Abstract
Japanese encephalitis (JE) is a major contributor for viral encephalitis in Asia. Vaccination programme has limited success for largely populated JE endemic countries like India and disease exposure is unavoidable. Involvement of chemokines and its co-receptors for adverse prognosis of JE have been documented both in vitro and in vivo. Identification of the genetic predisposing factor for JE infection in humans is crucial but not yet established. Therefore, we investigated the association of single nucleotide polymorphisms (SNPs) in chemokines (CCL2 and CCL5) and its co-receptors (CCR2 and CCR5) with their protein level for JE. The study enrolled 87 symptomatic JE cases (mild: severe = 24:63) and 94 asymptomatic controls. Our study demonstrated that CCL2 (rs1024611G), CCL5 (rs2280788G) and CCR2 (rs1799864A) significantly associated with JE (Odds ratio = 1.63, 2.95 and 2.62, respectively and P = 0.045, P = 0.05 and P = 0.0006, respectively). The study revealed that rs1024611G allele was associated with elevated level of CCL2. CCL5 elevation associated with JE mortality having a Cox proportional hazard of 1.004 (P = 0.033). In conclusion, SNPs of chemokine viz. CCL2 (rs1024611G) and its receptor CCR2 (rs1799864A) significantly associated with JE which may serve as possible genetic predisposing factor and CCL5 protein level may act as marker for disease survival.
Introduction
Japanese encephalitis (JE), a mosquito borne disease, is one of the major viral encephalitis in Asia and parts of Western Pacific1. It is estimated that globally 67,900 people are infected with Japanese encephalitis virus (JEV) per year. Among the JE endemic countries, India is one of the largest contributors of the disease burden2. Persistent JE outbreaks since the last four decades have been reported from the northeast (NE) India, particularly the state of Assam which confers around 30–40% of the country’s total JE burden (http://nvbdcp.gov.in/Doc/je-aes-cd-May16.pdf).
Several mechanisms have been postulated for JE related morbidity and mortality but exact aetiology still remains unclear. Differential phenotypic spectrum upon JE infection seems a unique feature for the disease morbidity where it varies from asymptomatic to symptomatic3. Epidemiological survey documented that JE associated mortality ranges from 20–30% among the symptomatic individuals4. Studies have shown that host innate and adaptive immune response activated by JEV infection, are involved in controlling viral loads in infected humans5. Beside other flaviviruses viz. West Nile virus (WNV), Tick borne encephalitis virus (TBEV) and Dengue, the differential expressions of cytokines and chemokines are observed in mice brain and spleen during JEV infection6–9. Altered homeostasis of chemokines including CCL2 (also known as MCP-1 [monocyte chemoattractant protein]), CCL3 (also known as MIP-1α [macrophage inflammatory protein 1-α]), CCL5 (also known as RANTES [regulated on activation, normal T cell expressed and secreted]), CXCL8 (IL-8) and cytokines viz. tumour necrosis factor α (TNFα), interleukin-6 (IL-6), IL-1β have been associated with the disease outcome10–14. In vitro studies demonstrated expression of cytokines and chemokines in neural cells after JEV infection11,15. Although chemokine cascade has been identified as a most potent contributor in JEV infected mice, reports have not yet been substantiated in humans6.
Differential phenotypic outcome among the individuals upon infection may hypothesize the aetiology of the disease to be multi-factorial, complex and dependant on host immune as well as genomic architecture. Recent genetic studies among varied ethnic groups have identified multiple genetic markers for several viral and bacterial diseases16,17 with a series of functional potency. Studies are extremely limited for identifying the genetic predisposing factors that may quantify the risk for JE. Therefore, detection of genetic variants of chemokines and their functional impact on disease status is imperative for deeper understanding of the disease mechanism.
Hence, in this present study, we have designed a case control association study to map functionally important single nucleotide polymorphisms (SNPs) of the candidate genes viz. CCL2, CCL5, CCR2 and CCR5 with morbidity of JE. Further, this study aims to evaluate the chemokine level and its interaction with genomic polymorphism to understand the differential phenotypic outcome of the disease.
Results
Vaccination status and gender were matched among the study subjects, though older individuals were predominantly affected by severity of the disease (P = 0.003) (Supplement Table S1). Incidence of fatal outcome was significantly higher among the severe JE cases compared to mild cases (P = 0.005).
Association of the SNPs of chemokines and their co-receptors for JE
Genotyping was done for the entire group of 87 cases (JE patients) and 94 controls. Electrophoretic gel images for genotyping each studied SNPs are shown in Fig. 1. No significant departure from Hardy Weinberg equilibrium (HWE) was observed at rs1024611A/G (CCL2-2518), rs2857656G/C (CCL2 -362), rs2280788C/G (CCL5 -28), rs2107538A/G (CCL5 -403), rs1799864G/A (CCR2 V64I) and rs1799987G/A (CCR5 59029) loci among the controls (Table 1).
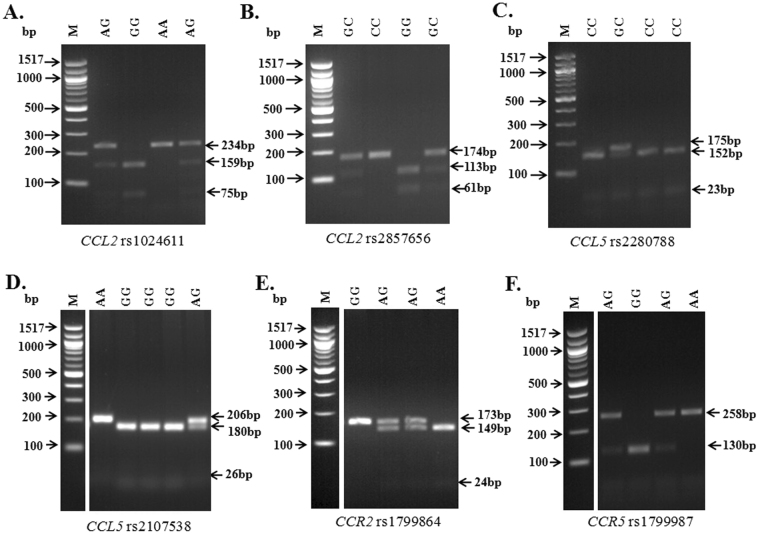
Figure 1.
