Abstract
Comparative protein structure modeling is a computational approach to build three-dimensional structural models for proteins using experimental structures of related protein family members as templates. Regular blind assessments of modeling accuracy have demonstrated that comparative protein structure modeling is currently the most reliable technique to model protein structures. Homology models are often sufficiently accurate to substitute for experimental structures in a wide variety of applications. Since the usefulness of a model for specific application is determined by its accuracy, model quality estimation is an essential component of protein structure prediction. Comparative protein modeling has become a routine approach in many areas of life science research since fully automated modeling systems allow also non-experts to build reliable models. In this manuscript, we describe practical approaches for automated protein structure modeling with SWISS-MODEL Workspace and the Protein Model Portal.
Keywords: protein structure prediction, molecular models, automation, homology modeling, comparative modeling, quality estimation, SWISS-MODEL, PMP, Protein Model Portal, QMEAN
1. Introduction
Knowing a protein’s three-dimensional structure is crucial for understanding its biological function at the molecular level. However, despite remarkable advances in protein structure determination by NMR and X-Ray crystallography, currently no experimental structural information is available for the vast majority of protein sequences resulting from large-scale genome sequencing and meta-genomics projects. To overcome this knowledge gap, over the past decades a wide variety of computational methods for predicting the structure of proteins have been developed. These methods differ significantly in their computational complexity, the range of proteins for which they can be applied, and the accuracy and reliability of the resulting models (1, 2). Here, we will focus on homology modeling (aka comparative or template based modeling), where a model for a protein of interest is constructed using structural information from homologous proteins (1–6). Regular blind assessment of prediction techniques has shown that comparative protein structure modeling is currently the only technique which is able to reliably provide models of high quality over a wide range of size, while de novo prediction methods are limited to small proteins and peptides (7). On the other side, comparative modeling techniques are limited to cases for which suitable template structures can be indentified. For example, this poses a major limitation when modeling membrane proteins, which are underrepresented in today’s structure databases but embody the majority of pharmaceutically interesting drug targets (8). The usefulness of protein structure models has been demonstrated in a variety of biological applications (9–11), such as rational design of mutagenesis experiments (12), providing receptor models for virtual screening (13, 14), to develop strategies for protein engineering, or to support experimental structure solution by crystallography (15, 16) or electron microscopy (17–19).
Computational modeling has become a valuable tool to complement experimental elucidation of protein structures. To make 3-dimesional information accessible to a broad community of biomedical researchers on a whole-genome scale, automated modeling pipelines had to be developed which were stable, reliable, accurate and easy to use. Almost two decades ago the first automated modeling server - SWISS-MODEL - was made available on the Internet (20). Since then, many more services have been developed to model the structures of proteins in an automated manner (21, 22), e.g. ModWeb (23), Robetta (24), HHpred (25), I-TASSER (26), Pcons (27), PHYRE (28), or M4T (29). Recent method developments aim to include additional experimental constraints into the modeling procedures (17–19, 30), and to establish methods specialized in certain protein families such as GPCRs (31, 32) or Antibodies (33, 34).
One main objective for automating the principal steps of comparative protein structure modeling – template selection, target-template alignment, model building and model quality evaluation (Fig. 1) – is the need of making these technologies accessible to an audience of non experts in bioinformatics. This includes facilitating the usage of computational tools which otherwise required highly specialized technical skills, maintaining up-to-date modeling software, and managing large amounts of sequence and structural data stored in biological databases, which are needed to complete the modeling tasks. Secondly, due to the huge number of protein sequences whose structure has not yet been experimentally characterized, automated procedures are essential to cope with this flood of data, e.g. to increase the coverage of structural information for proteomes of whole organisms or families of proteins (20, 35–37). Finally, from a theoretical perspective, automatic procedures ensure the reproducibility of the modeling methods by excluding individual human bias, which is a pre-requisite for the assessment and comparison of their reliability and accuracy (22, 38).
