Abstract
The extracellular matrix protein Emp of Staphylococcus aureus is a secreted adhesin that mediates interactions between the bacterial surface and extracellular host structures. However, its structure and role in staphylococcal pathogenesis remain unknown. Using multidisciplinary approaches, including circular dichroism (CD) and Fourier transform infrared (FTIR) spectroscopy, transmission electron (TEM) and immunogold transmission electron microscopy, functional ELISA assays and in silico techniques, we characterized the Emp protein. We demonstrated that Emp and its truncated forms bind to suprastructures in human skin, cartilage or bone, among which binding activity seems to be higher for skin compounds. The binding domain is located in the C-terminal part of the protein. CD spectroscopy revealed high contents of β-sheets (39.58%) and natively disordered structures (41.2%), and TEM suggested a fibrous structure consisting of Emp polymers. The N-terminus seems to be essential for polymerization. Due to the uncommonly high histidine content, we suggest that Emp represents a novel type of histidine-rich protein sharing structural similarities to leucine-rich repeats proteins as predicted by the I-TASSER algorithm. These new findings suggest a role of Emp in infections of deeper tissue and open new possibilities for the development of novel therapeutic strategies.
Introduction
Staphylococcus aureus is an important human pathogen that can cause serious invasive and persistent diseases, such as osteomyelitis or deep soft tissue infections1. Adhesion to the extracellular matrix of the host tissue is a critical step in S. aureus pathogenesis. This process is essentially mediated by adhesins, which are staphylococcal proteins that undergo strong interactions with the extracellular matrix. Adhesins can be divided into proteins that are covalently bound to bacterial cell wall peptidoglycans (microbial surface components recognizing adhesive matrix molecules, MSCRAMMs)2–5 and proteins that are only secreted but that re-bind to the bacterial cell surface (secretable expanded repertoire adhesive molecules, SERAMs)6. Fibronectin binding proteins (FnBPs) and collagen binding protein (Cna) belong to the group of MSCRAMMs, and their roles in bacterial adhesion to host structures and host cell invasion are well established3,7,8. The extracellular adhesion protein (Eap) and the extracellular matrix binding protein (Emp) are anchorless proteins and belong to the group of SERAMs9,10. Both proteins are essentially regulated by Sae, but their functions in bacterial adhesion remain unclear11. Whereas MSCRAMMs are mainly expressed in the exponential bacterial growth phase, Eap and Emp are expressed in the stationary growth phase and could therefore play roles in later infection processes10. Eap and Emp are not only found in bacterial supernatants but also efficiently bind to the bacterial wall via a neutral phosphatase or other unknown bacterial surface molecules12. PCR analysis revealed that the eap and emp genes are present in almost all S. aureus isolates, whereas they have not been found in any S. epidermidis isolates10,13.
The extracellular matrix (ECM) is composed of a variety of proteins and proteoglycans and glycoproteins that are secreted locally and assembled into an organized meshwork in close association with the surface of the cell that produced them14. Emp and Eap can interact with various ECM proteins, such as fibronectin, fibrinogen and some collagens9,10,15. However, Hansen et al. have demonstrated that the binding of Eap and Emp to extracellular proteins is strongly dependent on their incorporation into suprastructures16. For example, there is a striking binding specificity of Eap to different collagen types and structures. Collagen I, a major component of banded fibrils in skin, is a binding substrate in monomolecular form, but it is almost unrecognized by Eap when incorporated into banded fibrils16. This phenomenon can be explained by the aggregation of matrix macromolecules in situ into suprastructures such as fibrils, microfibrils, filaments, and networks. Furthermore, matrix suprastructures are usually composed of several molecular species, and their composition can be complexed with non-collagenous macromolecules attached to some fibril surfaces, forming a defined three-dimensional structure17. Conversely, the structure and function of Emp remain unknown. Surface plasmon resonance (SPR) spectroscopy has shown that Emp binds to different ECM components10. However, the structural adaptations and biochemical features of Emp that allow specific interactions with so many different ligands remain largely unexplored10. Kalinka et al. observed an increase in Emp expression in chronic osteomyelitis staphylococcal isolates18. Moreover, Emp is required for abscess formation and persistence19. Additionally, Emp is associated with endovascular disease6 and plays an important role in low-iron-induced biofilm formation20.
The conferring gene emp (1,023 nucleotides) encodes an extracellular mature protein with a calculated molecular mass of 35.5 kDa10. We blasted the Emp amino acid sequence against the database to identify relatives and investigated its structural properties by circular dichroism (CD), Fourier transform infrared (FTIR) spectroscopy, and transmission electron microscopy (TEM). The polydispersity was analysed using Nanosizer measurements. Furthermore, the role of individual Emp fragments (Fig. 1) and their interactions with suprastructures in human skin, cartilage or bone were studied in in vitro binding assays. Finally, a possible protein structure was predicted by I-TASSER. These results provide the first structural characterization of Emp, contributing to our understanding of the role of this protein in staphylococcal pathogenesis.
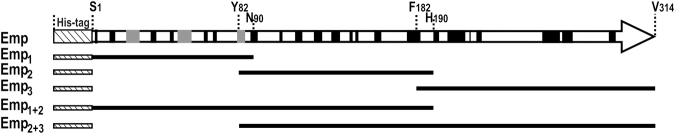
Figure 1.
Schematic illustration of the cloning strategy used for the Emp forms of S. aureus and the predicted secondary structure. The arrow indicates the primary structure of the His-tagged fusion products of the full-length Emp protein of 314 amino acids. The His-tag (shaded box) was excluded from continuous numbering of the protein sequence. The predicted secondary structure elements within the arrow are indicated as filled boxes: black for β-sheets; grey for α-helices. The black lines below the arrow indicate the different truncated Emp fragments, which all were fused to the His-tag (grey lines).
Results and Discussion
Emp is not related to any other protein family
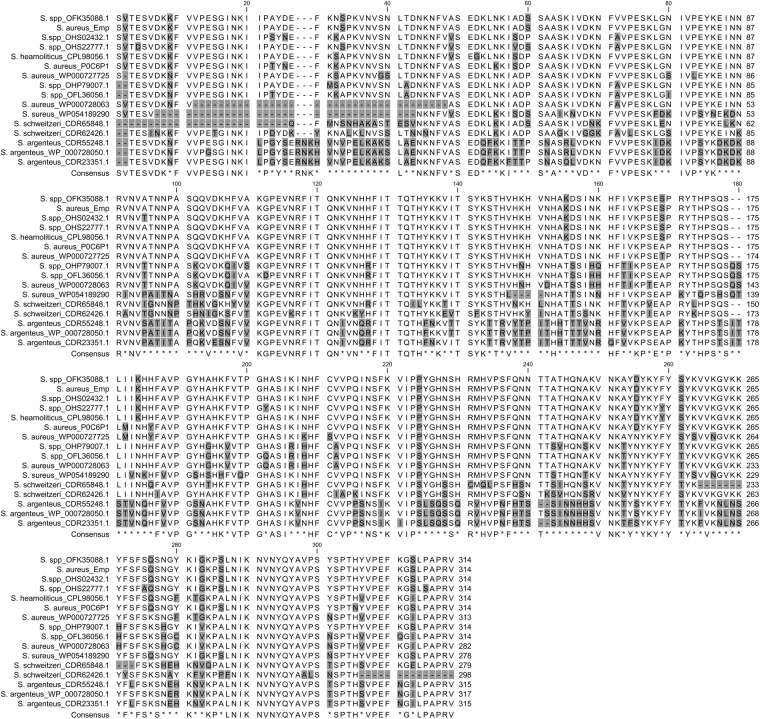
The blastp analysis of the Emp amino acid sequence did not reveal any relationship to other known or hypothetical proteins in the database, except for other Emp forms of a few Staphylococcus spp. (Fig. 2). Hundreds of Emp sequences were found in the protein database within various S. aureus strains, and the sequence identity to our Emp protein was almost higher than 90% (data not shown). Excluding the S. aureus from the blastp analysis, additional Emp proteins were found in other Staphylococcus species. High amino acid sequence similarities were also found to various Staphylococcus spp. strains (identity and positivity 90–100%) and S. heamoliticus (identity and positivity 92%). Whereas Emp proteins of S. schweitzeri and S. argenteus showed lower sequence similarities than S. aureus (identity 62–75% / positivity 74–84%, and identity 61–62% / positivity 73–75%, respectively), Emp of S. argenteus showed the strongest differences in sequence with the longest phylogenetic distance from the other species (Supplementary Figure S1). Our results indicate that Emp or analogous proteins are not present in poorly invasive and low-pathogenic staphylococci species like S. epidermidis21.
Figure 2.
Multiple alignment of Emp proteins from different Staphylococcus species. Only selected S. aureus Emp proteins that showed differences in their sequences were chosen. Similarly, only those Emp protein sequences of S. spp, S. haemolyticus, S. schweitzeri and S. argenteus were selected that showed different identity scores in the blastp analysis.
