Abstract
Aims: Organismal aging can be delayed by mutations that either activate stress responses or reduce the nutrient-sensing pathway signaling; thus, by using Drosophila melanogaster as an in vivo experimental screening platform, we searched for compounds that modulate these pathways.
Results: We noted that oral administration of the glycogen synthase kinase 3 (Gsk-3) inhibitor 6-bromoindirubin-3′-oxime (6BIO) in Drosophila flies extended healthy life span. 6BIO is not metabolized in fly tissues, modulated bioenergetic pathways, decreased lipid and glucose tissue load, activated antioxidant and proteostatic modules, and enhanced resistance to stressors. Mechanistically, we found that the effects on the stress-responsive pathways were largely dependent on the activity of the transcription factor nuclear factor erythroid 2-related factor (Nrf-2). Genetic inhibition of Gsk-3 largely phenocopied the 6BIO-mediated effects, while high levels of Gsk-3 expression and/or kinase activity suppressed proteostatic modules and reduced flies' longevity; these effects were partially rescued by 6BIO. Also, 6BIO was found to partially reduce the 3-phosphoinositide-dependent protein kinase-1 (Pdpk1) activity, a major effector of the insulin/insulin-like growth factor-1 cell signaling pathways.
Innovation: 6BIO exerts the unique property of increasing stress tolerance and in parallel partially suppressing the nutrient-sensing pathway signaling.
Conclusion: Our findings suggest that the 6BIO scaffold can be used for the development of novel antiaging compounds. Antioxid. Redox Signal. 27, 1027–1047.
Keywords: : 6-bromoindirubin-3′-oxime, aging, Gsk-3, Nrf-2, Pdpk1, proteostasis
Introduction
Organismal aging is a complex molecular process that relates to time-dependent decline of stress resistance and functional capacity, which leads to increased risk of morbidity and mortality; accordingly, age is the major risk factor for several diseases, including cancer, cardiovascular disease, neurodegeneration, and diabetes (31). It is nowadays evident that both healthspan (the disease-free period of life) and/or life span (maximum longevity) can be prolonged by genetic and/or diet interventions, suggesting that animals have the latent potential to live longer than they normally do. Specifically, several studies at model organisms have demonstrated that longevity can be increased by caloric restriction (CR) or gene mutations that reduce the activity of nutrient sensing, for example, the insulin/insulin-like growth factor-1 (InS/IGF-1) cell signaling pathways (1, 2, 15).
Alternatively, CR activates the AMP-activated kinase (AMPK) signaling pathway that functions as a nutrient and energy sensor, which represses anabolic pathways under conditions of reduced ATP cellular levels (1). It is assumed that CR induces a physiological state similar to that resulting from periods of food shortage triggering thus the mild (hormetic) activation of cellular defenses that eventually results in lower rates of stressors and/or damaged biomolecule accumulation in cells.
Innovation.
Life span can be increased by caloric restriction (CR) or by gene mutations that either activate stress responses or reduce the activity of nutrient signaling pathways. Nevertheless, it is challenging to implement CR in the population and genetic interventions cannot be applied in humans. We report herein that oral administration of the Gsk-3 inhibitor 6-bromoindirubin-3′-oxime (6BIO) in Drosophila flies prolonged healthy life span, enhanced stress tolerance, and modulated bioenergetic pathways; mechanistically, these effects were largely dependent on Nrf-2 activation. Thus, 6BIO (along with other indirubins found in edible mollusks) represents a scaffold for the development of novel antiaging dietary compounds.
The complex network of cellular defense pathways includes antioxidant molecules and enzymes, as well as several transcription factors that function as stress sensors and mobilize downstream genomic responses; these (among many others) include forkhead box O (Foxo), heat shock factor-1 (Hsf-1), and nuclear factor erythroid 2-related factor (Nrf-2). Nrf-2 plays a central role in the protection of cells against oxidative and/or xenobiotic damage by binding on genomic antioxidant response elements (AREs) and stimulating the expression of phase II and antioxidant genes (26, 50). Central to cell defense pathways are also the proteome damage responses (PDR) that ensure proteome stability during proteotoxic stress by activating the proteostasis network (PN); key components of PN are the molecular chaperones and the two main degradation machineries, namely the autophagy/lysosome pathway (ALP) and the ubiquitin/proteasome system (UPS) (28, 52).
ALP is mainly involved in the degradation of protein aggregates and damaged organelles and is (among others) subject to Tor regulation (29), while UPS is the main site of protein synthesis quality control and is involved in the recycling of both normal short-lived proteins and of nonrepairable misfolded or unfolded proteins (56). The 26S eukaryotic proteasome is constituted from a 20S core particle (CP) bound to 19S regulatory particles (RP). The 20S CP consists of four stacked heptameric rings (two of α type surrounding two of β type) that form a barrel-like structure; the caspase-, trypsin-, and chymotrypsin-like proteasome peptidase activities are located at the β1, β2, and β5 proteasomal subunits, respectively. The 19S RP is involved in substrate recognition, deubiquitination, unfolding, and translocation into the 20S CP (25).
Notably, the functionality of both the stress response module and of the PN declines during in vivo aging (40, 53, 55) and this fact has emerged as a key factor that fuels the appearance of aging and/or age-related diseases (20). In support, pharmacological disruption of proteasome functionality in young Drosophila flies or feeding of flies with diet-derived advanced glycation end products (AGEs) disrupts proteostasis and significantly reduces fly longevity (54, 55). On the contrary, activation of stress responsive pathways seems (under certain conditions) to delay the age-related accumulation of stressors and damaged biomolecules in cells, resulting in increased longevity (20).
Nonetheless, little of this knowledge had been translated to humans despite the fact that the need to increase healthspan is becoming urgent from both an economic and health perspective; this gap in the translational research continuum mostly relates to the fact that genetic interventions cannot be applied in humans and it is challenging to implement CR (2, 15). Thus, many studies have been devoted to the identification of synthetic or dietary natural products (NPs) (e.g., extracts or isolated compounds) that can prolong healthspan in model organisms. Preferred bioactivities in low (preferentially dietary relevant) doses could relate to either dampening the signaling from nutrient-sensing pathways, thus mimicking the CR systemic effect(s) (dietary restriction mimetics), or to mildly (hormetically) activating the stress responsive pathways. Indeed, several NPs have showed beneficial effects against aging or age-related diseases in model organisms; yet, in the majority of the reports, the underlying molecular mechanism(s) remain elusive (2).
We have been using Drosophila flies as an in vivo experimental screening platform for the isolation of dietary molecules with antiaging effects and for elucidation of the molecular basis of their function, since, apart from Drosophila powerful genetics, a number of key metabolic, antioxidant, proteostatic, and age-related pathways are conserved among flies and mammals. Our study was focused on the hemisynthetic cell-permeable indirubin derivative 6-bromoindirubin-3′-oxime (6BIO). Indirubins are a family of bis-indoles naturally occurring in edible gastropod mollusks and plants. Many indirubins by acting through competition with ATP binding are in fact dual inhibitors of both cyclin-dependent kinases (CDKs) and glycogen synthase kinase-3β (Gsk-3β) (34); a serine/threonine kinase originally identified for its role in phosphorylating, and negatively regulating, glycogen synthase (Gys), the rate-limiting enzyme in glycogen synthesis (5).
Mammals express two isoforms of Gsk-3 (-α and -β) with high homology and similar functions but distinct tissue distributions (5), while in Drosophila, only one Gsk-3 ortholog (named Shaggy) has been identified (37). Gsk-3 phosphorylates a wide range of substrates, including metabolic enzymes, signaling molecules, and transcription factors, and it thus influences several cellular functions, including cell survival, differentiation, proliferation, and metabolism; accordingly, Gsk-3 has been implicated in various pathologies, including carcinogenesis, neurodegeneration, and diabetes (5).
6BIO was developed as a potent and selective Gsk-3β inhibitor (38), while studies in cancer cells suggested that it may also inhibit 3-phosphoinositide-dependent protein kinase-1 (Pdpk1) (62), a major effector of the InS/IGF-1 pathway (18). We and others have previously shown that 6BIO exerts a wide range of interesting pharmacological properties, including modulation of mammalian stem cell pluripotency (46), as well as antineurodegenerative (3) and antitumor (8) activities. Nevertheless, the in vivo long-term effect(s) of dietary 6BIO and/or other indirubin derivatives in physiology and aging have not been studied. We report herein that orally delivered 6BIO in Drosophila flies bypasses the intestine barrier, it is not metabolized and increases flies' healthy life span by modulating bioenergetic pathways and activating cytoprotective modules.
Results
Oral administration of 6BIO in Drosophila flies modulated cytoprotective and metabolic pathways; it also extended flies' healthy life span
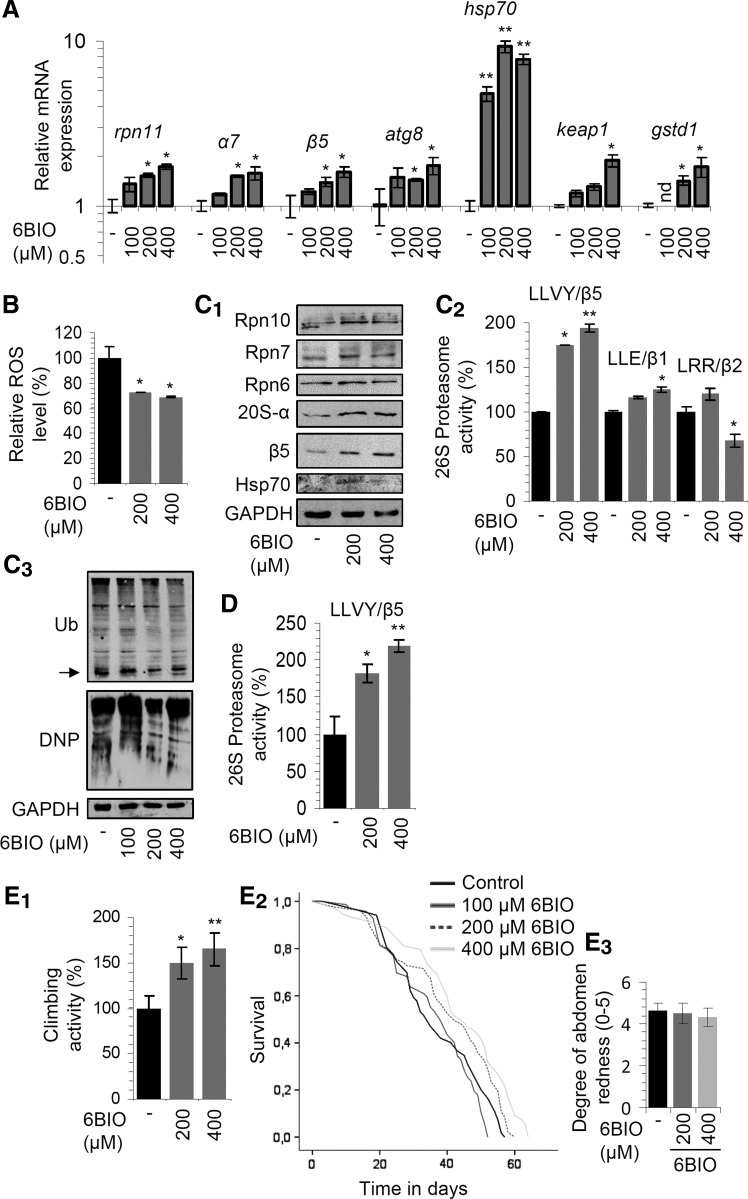
To identify bioactive dietary NPs that modulate antiaging pathways in higher metazoans, we used Drosophila flies as an in vivo experimental screening platform. Among the several NPs that positively regulated flies' longevity (data to be reported elsewhere), the indirubin derivative 6BIO was found to induce in young flies dose-dependent (100–400 μM) upregulation of main proteostatic genes, including 19S (rpn11) and 20S (α7, β5) proteasome genes, the autophagy-related gene 8 (atg8), and the hsp70 chaperone gene. We also noted the induction of genes involved in antioxidant responses, namely keap1 and gstd1 (Fig. 1A). In line with these findings, we found reduced levels of tissue reactive oxygen species (ROS) (Fig. 1B), upregulation of proteasomal protein subunits and of the Hsp70 chaperone (Fig. 1C1), as well as increased proteasome peptidase activities, namely chymotrypsin (LLVY/β5), caspase (LLE/β1), and trypsin like (LRR/β2) (Fig. 1C2); in support of the 6BIO-mediated increased proteasome functionality, we noted reduced levels of ubiquitinated and carbonylated proteins in the somatic tissues of 6BIO-treated flies (Fig. 1C3). Higher proteasome peptidase activities were also found in middle-aged 6BIO-fed flies (Fig. 1D), while feeding of flies with 6BIO delayed the age-related decline of locomotor activity (Fig. 1E1).
FIG. 1.
Oral administration of 6BIO in flies activated cytoprotective modules and increased healthy life span. (A) Relative expression of rpn11, α7, β5, atg8, hsp70, keap1, and gstd1 genes in young flies fed with the indicated concentrations of 6BIO for 6–9 days. (B) Relative (%) ROS levels in flies after 6BIO treatment with the shown 6BIO concentrations. (C1) Representative immunoblot analyses of tissue protein samples probed with antibodies against Rpn10, Rpn7, Rpn6, 20S-α, β5, and Hsp70. (C2) Relative (%) 26S proteasome activities in Drosophila tissues following treatment with the indicated doses of 6BIO. (C3) Immunoblot analyses of total protein ubiquitination (Ub; arrow denotes free ubiquitin) and carbonylation (DNP) in tissues of flies treated with increasing concentrations of 6BIO; treatment of flies in (B–C3) was as in (A). (D) Relative (%) chymotrypsin-like activity in tissues of middle-aged (at ∼45–55% of their life span) flies being exposed to the indicated concentrations of 6BIO. (E1) Climbing activity (%) of middle-aged flies constantly exposed or not (normal aging) to 200 or 400 μM 6BIO. (E2) Longevity curves of flies exposed to the indicated concentrations of 6BIO; flies' median life span and comparative statistics are reported in details in Supplementary Table S1. (E3) Quantitation of gustatory assays in flies fed (or not) with the indicated doses of 6BIO. Control samples always refer to treatment with the equivalent amount of the 6BIO solvent. Gene expression was plotted versus the respective control set to 1. (B, C2, D, E1) Control values were set to 100%; (E3) the absolute mean values are shown. The rp49 gene expression was used in (A) as a reference for total RNA input; probing with GAPDH was used in (C1, C3) as a total protein loading reference. Bars ± SD (n ≥ 2); *p < 0.05; **p < 0.01. See Western blot in Supplementary Figure S12. 6BIO, 6-bromoindirubin-3′-oxime; ROS, reactive oxygen species; SD, standard deviation.