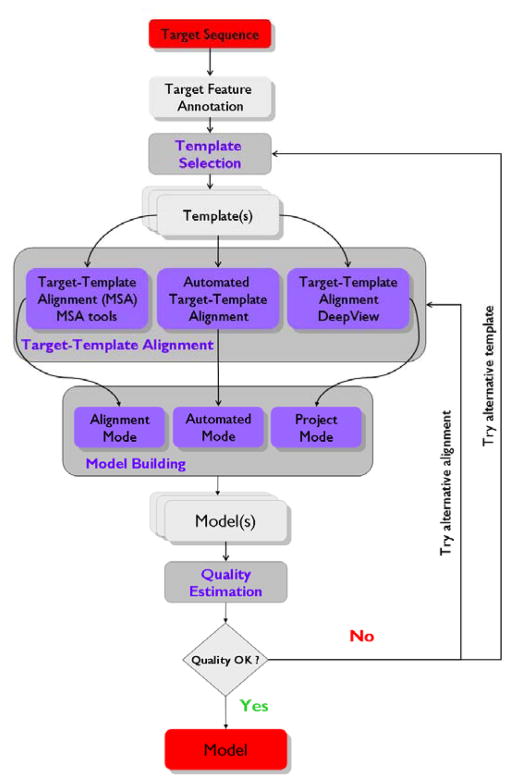
Fig. 1.
SWISS-MODEL workflow. The flowchart illustrates the classical steps to construct a homology model of a target sequence as they are implemented in SWISS-MODEL Workspace. Starting from the sequence of the protein of interest (target) one or more related structures (templates) are identified (template selection). Annotation of the target sequence (feature annotation) can guide the choice of appropriate template(s). Based on the evolutionary distance between target and template(s) sequences three different regimes of the target-template alignment step are available on the Swiss-Model Workspace: Automated, Alignment or Project Mode. Target and template(s) sequences are aligned (target-template alignment) either in a fully automated fashion, by using external alignment tools, and (optionally) adjusted visually with the help of the DeepView program. The model is then constructed based on these alignments. Finally the quality of the obtained model(s) can be estimated and verified and if necessary the procedure is repeated until a satisfactory result is obtained.
Validating the quality of the obtained models is a central aspect of protein structure modeling. The quality of models determines their usefulness for specific applications in life science research (9). Scoring functions which aim to estimate the expected accuracy of a protein model are therefore crucial to judge if it would be suitable to address a specific biomedical question. A well known first estimate for the expected quality of a structural model is the sequence identity between the target and the template sequences, where in general higher sequence similarity leads to more accurate models since the evolutionary structural divergence will be smaller (39) and alignment errors less likely to occur (40). However, sequence identity is only a first indicator and depending on the specific protein at hand, accurate models can be achieved based on very low sequence identity templates, while models based on medium sequence identity templates may contain significant errors. The development of more sophisticated scoring methods, taking into account various aspects of structural and sequence information in order to be able to judge the quality of obtained models (41–45), is currently matter of intensive research.
The SWISS-MODEL Server
Since the first release of the SWISS-MODEL server, the resource has evolved to reflect advances of modeling algorithms as well as Internet and web-technologies (46). The most recent version of the server is the SWISS-MODEL Workspace (47), a web based working environment, where users can easily compute and store the results of various computational tasks required to build homology models. In particular, the Workspace gives access to software and databases necessary to complete the four main steps of comparative modeling: (i) detection of experimental structures (templates) homologous to the protein of interest (target), (ii) alignment of the target and template(s) protein sequences, (iii) building of one or more models for the target protein (iv) and evaluation of the quality of the obtained model(s) (Fig. 1). In the fully “Automated” mode of SWISS-MODEL Workspace, the amino acid sequence (or the database accession code) of the protein of interest is sufficient as input to compute a structural model in a completely automated fashion. For non-trivial modeling cases, however, where the evolutionary distance between target and template is large, it is advisable to use the “Alignment” mode of the server, where a curated multiple sequence alignment of target, template and other family members of the protein can be submitted to compute the structural model. Similarly, the “Project” mode of the SWISS-MODEL Workspace allows the user to examine and manipulate the target-template alignment in its structural context within the DeepView (Swiss-Pdb Viewer) visualization and structural analysis tool (20). The server will then build the coordinates of the model according to the target-template alignment specified by the user.