The most extraordinary characteristic of the Emp proteins is their high content of histidines. Based on multiple alignments, at least 8 strongly conserved histidines were found in all aligned species that were primarily located within the core region of the protein (approximately from residue 123 to 240) (Fig. 2). Additional histidine residues seemed to be conserved in S. aureus, S. heamoliticus, S. schweitzeri and related species (up to 7), whereas some other His residues were conserved in S. argenteus (up to 5), indicating possible differences in functionality or substrate specificity.
Histidine-rich proteins (HRPs) have been observed in many prokaryotic and eukaryotic organisms but are not phylogenetically related to each other and do not form a structural superfamily. Due to the chemical properties of histidine, which play multiple roles in inter- and intramolecular protein interactions, they also exhibit different functions22. The HRPs known as filaggrins in mammals interact with the keratin cytoskeleton of the epidermis to form insoluble microfibers in the stratum corneum responsible for skin barrier functions23. In Plasmodium falciparum, the HRP-2 protein is a virulence factor that binds to the cytoskeletal components actin and phosphatidylinositol 4,5-bisphosphate24. Other HRPs, such as the Hpn of Helicobacter pylori, are responsible for Ni2+ storage and detoxification25. Moreover, human histidine-rich glycoproteins (HRGs) interact with many ligands, such as Zn2+, heme, fibrinogen, IgG or the complement system. HRG is involved in different processes associated with tissue injury and tumor growth to regulate cell adhesion and migration, fibrinolysis and coagulation, and the immune response26. Similarly, Emp binds to proteins such as fibronectin, fibrinogen and some collagens9,10,15; therefore, we suggest that Emp is a novel bacterial HRP.
The Emp was divided in three different constructs to be analysed (Fig. 1). The Emp1 is localized at the N-terminal, followed by Emp2 and Emp3 at the C-terminal. Combination of Emp1 and Emp2 was named Emp1+2 as well as the combination between Emp2 and Emp3 was named Emp2+3. The unique strongly conserved methionine (M229) and cysteine (C208) residues are located at the C-terminus of Emp3. For all Emp fragments, high isoelectric points (IEPs) were predicted, with an IEP of 9.99 for full-length EmpFL, an IEP of 8.89 for Emp1, an IEP of 10.38 for Emp2 and an IEP of 10.04 for Emp3. The high IEPs of fragments Emp2 and Emp3 (positively charged) suggest that those fragments might be primarily involved in interactions with negatively charged compounds or polymers.
Based on these protein sequence alignments, the most conserved region of Emp seems to be located at the C-terminus (residues 284 to 314), indicating that the C-terminus might also play a crucial role in Emp function, folding or polymerization.
Emp binds preferentially to skin and cartilage ECMs
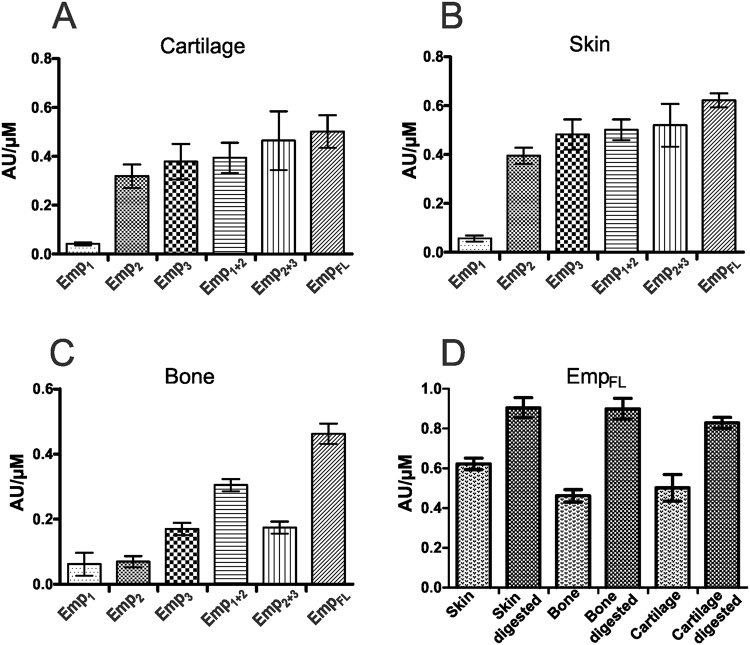
To investigate the capacity of Emp to interact with host ECMs, the soluble recombinant protein forms EmpFL, Emp1, Emp2, Emp3, Emp2+3 and Emp1+2 were assessed for binding to human cartilage, skin and bone, which were homogenized from the target tissues and immobilized on ELISA plates as it was described in material and methods. The EmpFL and all Emp forms, excluding Emp1, bound to skin and cartilage in a similar manner (Fig. 3A and B), whereas bone material was preferably bound by EmpFL, followed by Emp1+2 and, to similar extents, Emp3 and Emp2+3. Emp1 and Emp2 showed the lowest binding activity to the bone material (Fig. 3C). The specificity of the target might therefore depend on the C-terminal sequence because variations seemed to modulate the binding properties. Interactions between Emp and ECMs were higher with the C-terminal part of the protein (Emp2 and Emp3) but were almost absent within the N-terminus (Emp1), suggesting that Emp might consist of two domains: the C-terminal domain, which is responsible for selective binding to target structures; and the N-terminal domain, the function of which is not related to target interactions.
Figure 3.
The binding specificities of EmpFL and Emp fragments to different ECMs determined by ELISA assays: cartilage (A), skin (B), bone (C). (D) Binding of EmpFL to enzymatically digested ECMs in comparison to non-digested ECMs. The binding specificity was expressed as absorption units (AU) per µM protein. The ECMs were immobilized on ELISA plates at saturated concentrations and were exposed to 1 µM concentrations of Emp forms.
The binding activity of EmpFL was compared between enzymatically digested and non-digested suprastructures (Fig. 3D). Thereby, Emp bound more efficiently to digested material, which offers more binding sites following the enzymatic release of matrix components.
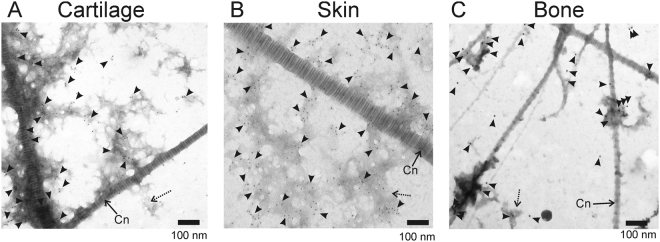
The specific binding of Emp to ECMs was analysed by immunogold electron microscopy (Fig. 4). The ECMs were and immobilized to the formvar/carbon coated nickel grids, enzymatically digested and exposed to EmpFL. As controls, ECMs were treated with PBS. Subsequently, the grids were incubated with polyclonal rabbit antibodies against Emp and anti-rabbit IgG conjugated to colloidal gold particles. Non-specific binding was not detected in negative controls without Emp or anti-Emp treatment (data not shown). EmpFL adhered to all tested matrices but was primarily found in the enzymatic-treated regions because more epitopes are available (indicated by arrowheads in Fig. 4). These experiments also showed that more Emp particles were found in cartilage and skin matrix compartments than in the bone matrix.
Figure 4.
Immunogold electron microscopic images of the digested ECMs exposed to EmpFL. The Emp proteins are visualized by antibodies against Emp and secondary gold-labelled antibodies visible as dark spherical particles (indicated by arrowheads) located predominantly in the destroyed ECM structural formations (dashed arrows). The fibers of extracellular collagen (Cn) are indicated by arrows. The scale bars indicate 100 nm.
These results suggested that Emp preferably interacts with matrix components usually buried within matrix suprastructures which becomes available after proteolytic treatment. This situation might take place during infection, when cytotoxic factors of pathogens and the host immune system damages most host matrices (i.e., inflammation) and internal matrix components are exposed. Emp might be used by S. aureus to bind to these structures for internalization into different host cells and tissues27–29.
