Abstract
Macrophages act as the first line of host immune defense against Mycobacterium tuberculosis (Mtb). Recent studies have demonstrated circular RNAs (circRNAs) are implicated in a variety of physiological and pathological processes; however, the role of circRNAs in macrophages response to Mtb infection remain unknown. To address this issue, here we characterized circRNAs expression profiles in human monocyte derived macrophages (MDMs) response to Mtb infection using microarray assay. Our results revealed that many circRNAs were differentially expressed in human MDMs after Mtb infection; of these, 32 circRNAs were up-regulated and 110 were down-regulated. Real time PCR results were generally consistent with the microarray data. Furthermore, we found that hsa_circ_0043497 and hsa_circ_0001204 may be effective diagnostic biomarkers for TB. This study provides the first evidence that circRNAs alterations are involved in human MDMs response to TB infection and reveal potential targets for diagnostics and the treatment of TB.
Introduction
Tuberculosis (TB), an infectious disease caused by Mycobacterium tuberculosis (Mtb) infection, remains a leading cause of morbidity and mortality worldwide1. In 2015, World Health Organization (WHO) reported that an estimated 10.3 million cases of TB occurred and that 1.4 million died of TB2. Despite the high rate of Mtb infection in humans, especially in developing countries, only 5–10% of infected people develop active TB in their lifetime3. Interactions between Mtb and the host largely determine the development and outcome of TB infection.
Macrophages play a critical role in the host immune response against mycobacterial infection4. They serve as the major host cell niche for intracellular growth and persistence of Mtb during all phases of TB, from primary infection with bacillary dissemination, through latency and reactivation TB5. Despite several studies that have been conducted on Mtb-macrophages interaction, however, the underlying molecular regulation is not fully understood.
Circular RNAs (circRNAs) are a special type of non-coding RNA that are formed from the covalent linkage of the 3′ and 5′ ends to form a closed loop6. Recent evidence has suggested that circRNAs play essential roles in various physiological and pathological processes7,8. Many circRNAs are abundant, stable, conserved and potentially function as competing endogenous RNAs9. Due to a lack of 3′ and 5′ ends and resistance to RNases, circRNAs might be used as potential biomarkers and treatment targets for human diseases10. With the increasing studies about circRNAs, researchers have reported that circRNAs are involved in the development of several types of diseases, such as cancer11, cardiovascular disease12, and neurological disorders13. However, the role of circRNAs within macrophages innative response to TB infection has yet to be explored.
In this study, we analysed the expression patterns of circRNAs in Mtb-infected human monocyte derived macrophages (MDMs) using microarray assay. Our data revealed that a number of circRNAs were consistently altered under Mtb infection. And then, we demonstrated that hsa_circ_0043497 and hsa_circ_0001204 may be effective diagnostic biomarkers for TB. These findings provided novel insight into the pathogenesis of TB and provide a basis for the diagnosis and therapy of TB.
Materials and Methods
Ethics Statement
This study was approved by the ethical committee of the First Affiliated Hospital of Nanchang University and conducted in accordance with the Declaration of Helsinki. All participants provided informed consent before commencement of the study.
Sample collection
The peripheral blood samples (5 mL) used to validate candidate biomarkers were collected from 96 patients with active pulmonary TB and 85 healthy controls. Patients with pulmonary TB were recruited from the Jiangxi Chest Hospital and the First Affiliated Hospital of Nanchang University from January 2016 to July 2017. All of these patients were diagnosed with TB on the basis of the typical TB clinical symptoms, bacterial culture, and imaging examinations, and in accordance with the Health criteria in People’s Republic of China: The legal diagnostic criteria of infectious diseases (WS288–2008) and the revised international definitions in TB control of the WHO. Individuals with malignant tumor, HIV infections were excluded. Subsequently 12 active pulmonary TB (TB naive) inpatients were treated according to prevailing China National Tuberculosis Program guidelines (2HRZE/6HE). peripheral blood samples were obtained at diagnosis prior to treatment initiation and after completion of treatment (six months). After sample collection, peripheral blood mononuclear cells (PBMCs) were freshly isolated by density gradient centrifugation on Ficoll-Paque (Sigma, USA) according to the manufacturer’s protocol. Then, the PBMCs samples were lysed with TRIzol reagent (Invitrogen, USA) and stored at −80 °C.
Cell purification and culture
The peripheral blood samples (5 mL) were collected from 22 healthy donors. Individuals with cancer, allergic diseases, immune-compromised conditions, diabetes or other infectious diseases such as HBV, HCV and HIV infection were excluded. After sample collection, PBMCs were isolated by density gradient centrifugation. To develop MDMs, the monocytes were purified using anti-CD14 magnetic beads (Miltenyi Biotec, USA) and the purified monocytes were cultured in RPMI 1640 medium contains 10% Human serum and 0.05% Glutamine (Sigma, USA) for 7 days at 5% CO2 and 37 °C14. The MDMs were identified by morphologic observation and flow cytometric analysis followed by anti-CD68 staining.
Bacterial strains and culture
Mtb H37Rv strain 27294 was purchased from the American Type Culture Collection (ATCC). They were grown in Middlebrook 7H9 broth containing 10% oleic acid-albumin-dextrose-catalase (OADC) and 0.05% Tween-80 at 37 °C. They were washed with PBS and passed through a syringe, to disrupt the clumps, before infecting human MDMs.
Cell infection
The human MDMs were plated in 12-well plates, and incubated overnight. The cells were infected at a multiplicity of infection (MOI) of 5. Uninfected cells which received only PBS served as controls. After 4–6 h incubation at 37 °C, non-phagocytosed bacteria were washed off using PBS. MDMs were replenished with fresh RPMI 1640 and incubated for 24 h at 37 °C with 5% CO2. After 24 h, cells were harvested and RNA was isolated.