Programs like SWISS-MODEL generate the structural coordinates of the model based on the mapping between the target residues and the corresponding amino acids of the structural template(s). Regions of the protein, for which no template information is available, typically insertions and deletions in loop regions, are built by using libraries of backbone fragments (48) or by constraint space de novo reconstruction of these backbone segments (49). Local suboptimal geometry of the obtained model, e.g. distorted bonds, angles and close atomic contacts due to imperfect combination of fragments from structural templates, is regularized by limited energy minimization using the Gromos96 force field (50). Finally the quality of the overall model is validated using specialized model quality estimation tools (MQE) such as ANOLEA (44) or QMEAN (51). Often when building a structural model for a specific protein, it is useful to produce several models based on alternative target-template alignments, especially if the sequences are only distantly related. The expected quality of the produced models can then be predicted to identify which has(have) the highest probability of being the most accurate. Moreover, based on hypotheses about the functional mechanisms of a protein, the visualization of key residues in their structural context may facilitate deciding which models are the most useful for the biochemical application of interest. Workspace offers additional tools to support the building of protein 3D-model(s) such as programs for functional and domain annotation, template identification, and structure assessment. Please see “Materials and on Methods” section for details.
Protein Model Portal
The goal of Protein Model Portal (PMP) (52) of the Nature PSI Structural Biology Knowledgebase (53) is to promote the efficient use of molecular models in biomedical research. PMP provides a comprehensive view of structural information for proteins by combining information on experimental structures and theoretical models from various modeling resources. When searching the Protein Model Portal, data about experimental structures are derived from the latest version of the PDB databank (54), whereas comparative models are obtained from repositories of precompiled models (36, 37). It is not feasible to regularly pre-compute models for all protein sequences known today, and a more suitable template may have become available for a given protein of interest since it was initially modeled. Therefore, PMP provides an interface to simultaneously submit a modeling request to several state-of-the-art modeling resources (29, 47, 55, 56) to receive a set of up-to-date models by different homology modeling programs. Using different independent methods for modeling may indicate which parts of the protein structure model are expected to be more and which to be less reliable. In other words, regions of the protein which are consistently predicted to be similar by different independent methods are considered more likely to be correct (57). Finally to estimate the quality of the obtained models, PMP provides an interface to submit models in parallel to several model quality estimation tools, e.g. ModEval (43), ModFold (58), and QMEAN (41, 51).
In this manuscript, we will illustrate the use of SWISS-MODEL and Protein Model Portal for automated comparative protein structure modeling for a selection of examples.
2. Material
2.1 Swiss-Model Workspace
2.1.1 Access to the service
A computer with a web browser and connection to the Internet to access the web address of the server: http://swissmodel.expasy.org/workspace/.
The Java runtime environment (JRE) installed on the computer to run Astex (59) a molecular graphics program accessible on the server web site. Java is typically installed on most computers. You can get the latest version at http://java.com.
2.1.2 Software
The DeepView (Swiss-PdbViewer) software (v4.0) (20), downloaded and installed from http://spdbv.vital-it.ch/. Microsoft Windows and Mac versions of the program are available.
To learn the basic handling of the program DeepView, we recommend following Gale Rhodes’ tutorial at: http://spdbv.vital-it.ch/TheMolecularLevel/SPVTut/index.html.
2.1.3 Programs accessible through the server
Several tools necessary to complete the modeling task are accessible through the server, i.e. they do not require local installation on the computer:
Protein sequence structure and function annotation programs: InterProScan (60) for protein domain motifs and families recognition, PsiPred (61) for secondary structure prediction, DisoPred (62) for disorder prediction and MEMSAT (63) to predict transmembrane segments.
Database search programs for template selection: Blast (64), Iterative Profile Blast (64) and HHsearch (65).
Programs for protein structure and model quality evaluation: QMEAN (41), Gromos (50) and Anolea (44) to estimate the local (per residue) accuracy of the models; DFire (45) to estimate the global quality of the models; Whatchek (66) and Procheck (67) to verify the stereochemistry of protein structures and molecular models; DSSP (68) and Promotif (69) to evaluate structural features, such as secondary and super-secondary structures elements.
2.2 PMP
2.2.1 Access to the service
A computer with a web browser installed and a connection to the internet to access the web address of the server: http://proteinmodelportal.org/.
The Java runtime environment (JRE) installed on the computer to run Jmol (70), a viewer for chemical structures embedded in the web site. Java is typically installed on most computers. You can get the latest version at http://java.com.
2.2.2 Participating resources
Following resources are currently participating to the Protein Model Portal:
The PDB (54) protein structure database.
Comparative models providers: CSMP - Center for Structures of Membrane Proteins (71), JCSG - Joint Center for Structural Genomics (72), MCSG - Midwest Center for Structural Genomics (73), NESG - Northeast Center for Structural Genomics (74), NYSGXRC - NYSGX Research Center for Structural Genomics (75), JCMM - Joint Center for Molecular Modeling (76), ModBase (37) and SWISS-MODEL Repository (36) databases of comparative protein structure models.