Emp binds host tissue components
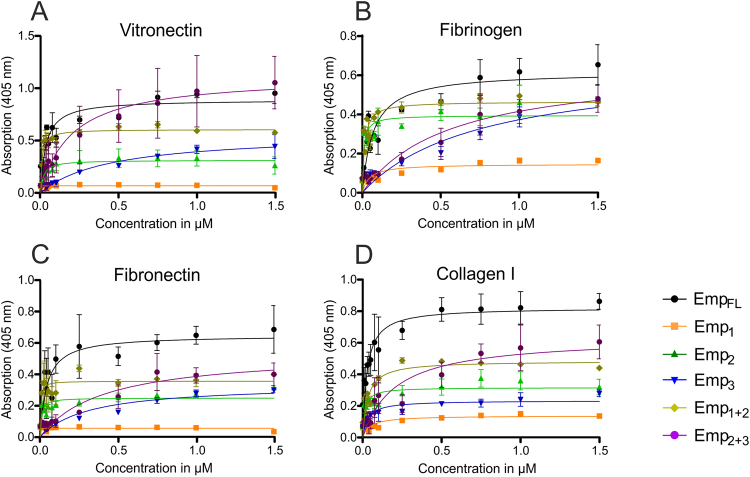
S. aureus adheres to ECM substrates and eukaryotic cells to initiate invasive infection15,30. Each ECM contains a special arrangement of components like fibrinogen (Fg), fibronectin (Fn), vitronectin (Vn) and collagen I (Cn I), which form a three-dimensional structural complex30. ELISA was performed to study the specific interactions of the Emp forms and either Fg, Fn, Vn or Cn I. All matrix components were immobilized on ELISA plates and incubated with increasing concentrations of EmpFL and the different Emp forms. The mean equilibrium dissociation constant (KD), which represents a measure of affinity, was calculated from the steady-state binding curves of the different Emp forms at different ligand concentrations (Fig. 5). Thereby, the lower the KD value indicates a higher affinity.
Figure 5.
Binding kinetics of the Emp forms to different matrix components: vitronectin (A), fibrinogen (B), fibronectin (C) and collagen I (D). The legend (in D) corresponds to all diagrams. The curves represent the average values with the standard deviation of at least three independent measurements.
EmpFL and all Emp forms, except for Emp1, bound to all components in a concentration-dependent manner (Fig. 5). Although the KD values for Emp1 binding to Fg and Cn I could be calculated, it remained an artefact of the assay because the maximal signal was very low compared with the other Emp forms, indicating that Emp did not interact with the compounds. The affinity of the other Emp forms varied strongly depending on the Emp fragment and the substrate.
The KD values of EmpFL were 27 nM for Cn I, 34 nM for Vn, 43 nM for Fn, and 70 nM for Fg (Table 1). In previous surface plasmon resonance (SPR) experiments, similar results were obtained for these ligands10. However, the protein was immobilized on the chip surface and the ligands were flushed through the surface. The KD values obtained were: 21 nM for Fn and 91 nM for Fg, but even much lower KD values were observed for Vn (0.122 nM however, a molecular mass of 1200 kDa was assumed that might cause the difference)10. The KD for Cn I has not been calculated due to unknown molecular mass of the Cn-polymer but the SPR sensograms indicated also high affinity to EmpFL. These results suggest stronger binding of Emp to Cn I and Vn than to Fg and Fn. According to these results, Emp has an affinity similar to Eap for host components of ECMs16,31. To investigate the region responsible for the binding activity of Emp, the different Emp forms were evaluated. Emp2 and Emp1+2 showed the lowest KD values, indicating the highest affinity to all tested host components with a tendency towards a higher affinity to Fg and Fn. Because Emp1 showed nearly no activity to the matrix components, it can be assumed that most of the amino acid residues involved in the binding of Emp to host components must be located in the central part between residues 100 to 200 (within Emp2). The C-terminal part of Emp (Emp3) showed relatively high KD values for all ligands but not for Cn I, suggesting that this Emp part do not participate in the ligand-binding process. Interestingly, the presence of the C-terminus reduced the affinity of Emp2+3 for all ligands, suggesting that the C-terminus might be even necessary for the dissociation of Emp from host components.
Table 1.
Binding affinities of the Emp protein forms to various substrates, expressed as dissociation constants (KD) in nM.
| Substrate | EmpFL | Emp1 | Emp2 | Emp3 | Emp1+2 | Emp2+3 |
|---|---|---|---|---|---|---|
| Vn | 34 ± 6 | / | 18 ± 4 | 449 ± 103 | 9 ± 3 | 200 ± 50 |
| Fg | 70 ± 1 | 44 ± 12 | 9 ± 2.6 | 756 ± 224 | 14 ± 3 | 556 ± 115 |
| Fn | 43 ± 11 | / | 8 ± 3 | 311 ± 79 | 3 ± 2 | 404 ± 95 |
| Cn l | 27 ± 5 | 47 ± 12 | 13 ± 6 | 32 ± 7 | 31 ± 5 | 207 ± 44 |
/ = no binding.
The secondary structure of Emp is dominated by β-sheets and natively disordered structures
The predicted secondary structure of Emp revealed a high proportion of β-sheets; only 3 α-helices have been predicted in the N-terminal fragment, Emp1. Most of the β-sheets seem to be separated by longer sequences that can be expected to form unstructured turns. To confirm the predicted secondary Emp structure, two methods have been applied that demand different light spectra and optical properties of the protein: FTIR and CD spectroscopy.
Using FTIR spectroscopy, the amide I bands in the IR spectra of EmpFL were compared to the amide I bands of concanavalin A (14 β-sheets), myoglobin (8 α-helices), lysozyme (28% α-helices, 12% β-sheets and 60% other structures) (Supplementary Figure S2). It is well accepted that the amide I band depends on the secondary structure of proteins and can be used to estimate secondary structures32,33. The amide I band of the Emp protein showed a higher maximum near 1655cm−1, a low wavenumber slope similar to lysozyme and an unusually extended high wavenumber shoulder. Thus, the spectra could not be reliably evaluated. The deconvolution routine provides reasonable results only if the reference proteins have similar structures to the unknown protein33,34. However, this is not the case for the Emp protein: the broad high wavenumber shoulder in its amide I band has not been observed in IR spectra of common reference proteins, indicating an unusual and unknown conformation.
Because the FTIR analysis remained unsatisfactory, CD spectroscopy was conducted. CD spectroscopy is based on the optical activity of chiral residues that differentially absorb circular polarized light depending on the circular polarization direction35. Because the specific peptide bonds absorb in near-ultraviolet (UV) light (approximately 180 nm to 240 nm), the experiments were performed in NaF-containing buffer instead of NaCl, which interferes with the signal.
Based on the CD spectra (Supplementary Figure S3), proportions of 39.58% of β-sheets and 41.2% of natively disordered structures, but only 4.8% of α-helices and 13.75% of α-turns, were predicted for EmpFL, which confirmed the in situ prediction. A more detailed evaluation of the CD data further indicated that most of the helical structures were rather distorted (αD = 4.58%) than regular (αR = 0.23%), while the β- sheets seemed to exhibit predominantly regular conformations (βR = 25.95% vs βD = 13.63%). As distorted helices, all helical structures of less than four residues are assigned36. The deconvolution offers additional information about the averaged lengths and segment numbers of the secondary structures. The estimated segment numbers were 1.25 per 100 residues for the α-helices and 6.78 per 100 residues for the β-sheets. The predicted average lengths per segment were 3.98 residues per α-helix and 5.88 residues per β-sheet. The normalized root mean square deviation (NRMSD) fit parameter, which is a measure of the difference between the experimental ellipticities and back-calculated spectra for the derived structure based on the reference databases, was only 0.49 due to the strong scattering of the signals below 200 nm. The noisy CD data below 200 nm can be a sign of protein aggregation suggesting that Emp forms a complex quaternary structure37.
The tertiary structure of Emp indicates a fibrous structure
The idea of a fibrous structure of Emp was supported by the observation that the attempt to purify mono-dispersed EmpFL for structural analyses by preparative size exclusion FPLC, using both Sephadex 75 and Sephadex 200 columns, failed because we could not separate single peaks (data not shown). The protein yielded a plateau that indicated a poly-dispersed solution of polymers of different lengths. It is also possible that the protein interacted with the matrix due its adhesive characteristics, hampering the separation.
