Since the start of this century, a handful of research groups have pursued the synthesis and large-scale engineering of genomes. Work on synthetic genomes has seen the field scale-up from the full synthesis of the small poliovirus genome (2002) (1), to a complete working synthetic bacterial genome (2010) (2), and more recently to the construction and validation of multiple rewritten eukaryote chromosomes for the model organism Saccharomyces cerevisiae (2014, 2017) (3–8). The costs and time-scales for assembling entire bacterial genomes and eukaryotic chromosomes mean that synthetic genome engineering is not yet a routine approach to manipulating cells for research or biotechnology. However, by stepping down a scale from bacteria to viruses, opportunities quickly arise, even for those viruses with comparatively large genomes, like the double-stranded DNA herpes simplex virus (HSV) type 1 genome, over 150 kb in length. In PNAS, Oldfield et al. (9) engineer the HSV KOS strain genome, leveraging synthetic genomic cloning approaches to rapidly construct HSV variants with combinatorial mutations for functional evaluation.
Large-scale genomic engineering has been achieved by a handful of groups taking different approaches, but broadly the strategies employed fall into two categories: multiplexed editing and hierarchical assembly. For editing, new technologies, such as multiplex automated genome engineering-based targeted mutation (10) and the new genome editing tools of CRISPR-Cas9 allow existing genomes to be extensively modified toward a target sequence over several generations within their host cells (11). This can be an efficient approach if the cell grows fast and is easy to manipulate with molecular biology methods. For the alternative hierarchical assembly strategy, a designed or modified target genome sequence is instead put together gradually from smaller subgenomic fragments that are linked together by various DNA assembly methods. Depending on the size of the genome or chromosome being built, this may require many rounds of assembly, as the typical starting material for DNA assembly projects is almost always fragments of DNA smaller than 15 kb, with these obtained either from commercial synthesis or from PCR amplification of natural DNA regions.
While the assembly strategy is typically more costly and time-consuming than the editing strategy, it allows for many more design changes throughout the genome, including large-scale rearrangements, and its efficiency is not determined by our ability to work with the target organism. So long as the target organism can be transformed with the assembled DNA, then the rest of the work can be done in model organisms that grow rapidly. Indeed, over the last decade a typical path for assembly-based synthetic genomics has emerged, where starting DNA fragments from 1 to 15 kb are first assembled by in vitro reactions into bigger pieces (10–100 kb) using Escherichia coli as the initial host that accepts and amplifies successful assembly products. The larger assemblies from this first round are then assembled into chromosome-scale pieces using S. cerevisiae as the host and exploiting yeast’s remarkable talent for accurate large-scale homologous recombination.
Through this route in 2010, the J. Craig Venter Institute constructed the first completely synthetic bacterial genome (2). It was extracted from its yeast host and used to transform Mycoplasma cells, resulting in a bacteria growing and dividing with the accepted synthetic genome. In 2016, the same strategy allowed the Venter Institute team to construct a working, rationally reduced Mycoplasma genome with large-scale changes from the natural sequence, including removal of hundreds of genes (12). The many changes in this new genome were facilitated not just by the DNA being synthesized, but also by the fact that the synthetic DNA was assembled and hosted in yeast. While in the yeast host cells, the synthetic genome can be extensively modified due to the ease and high efficiency of recombination and genome-editing methods in S. cerevisiae. Genome assemblies hosted in yeast are effectively still “in the workshop” and if desired can be customized further as and when is needed.
Oldfield et al. (9) used a hierarchical assembly strategy in their work, differing in one important respect from published bottom-up synthetic genomics projects. Rather than synthesizing from scratch, Oldfield et al. first isolated 11 overlapping subsections of the HSV genome as amplification fragments, each ∼14 kb in length, and individually cloned these into yeast vectors using transformation-associated recombination cloning with S. cerevisiae. Next Oldfield et al. reconstructed the complete genome in yeast using either the wild-type or specifically mutated cloned fragments (Fig. 1). Encoding both yeast and E. coli parts for selection, replication, and segregation on the vectors enabled shuttling of the constructs between these two organisms and leveraged their individual strengths: efficient homologous recombination and CRISPR-Cas9 editing in yeast, and rapid amplification and easy isolation of high quantities of DNA from E. coli. In abstraction, this work provides a blueprint for the deconstruction and reconstruction in model organisms of virtually any extant DNA sequence with the additional capability to rapidly generate engineered variants.