Interactive services for model building: ModWeb (37), M4T (29), SWISS-MODEL (47) and I-Tasser (56).
Model quality estimation tools: ModFOLD (58), QMEAN (51) and ModEval (43).
3. Methods
Please note that the examples used in this section to describe the usage and the results obtainable from the SWISS-MODEL Workspace and Protein Model Portal represent the status of the these resources at the time of writing. Different results, in general better, may be obtained at a later point since more closely related experimental template structures might become available.
3.1 Swiss-Model Workspace
We will use the Caulobacter crescentus protein PopA (UniProt accession code Q9A784 (77)) to demonstrate how to use the SWISS-MODEL Workspace to generate and analyze comparative models. PopA is a paralog in C. crescentus of PleD, a response regulator protein which is a component of the signal transduction pathway controlling transitions between motile and sessile lifestyles in eubacteria (78). PleD catalyzes the condensation of two GTP molecules to the cyclic dinucleotide di-GMP (c-di-GMP), an ubiquitous second messenger in bacteria (79). The diguanylate cyclase activity is harbored by the GGDEF (or DGC) domain of the protein. PleD also contains two response regulatory domains, CheY-like response regulator receiver (Rec, also called D1) domains.
3.1.1 User account
The SWISS-MODEL Workspace is freely accessible at http://swissmodel.expasy.org. For each user, the results of their computations are organized in a personal account, a workspace. Each calculation is stored as a “work unit” of the Workspace, displaying title and status of the computation. Work units are automatically deleted after a week, unless the storage of the results is prolonged by the user.
Alternatively, occasional users have the possibility to use SWISS-MODEL without the need to create a personal account by bookmarking the results pages for future reference.
3.1.2 Target sequence feature annotation
Tools to analyze the sequence of a protein and predict its functional and structural characteristics can be very useful in identifying the most probable structural template(s) (see paragraph 3.1.3). These programs are accessible in the “Domain Annotation” Tools section on the Workspace (Fig. 2). It is sufficient to provide the sequence or the UniProt accession code (80) of the protein of interest and select among a list of available tools:
Fig. 2.
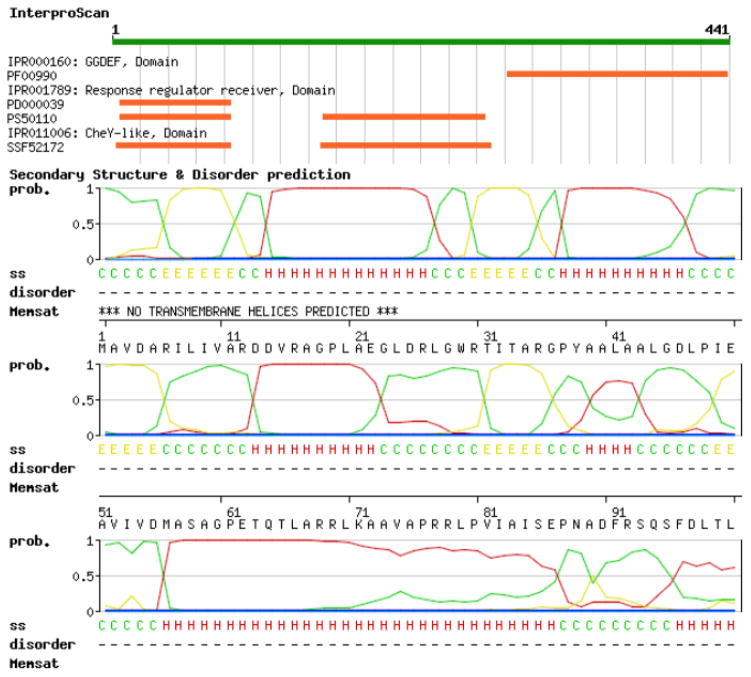
SWISS-MODEL Workspace target sequence feature annotation. To predict functional and structural features of the target proteins, several annotation tools are available on the SWISS-MODEL Workspace. In this example, the C. crescentus PopA protein (represented as a green bar on the top) is predicted to contain a C-terminal GGDEF domain and two N-terminal receiver domains. The likelihood (between 0 and 1, where 1 means highest probability) of the occurrence of secondary structure elements are depicted as curves (red for alpha-helices, yellow for β-strands and green for coiled regions). Prediction of disordered regions and transmembrane domains is also available. In particular, for PopA neither intrinsically unstructured regions nor portions of the protein spanning the membrane are detected.