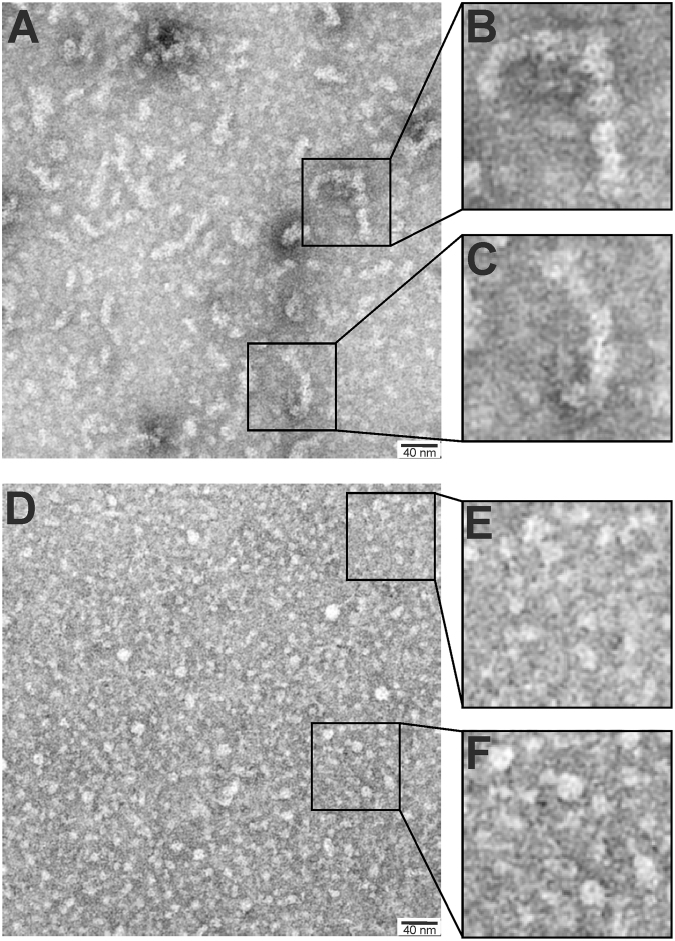
Thus, we analysed EmpFL and Emp2+3 by TEM. The EmpFL protein formed bead-like fibril structures of approximately 8 to 180 nm in thickness and 20 to 180 nm in length, which might be even longer due to coiling that was not clearly visible on the images (Fig. 6A). Zooming into such structures (Fig. 6B and C) revealed packages of smaller ‘donut’-like structures of approximately 8 nm in diameter (80 Å), which might represent the monomers. The N-truncated Emp2+3 protein did not form fibrils (Fig. 6D), but the ‘donut’-like structures could be observed in small packages of a few (2–4) units (Fig. 6E and F), indicating that the N-terminus is necessary for fibrillation.
Figure 6.
TEM images of EmpFL (A to C) and Emp2+3 (D to F). The proteins are visible as light structures.
The hydrodynamic diameter, as determined by the Nanosizer, was approximately 190 nm (±45 nm) for the main peak and 10.1 nm (±3.5 nm) for a low-molecular-mass fraction (Supplementary Figure S4). This finding supports the TEM results, but the polydispersity index of 0.62 ± 0.12 indicated a broad distribution of the Emp particles. Both parameters support the idea of a polymeric and fibrous tertiary structure of Emp.
The protein model of Emp revealed structural similarities to leucine-rich repeat proteins
Based on the Emp sequence, a possible protein structure of Emp was predicted using the I-TASSER on-line server. The analysis yielded four models, among which only one structure (Fig. 7) exhibited eligible scores: C-score = −2.95, TM-score = 0.38 ± 0.13 and root mean square deviation of atomic positions (RMSD) = 13.4 ± 4.1 Å. This predicted Emp structure (Fig. 7) showed a nearly 90° angled shape, with two arms of approximately 50 Å (5 nm, N-terminal) and 64 Å (6.4 nm, C-terminal) and a diagonal between both arms 73 Å (7.3 nm) in length. The primary structure contained short β-sheets and disordered structures, as predicted by CD spectroscopy. However due to the low C- and TM-scores, the overall folding of Emp remains unclear. The C-score is a confidence score for estimating the quality of predicted models by I-TASSER and typically ranges between −5 and 2; the higher the C-score, the higher is the confidence of the model. The template modelling (TM)-score and the RMSD are known standards for measuring structural similarity between two proteins with different tertiary structures. The TM-score ranges between 0 and 1, where 1 indicates a perfect match between two structures; the likelihood of similar folding increases when the TM-score exceeds 0.538.
Figure 7.
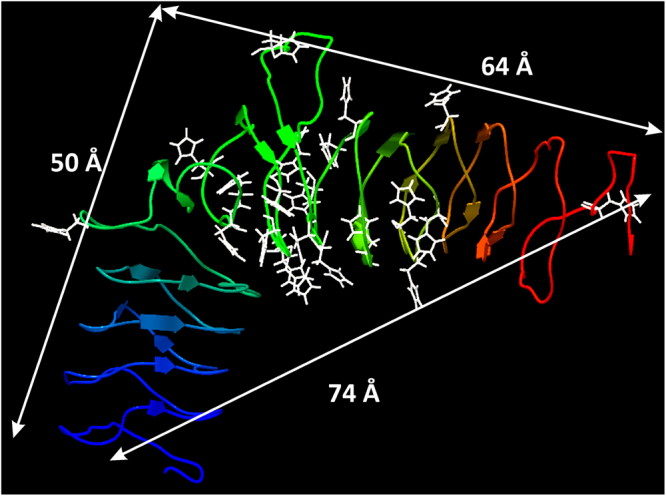
Predicted protein structure of Emp. The analysis was performed using the I-TASSER on-line server. The backbone is shown as a cartoon coloured from blue (N-terminus) to red (C-terminus); the histidine residues are represented as white sticks. The distances of the arms and the diagonal are indicated by arrows in Angstroms (Å).
The top ten identified structural analogues, all exhibit an arc or horseshoe shape, were primarily different leucine-rich repeat (LRR) proteins and some eukaryotic receptors containing LRR motifs (for details, see Supplementary Table S1). The LRR domains are often associated with innate immunity in higher eukaryotes and can be found in mammalian Toll-like receptors (TLRs) and NOD-like receptors (NLRs), which sense pathogen-associated molecular patterns (PAMPs)39. Interestingly, the known Emp proteins do not contain noticeable amounts of leucine and no leucine repeats such as LRRs, but most of the histidines seem to be located in the concave face of the Emp-arc and might be co-responsible for the predicted shape. Histidines are known to be involved in interactions with different ligands, most often with cationic metals. However, due to very low C-scores, no eligible predictions can be made regarding the ligands based on the predicted structure.
Conclusions
Our results suggested similar behaviours for Emp and Eap. S. aureus has a greater capacity to adhere to host tissue during inflammation. However, the roles of both proteins seem to be related to the late stage of infection. During the early stage of infection, many staphylococcal virulence factors, e.g. the staphylococcal protein A (SpA), interact with immunoglobulins inhibiting the adaptive host immune response. Staphylococcal virulence factors have also been shown to recruit neutrophils simultaneously inducing their apoptosis that triggers damage to host tissue due to enzymatic and acidic degradation of the ECMs40,41. This might enable interactions of Eap and Emp with matrix compounds that facilitate the access to deeper tissue for S. aureus. Furthermore, Eap contributes to the anti-inflammatory functions by blocking adhesion receptors of the leukocytes required for inflammatory infiltration of infected tissues protecting S. aureus from killing by leucocytes42. Thus, disintegrating suprastructures of the ECM are less suitable for infiltration by leukocytes, the directional migration of which depends on specific signaling molecules exposed by intact ECM.
Emp exhibits higher expression in chronic osteomyelitis isolates18. Moreover, S. aureus requires Emp for staphylococcal accumulation which is the prerequisite for abscess formation and persistence in host tissues19. During infection, S. aureus induces the expression of proteolytic enzymes like V8, aureolysin to damage the ECM that favours the ability of S. aureus to develop deep tissue infections43,44. This mechanism facilitates the interaction between Emp and damaged ECM to promote S. aureus persistence (Fig. 8).
Figure 8.
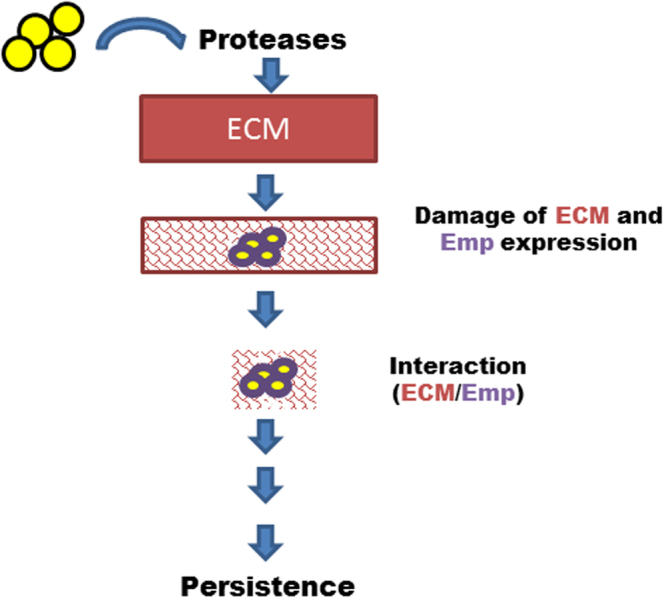
Hypothetical role of Emp during infection. S. aureus secretes proteases which disrupt the integrity of ECM. This process favours the exposition of epitopes and induces an interaction between Emp and ECM that contributes to staphylococcal persistence.