RNA isolation
Total RNA was extracted using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s protocol. The integrity of the RNA was assessed by electrophoresis on a denaturing agarose gel. A NanoDrop ND-1000 spectrophotometer was used for the accurate measurement of RNA concentration.
CircRNAs microarray and computational analysis
Human CircRNA Array V2.0 (8 × 15 K) is manufactured by Arraystar Technologies (Rockville, MD, USA). Six MDMs samples, including three infected samples and three uninfected samples, were sent to KangChen Bio-tech (Shanghai, China) for the Arraystar circRNA microarray analysis. Microarray hybridization were performed according to the protocols of Arraystar. The scatter plot is a visualization method used for assessing the circRNA expression variation. Differentially expressed circRNAs with statistical significance (fold changes >2.0 and P < 0.05) between groups were identified using fold change cut-off or volcano plot filtering, respectively. The circRNAs/microRNAs (miRNAs) interaction was predicted using Arraystar’s home-made miRNA target prediction software based on TargetScan and miRanda.
Quantitative real-time PCR
Total RNA (2 μg) was reversely transcribed into cDNA using the Reverse Transcription System Kit (Takara, Dalian, China). The expression levels of circRNAs and miRNAs were determined by quantitative real-time PCR (RT-qPCR) using SYBR Master Mix (Applied Biosystems, Foster City, CA, USA) and mirVanaTM RT-qPCR miRNA Detection Kit (Ambion, Austin, USA) on Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems, Foster City, USA), respectively. Primers used in this study were listed in Supplementary Table 1. The PCR primers of U6 and miRNAs were purchased from RIBOBIO. All RT-qPCR experiments were performed in triplicate. The relative expression levels of circRNAs and miRNAs were calculated by the 2−ΔΔCt method. The the relative expression levels of circRNAs were normalized to GAPDH and the relative expression levels of miRNAs were normalized to U610. PCR product was examined by agarose gel electrophoresis using 2% (w/v) LE agarose (Seakem) stained with ethidium bromide.
Statistics
Numerical data were shown as the mean ± standard error of the mean (SEM). A one-way ANOVA test, Mann-Whitney test or Student t-test was used for statistical analysis. Receiver operating characteristic (ROC) analysis was used to evaluate the power of candidate circRNAs. All statistical tests were performed with GraphPad Prism 5.0 (GraphPad Software, San Diego, USA). P < 0.05 was considered statistically significant.
Results
Overview of circRNA profiles
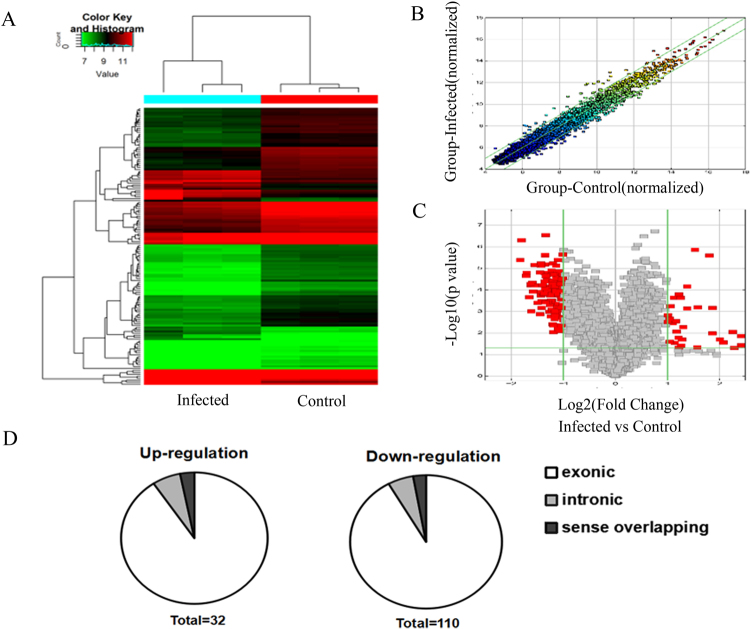
To explore the potential circRNAs involved in human MDMs response to TB infection, we examined the circRNAs expression profiles through microarray analysis. In total, 142 circRNAs were identified with differential expression between Mtb-infected group and uninfected group (fold change >2.0, P < 0.05), among which 32 circRNAs were up-regulated while 110 circRNAs were down-regulated (Fig. 1 and Supplementary Table 2). Specifically, the most up-regulated circRNAs were: hsa_circ_0003528, hsa_circ_0001417, hsa_circ_0043497 and hsa_circ_0038929, of which hsa_circ_0003528 was the highest. The most highly down-regulated were: hsa_circ_0068784, hsa_circ_0057090, hsa_circ_0056247 and hsa_circ_0001204, of which hsa_circ_0068784 showed the largest down-regulation. In present study, the top 40 up- and down-regulated circRNAs are listed in Table 1 by fold change. We summarized the classification of dysregulated circRNAs (Fig. 1). Among the up-regulated circRNAs, there were 29 exonic, 2 intronic and 1 sense overlapping. Among the down-regulated circRNAs, there were 101 exonic, 6 intronic and 3 sense overlapping.
Figure 1.
Analysis of differentially expressed circRNAs in Mtb-infected human MDMs and uninfected MDMs. (A) Hierarchical clustering results of circRNAs expression profiles among the Mtb-infected group and control group. “Red” indicated high relative expression and “green” indicated low relative expression. (B) CircRNAs in the Scatter-Plot above the top green line and below the bottom green line indicated more than 2.0 fold change of circRNAs between the two groups. (C) Volcano-Plot of circRNAs expression profile. The vertical lines correspond to 2.0-fold up and down and the horizontal line represents a P-value of 0.05. The red point in the plot represents the differentially espressed circRNAs with statistical significance. (D) classification of dysregulated circRNAs.