Furthermore, 6BIO extended median and maximum life span at concentrations of 200 and 400 μM (Fig. 1E2; statistical analyses of all longevity assays are presented in Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/ars) in a sex-independent manner with no effects (at the doses used) on development or fecundity (not shown) and the rate of food consumption (Fig. 1E3). We also studied the effects of 6BIO at another well-established biomarker of in vivo aging-related biomolecule damage, namely AGEs (23). To this end, the rate of AGE accumulation was studied by confocal laser scanning microscopy (CLSM) in isolated fly brain tissues, as well as by immunoblotting in somatic tissue lysates. As shown in Supplementary Figure S1A by both assays, we observed reduced levels of AGEs in the tissues of 6BIO-treated flies. Furthermore, we tested the effects of 6BIO in a fly model of Alzheimer's disease; that is, in double transgenic flies expressing the 695 amino acid isoform of human amyloid precursor protein (hAPP) and β-secretase (hBACE1). We found that treatment with 6BIO alleviated the disease-related decrease in middle-aged fly locomotion (climbing) activity (Supplementary Fig. S1B).
Given the fact that the genetic or dietary interventions that extend life span often impact on bioenergetic and/or nutrient-sensing pathways, we then assayed mitochondria functionality as well as sugar and lipid metabolism. We found that 6BIO upregulated the fusion/fission mitochondrial dynamics-related genes, namely mitochondrial assembly regulatory factor (marf), optic atrophy 1 (opa1), and dynamin-related protein (drp1), while it did not affect mitochondrial lon protease or the succinate dehydrogenase subunit A (sdhA) gene expression (Supplementary Fig. S2A); these effects were more prominent at 200 μM of 6BIO, indicating that the 6BIO concentration that activates antioxidant and proteostatic modules is likely higher than that which modulates mitostatic genes. In line with potential effects in mitochondria dynamics, we noted a trend (yet not statistically significant) for increased mitochondria numbers and mitochondrial network density in 6BIO-fed flies' tissues (Supplementary Fig. S2B). Furthermore, 6BIO increased the mitochondrial respiratory ST3/ST4 ratio (Supplementary Fig. S2C1) indicating increased coupling of the mitochondrial respiratory chain activity to oxidative phosphorylation and, likely, increased substrate usage; these findings coincided with increased phosphorylation and expression of AMPKα (Supplementary Fig. S2C2), which (among others) repress anabolic pathways (1).
Interestingly, prolonged treatment with 6BIO increased levels of trehalose and glycogen in middle-aged fly somatic tissues (Supplementary Fig. S3A1) and reduced glucose levels in both fly tissues and in the hemolymph (Supplementary Fig. S3A1, A2). Also, CLSM analyses of 6BIO-fed larvae fat bodies stained with the BODIPY dye (stains neutral lipids and lipid droplets) revealed significant reduction in lipid droplet number and staining intensity (Supplementary Fig. S3B), indicating a 6BIO-mediated lipolytic effect; in support, 6BIO was found to mildly sensitize flies to starvation conditions (data not shown).
Overall, orally delivered 6BIO promoted pleiotropic effects, including activation of the main cellular proteostatic and antioxidant modules, upregulation of mitochondrial energetics, reduction of glucose and lipid tissue levels, and extension of flies' healthy life span.
6BIO bypasses the intestine barrier, it is not metabolized in fly tissues, and can dock in the substrate binding pocket of Shaggy (Sgg; the fly Gsk-3 ortholog); 6BIO analogues with high selectivity against Sgg/Gsk-3 partially phenocopy the 6BIO effects
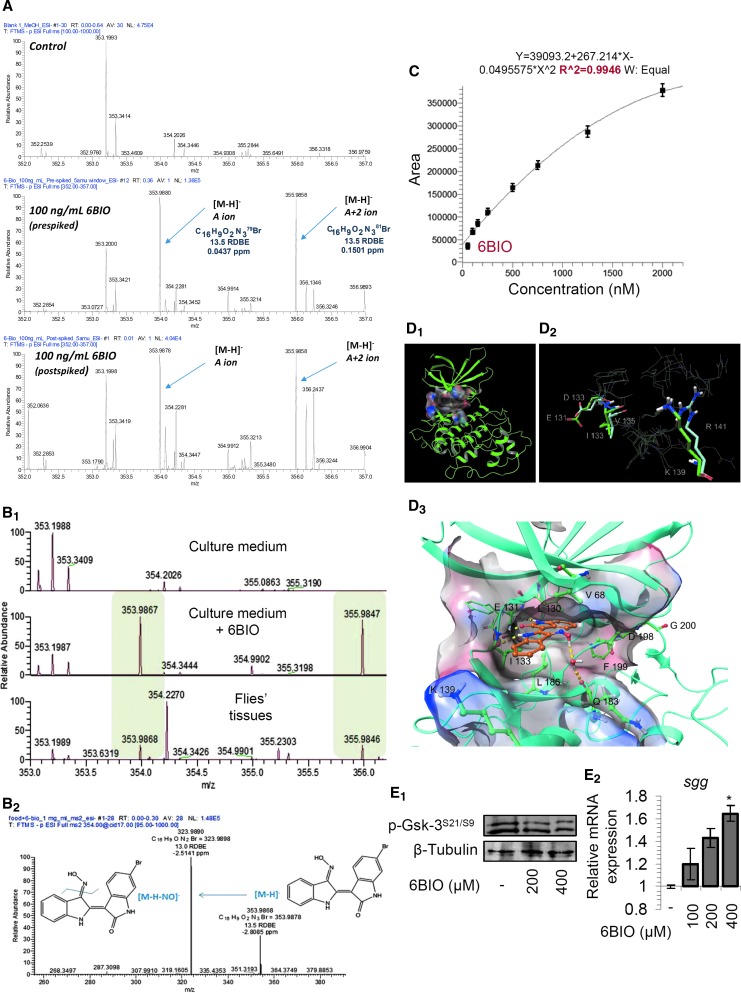
We then addressed the issue of whether 6BIO can pass the intestine barrier, as well as whether it is metabolized in fly tissues. To this end, we performed liquid chromatography/high-resolution mass spectroscopy (LC-HRMS) analyses in flies fed with 6BIO-supplemented culture medium (Fig. 2A–C). We first developed a 6BIO detection method using LC-HRMS and HRMS/MS analyses in tissue extracts after not-spiking, prespiking, or postspiking 400 μM 6BIO (Fig. 2A). Following the verification of the method accuracy and specificity (HRMS full scan and HRMS/MS) and the identification of spiked 6BIO in tissue extracts (Fig. 2A), we analyzed samples derived from either culture medium containing (or not) 400 μM 6BIO or from samples derived from flies fed (or not) with 400 μM 6BIO. As shown in Figure 2B, we noted the characteristic isotopic peaks of 6BIO in both the 6BIO-containing culture medium and in the treated fly tissue samples. With the use of a calibration curve, we also found that the actual 6BIO concentration in fly tissues ranges from 514.4 ng/ml (feeding with 400 μM 6BIO) up to 1849.2 ng/ml (feeding with 4000 μM 6BIO) (Fig. 2C). Given the fact that at low concentrations there is an almost linear correlation between the 6BIO concentration in culture medium and the HRMS-derived 6BIO concentration in tissue extracts, we concluded that the 6BIO concentration in fly tissues fed with 200 or 400 μM 6BIO should be in the range of ∼720 nM–1.44 μM, respectively. Reportedly, as little as 1 μM of 6BIO significantly inhibited Gsk-3 in human cells (34), and thus, the levels of 6BIO found in fly tissues are in the reported inhibitory range of 6BIO on Gsk-3.
FIG. 2.
6BIO is not metabolized in fly tissues and can effectively dock in the substrate binding pocket of Sgg/Gsk-3. (A) Full scan LC-HRMS analyses of fly tissue samples after no spiking (upper panel), prespiking (middle panel), or postspiking (bottom panel) of 400 μM 6BIO. The 6BIO ion is evident whether in pre- or postspiking as demonstrated by its characteristic isotope patterns. Additional information related to RDBE value and measurement accuracy (Δm in ppm) is also annotated verifying the identity of the molecule and the accuracy of the method. (B1) Comparative LC-HRMS spectra from standard culture medium (upper panel), culture medium plus 400 μM 6BIO (middle panel), and from tissue samples of flies fed with 400 μM 6BIO (lower panel); the isotopic peaks of 6BIO are detected in middle and lower panels at m/z 353.986 and 355.984. (B2) LC-HRMS/MS spectra of 6BIO where the main fragment [M-H-NO]− is detected confirming its accurate detection and identification. Additional spectrometric features such as suggested molecular formula, RDBE value, and Δm are also annotated. (C) Calibration curve used to determine 6BIO concentration in fly tissue samples; standard solutions were prepared at 50–2000 ng/ml. The curve showed a fitting of r2 > 0.99. (D1) Homology model of Sgg/Gsk-3 represented in ribbons; the surface of the catalytic site is displayed. (D2) Superimposition of the binding sites of Sgg/Gsk-3 (green) and mammalian Gsk-3 (turquoise); the differences in amino acid sequence and ligand (6BIO) are highlighted. (D3) 6BIO bound in Sgg/Gsk-3; the binding cavity has been surfaced and key residues are displayed. Ligands and residues are represented as ball and sticks and H-bonds are displayed in yellow dashed lines. Representative immunoblot analyses of protein samples probed with an antibody against p-Gsk-3S21/S9 (E1), and relative expression of sgg/gsk-3 gene (E2) in somatic tissues of young flies treated with the indicated 6BIO concentrations for 6–9 days. β-tubulin probing (E1) and rp49 gene expression (E2) were used as reference for total protein and RNA input, respectively; bars ± SD (n = 2); *p < 0.05. See Western blot in Supplementary Figure S13. Gsk-3, glycogen synthase kinase-3; LC-HRMS, liquid chromatography/high-resolution mass spectroscopy; RDBE, ring double bond equivalent.
We also synthesized a number of additional 6BIO derivatives, namely, 6,5-dichloroindirubin (794), 6BIO-piperazine (6BIO-Pip), and 6-bromo-5-nitroindirubin-3′-acetoxime (796) (Supplementary Fig. S4). All potent inhibitors of mammalian Gsk-3 that show minimal inhibitory effects on CDKs (38). We also used in our analyses 7-bromoindirubin-3′-oxime (7BIO) (38) that has only a marginal inhibitory activity toward CDKs and Gsk-3 (41). We found that feeding of flies with 400 or 800 μM of 794 increased proteasome and antioxidant gene expression (Supplementary Fig. S5A1), and it upregulated proteasomal protein subunit expression (Supplementary Fig. S5A2) and peptidase activities (Supplementary Fig. S5A3); moreover, all three concentrations of 794 tested (400, 600, or 800 μM) increased flies' life span (Supplementary Fig. S5A4). Similarly, 6BIO-Pip increased proteasome subunits and Hsp70 expression, reduced ubiquitinated proteins (Supplementary Fig. S5B1), and enhanced (at 150 μM) proteasome peptidase activities (Supplementary Fig. S5B2); notably, however, although 6BIO-Pip showed a trend to increase mean and median life span of flies up to 150 μM, it did not increase maximum longevity and differences were significant only at 50 μM of added 6BIO-Pip (Supplementary Fig. S5B3). The compound 796 only slightly induced proteasome expression and activities, while 7BIO did not significantly affect proteasome functionality (data not shown) or fly longevity (Supplementary Fig. S5C).
To gain insight on the possible mode of 6BIO (or the synthesized analogues) interaction with Sgg/Gsk-3, we then performed docking scoring calculations. We first built a homology model based on the amino acid sequence of Sgg/Gsk-3, which shows very high homology (∼84%) with the mammalian Gsk-3α/β orthologs; minor differences occurred mainly in the secondary structures of the protein, while the catalytic sites presented ∼98% homology (see Materials and Methods section). Homology model construction was performed using Prime Schrödinger® and the cocrystal structure of Gsk-3β with 6BIO (PDB 1UV5) (38) (Fig. 2D1). After minimization and loop refinement, some minor differences have been noted in the binding cavity (Fig. 2D2). Specifically, Val135GSK-3β is replaced by Ile133Sgg/Gsk-3, Asp133GSK-3β is substituted by Glu131Sgg/Gsk-3, and Arg141GSK-3β is exchanged by Lys139Sgg/Gsk-3.
The refined model was further subjected to molecular dynamics (10 ns) using Desmond, Schrödinger. Induced fit docking calculations (IFD; Schrödinger) were used to define the interaction modes of 6BIO, 794, 6BIO-Pip, and 796 with Sgg/Gsk-3. As it was anticipated due to high homology between the fly and human orthologs, the studied indirubins kept their characteristic binding mode. Concerning 6BIO, the interactions within the cavity implied the formation of a network of hydrogen bonds between N-1, O-2 with Ile133, and N′-1 with Glu131 (Fig. 2D3 and Supplementary Fig. S5D). The planar aromatic system of the bis-indole molecule interacted with hydrophobic residues through Van der Waals interactions (Leu186 and Val68), while the 3′-oxime moiety of 6BIO and of compound 796 could eventually bring further stabilization of the molecule through H-bond with surrounding water molecules. The ketone analogue 794 is expected to exhibit relatively lower affinity, while in agreement to previous findings in mammalian systems, 6BIO-Pip forms an additional H-bond with Asp200GSK-3β in the catalytic site (59). Overall, 6BIO and the synthesized analogues 6BIO-pip and 796 showed high predicted interaction energy against Sgg/Gsk-3.
We then studied the 6BIO-mediated effects on Sgg/Gsk-3 phosphorylation and gene expression status and observed reduced levels of the inhibitory Akt-mediated Sgg/Gsk-3 phosphorylation on Ser21/9 (37) (Fig. 2E1) and a dose-dependent induction of the sgg/gsk-3 gene expression (Fig. 2E2). These findings indicate the existence of a regulatory feedback loop aiming to restore physiological Sgg/Gsk-3 kinase activity levels; consistently, 6BIO-mediated inhibition of Gsk-3 in SH-SY5Y cells resulted in increased Gsk-3β expression levels (34).
Taken together, these observations suggest that most indirubin derivatives with high selectivity against Sgg/Gsk-3 exert (at the doses used) beneficial effects on fly physiology and longevity. The noted differences of the in vivo bioactivities of the compounds tested could be due to compound-specific inner physicochemical properties (e.g., solubility, cell permeability, metabolization, or intestinal barrier crossing) or to cross-selectivity against other kinases.
