Significance
Type I restriction-modification (R-M) enzymes are large molecular machines found in the majority of bacterial species. They can add methylation modifications to the self-DNA and degrade the invading unmodified DNA. The lack of high-resolution structures of type I R-M complexes impairs our understanding of the mechanism of subunit assembly and conformational transition. Here we report the first high-resolution structure of the type I MTase complex in its “open” conformation, including one DNA-recognition subunit, two DNA-modification subunits, one bound DNA, and two S-adenosyl methionine cofactors. We propose an updated model for the complex assembly and conformational transition. The structural and biochemical characterization of the type I R-M system reported in this study provides guidelines for future applications in molecular biology.
Keywords: type I R-M system, EcoKI, MTase, crystal structure
Abstract
Type I restriction-modification (R-M) systems are multisubunit enzymes with separate DNA-recognition (S), methylation (M), and restriction (R) subunits. Despite extensive studies spanning five decades, the detailed molecular mechanisms underlying subunit assembly and conformational transition are still unclear due to the lack of high-resolution structural information. Here, we report the atomic structure of a type I MTase complex (2M+1S) bound to DNA and cofactor S-adenosyl methionine in the “open” form. The intermolecular interactions between M and S subunits are mediated by a four-helix bundle motif, which also determines the specificity of the interaction. Structural comparison between open and previously reported low-resolution “closed” structures identifies the huge conformational changes within the MTase complex. Furthermore, biochemical results show that R subunits prefer to load onto the closed form MTase. Based on our results, we proposed an updated model for the complex assembly. The work reported here provides guidelines for future applications in molecular biology.
To fend off the constant challenge of phage infection, bacteria have evolved a set of defense systems. Central among them are the restriction-modification (R-M) and CRISPR-Cas (clustered regularly interspaced short palindromic repeats-CRISPR associated) systems, which provide innate and adaptive immune protection, respectively, by directly targeting the incoming viral DNA (1, 2). Recently, it has been shown that the R-M and CRISPR-Cas systems are compatible and act together to increase the overall phage resistance of the host cells (1). The R-M systems can add methylation modifications to the self-DNA and degrade the invader DNA, which lacks the modifications. The R-M systems were also found to be able to regulate the gene expression that leads to pathogenicity through the epigenetic effect of their DNA methylation (3). The R-M systems are almost universal and exist in ∼90% of bacterial and archaeal genomes (4). Based on the subunit composition, cofactor requirements, and DNA cleavage properties, the R-M systems can be broadly divided into four types: type I, type II, type III, and type IV (5, 6).
The creation of modification (MTases) and restriction enzymes (REases) with programmable DNA-binding, -methylation, and -cleavage specificities has long been a goal of modern biology. Type I restriction enzymes were the first REases to be purified and have been studied for five decades (7–10), which has helped launch the molecular biology revolution (6, 11). Type I R-M systems are encoded by three genes, termed hsd genes, for host-specific determinant: hsdS encodes the specific DNA-recognition subunit (S), hsdM the modification or methyltransferase subunit (M), and hsdR the restriction or endonuclease subunit (R). The S subunit determines the DNA recognition sequence, which consists of two independent DNA-binding domains (Target Recognition Domains, or TRDs) connected by two long alpha helices (Conserved Regions, or CRs). This results in a characteristic asymmetric, bipartite recognition sequence consisting of two half-sequences, each containing 2–5 base pairs separated by a gap with a fixed number of nonspecific DNA bases (6, 12). For example, the EcoKI system recognizes the DNA sequence of 5′-AACNNNNNNGTGC-3′. It has been shown that the TRD domains from different type I R-M systems could be exchanged to gain the hybrid specificities (6), which greatly extends the potential of molecular biology applications of the type I R-M enzymes.
The S/M/R subunits can assemble into two types of complexes: 2M+1S as the MTase complex with only methyltransferase activity and 2R+2M+1S as the REase complex with both methyltransferase and endonuclease activities (6). In this way, the type I R-M systems can change both REase and MTase specificities harmoniously by using a common S subunit (6). This feature is critical for the host cells to periodically change the REase specificity to overcome the threat of the viruses with modified target sequences, while changing the MTase specificity in exactly the same way to modify and protect the host DNA (6). It has been proposed that the M subunits bind mainly to the TRD domains (12, 13) and the conserved “IPLPPL” regions connecting CRs and TRDs (14) in the S subunit to form the MTase complex. However, if this is the case, it is hard to understand why the type I R-M enzymes can still be properly assembled after exchanging the TRD domains among the different enzymes (6). Atomic structures of type I R-M enzyme complexes have been difficult to obtain. Until now, only the crystal structures of individual subunits have been solved (15–20), but those of complexes have not. The low-resolution negative stain electron microscopy (EM) studies revealed that type I R-M enzymes will undergo an “open” to “closed” conformational change upon binding to the target DNA (11, 13). However, despite a half century of efforts, the molecular details of the interactions among different subunits and the dynamic conformational transitions are still unclear due to the lack of high-resolution structures.
To better understand how the type I MTase assembles, and how the complex dynamically changes conformation from open to closed upon target DNA binding, we determined the crystal structure of the type I MTase complex from Thermoanaerobacter tengcongensis (termed Tte-MTase) bound to a random DNA in the open conformation. Here we report the atomic structure of the type I R-M enzyme complex. The intermolecular interactions between the S and M subunits are mediated by a newly identified four-helix bundle formed by the CRs of the S subunit and the C-terminal alpha helices of the two M subunits. The four-helix bundle, rather than the TRD domains, determines the specificity of the interaction between the S and M subunits. The linker region between the C-terminal alpha helix and the N-terminal globular domain of the M subunit is essential to control the conformational transition of the MTase complex. Furthermore, biochemical results show that the R subunits prefer to load onto the closed form MTase rather than the open form. Based on our crystal structure, the previous low-resolution EM model (11, 13), and the biochemical data, we propose an updated model for the complex assembly and conformational transition of the type I R-M system. The structural and biochemical characterization of the type I R-M system reported in this study provides guidelines for future applications in molecular biology.
Results
Overall Structure of the Type I MTase Complex in the Open Form.