Genotyping of all the studied SNPs in agarose gel electrophoresis. (A) represents CCL2 rs1024611A/G where AA homozygote was identified by a single 234 bp fragment, GG homozygote was characterized by 159 bp and 75 bp while AG heterozygote was identified by 234 bp, 159 bp and 75 bp fragments; (B) represents CCL2 rs2857656G/C where CC genotype was identified by uncut 174 bp band, GG was identified by two fragments of 113 and 61 while CG heterozygote was characterized by 174 bp, 113 bp and 61 bp; (C) represents CCL5 rs2280788C/G where CC genotype was identified by 152 bp and 23 bp fragments while for GC heterozygote, three bands at 175 bp, 152 bp and 23 bp was observed; (D) represents CCL5 rs2107538G/A where an uncut single band of 206 bp corresponded to AA homozygote, 180 bp and 26 bp corresponded to GG homozygote while presence of three bands at 206 bp, 180 bp and 26 bp was identified as AG heterozygote; (E) denotes CCR2 rs1799864A/G where an uncut single band of 173 bp was identified as GG, 149 bp and 24 bp was characterized by AA homozygote while AG heterozygote was identified by 173 bp, 149 bp and 24 bp fragments; (F) represents CCR5 rs1799987A/G where AA genotype was characterized by 258 bp fragment, GG genotype was identified by 130 bp fragment while for AG heterozygote bands at 258 bp and 130 bp was observed. Images (D), (E) and (F) are vertically sliced images of juxtaposed lanes that were non-adjacent to the molecular marker and are delineated with white space. Gel images shown here are cropped images and full length gels are presented in Supplementary Fig. S7. Abbreviations: M, molecular size marker; bp, base pairs.
Table 1.
Genotype and allele distribution among cases and controls.
| Gene rs id | HWE for controls χ2 (P value) | Genotype/Allele | Case (n = 87) | Control (n = 94) | Odds ratio (95% CI) | P value |
|---|---|---|---|---|---|---|
| CCL2 rs1024611 | 1.13 (P = 0.287) | AA | 47 | 57 | 0.762 (0.42–1.37) | 0.452 |
| AG | 22 | 30 | 0.722 (0.37–1.38) | 0.411 | ||
| GG | 18 | 7 | 3.24 (1.28–8.2) | 0.016* | ||
| A | 116 | 144 | 0.611 (0.38–0.96) | 0.046 | ||
| G | 58 | 44 | 1.636 (1.03–2.59) | 0.046* | ||
| CCL2 rs2857656 | 2.34 (P = 0.126) | GG | 56 | 60 | 1.023 (0.55–1.88) | 1 |
| GC | 21 | 27 | 0.789 (0.40–1.53) | 0.505 | ||
| CC | 9 | 7 | 1.434(0.51–4.03) | 0.603 | ||
| G | 133 | 147 | 0.951 (0.57–1.56) | 0.899 | ||
| C | 39 | 41 | 1.051 (0.63–1.72) | 0.899 | ||
| CCL5 rs2280788 | 0.07 (P = 0.791) | CC | 75 | 89 | 0.35 (0.118–1.041) | 0.072 |
| GC | 11 | 5 | 2.57 (0.857–7.744) | 0.115 | ||
| GG | 1 | 0 | — | |||
| C | 161 | 183 | 0.33 (0.118–0.969) | 0.05 | ||
| G | 13 | 5 | 2.95 (1.031–8.47) | 0.05* | ||
| CCL5 rs2107538 | 3.10 (P = 0.078) | GG | 39 | 46 | 0.84 (0.472–1.521) | 0.655 |
| AG | 36 | 34 | 1.24 (0.684–2.268) | 0.541 | ||
| AA | 11 | 14 | 0.82 (0.353–1.934) | 0.673 | ||
| G | 114 | 126 | 0.96 (0.623–1.499) | 0.911 | ||
| A | 58 | 62 | 1.03 (0.666–1.603) | 0.911 | ||
| CCR2 rs1799864 | 2.34 (P = 0.125) | GG | 48 | 74 | 0.33 (0.173–0.637) | 0.0008* |
| AG | 27 | 17 | 2.03 (1.017–4.081 | 0.05 | ||
| AA | 9 | 3 | 3.5 (0.915–13.382) | 0.072 | ||
| G | 123 | 165 | 0.40 (0.231–0.702) | 0.001* | ||
| A | 45 | 23 | 2.62 (1.508–4.567) | 0.0006* | ||
| CCR5 rs1799987 | 3.60 (P = 0.057) | GG | 31 | 35 | 0.93 (0.509–1.710) | 0.877 |
| AG | 34 | 35 | 1.08 (0.593–1.971) | 0.878 | ||
| AA | 22 | 20 | 1.25 (0.627–2.499) | 0.598 | ||
| G | 96 | 105 | 0.97 (0.642–1.473) | 0.916 | ||
| A | 78 | 83 | 1.02 (0.678–1.556) | 0.916 |
P values were calculated by chi square test and * denotes statistical significance at P < 0.05. Abbreviations: rs ID, reference SNP cluster ID; HWE, Hardy Weinberg Equilibrium; CI, confidence interval; P value, level of significance.
For three tested SNPs, significant differences in allele frequencies were observed across JE cases and control groups. In JE cases, the frequency of rs1024611G allele (CCL2) was significantly higher than the control group (case: 36.78% vs. control: 23.4%; OR = 1.636, 95%CI = 1.03–2.59, P = 0.04; Table 1). In case of CCL5 rs2280788, our analysis revealed an increased frequency of G allele among cases than controls (JE cases: 7.47% vs. control: 2.65%; OR = 2.95, 95%CI = 1.031–8.47, P = 0.05; Table 1). The data revealed that the CCL2 rs1024611G (OR = 2.337, 95%CI = 1.2–4.54, P = 0.017) and CCL5 rs2280788G (OR = 5.22, 95%CI = 1.523–17.947, P = 0.01) significantly associated with mild phenotype compared to controls (Supplementary Table S2).