The 6BIO-mediated effects on cytoprotective modules and longevity largely depend on the functionality of the transcription factor cap'-n'-collar isoform-C; the Nrf-2 ortholog in Drosophila
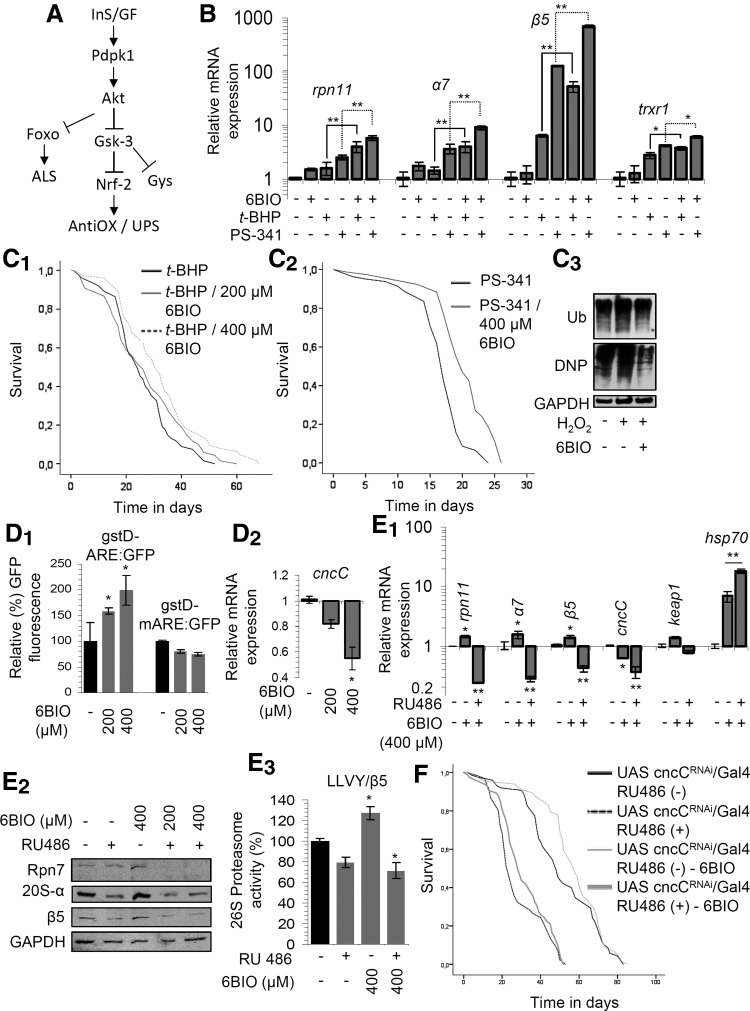
Given the activating effect of 6BIO on proteasome activities, we initially asked whether 6BIO could directly activate proteasome peptidases. In experiments where 6BIO was directly added (at concentrations ranging from 0.5 to 20 μM) in fly tissue lysates, we found no effect (or even a slight decrease) on proteasome activities (data not shown), and thus, 6BIO effects are mediated by indirect means (e.g., modulation of a proteasome regulatory mechanism). Considering the 6BIO inhibitory effect on Sgg/Gsk-3, the fact that Sgg/Gsk-3 is negatively regulated by nutrient-sensing pathways (mainly Akt), a number of reports showing that Gsk-3 exerts an inhibitory effect on Nrf-2 (24, 39), as well as that according to our recent findings cap'-n'-collar isoform-C (CncC)/Nrf-2 is a main regulator of antioxidant responses and proteasome functionality in fly somatic tissues (55) (Fig. 3A), we hypothesized that the 6BIO-mediated effects largely depend on CncC/Nrf-2 activation.
FIG. 3.
The 6BIO-mediated effects on antioxidant responses, proteasome activation, and longevity largely depend on the functionality of CncC/Nrf-2. (A) Schematic presentation of the InS/GF signaling pathway that culminates in Sgg/Gsk-3 suppression; additional modules (e.g., Foxo, Nrf-2, and Gys) and their reported regulatory inter-relationships are shown (→ denotes positive regulation and ⊣a negative regulatory effect). (B) Relative expression of the rpn11, α7, β5, and trxr1 genes in somatic tissues of young flies following exposure (for 3 days) to 400 μM 6BIO, 20 mM of the oxidant t-BHP, 5 μM of the specific proteasome inhibitor PS-341, or to shown combinations; as is evident, 6BIO significantly enhances genomic responses to oxidative (t-BHP) or to proteotoxic (PS-341) stress. Longevity curves of flies exposed to 4 mM t-BHP (C1) or to 1 μM PS-341 (C2) with or without the addition of the indicated doses of 6BIO. (C3) Immunoblotting analyses of ubiquitinated (Ub) or carbonylated (DNP) protein levels in fly tissues treated with 0.8% H2O2 with or without the addition of 400 μM 6BIO. (D1) Relative (%) GFP levels in the somatic tissues of young transgenic gstD-ARE:GFP or gstD-mARE:GFP flies after treatment with the indicated concentrations of 6BIO. (D2) Relative expression of the cncC/nrf-2 gene following exposure to the shown concentrations of 6BIO; in both (D1, D2), flies were treated with 6BIO for 6–9 days. Relative expression of the rpn11, α7, β5, cncC/nrf-2, keap1, and hsp70 genes (E1), representative immunoblot analyses of flies' protein samples probed with antibodies against Rpn7, 20S-α, and β5 (E2), and relative proteasome activity (E3) in somatic tissues of young flies after RNAi-mediated CncC/Nrf-2 knockdown; transgenic flies were treated (or not) for 5 days with the transgene inducer RU486 and the shown 6BIO doses. (F) Longevity curves of flies after CncC/Nrf-2 knockdown with or without the addition of 400 μM 6BIO. GAPDH probing (E2) and rp49 gene expression (B, D2, E1) were used as reference. Control sample values in (D1, E3) were set to 100%. Statistics of the longevity assays is shown in Supplementary Table S1. Bars ± SD (n ≥ 2); *p < 0.05; **p < 0.01. See Western blot in Supplementary Figure S13. ARE, antioxidant response element; CncC, cap'-n'-collar isoform-C; Foxo, forkhead box O; GFP, green fluorescent protein; H2O2, hydrogen peroxide; Nrf-2, nuclear factor erythroid 2-related factor.
We found that 6BIO significantly amplified the magnitude of CncC/Nrf-2-mediated antioxidant and proteasome gene expression responses (Fig. 3B) and it extended the longevity of flies exposed to either oxidative (Fig. 3C1) or proteotoxic (Fig. 3C2) stress; consistently, it suppressed proteome instability (i.e., the rate of ubiquitinated and carbonylated protein accumulation) after exposure of flies to oxidative stress (Fig. 3C3). Also, treatment of flies with the antioxidant tiron could not increase the longevity of 6BIO-treated cells (Supplementary Fig. S6), suggesting that antioxidant protection and 6BIO treatment are largely epistatic, indicating thus a common molecular pathway.
Furthermore, 6BIO activated the CncC/Nrf-2-regulated AREs when administrated in gstD-ARE:GFP reporter lines (Fig. 3D1) while, interestingly enough, it also promoted the downregulation of cncC/nrf-2 gene expression (Fig. 3D2) indicating, as in the case of the sgg/gsk-3 gene (Fig. 2E2), the existence of regulatory feedback loops that aim to restore normal activity in the InS/GFs→Sgg/Gsk-3→CncC/Nrf-2 signaling axis (Fig. 3A). Finally, we noted that RNAi-mediated knockdown of CncC/Nrf-2 abolished the 6BIO-induced upregulation of proteasomal genes and the CncC/Nrf-2 target gene keap1 (Fig. 3E1), as well as of proteasome protein subunits (Fig. 3E2) and peptidase activities (Fig. 3E3). Interestingly, CncC/Nrf-2 knockdown further enhanced 6BIO-mediated hsp70 gene upregulation (Fig. 3E1), suggesting the involvement of additional CncC/Nrf-2 mediators in 6BIO-promoted genomic effects. We next assessed whether CncC/Nrf-2 activity is required for the prolongevity effect of 6BIO and found that RNAi-mediated knockdown of CncC/Nrf-2 expression largely abolished the prolongevity effect of 6BIO (Fig. 3F). Notably, we observed a tendency for 6BIO to increase the mean (∼10%) and median (∼20%) life span of CncC/Nrf-2 RNAied flies (not reaching statistical significance though); thus, we cannot exclude the possibility that, as in the case of the hsp70 gene, 6BIO also modulates additional to CncC/Nrf-2 signaling pathways.
To further investigate the effect of 6BIO on proteolytic pathways, we also assayed autophagy in larvae muscles after exposing transgenic larvae expressing an Atg8mcherry reporter to 6BIO. In accordance to recent findings for lithium (a therapeutic Gsk-3 inhibitor) (9), 6BIO tended to suppress ALP (Supplementary Fig. S7A) activity; yet, at the doses (or the duration of the treatment) used herein, this effect did not reach statistical significance. Consistently, 6BIO downregulated the enzymatic activity of lysosomal cathepsins B, L (Supplementary Fig. S7B). Our findings thus do not support a role for autophagy in the prolongevity effects of 6BIO. Finally, we found that the 6BIO-mediated induction of UPS genes is independent of Foxo activity (the other arm of cellular antioxidant responses; see Fig. 3A) since RNAi-mediated KD of foxo gene expression did not abolish proteasomal gene upregulation (Supplementary Fig. S7C); this finding coincided with higher foxo (Supplementary Fig. S7C) and atg8 (Fig. 1A) gene expression on 6BIO treatment, likely due to a compensatory response to lower Foxo and ALP activities.
Overall, these data suggest that the 6BIO-mediated effect on antioxidant and proteostatic (i.e., proteasome) modules, as well as on healthy life span extension, largely (but not entirely) depends on CncC/Nrf-2 activation.
Genetic inhibition of Sgg/Gsk-3 in adult flies largely phenocopied the 6BIO effects and increased flies' healthy life span
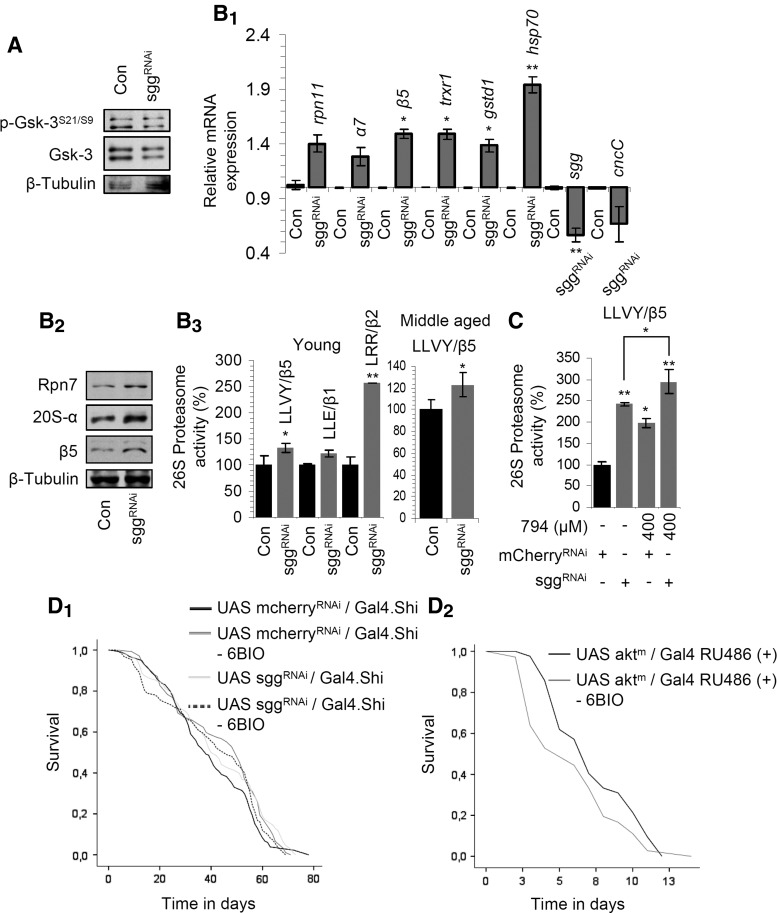
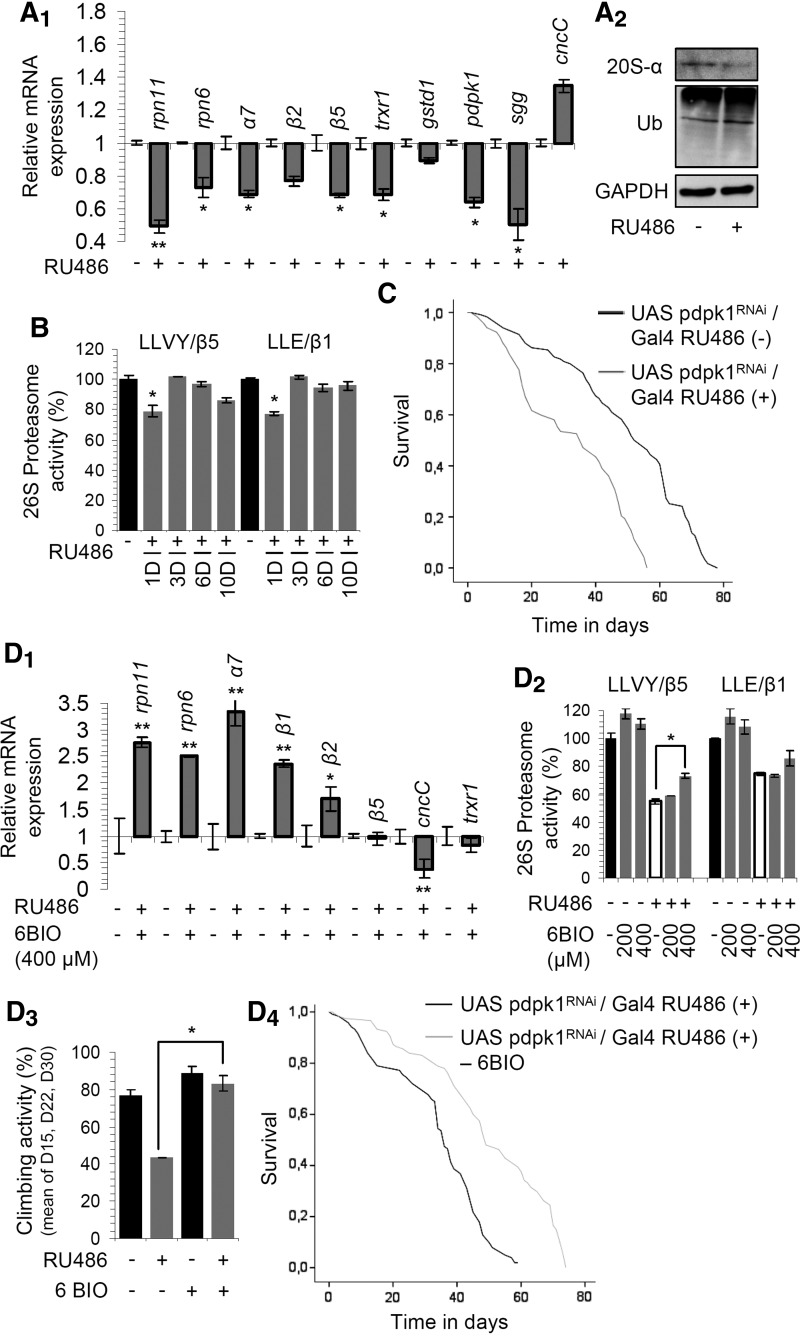
We then sought to phenocopy the effects of 6BIO by genetic approaches. Since we had previously found that high levels of CncC/Nrf-2 expression (55) reduced fly longevity, and it was reported that Sgg/Gsk-3 mutants are defective for larval growth (37), we used for these experiments a ubiquitous weak (shi-GAL4.S) driver. Similarly to 6BIO treatment, RNAi of Sgg/Gsk-3 resulted in reduced levels of the inhibitory Sgg/Gsk-3 Ser21/9phosphorylation (Fig. 4A) and triggered the upregulation of proteasome (rpn11, α7, β5) and antioxidant (trxr1, gstd1) genes, as well as of hsp70, while, as in the case of 6BIO treatment, the cncC/nrf-2 gene expression was downregulated (Fig. 4B1). Furthermore, we found increased levels of proteasome protein subunit expression (Fig. 4B2) and of proteasomal peptidase activities in both young and middle-aged flies (Fig. 4B3). Notably, proteasomal activities were further increased (compared to Sgg/Gsk-3 RNAied flies) when Sgg/Gsk-3 RNAi was combined with 794 treatment (Fig. 4C), likely due to inhibition of the remaining enzyme; this observation highlights the molecular differences between genetic interventions and treatment with selective inhibitors.