InterProScan (60) queries protein sequences against the InterPro database (81) (see Note 1). In our example, InterProScan predicts the presence of a GGDEF domain in the C-terminal region of the PopA protein and two receiver domains in the N-terminal, respectively. Details about the location in the protein of different domains and signatures are graphically displayed and links to the InterPro database provide additional information about the protein classification and documentation about the signature annotations.
DISOPRED (62) detects intrinsically unstructured regions in protein, i.e. segments of protein with no defined 3-dimensional structure in solution (see Note 2). Disordered residues are represented by asterisks (*), whereas ordered are shown with dots (.). PopA is predicted to contain no intrinsically disordered regions.
MEMSAT (63) predicts regions of proteins spanning cellular membranes, indicated with “X” in the output of the program. PopA appears to not contain any trans-membrane segments.
PsiPred (61) predicts the occurrence of secondary structure elements, such as alpha helixes, extended beta strands or coil regions, which are graphically indicated by a letter H, E and C respectively.
Comparing the functional annotations of the target protein with the protein features of possible templates can help deciding if a given structure can be used as scaffold to build a comparative model. A protein with a known 3D-structure sharing the same type of domains, or having a similar secondary structure elements arrangement can indicate an evolutionary relationship to the target protein. Indications about the presence of trans-membrane domains or disordered regions are also valuable hints regarding the function and the domain architecture of the target protein and can be taken into account when evaluating if templates are available and for which region(s) of the protein of interest.
3.1.3 Template detection
A prerequisite for building a homology model is the availability of one or more evolutionary related proteins whose structure has been elucidated experimentally (see Note 3). For this purpose the target protein sequence can be queried against a sequence library (SMTL – SWISS-MODEL Template Library) extracted from known structures using increasingly sensitive search methods. The sequence (in FASTA or raw sequence format) or the corresponding UniProt AC can be submitted to the following search tools available in the Workspace “Template identification” tools section:
Blast (64), to detect evolutionarily closely related protein structures. Basic Blast standard parameters can be adjusted to regulate the sensitivity and the selectivity of the program (see Note 4).
Iterative Profile Blast (64), is used to identify more distantly related proteins (see Note 5).
HHSearch (65), an HMM based profile-profile comparison tool, is a very sensitive search method to detect remotely related sequences (see Note 6).
A graphical synopsis of the search results is presented showing the region(s) of the related target protein(s) aligned to the query sequence. The matches are colored according to their statistical significance (Expectation- and/or Probability-values, for details see Note 7), green color indicating more reliable hits. Domain boundaries according to InterPro annotations are also shown to guide the choice of suitable template with respect to functional domains. Details about the detected templates are accessible below the graphical representation, alongside with the alignment of the template sequence to the protein of interest.
In this example, Blast and Profile Blast template recognition tools detect three structures (PDB ID 1w25, 2wb4, and 2v0n) as possible templates for PopA. They represent structures of the paralog PleD protein in C. crescentus in complex with c-di-GMP, the activated form in complex with c-di-GMP and the activated form in complex with c-di-GMP and GTP-alpha-S, respectively (82, 83). HHsearch additionally detect the Pseudomonas aeruginosa diguanylate cyclase WspR (84) as potential template. All four structures span the full length of the target protein (see Note 8) ; three of them are paralogs whereas the WspR protein is an ortholog protein. Since all structures represent statistically significant hits (very low E-values), users should decide based on templates annotations which is(are) the most suitable template(s) for building the comparative model for PopA. Typically, one would select a template with high sequence similarity (PDB IDs 1w25, 2wb4 or 2v0n (82, 83)), unless specific features are considered important for the planned application: i.e. using templates in active or inactive forms, bound to specific ligands, etc. (see Note 9).
If clustered versions of the templates library are searched using the template detection tools, all the structures of the same cluster can be retrieved by clicking the corresponding “show template cluster” link of the results list.
3.1.4 Target-template Alignment
The target-template alignment generated by the template search tools can be used as starting point to create the correspondence between the residues of the target protein and the structure of the template, to ultimately produce the homology model. This is a critical step since standard homology modeling techniques will not recover from an incorrect input alignment, therefore special care should be addressed to this step.