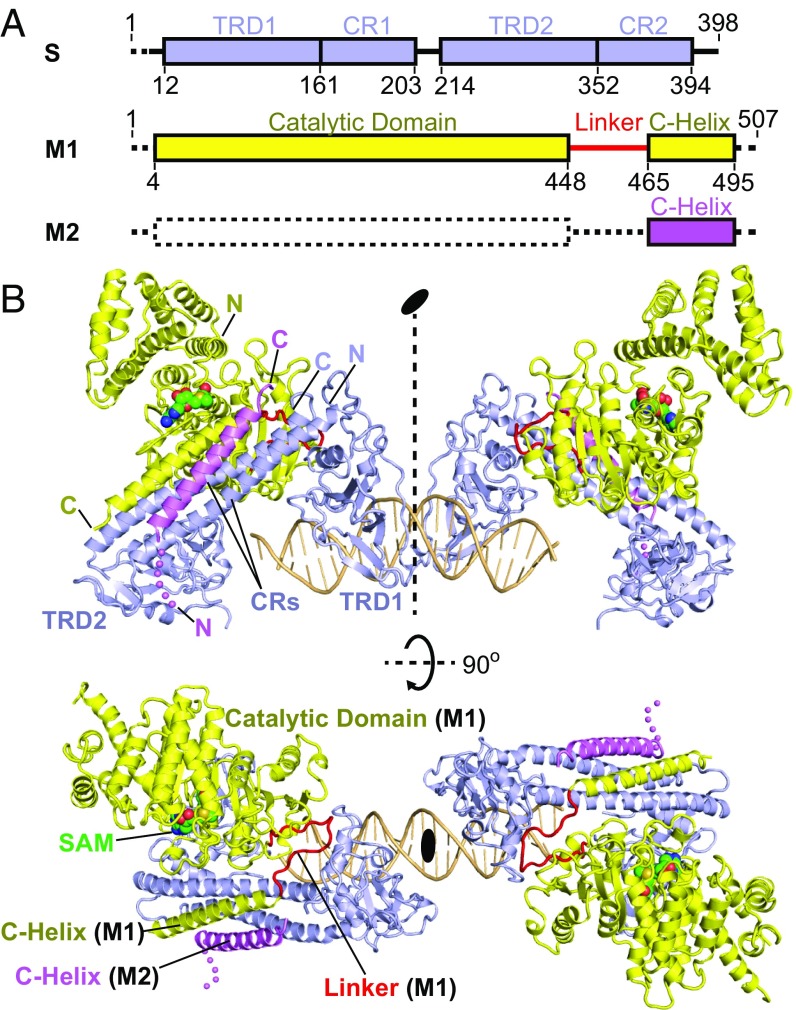
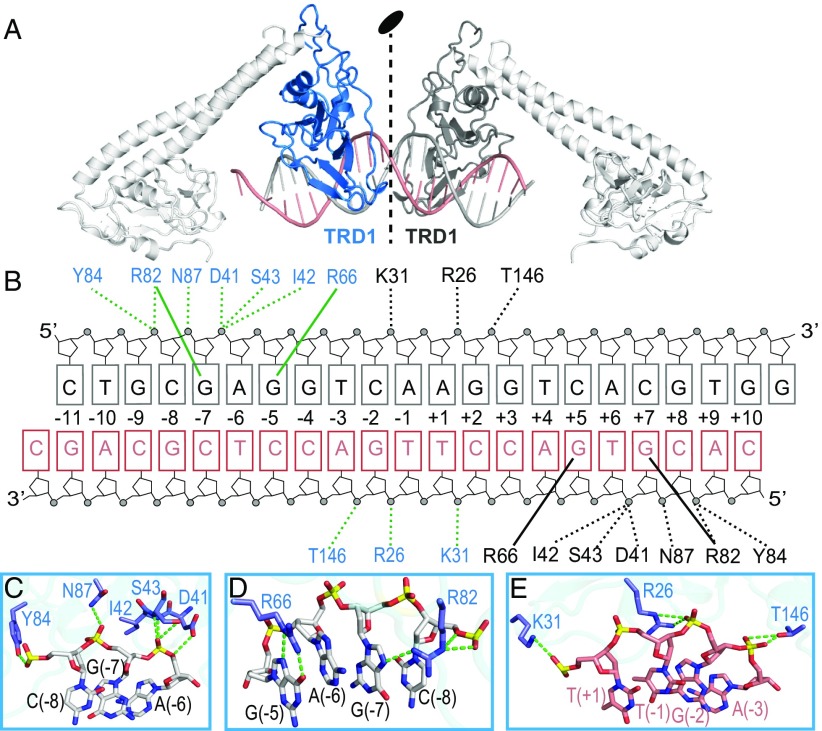
It has been a long-term challenge to determine the atomic structure of type I R-M complexes. Due to its dynamic nature, the open form structure is considerably more difficult to achieve than the closed form. To obtain the diffraction-quality crystals, we screened several type I enzymes from different species in combination with DNAs of various sequences and lengths. Here, we report the 3.2-Å crystal structure of the full-length type I MTase complex (including one S and two M subunits) from T. tengcongensis bound to a nontarget DNA and the cofactor S-adenosyl methionine (SAM) (Fig. 1 and Fig. S1; X-ray statistics in Table S1). The asymmetric unit contains two Tte-MTase complexes, one DNA duplex, and two SAM molecules (Fig. 1B). Related by a noncrystallographic symmetry, the two Tte-MTase complexes in the asymmetric unit are almost identical (r.m.s.d. ∼0.17 Å, Fig. S1A). This assembly appeared to be a result of crystal packing but may also represent the structure of two collision type I R-M complexes bound to the same DNA. One of the two M subunits in each Tte-MTase complex in the asymmetric unit lacks the N-terminal catalytic portion due to the lack of electron density, indicating the dynamic nature of this region (Fig. 1). The overall structure of the complex clearly shows an open conformation with loose interactions among S subunit, M subunits, and DNA in contrast to the compact organization observed in the closed-form, low-resolution EM models (11, 13). The S subunit is composed of four continuous structural regions (TRD1-CR1-TRD2-CR2) and displays a pseudo twofold symmetry and cyclic topology organization (Fig. 1) similar to the structure of the S subunit in the free state (15, 16, 20). The M subunit contains an N-terminal catalytic domain, a flexible linker, and a C-terminal alpha helix, with a SAM molecule bound within the catalytic pocket (Fig. 1). The catalytic domain of the M subunit adopts a bilobal scaffold characteristic of members of the γ-class N6-adenine MTase family. The DNA duplex interacts only with TRDs of the S subunit and not with the M subunits (Fig. 1B), further confirming the open conformation of the complex.
Fig. 1.
Overall structure of the Tte-MTase complex bound to DNA and SAM. (A) Schematics of the domain architectures of the S and M subunits of the type I R-M system from T. tengcongensis. The dashed lines indicate the disordered regions in the crystal structure. (B) Ribbon diagram of the structure of two Tte-MTase complexes in complex with one DNA and two SAM molecules in the asymmetric unit shown in two different views. Same color code as in A. The two MTase complexes are related by a noncrystallographic twofold symmetry. The individual domains of S and M subunits have been labeled in the Upper and Lower panels, respectively. The SAM molecules are shown in space-filling presentation and colored in green.
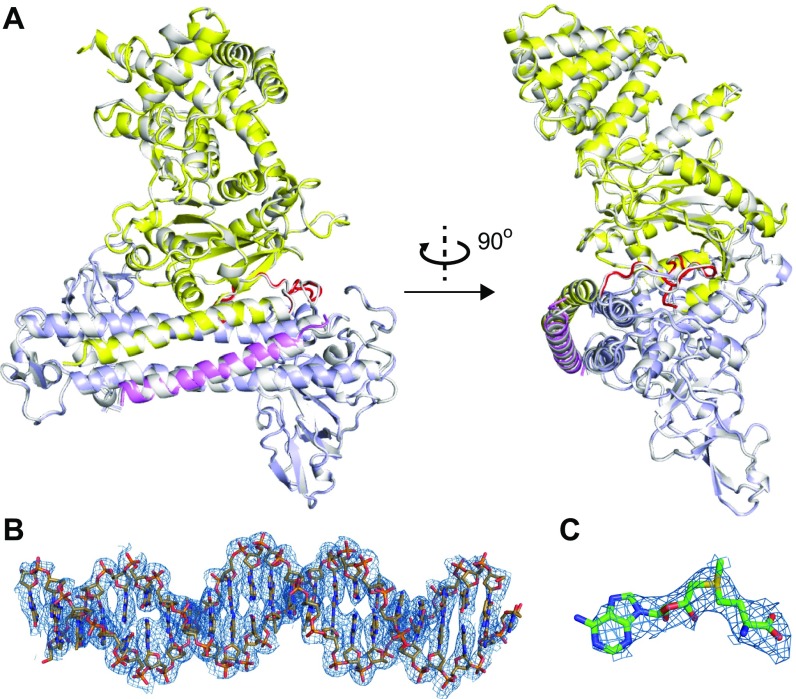
Fig. S1.