FIG. 4.
Genetic inhibition of Sgg/Gsk-3 in adult flies largely phenocopy the 6BIO-induced effects. (A) Representative immunoblot analyses of tissue protein samples probed with antibodies against p-Gsk-3S21/S9 and Gsk-3 after Sgg/Gsk-3 RNAi. Relative expression of the rpn11, α7, β5, trxr1, gstd1, hsp70, sgg, and cncC/nrf-2 genes (B1) and representative immunoblot analyses of fly tissue protein samples probed with antibodies against Rpn7, 20S-α, and β5 (B2) after RNAi-mediated Sgg/Gsk-3 knockdown. (B3) Relative (%) proteasome activities in somatic tissues of young (left) or middle-aged (right) flies after Sgg/Gsk-3 RNAi; controls in (A), (B1–B3) refer to flies expressing mcherry RNAi. (C) Relative (%) proteasome activities (vs. respective controls) after RNAi-mediated Sgg/Gsk-3 knockdown and treatment (or not) of flies for 5 days with the indicated concentration of 794. Longevity curves of flies expressing Sgg/Gsk-3 RNAi (D1) or a constitutively active mutated Akt (D2) cotreated (or not) with 400 μM 6BIO. Statistics of longevity curves is reported in Supplementary Table S1. β-tubulin (A, B2) or rp49 gene expression (B1) was used as reference. Control sample values in (B3, C) were set to 100%. Bars ± SD (n ≥ 2); *p < 0.05; **p < 0.01. See Western blot in Supplementary Figure S14.
RNAi-mediated knockdown of Sgg/Gsk-3 mildly increased mean and median life span of flies (Fig. 4D1) while in support to the notion that 6BIO extends life span mainly through Sgg/Gsk-3 inhibition, either addition of 400 μM 6BIO in Sgg/Gsk-3 RNAied transgenic flies or reduction of the endogenous Sgg/Gsk-3 activity by overexpressing its upstream suppressor Akt did not increase overall longevity (Fig. 4D1, D2). The noted increase (∼15%) in the median life span of 6BIO-treated Sgg/Gsk-3 RNAied flies may relate to either the 6BIO inhibitory effect on the still expressed enzyme or to 6BIO effects on other signaling pathways (see below the 6BIO effects on Pdpk1 kinase activity). Thus, genetic downregulation of Sgg/Gsk-3 and 6BIO treatment are largely epistatic, indicating a common molecular pathway.
High expression and/or activity levels of Sgg/Gsk-3 in flies' somatic tissues reduce longevity; this effect can be partially rescued by 6BIO or indirubin analogues with high selectivity against Sgg/Gsk-3
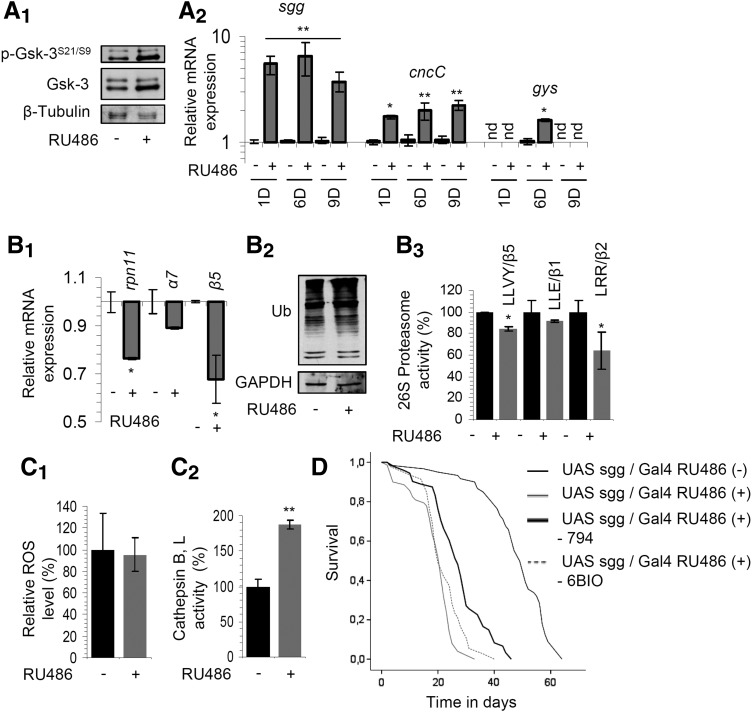
We then sought to study the long-term effects of increased Sgg/Gsk-3 expression and/or kinase activity levels on fly physiology and longevity. Inducible overexpression of Sgg/Gsk-3 in adult flies increased the inhibitory Sgg/Gsk-3 Ser21/9phosphorylation (Fig. 5A1), as well as the gene expression of Sgg/Gsk-3 downstream targets, namely cncC/Nrf-2 and gys genes (Fig. 5A2). It also suppressed proteasomal gene expression (Fig. 5B1), increased ubiquitinated proteome (Fig. 5B2) and reduced proteasome peptidase activities (Fig. 5B3). Sgg/Gsk-3 overexpression caused no significant effect on total ROS levels (Fig. 5C1) and increased the enzymatic activities of lysosomal cathepsins (Fig. 5C2), suggesting activation of the ALP pathway. Finally, sustained overexpression of Sgg/Gsk-3 promoted the significant shortening of flies' life span; this effect was partially rescued by 6BIO or 794 (Fig. 5D).
FIG. 5.
High expression levels of Sgg/Gsk-3 in flies' somatic tissues suppressed proteasome functionality, enhanced activities of cathepsins B, L, and reduced longevity. (A1, A2) Representative protein (blots were probed with antibodies against p-Gsk-3S21/S9 and Sgg/Gsk-3) and gene (sgg/gsk-3, cncC/nrf-2, gys) expression analyses in fly tissues following inducible [RU486 (+)] Sgg/Gsk-3 overexpression for 5 days (A1) or for 1–9 days (1D, 6D, 9D) (A2). Relative expression of rpn11, α7, and β5 genes (B1); total protein ubiquitination (B2) and (%) proteasome activities (B3) in tissues of young flies after inducible Sgg/Gsk-3 overexpression for 5 days. Relative (%) ROS (C1) or cathepsin B, L activity (C2) levels in young fly tissues after overexpression of Sgg/Gsk-3 for 15 and 7 days, respectively. (D) Longevity curves of Sgg/Gsk-3 overexpressing flies (EtOH vs. RU486) in the presence (or not) of 400 μM 794 or 400 μM 6BIO; the statistical analyses of the longevity experiments are reported in Supplementary Table S1. β-tubulin (A1), GAPDH (B2), and rp49 gene expression (A2, B1) were used as reference. Control samples in (B3, C1, C2) were set to 100%. Bars indicate ± SD (n ≥ 2); *p < 0.05; **p < 0.01. See Western blot in Supplementary Figure S14.
Then, we studied the effects mediated by overexpression of two Sgg/Gsk-3 mutants, namely Sgg/Gsk-3S9E and Sgg/Gsk-3S9A; these mutants when expressed in human cells exhibited a higher kinase activity of ∼1.9 and ∼2.6, respectively (13). Inducible overexpression of Sgg/Gsk-3S9E in fly tissues increased the inhibitory Sgg/Gsk-3 Ser21/9 phosphorylation (Supplementary Fig. S8A1), downregulated proteasome gene expression, and upregulated the cncC/nrf-2 gene expression (Supplementary Fig. S8A2). It also increased proteome ubiquitination (Supplementary Fig. S8B1) and suppressed proteasome activities (Supplementary Fig. S8B2), while, similarly to Sgg/Gsk-3WT overexpression, it had no effect on tissue ROS levels (Supplementary Fig. S8C1) and it increased cathepsin enzymatic activities (Supplementary Fig. S8C2). Moreover, it reduced flies' longevity and this effect was partially rescued by addition of 6BIO in fly culture medium (Supplementary Fig. S8D). Similarly to the effects noted for Sgg/Gsk-3S9E, inducible overexpression of the highly active Sgg/Gsk-3S9A mutant (Supplementary Fig. S9A1) increased Sgg/Gsk-3 Ser21/9 inhibitory phosphorylation (Supplementary Fig. S9A2), suppressed proteasome genes (Supplementary Fig. S9B1) and 20S-α protein subunit (Supplementary Fig. S9B2) expression, and it induced the expression of the cncC/nrf-2 and gys genes (Supplementary Fig. S9B1). Furthermore, high expression levels of Sgg/Gsk-3S9A increased total cellular ubiquitination (Supplementary Fig. S9B2), suppressed proteasome activities (Supplementary Fig. S9B3), increased tissue ROS (Supplementary Fig. S9C), and significantly reduced flies' longevity (Supplementary Fig. S9D).
Finally, we sought to study the effects of sustained activation of endogenous Sgg/Gsk-3. To this end, we knocked down by RNAi the Akt activating kinase Pdpk1 (Fig. 3A). Pdpk1 knockdown suppressed proteasome gene (Fig. 6A1) and 20S-α protein subunit (Fig. 6A2) expression, increased proteome ubiquitination (Fig. 6A2), and in the short term downregulated proteasome peptidase activities (Fig. 6B); thus, decreased signaling from the InS/IGF-1 pathway culminates in lower proteasome functionality. Also, Pdpk1 RNAi suppressed antioxidant genes (trxr1, gstd1) and sgg/gsk-3 gene expression; upregulated cncC/nrf-2 gene (Fig. 6A1) and reduced flies' median and maximum life span (Fig. 6C). The Pdpk1 RNAi-mediated effects on gene expression patterns and proteasome activities were inverted by 6BIO treatment (Fig. 6D1, D2). Also, 6BIO increased the locomotor activity of Pdpk1 RNAied middle-aged flies (Fig. 6D3) and it significantly increased flies' longevity (Fig. 6D4); the beneficial effect of 6BIO on the physiology and longevity of flies with high Sgg/Gsk-3 expression and kinase activity levels was further confirmed in double transgenic flies where Pdpk1 RNAi was combined with Sgg/Gsk-3 overexpression (data not shown).
FIG. 6.
Activation of endogenous Sgg/Gsk-3 in Drosophila somatic tissues decreased proteasome functionality and reduced longevity; these effects were largely rescued by 6BIO. Relative expression of rpn11, rpn6, α7, β2, β5, trxr1, gstd1, pdpk1, sgg/gsk-3, and cncC/nrf-2 genes (A1), and representative blots showing 20S-α proteasome subunit expression and total protein ubiquitination (A2) in young fly tissues after inducible RNAi-mediated Pdpk1 knockdown for 6 days. (B) Relative proteasome activities in young fly tissues following Pdpk1 RNAi for 1, 3, 6, and 10 (1D–10D) days. (C) Longevity curves after induced (or not) Pdpk1 RNAi. (D1, D2) Relative expression of the rpn11, rpn6, α7, β1, β2, β5, cncC/nrf-2, and trxr1 genes (D1) and relative (%) proteasome activities (D2) after Pdpk1 RNAi in flies treated (or not) for 6 days with the shown 6BIO doses. (D3) Mean climbing activity (%) of Pdpk1 RNAied middle-aged flies cultured (or not) in the presence of 400 μM 6BIO. (D4) Longevity curves after inducible Pdpk1 RNAi in flies treated (or not) with 400 μM 6BIO; statistical analyses of life span experiments (C, D4) are reported in Supplementary Table S1. GAPDH (A2) and rp49 gene expression (A1, D1) were used as reference. Control samples in (B, D2, D3) were set to 100%. Bars indicate ± SD (n ≥ 2); *p < 0.05; **p < 0.01. See Western blot in Supplementary Figure S14. Pdpk1, 3-phosphoinositide-dependent protein kinase-1.
In conclusion, sustained high Sgg/Gsk-3 expression levels and/or kinase activity are toxic; 6BIO (or other Sgg/Gsk-3 selective 6BIO analogues) can partially rescue these detrimental effects.
6BIO suppresses the nutrient-sensing pathway signaling by reducing the Pdpk1 kinase activity
It was previously reported that 6BIO also inhibits Pdpk1 with an IC50 in human endothelial cells of less than 1 μM (62); this concentration is in the range of 6BIO concentration found in fly tissues (see HRMS-derived 6BIO concentration in tissue extracts). Considering the high homology between mammalian Pdpks and the fly ortholog (data not shown), the effects of 6BIO in fly tissue bioenergetic and metabolic pathways (Supplementary Figs. S2 and S3), as well as that the 6BIO effects were seemingly not entirely dependent on Sgg/Gsk-3 inhibitory effect (e.g., Figs. 3F and 4D1), we also investigated the 6BIO effects on Pdpk1 activity. We noted that 6BIO caused the upregulation of the phosphoenolpyruvate carboxykinase (pepck) and glucose-6-phosphatase (g6p) genes (Supplementary Fig. S10A); this effect correlates with reduced signaling of the nutrient pathway. Notably, Sgg/Gsk-3 knockdown suppressed pepck and g6p expression, while g6p gene was induced during Pdpk1 knockdown (that results in Sgg/Gsk-3 activation) or Sgg/Gsk-3 overexpression (Supplementary Fig. S10A). Furthermore, treatment of wild-type flies with 6BIO upregulated pdpk1 gene expression (Supplementary Fig. S10B1) and this response is reminiscent of 6BIO-mediated sgg gene induction (Fig. 2E2); it also reduced Pdpk1 Ser/Thr-dependent phosphorylation and upregulated Akt (Supplementary Fig. S10B2). These findings suggest that 6BIO likely reduces the nutrient pathway signaling by partially inhibiting Pdpk1 kinase activity.