The alignments in the output of the template identification tools can be retrieved as DeepView format file for further inspection. The file contains the target sequence aligned to the structure of the template. This allows the users to inspect the occurrence of amino acid insertions/deletions in the alignment in their structural context. For instance it is more likely that during evolution an insertion/deletion has occurred in a flexible surface loop rather than in a well structured secondary structure element such as an alpha helix or a beta strand in the core of the structure. The alignment between target and template sequences can be modified using the DeepView program’s “alignment window” and the changes visualized in the 3D-environment of the structure. The “alignment window” also allows verifying if important residues of both target and template sequences (i.e. amino acids belonging to active sites) are correctly aligned. For this purpose the DeepView function “scan for Prosite Patterns” (85) of the “Edit” menu can be applied.
Alternatively, pair-wise or multiple sequence alignment between the target, the template and preferably related sequences, can be generated with other state-of-the-art alignments tools (see Note 10) and submitted to the server for computation of models (see next paragraph 3.1.5 Model building).
3.1.5 Model building
Three variations of the model generation step are available in Workspace: “Automated”, “Alignment” and “Project” Modes. These are accessible in the “Modelling” section of the server:
The Automated mode is recommended when the sequence similarity between target and template proteins is high, i.e. larger than 60%. It is sufficient to submit the target sequence (either in raw or Fasta format) and the Swiss-Model pipeline will select the template(s) based on a hierarchical procedure to search and select the most suitable structures (36). If several templates are available or a custom-made structure is required, the user can additionally specify to use a particular template by either indicating its PDB ID code or by uploading a file in PDB format of the structure (see Note 11).
The Alignment method is appropriate for more distantly related target and template sequences. Multiple sequence alignment algorithms and PSSM or HHM based profile-profile methods (86) will generate the reasonable alignments. However, often these alignments can be verified manually and improved using for instances sequence alignment editors like JalView (87). The alignment in one of the supported formats (FASTA, MSF, ClustalW, PFAM and SELEX) can be subsequently submitted to the Workspace server. The alignment is checked for format compatibility and the user is required to indentify the sequences of the target and of the template protein and the PDB protein chain ID of the template structure (see Note 12) when submitting the alignment for the computation of models.
If the protein target-template sequence identity is close to the twilight-zone (i.e. sequence identity below 20%) (88), particular care should be taken in manually curating the alignment between the target protein and the template structure prior computation of the comparative model. This is facilitated by the DeepView program (see paragraph 3.1.4 Target-Template alignment - point 2.). The target-template alignment is saved as DeepView “project file” and submitted for computation to the “Project Mode” of the server. The DeepView program also enables calculation of models using structures which are not part of the SMTL library (see Note 12).
Modeling of oligomeric proteins, i.e. a group of two or more associated polypeptide chains, is possible using DeepView and the “Project Mode” of the server. The prerequisite is to determine the correct quaternary structure of the template protein - which is typically not identical with the coordinates representing the asymmetric unit of a PDB entry. Prediction of the most likely biological assembly for a particular protein can be retrieved from the PISA database (89). A DeepView project file with the sequences of the homo-multimeric or hetero-multimeric protein target sequences and template structure is then created (for details please see Note 13) and submitted to the server to obtain a model for the oligomeric complex.
After the computation of the structure for the macromolecule of interest is completed, the results are stored in a summary page of the workspace (Fig. 3) and users are notified by email.
Here we model the structure of PopA based on the structure of the activated diguanylate cyclase PleD in complex with c-di-GMP (PDB ID 2wb4). Activation of the PleD protein occurs upon phosphorylation-induced dimerization (90). For this reason we model the structure of PopA based on the homodimer activated form of PleD. The most likely biological assembly of the template is downloaded from the PISA database (89). A DeepView project file of the target sequence aligned to the homodimeric template is created and the alignment carefully inspected. Particular attention is devoted in correctly aligning residues which constitute important functional sites, i.e. the catalytic A-site and the inhibitory I-site of the diguanylate cyclase (DGC or GGDEF) domain and the phosphor acceptor P-site in the receiver domain of both proteins (82, 91). Insertions and deletion in the target-template alignment are visually assessed in the context of the template PleD structure and also guided by the secondary structure element predictions of the target PopA sequence (see paragraph 3.1.2 Target sequence feature annotation). Finally, the “Project file” containing the target-template alignment and the structure of the template is submitted to the server to calculate the comparative model for PopA.