Table 1.
Top 40 differently expressed circRNAs in M. tuberculosis-infected human monocyte derived macrophages.
| CircRNA ID | P-value | FDR | FC | Regulation | CircRNA_type | Chrom | Strand | Best_transcript | GeneSymbol |
|---|---|---|---|---|---|---|---|---|---|
| hsa_circ_0003528 | 0.037466755 | 0.115427239 | 5.341549 | up | exonic | chr5 | + | NM_021982 | SEC. 24 A |
| hsa_circ_0001417 | 0.013874737 | 0.062154272 | 5.3239089 | up | exonic | chr4 | — | NM_032217 | ANKRD17 |
| hsa_circ_0043497 | 0.049072061 | 0.137777454 | 4.866483 | up | exonic | chr17 | — | NM_014815 | MED24 |
| hsa_circ_0038929 | 0.008851434 | 0.04811339 | 4.7593614 | up | exonic | chr16 | + | NM_007317 | KIF22 |
| hsa_circ_0000994 | 0.02769979 | 0.094239935 | 4.2102057 | up | exonic | chr2 | — | NM_021097 | SLC8A1 |
| hsa_circ_0001216 | 0.019989045 | 0.076864421 | 4.0183976 | up | sense overlapping | chr22 | — | NM_005080 | XBP1 |
| hsa_circ_0007705 | 0.000698384 | 0.012309287 | 3.6367029 | up | exonic | chr15 | + | NM_016396 | CTDSPL2 |
| hsa_circ_0030045 | 0.010558813 | 0.053165131 | 3.6256505 | up | exonic | chr13 | + | NM_014252 | SLC25A15 |
| hsa_circ_0006682 | 2.5579E-06 | 0.002191928 | 3.4860902 | up | intronic | chr17 | + | ENST00000439730 | GOSR2 |
| hsa_circ_0005108 | 0.000648282 | 0.011824622 | 2.9961246 | up | exonic | chr14 | — | NM_203301 | FBXO33 |
| hsa_circ_0028152 | 0.046309846 | 0.132935223 | 2.9723266 | up | exonic | chr12 | — | NM_014776 | GIT2 |
| hsa_circ_0003748 | 1.35271E-06 | 0.002016717 | 2.8810825 | up | exonic | chr3 | — | NM_016291 | IP6K2 |
| hsa_circ_0076260 | 6.90949E-05 | 0.005050619 | 2.5996407 | up | exonic | chr6 | + | NM_001206927 | DNAH8 |
| hsa_circ_0083157 | 0.000732236 | 0.012499195 | 2.5949787 | up | exonic | chr7 | + | NM_014671 | UBE3C |
| hsa_circ_0008157 | 0.005409066 | 0.035950336 | 2.4669575 | up | exonic | chr11 | — | NM_020153 | IFT46 |
| hsa_circ_0007209 | 0.020193068 | 0.077300669 | 2.4254376 | up | exonic | chr8 | — | NM_020863 | ZFAT |
| hsa_circ_0002824 | 0.037701504 | 0.115881588 | 2.392587 | up | exonic | chr9 | + | NM_004972 | JAK2 |
| hsa_circ_0022382 | 0.000235706 | 0.007462334 | 2.376217 | up | exonic | chr11 | + | NM_004265 | FADS2 |
| hsa_circ_0091994 | 0.000162781 | 0.006608264 | 2.3325785 | up | exonic | chrX | — | NM_001456 | FLNA |
| hsa_circ_0026616 | 0.047782317 | 0.135397186 | 2.3247835 | up | exonic | chr12 | — | NM_015665 | AAAS |
| hsa_circ_0068784 | 4.17569E-06 | 0.002492284 | 3.5480508 | down | exonic | chr4 | + | NM_003441 | ZNF141 |
| hsa_circ_0057090 | 4.9948E-07 | 0.00147459 | 3.4927401 | down | exonic | chr2 | + | NM_002610 | PDK1 |
| hsa_circ_0056247 | 2.22076E-05 | 0.003841024 | 3.2888045 | down | exonic | chr2 | + | NM_002830 | PTPN4 |
| hsa_circ_0001204 | 1.64596E-05 | 0.003817991 | 3.2644161 | down | intronic | chr22 | + | ENST00000351989 | DGCR8 |
| hsa_circ_0001747 | 0.000218714 | 0.007377021 | 3.149758 | down | exonic | chr7 | + | NM_013255 | MKLN1 |
| hsa_circ_0017311 | 5.30031E-05 | 0.004765914 | 3.0933966 | down | exonic | chr1 | + | NM_152609 | CNST |
| hsa_circ_0047744 | 5.99584E-05 | 0.004765914 | 3.0829551 | down | exonic | chr18 | + | NM_015285 | WDR7 |
| hsa_circ_0030569 | 0.001000539 | 0.014333503 | 3.0627677 | down | exonic | chr13 | — | NM_005845 | ABCC4 |
| hsa_circ_0089866 | 4.7015E-05 | 0.004765914 | 3.0233465 | down | exonic | chrX | + | NM_015691 | WWC3 |
| hsa_circ_0023748 | 0.000120414 | 0.006050795 | 2.9884338 | down | exonic | chr11 | — | NM_033547 | INTS4 |
| hsa_circ_0008153 | 6.89986E-06 | 0.002809671 | 2.92489 | down | exonic | chr15 | + | NM_006537 | USP3 |
| hsa_circ_0004893 | 6.21515E-05 | 0.004860577 | 2.9098745 | down | exonic | chr1 | — | NM_080391 | PTP4A2 |
| hsa_circ_0012425 | 0.000394458 | 0.009352714 | 2.8215173 | down | exonic | chr1 | — | NM_001981 | EPS15 |
| hsa_circ_0039261 | 1.57454E-05 | 0.003817991 | 2.7291055 | down | exonic | chr16 | + | NM_000293 | PHKB |
| hsa_circ_0037680 | 0.001355771 | 0.016823871 | 2.72776 | down | exonic | chr16 | — | NM_004380 | CREBBP |
| hsa_circ_0000028 | 7.56027E-06 | 0.002879975 | 2.696051 | down | exonic | chr1 | — | NM_032236 | USP48 |
| hsa_circ_0008205 | 0.000125451 | 0.00607151 | 2.6958097 | down | exonic | chr8 | — | NM_144649 | TMEM71 |
| hsa_circ_0003615 | 6.03401E-06 | 0.002727301 | 2.6943212 | down | exonic | chr11 | — | NM_024079 | ALG8 |
| hsa_circ_0002383 | 0.000178628 | 0.006792563 | 2.6911006 | down | exonic | chr11 | — | NM_006645 | STARD10 |
| hsa_circ_0003768 | 0.000673145 | 0.0120992 | 2.6750548 | down | exonic | chr11 | + | NM_003477 | PDHX |
Note: CircRNA ID: The circRNA ID is in circBase (http://www.circbase.org/). P-value: P-value calculated from paired t-test. FDR: FDR is calculated from Benjamini Hochberg FDR. Fold Change: Te absolute ratio (no log scale) of normalized intensities between two conditions.