We attempted to verify these observations genetically by inducible Pdpk1 overexpression. Pdpk1 is expressed in relatively low levels in adult fly tissues (data not shown) and we noted lethality at pupal stages even at conditions of non-RU486 addition in Pdpk1 transgenic flies' culture medium; this effect likely relates to mild transgene overexpression due to leakage effects (55). In an alternative strategy, we inducibly overexpressed a mutated constitutively active insulin receptor InRA1325D, which is expected to increase the activity of the endogenous Pdpk1 (and at the same time suppress Sgg/Gsk-3 kinase activity; Fig. 3A). We observed that 6BIO could not rescue the InRA1325D overexpression-mediated shortening of fly longevity despite some nonsignificant increase in median life span (Supplementary Fig. S10C). Nevertheless, feeding of noninduced (EtOH treated) Pdpk1 transgenic larvae with 6BIO resulted in higher larvae survival rates and increased rates of pupation indicating a partial rescue (Supplementary Fig. S10D); however, even the 6BIO-fed Pdpk1 overexpressing transgenic larvae/pupae did not hatch. These findings support the notion that, as proposed in human cells, 6BIO likely exerts mild inhibitory effects to Pdpk1 and the nutrient pathway signaling activity.
Discussion
By screening for NPs that modulate antiaging pathways, we found that oral administration in Drosophila flies of 6BIO or other indirubin analogues that act as selective Gsk-3 inhibitors activated cytoprotective mechanisms, increased resistance to stressors, delayed age-related phenotypes, and extended healthy life span. In support to these findings, it was very recently reported that lithium extended flies' life span and increased resistance to xenobiotics (9); also, lithium was found to exert beneficial effect in worms' longevity (33) and it also ameliorated Aβ pathology in an adult-onset Drosophila model of Alzheimer's disease (48). Gsk-3 is a pleiotropic serine/threonine kinase that functions as a negative regulator of Gys1 activity and hence glycogen synthesis, while it seems to also be central in a number of additional diverse signaling pathways, including Wnt signaling, protein synthesis, the circadian clock, cell cycle regulation, as well as cell survival (5) and death (11, 45); the latter may (among others) correlate with the Sgg/Gsk-3 overexpression-mediated suppression of proteasomal activity by ∼20%, which, as shown before (55), decreases fly longevity by ∼50%.
Gsk-3 activity is suppressed by stress as well as by nutrient-sensing pathways and specifically the InS/PI3K/Akt axis (16); this mode of inhibitory regulation by the InS/PI3K/Akt pathway is also conserved in flies (37). Accordingly, Gsk-3 has been implicated in the pathogenesis of several diseases, including neurodegeneration, diabetes, and cancer, as well as neuropsychiatric and mood disorders (5). It is not thus surprising that indirubins, and particularly 6BIO, have demonstrated a wide range of bioactivities, including anticancer (8) and antineurodegenerative (3) effects, as well as regenerative properties in mammalian stem cells (46); our finding that 6BIO also exerts in vivo antiaging effects further expands the reported bioactivities of this molecule. In support to Gsk-3 functional involvement in higher metazoan longevity, we noted that genetic inhibition of Sgg/Gsk-3 expression mildly enhanced fly longevity, whereas increased expression and/or kinase activity of Sgg/Gsk-3 severely reduced life span; this latter effect was partially rescued by 6BIO indicating that the 6BIO inhibitory effect on Gsk-3 activity is central to its antiaging properties. Nonetheless, complete absence of Gsk-3 in various model organisms shortens life span or prevents development (6, 21, 37), while homozygous knockout of Gsk-3β in mice yields embryonic lethality (21, 27). Thus, only moderate Gsk-3 inhibition would be beneficial in the long term; in support, we found that high 6BIO doses reduced longevity despite high activities of the antioxidant and proteostatic modules (data not shown).
We noted that the 6BIO-mediated effects on antioxidant responses, proteasome functionality, and longevity were largely dependent on the activity of CncC/Nrf-2. Indeed, Nrf-2 functionality is regulated by Gsk-3 since it is directly phosphorylated at its Neh6 domain by Gsk-3 and this modification leads to its SCF/β-TrCP-dependent degradation (39). Also, Gsk-3β phosphorylates Fyn kinase at threonine residue(s), which then accumulates in the nucleus and by phosphorylating Nrf-2 triggers its nuclear export, ubiquitination, and degradation (24). Thus, as we found herein, it is anticipated that Gsk-3 inhibition will promote the stabilization and activation of Nrf-2. Supportively, Nrf-2 was stabilized by Gsk-3 inhibitors in Keap1-null mouse embryo fibroblasts (39), while chemical (including lithium) or RNAi-mediated inhibition of Gsk-3β led to Nrf-2 activation and protection against oxidants in mammalian cells (43, 47). Interestingly, as we and others recently showed, there is a threshold in the Nrf-2 activation levels, correlating with beneficial long-term effects since high levels of CncC/Nrf-2 activity in flies result in decreased longevity despite the underlying activation of antioxidant responses and proteasome (9, 55). These data suggest that only mild Nrf-2 activation would be beneficial in the long term, while higher levels ensure transient protection from stressors.
Part of the 6BIO protective effects obviously relate to activation of main proteostatic mechanisms, namely the CncC/Nrf-2-dependent proteasome activation and the upregulation of molecular chaperones. Higher activity of the PN modules seems to delay the age-related accumulation of stressors in cells, resulting in increased life span (20). This assumption is supported by findings indicating that forced reinvestment of resources from the germ line to the soma of Caenorhabditis elegans promotes higher somatic proteasome activities, clearance of damaged proteins, and increased longevity (58), while, as we and others showed, Drosophila reproductive tissues age at significantly lower rates (compared to the soma) since they exhibit higher capacity to prevent accumulation of damaged proteins due to increased intrinsic proteasome activities (17, 53).
Interestingly, the 6BIO-mediated effect on chaperone (e.g., hsp70 gene) upregulation was not dependent on CncC/Nrf-2 and in fact CncC/Nrf-2 knockdown further increased hsp70 upregulation. In support, Gsk-3 exerted a repressive action on the transcriptional activation of Hsf-1 (10, 61), and Gsk-3β inhibition protected mesothelial cells during experimental peritoneal dialysis through upregulation of the heat shock response (44). Notably, Gsk-3 also enhanced the transactivation activity of Foxo in both mammalian (22) and in Drosophila S2 (32) cells; and it also positively regulated autophagic flux in hippocampal neural stem cells (19) and in fibroblasts (63). In line with these reports, we noted reduced autophagic and cathepsin B, L activity in tissues of 6BIO-treated flies; on the contrary, high expression and/or kinase activity of Sgg/Gsk-3 levels activated cathepsins B, L indicating increased lysosomal activity. Thus, the Gsk-3 inhibition-mediated UPS and chaperone activation may occur at the cost of reduced ALP activity.
The dynamic process of extensive wiring and functional crosstalk among proteostatic and genomic modules is particularly evident in our study since 6BIO-mediated Sgg/Gsk-3 inhibition triggers counteracting genomic (e.g., sgg upregulation, cncC/nrf-2 downregulation, or enhanced hsp70 upregulation in CncC/Nrf-2 RNAied flies) and proteomic (e.g., reduced levels of Sgg/Gsk-3 inhibitory Ser21/9 phosphorylation) responses; the latter likely relates to reduced upstream Akt activity indicating that cells “attempt” to adapt to the sudden condition of reduced Sgg/Gsk-3 activity. These findings are particularly significant for the drug development research pipeline since they indicate that even in relative simple organisms such as the fly, the “system” is equipped with numerous sensors and responses to counteract disturbances and return to an evolutionary preset species-specific “ideal” equilibrium status.
Intriguingly, 6BIO was also found to modulate cellular bioenergetic and metabolic pathways, and to decrease lipid and glucose tissue load. In support, lithium improved mitochondrial energetic function in C. elegans (51), while it has been shown that Nrf-2 regulates mitochondria functionality and dynamics (36). The increased mitochondria respiration that we found in 6BIO-treated flies, along with AMPK activation, suggests efficient substrate usage and increased AMP/ATP ratio. In line with this finding, Gsk-3 was found to directly inhibit the AMPK kinase activity, while disrupting Gsk-3 function within the AMPK complex sustains higher AMPK activity and cellular catabolic processes; thus, Gsk-3 acts as a critical sensor for anabolic signaling to regulate AMPK (49). In addition to these effects on cellular energetics, Gsk-3 also acts as a negative regulator in the hormonal control of glucose homeodynamics through Gys1 and hence glycogen synthesis inhibition (16); thus, the shown effects of 6BIO on metabolic pathways are not surprising. In line with our findings, Gsk-3 inhibitors improved glucose disposal in prediabetic insulin-resistant rat skeletal muscle (12), while our observation that 6BIO affected lipid metabolism is consistent with recent findings showing that lithium exerts lipolytic effects (9); nevertheless, the mechanism behind the Gsk-3 inhibitor-mediated lipolytic effects remains to be clarified.
Finally, according to our presented data and recent findings in human cells, 6BIO likely also mildly inhibits Pdpk1 kinase activity (62); this action may also explain the 6BIO impact on metabolic pathways. Pdpk1 is downstream to PI3K and controls many central signaling pathways by phosphorylating (among others) Akt, thereby regulating cell growth and survival in both mammalian cells (18) and flies (42); notably, serine- and threonine/valine-dependent activation of Pdpk1 and Tor orthologs converges on Sch9 to promote Drosophila aging (35). The in vivo inhibitory effect of 6BIO on Drosophila Pdpk1 is further supported by our finding that Sgg/Gsk-3 RNAi could not phenocopy the 6BIO-mediated increased expression of the pepck and g6p metabolic genes, which rather relates to relaxation of Foxo suppression through the 6BIO-mediated Pdpk1→Akt axis inhibition. Indeed, hepatic expression of Pepck and G6pase is rapidly and completely inhibited by insulin (14), while, as we found herein, genetic inhibition of Gsk-3 selectively reduced pepck and g6p gene expression (30). Finally, in line with findings in human cells, 6BIO reduced Pdpk1 substrate phosphorylation, increased Akt protein and pdpk1 gene expression, and conferred a partial rescue in Pdpk1 overexpression-mediated larval lethality. These findings suggest that 6BIO likely reduces the nutrient-sensing pathway signaling and thus also acts as a CR mimetic.
Taken together our findings indicate that Gsk-3 is a central hub in the functional wiring of nutrient-sensing, antioxidant, and proteostatic signaling pathways (Supplementary Fig. S11). Considering that 6BIO activates cytoprotective modules and in parallel suppresses anabolic pathways (Supplementary Fig. S11), as well as that, according to a very recent report, CR mimetics can be used as therapeutics in DNA damage-related progeroid diseases (57), 6BIO has the potential to enter the challenging field of translational medicine. Moreover, and given the fact that indirubins are found in edible gastropod mollusks and plants representing thus natural sources of Gsk-3 and, likely, Pdpk1 inhibitors, we suggest that the 6BIO scaffold can be used for the synthesis of novel antiaging compounds.
Materials and Methods
Chemicals and synthesis procedures
All chemicals were purchased from Sigma. Microwave-assisted reactions were performed in a single-mode CEM apparatus. NMR spectra were recorded on the Bruker Avance 600 spectrometer (600 MHz). HRMS spectra were determined on an MSQ Orbitrap Thermo Finnigan spectrometer. Purification of the synthesized products was conducted using flash silica gel 60 (40–63 μm) from Merck. Purity of the compounds has been determined by high-performance liquid chromatography (HPLC) and was above 95%.
Synthesis of the isatins and indirubins
The isatins were synthesized as previously described (38); the synthesis scheme is summarized in Supplementary Figure S4. The mixture of 6-bromoisatin and 4-bromoisatin has been separated by pH-dependent precipitation (38). The mixture of 5,6-dichloroisatin and 4,5-dichloroisatin has been separated on silica gel (CH2Cl2 to CH2Cl2/AcOEt 7/3) (Supplementary Fig. S4).
The corresponding isatin was dissolved in 30 ml of anhydrous MetOH and the corresponding 3-acetoxyindole was introduced. The mixture was stirred for 5 min prior the addition of sodium carbonate. The solution was then stirred for 3.5 h at room temperature protected from light. After completion, an aqueous methanolic solution (1/1) was added. The precipitate was filtered and washed successively with H2O/MeOH (1/1) and water. The resulting purple solid is dried to afford the corresponding indirubin with 80–90% yield.
Synthesis of indirubin derivatives, oximes, 796 and 6BIO-Pip
The targeted compounds have been synthesized as previously described (38). This approach consists in the coupling of a correctly substituted isatin and the 3-acetoxyindole in alkaline methanol to afford the indirubin skeleton. Then, the 3′-position reacts with hydroxylamine in refluxing pyridine leading to the corresponding indirubin-3′-oxime. To a solution of the corresponding indirubin in pyridine, hydroxylamine hydrochloride was added. The mixture was warmed in reflux for 1.5 h. After completion and cooling to 70°C, water was added. The resulting precipitate was filtered, washed with water and cyclohexane, and dried to afford the corresponding oxime as a red solid with 100%.
796 was synthesized after acetylation of the oxime as was previously described (38) and was obtained with 100% yield.
For synthesis of 6BIO-Pip, the corresponding indirubin-3′-oxime was dissolved in anhydrous dimethylformamide (DMF). Then, 1,2-dibromoethane and a catalytic amount of triethylamine were added and the mixture was stirred at 50°C for 24 h. After completion, water was added and the precipitate filtered, washed with water, and dried. The ether was dissolved in anhydrous DMF. The corresponding amine was then added and the resulting mixture was warmed at 80°C under a radiation of 150 W for 20 min. After completion, water is added and the resulting precipitate is filtered, washed with water, and dried. The corresponding derivatives are obtained as purple solid.
In all cases, analytical data (NMR and MS) were conformed to literature values and were as follows:
5,6-dichloroisatin: 1H NMR (CDCl3, 400 MHz, δ ppm, J in Hz): 7.84 (1H, bs, NH), 7.74 (1H, s, H-4), 7.10 (1H, s, H-7).