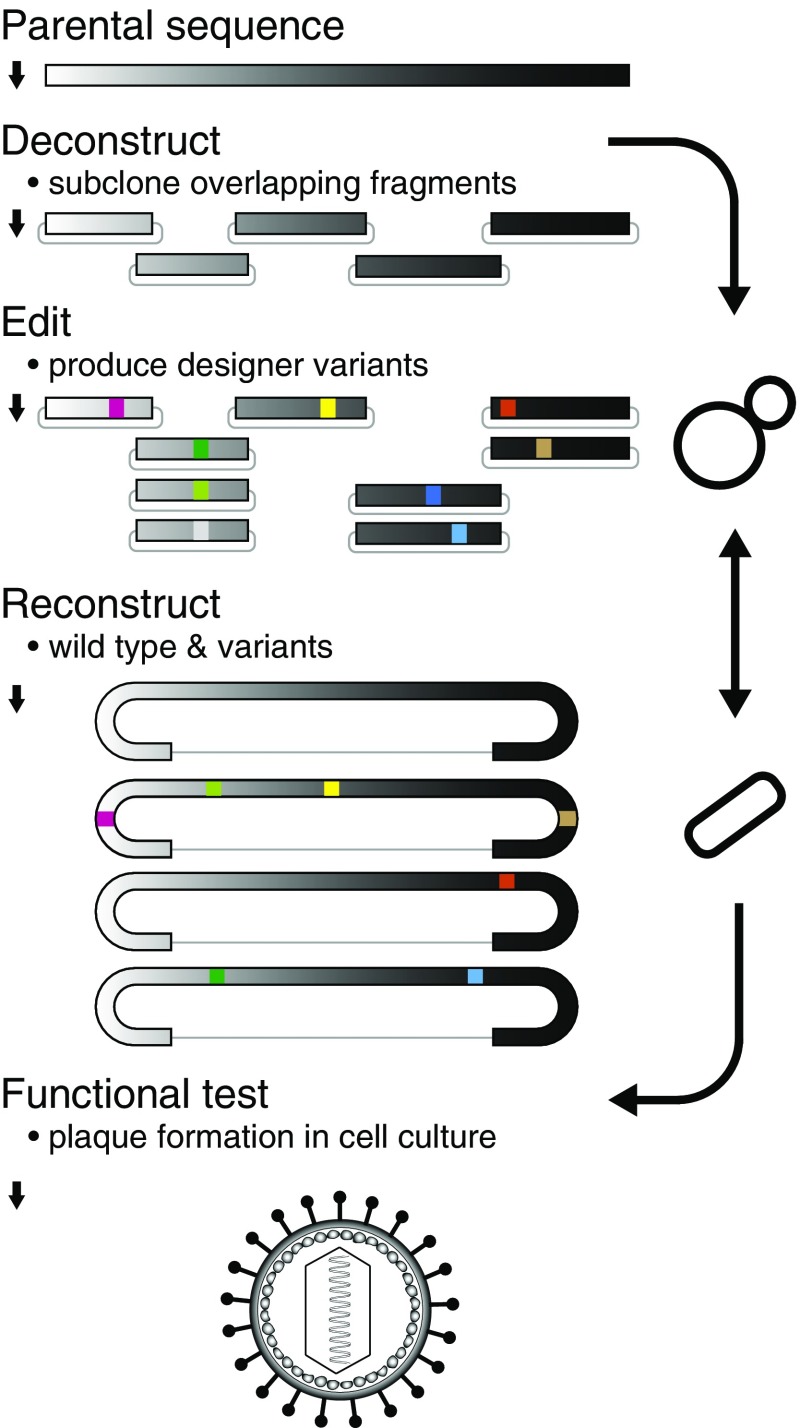
Fig. 1.
A synthetic genomic assembly workflow for DNA viruses. A double-stranded DNA viral genome like HSV-1 can be produced synthetically using a generic workflow. Deconstruct: the parental genome sequence is subcloned into fragments with terminal sequence homology (overlaps), using a vector that shuttles between yeast and E. coli. Edit: specific variants are introduced into the fragments using editing tools available in yeast or E. coli. Reconstruct: wild-type and variant genomes are reassembled by mixing and matching subcloned parts. Functional test: infectivity of synthetic viral genomes is tested in cell culture.
Having assembled the complete and several modified versions of the HSV-1 genome, Oldfield et al. (9) then tested their functionality by purifying the constructed DNA and transfecting it into Vero mammalian cells. The complete reconstructed viral genome triggered plaque formation and quickly produced viable HSV-1 viruses capable of infection within the cell culture. Infection was also seen from all versions of the genome that had deletions of individual genes encoding the tegument proteins that surround the virus capsid. However, most versions of the genome that had two or more of these tegument genes deleted in combination were not able to spread. This confirmed the known redundancy of these genes and provides valuable new information on their genetic interactions and their role in mammalian cell infection.