Validation of circRNAs expression
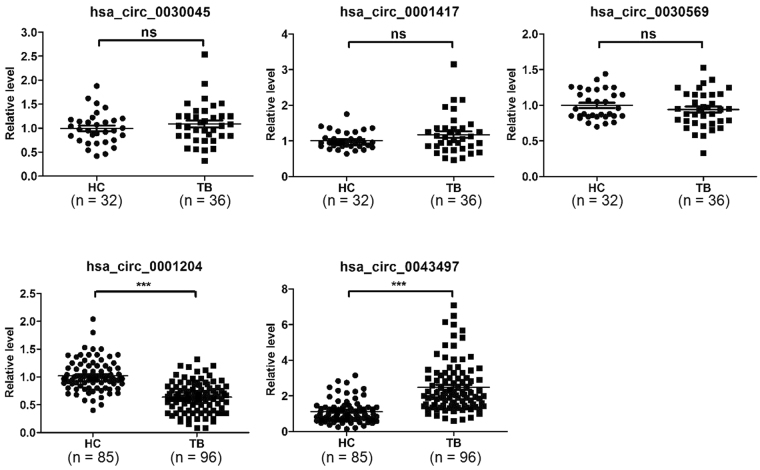
To validate microarray analysis findings, we randomly selected 5 circRNAs from the differentially expressed circRNAs with fold change >3.0 and analyzed their expression by RT-qPCR in expanded MDMs samples. The results showed that these circRNAs were significantly differentially expressed. Particularly, hsa_circ_0030045, hsa_circ_0001417 and hsa_circ_0043497 are significantly up-regulated in Mtb-infected group, while hsa_circ_0030569 and hsa_circ_0001204 are significantly down-regulated in Mtb-infected group (Fig. 2). Our data confirmed the findings of the microarray analysis.
Figure 2.
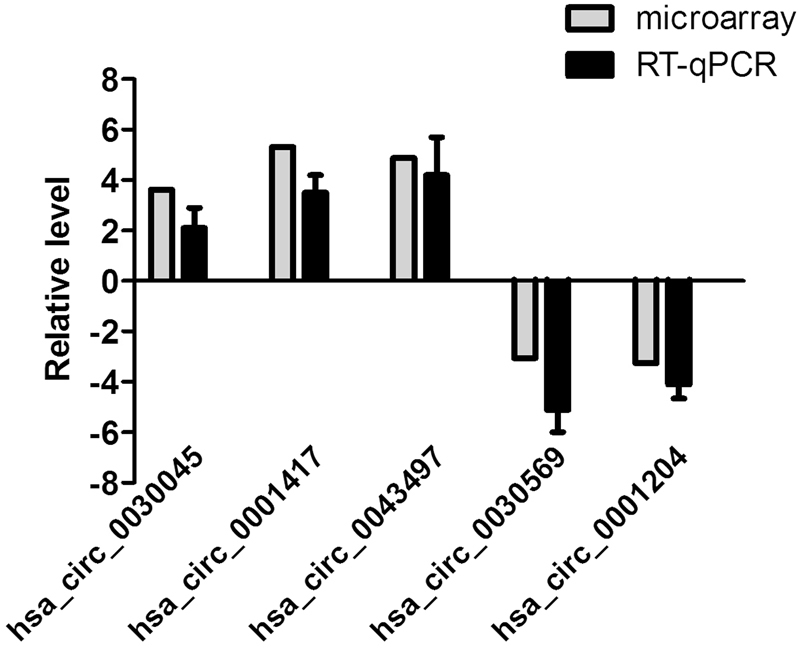
Confirmation of the differential expression of circRNAs by RT-qPCR. Five differentially expressed circRNAs were validated by RT-qPCR. Individual RT-qPCR assays were performed using samples from six additional human MDMs after 24 h of infection with H37Rv. Data are expressed as the means ± SEM. The relative expression levels of circRNAs were normalized to levels of GAPDH.