Genotype of CCR2 rs1799864 documented as protected allele where the frequency of rs1799864G significantly reduced among the cases with an odds ratio of 0.40 (95% CI: 0.231–0.702, P = 0.001). Study further revealed that the risk of the disease significantly increased by 2.62 fold for rs1799864A of CCR2 (95%CI = 1.508–4.567, P = 0.0006). Moreover, CCR2 rs1799864A allele frequency was significantly higher among mild and severe JE cases than controls (mild JE cases: 54.16% vs. control: 36.50%, P = 0.01; severe JE cases: 26.22% vs. controls: 12.23%, P = 0.002) (Supplementary Table S2).
Among the six tested SNPs, the study did not reveal significant association for rs2857656G/C of CCL2, rs2107538G/A of CCL5 and rs1799987G/A of CCR5 genotypes. Moreover, genotype and allele frequencies of all the studied SNPs did not differ significantly among mild and severe JE cases.
Combined allelic association between JE cases and controls
To evaluate synergic effect for alleles of associated SNPs, χ2 test was used. The study revealed that the individuals lacking the variant allele of CCL2 rs1024611, CCL5 rs2280788 and CCR2 rs1799864, in combination, decreased the risk of the disease by 0.47 fold (95%CI = 0.253–0.878; P = 0.02) (Table 2). However, no significant difference of combined SNPs was observed across mild and severe JE cases.
Table 2.
Frequency distribution of combined alleles among cases and controls.
| Combination allele | Case n = 87 (%) | Control n = 94 (%) | OR (95% CI) | P value |
|---|---|---|---|---|
| All 3 SNPs are dominant | 4 (4.59) | 0 | — | — |
| Two SNPs have dominant allele | 19 (21.83) | 11 (11.70) | 2.108 (0.939–4.733) | 0.074 |
| One SNP has dominant allele | 38 (43.67) | 42 (44.68) | 0.960 (0.533–1.727) | 1 |
| None of the SNP has dominant allele | 24 (27.58) | 42 (44.68) | 0.471 (0.253–0.878) | 0.020* |
P values were calculated by chi square test and * denotes statistical significance at P < 0.05. Abbreviations: CI, confidence interval; P value, level of significance.
Serum protein levels and its relation with genotype
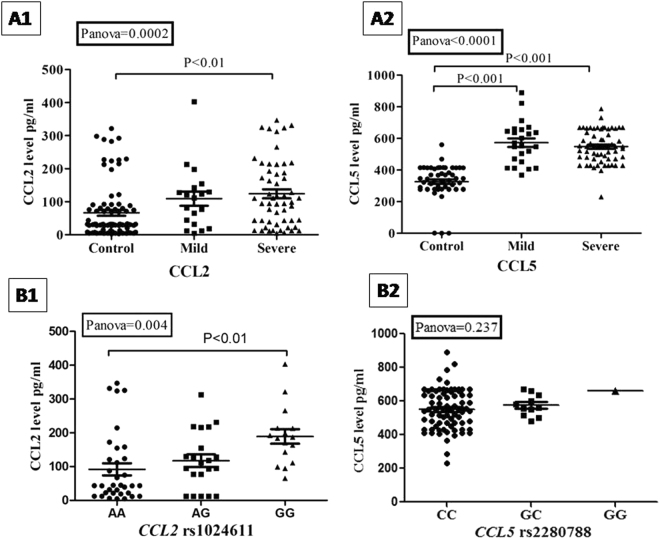
Serum concentration of CCL2 (sCCL2) and CCL5 (sCCL5) was significantly elevated among JE cases compared to controls (P = 0.0002 and P < 0.0001, respectively) (Fig. 2). Tukey’s test demonstrated that the mean concentration of sCCL2 was significantly elevated among severe JE cases than controls (P < 0.01) where the difference is insignificant between mild and severe phenotype of JE. In addition, the concentration of sCCL5 was significantly elevated among the cases compared to control group (P < 0.0001). We also performed non-parametric test for the data which showed similar trend of result (Supplementary Table S3). Figure 2 shows the P- values for parametric tests only.
Figure 2.
Summary of serum chemokine protein concentration among study groups (A1 and A2) and according to genotypes of CCL2 rs1024611 as well as CCL5 rs2280788 (B1 and B2) in cases. P values were calculated by one way ANOVA with Tukey’s multiple comparison test.
The study further examined the differential distribution pattern of serum chemokine viz.CCL2 and CCL5 with respect to their genotypes among the JE cases. The study found concomitant elevation of sCCL2 concentration level with the increase of rs1024611G allele in a significant manner (P < 0.01) (Fig. 2). Additionally, Tukey’s test revealed that levels of sCCL2 was significantly higher (P = 0.004) among the cases with rs1024611GG genotype (189.10 ± 21.6 pg/ml) and lower among the rs1024611AA genotype (91.96 ± 16.29 pg/ml). However, we did not find any significant distributional difference of sCCL5 in context of rs2280788 genotype (550.8 ± 120.2 pg/ml in CC, 574.4 ± 70.3 pg/ml in GC and 661.6 pg/ml in GG; P = 0.237).
Independent effect of SNPs and their interaction with JE
Binary logistic regression analysis of case- control data revealed that rs1799864A allele of CCR2 significantly increased the disease risk with a log odds of 2.28 (P = 0.022) (Table 3). Concentration of sCCL2 and sCCL5 increased significantly the disease risk by 2.752 (P = 0.005) and 2.748 (P = 0.005) fold, respectively. Gene and protein interaction i.e. effect of corresponding variant allele and its subsequent elevation of sCCL2 levels significantly increased the disease risk with a log odds of 2.024 (P = 0.043). However, interactions among the other SNPs did not reveal any significant effect on the disease status.
Table 3.