794: 1H NMR (400 MHz, DMSO, δ ppm, J in Hz): 11.17 (2H, s, N-H, N¢-H) 8.94 (1H, s, H-4), 7.67 (1H, d, J = 7.5 Hz, H-4′), 7.60 (1H, t, J = 7.5 Hz, H-6′), 7.43 (1H, d, J = 7.5, Hz, H-7′), 7.08 (1H, s, H-7), 7.06 (1H, t, J = 7.5 Hz, H-5′).
6BIO: 1H NMR (DMSO-d6, 400 MHz, δ ppm, J in Hz): 13.61 (1H, brs, NOH), 11.72 (1H, s, N′-H), 10.85 (1H, s, N-H), 8.53 (1H, d, J = 8.2 Hz, H-4), 8.19 (1H, d, J = 7.5 Hz, H-4′), 7.39 (2H, brs, H-7′, 6′), 7.07 (1H, d, J = 8.2, Hz, H-5), 7.01 (2H, brs, H-7, 5′).
796: 1H NMR (DMSO-d6, 400 MHz, δ ppm, J in Hz): 11.73 (1H, s, N′-H), 11.41 (1H, s, N-H), 9.56 (1H, s, H-4), 8.22 (1H,d, J = 7.8 Hz, H-4′), 7.53 (2H, m, H-6′, 7′), 7.21 (1H, s, H-7), 7.13 (1H, t, J = 7.8 Hz, H-5′) 2.46 (3H, s, OCOCH3).
6BIO-Pip: 1H NMR (400 MHz, DMSO-d6, δ ppm, J in Hz) 11.69 (1H, s, H-1′), 10.92 (1H, s, H-1), 8.53 (1H, d, J = 8.5 Hz, H-4), 8.15 (1H, d, J = 7.4 Hz, H-4′), 7.43 (2H, m, H-6′, H-7′), 7.14 (1H, d, J = 8.5 Hz, H-5), 7.03 (2H, m, H-5′, H-7), 4.69 (2H, br t, H-1′′), 2.83 (2H, br t, H-2′′), 2.71 (4H, brs, H-4′′, H-5′′), 2.46 (4H, brs, H-3′′, H-6′′, partially overlapped with DMSO).
Sample preparation and ultra-HPLC; high-resolution mass spectrometry for the identification of 6BIO in fly culture medium and tissues
Working samples consisting of 20, 50, and 100 flies (50% males/50% females) were initially tested to obtain the required mass for the analyses. In parallel, two different concentrations of 400 and 4000 μM 6BIO were added in fly culture medium; both concentrations were found adequate for 6BIO detection and quantification. A stock solution of 1 mg/ml 6BIO (diluted in DMSO) was further diluted to give a series of calibration solutions with the concentrations of 50, 100, 150, 250, 500, 750, 1250, and 2000 ng/ml. The quality control solutions were prepared in triplicate at 4 different concentrations of 75, 400, 1000, and 1750 ng/ml.
For sample preparation, the protein precipitation technique was used in fly tissue samples before the LC-MS analysis. Somatic tissues (head/thorax) from 100 flies were placed into an Eppendorf with 50 μl of cold MetOH and homogenized with a pestle. The volume of cold MetOH was slowly increased to 500 μl to attain complete homogenization. Following a centrifugation step at 12,000 g for 3 min (4°C), the supernatant was collected and 10 μl of the isolated supernatant was injected into the UHPLC-LTQ-Orbitrap system. For analysis of fly culture medium, a blank and a sample containing 400 μM were used; both samples were extracted with MetOH to a final concentration of 100 μg/ml.
Analysis of tissue samples with high-resolution mass spectrometry
Samples were analyzed on a UHPLC-LTQ-Orbitrap Discovery system (Thermo Scientific, Inc.) consisting of an Accela UHPLC system coupled to a hybrid LTQ-Orbitrap Discovery mass spectrometer. An aliquot of 10 μl of sample was injected into a C18 Hypersil GOLD column (50 × 2.1 mm i.d., 1.9 μm particle size; Thermo Scientific, Inc.) for separation (at room temperature), with a flow rate of 500 μl/min. An LC gradient method was used; initiated from 10% B to 55% B in 2 min (A, water with 0.1% acetic acid; B, acetonitrile), hold at 55% B for 0.3 min, and then increased to 100% B in 1 min, restored to 10% in 0.1 min, and hold at 10% B for 1 min. The total run time was 4.4 min.
Mass spectrometric detection was carried out on LTQ-Orbitrap Mass Spectrometer (Thermo Finnigan) with an electrospray ionization (ESI) interface. The ESI source was set in a negative ionization mode. LC-MS based quantitation was performed using multiple reaction monitoring of the transitions of m/z 354.0–324.0. The optimized ionization parameters were as follows: ion spray voltage of 3.5 kV, source current 100 μA, capillary temperature 300°C, capillary voltage −50 V, and tube lens −50 V. Two microscans were applied with a maximum injection time of 100 ms. Nitrogen was used as sheath gas (flow rate 50 arb) and auxiliary gas (flow rate of 10 arb). For direct infusion experiments, the ionization parameters were slightly modified (ion spray voltage of 4.5 kV, source current 100 μA, capillary temperature 270°C, capillary voltage −50 V, and tube lens −110 V) except the gasses that were adjusted accordingly; sheath gas 15 arb and auxiliary gas 6 arb. All samples were analyzed in duplicate and data were acquired and processed using the Xcalibur 2.0 software provided with the system.
Fly stocks
Oregon R Drosophila flies were used as the wild-type strain. The w1118 stock and the transgenic strains UAS sgg, UAS sggS9E (expresses a mutated constitutively active sgg), UAS sggS9A (expresses a mutated constitutively active sgg with higher kinase activity than sggS9E), UAS sggRNAi, UAS inrA1325D (expresses a mutated constitutively active insulin receptor), UAS akt1m (expresses a mutated constitutively active Akt protein with a myristoylation motif), UAS pdpk1, UAS pdpk1RNAi, UAS mcherryRNAi, UAS foxoRNAi; the double transgenic lines UAS hAPP, UAS hBACE1, along with the Gal4 ubiquitous driver (shi-Gal4.S), were obtained from the Bloomington Drosophila Stock Center. The gstD-ARE:GFP/II (ARE of the gstd gene) and the gstD-mARE:GFP/III (mutated version of gstD-ARE) reporter transgenic lines, along with the transgenic lines UAS cncCRNAi and the tubulin-GeneSwitch-Gal4 (tubGSGal4) driver were a gift from Prof. D. Bohmann (University of Rochester). The conditional driver (tubGSGal4) is ubiquitously activated on dietary administration of RU486 (320 μM) as reported previously (55). The transgenic lines UAS mitoGFP, Gal4 Mef2 (muscle targeted transgene expression), and the double transgenic line UAS atg8mcherry, Gal4-Mef2 were a gift from Prof. A. Daga (University of Padua, Padova, Italy). Since we recently reported that gonads display distinct aging rates and regulation of proteostatic mechanisms compared to adult somatic tissues (53, 55), in all presented experiments referring to adult flies, only microdissected somatic tissues (head and thorax; equal numbers from mated male and female flies) were analyzed.
Fly culture and exposure to compounds
Fly stocks were maintained at 25°C, 60% relative humidity on a 12-h light:12-h dark cycle, and were fed standard medium (unless otherwise indicated). All used compounds (indirubins, PS-341, hydrogen peroxide [H2O2], 4,5-dihydroxy-1,3-benzenedisulfonic acid [Tiron], and tert-butyl hydroperoxide [t-BHP]) were added in fly culture medium; doses and duration of fly exposure to compounds are indicated in figure legends. The proteasome inhibitor PS-341 was obtained from Santa Cruz, while H2O2, Tiron, and t-BHP from Sigma. Synthesized indirubin molecules were dissolved in DMSO, aliquoted, and stored at −20°C.
Climbing, gustatory and longevity assays
The mobility of flies was assayed as described previously (55). Thirty flies (15 males and 15 females) were placed in a 100-ml cylinder with a line drawn at the 66 ml mark. Flies were gently tapped to the bottom of the cylinder and the number of flies that climbed above the 66 ml mark after 20 s was recorded.
Gustatory assay was done as described previously (55). Briefly, young flies were starved for 20 h on water-soaked Whatman paper and were then transferred for 2 h to vials containing standard food mixed with 0.2% sulforhodamine B sodium salt (Acid-Red) (control) or standard food containing 6BIO and 0.2% sulforhodamine B sodium salt. The degree of abdominal redness for each fly was scored from grade 0 (colorless abdomen) to grade 5 (fully red abdomen); at least 30 flies per treatment were scored.
For longevity assays (performed in duplicates at two experimental replicates), female and male flies (equal numbers per sex) were collected and cultured in vials; flies were transferred to vials with fresh food every 3–4 days and deaths were scored every day. For survival curves and statistical analyses, the Kaplan–Meier procedure and log-rank (Mantel–Cox) test were used; significance was accepted at p < 0.05. Statistical analyses of flies' survival curves are presented in Supplementary Table S1.
RNA extraction and quantitative real-time polymerase chain reaction analyses
Total RNA was extracted from flies' somatic or larvae tissues by using RNAzol (Molecular Research Center, Inc.). Extracted total RNA was converted to cDNA with the Maxima First-Strand cDNA Synthesis Kit of Thermo scientific, Inc. (#K1642). cDNA was then subjected to quantitative real-time polymerase chain reaction (Q-RT-PCR) analysis using the SYBR Green/ROX qPCR Master Mix of Thermo Scientific, Inc. (#K0223). Primers were designed using the primer-BLAST tool (www.ncbi.nlm.nih.gov/tools/primer-blast/) and were the following:
rpn11-F: ACAACAAGTCACTGGAGGACG, rpn11-R: TGCTTGCCCACGTTCTTGAT; rpn10-F: TGCCTTCGCTATGCAGATGT, rpn10-R: GTTTGCCTCGTCCGTCTTTG; rpn6-F: TCTACTGTCCGCCAAAGGTG, rpn6-R: TTCCACTGACGAGCTGGTTG; α7-F: ACCGACGAATTGGTGGAGAG, α7-R: ACCCATTTCGAAGCGGAAGT; β1-F: GCGACGCATCTCTACAACAC, β1-R: CGAGGAAATGAAGCTGGGAGT; β2-F: AGCCACCGACCACCACCAAGA, β2-R: CCACAACGCGCACCTCACGA; β5-F: GCCATCTACCATGCCACCTT, β5-R: TTACCCAGCCGTCCTCCTTA; atg8-F: ACGCCTTCGAGAAGCGTCGC, atg8-R: CCAAATCACCGATGCGCGCC; hsp70-F: AAGAACCTCAAGGGTGAGCG, hsp70-R: CGTCGATGGTCAGGATGGAG; cncC-F: CCAACGAGGTGGAAATCGGA, cncC-R: ACTTGACATTGGTAGCCGCA; keap1-F: GCGCTCGTCAGCCCATTTT, keap1-R: GGATGCGCATAATTCCTCTTCTT; gstD1-F: TGATCAATCAGCGCCTGTACT, gstD1-R: GCAATGTCGGCTACGGTAAG; trxr1-F: GACACCAGTGGAACTACCCG, trxr1-R: ATTTTGAGTGCATGTCGGCG; pdpk1-F: TTCAAACCAGTCGCTTCGTG, pdpk1-R: TGTGTATGTATATCAATGCCAGCG; g6p-F: GTCGGTGCGAGAGGCATTTA, g6p-R: TATAGTGGCGAGGCCAAAGC; pepck-F: TGAACGCAAAGTCCTCGACA, pepck-R: ACATTGCCGGAGATAATTTTGC; sgg-F: TGCGCAAATTGGAGCATTGT, sgg-R: TGTTCTCTTGTCGGTGTGCC; gys-F: TTCCAGTTGTCTTGTTCCACCA, gys-R: AGTTTCGAAGGCATCCGGG; marf-F: CCGCTATCCCGGTTCAACTC, marf-R: AGCGGTGATTTGTCGTTGGA; drp1-F: CAACGCACGTGGTCAACCTA, drp1-R: GATCTGCGCCTCGATATCCTC; opa1-F: CGAGGAGTTCCTACTTGC, opa1-R: TGAGATTCCGCGAGAACTGG; lon-F: TCTCGCGAGTGCTCTTCATC, lon-R: ATTCCCGGCAATAACTGCGA; sdhA-F: CACCGGTGGATATGGACGAG, sdhA-R: CCATAGCAGTACCGTCACCG; and foxo-F: CAGTGCCGGATGGAAGAACT, foxo-R: ATCCACCAGGATGACTTGCC. The ribosomal gene rp49 (rp49-F: AGCACTTCATCCGCCACC and rp49-R: ATCTCGCCGCAGTAAACG) was used as a normalizer as described before (54).
Isolation of adult flies' hemolymph
Hemolymph was isolated from equal numbers of young male and female flies according to standard procedures as described previously (53).
Mitochondria isolation and measurement of mitochondrial respiration
For mitochondria isolation, somatic tissues of flies were homogenized in ice-cold isolation buffer (0.32 M sucrose, 10 mM EDTA, 10 mM Tris/HCl, pH 7.3) containing 2% (w/v) bovine serum albumin (BSA). Samples were filtered through a layer of gauze, which was then washed with additional isolation buffer up to a final volume of 1.5 ml. Following centrifugation for 10 min at 2200 g, the pellet was washed with BSA-free isolation buffer and resuspended in 200 μl of the same buffer. The protein content of isolated mitochondria was measured by the Bradford method.
Respiration of isolated mitochondria was determined using a Clark-type oxygen electrode connected to the computer-operated Oxygraph control unit (Hansatech Instruments). Freshly isolated mitochondria (150 μg of protein) were added to the respiration buffer (120 mM KCl, 5 mM KH2PO4, 3 mM HEPES, 1 mM EGTA, 1 mM MgCl2, and 0.2% BSA, pH 7.2) in which 5 mM glutamate/2.5 mM malate have been added. Basal O2 consumption was recorded (State 2) and after 2 min, 500 μM of ADP was added (State 3), followed by 6 μM of oligomycin (State 4) and 100 nM of the uncoupler carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP) (State FCCP). For all the experiments, the temperature was maintained at 25°C and the total reaction volume was 300 μl; the respiratory control ratio was calculated as the ratio of State 3/State 4 (ST3/ST4).
Preparation of tissue protein extracts, immunoblot analysis, and detection of protein carbonyl groups
Tissue extract preparation, adjustment of protein content of each sample lysate by Bradford assay (Bio-Rad Laboratories), and immunoblotting were essentially performed as described previously (53–55). Analysis of blot quantification was done by scanning densitometry.