(A) Two views of the structural superimposition of the two MTases complexes (one in different colors; the other one in silver) in the asymmetric unit. (B and C) The 2Fo − Fc electron density maps around DNA (B) and SAM (C) are shown as the blue meshes (contoured at 1.2σ).
Table S1.
X-ray statistics of Tte-MTase + DNA
| Beamline | SSRF-BL19U1/BL17U |
| Wavelength | 0.9800 |
| Space group | P41 |
| Unit cell | |
| a, b, c, Å | 121.6, 121.6, 280.4 |
| α, β, γ, ° | 90.0, 90.0, 90.0 |
| Resolution, Å | 47–3.2 (3.28–3.2)* |
| Rmerge | 0.295 (0.862) |
| CC (1/2) | 0.906 (0.586) |
| I/σ (I) | 27.8 (2.4) |
| Completeness, % | 95.3 (100.0) |
| Redundancy | 2.8 (2.8) |
| No. of unique reflections | 63,642 |
| Rwork/Rfree, % | 22.2/27.2 |
| r.m.s.d. | |
| Bond lengths, Å | 0.015 |
| Bond angles, ° | 2.010 |
Highest resolution shell (in Å) is shown in parentheses.
Four-Helix Bundle Interface Between S and M Subunits.
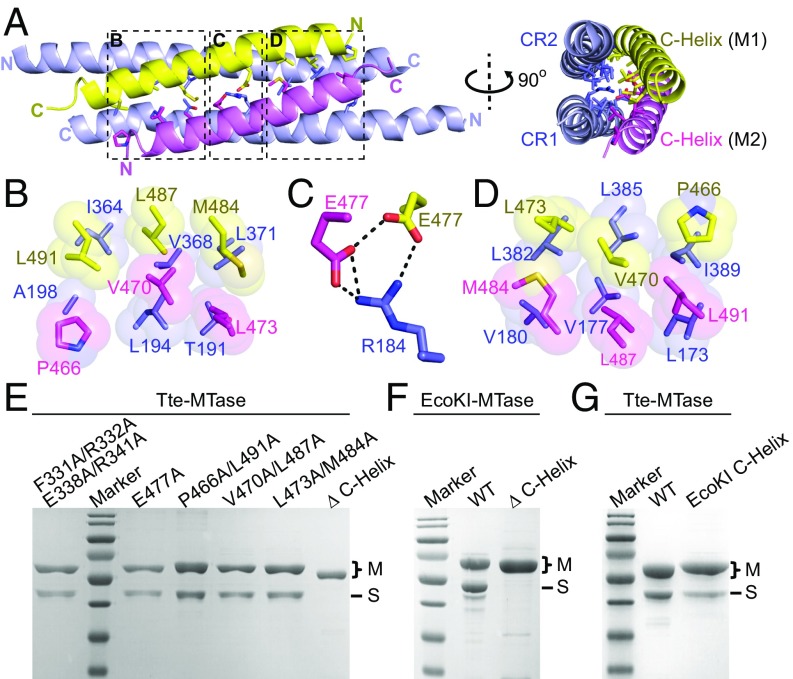
To our surprise, the majority of intermolecular interactions are mediated by the two CRs of the S subunit and the C-terminal alpha helices of the two M subunits (Figs. 1A and 2A). This is in contrast to the previous understanding that the TRD domains (12, 13) or the conserved IPLPPL motifs are the major interacting regions (14). The two helices of the M subunits adopt an antiparallel arrangement and form a four-helix bundle structure together with the coiled-coil region of the S subunit (Fig. 2A). Although it was found to be primarily a monomer in solution (21), the M subunit was forced into a dimer organization in the MTase complex through the formation of the four-helix bundle. The interacting surface within the four-helix bundle between the S and M subunits is ∼1,465 Å2 and is formed by both hydrogen bond interactions and van der Waals contacts (Fig. 2 B–D). Both the antiparallel arrangement of each alpha helix and the detailed intermolecular interactions represented a pseudo twofold symmetry of the four-helix bundle structure. In the middle of the four-helix bundle is a hydrogen-bonding network formed by Arg184 from CR1 of the S subunit and Glu477 from each M subunit (Fig. 2C). There are a total of six groups of hydrophobic contacts within the four-helix bundle (Fig. 2 B and D), which can be divided into three pairs based on the pseudo twofold symmetry: Leu173 (CR1)-Ile389 (CR2)-Pro446 (M1)-Leu491 (M2) and Ala198 (CR1)-Ile364 (CR2)-Leu491 (M1)-Pro466 (M2); Val177 (CR1)-Leu385 (CR2)-Val470 (M1)-Leu487 (M2) and Leu194 (CR1)-Val368 (CR2)-Leu487 (M1)-Val470 (M2); and Val180 (CR1)-Leu382 (CR2)-Leu473 (M1)-Met484 (M2) and Thr191 (CR1)-Leu371 (CR2)-Met484 (M1)-Leu473 (M2). We generated mutations to break either the hydrogen-bonding interactions (E477A) or each pair of the hydrophobic contacts (P466A/L491A or V470A/L487A or L473A/M484A) and then examined the Tte-MTase complex formation. As expected, none of these separated mutations could abolish the assembly of the complex by themselves (Fig. 2E). However, the complex cannot be formed if the whole alpha helix of the M subunit is deleted (Fig. 2E), indicating the critical role of the four-helix bundle in complex assembly. We also checked the complex formation by deleting the C-helix in the well-characterized EcoKI system and observed similar results to the Tte system (Fig. 2F). Our structural and mutational observation was also supported by the previous biochemical results, which showed that partial deletions of the C-terminal helix of the M subunit would impact the MTase complex formation (20, 21).
Fig. 2.
Four-helix bundle interface between S and M subunits. (A) Ribbon diagram of the four-helix bundle structure consisting of the CR regions (light blue) from the S subunit and two C-helices (yellow and violet) from the M subunits. The key residues are shown in stick representation. (B–D) Detailed interactions within the four-helix bundle interface as highlighted by the dashed boxes in A. The hydrophobic interactions are shown in space-filling presentation (B and D). The hydrogen bonds are shown as thick black dashed lines (C). Same color code as in A. (E–G) SDS/PAGE results of the coexpression of the wild-type S subunit (no tag) and different M subunits (His tagged) as indicated for both Tte (E and G) and EcoKI (F) systems. The SDS/PAGE gels were stained with Coomassie blue.
The C-terminal alpha helix is predicted to exist in the M subunits of other species (Fig. S2), suggesting that the four-helix bundle structure is structurally conserved among different type I R-M systems. To find out whether the C-helix motif can be exchanged between different M subunits in a way similar to the TRD domains of S subunits, we substituted the C-helix of Tte-M for the C-helix of EcoKI-M and then checked the complex formation. Our results showed that, although a small amount of Tte-S could still be copurified with the hybrid Tte-M, the binding affinity between these subunits was significantly decreased compared with the wild-type proteins (Fig. 2G). This indicated that the four-helix bundle, rather than the TRD domains, determines the specificity of S–M interaction.