Association of chemokine proteins, SNP genotypes and its interaction to explain the disease status.
| Independent variable | Standard error | Z value | P value | |
|---|---|---|---|---|
| Independent effect of serum protein level on disease status | CCL2 | 0.01883 | 2.752 | 0.00593* |
| CCL5 | 0.03439 | 2.748 | 0.00599* | |
| Independent effect of genotype on disease status | rs1024611 | 0.29991 | 1.053 | 0.292431 |
| rs2857656 | 0.27033 | 0.101 | 0.919405 | |
| rs2107538 | 0.29961 | 1.442 | 0.149178 | |
| rs2280788 | 1.30589 | 1.273 | 0.203187 | |
| rs1799864 | 0.38339 | 2.284 | 0.022391* | |
| rs1799987 | 0.25315 | −0.437 | 0.661915 | |
| Effect of genotype-genotype interaction on disease status | rs1024611: rs2280788 | 0.79428 | 0.444 | 0.65703 |
| rs1024611: rs1799864 | 0.41118 | 0.145 | 0.88486 | |
| rs2280788: rs1799864 | 1.03252 | −1.360 | 0.17398 | |
| Effect of genotype-protein interaction on disease status | rs1024611:CCL2 | 0.00341 | 2.024 | 0.0430* |
| rs1799864:CCL2 | 0.00389 | 0.300 | 0.76408 | |
| rs2280788:CCL5 | 1.151e + 01 | 0.004 | 0.99696 |
*Denotes P value significant at <0.05 level. Logistic regression was performed on case (n = 87) and control (n = 94) as dependant variable while the selected SNPs and protein were taken as the independent variable. It was used to fit a regression model to examine the effect of a protein level and genotype on the disease status. Abbreviations: Z value, regression coefficients divided by standard error; P value, level of significance.
Survival analysis
The cases were followed up for one year post diagnosis. The overall survival rate at one year was 64.36% and the mean age of the fatal cases were 43 years (SD ± 19). The Kaplan Meier survival analysis did not reveal any significant difference among the genotypes of the associated SNPs (see Supplementary Fig. S4). The proportional death hazard significantly increased (P = 0.033) by 0.4% with increase of CCL5 protein even after considering together with other variables (age, gender, CCL2 protein, rs1024611, rs2280788 and rs1799864) in Cox proportional hazard model (Table 4).
Table 4.
Cox proportional hazard analysis with putative prognostic factors for survival.
| Selected factor | Hazard ratio for death (95% CI) | P value |
|---|---|---|
| CCL5 | 1.004 (1.0003–1.008) | 0.033* |
| CCL2 | 0.998 (0.994–1.003) | 0.557 |
| Age | 0.990 (0.969–1.011) | 0.358 |
| Gender | 1.155 (0.485–2.749) | 0.744 |
| SNP1 (rs1024611) | 1.035 (0.446–2.405) | 0.934 |
| SNP4 (rs2280788) | 0.710 (0.146–3.445) | 0.671 |
| SNP5 (rs1799864) | 1.297 (0.574–2.934) | 0.530 |
P values were calculated by chi square test and * denotes statistical significance at P < 0.05. Abbreviations: CI, confidence interval; P value, level of significance.
Discussion
Intra-cellular chemokine cascade may play a pivotal role for anti viral host defence18. Murine model and in-vitro studies have potentially demonstrated the activation of chemokine on JEV infection6,15. Identification of genomic markers for JE endemic area seems to be crucial but studies are substantially limited. Present study reveals that the variant allele of chemokines viz. CCL2 and CCL5 along with CCR2 receptor are significantly associated with the disease.
Previous study demonstrated that chemokines are crucially involved in trafficking of peripheral monocytes, lymphocytes and neutrophils into the site of infection18. Among the varied group of chemokines, CCL2 is recognized as the most effective regulator of monocyte infiltration but its impact on JEV infection and its severity is still unexplored. During the severity of JEV infection, the virus usually infiltrates blood brain barrier (BBB) to CNS leading to influx of leukocytes. The infected cells in response trigger the production of cytokine and chemokine cascade which ultimately causes neuroinflammation. It has been also postulated that the elevated level of inflammatory cytokines viz. IL-6 and TNFα and chemokines viz. CCL5 and IL-8 may crucially be associated with adverse prognosis of JE12,14. As observed in our study, rs1024611G and rs2280788G at the promoter of CCL2 and CCL5 respectively, increases risk of the disease 1.63 and 2.95 folds significantly (Table 1). The study also demonstrated that chemokine concentration of sCCL2 and sCCL5 level significantly increased among the cases compared to controls (P = 0.0002 and P < 0.0001, respectively) (Fig. 2). Previous study demonstrated that the level of serum CCL2 significantly elevated upon administration of live attenuated JE vaccine in humans19. Recently, it was documented that expression of CCL2 elevated 1387.79 fold in JEV infected mice brain as one of the topmost hits from microarray dataset6. Elevated plasma concentration of CCL5 has been documented on JEV infected cases14. Repeated intra and inter species studies documented the crucial involvement of chemokine and cytokines for the adverse prognosis of JE. Single nucleotide polymorphisms (SNPs) found in CCL2 and CCL5 gene (chromosome 17), in CCR2 and CCR5 gene (chromosome 3) have been related to infectious diseases including West Nile virus20,21, dengue22,23, HIV24,25, Hepatitis C26 and tuberculosis27. However, there are no available reports which relate chemokine and its co-receptor SNPs to JE. Till date, genetic studies associating host genotype and JE are limited only to the analysis of TNFα and TLR3 polymorphisms28,29. rs1024611G and rs2280788G are associated with communicable diseases like pulmonary tuberculosis30,31 and Hepatitis C32,33. Not only in infectious diseases, transition of adenine (A) to guanine (G) at CCL2 (−2518) locus i.e. rs1024611G signify important risks for variety of non communicable diseases. G allele of the locus is associated with coronary artery disease risk (OR = 2.2, 95% CI 1.25–3.92, P < 0.005)34 as well as systemic lupus erythematosus (OR = 4.2, 95% CI 1.8–9.6, P < 0.0001)35. Multi factorial genetic model reveals that rs2280788G may act as a predicting marker for the adverse prognosis of diabetic nephropathy36.