That Oldfield et al. (9) have chosen to engineer a human pathogen in this work raises questions regarding the potential dual use of synthetic genomic approaches and prompts deep consideration of the benefits and risks that result from developing a method to engineer infectious viruses. Importantly, the lack of synthesis in the workflow presented by Oldfield et al. makes it possible to circumvent the biosecurity efforts of the International Gene Synthesis Consortium (www.genesynthesisconsortium.org), who screen gene synthesis orders and aim to prevent misuse by restricting production of sequences associated with dangerous pathogens. Although this screening mechanism should keep in check the de novo synthesis of viral genomes, the system is clearly not yet foolproof. Recently it was reported that the ∼212-kb horsepox virus was synthesized from scratch using mail-order synthetic DNA (13). While horsepox virus is not known to harm humans or agriculture, its relative smallpox, which was declared eradicated in 1980, causes horrific disease; a similar de novo synthesis strategy could readily be employed to construct the smallpox genome, likely to cost around ∼$100K USD for DNA (based on today’s gene synthesis prices), although requiring significant scientific expertise. Together with the work of Oldfield et al. (9), there now exist clear pathways to construct both extinct and extant viruses using synthetic genomic technologies.
Notably, Oldfield et al. (9) include a measured critical discussion of dual-use concerns in their work, arguing that the possible negative uses of this technology are not as realistic as other options already available to potential bad actors. The authors also stress that this approach to viral research is likely to yield many positive benefits. Being able to comprehensively modify HSV-1 and other DNA viral genomes could have wide-ranging applications, such as in the accelerated development of vaccines or for developing oncolytic viruses as cancer therapies, designing them to selectively kill tumorgenic cells. However, most importantly, it gives a new tool for probing knowledge of viral and genome biology. For HSV type 1, this is important as it is the causative agent of a number of human pathologies, ranging in severity from cold sores to encephalitic infection (14). In the past, studying the virus at the genomic level has been hampered by its size and difficulty to manipulate in vitro. The synthetic genomics cloning strategy here enabled Oldfield et al. (9) to fluorescently tag individual HSV-1 genes, as well as analyze the interactions between genes of the HSV-1 genome. These initial studies with their method provide an important proof-of-concept for more complex combinatorially engineered versions of the HSV-1 genome in the future.
While the handful of high-profile synthetic genomics projects undertaken so far have been mostly motivated by a desire to push the boundaries of synthetic biology and take early advantage of the falling cost of synthetic DNA, the work of Oldfield et al. (9) demonstrates that the approach can also be employed as a method to probe genome function, and without having to rely on costly synthetic DNA as well. Enabled by the new methods for large-scale DNA assembly and efficient genome editing in model organisms, synthetic genomics approaches offer a new tool for bioscience research.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
See companion article on page E8885.
References
- 1.Cello J, Paul AV, Wimmer E. Chemical synthesis of poliovirus cDNA: Generation of infectious virus in the absence of natural template. Science. 2002;297:1016–1018. doi: 10.1126/science.1072266. [DOI] [PubMed] [Google Scholar]
- 2.Gibson DG, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell LA, et al. Synthesis, debugging, and effects of synthetic chromosome consolidation: synVI and beyond. Science. 2017;355:eaaf4831. doi: 10.1126/science.aaf4831. [DOI] [PubMed] [Google Scholar]
- 4.Shen Y, et al. Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome. Science. 2017;355:eaaf4791. doi: 10.1126/science.aaf4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu Y, et al. Bug mapping and fitness testing of chemically synthesized chromosome X. Science. 2017;355:eaaf4706. doi: 10.1126/science.aaf4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie ZX, et al. “Perfect” designer chromosome V and behavior of a ring derivative. Science. 2017;355:eaaf4704. doi: 10.1126/science.aaf4704. [DOI] [PubMed] [Google Scholar]
- 7.Zhang W, et al. Engineering the ribosomal DNA in a megabase synthetic chromosome. Science. 2017;355:eaaf3981. doi: 10.1126/science.aaf3981. [DOI] [PubMed] [Google Scholar]
- 8.Annaluru N, et al. Total synthesis of a functional designer eukaryotic chromosome. Science. 2014;344:55–58. doi: 10.1126/science.1249252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oldfield LM, et al. Genome-wide engineering of an infectious clone of herpes simplex virus type 1 using synthetic genomic assembly methods. Proc Natl Acad Sci USA. 2017;114:E8885–E8894. doi: 10.1073/pnas.1700534114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isaacs FJ, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang L, et al. Genome-wide inactivation of porcine endogenous retroviruses (PERVs) Science. 2015;350:1101–1104. doi: 10.1126/science.aad1191. [DOI] [PubMed] [Google Scholar]
- 12.Hutchison CA, 3rd, et al. Design and synthesis of a minimal bacterial genome. Science. 2016;351:aad6253. doi: 10.1126/science.aad6253. [DOI] [PubMed] [Google Scholar]
- 13.Koblentz GD. The de novo synthesis of horsepox virus: Implications for biosecurity and recommendations for preventing the reemergence of smallpox. Health Secur. August 24, 2017 doi: 10.1089/hs.2017.0061. [DOI] [PubMed] [Google Scholar]
- 14.Heming JD, Conway JF, Homa FL. Herpesvirus capsid assembly and DNA packaging. Adv Anat Embryol Cell Biol. 2017;223:119–142. doi: 10.1007/978-3-319-53168-7_6. [DOI] [PMC free article] [PubMed] [Google Scholar]