Fig. S2.
The secondary structure prediction for the C-terminal regions of the M subunits from different type I R-M systems. The linker and C-Helix motifs are colored in red and pink, respectively. The Jpred4 server (www.compbio.dundee.ac.uk/jpred/) was used for the secondary structure prediction.
Other Intermolecular Interactions Between S and M Subunits.
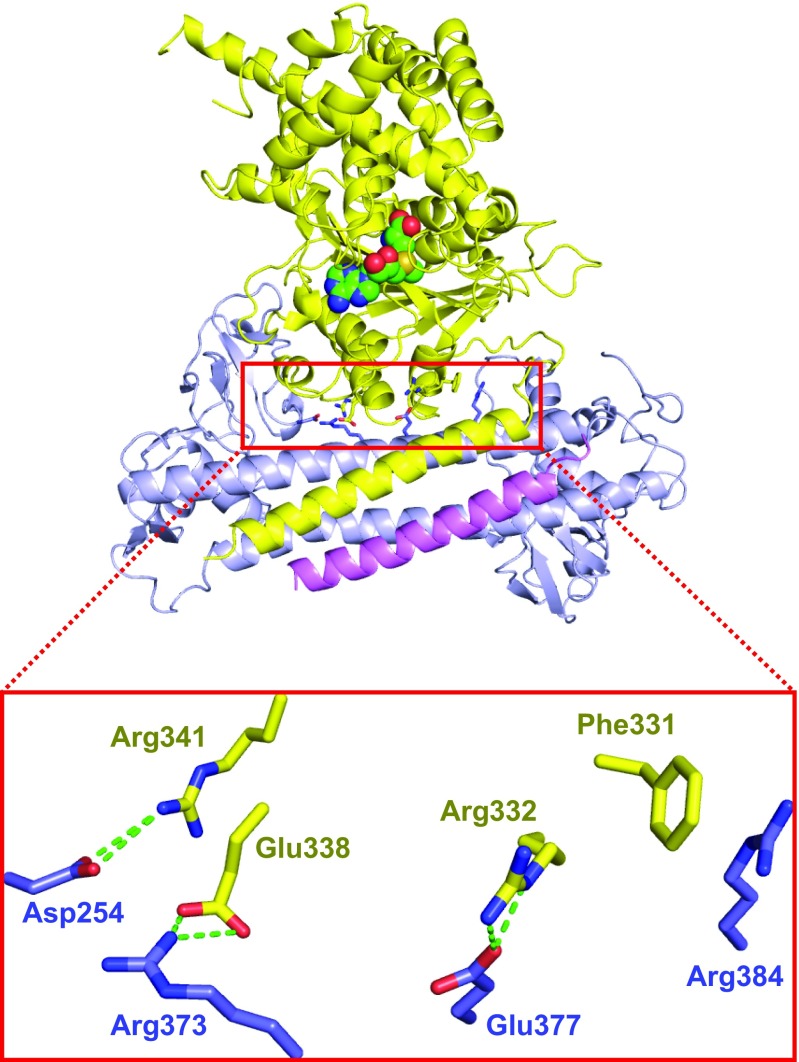
In addition to the four-helix bundle interface, the N-terminal catalytic domain of the M1 subunit also interacts with the S subunit through three hydrogen-bonding interactions [Arg332(M1)-Glu377(CR2), Glu338(M1)-Arg373(CR2), and Arg341(M1)-Asp254(TRD2)] and one stacking interaction [Phe331(M1)-Arg(Arg384)] (Fig. S3). Due to the dynamic nature of the M2 catalytic domain in the structure, we speculated that the current conformation of the M1 subunit was just one of its possible states. Thus, the above contacts between the M1 catalytic domain and the S subunit may not be functionally specific. To prove our speculation, we generated a group mutation removing all of the four interactions (F331A/R332A/E338A/R341A) and found that the Tte-MTase complex assembly was not affected (Fig. 2E). Together, these results further confirmed that the newly identified four-helix bundle structure is the major interacting interface between the S and M subunits in type I R-M systems.
Fig. S3.
The intermolecular interactions between the S and M subunits in addition to the four-helix bundle interface. Same color code as in Fig. 1. The hydrogen bonds are shown by green dashed lines.
Intermolecular Interactions Between DNA and TRD.
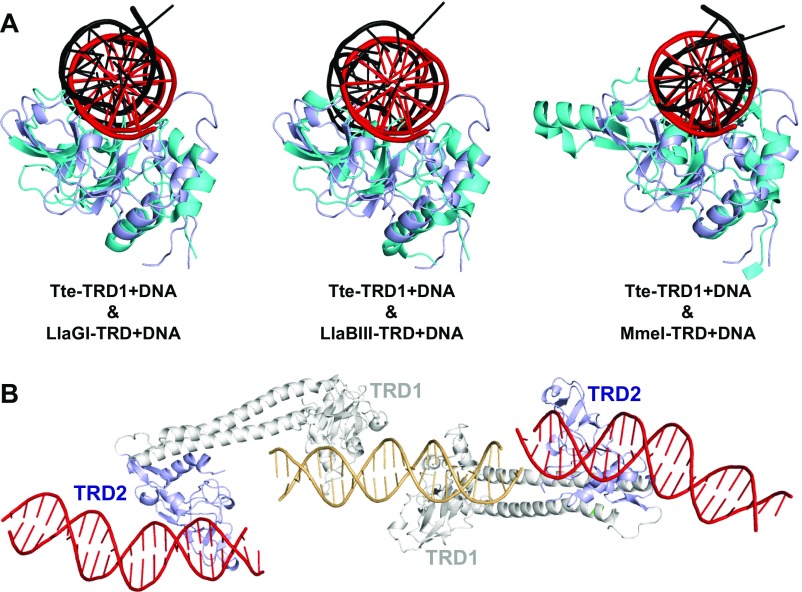
The specific recognition sequence of the R-M enzymes from T. tengcongensis has not been characterized, and thus we employed a random nonspecific DNA to determine the structure of the open-form MTase. In the asymmetric unit, the TRD1 domains from two S subunits bind to one DNA molecule (Fig. 3A). Because of the pseudo twofold symmetry, the two TRD1 domains bind to DNA in a very similar way (Fig. 3B). The DNA binds to the positively charged surface of the TRD1 domain, with the majority of the intermolecular contacts being mediated by the phosphate backbone of the DNA and TRD1 (including the side chains of Arg26, Lys31, Asp41, Ser43, Arg82, Tyr84, Asn87, and Thr146; and the main chains of Ile42 and Ser43) (Fig. 3 B–E). The residues Arg66 and Arg82 also form the base-specific hydrogen bonds with the Hoogsteen edges of guanines at positions +5 and +7 (or positions −5 and −7 for the pseudo symmetrical TRD1), respectively (Fig. 3 B and D). Given that the DNA in our structure is not the actual target sequence of Tte-MTase and the open conformation of the complex, no adenine base flipping was observed. This is the DNA-bound structure for the TRD domain of the classical type I enzymes. The TRD of the type I R-M system has a similar DNA-binding pattern to the TRD of LlaGI (22)/LlaBIII (23) (type ISP) and MmeI (24) (type IIL) (Fig. S4A). The TRD2 domains of the two S subunits in the structure bind to neighboring symmetric DNA molecules due to the crystal packing (Fig. S4B). Although the binding surface of TRD2 was not fully covered by the bound DNA, several similar interactions to TRD1 were observed.