Our study revealed that G allele of CCL2 promoter i.e. rs1024611 was significantly associated with elevated serum levels of the CCL2 where rs1024611AA individuals showed significantly reduced level of the same (91.96 ± 16.29 pg/ml) compared to variant homozygous genotype i.e. rs1024611GG (P < 0.01). Transition from A to G which is associated with elevated sCCL2 concentration may postulate its functional potentiality on transcription regulation. Furthermore, ChipSeq data reveals that the transcription factor viz. Myb and NFκB binds to CCL2 promoter locus (rs1024611) and may modulate the gene regulation. A functional study reported that rs1024611, located in the promoter region, has been implicated with increased CCL2 expression37. Persistent replication of studies including the present observation suggests that rs1024611G could possibly result in an up-regulation of CCL2 and might boost leukocyte trafficking into CNS. This could trigger massive inflammation and may be related to adverse prognosis of the disease. On the other hand, very few functional studies on rs1799864 are available and report its involvement on the stability of CCR2 by a missense mutation from valine to isoleucine at the 64th position of the protein38. Studies have documented that CCR2 genetic deficiency may result in partial or entire absence of leukocytes in inflamed tissue in WNV and JEV in mice model39,40. Repeated observations postulate that rs1024611G, with 8 regulatory motifs, may act as a genomic marker with its strong functional potentiality.
The logistic regression analysis further revealed that rs1799864A allele significantly increases the disease risk 2.284 fold independently and the gene- protein interaction of rs1024611G and CCL2 protein level significantly associated with the odds of 2.024 (P = 0.043) for JE (Table 3). Further multinomial regression revealed rs1799864A independently associated with the mild phenotype of JE (P = 0.028) and gene-protein interaction for rs1024611G and CCL2 significantly explain the mild phenotype (P = 0.015) (Supplementary Table S5).
Our study also observed that rs1799864GG of CCR2 was significantly less among the cases compared to controls that may provide protection by reducing the risk of JE by 0.33 fold for the present JE endemic population. Present study also reveals that the absence of variant allele of rs1024611, rs2280788 and rs1799864 synergistically reduces the disease risk 0.47 fold, P = 0.02. However, a previous study documented rs1799864A allele to provide protective effect among the human Pappiloma virus infected Brazilian patients41. Contradictory events for rs1799864 may implicate the heterogeneity among the population architecture (Supplementary Table S6). The allelic and genotype distribution in our study population were evidently different from other non JE endemic African and American population (Supplementary Table S6). Although the complete genomic architecture is still enigmatic for the population, the genotype frequencies of the tested associated SNPs were similar to the South Asian population reported in the 1000 Genomes Project Phase III database except rs1799864G that significantly decreased the disease risk. Additionally, it must be noted that the study population has close geographical proximity with the other highly JE endemic Southeast Asian countries. Fatality is the most adverse outcome that associated with mild and severe phenotype. When JEV evades the host’s primary immune response, it enters into the CNS and damages the non renewable neuronal cells. This may further induce severe inflammation causing encephalitis and death. According to the WHO reports, the case fatality has been recorded as 30% which is similar to our study where case fatality in JE patients is 35.63% (Supplementary Table S1). The study observed that 32% of mild JE cases and 67.7% from severe cases had a fatal outcome which is significantly high in severe cases (P = 0.005) (Supplementary Table S1). The study demonstrated that serum CCL5 significantly associated with the mortality of JE infection with Cox proportional hazards (P = 0.03) (Table 4).
In the present study, adults were observed to be predominantly suffering from JE. This could be attributed to regular childhood immunization programmes for the past 10 years in the study population. This phenomenon of changing age specificities of JE has been previously documented in other JE endemic areas with child immunizations4. Although a definite explanation is dubious, male preponderance among severe JE cases is well reflected in this study as well as previous studies42,43.
No significant difference was observed when SNPs were compared among mild and severe JE cases. This could be probably explained with further longitudinal studies on respective chemokine and its receptor gene expression. Though the present pilot study is limited with its sample size, but replication of findings in a large cohort is extremely important for validation. As accepted, gene expression is governed by many regulatory factors rather than exclusively by promoter. Thereby, it is possible that other genetic influences may have a role in the prognosis of the disease which was not evaluated in our study.
In conclusion, the present findings suggests that CCL2 (rs1024611G), CCL5 (rs2280788C) and CCR2 (rs1799864A) is associated with JEV infection and may act as a predictive marker for the adverse prognosis of JE in the population from northeast India. Moreover, rs1024611G (risk allele) is associated with increased serum CCL2 and may play a critical role in its adverse prognosis. Notably, CCL5 protein levels can be crucial marker for survival of JE as it is significantly associated with the fatal outcome.
Materials and Methods
Study cohort and diagnosis
Present cross sectional study comprised of 181 subjects (Case: 87 and Controls: 94). Cases were recruited from Assam Medical College & Hospital and other hospitals in Dibrugarh, India. The diagnosis of acute JE infection was based on clinical symptoms and serological positivity in accordance with WHO criteria44. The likelihood of cross reactivity with other circulating flavivirus viz. West Nile (WN) virus was considered. As a result, JE infected individuals nonreactive to WN specific IgM antibodies were confirmed as case. Cross-reactivity with dengue was not tested as the clinical symptoms are distinctly different from JE.
Cases were further stratified as mild and severe JE on the basis of their clinical symptoms as diagnosed by clinicians. Mild JE (n = 24) was characterized as JE patients who had acute fever, irritability, headache and muscle pain while patients with paralysis and encephalitis (altered mental status varying from confusion to coma) were considered as severe JE (n = 63). Asymptomatic serologically confirmed JE positive individuals from JE endemic area were considered as controls. People with other infections within one month, chronic inflammatory and autoimmune diseases were excluded from the study. Each case was followed up for one year and health status was obtained through structured questionnaire. The study was approved by the Institutional Ethics Committee of Regional Medical Research Centre, Indian Council of Medical Research (ICMR), Assam, India. Informed written consent from each participant was obtained prior to the study. Additionally, the study was carried out in accordance with Declaration of Helsinki and all methods were performed according to the recommended guidelines and regulations.