Diagnostic value of hsa_circ_0043497 and hsa_circ_0001204 expression levels in TB
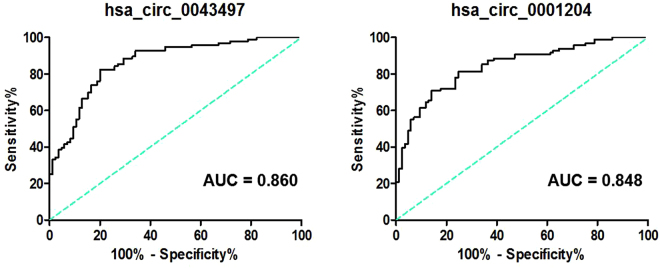
Recently, many studies have demonstrated that circRNAs can serve as novel diagnostic markers for diseases. Thus, we sought to determine whether the circRNAs identified in our microarray analysis could potentially serve as diagnostic biomarkers for TB. The levels of 5 circRNAs (hsa_circ_0030045, hsa_circ_0001417, hsa_circ_0043497, hsa_circ_0030569 and hsa_circ_0001204) were measured in PBMCs samples from 96 patients with active pulmonary TB and 85 healthy controls using RT-qPCR. The demographic characteristics of the participants are showed in Table 2. Of the five circRNAs studied, the levels of hsa_circ_0043497 were significantly elevated and hsa_circ_0001204 were significantly reduced in PBMCs of TB patients as compared to healthy controls. There were no significant differences in the level of hsa_circ_0030045, hsa_circ_0001417 and hsa_circ_0030569 between TB patients and controls (Fig. 3). To compare the diagnostic value of hsa_circ_0043497 and hsa_circ_0001204 as candidate biomarkers of TB, we performed ROC curve analysis for hsa_circ_0043497 and hsa_circ_0001204. ROC curve analysis exhibited a significant distinguishing efficiency with an area under the curve (AUC) value of 0.860 for hsa_circ_0043497 and 0.848 for hsa_circ_0001204 (Fig. 4). These results suggest that altered expression of hsa_circ_0043497 and hsa_circ_0001204 in PBMCs are potential novel biomarkers for the diagnosis of TB.
Table 2.
Demographic and clinical characteristics of TB patients and healthy controls.
| Active TB patients (n = 96) | Healthy controls (n = 85) | |
|---|---|---|
| Sex (female/male) | 96 (40/56) | 85 (33/52) |
| Age (mean ± SD) | 34.57 ± 4.83 | 36.19 ± 5.06 |
| ELISPOT-positive | 96/96 | 0/85 |
| M. tuberculosis culture-positive | 58/96 | N/A |
| Lung cavity | 52/96 | 0/85 |
| New/retreatment | 80/16 | N/A |
Note: N/A: not applicable.
Figure 3.
Comparison of circRNAs levels in active TB patients and healthy control. The expression of five circRNAs in 96 pulmonary TB patients and 85 healthy controls were measured by RT-qPCR. Statistical analysis was performed using the nonparametric Mann-Whitney test. The relative expression levels of circRNAs were normalized to levels of GAPDH. ***P < 0.001.
Figure 4.
Receiver operating characteristic (ROC) curves of hsa_circ_0043497 and hsa_circ_0001204 between active TB patients and healthy control. ROC curves of hsa_circ_0043497 and hsa_circ_0001204 showed a moderate distinguishing efficiency.
Effects of anti-TB therapy on level of hsa_circ_0043497 and hsa_circ_0001204
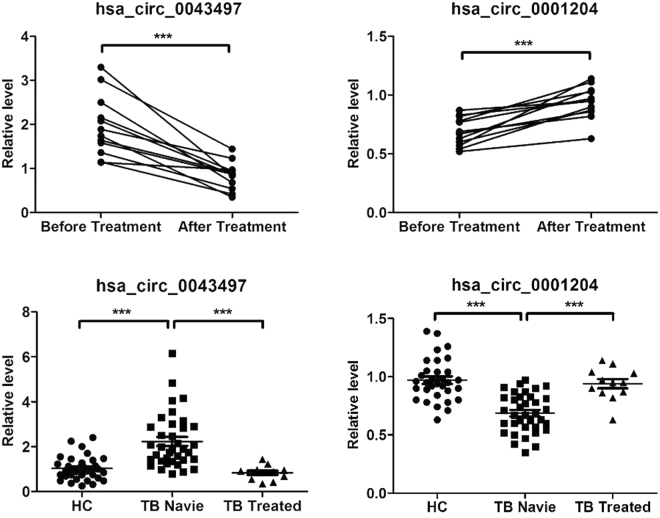
Furthermore, we compared the expression of hsa_circ_0043497 and hsa_circ_0001204 in 12 patients before and after TB therapy. As compared to their pre-treatment, the levels of hsa_circ_0043497 decreased after anti-TB treatment. As compared to controls, the mean levels of hsa_circ_0043497 which was higher in TB infected group came to near normal after therapy. The mean hsa_circ_0043497 levels did not differ significantly between healthy control and TB treated group. As compared to healthy controls, the mean levels of hsa_circ_0001204 were significantly lower in TB infected group; the levels increase post therapy. The mean hsa_circ_0001204 levels did not differ significantly between healthy control and TB treated group (Fig. 5).
Figure 5.
Change in the levels of hsa_circ_0043497 and hsa_circ_0001204 in response to therapy. The levels of hsa_circ_0043497 and hsa_circ_0001204 were measured by RT-qPCR. Dot plot shows relative levels of hsa_circ_0043497 and hsa_circ_0001204 in the same patients before and after completion of therapy (n = 12). Relative levels of hsa_circ_0043497 and hsa_circ_0001204 in healthy controls (n = 32), pulmonary TB naive patients (n = 36) and TB treated group (n = 12). The value between the healthy control and TB treated group is not significantly different. ***P < 0.001.