For the detection of protein carbonyl groups, the OxyBlot protein oxidation detection kit (Millipore; #s7150) was used, as per the manufacturer's instruction.
Measurement of ROS, green fluorescent protein, proteasome, and cathepsin B, L enzymatic activities in tissue extracts
Tissue extract preparation and measurement by fluorometry (using a Versa Fluor Fluorometer System; Bio-Rad Laboratories) of ROS, green fluorescent protein (GFP), proteasome, and cathepsin B, L enzymatic activities were done as described previously (54, 55). Fluorescence intensity was normalized to the total protein level per sample and expressed as the relative percentage versus the corresponding control; in adult flies, equal numbers of male and female flies were used.
Measurement of glucose, trehalose, and glycogen levels
Glucose, trehalose, and glycogen tissue levels were measured as described previously (4) with minor modifications. More specifically, somatic tissues of six flies (three males and three females) were homogenized either in cold phosphate-buffered saline (PBS) for glucose or glycogen measurement, or in Trehalose buffer (5 mM Tris pH 6.6, 137 mM NaCl, 2.7 mM KCl) for trehalose measurement. Extracts were cleared by centrifugation at 1200 g and the supernatant was incubated for 10 min at 70°C; a small amount of the cleared extract was used for protein quantification by Bradford assay. After centrifugation at maximum speed for 3 min, 30 μl of diluted (1/4) (no sample dilution was done in the case of trehalose measurement) supernatant was transferred to a 96-well plate. Glucose measurement was performed by adding 100 μl of Glucose Reagent (Sigma; GAGO-20) followed by a 30-min incubation at 37°C. For glycogen measurement, the same procedure as for the glucose assay was followed except that the samples were incubated with or without 1 U of amyloglucosidase (Sigma; A7420). For trehalose measurement, 100 μl of glucose reagent was added and samples were incubated with or without 0.05 U/ml of trehalose (Sigma; T8778) for 18 h at 37°C. Absorbance was recorded at 540 nm and the trehalose or glycogen levels were calculated after the subtraction of the glucose measured at this step from the total amount of free glucose measured after trehalose or amyloglucosidase digestion. Glucose measurement in hemolymph collected from 20 flies was done by the addition of diluted (1:8) hemolymph in a 96-well plate followed by the addition of the glucose reagent. At least three replicates per genotype or experimental condition were performed.
Larvae body wall or fat body preparations for CLSM viewing
Third instar larvae were collected and dissected in PBS. Larvae were placed into a Petri anatomy dish and immobilized with two small pins (one to the top and one to the edge of larvae) so that the abdominal region was upward. Larvae were cut just above the edge and then a slit was made until the larvae mouth to enable removal of the internal organs. The body of the larvae was then opened and stretched with small pins.
For boron–dipyrromethene fluorescent dye (BODIPY; Thermo Fischer Scientific) staining of larvae fat body, the procedure of larvae body wall preparation was followed without removing the fat body. Dissected larvae were fixed in 4% formaldehyde for 15 min, washed in PBS containing 0.3% Triton X-100, and incubated with the BODIPY dye or DAPI (Thermo Fischer Scientific) for 15 min at room temperature. After three washes with PBS, samples were mounted in Mowiol® 4–88 (Sigma) and viewed in a Digital Eclipse C1 (Nikon) CLSM equipped with 40 × 1.0 NA differential interference contrast (DIC) and 60 × 1.4 NA DIC Plan Apochromat objectives; image capturing was done using the EZC1 acquisition and images were analyzed with the CLSM software (Nikon, Inc.); at least 10 animals per treatment were viewed and representative captures are shown.
Immunofluorescence staining of adult fly brain and CLSM visualization
Immunostaining of isolated adult fly brains was performed as described previously (60) with minor modifications. Briefly, isolated fly brains were fixed in 4% formaldehyde for 30 min, washed in PBS containing 0.3% Triton X-100, and incubated (overnight at 4°C) with the anti-AGEs antibody. Secondary antibody or DAPI (Thermo Fischer Scientific; for nuclear visualization) staining was applied for 1 h at RT. Following three washes with PBS, samples were mounted in Mowiol 4–88 (Sigma) and viewed in a Digital Eclipse Nikon C1 CLSM. Z-stacks with a step size of 0.5 μm were taken using identical settings; each stack consisted of 25–30 plane images (representative captures are shown).
Alignment of mammalian Gsk-3 protein isoforms with their fly ortholog
Alignment of protein sequences was done with CLUSTAL W (1.83) multiple sequence alignment. The mammalian Gsk-3 sequences used were Gsk-3β (NP_002084.2) and the Gsk-3β isoform 2 from Homo sapiens (NP_001139628.1) along with Gsk-3β from Mus musculus (NP_062801.1). The Drosophila melanogaster Gsk-3 isoform J (NP_996336.1) that was used for the molecular docking analyses and the “canonical” Gsk-3 D. melanogaster isoform B (NP_476715.1) were also used in the alignment analyses; bold residues denote the kinase ATP binding site (see alignment Table 1).
Table 1.
Mammalian Gsk-3 Versus Fly Sgg/Gsk-3 Alignment
 |
Molecular dynamics
The protein was prepared for molecular simulation studies using the Protein Preparation Workflow (Schrödinger Suite 2015 Protein Preparation Wizard) implemented in the Schrödinger suite and accessible from within the Maestro program (Maestro, version 10; Schrödinger, LLC). The hydrogen atoms were added and the orientation of hydroxyl groups, Asn, Gln, and the protonation state of His were optimized to maximize hydrogen bonding. Finally, the ligand−protein complex was refined with a restrained minimization performed by the impref utility, based on the Impact molecular mechanics engine (Impact version 6.6; Schrödinger, LLC) and the OPLS2001 force field, setting a maximum root mean square difference (RMSD) of 0.30. Ligand preparation for docking was performed with the LigPrep (LigPrep, version 2.5; Schrödinger, LLC) application, which consists of a series of steps that perform conversions, apply corrections to the structure, generate ionization states and tautomers, and optimize the geometries. The homology model created from human Gsk-3 crystal structure with 6BIO present in the substrate binding site (PDB 1UV5) was utilized as the initial input structure. For the MD simulations, Desmond v.3 software was applied (Desmond Molecular Dynamics System, version 3.8, D. E. Shaw Research) (7). The system was prepared by embedding the protein in a POPC lipid bilayer, solvating the membrane by TIP3P explicit water, neutralizing with counter ions, and adding 150 mM salt. The stepwise equilibration protocol for membrane proteins as developed by Desmond has been followed on eight steps. No driving forces were applied to the ligand. The atomic Cα RMSD of the two monomeric units following equilibration was 1.05 Å. Constant pressure and constant temperature (NPT) runs were performed for 10 ns simulations in three independent experiments with Langevin thermostat and barostat and semi-isotropic pressure restraints for the substrate studied (6BIO). RMS fluctuation calculations were based on frame 0 as a reference while Cα atoms of all residues were selected for RMSF calculation and fitting. Whole residue RMSF ranged from 1 Å for rigid secondary structural elements to 6 Å for mobile loops in both monomers similar to what has been observed for membrane proteins. Figures were created with Maestro v10.1 (Schrödinger, LLC).
Induced fit docking
All selected molecules passed through exhaustive molecular docking calculations using the IFD protocol (Induced Fit Docking protocol 2015-2, Glide version 6.4, Prime version 3.7, Schrödinger, LLC, 2015), which is intended to circumvent the inflexible binding site and accounts for the side-chain or backbone movements, or both, upon ligand binding. In the first stage of the IFD protocol, softened-potential docking step, 20 poses per ligand were retained. In the second step, for each docking pose, a full cycle of protein refinement was performed, with Prime 3.7 (Prime, version 3.7; Schrödinger, LLC) on all residues having at least one atom within 8 Å of an atom in any of the 20 ligand poses. The Prime refinement starts with a conformational search and minimization of the side-chains of the selected residues and after convergence to a low-energy solution, an additional minimization of all selected residues (side-chain and backbone) is performed with the truncated Newton algorithm using the OPLS parameter set and a surface Generalized Born implicit solvent model. The obtained complexes are ranked according to Prime calculated energy (molecular mechanics and solvation), and those within 30 kcal/mol of the minimum energy structure are used in the last step of the process, redocking with Glide 6.4 (Glide, version 6.4, Schrödinger, LLC, 2015) using standard precision and scoring. In the final round, the ligands used in the first docking step are redocked into each of the transporter structures retained from the refinement step. The final ranking of the complexes is done by a composite score, which accounts for the transporter–ligand interaction energy (GlideScore) and solvation energies (Prime energy).
Antibodies used
The polyclonal antibody against the Drosophila β5 proteasome subunit was a gift from Prof. M. Figueiredo-Pereira (Department of Biological Sciences, Hunter College of the City University of New York, NY). Primary antibodies against the 20S-α (sc-65755), Rpn10 (sc-65748), and Rpn7 (sc-65750) Drosophila proteasome subunits; the anti-Hsp70 (sc-25837), anti-ubiquitin (Ub) (sc-8017), anti-AMPKα (sc-25792), anti-GAPDH (sc-25778); anti-β-tubulin (sc-20852) and the HRP-conjugated secondary antibodies were from Santa Cruz Biotechnology (Heidelberg, Germany). The antibodies against Gsk-3 (05-412) and the Rpn6 (NBP1-46191) proteasome subunit were obtained from Millipore S.A. and Novus Inc. respectively. The antibodies against phospho-Gsk-3S21/S9 (#9331), phospho-AktS505 (#4054), Akt (#9272), phospho-AMPKαT172 (#2535), and the phosphoSer/Thr Pdpk1 docking motif (#9634) were purchased from Cell Signaling; the anti-AGE antibody was from Cosmo Bio (KAL-KH001-01).
Statistical analyses
Experiments were performed at least in duplicate (for each biological replicate, n ≥ 2). Assays were done after pooling isolated male and female somatic tissues from 10 to 20 flies (see above), unless otherwise stated. For statistical analyses, the MS Excel and the Statistical Package for Social Sciences (IBM SPSS; version 19.0 for Windows) were used. Statistical significance was evaluated using one-way analysis of variance (ANOVA). Data points correspond to the mean of the independent experiments and error bars denote standard deviation (SD); significance at p < 0.05 or p < 0.01 is indicated in graphs by one or two asterisks, respectively.
Supplementary Material
Abbreviations Used
- 6BIO
6-bromoindirubin-3′-oxime
- AGEs
advanced glycation end products
- ALP
autophagy lysosome pathway
- AMPK
AMP-activated kinase
- ARE
antioxidant response element
- BSA
bovine serum albumin
- CDKs
cyclin-dependent kinases
- CLSM
confocal laser scanning microscopy
- CncC
cap'-n'-collar isoform-C
- CP
core particle
- CR
caloric restriction
- DIC
differential interference contrast
- DMF
dimethylformamide
- ESI
electrospray ionization
- FCCP
p-trifluoromethoxyphenylhydrazone
- Foxo
forkhead box O
- GFP
green fluorescent protein
- Gsk-3
glycogen synthase kinase-3
- Gys
glycogen synthase
- H2O2
hydrogen peroxide
- hAPP
human amyloid precursor protein
- Hsf-1
heat shock factor-1
- Hsp
heat shock protein
- IFD
induced fit docking
- InS/IGF-1
insulin/insulin-like growth factor-1
- Keap1
Kelch-like ECH-associated protein 1
- LC-HRMS
liquid chromatography/high-resolution mass spectroscopy
- NPs
natural products
- Nrf-2
nuclear factor erythroid 2-related factor
- PBS
phosphate-buffered saline
- Pdpk1
3-phosphoinositide-dependent protein kinase-1
- PDR
proteome damage responses
- PN
proteostasis network
- Q-RT-PCR
quantitative real-time polymerase chain reaction
- RDBE
ring double bond equivalent
- RMSD
root mean square difference
- ROS
reactive oxygen species
- RP
regulatory particle
- SD
standard deviation
- t-BHP
tert-butyl hydroperoxide
- UHPLC
ultrahigh-performance liquid chromatography
- UPS
ubiquitin/proteasome system
Acknowledgments
We thank Profs M. Figueiredo-Pereira (Hunter College, NY) for the β5 antibody, as well as A. Daga (University of Padua, Padova, Italy) and D. Bohmann (University of Rochester, NY) for the fly lines. Molecular dynamics and Induced Fit Docking studies were supported by computational time granted from the Greek Research & Technology Network in the National HPC facility (ARIS) under project ID emikros-pr001046; computational analyses were also supported by the Cy-Tera Project (NEAYPODOMI/STRATI/0308/31) cofunded by the European Regional Development Fund and the Republic of Cyprus through the Research Promotion Foundation. We also acknowledge funding for the support of this work to IPT and ALS from the EU grant TASCMAR (EU-H2020/634674) and the Hellenic GSRT grant NatProt (3207).
Authors Disclosure Statement
No competing financial interests exist.