Emp seems to form fibrous structures at least 200 nm in length. The pili and fibers are typically found in bacterial adhesins45. Most fimbriae adhesins were found in Gram-negative bacteria, but many of them were also found in Gram-positive bacteria46. Pilin proteins serve a variety of functions, including adhesion to eukaryotic host cells, biofilm formation and horizontal gene transfer. Their polymerization permits extension and lengthening of the fiber, and their depolymerization results in pilus retraction47. However, the mechanisms known for fimbriae and pilins, which are multi-modular and involve several specific proteins in an energy-dependent manner, do not seem to apply to Emp. Emp correlates better with the protein ‘curli’ described for Gram-negative bacteria, which is also involved in adhesion to host components and invasion, cell aggregation and biofilm formation, and it may induce the inflammatory response48. In E. coli, the protein CsgA is also secreted into the environment and re-binds to its specific CsgB protein. Both proteins are encoded by the operon csgAB, which is divergently located in the csgDEFG operon; the second operon encodes additional proteins that are involved in curli assembling48. In contrast, Emp is encoded by a monocistronic gene and seems to directly re-bind to the S. aureus surface. A specific protein that is recognized by Emp has not been identified.
The adhesive properties of Emp are located in the C-terminus, which seems to be highly conserved within different staphylococcal species. The C-terminus also bears all of the conserved histidines that might be key residues for the adhesive characteristics of Emp. The N-terminus seems to be related to the polymerization of Emp to form a fibrillary structure. Emp showed higher adhesion to skin ECM than to cartilage or bone. These results suggest a role for Emp in wound infections, as previously described for Eap49. The fibre-like structure of Emp might also facilitate adherence of S. aureus to catheters and foreign material, resulting in biofilm and abscess formation19,20.The Emp monomer seems to exhibit a novel protein structure that is not related to any other known structure, but might be analogous to the LRR domains. We hypothesize that the high histidine content of the Emp-homologs could play a key role in their structure and function which is still elusive. It is also unclear how the fibre-like polymers are formed by these monomers and which lengths they may achieve in vivo. But this study provides the first insights into the structural properties of Emp, which might be a novel immune evasion mechanism of S. aureus and a potential target for the development of more efficient therapeutic alternatives to prevent or resolve staphylococcal infection.
Materials and Methods
Cloning of the Emp expression plasmids
The sequence encoding the full-length Emp protein (without the signal peptide) of S. aureus was amplified and cloned into the expression plasmid pQ30-UA (Qiagen, Hilden, Germany) to generate plasmid pQemp10. The truncated and recombined fragments of Emp (Emp1, Emp2, Emp3, Emp1+2, Emp2+3) were amplified by PCR using the sets of primers listed in Table 2. The PCR products were ligated into plasmid pQ30-UA (Qiagen) (Fig. 1) using T4 DNA Ligase (Thermo Fisher Scientific, Massachusetts, USA) according to the manufacturer’s protocol. The ligation mixture was transformed into freshly prepared competent E. coli TG1 cells, and transformants were selected on LB agar plates supplemented with 100 µg/ml ampicillin as described previously10. The plasmids were isolated using the QIAGEN Plasmid Mini Kit (Qiagen, Hilden, Germany) and were confirmed by sequencing at Eurofins Genomics (Ebersberg, Germany). Representative plasmids containing the different emp fragments were designated pQemp1, 2, 3, 1 + 2, and 2 + 3.
Table 2.
Primers used in this study.
| Name | Sequence 5′→3′ | Position relative to ATG of Emp |
|---|---|---|
| EmpFor1UA | GCATCAGTGACAGAGAGTGTTGACAAA | +76 |
| EmpRev1UA | ATTCACGCGATTATTGATTTCTTTGTA | +348 |
| EmpFor2UA | TACAAAGAAATCAATAATCGCGTGAAT | +322 |
| EmpRev2UA | ATGCGCGTGATATCCTGGAACTGCAAA | +648 |
| EmpFor3UA | TTTGCAGTTCCTGGATATCACGCGCAT | +622 |
| EmpRev3UA | TTATACTCGTGGTGCTGGTAAGCTACC | +1024 |
Expression and purification of recombinant Emp protein forms
His-tagged Emp forms were expressed for 24 h in 2 L of LB broth supplemented with 1% lactose. The cells were harvested by centrifugation at 5,600 rpm and 4 °C for 10 min. The pellets were resuspended in 10 ml of lysis buffer (8 M urea, 0.5 M NaCl, 10 mM β-mercaptoethanol, 5% glycerol, and 1% Triton X-100 in 50 mM sodium phosphate buffer, pH8.0) and were sonicated using a Bandelin Sonopuls GM 2200 or a Bandelin Electronic UW 2200 (Bandelin Electronic GmbH & Co. KG, Berlin, Germany) for 6 min on ice. After centrifugation at 13,000 rpm at 4 °C for 30 min, the supernatants were mixed with 3 ml of Ni-nitrilotriacetic acid (NTA) affinity resin (Qiagen, Hilden, Germany) and shaken at 33 rpm for 2 h at 4 °C on a Stuart™ SRT6 roller mixer (Sigma, Missouri, USA). Subsequently, the mixtures were applied to columns, and the flowthroughs were discarded. Each column was washed three times with 10 ml of lysis buffer containing 30 mM imidazole and eluted with 3 ml of lysis buffer containing 250 mM imidazole. The purity of the eluted proteins was analysed by SDS-electrophoresis on 12% acryl amid gels and western blot analysis as described previously10. The proteins were solubilized sequentially by dialysis against buffer A1 (4 M urea, 0.5 M NaCl, and 10 mM β-mercaptoethanol in 50 mM phosphate buffer, pH8.0), buffer B1 (1 M urea, 0.5 M NaCl, and 10 mM β-mercaptoethanol in 50 mM sodium phosphate buffer, pH8.0) and finally buffer C1 (0.25 M NaCl in 50 mM sodium phosphate buffer, pH8.0). The protein was quantified using a NanoDrop (PeqLab, Erlangen, Germany) at 280 nm by applying the specific extinction coefficient (Supplementary Table S2).
Sequence analysis
All sequence analyses were performed using CLC Main Workbench 7.0.2 (Qiagen). The secondary structure of the recombinant (His-tagged) Emp protein sequence was predicted using the Hidden Markov Model (HMM) provided by the CLC Main Workbench. Multiple alignments were performed by applying the Clustal Omega algorithm50 for multiple sequence alignments. Phylogenetic analysis was performed based on maximum likelihood phylogeny tree construction with 100 bootstrap replicates.
The Emp sequence was compared against the NCBI database using the BLAST platform and the blastp tool to identify related proteins.
Extraction of matrix suprastructures
Fragments of suprastructural aggregates from human skin, bone and articular cartilage were obtained as described previously16,18. Briefly, slices of cartilage were homogenized twice using a Polytron in 15 volumes of PBS (150 mM NaCl in 2 mM sodium phosphate buffer, pH7.4) containing a mixture of protease inhibitors (0.1 mM phenylmethylsulforyl fluoride (PMSF), 0.1 M 6-aminocaproic acid, 5 mM benzamidine and 5mM N-ethylmaleimide (NEM)), and the homogenates were cleared by centrifugation at 4800 rpm for 6 min. This procedure was repeated twice. The supernatants containing suprastructural cartilage aggregates were used for adherence analysis. The epidermal layer of the skin samples was removed after treatment with 0.05 M Tris-HCl, pH7.4, containing 1 M NaCl for 4 h, and the dermal layer was frozen in liquid nitrogen. A 200-µm-thick layer was removed from the dermal surface by a dermatome and was repeatedly homogenized in PBS containing protease inhibitors (0.1 mM PMSF, 0.1 M 6-aminocapron acid, 5 mM benzamidine and 5 mM NEM). Between each homogenization step, tissue debris was removed by centrifugation at 4,800 rpm for 6 min. The supernatants containing supramolecular dermal aggregates were used for adherence analysis.
Fragments of suprastructural aggregates from bone were extracted from human hips recovered from joint replacements. The hips were carefully cleaned of adhering connective tissue and bone marrow followed by demineralization using sodium-EDTA (20 g/l, pH7.4) for four weeks, changing the demineralizing buffer every 3 days. After demineralization, the bone was mechanically disrupted into small pieces and homogenized in PBS (150 mM NaCl in 2 mM sodium phosphate buffer, pH7.4) containing a mixture of protease inhibitors (0.1 mM PMSF, 0.1 M 6-aminocaproic acid, 5 mM benzamidine and 5 mM NEM) using a Polytron homogenizer (Kinematica, Luzern, Switzerland). The obtained homogenates were clarified by centrifugation at 4,800 rpm for 6 min and were used for the experiments.