Fig. 3.
Intermolecular interactions between DNA and TRD. (A) Ribbon diagram of the DNA molecule bound to the TRD1 domains from the two S subunits in the asymmetric unit. The TRD1 domains are colored in blue and black, respectively. The two DNA strands are colored in black and salmon. (B) Schematic of the detailed interactions between TRD1s and DNA. Same color code as in A. The hydrogen bonds between protein and the sugar-phosphate backbone or the bases are shown in dashed and solid lines, respectively. (C–E) Interactions between one TRD1 domain and the bound DNA in three different regions. The hydrogen bonds are shown by green dashed lines.
Fig. S4.
(A) Structure superimposition of the Tte-TRD1+DNA with the TRD domains from other R-M systems: LlaGI-TRD+DNA (Left), LlaBIII-TRD+DNA (Middle), and MmeI-TRD+DNA (Right). The Tte-TRD1 and the bound DNA are in light blue and red, respectively. The other TRDs and bound DNAs are in cyan and black, respectively. (B) The TRD2 domains (light blue) in the asymmetric unit bind to neighboring symmetric DNA molecules (red) by using the similar DNA-binding surface to TRD1.
Conformational Change from Open to Closed.
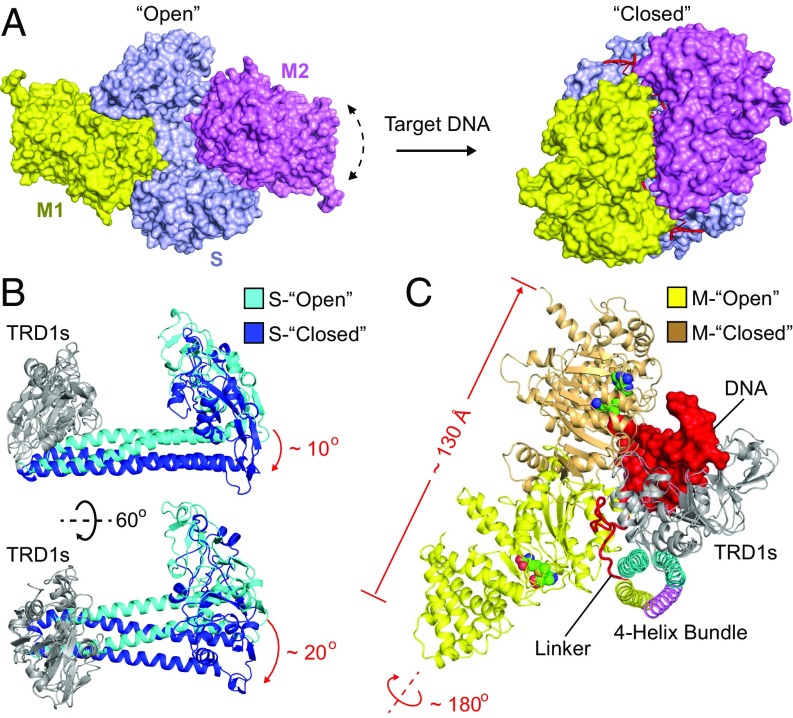
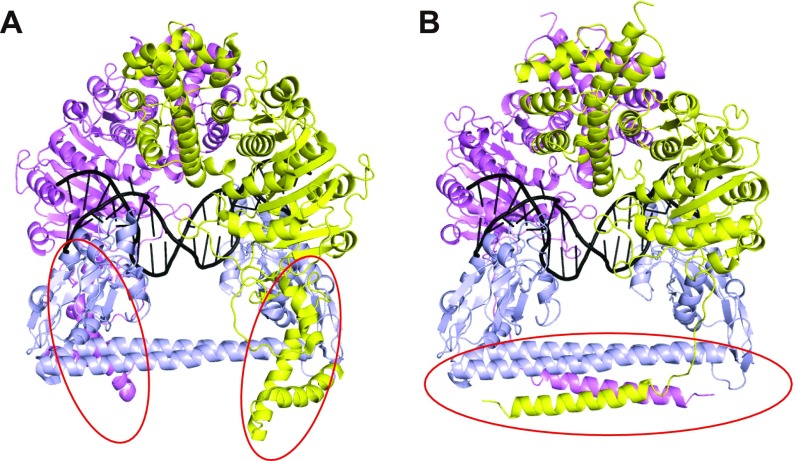
Based on our structural and biochemical results, we re-examined the previous low-resolution EM model of the closed-form type I MTase complex (PDB ID code 2Y7H) and made several corrections to generate a more accurate model (Fig. 4A and Fig. S5). The major changes were for the conformation of the C-terminal alpha helix of the M subunit and the four-helix bundle structure (Fig. S5). To better compare the open and closed structures of the MTase complex, we superimposed the TRD1 domains of the S subunits as the reference point. The CRs and TRD2 domains of the S subunit in the open structure undergo significant rotation and movement toward the bound target DNA to form the compact closed structure (Fig. 4B). The more dramatic conformational changes occur within the M subunits. The N-terminal catalytic domain of the M subunit is far from the bound DNA in the open structure, but it will move more than 130 Å to interact with the target DNA (Fig. 4C). It should be noted that the catalytic pocket of the M subunit is facing opposite to the TRD domain and the bound DNA in the open structure, indicating that a significant rotation (∼180°) is needed to transit from open to closed (Fig. 4C). The significant conformational changes of the M subunits rely on the flexible linker between the C-terminal helix and the N-terminal global domain (Fig. 4A). Similar to the C-helix region, this flexible linker is also predicted to exist in the M subunits of other type I R-M systems (Fig. S2), indicating a conserved and important function of this region. The previous ∼35-Å EM map of the open form EcoR124I system had shown the very thin connections between S and M subunits (11), which is consistent with our finding that the linker region is the pivot point for flexing to allow the enzyme to undergo large conformational changes.
Fig. 4.
Conformational transition from open to closed. (A) Surface views of the open (Left) and closed (Right) structures of the type I MTases. The color code for proteins is the same as in Fig. 1. The DNA is shown in ribbon and red. Given the low-quality electron density, the M2 subunit of the open complex was modeled in a similar conformation to M1 by superimposing the C-helices of the two M subunits. The dashed arrow line indicates the dynamic nature of this subunit in the structure. (B) Two views of the conformational changes in the S subunit between open (cyan) and closed (blue). The TRD1 domains (gray) are superimposed as the reference point. (C) The conformational changes in the M subunit between open (yellow) and closed (brown). The DNA (red) is shown in surface presentation. The TRD1 domains are superimposed as the reference point.
Fig. S5.
(A) The negative-stain EM closed model of the EcoKI-MTase. Same color code for the proteins as in Fig. 1. The DNA is shown in black ribbon. The red circles indicate the C-helix regions in the wrong conformation. (B) The updated closed model of the Tte-MTase. The S and M subunits of the EcoKI enzyme in A were replaced by their Tte counterparts by superimposition. The red circle indicates the revised four-helix bundle region based on the crystal structure.
R Subunit Prefers to Load onto the Closed MTase.