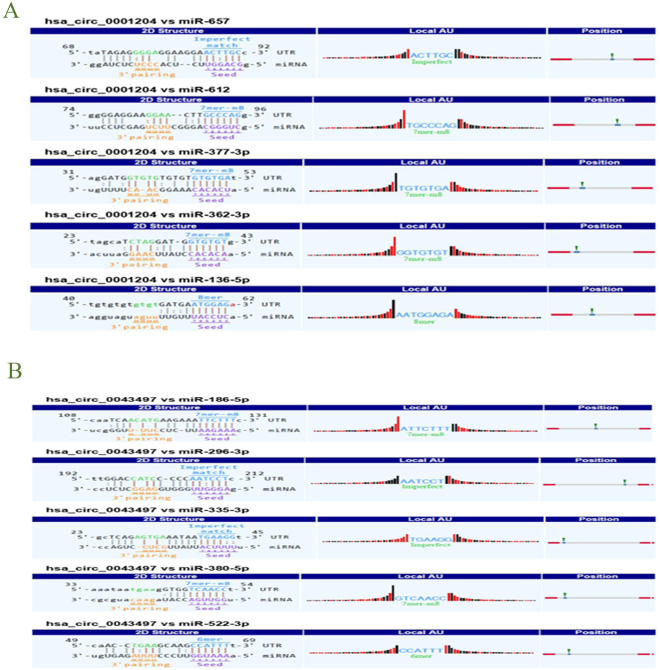
Target gene prediction
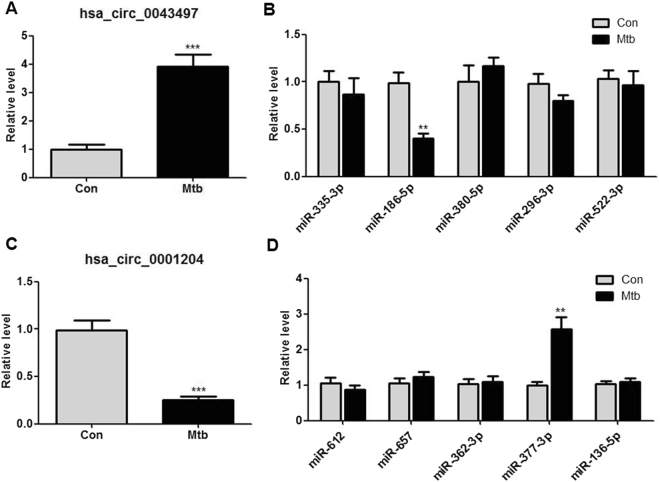
To evaluate circRNAs potential functions, we investigated potential miRNAs binding with circRNAs using Arraystar’s home-made miRNA target prediction software. We selected 18 differentially expressed circRNAs with the highest fold-change (fold-change > 3.0; P < 0.05) to predict their miRNA response elements (MREs), including 9 up-regulated circRNAs and 9 down-regulated circRNAs. Five MREs with good mirSVR scores for each circRNA are shown in Table 3. In the study, two confirmed circRNAs (hsa_circ_0043497 and hsa_circ_0001204) and their target miRNAs were focused (Fig. 6). Then, the expression levels of hsa_circ_0043497-target miRNAs and hsa_circ_0001204-target miRNAs were validated by RT-qPCR in ten additional human MDMs after 24 h of infection with Mtb. Of the ten miRNAs studied, the levels of miR-186–5p were significantly reduced and the levels of miR-377-3p were significantly elevated in Mtb-infected MDMs (Fig. 7).
Table 3.
Annotation for differentially expressed circRNAs/miRNAs interaction.
| CircRNA ID | P-value | FDR | FC | Regulation | MRE1 | MRE2 | MRE3 | MRE4 | MRE5 |
|---|---|---|---|---|---|---|---|---|---|
| hsa_circ_0003528 | 0.037466755 | 0.115427239 | 5.341549 | up | miR-421 | miR-181a-2-3p | miR-505-3p | miR-429 | miR-380-5p |
| hsa_circ_0001417 | 0.013874737 | 0.062154272 | 5.3239089 | up | miR-298 | miR-766-5p | miR-323a-5p | miR-138-5p | miR-486-3p |
| hsa_circ_0043497 | 0.049072061 | 0.137777454 | 4.866483 | up | miR-335-3p | miR-186-5p | miR-380-5p | miR-296-3p | miR-522-3p |
| hsa_circ_0038929 | 0.008851434 | 0.04811339 | 4.7593614 | up | miR-485-5p | miR-221-5p | miR-492 | miR-182-5p | miR-545-3p |
| hsa_circ_0000994 | 0.02769979 | 0.094239935 | 4.2102057 | up | miR-27b-3p | miR-27a-3p | miR-373-5p | miR-335-3p | miR-628-5p |
| hsa_circ_0001216 | 0.019989045 | 0.076864421 | 4.0183976 | up | miR-326 | miR-3154 | miR-4747-5p | miR-676-5p | miR-330-5p |
| hsa_circ_0007705 | 0.000698384 | 0.012309287 | 3.6367029 | up | miR-103a-2-5p | miR-190a-5p | miR-578 | miR-22-5p | miR-190b |
| hsa_circ_0030045 | 0.010558813 | 0.053165131 | 3.6256505 | up | miR-650 | miR-605-3p | miR-876-5p | miR-526b-5p | miR-569 |
| hsa_circ_0006682 | 2.5579E-06 | 0.002191928 | 3.4860902 | up | miR-485-5p | miR-6884-5p | miR-377-5p | miR-6848-5p | miR-6866-5p |
| hsa_circ_0068784 | 4.17569E-06 | 0.002492284 | 3.5480508 | down | miR-4673 | miR-3692-5p | miR-6754-5p | miR-23a-3p | miR-7113-5p |
| hsa_circ_0057090 | 4.9948E-07 | 0.00147459 | 3.4927401 | down | miR-1193 | miR-548k | miR-643 | miR-3064-5p | miR-4299 |
| hsa_circ_0056247 | 2.22076E-05 | 0.003841024 | 3.2888045 | down | miR-450a-2-3p | miR-23b-5p | miR-23a-5p | miR-511-5p | miR-7-5p |
| hsa_circ_0001204 | 1.64596E-05 | 0.003817991 | 3.2644161 | down | miR-612 | miR-657 | miR-362-3p | miR-377-3p | miR-136-5p |
| hsa_circ_0001747 | 0.000218714 | 0.007377021 | 3.149758 | down | miR-616-5p | miR-30d-3p | miR-320b | miR-320a | miR-302c-5p |
| hsa_circ_0017311 | 5.30031E-05 | 0.004765914 | 3.0933966 | down | miR-767-3p | miR-412-3p | miR-335-3p | miR-191-3p | miR-421 |
| hsa_circ_0047744 | 5.99584E-05 | 0.004765914 | 3.0829551 | down | miR-503-5p | miR-424-5p | miR-15a-5p | miR-16-5p | miR-497-5p |
| hsa_circ_0030569 | 0.001000539 | 0.014333503 | 3.0627677 | down | miR-30d-3p | miR-29a-5p | miR-584-3p | miR-30a-3p | miR-30e-3p |
| hsa_circ_0089866 | 4.7015E-05 | 0.004765914 | 3.0233465 | down | miR-1914-5p | miR-556-5p | miR-3692-5p | miR-4268 | miR-4640-5p |
Note: We selected 18 differentially expressed circRNAs with the highest fold-change (fold-change >3.0; P < 0.05) to predict their miRNAs response elements (MREs), including 9 up-regulated circRNAs and 9 down-regulated circRNAs. The circRNAs/miRNAs interaction was predicted with Arraystar’s home-made miRNA target prediction software based on TargetScan and miRanda.
Figure 6.