References
- 1.Alic N. and Partridge L. Death and dessert: nutrient signalling pathways and ageing. Curr Opin Cell Biol 23: 738–743, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Argyropoulou A, Aligiannis N, Trougakos IP, and Skaltsounis AL. Natural compounds with anti-aging activity. Nat Prod Rep 30: 1412–1437, 2013 [DOI] [PubMed] [Google Scholar]
- 3.Avrahami L, Farfara D, Shaham-Kol M, Vassar R, Frenkel D, and Eldar-Finkelman H. Inhibition of glycogen synthase kinase-3 ameliorates β-amyloid pathology and restores lysosomal acidification and mammalian target of rapamycin activity in the Alzheimer disease mouse model: in vivo and in vitro studies. J Biol Chem 288: 1295–1306, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrio L, Dekanty A, and Milán M. MicroRNA-mediated regulation of Dp53 in the Drosophila fat body contributes to metabolic adaptation tonutrient deprivation. Cell Rep 8: 528–541, 2014 [DOI] [PubMed] [Google Scholar]
- 5.Beurel E, Grieco SF, and Jope RS. Glycogen synthase kinase-3 (GSK3): regulation, actions, and diseases. Pharmacol Ther 148: 114–131, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourouis M. Targeted increase in shaggy activity levels blocks wingless signaling. Genesis 34: 99–102, 2002 [DOI] [PubMed] [Google Scholar]
- 7.Bowers KJ, Chow E, Xu H, Dror RO, Eastwood MP, Gregersen BA, Klepeis JL, Kolossvary I, Moraes MA, Sacerdoti FD, Salmon JK, Shan Y, and Shaw DE. Scalable algorithms for molecular dynamics simulations on commodity clusters. Proceedings of the 2006 ACM/IEEE Conference on Supercomputing (SC06), Tampa, FL, ACM Press, 2006 [Google Scholar]
- 8.Braig S, Kressirer CA, Liebl J, Bischoff F, Zahler S, Meijer L, and Vollmar AM. Indirubin derivative 6BIO suppresses metastasis. Cancer Res 73: 6004–6012, 2013 [DOI] [PubMed] [Google Scholar]
- 9.Castillo-Quan JI, Li L, Kinghorn KJ, Ivanov DK, Tain LS, Slack C, Kerr F, Nespital T, Thornton J, Hardy J, Bjedov I, and Partridge L. Lithium promotes longevity through GSK3/NRF2-dependent hormesis. Cell Rep 15: 638–650, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu B, Soncin F, Price BD, Stevenson MA, and Calderwood SK. Sequential phosphorylation by mitogen-activated protein kinase and glycogen synthase kinase 3 represses transcriptional activation by heat shock factor-1. J Biol Chem 271: 30847–30857, 1996 [DOI] [PubMed] [Google Scholar]
- 11.Crowder RJ. and Freeman RS. Glycogen synthase kinase-3β activity is critical for neuronal death caused by inhibiting PI 3-kinase or Akt but not for death caused by NGF withdrawal. J Biol Chem 275: 34266–34271, 2000 [DOI] [PubMed] [Google Scholar]
- 12.Dokken BB, Sloniger JA, and Henriksen EJ. Acute selective glycogen synthase kinase-3 inhibition enhances insulin signaling in prediabetic insulin-resistant rat skeletal muscle. Am J Physiol Endocrinol Metab 288: E1188–E1194, 2005 [DOI] [PubMed] [Google Scholar]
- 13.Eldar-Finkelman H, Argast GM, Foord O, Fischer EH, and Krebs EG. Expression and characterization of glycogen synthase kinase-3 mutants and their effect on glycogen synthase activity in intact cells. Proc Natl Acad Sci U S A 93: 10228–10233, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finlay D, Patel S, Dickson LM, Shpiro N, Marquez R, Rhodes CJ, and Sutherland C. Glycogen synthase kinase-3 regulates IGFBP-1 gene transcription through the thymine-rich insulin response element. BMC Mol Biol 5: 15, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fontana L. and Partridge L. Promoting health and longevity through diet: from model organisms to humans. Cell 161: 106–118, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forde JE. and Dale TC. Glycogen synthase kinase 3: a key regulator of cellular fate. Cell Mol Life Sci 64: 1930–1944, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fredriksson Å, Johansson Krogh E, Hernebring M, Pettersson E, Javadi A, Almstedt A, and Nyström T. Effects of aging and reproduction on protein quality control in soma and gametes of Drosophila melanogaster. Aging Cell 11: 634–643, 2012 [DOI] [PubMed] [Google Scholar]
- 18.Gagliardi PA, di Blasio L, and Primo L. PDK1: A signaling hub for cell migration and tumor invasion. Biochim Biophys Acta 1856: 178–188, 2015 [DOI] [PubMed] [Google Scholar]
- 19.Ha S, Ryu HY, Chung KM, Baek SH, Kim EK, and Yu SW. Regulation of autophagic cell death by glycogen synthase kinase-3β in adult hippocampal neural stem cells following insulin withdrawal. Mol Brain 8: 30, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haigis MC. and Yankner BA. The aging stress response. Mol Cell 40: 333–344, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoeflich KP, Luo J, Rubie EA, Tsao MS, Jin O, and Woodgett JR. Requirement for glycogen synthase kinase-3 beta in cell survival and NF-kappaB activation. Nature 406: 86–90, 2000 [DOI] [PubMed] [Google Scholar]
- 22.Huo X, Liu S, Shao T, Hua H, Kong Q, Wang J, Luo T, and Jiang Y. GSK3 protein positively regulates type I insulin-like growth factor receptor through forkhead transcription factors FOXO1/3/4. J Biol Chem 289: 24759–24770, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobson J, Lambert AJ, Portero-Otín M, Pamplona R, Magwere T, Miwa S, Driege Y, Brand MD, and Partridge L. Biomarkers of aging in Drosophila. Aging Cell 9: 466–477, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain AK. and Jaiswal AK. GSK-3beta acts upstream of Fyn kinase in regulation of nuclear export and degradation of NF-E2 related factor 2. J Biol Chem 282: 16502–16510, 2007 [DOI] [PubMed] [Google Scholar]
- 25.Kästle M. and Grune T. Interactions of the proteasomal system with chaperones: protein triage and protein quality control. Prog Mol Biol Transl Sci 109: 113–160, 2012 [DOI] [PubMed] [Google Scholar]
- 26.Kensler TW. and Wakabayashi N. Nrf2: friend or foe for chemoprevention? Carcinogenesis 31: 90–99, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerkela R, Kockeritz L, Macaulay K, Zhou J, Doble BW, Beahm C, Greytak S, Woulfe K, Trivedi CM, and Woodgett JR. Deletion of GSK-3beta in mice leads to hypertrophic cardiomyopathy secondary to cardiomyoblast hyperproliferation. J Clin Invest 118: 3609–3618, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Labbadia J. and Morimoto RI. The biology of proteostasis in aging and disease. Ann Rev Biochem 84: 435–464, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levine B. and Kroemer G. Autophagy in the pathogenesis of disease. Cell 132: 27–42, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lochhead PA, Coghlan M, Rice SQ, and Sutherland C. Inhibition of GSK-3 selectively reduces glucose-6-phosphatase and phosphatase and phosphoenolypyruvate carboxykinase gene expression. Diabetes 50: 937–946, 2001 [DOI] [PubMed] [Google Scholar]
- 31.López-Otín C, Blasco MA, Partridge L, Serrano M, and Kroemer G. The hallmarks of aging. Cell 153: 1194–1217, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mattila J, Kallijärvi J, and Puig O. RNAi screening for kinases and phosphatases identifies FoxO regulators. Proc Natl Acad Sci U S A 105: 14873–14878, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McColl G, Killilea DW, Hubbard AE, Vantipalli MC, Melov S, and Lithgow GJ. Pharmacogenetic analysis of lithium-induced delayed aging in Caenorhabditis elegans. J Biol Chem 283: 350–357, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meijer L, Skaltsounis AL, Magiatis P, Polychronopoulos P, Knockaert M, Leost M, Ryan XP, Vonica CA, Brivanlou A, Dajani R, Crovace C, Tarricone C, Musacchio A, Roe SM, Pearl L, and Greengard P. GSK-3-selective inhibitors derived from Tyrian purple indirubins. Chem Biol 10: 1255–1266, 2003 [DOI] [PubMed] [Google Scholar]
- 35.Mirisola MG, Taormina G, Fabrizio P, Wei M, Hu J, and Longo VD. Serine- and threonine/valine-dependent activation of PDK and Tor orthologs converge on Sch9 to promote aging. PLoS Genet 10: e1004113, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palikaras K, Lionaki E, and Tavernatakis N. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature 521: 525–528, 2015 [DOI] [PubMed] [Google Scholar]
- 37.Papadopoulou D, Bianchi MW, and Bourouis M. Functional studies of shaggy/glycogen synthase kinase 3 phosphorylation sites in Drosophila melanogaster. Mol Cell Biol 24: 4909–4919, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polychronopoulos P, Magiatis P, Skaltsounis AL, Myrianthopoulos V, Mikros E, Tarricone A, Musacchio A, Roe SM, Pearl L, Leost M, Greengard P, and Meijer L. Structural basis for the synthesis of indirubins as potent and selective inhibitors of glycogen synthase kinase-3 and cyclin-dependent kinases. J Med Chem 47: 935–946, 2004 [DOI] [PubMed] [Google Scholar]
- 39.Rada P, Rojo AI, Chowdhry S, McMahon M, Hayes JD, and Cuadrado A. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol 31: 1121–1133, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rahman MM, Sykiotis GP, Nishimura M, Bodmer R, and Bohmann D. Declining signal dependence of Nrf2-MafS-regulated gene expression correlates with aging phenotypes. Aging Cell 12: 554–562, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ribas J, Bettayeb K, Ferandin Y, Knockaert M, Garrofé-Ochoa X, Totzke F, Schächtele C, Mester J, Polychronopoulos P, Magiatis P, Skaltsounis AL, Boix J, and Meijer L. 7-Bromoindirubin-3′-oxime induces caspase-independent cell death. Oncogene 25: 6304–6318, 2006 [DOI] [PubMed] [Google Scholar]
- 42.Rintelen F, Stocker H, Thomas G, and Hafen E. PDK1 regulates growth through Akt and S6K in Drosophila. Proc Natl Acad Sci U S A 98: 15020–15025, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rizak J, Tan H, Zhu H, and Wang JF. Chronic treatment with the mood-stabilizing drug lithium up-regulates nuclear factor E2-related factor 2 in rat pheochromocytoma PC12 cells in vitro. Neuroscience 256: 223–229, 2014 [DOI] [PubMed] [Google Scholar]
- 44.Rusai K, Herzog R, Kuster L, Kratochwill K, and Aufricht C. GSK-3β inhibition protects mesothelial cells during experimental peritoneal dialysis through upregulation of the heat shock response. Cell Stress Chaperones 18: 569–579, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanchez JF, Sniderhan LF, Williamson AL, Fan S, Chakraborty-Sett S, and Maggirwar SB. Glycogen synthase kinase 3β-mediated apoptosis of primary cortical astrocytes involves inhibition of nuclear factor κB signaling. Mol Cell Biol 23: 4649–4662, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sato N, Meijer L, Skaltsounis L, Greengard P, and Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med 10: 55–63, 2004 [DOI] [PubMed] [Google Scholar]
- 47.Schäfer M, Goodenough S, Moosmann B, and Behl C. Inhibition of glycogen synthase kinase 3 beta is involved in the resistance to oxidative stress in neuronal HT22 cells. Brain Res 1005: 84–89, 2004 [DOI] [PubMed] [Google Scholar]
- 48.Sofola-Adesakin O, Castillo-Quan JI, Rallis C, Tain LS, Bjedov I, Rogers I, Li L, Martinez P, Khericha M, Cabecinha M, Bähler J, and Partridge L. Lithium suppresses Aβ pathology by inhibiting translation in an adult Drosophila model of Alzheimer's disease. Front Aging Neurosci 6: 190, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzuki T, Bridges D, Nakada D, Skiniotis G, Morrison SJ, Lin JD, Saltiel AR, and Inoki K. Inhibition of AMPK catabolic action by GSK3. Mol Cell 50: 407–419, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sykiotis GP. and Bohmann D. Stress-activated cap'n'collar transcription factors in aging and human disease. Sci Signal 3: re3, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tam ZY, Gruber J, Ng LF, Halliwell B, and Gunawan R. Effects of lithium on age-related decline in mitochondrial turnover and function in Caenorhabditis elegans. J Gerontol A Biol Sci Med Sci 69: 810–820, 2014 [DOI] [PubMed] [Google Scholar]
- 52.Trougakos IP, Sesti F, Tsakiri E, and Gorgoulis VG. Non-enzymatic post-translational protein modifications and proteostasis network deregulation in carcinogenesis. J Proteomics 92: 274–298, 2013 [DOI] [PubMed] [Google Scholar]
- 53.Tsakiri EN, Sykiotis GP, Papassideri IS, Gorgoulis VG, Bohmann D, and Trougakos IP. Differential regulation of proteasome functionality in reproductive vs. somatic tissues of Drosophila during aging or oxidative stress. FASEB J 27: 2407–2420, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsakiri EN, Iliaki KK, Höhn A, Grimm S, Papassideri IS, Grune T, and Trougakos IP. Diet-derived advanced glycation end products or lipofuscin disrupts proteostasis and reduces life span in Drosophila melanogaster. Free Radic Biol Med 65: 1155–1163, 2013 [DOI] [PubMed] [Google Scholar]
- 55.Tsakiri EN, Sykiotis GP, Papassideri IS, Terpos E, Dimopoulos MA, Gorgoulis VG, Bohman D, and Trougakos IP. Proteasome dysfunction in Drosophila signals to an Nrf2-dependent regulatory circuit aiming to restore proteostasis and prevent premature aging. Aging Cell 12: 802–813, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsakiri EN. and Trougakos IP. The amazing ubiquitin-proteasome system: structural components and implication in aging. Int Rev Cell Mol Biol 314: 171–237, 2015 [DOI] [PubMed] [Google Scholar]
- 57.Vermeij WP, Dollé ME, Reiling E, Jaarsma D, Payan-Gomez C, Bombardieri CR, Wu H, Roks AJ, Botter SM, van der Eerden BC, Youssef SA, Kuiper RV, Nagarajah B, van Oostrom CT, Brandt RM, Barnhoorn S, Imholz S, Pennings JL, de Bruin A, Gyenis Á, Pothof J, Vijg J, van Steeg H, and Hoeijmakers JH. Restricted diet delays accelerated ageing and genomic stress in DNA-repair-deficient mice. Nature 2016. DOI: 10.1038/nature19329 [DOI] [PMC free article] [PubMed]
- 58.Vilchez D, Morantte I, Liu Z, Douglas PM, Merkwirth C, Rodrigues AP, Manning G, and Dillin A. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature 489: 263–268, 2012 [DOI] [PubMed] [Google Scholar]
- 59.Vougogiannopoulou K, Ferandin Y, Bettayeb K, Myrianthopoulos V, Lozach O, Fan Y, Johnson CH, Magiatis P, Skaltsounis AL, Mikros E, and Meijer L. Soluble 3′,6-substituted indirubins with enhanced selectivity toward glycogen synthase kinase-3 alter circadian period. J Med Chem 51: 6421–6431, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.White KE, Humphrey DM, and Hirth F. The dopaminergic system in the aging brain of Drosophila. Front Neurosci 4: 205, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wigmore SJ, Sangster K, McNally SJ, Harrison EM, Ross JA, Fearon KC, and Garden OJ. De-repression of heat shock transcription factor-1 in interleukin-6- treated hepatocytes is mediated by downregulation of glycogen synthase kinase 3beta and MAPK/ERK-1. Int J Mol Med 19: 413–420, 2007 [PubMed] [Google Scholar]
- 62.Zahler S, Tietze S, Totzke F, Kubbutat M, Meijer L, Vollmar AM, and Apostolakis J. Inverse in silico screening for identification of kinase inhibitor targets. Chem Biol 14: 1207–1214, 2007 [DOI] [PubMed] [Google Scholar]
- 63.Zhou J, Freeman TA, Ahmad F, Shang X, Mangano E, Gao E, Farber J, Wang Y, Ma XL, Woodgett J, Vagnozzi RJ, Lal H, and Force T. GSK-3α is a central regulator of age-related pathologies in mice. J Clin Invest 123: 1821–1832, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.