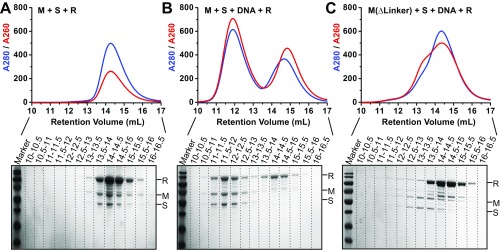
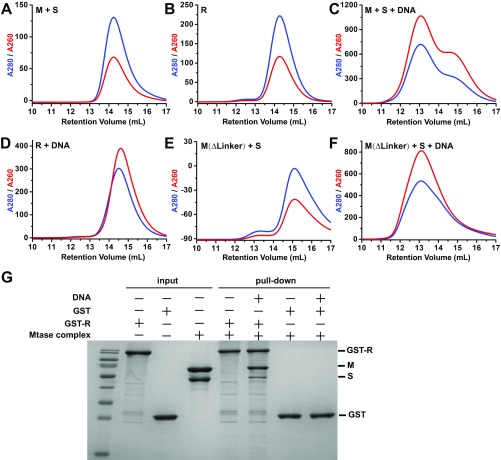
To better understand whether the huge conformational transition from open to closed would affect the assembly of the R subunits, we checked the REase complex formation by using the well-characterized EcoKI system. We first purified the separated R subunit and MTase complex (2M+1S) of the EcoKI system to homogeneity (Fig. S6) and then incubated the R subunit with either the apo-form MTase (Fig. S6A) or the MTase bound to a target DNA (Fig. S6C). By using size-exclusion chromatography, we found that, after incubation, the R subunit and apo-form MTase were eluted in a similar retention volume to their individual forms (Figs. S6A and S7 A and B), suggesting that no larger complex was formed. When we added the target DNA, a stable larger complex was formed and eluted earlier on the column than both the MTase–DNA complex and the R subunit (Figs. S6B and S7 B and C). The complex contains all of the three subunits as well as the target DNA (Fig. S6B). It should be noted that the R subunit by itself could not form a stable complex with DNA under the same condition (Fig. S7D). These results indicate that the R subunit prefers to load onto the closed-form MTase rather than the open form. The GST pull-down assay showed similar results to the size-exclusion chromatography (Fig. S7G). To further confirm this conclusion, we generated the deletion mutation of the linker region for the EcoKI M subunits (∆461–479) and purified the 2M(∆461–479)+1S MTase complex to homogeneity (Fig. S7E). We then incubated this mutated MTase with the target DNA and the R subunits and checked the larger REase complex formation using size-exclusion chromatography (Fig. S6C). As expected, although the linker-deleted MTase can still bind to DNA (largely due to the S subunit, Fig. S7F), it cannot form the larger REase complex with R subunits (Fig. S6C). This indicated that the failure to form the correct closed MTase (due to the linker deletion) will abolish the assembly of R subunits.Taken together, these results indicate that the closed conformation of the MTase is critical for R subunit assembly.
Fig. S6.
(A–C) Size-exclusion chromatography curves (Upper) of different incubation samples and the related SDS/PAGE results (Lower) for the fractions from 10 mL to 16.5 mL (0.5 mL for each fraction).
Fig. S7.
(A–F) Size-exclusion chromatography curves of different incubation samples of the EcoKI system. (G) GST pull-down results showing that the R subunit bound to the Mtase complex in its closed state (with DNA) but not in the open state (no DNA).
Discussion
We present here the atomic structure of a type I R-M MTase complex bound to DNA, a structure that provides detailed insights into the complex assembly and the dynamic conformational changes between open and closed states. As the first-discovered R-M systems, the type I R-M enzymes have been extensively studied genetically, biochemically, and biophysically for five decades (6, 25–27). However, in contrast to the enormously useful type II enzymes, the type I enzymes have not been fully developed as convenient molecular biological tools. One reason is that the characterization of type I systems has been relatively difficult because the position of DNA cutting is nonspecific. However, this situation has changed with the new single-molecule real-time DNA sequencing technology, which can directly detect the DNA methylations (6, 12). Another reason is the lack of high-resolution structural information about the complex assembly, intermolecular interaction, and the conformational transition between different states. The first insights into the molecular organization of the type I R-M enzymes came from low-resolution models using a combination of negative-stain EM, neutron scattering, and structural modeling (11, 13). Although these models have greatly advanced our understanding of the overall organization of these complexes, there are still many ambiguous and unsolved questions at the resolution of 20–35 Å.
One advantage of type I enzymes is that the sequence specificity can be easily changed through TRD recombination, which simultaneously couples the M and R activities (6). However, this phenomenon has long been confusing, given that the TRD domains were considered to be the major contacting region with the M subunits. The identification of a four-helix bundle interface formed by the CRs from the S subunit and the C-terminal helices from the M subunits provides a perfect explanation for this question. Our structure also showed that the TRD domain of the type I R-M system bound to DNA in a manner similar to other R-M systems (Fig. S4A), suggesting that one could design new R-M enzymes with hybrid activities by exchanging TRD domains among different R-M systems. It has also been shown that the gap length between the two recognition sequences can be changed by adjusting the length of the CR regions of the S subunit (6). The identification of the four-helix bundle structure, however, raises concerns that the C-helix motifs of the M subunits may need to be changed accordingly along with the CR regions.
The formation of the four-helix bundle structure definitely requires the C-terminal helices from two M subunits, indicating that the previously proposed 1M+1S complex (21) is unlikely to be functionally relevant. We also found that a flexible linker between the C-terminal helix and N-terminal catalytic domain of the M subunit is critical for the transition between open and closed states (Figs. 1B and 4C), which confirmed the findings from previous low-resolution negative-stain EM models (11). In addition, our results showed that the four-helix bundle, but not the TRD domains, determines the specificity of the combination between different S and M subunits. This observation greatly extends our current understanding of the approaches to designing hybrid type I enzymes with various specificities. Theoretically, the TRDs of S subunits and the catalytic domains of M subunits can be freely exchanged as long as the intermolecular interactions of the four-helix bundle are maintained.