Adherence assays with the Emp forms
Polystyrene microtiter plates were coated with 5 μg/ml of human skin, cartilage or bone fragment or with 0.25 μg/well of ECM components collagen I (Cn I), fibronectin (Fn), fibrinogen (Fg) or vitronectin (Vn) in 0.1 M Na2CO3 overnight at 4 °C. After washing with PBST (137.93 mM NaCl, 2.67 mM KCl, 1.47 mM KH2PO4, 8.06 mM Na2HPO4 × 7 H2O and 0.2% Tween 20, pH7), the wells were blocked by incubation with protein-free blocking buffer (Pierce, Rockford, IL USA) for 1 h at room temperature (RT). The wells were washed with PBST, and 50 µl of different concentrations of EmpFL and Emp fragments (0, 0.01, 0.02, 0.03, 0.04, 0.05, 0.075, 0.1, 0.25, 0.5, 0.75, 1, and 1.5 µM) were added to each well for the concentration-dependent assays. Other binding assays were performed with 1 µM of the EmpFL and Emp forms. The plate was incubated at RT for 1 h with shaking at 70 rpm. The wells were washed with PBST, and 50 µl of a 1:1000 dilution of polyclonal anti-Emp IgG (Squarix Biotechnology, Germany) was added to each well followed by re-incubation of the plate at RT for 1 h with shaking at 70 rpm. The wells were washed with PBST, and 50 µl of a 1:2000 dilution of alkaline phosphatase (AP)-conjugated anti-rabbit goat IgG (Dako, Eching, Germany) was added to each well followed by shaking at 70 rpm for 1 h at RT. The plates were washed three times with PBST, and 50 µl of AP-buffer (1 mM MgCl2, in 0.1 M Tris/HCl pH9.5) supplemented with ready-to-use Alkaline Phosphatase Yellow (pNPP) substrate (Sigma-Aldrich, Missouri, USA) was added to each well. The colour changes were measured at 405 nm after a 30 min incubation at RT using an iMark microplate reader (Bio-Rad, Hercules, California, USA).
Immunogold transmission electron microscopy
Aliquots of suprastructural fragments were absorbed to formvar/carbon nickel grids, washed with PBS and treated for 30 min with 2% (w/v) dried skim milk in PBS for cartilage and bone or 2% (w/v) bovine serum albumin (BSA) in PBS for skin. This procedure was followed by incubation with recombinant Emp (full-length or fragments) in PBS at different concentrations for 1 h at RT. After several steps of washing with PBS, the grids were allowed to react with a rabbit polyclonal antibody against Emp diluted 1:100 in 0.2% (w/v) dried skim milk or PBS containing 2% (v/v) BSA-c (Aurion, Wageningen, Netherland) and 0.025% (v/v) Tween 20 (blocking solution). After washing with PBS, the grids were placed on drops of PBS containing goat antibodies against rabbit immunoglobulins conjugated to colloidal gold particles (Jackson Immuno Research Laboratories) diluted 1:30 in 0.2% dried skim milk or blocking solution. In some experiments, the suprastructural fragments were treated with purified collagenase (CLSPA; Worthington, New Jersey, USA) for 2 h at 37 °C. Finally, the grids were washed five times with distilled water and negatively stained with 2% (w/v) uranyl acetate for 10 min. Electron micrographs were obtained with a Philips EM-410 electron microscope at 60 kV.
Transmission electron microscopy (TEM)
Carbon-coated grids (400 meshes) (Quantifoil, Großlöbichau, Germany) were hydrophilized by glow discharge at low pressure in air. Subsequently, 20 μl a solution of purified EmpFL (3.1nmol) or Emp2+3 (2.4nmol), was adsorbed onto the hydrophilic grids for 1 min. The grids were washed twice with drops of distilled water and were stained with a drop of 2% uranyl acetate in distilled water. The samples were analysed using a Zeiss EM902A electron microscope (Carl Zeiss AG, Oberkochen, Germany) operated at an acceleration voltage of 80 kV. Images were recorded with a FastScan-CCD camera at 1024 × 1024 pixels (TVIPS, Munich, Germany).
Circular dichroism spectroscopy (CD)
Chiral macromolecules, such as proteins differentially absorb left- and right-handed circularly polarized light (L-CPL or R-CPL). These differences can be visualized by circular dichroism (CD) spectroscopy. Depending on whether L-CPL or R-CPL is absorbed to a greater or to a lesser extent, the CD signal can be positive or negative. The electron transitions events of the chiral amide groups, which can be observed at the far ultraviolet (UV) region (below 260 nm), are highly influenced by their surrounding geometries (ϕ and ψ torsion angles) of the polypeptide backbone. Thus, the resulting CD spectra are specific for the secondary structures of the protein (α-helices, β-turns, and natively disordered)51.
For CD spectroscopy, the proteins were finally dialysed against 250 mM NaF in 50 mM sodium phosphate buffer, pH7. CD measurements were performed on a Jasco J-815 spectropolarimeter in 1mm quartz cuvettes (Hellma, Müllheim, Germany). CD spectra were recorded between 185 and 320 nm. Twenty scans for the analysis of protein secondary structure were accumulated and averaged. The buffer reference spectra (250 mM NaF in 50 mM sodium phosphate buffer, pH7) were used as baselines and were subtracted from the CD spectra of the samples. Signal intensities were expressed in millidegrees [θ] and converted to the mean molar ellipticity per residue [θ]MRE in (deg cm2 dmol−1 residue−1) according to equation (1), where ‘N’ represents the number of amino acid residues, ‘d’ is the path length of the cell in cm, ‘c’ is the concentration in g/L of protein, and M is the molecular mass in Da.
| 1 |
The analysis of protein secondary structures was performed on the DichroWeb server52 using SELCON353–55, CONTIN56,57, CDSSTR55,58 algorithms and different reference datasets. As CD data were reliable only above 190 nm, datasets 3, 4, 6, 7, SP175 and SMP180 could be used. The normalized root mean square deviation (NRMSD) was used as a goodness-of-fit parameter for each deconvolution output52.
Hydrodynamic protein size measurements
The photon correlation spectroscopy (PCS) uses the quasi-elastic/dynamic light scattering of small particles in suspension or polymers in solution based on the principle that smaller particles move with higher velocity than larger particles. Thereby, the time-dependent Brownian motion of the particles/polymers is detected as speckle patterns and the hydrodynamic diameter information of the particles is derived from the autocorrelation of the intensity traces of each spot, where is the decay rate, q is the wave vector and Dt is the diffusion coefficient that can be used to calculate the hydrodynamic diameter by Stokes-Einstein equation.
Applying the PCS the hydrodynamic diameter distribution and the polydispersity index of 12.94 µM EmpFL in dialysis buffer (50 mM sodium phosphate, pH7, 250 mM NaF) were measured. The experiments were performed in a low-volume cuvette (ZEN 0112, Brand, Wertheim, Germany) using a Zetasizer Nano ZS (Malvern Instruments, Herrenberg, Germany) at 25 °C and at a scattering angle of 173°. It total, six measurements were collected and evaluated using Zetasizer software (version 5.03, Malvern Instruments) and applying the refractive index (1.33) and viscosity (0.9872mPa*s) of the dialysis buffer. Data are expressed as the means with standard deviations.
Fourier transform infrared spectroscopy (FTIR)
In the Fourier transform infrared spectroscopy (FTIR), all of the infrared (IR) frequencies are assessed simultaneously by splitting the IR light into two beams; one with a fixed wave length, the other with is constantly length changing wave length. Both beams are merged to an interfering beam to with the sample is exposed resulting in an interferogram. The infrared frequencies encoded in an interferogram must be deconvoluted by the Fourier transformation yielding a frequency spectrum. The IR spectrum is a kind of molecular finger print of biomacromolecules. In contrast to other techniques, FTIR requires only small amounts of proteins (1 mM) in variable deuterium water allowing high quality spectra independent of the background fluorescence or light scattering. Additionally, FTIR is not limited to specific protein sizes. Because of the repeating units in proteins, nine characteristic IR absorption bands (amides A, B and I–VII) can be measured. Thereby, the amide I bands (1,700–1,600 cm−1) are the most sensitive vibrational bands of the polypeptide backbone, which gives specific peaks depending of the secondary structural components59.
For FTIR spectroscopy, the proteins were dialyzed stepwise against buffer A2 (50% D2O and 0.25 M NaCl in 50 mM sodium phosphate buffer, pH8.0), buffer B2 (75% D2O and 0.25 M NaCl in 50 mM sodium phosphate buffer, pH8.0) and finally buffer C2 (100% D2O and 0.25 M NaCl in 50 mM sodium phosphate buffer, pH8.0). Infrared (IR) spectra were recorded with a Nicolet Magna 750FTIR spectrometer (Thermo Fisher Scientific, Waltham, Massachusetts, USA) equipped with a DTGS detector. The standard proteins myoglobin, concanavalin A and lysozyme were prepared in buffer C2 at concentrations of 3 µg/µl, which corresponded to the estimated concentration of Emp. Forty-five microliters of the sample solution was placed in a demountable press-on cell for analysis of the liquids with calcium fluoride windows and a 100 µm Teflon spacer (Pike Technologies, Wisconsin, USA). A total of 256 scans were averaged per interferogram at a spectral resolution of 4 cm−1. The total spectral range was 950–4000 cm−1. The IR spectra were corrected for water vapour with OMNIC software version 9 (Thermo Fisher Scientific). The IR spectra were baseline-corrected and normalized in the spectral range from 1780–1540 cm−1 for comparison of amide I bands.