A snippet of the detailed annotation for circRNAs/miRNAs interaction. (A) hsa_circ_0001204. (B) hsa_circ_0043497. The circRNAs/miRNAs interaction was predicted with Arraystar’s home-made miRNA target prediction software based on TargetScan and miRanda. Binding sites of conserved miRNAs with good mirSVR scores are represented.
Figure 7.
Expression levels of hsa_circ_0043497 and hsa_circ_0001204 and their target miRNAs in Mtb-infected human MDMs. RT-qPCR was performed to evaluate the expression levels of hsa_circ_0043497 (A) and its top 5 predicted miRNA targets (miR-335-3p, miR-186-5p, miR-380-5p, miR-296-3p and miR-522-3p) (B), hsa_circ_0001204 (C) and its top 5 predicted miRNA targets (miR-612, miR-657, miR-362-3p, miR-377-3p and miR-136-5p) (D) in ten human MDMs after 24 h of infection with H37Rv. Data are expressed as the means ± SEM. The relative expression levels of circRNAs were normalized to levels of GAPDH and the relative expression levels of miRNAs were normalized to levels of U6. **P < 0.01, ***P < 0.001 compared to the control group.
Discussion
CircRNAs are a new class of long non-coding RNAs which do not have 5′ or 3′ ends but are covalently linked to form a closed circular structure. Recent evidence has suggested that circRNAs play essential roles in various physiological and pathological processes15. The dysregulation of circRNAs have also been implicated in the occurrence and progression of many human diseases16. However, the relationship between circRNAs and TB infection has not been reported.
In this study, we comprehensively profiled the circRNAs expression of human MDMs response to Mtb infection in order to improve our understanding of the pathogenesis of TB infection as well as identify potential circRNAs biomarkers for TB. We demonstrated that the expression of 142 circRNAs was significantly different in the Mtb-infected group compared with uninfected group. Among these, 32 circRNAs were up-regulated, while 110 circRNAs were down-regulated. Most of differentially expressed circRNAs originate from the exons. Some are from introns, while, a few are other sources. We then randomly selected five circRNAs from the differentially expressed cricRNAs with a fold change >3.0 and performed a quantitative real time PCR to examine these circRNAs’ expression levels in Mtb-infected MDMs. RT-qPCR results were generally consistent with the microarray data. To our knowledge, this is the first report on the expression of circRNAs in the human MDMs response to TB infection.
Early diagnosis of TB infection is essential for controlling the spread of the disease and providing early therapy for TB epidemic17. Several groups have reported the potential use of circRNAs as promising biomarkers for detection of human diseases18,19. For example, recently, Cui et al.20 showed that hsa_circRNA_103636 in PBMCs can be used as a novel biomarker for the diagnosis and treatment of major depressive disorder. Chen et al.21 reported that hsa_circ_0000190 yields a diagnostic characteristic with a sensitivity of 71.2% and a specificity of 75.0% for detection of gastric cancer. However, there are no reports regarding the use of circRNAs as biomarkers for TB. In this study, there were many differentially expressed circRNAs induced by Mtb infection, which indicated that circRNAs have the potential to be novel biomarkers for TB. To identify the clinically applicable biomarkers, 5 significant difierentially expressed circRNAs were selected for further analysis. We found the expression of hsa_circ_0043497 and hsa_circ_0001204 were significantly elevated in TB infected patients compared to healthy subjects. In ROC plots, the AUC was 0.856 for hsa_circ_0043497 and 0.882 for hsa_circ_0001204. Next, we investigated the effect of anti-TB drug treatment on hsa_circ_0043497 and hsa_circ_0001204 expression. In the present study, we observed that the levels of hsa_circ_0043497 and hsa_circ_0001204 which were altered in naive TB patients reverted to normal after completion of anti-TB therapy and proven to be acid-fast bacillius (AFB) negative in sputum. However, a larger sample size is needed to confirm our results.