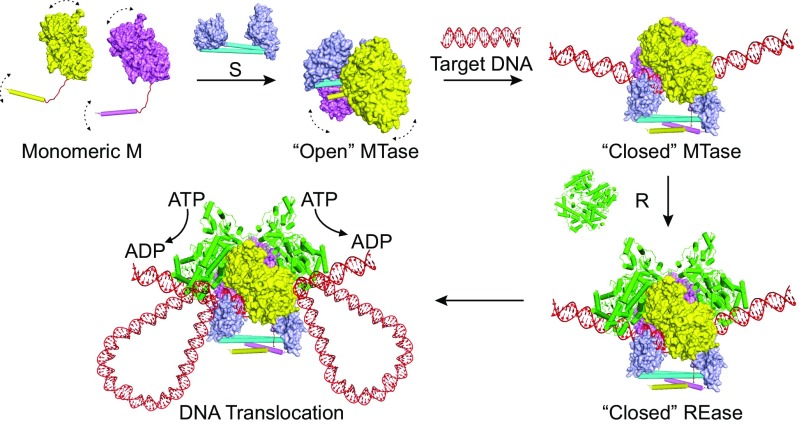
It has been proposed that the type I R-M enzymes reach the “closed initiation” complex via two possible routes (11): (i) the enzyme stays in the closed form and just opens up transiently to allow the DNA to bind into the MTase core, followed by closing again and diffusion of the complex on the DNA until it meets the recognition site; and (ii) the enzyme stays in the open form and nonspecifically binds to the DNA using its R subunits, followed by diffusion along the DNA until the MTase core recognizes the target sequence. However, none of the above speculated models has solid evidence to support them. Regarding the first model, the previous EM model (11) has clearly shown that the type I enzymes adopt the open or “relaxation” conformation in the apo form. For the second model, the crystal structure presented here provides evidence that the TRD domains can directly bind to the nontarget DNA without the help of M and R subunits (Fig. 1B). Furthermore, the R subunits cannot form a stable complex with the open-form MTase (Fig. S6A). In addition, we also observed that the MTase core binds to DNA (either target or nontarget) more strongly than the R subunits (Fig. S7). Together, these results indicate that the open form MTase core nonspecifically binds to the DNA by itself rather than through its R subunits. Based on the results from both the current and previous studies, we propose an updated model for the assembly of the type I R-M enzymes (shown in Fig. 5): (i) two monomeric M subunits bind to one S subunit through the four-helix bundle interactions and form an open MTase complex; (ii) the open MTase complex dynamically binds to (and dissociates from) the DNA and reads the sequence by using its TRD domains; (iii) when reaching the recognition site, the M subunits undergo large conformational changes and move toward the DNA to form the compact closed complex; (iv) the two R subunits load onto the closed MTase complex through the interactions with both M subunits; and (v) the R subunits initiate the DNA translocation by hydrolyzing the ATP molecules.
Fig. 5.
The working model. The proposed model for the complex assembly and conformational changes of the type I R-M systems. The REase structure is modeled based on the crystal structure of the EcoR124I-R subunit (18) and the low-resolution EM model of EcoKI REase (11).
In summary, we present the atomic structure of the type I MTase complex and reveal the molecular mechanisms underlying the subunit assembly and conformational transition. The results presented here rationalize a large body of experimental data obtained by many different methods over the past 50 y and provide guidelines for future applications of type I R-M systems in molecular biology.
Materials and Methods
Expression and Purification of the MTase Complexes.
The Tte-hsdS and Tte-hsdM genes were amplified by PCR from T. tengcongensis genomic DNA. The EcoKI-hsdS and EcoKI-hsdM genes were amplified by PCR from the Escherichia coli K-12 substrain MG1655 genomic DNA. The PCR products of hsdM and hsdS genes were subcloned into the modified pet-Duet vector (Novagen) at cloning sites 1 (with N-terminal 6XHis tag) and 2 (without tag), respectively. The E. coli BL21 (DE3) cell strain was used to express the recombinant proteins. The cells were grown at 37 °C until OD600 reached ∼0.8. The temperature was then shifted to 28 °C, and the cells were induced by addition of isopropyl β-d-1-thiogalactopyranoside to the culture medium at a final concentration of 0.4 mM. After induction, the cells were grown overnight. The fusion protein was purified over a Ni-NTA affinity column. After dialysis, the protein sample was further fractionated over a Resource Q column, followed by gel filtration on a Superdex 200 10/300 GL column. The final sample of MTase complexes contains about 20 mg/mL protein, 8 mM Hepes (pH 7.0), 100 mM KCl, 5% glycerol, and 1 mM DTT. Based on the coexpression vectors of wild-type HsdM and HsdS, we also constructed several coexpression vectors of the mutant HsdM and HsdS. The mutant M–S complexes were expressed and purified by a protocol similar to that of wild-type proteins.
Crystallization for the MTase-DNA-SAM Complex.
The double-strand DNA used for crystallization is 21 bp with a 1-nt 3′-overhang at either end (upper strand 5′-CTGCGAGGTCAAGGTCACGTGG-3′; lower strand 5′-CACGTGACCTTGACCTCGCAGC-3′). All of the DNA oligos were synthesized from Invitrogen. The two DNA strands were dissolved in H2O and mixed together with a molar ratio of 1:1 and then heated at 95 °C for 5 min and annealed by slow cooling to room temperature. The M-S-DNA-SAM quaternary complex was prepared by first incubating the protein and DNA at a molar ratio of 1:1.3 at 20 °C for 30 min, followed by gel filtration purification in a buffer containing 1 mM SAM, 8 mM Hepes (pH 7.0), 100 mM KCl, 5% glycerol, and 1 mM DTT. The purified M–S-DNA-SAM quaternary complex was then concentrated to ∼30 mg/mL before crystallization.
The crystals of M-S-DNA-SAM were generated by hanging drop vapor diffusion method at 12 °C from drops mixed from 1.5 μL of the complex solution and 1.5 μL of reservoir solution [0.1 M Tris⋅HCl, 54% 2-methyl-2,4-pentanediol, 0.2 M NH4H2PO4 (pH 8.8)].
Structure Determination.
The diffraction data sets were collected from crystals cooled to 90 K using 0.3° oscillations in beamline BL19U1 in Shanghai Synchrotron Radiation Facility, China. The data were indexed, integrated, and scaled using HKL2000 (28). Molecular replacement solutions were obtained by Phaser using the two models, including HsdM (PDB ID code 3UFB) from Vibrio vulnificus YJ016 (29) and HsdS (PDB ID code 3OKG) from T. tengcongensis (20). After a density modification process using RESOLVE (30), we could build an ∼80% sequence of the protein and most of the DNA into the electron density. The model building was mostly carried out using the program COOT (31), and final structural refinement was carried out using the program PHENIX (32). The statistics of the data collection and refinement are listed in Table S1.
Expression and Purification of the EcoKI-R Subunit.
The EcoKI-hsdR gene was amplified by PCR from the E. coli K12 genomic DNA and subcloned into the Phat2 vector (with N-terminal 6XHis tag). The protein was expressed in E. coli BL21 (DE3) cells under a condition similar to that of the MTase complexes. The His-tagged protein was purified over Ni-NTA, Resource Q, and gel filtration columns. The final sample contains about 20 mg/mL protein, 8 mM Hepes (pH 7.0), 300 mM KCl, 5% glycerol, and 1 mM DTT.
GST Pull-Down Assay.
GST pull-down assay was employed to identify the interactions between EcoKI-HsdR and EcoKI-MTase with or without DNA. Purified GST (20 μg) or GST-tagged protein HsdR (60 μg) was incubated with MTase (30 μg) or MTase (30 μg)/target DNA (20 μg) complex for 2 h at 4 °C with 500 μL binding buffer [10 mM Hepes (pH 7.0) 50 mM NaCl, 1 mM DTT, and 1% Triton X-100]. Then the samples were incubated with glutathione-Sepharose 4B (GE Health) beads (50 μL) at 4 °C for 2 h. After incubation, beads were washed four times with binding buffer containing 400 mM NaCl, and the bound proteins were eluted and subsequently boiled with sample loading buffer. All samples were analyzed with SDS/PAGE.
In Vitro Assembly of the EcoKI REase Complex.