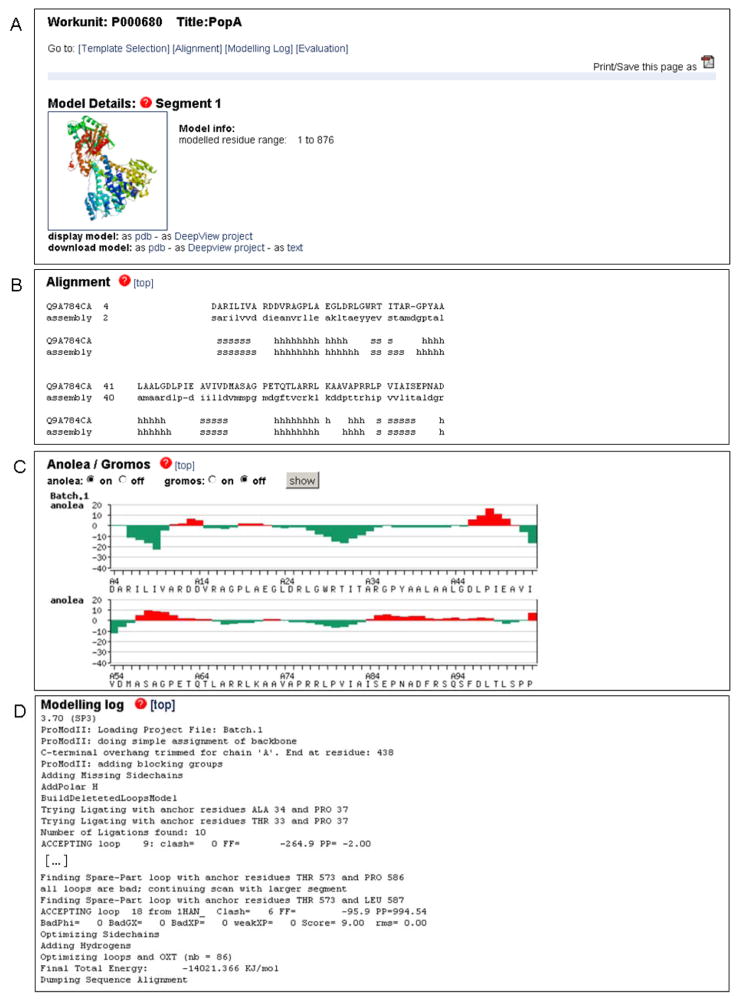
The Swiss-Model Workspace’s modeling results page is composed of different sections (Fig. 3). 1) In the “Model details” section the structure of the computed macromolecule is available for download as PDB file or DeepView “Project file” for further analysis. The model can be also displayed directly from the web site by clicking on the model image which will launch the molecular graphics program Astex Viewer (59). In the fully automated mode, additional details are provided, i.e. the template the model was based on (with a link to the corresponding PDB entry), the sequence identity and statistical significance of the target-template alignment (see Note 7). 2) The “Alignment” section contains the details of target-template alignment including secondary structure elements assignments. 3) Estimation of model quality based on Anolea (44) and Gromos (50) is available as residue based graphical plot, to indicate parts of the model with unfavorable interactions. 4) Technical modeling details are accessible in the “Modeling Log” section. 5) If the “Automated” mode is applied, an additional “Template Selection Log” is present in the results section, providing information about the template selection step performed to search the SWISS-MODEL Template library for suitable templates.
Fig. 3.
Typical representation SWISS-MODEL Workspace modeling results. In this example the C. crescentus PopA protein was modeled based on the structure of the paralog protein PleD (PDB ID 2wb4) using the Project Mode of the server. (A) The comparative model for PopA can be downloaded as PDB or DeepView project file. The model can be visualized directly on the web-page by clinking on the ribbon plot which will launch a java based visualization tool. In the Automated Mode, additional information about the template and the statistical significance of the target-template alignment would be shown in this section. (B) Details of the target-template alignment are provided together with the secondary structure elements assignments. (C) Anolea (44) and Gromos energy (50) plots provide residue based quality estimates of the model. Regions with positive energy values (red bars) indicate unfavorable interactions and regions of likely modeling errors. (D) Details about the modeling procedure are available at the end of the results. In the Automated Mode an additional section regarding the template selection step will be shown.