Sampling
Venous blood was collected from study participants in EDTA vials (for genomic DNA isolation) and sterile plain vials for serum. The samples were stored at −80 °C until further processing.
In silico annotation for SNP selection in promoter and coding region
The study used the UCSC Genome Browser and ChipSeq data to predict the functional potentiality of single nucleotide polymorphisms (SNPs) that are present in CCL2, CCL5, CCR2 and CCR5 promoter and coding regions in 1000 Genome datasets (http://www.1000genomes.org/home). Predictions about the functional consequences of its variant region and impact were assessed using HaploregV345.
Genotyping
Genomic DNA was isolated by QIAamp Blood DNA kit (Qiagen, USA; Cat no.: 51104) from each study subject as per manufacturer’s instructions from venous blood. Genotyping for the candidate SNPs of chemokine and chemokine receptor genes viz. rs1024611A/G (-2518) and rs2857656G/C (-362) of CCL2, rs2107538G/A (-403) and rs2280788C/G (-28) of CCL5, rs1799864A/G (V64I/+190) of CCR2 and rs1799987G/A (-59029) of CCR5 was performed by PCR-restriction fragment length polymorphism (RFLP)46–48. Details of SNP gene location, primer sequences, cycling conditions, and restriction enzymes (RE) with the amplicon size are given in Table 5.
Table 5.
Details of PCR primers and conditions for genotyping of study SNPs.
| Gene | rsID (location) | Primer (5′-3′) | Annealing temperature (°C) | Amplicon size (bp) | Restriction Enzyme | Allele size (bp) |
|---|---|---|---|---|---|---|
| CCL2 | rs1024611 (−2518A/G) | F-TCTCTCACGCCAGCACTGACC | 56 | 238 | PvuII | G = 159,75 A = 234 |
| R-GAGTGTTCACATAGGCTTCTG | ||||||
| rs2857656 (−362G/C) | F-GAGCCTGACATGCTTTCATCTA | 58 | 174 | Hpy188I | G = 113,61 C = 174 | |
| R-TTTCCATTCACTGCTGAGAC | ||||||
| CCL5 | rs2280788 (−28C/G) | F-ACTCCCCTTAGGGGATGCCCG | 55 | 175 | HincII | C = 152,23 G = 175 |
| R-GCGCAGAGGGCAGTAGCAAT | ||||||
| rs2107538 (−403G/A) | F-CACAAGAGGACTCATTCCAACTCA | 50 | 206 | RsaI | G = 180,26 A = 206 | |
| R-GTTCCTGCTTATTCATTACAGATCGTA | ||||||
| CCR2 | rs1799864 (V64I) | F-TTGGTTTTGTGGGCAACATGATGG | 56 | 173 | BsaBI | A = 149,24 G = 173 |
| R-CATTGCATTCCCAAAGACCCACTC | ||||||
| CCR5 | rs1799987 (59029 G/A) | F-CCCGTGAGCCCATAGTTAAAACTC | 65 | 258 | Bsp1286I | G = 130 A = 258 |
| R-TCACAGGGCTTTTCAACAGTAAGG |
Abbreviations: rs ID, reference SNP cluster ID; bp, base pair.
All PCRs were done in 20 μl reaction volumes using 2X Master Mix (Promega, Wisconsin, USA; Cat no.: M7505). PCR was performed using Applied Biosystems Veriti 96 well Thermal Cycler (California, USA). The amplified products were digested with 1.5U of specific restriction enzyme (New England Biolabs, Massachusetts, USA) as per manufacturer’s instructions and subsequently separated by electrophoresis on 3% agarose gel stained with 1 mg/ml ethidium bromide.
Serum CCL2 and CCL5 protein assay
Protein concentration of CCL2 and CCL5 were determined from serum using commercially available ELISA kits as per manufacturer’s instruction (RayBio®, USA; Cat no.: ELH-MCP1-001 and ELH-RANTES-1). The minimum detectable quantity of CCL2 and CCL5 as per the used kit is 2 pg/ml and 3 pg/ml respectively.
Statistical analysis
Age, gender, vaccination status and fatality were compared between JE (mild and severe) cases and controls using chi square (χ2) and two-tailed Student’s t-test. Genotypic frequency among cases and controls were compared byχ2 test. Hardy Weinberg equilibrium was analysed byχ2 test with P > 0.05 considered as equilibrium. Odds ratio (OR) was calculated using Fisher’s exact test at 95% confidence interval (95% CI). Protein level was analyzed by one way ANOVA and Tukey’s multiple comparison tests among the study groups viz. mild, severe and control. Data were presented in mean ± standard deviation of mean (SD). Logistic regression model was used to fit a regression model to examine the effect of chemokine protein level and genotypes on the disease (cases vs. control). The dependant variable i.e. cases and control was coded as 1 and 0 respectively. In addition to the independent SNP effect, combined or synergic effect was also tested to examine if risk alleles of the three associated loci could magnify the genetic risk usingχ2 test. The survival analysis for surviving and fatal JE cases was estimated using Kaplan Meier method. The possible predictive significance of chemokine proteins and SNPs on JE survival was assessed using Cox proportional-hazards modelling. The p-value < 0.05 was considered as significant. All the statistical analysis was conducted in R package (version 3.2) and GraphPad Prism 5.
Data availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).
Electronic supplementary material
Acknowledgements
The authors thank the Director and the entire arbovirology division of Regional Medical Research Centre (ICMR), Assam, India. The authors are also grateful to Niranjan K. Baruah for his excellent field support. The fund for this study was provided at Regional Medical Research Institute, Assam, India and supported by Indian Council of Medical Research, New Delhi, India.