The size-exclusion chromatography method was used to check the REase complex formation. Before loading onto the column, the R subunit was first incubated with either the apo-form MTase complex (open form) or the MTase complex bound to target DNA (closed form) at 4 °C for 30 min in the buffer containing 8 mM Hepes (pH 7.0), 50 mM KCl, 5% glycerol, and 1 mM DTT. The size-exclusion chromatography running buffer is the same as that of the incubation buffer. The target DNA used in this study is a 40-bp DNA containing the EcoKI specific recognition sequences (shown as underlined): 5′-AAAAATCTAAAATCAACGTCGACGTGCAAAAAGAGAAAAA-3.
Acknowledgments
We thank Professor Dong-Cai Liang for his advice and support throughout the project and the staff of beamline BL19U1/BL17U at the National Facility for Protein Science Shanghai and the Shanghai Synchrotron Radiation Facility, China, for assistance in data collection. This work was supported by the National Natural Science Foundation of China Grants 31371310, 31570794, and 31670903; the Chinese Academy of Sciences Pilot Strategic Science and Technology Projects B Grants XDB08010301, XDB08010303, and XDB08020204; the Starting-Up Funding of the Institute of Biophysics; and the Thousand Young Talents Program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 5YBB).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711754114/-/DCSupplemental.
References
- 1.Dupuis ME, Villion M, Magadán AH, Moineau S. CRISPR-Cas and restriction-modification systems are compatible and increase phage resistance. Nat Commun. 2013;4:2087. doi: 10.1038/ncomms3087. [DOI] [PubMed] [Google Scholar]
- 2.Wright AV, Nuñez JK, Doudna JA. Biology and applications of CRISPR systems: Harnessing nature’s toolbox for genome engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 3.Seib KL, et al. Specificity of the ModA11, ModA12 and ModD1 epigenetic regulator N(6)-adenine DNA methyltransferases of Neisseria meningitidis. Nucleic Acids Res. 2015;43:4150–4162. doi: 10.1093/nar/gkv219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stern A, Sorek R. The phage-host arms race: Shaping the evolution of microbes. BioEssays. 2011;33:43–51. doi: 10.1002/bies.201000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts RJ, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loenen WA, Dryden DT, Raleigh EA, Wilson GG. Type I restriction enzymes and their relatives. Nucleic Acids Res. 2014;42:20–44. doi: 10.1093/nar/gkt847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arber W, Linn S. DNA modification and restriction. Annu Rev Biochem. 1969;38:467–500. doi: 10.1146/annurev.bi.38.070169.002343. [DOI] [PubMed] [Google Scholar]
- 8.Bickle T, Arber W. Host-controlled restriction and modification of filamentous 1- and F-specific bacteriophages. Virology. 1969;39:605–607. doi: 10.1016/0042-6822(69)90112-3. [DOI] [PubMed] [Google Scholar]
- 9.Meselson M, Yuan R. DNA restriction enzyme from E. coli. Nature. 1968;217:1110–1114. doi: 10.1038/2171110a0. [DOI] [PubMed] [Google Scholar]
- 10.Linn S, Arber W. Host specificity of DNA produced by Escherichia coli, X. In vitro restriction of phage fd replicative form. Proc Natl Acad Sci USA. 1968;59:1300–1306. doi: 10.1073/pnas.59.4.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kennaway CK, et al. Structure and operation of the DNA-translocating type I DNA restriction enzymes. Genes Dev. 2012;26:92–104. doi: 10.1101/gad.179085.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan RD, et al. Novel m4C modification in type I restriction-modification systems. Nucleic Acids Res. 2016;44:9413–9425. doi: 10.1093/nar/gkw743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kennaway CK, et al. The structure of M.EcoKI Type I DNA methyltransferase with a DNA mimic antirestriction protein. Nucleic Acids Res. 2009;37:762–770. doi: 10.1093/nar/gkn988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abadjieva A, Webb M, Patel J, Zinkevich V, Firman K. Deletions within the DNA recognition subunit of M.EcoR124I that identify a region involved in protein-protein interactions between HsdS and HsdM. J Mol Biol. 1994;241:35–43. doi: 10.1006/jmbi.1994.1471. [DOI] [PubMed] [Google Scholar]
- 15.Calisto BM, et al. Crystal structure of a putative type I restriction-modification S subunit from Mycoplasma genitalium. J Mol Biol. 2005;351:749–762. doi: 10.1016/j.jmb.2005.06.050. [DOI] [PubMed] [Google Scholar]
- 16.Kim JS, et al. Crystal structure of DNA sequence specificity subunit of a type I restriction-modification enzyme and its functional implications. Proc Natl Acad Sci USA. 2005;102:3248–3253. doi: 10.1073/pnas.0409851102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Obarska A, et al. Structural model for the multisubunit Type IC restriction-modification DNA methyltransferase M.EcoR124I in complex with DNA. Nucleic Acids Res. 2006;34:1992–2005. doi: 10.1093/nar/gkl132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lapkouski M, et al. Structure of the motor subunit of type I restriction-modification complex EcoR124I. Nat Struct Mol Biol. 2009;16:94–95. doi: 10.1038/nsmb.1523. [DOI] [PubMed] [Google Scholar]
- 19.Uyen NT, et al. The fragment structure of a putative HsdR subunit of a type I restriction enzyme from Vibrio vulnificus YJ016: Implications for DNA restriction and translocation activity. Nucleic Acids Res. 2009;37:6960–6969. doi: 10.1093/nar/gkp603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao P, Tang Q, An X, Yan X, Liang D. Structure of HsdS subunit from Thermoanaerobacter tengcongensis sheds lights on mechanism of dynamic opening and closing of type I methyltransferase. PLoS One. 2011;6:e17346. doi: 10.1371/journal.pone.0017346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dryden DT, Cooper LP, Thorpe PH, Byron O. The in vitro assembly of the EcoKI type I DNA restriction/modification enzyme and its in vivo implications. Biochemistry. 1997;36:1065–1076. doi: 10.1021/bi9619435. [DOI] [PubMed] [Google Scholar]
- 22.Kulkarni M, Nirwan N, van Aelst K, Szczelkun MD, Saikrishnan K. Structural insights into DNA sequence recognition by Type ISP restriction-modification enzymes. Nucleic Acids Res. 2016;44:4396–4408. doi: 10.1093/nar/gkw154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chand MK, et al. Translocation-coupled DNA cleavage by the Type ISP restriction-modification enzymes. Nat Chem Biol. 2015;11:870–877. doi: 10.1038/nchembio.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Callahan SJ, et al. Structure of type IIL restriction-modification enzyme MmeI in complex with DNA has implications for engineering new specificities. PLoS Biol. 2016;14:e1002442. doi: 10.1371/journal.pbio.1002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray NE. Type I restriction systems: Sophisticated molecular machines (a legacy of Bertani and Weigle) Microbiol Mol Biol Rev. 2000;64:412–434. doi: 10.1128/mmbr.64.2.412-434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loenen WA. Tracking EcoKI and DNA fifty years on: A golden story full of surprises. Nucleic Acids Res. 2003;31:7059–7069. doi: 10.1093/nar/gkg944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tock MR, Dryden DT. The biology of restriction and anti-restriction. Curr Opin Microbiol. 2005;8:466–472. doi: 10.1016/j.mib.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.Park SY, et al. Structural characterization of a modification subunit of a putative type I restriction enzyme from Vibrio vulnificus YJ016. Acta Crystallogr D Biol Crystallogr. 2012;68:1570–1577. doi: 10.1107/S0907444912038826. [DOI] [PubMed] [Google Scholar]
- 30.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams PD, et al. PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]