Prediction of the Emp structure
The I-TASSER (Iterative Threading ASSEmbly Refinement) on-line server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) was used to predict the Emp structure without any specific assumptions. In I-TASSER, full-length structural models are constructed by iterative fragment assembly simulations utilizing structural templates identified in the protein data bank using multiple threading alignments60,61. Furthermore, the constructed model is compared to structures of known proteins (top ten structural analogues), offering insights into funicular properties62. The most fitting model was visualized and further analysed using CLC Main Workbench.
Curve fittings and statistics
Statistical analyses and curve fittings were performed using GraphPad Prism version 4.00 (GraphPad Software, San Diego California USA, www.graphpad.com). The binding constants of the Emp forms were calculated by applying the one-site binding model according to equation (2); where X represents the concentration of the ligand and Y is the specific binding signal that increases to a maximum plateau value Ymax. The KD is the equilibrium dissociation constant, expressed in the same units as the X-axis (concentration in µM). When the drug concentration equals KD, half the binding sites are occupied at equilibrium.
| 2 |
Differences in the binding of the EmpFL and Emp fragments to the suprastructures were assessed by one-way ANOVA and Dunnett’s multiple comparison test or by two-way ANOVA and the Bonferroni post-test (two-tailed, 95% confidence intervals). Significance was assumed with p-values ≤ 0.05.
Ethical statements
ECM preparations and experimental protocols from human skin and cartilage and human bone tissue were approved by the ethics committee of the University of Muenster (Ethikkomission der Ärztekammer Westfalen-Lippe und der Medizinischen Fakultät der Wesfälischen Wilhelms-Universität, Münster), Az. 2009-442-f-s and Az. 2010-155-f-s, respectively. All experiments were performed in accordance with relevant guidelines and regulations and approved by the universities of Muenster and Jena, Germany. The informed consent was obtained from all participants.
Electronic supplementary material
Acknowledgements
We would like to thank to Prof. Dr. Ravoo from Organic Chemistry Institute and CeNTech, Westfälische Wilhelms-Universität Münster, Münster, Germany to allow us to use the CD instrument.
Author Contributions
J.G.: Performed all experiments with E.C.M., prepared the samples to measure by different tools, contributed to the C.D. measurement, designed and generated figures and drafted the manuscript. S.N.: Contributed to the measurements and protein structure analysis (CD, Nanosizer, TEM, FTIR) and designed the figures for these applications. C.P.: Contributed to the protein preparation and analysis (Nanosizer, FPLC). U.H.: Performed the extraction of matrices and immunogold transmission electron microscope. F.R.: Performed the C.D. C.K.: Performed the FTIR. M.W.: Performed the TEM. M.H.: Designed and created the Emp forms. G.P.: Participated in writing the manuscript. M.W.P.: Participated in writing the manuscript. B.T.: Participated in writing the manuscript. O.M.: Participated in planning of the experiments, writing of the manuscript, participate in design of the figures, performed the protein structure analysis (I-TASSER) and calculated the binding constants and statistics. L.T.: Planned experiments, coordinated the project, analysed the data, participated in design of the figures and writing of the manuscript. Corresponding author. O.M. and L.T.: wrote the main manuscript text and prepared figures.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Oliwia Makarewicz and Lorena Tuchscherr contributed equally to this work.
A correction to this article is available online at https://doi.org/10.1038/s41598-018-26315-6.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-14168-4.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
5/25/2018
A correction to this article has been published and is linked from the HTML and PDF versions of this paper. The error has been fixed in the paper.
Contributor Information
Oliwia Makarewicz, Email: Oliwia.Makarewicz@med.uni-jena.de.
Lorena Tuchscherr, Email: Lorena.tuchscherrdehauschopp@med.uni-jena.de.
References
- 1.Lowy FD. Staphylococcus aureus infections. The New England journal of medicine. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Foster TJ, Hook M. Surface protein adhesins of Staphylococcus aureus. Trends in microbiology. 1998;6:484–488. doi: 10.1016/S0966-842X(98)01400-0. [DOI] [PubMed] [Google Scholar]
- 3.Patti JM, et al. The Staphylococcus aureus collagen adhesin is a virulence determinant in experimental septic arthritis. Infection and immunity. 1994;62:152–161. doi: 10.1128/iai.62.1.152-161.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sinha B, Herrmann M. Mechanism and consequences of invasion of endothelial cells by Staphylococcus aureus. Thrombosis and haemostasis. 2005;94:266–277. doi: 10.1160/TH05-04-0235. [DOI] [PubMed] [Google Scholar]
- 5.Speziale P, Pietrocola G, Foster TJ, Geoghegan JA. Protein-based biofilm matrices in Staphylococci. Frontiers in cellular and infection microbiology. 2014;4:171. doi: 10.3389/fcimb.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chavakis T, Wiechmann K, Preissner KT, Herrmann M. Staphylococcus aureus interactions with the endothelium: the role of bacterial “secretable expanded repertoire adhesive molecules” (SERAM) in disturbing host defense systems. Thrombosis and haemostasis. 2005;94:278–285. doi: 10.1160/TH05-05-0306. [DOI] [PubMed] [Google Scholar]
- 7.Hauck CR, Ohlsen K. Sticky connections: extracellular matrix protein recognition and integrin-mediated cellular invasion by Staphylococcus aureus. Current opinion in microbiology. 2006;9:5–11. doi: 10.1016/j.mib.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Sinha B, et al. Fibronectin-binding protein acts as Staphylococcus aureus invasin via fibronectin bridging to integrin alpha5beta1. Cellular microbiology. 1999;1:101–117. doi: 10.1046/j.1462-5822.1999.00011.x. [DOI] [PubMed] [Google Scholar]
- 9.Harraghy N, et al. The adhesive and immunomodulating properties of the multifunctional Staphylococcus aureus protein Eap. Microbiology. 2003;149:2701–2707. doi: 10.1099/mic.0.26465-0. [DOI] [PubMed] [Google Scholar]
- 10.Hussain M, et al. Identification and characterization of a novel 38.5-kilodalton cell surface protein of Staphylococcus aureus with extended-spectrum binding activity for extracellular matrix and plasma proteins. Journal of bacteriology. 2001;183:6778–6786. doi: 10.1128/JB.183.23.6778-6786.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harraghy N, et al. sae is essential for expression of the staphylococcal adhesins Eap and Emp. Microbiology (Reading, England) 2005;151:1789–1800. doi: 10.1099/mic.0.27902-0. [DOI] [PubMed] [Google Scholar]
- 12.Flock M, Flock JI. Rebinding of extracellular adherence protein Eap to Staphylococcus aureus can occur through a surface-bound neutral phosphatase. Journal of bacteriology. 2001;183:3999–4003. doi: 10.1128/JB.183.13.3999-4003.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hussain M, et al. eap Gene as novel target for specific identification of Staphylococcus aureus. Journal of clinical microbiology. 2008;46:470–476. doi: 10.1128/JCM.01425-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alberts B, J. A. & Lewis, J. et al. In molecular Biology of the Cell. 4th edition. Ch. The Extracellular Matrix of Animals., (Garland Science; 2002., 2002).