Author Contributions
P.C. and S.A.K. designed the study and wrote the manuscript. P.C. performed all the experiments and interpreted the data. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-14091-8.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lobo DA, Velayudhan R, Chatterjee P, Kohli H, Hotez PJ. The Neglected Tropical Diseases of India and South Asia: Review of heir Prevalence, Distribution, and Control or Elimination. PLoS Neglected Tropical Diseases. 2012;5:e1222. doi: 10.1371/journal.pntd.0001222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang H, Liang G. Epidemiology of Japanese encephalitis: past, present, and future prospects. Therapeutics and Clinical Risk Management. 2015;11:435–448. doi: 10.2147/TCRM.S51168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Solomon T, et al. Japanese encephalitis. Journal of Neurology, Neurosurgery, and Psychiatry. 2000;68:405–415. doi: 10.1136/jnnp.68.4.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischer, M. et al. Japanese encephalitis prevention and control: advances, challenges, and new initiatives in Emerging Infections (ed. Scheld, W.M., Hammer, S.M., Hughes, J.M.) 93-124 (ASM Press, 2008)
- 5.Abraham, S., Shwetank, G.K. & Manjunath, R. Japanese encephalitis virus: innate and adaptive immunity. FLAVIVIRUS ENCEPHALITIS, 339, 10.5772/22083 (2011).
- 6.Yang Y, et al. Japanese encephalitis virus infection induces changes of mRNA profile of mouse spleen and brain. Virology Journal. 2011;8:80. doi: 10.1186/1743-422X-8-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zajkowska J, et al. Evaluation of CXCL10, CXCL11, CXCL12 and CXCL13 chemokines in serum and cerebrospinal fluid in patients with tick borne encephalitis (TBE) Advances in medical sciences. 2011;56:311–317. doi: 10.2478/v10039-011-0033-z. [DOI] [PubMed] [Google Scholar]
- 8.Shirato K, Kimura T, Mizutani T, Kariwa H, Takashima I. Different chemokine expression in lethal and non‐lethal murine west nile virus infection. Journal of medical virology. 2004;74:507–513. doi: 10.1002/jmv.20205. [DOI] [PubMed] [Google Scholar]
- 9.Rathakrishnan A, et al. Cytokine expression profile of dengue patients at different phases of illness. PloS one. 2012;7:e52215. doi: 10.1371/journal.pone.0052215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen CJ, et al. Glial activation involvement in neuronal death by Japanese encephalitis virus infection. Journal of General Virology. 2010;91:1028–1037. doi: 10.1099/vir.0.013565-0. [DOI] [PubMed] [Google Scholar]
- 11.Ghoshal A, et al. Proinflammatory mediators released by activated microglia induces neuronal death in Japanese encephalitis. Glia. 2007;55:483–496. doi: 10.1002/glia.20474. [DOI] [PubMed] [Google Scholar]
- 12.Ravi V, et al. Correlation of tumor necrosis factor levels in the serum and cerebrospinal fluid with clinical outcome in Japanese encephalitis patients. Journal of Medical Virology. 1997;51:132–136. doi: 10.1002/(SICI)1096-9071(199702)51:2<132::AID-JMV8>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 13.Saxena V, Mathur A, Krishnani N, Dhole TN. Kinetics of cytokine profile during intraperitoneal inoculation of Japanese encephalitis virus in BALB/c mice model. Microbes and Infection. 2008;10:1210–1217. doi: 10.1016/j.micinf.2008.06.015. [DOI] [PubMed] [Google Scholar]
- 14.Winter PM, et al. Proinflammatory Cytokines and Chemokines in Humans with Japanese Encephalitis. The Journal of Infectious Diseases. 2004;190:1618–26. doi: 10.1086/423328. [DOI] [PubMed] [Google Scholar]
- 15.Chen CJ, Chen JH, Chen SY, Liao SL, Raung SL. Upregulation of RANTES Gene Expression in Neuroglia by Japanese Encephalitis Virus Infection. Journal of Virology. 2004;78:12107–12119. doi: 10.1128/JVI.78.22.12107-12119.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapman SJ, Hill AV. Human genetic susceptibility to infectious disease. Nature Reviews Genetics. 2012;13:175–88. doi: 10.1038/nrg3114. [DOI] [PubMed] [Google Scholar]
- 17.Asner SA, Morré SA, Bochud PY, Greub G. Host factors and genetic susceptibility to infections due to intracellular bacteria and fastidious organisms. Clinical Microbiology and Infection. 2014;20:1246–53. doi: 10.1111/1469-0691.12806. [DOI] [PubMed] [Google Scholar]
- 18.Larena, M. & Lobigs, M. Immunobiology of Japanese encephalitis virus. Flavivirus Encephalitis, 317–338, 10.5772/21777 (2011).