Some studies have revealed that circRNAs could function as miRNAs sponges, modulate alternative splicing or transcription, and regulate the expression of parental genes7,8. To evaluate hsa_circ_0043497 and hsa_circ_0001204 potential functions, we investigated potential miRNAs binding with hsa_circ_0043497 and hsa_circ_0001204. The potential miRNAs targets of hsa_circ_0043497 include miR-335-3p, miR-186-5p, miR-380-5p, miR-296-3p and miR-522-3p. For hsa_circ_0001204, the potential miRNAs targets include miR-612, miR-657, miR-362-3p, miR-377-3p and miR-136-5p. These potential target miRNAs were verified in MDMs with or without Mtb infection using RT-qPCR. And the data of RT-qPCR showed that the expression levels of miR-377-3p were markedly elevated and the expression levels of miR-186-5p were significantly reduced in Mtb-infected MDMs. However, due to the limited known function of circRNAs and miRNAs, a lot of circRNAs/miRNAs interactions should be analyzed in the future.
In conclusion, in the study, we described differentially expressed circRNAs in human MDMs response to TB infection. Furthermore, hsa_circ_0043497 and hsa_circ_0001204 were identified as potential non-invasive molecular markers for rapid diagnosis of TB. Our findings shed a novel light on our understandings of the pathogenesis of TB infection. To our knowledge, this is the first research addressing circRNAs expression profiles in macrophages response to TB infection. Further studies should focus on the function of circRNAs involved in TB infection, which may lead to new theories for TB pathogenesis and give new potentially therapeutic targets in active TB.
Electronic supplementary material
Acknowledgements
We thank all the donors who participated in work. This work was supported by the grants from the National Natural Science Foundation of China (81560001, 81560344), the Province Natural Science Foundation of Jiangxi Province (20171BAB205111), the Foundation for Distinguished Young Scientists of Jiangxi Province (20171BCB23087), the Foundation of Scientific & Technical Research Project of Jiangxi Province (20151BBG70208, 2015BBG70227), and the Science and Technology Project of Health and Family Planning Commission of Jiangxi Province of China (20161018).
Author Contributions
Z.H. and J.L. conceived and designed the experiments. Z.H., R.S., Z.D., J.X. and Q.L. performed the experiments. Z.H., Q.L. and J.L. analyzed data, prepared the figures and drafted the manuscript. All authors reviewed and agreed on the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-13885-0.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Qing Luo, Email: lxc042@163.com.
Junming Li, Email: lisir361@163.com.
References
- 1.Dheda K, Barry CE, 3rd, Maartens G. Tuberculosis. Lancet. 2016;387:1211–1126. doi: 10.1016/S0140-6736(15)00151-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Global tuberculosis report. World Health Organization (2016).
- 3.Peddireddy V, Doddam SN, Ahmed N. Mycobacterial Dormancy Systems and Host Responses in Tuberculosis. Front Immunol. 2017;8:84. doi: 10.3389/fimmu.2017.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Z, et al. Mycobacterium tuberculosis-Induced Polarization of Human Macrophage Orchestrates the Formation and Development of Tuberculous Granulomas In Vitro. PLoS One. 2015;10:e0129744. doi: 10.1371/journal.pone.0129744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korb VC, Chuturgoon AA, Moodley D. Mycobacterium tuberculosis: Manipulator of Protective Immunity. Int J Mol Sci. 2016;17:131. doi: 10.3390/ijms17030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Memczak S, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 7.Ashwal-Fluss R, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 8.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer JW, Leung AK. CircRNAs: a regulator of cellular stress. Crit Rev Biochem Mol Biol. 2017;52:220–233. doi: 10.1080/10409238.2016.1276882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Z, et al. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci Rep. 2017;7:39918. doi: 10.1038/srep39918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He J, Xie Q, Xu H, Li J, Li Y. Circular RNAs and cancer. Cancer Lett. 2017;396:138–144. doi: 10.1016/j.canlet.2017.03.027. [DOI] [PubMed] [Google Scholar]
- 12.Fan X, et al. Circular RNAs in Cardiovascular Disease: An Overview. Biomed Res Int. 2017;2017:5135781. doi: 10.1155/2017/5135781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lukiw W. J. Circular RNA (circRNA) in Alzheimer’s disease (AD). Front Genet. 2013;4:307. doi: 10.3389/fgene.2013.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Z, et al. Identification of Differentially Expressed Long Non-coding RNAs in Polarized Macrophages. Sci Rep. 2016;6:19705. doi: 10.1038/srep19705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 16.Salzman J. Circular RNA Expression: Its Potential Regulation and Function. Trends Genet. 2016;32:309–316. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goletti D, Petruccioli E, Joosten SA, Ottenhoff TH. Tuberculosis Biomarkers: From Diagnosis to Protection. Infect Dis Rep. 2016;8:6568. doi: 10.4081/idr.2016.6568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang F, Nazarali AJ, Ji S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am J Cancer Res. 2016;6:1167–1176. [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Z, et al. Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol. 2017;54:237–245. doi: 10.1007/s00592-016-0943-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui X, et al. hsa_circRNA_103636: potential novel diagnostic and therapeutic biomarker in Major depressive disorder. Biomark Med. 2016;10:943–952. doi: 10.2217/bmm-2016-0130. [DOI] [PubMed] [Google Scholar]
- 21.Chen S, Li T, Zhao Q, Xiao B, Guo J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta. 2017;466:167–171. doi: 10.1016/j.cca.2017.01.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.