- 15.Patti JM, Hook M. Microbial adhesins recognizing extracellular matrix macromolecules. Current opinion in cell biology. 1994;6:752–758. doi: 10.1016/0955-0674(94)90104-X. [DOI] [PubMed] [Google Scholar]
- 16.Hansen U, et al. The anchorless adhesin Eap (extracellular adherence protein) from Staphylococcus aureus selectively recognizes extracellular matrix aggregates but binds promiscuously to monomeric matrix macromolecules. Matrix biology: journal of the International Society for Matrix Biology. 2006;25:252–260. doi: 10.1016/j.matbio.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 17.Myllyharju J, Kivirikko KI. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends in genetics: TIG. 2004;20:33–43. doi: 10.1016/j.tig.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Kalinka J, et al. Staphylococcus aureus isolates from chronic osteomyelitis are characterized by high host cell invasion and intracellular adaptation, but still induce inflammation. International journal of medical microbiology: IJMM. 2014;304:1038–1049. doi: 10.1016/j.ijmm.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 19.Cheng AG, et al. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2009;23:3393–3404. doi: 10.1096/fj.09-135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson M, Cockayne A, Morrissey JA. Iron-regulated biofilm formation in Staphylococcus aureus Newman requires ica and the secreted protein Emp. Infection and immunity. 2008;76:1756–1765. doi: 10.1128/IAI.01635-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campoccia, D. et al. Orthopedic implant-infections. Incompetence of Staphylococcus epidermidis, Staphylococcus lugdunensis and Enterococcus faecalis to invade osteoblasts. Journal of biomedical materials research. Part A, 10.1002/jbm.a.35564 (2015). [DOI] [PubMed]
- 22.Liao SM, Du QS, Meng JZ, Pang ZW, Huang RB. The multiple roles of histidine in protein interactions. Chemistry Central journal. 2013;7:44. doi: 10.1186/1752-153X-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandilands A, Sutherland C, Irvine AD, McLean WH. Filaggrin in the frontline: role in skin barrier function and disease. Journal of cell science. 2009;122:1285–1294. doi: 10.1242/jcs.033969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benedetti CE, et al. Plasmodium falciparum histidine-rich protein II binds to actin, phosphatidylinositol 4,5-bisphosphate and erythrocyte ghosts in a pH-dependent manner and undergoes coil-to-helix transitions in anionic micelles. Molecular and biochemical parasitology. 2003;128:157–166. doi: 10.1016/S0166-6851(03)00057-4. [DOI] [PubMed] [Google Scholar]
- 25.Ge R, et al. Expression and characterization of a histidine-rich protein, Hpn: potential for Ni2 + storage in Helicobacter pylori. The Biochemical journal. 2006;393:285–293. doi: 10.1042/BJ20051160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones AL, Hulett MD, Parish CR. Histidine-rich glycoprotein: A novel adaptor protein in plasma that modulates the immune, vascular and coagulation systems. Immunology and cell biology. 2005;83:106–118. doi: 10.1111/j.1440-1711.2005.01320.x. [DOI] [PubMed] [Google Scholar]
- 27.Grundmeier M, et al. Staphylococcal strains vary greatly in their ability to induce an inflammatory response in endothelial cells. The Journal of infectious diseases. 2010;201:871–880. doi: 10.1086/651023. [DOI] [PubMed] [Google Scholar]
- 28.Tuchscherr L, et al. Staphylococcus aureus small-colony variants are adapted phenotypes for intracellular persistence. The Journal of infectious diseases. 2010;202:1031–1040. doi: 10.1086/656047. [DOI] [PubMed] [Google Scholar]
- 29.Tuchscherr L, et al. Staphylococcus aureus phenotype switching: an effective bacterial strategy to escape host immune response and establish a chronic infection. EMBO molecular medicine. 2011;3:129–141. doi: 10.1002/emmm.201000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patti JM, Allen BL, McGavin MJ, Hook M. MSCRAMM-mediated adherence of microorganisms to host tissues. Annual review of microbiology. 1994;48:585–617. doi: 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- 31.Hussain M, et al. More than one tandem repeat domain of the extracellular adherence protein of Staphylococcus aureus is required for aggregation, adherence, and host cell invasion but not for leukocyte activation. Infection and immunity. 2008;76:5615–5623. doi: 10.1128/IAI.00480-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cooper EA, Knutson K. Fourier transform infrared spectroscopy investigations of protein structure. Pharmaceutical biotechnology. 1995;7:101–143. doi: 10.1007/978-1-4899-1079-0_3. [DOI] [PubMed] [Google Scholar]
- 33.Jackson M, Mantsch HH. The use and misuse of FTIR spectroscopy in the determination of protein structure. Critical reviews in biochemistry and molecular biology. 1995;30:95–120. doi: 10.3109/10409239509085140. [DOI] [PubMed] [Google Scholar]
- 34.Surewicz WK, Mantsch HH, Chapman D. Determination of protein secondary structure by Fourier transform infrared spectroscopy: a critical assessment. Biochemistry. 1993;32:389–394. doi: 10.1021/bi00053a001. [DOI] [PubMed] [Google Scholar]
- 35.Wallace, B. A. & Janes, R. W. Modern techniques for circular dichroism and synchrotron radiation circular dichroism spectroscopy. Vol. 1 (IOS Press, 2009).
- 36.Lees JG, Miles AJ, Janes RW, Wallace BA. Novel methods for secondary structure determination using low wavelength (VUV) circular dichroism spectroscopic data. BMC bioinformatics. 2006;7:507. doi: 10.1186/1471-2105-7-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitmore L, Wallace BA. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic acids research. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng A, Xavier RJ. Leucine-rich repeat (LRR) proteins: integrators of pattern recognition and signaling in immunity. Autophagy. 2011;7:1082–1084. doi: 10.4161/auto.7.9.16464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beavers, W. N. & Skaar, E. P. Neutrophil-generated oxidative stress and protein damage in Staphylococcus aureus. Pathogens and disease74, 10.1093/femspd/ftw060 (2016). [DOI] [PMC free article] [PubMed]
- 41.Koymans, K. J. et al. Staphylococcal Superantigen-Like Protein 1 and 5 (SSL1 & SSL5) Limit Neutrophil Chemotaxis and Migration through MMP-Inhibition. International journal of molecular sciences17, 10.3390/ijms17071072 (2016). [DOI] [PMC free article] [PubMed]
- 42.Chavakis T, et al. Staphylococcus aureus extracellular adherence protein serves as anti-inflammatory factor by inhibiting the recruitment of host leukocytes. Nature medicine. 2002;8:687–693. doi: 10.1038/nm728. [DOI] [PubMed] [Google Scholar]
- 43.Koziel J, Potempa J. Protease-armed bacteria in the skin. Cell and tissue research. 2013;351:325–337. doi: 10.1007/s00441-012-1355-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pietrocola G, Nobile G, Rindi S, Speziale P. Staphylococcus aureus Manipulates Innate Immunity through Own and Host-Expressed Proteases. Frontiers in cellular and infection microbiology. 2017;7:166. doi: 10.3389/fcimb.2017.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klemm P, Schembri MA. Bacterial adhesins: function and structure. International journal of medical microbiology: IJMM. 2000;290:27–35. doi: 10.1016/S1438-4221(00)80102-2. [DOI] [PubMed] [Google Scholar]
- 46.Kline KA, Falker S, Dahlberg S, Normark S, Henriques-Normark B. Bacterial adhesins in host-microbe interactions. Cell host & microbe. 2009;5:580–592. doi: 10.1016/j.chom.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 47.Piepenbrink KH, Sundberg EJ. Motility and adhesion through type IV pili in Gram-positive bacteria. Biochemical Society transactions. 2016;44:1659–1666. doi: 10.1042/BST20160221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barnhart MM, Chapman MR. Curli biogenesis and function. Annual review of microbiology. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bur S, Preissner KT, Herrmann M, Bischoff M. The Staphylococcus aureus extracellular adherence protein promotes bacterial internalization by keratinocytes independent of fibronectin-binding proteins. The Journal of investigative dermatology. 2013;133:2004–2012. doi: 10.1038/jid.2013.87. [DOI] [PubMed] [Google Scholar]
- 50.Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular systems biology7, 539, http://www.ncbi.nlm.nih.gov/pubmed/21988835 (2011). [DOI] [PMC free article] [PubMed]
- 51.Greenfield, N. J. Using circular dichroism spectra to estimate protein secondary structure. Nature protocols1, 2876–2890, 10.1038/nprot.2006.202 (2006). [DOI] [PMC free article] [PubMed]
- 52.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic acids research. 2004;32:W668–673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sreerama N, Woody RW. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Analytical biochemistry. 1993;209:32–44. doi: 10.1006/abio.1993.1079. [DOI] [PubMed] [Google Scholar]
- 54.Sreerama N, Venyaminov SY, Woody RW. Estimation of the number of alpha-helical and beta-strand segments in proteins using circular dichroism spectroscopy. Protein science: a publication of the Protein Society. 1999;8:370–380. doi: 10.1110/ps.8.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Analytical biochemistry. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 56.Provencher SW, Glockner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 1981;20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- 57.Van Stokkum IH, Spoelder HJ, Bloemendal M, van Grondelle R, Groen FC. Estimation of protein secondary structure and error analysis from circular dichroism spectra. Analytical biochemistry. 1990;191:110–118. doi: 10.1016/0003-2697(90)90396-Q. [DOI] [PubMed] [Google Scholar]
- 58.Compton LA, Johnson WC., Jr. Analysis of protein circular dichroism spectra for secondary structure using a simple matrix multiplication. Analytical biochemistry. 1986;155:155–167. doi: 10.1016/0003-2697(86)90241-1. [DOI] [PubMed] [Google Scholar]
- 59.Yang, H., Yang, S., Kong, J., Dong, A. & Yu, S. Obtaining information about protein secondary structures in aqueous solution using Fourier transform IR spectroscopy. Nature protocols10, 382–396, 10.1038/nprot.2015.024 (2015). [DOI] [PubMed]
- 60.Yang J, Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucleic acids research. 2015;43:W174–181. doi: 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nature protocols. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang J, Roy A, Zhang Y. Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics. 2013;29:2588–2595. doi: 10.1093/bioinformatics/btt447. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.