- 19.Zhang JS, Zhao QM, Zuo SQ, Jia N, Guo XF. Cytokine and chemokine responses to Japanese encephalitis live attenuated vaccine in a human population. International Journal of Infectious Diseases. 2012;16:e285–e288. doi: 10.1016/j.ijid.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 20.Bigham AW, et al. Host genetic risk factors for West Nile virus infection and disease progression. PloS one. 2011;6:e24745. doi: 10.1371/journal.pone.0024745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durrant DM, Daniels BP, Pasieka T, Dorsey D, Klein RS. CCR5 limits cortical viral loads during West Nile virus infection of the central nervous system. Journal of neuroinflammation. 2015;12:p.233. doi: 10.1186/s12974-015-0447-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guabiraba R, et al. Role of the chemokine receptors CCR1, CCR2 and CCR4 in the pathogenesis of experimental dengue infection in mice. PLoS One. 2010;5:e156802010. doi: 10.1371/journal.pone.0015680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alagarasu K, Bachal RV, Damle I, Shah PS, Cecilia D. Association of FCGR2A p. R131H and CCL2 c.-2518 A > G gene variants with thrombocytopenia in patients with dengue virus infection. Human immunology. 2015;76:819–822. doi: 10.1016/j.humimm.2015.09.042. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez E, et al. HIV-1 infection and AIDS dementia are influenced by a mutant MCP-1 allele linked to increased monocyte infiltration of tissues and MCP-1 levels. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:13795–13800. doi: 10.1073/pnas.202357499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ometto L, et al. Polymorphisms in the CCR5 promoter region influence disease progression in perinatally human immunodeficiency virus type 1–infected children. Journal of Infectious Diseases. 2001;183:814–8. doi: 10.1086/318828. [DOI] [PubMed] [Google Scholar]
- 26.Montes-Cano MA, et al. CCL2-2518 A/G and CCR2 190 A/G do not influence the outcome of hepatitis C virus infection in the Spanish population. World journal of gastroenterology. 2007;13:2187–2192. doi: 10.3748/wjg.v13.i15.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng WX, et al. Tag SNP polymorphism of CCL2 and its role in clinical tuberculosis in Han Chinese pediatric population. PLoS One. 2011;6:e14652. doi: 10.1371/journal.pone.0014652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pujhari SK, Ratho RK, Prabhakar S, Mishra B, Modi M. TNF-α promoter polymorphism: a factor contributing to the different immunological and clinical phenotypes in Japanese encephalitis. BMC infectious diseases. 2012;12:23. doi: 10.1186/1471-2334-12-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biyani S, et al. Toll-like receptor-3 gene polymorphism in patients with Japanese encephalitis. Journal of neuroimmunology. 2015;286:71–6. doi: 10.1016/j.jneuroim.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 30.Ganachari M, et al. Joint effect of MCP-1 genotype GG and MMP-1 genotype 2G/2G increases the likelihood of developing pulmonary tuberculosis in BCG-vaccinated individuals. PLoS One. 2010;5:e8881. doi: 10.1371/journal.pone.0008881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alqumber MA, et al. A genetic association study of CCL5-28 C > G (rs2280788) polymorphism with risk of tuberculosis: a meta-analysis. PloS one. 2013;8:e83422. doi: 10.1371/journal.pone.0083422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huik K, et al. A CCL5 Haplotype Is Associated with Low Seropositivity Rate of HCV Infection in People Who Inject Drugs. PloS one. 2016;11:e0156850. doi: 10.1371/journal.pone.0156850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mühlbauer M, et al. A novel MCP-1 gene polymorphism is associated with hepatic MCP-1 expression and severity of HCV-related liver disease. Gastroenterology. 2003;125:1085–1093. doi: 10.1016/S0016-5085(03)01213-7. [DOI] [PubMed] [Google Scholar]
- 34.Szalai C, et al. Involvement of polymorphisms in the chemokine system in the susceptibility for coronary artery disease (CAD). Coincidence of elevated Lp(a) and MCP-1–2518 G/G genotype in CAD patients. Atherosclerosis. 2001;158:233–239. doi: 10.1016/S0021-9150(01)00423-3. [DOI] [PubMed] [Google Scholar]
- 35.Brown KS, et al. Monocyte chemoattractant protein-1: plasma concentrations and A (-2518)G promoter polymorphism of its gene in systemic lupus erythematosus. Journal of Rheumatology. 2007;34:740–746. [PubMed] [Google Scholar]
- 36.Blech I, et al. Predicting diabetic nephropathy using a multifactorial genetic model. PloS one. 2011;6:e18743. doi: 10.1371/journal.pone.0018743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rovin BH, Lu L, Saxena R. A novel polymorphism in the MCP-1 gene regulatory region that influences MCP-1 expression. Biochemical and Biophysical Research Communications. 1999;259:344–348. doi: 10.1006/bbrc.1999.0796. [DOI] [PubMed] [Google Scholar]
- 38.Nakayama EE, Tanaka Y, Nagai Y, Iwamoto A, Shioda T. A CCR2-V64I polymorphism affects stability of CCR2A isoform. AIDS. 2004;18:729–38. doi: 10.1097/00002030-200403260-00003. [DOI] [PubMed] [Google Scholar]
- 39.Kim JH, et al. CCL2, but not its receptor, is essential to restrict immune privileged central nervous system‐invasion of Japanese encephalitis virus via regulating accumulation of CD11b + Ly‐6Chi monocytes. Immunology. 2016;149:186–203. doi: 10.1111/imm.12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim JK, et al. Chemokine receptor Ccr2 is critical for monocyte accumulation and survival in West Nile virus encephalitis. The Journal of Immunology. 2011;186:471–478. doi: 10.4049/jimmunol.1003003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santos EU, et al. CCR2 and CCR5 genes polymorphisms in women with cervical lesions from Pernambuco, Northeast Region of Brazil: a case-control study. Memórias do Instituto Oswaldo Cruz. 2016;111:174–80. doi: 10.1590/0074-02760150367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borah J, Dutta P, Khan SA, Mahanta J. A comparison of clinical features of Japanese encephalitis virus infection in adult and pediatric age group with Acute Encephalitis Syndrome. Journal of Clinical Virology. 2011;52:45–49. doi: 10.1016/j.jcv.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 43.Saxena SK, Mishra N, Saxena R, Singh M, Mathur A. Trend of Japanese encephalitis in North India: evidence from thirty-eight acute encephalitis cases and appraisal of niceties. The Journal of Infection in Developing Countries. 2009;3:517–30. doi: 10.3855/jidc.470. [DOI] [PubMed] [Google Scholar]
- 44.Solomon T, et al. A cohort study to assess the new WHO Japanese encephalitis surveillance standards. Bulletin of the World Health Organization. 2008;86:178–86. doi: 10.2471/BLT.07.043307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic acids research. 2012;40:D930–4. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye DQ, et al. The correlation between monocyte chemoattractant protein-1and the arthritis of systemic lupus erythematosus among Chinese. Archives of Dermatological Research. 2005;296:366–371. doi: 10.1007/s00403-004-0531-y. [DOI] [PubMed] [Google Scholar]
- 47.Thye T, et al. MCP-1 promoter variant 2362C associated with protection from pulmonary tuberculosis in Ghana, West Africa. Human Molecular Genetics. 2009;18:381–388. doi: 10.1093/hmg/ddn352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abdi R, et al. Chemokine receptor polymorphism and risk of acute rejection in human renal transplantation. Journal of the American Society of Nephrology. 2002;13:754–758. doi: 10.1681/ASN.V133754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).