3.1.6 Model quality estimation
Finally the quality of the obtained model(s) can be assessed and estimated using the programs available in the “Structure assessment” tools section of the Workspace. A list of quality estimation algorithms and programs to verify the structural quality of proteins can be applied to the obtained models. We distinguish between programs to predict the local (per residue) and the global expected accuracy of the computed models (see paragraph 2.1.3 Programs accessible trough the server) and tools to verify the structure of the calculated models, e.g. structure geometries, packing quality, most probable side chain conformations, etc....
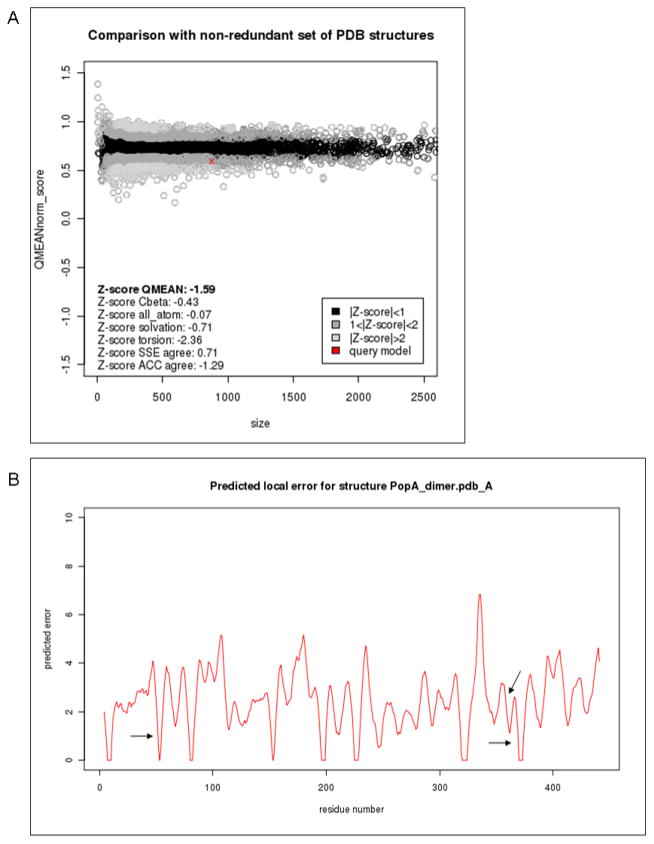
We analyze the quality of the homology model for PopA using QMEAN (41, 51) and Anolea (44) tools. The QMEAN scoring function estimates the local structural error at a given position in the protein. Regions in the model with low associated values are expected to be more reliably predicted. Anolea calculates pseudo energies based on potentials of mean force. Negative energy values indicate regions of the protein with favorable interatomics interactions. The sequence identity (~22%) between PopA and the template structure of PleD is close to the twilight zone of sequence alignments. For this reason is not surprising that the expected quality of some regions of the model is not high. However we verified that functional important sites of the protein, e.g. the P- A- and I- sites were better modeled than other loop regions of the protein (Fig. 4B).
The QMEAN Z- score is a quality estimate which relates structural features observed in a model to their expected distributions based on statistics for experimental protein structures of comparable size (54, 92). QMEAN Z- scores are normalized such that more positive values represent better model quality. Based on this measure, the quality of the obtained model for PopA of -1.59 lies within the expected range and is comparable to a medium resolution experimental structure (Fig. 4A).
We validate the predicted structure of PopA using the program Procheck (67). The analysis reveals a satisfactory quality of the model structure, e.g. in the Ramachandran plot (93) 91.1% of the PopA residues occupy the most favored regions, with only 7 residues in disallowed areas of the plot.
Finally regions of the comparative models, containing errors or of low quality can be further inspected and the corresponding segments in the target-template alignment adjusted to create a new model. The process (see Fig. 1) can be iterated until satisfactory results are obtained. This is facilitated by the use of the DeepView project files downloadable from the modeling results web site.
Fig. 4.
Examples of SWISS-MODEL Workspace model quality estimation plots calculated using QMEAN. (A) The global estimated energy of the PopA model (red cross) is compared to the QMEAN energy estimates (51, 92) for a non-redundant set of high-quality experimental protein crystal structures of similar length, and their deviation from the expected distributions is represented as Z-scores. The QMEAN quality estimate for PopA lies within the expected range for models of this type and is comparable to a medium resolution experimental structure. (B) Local (per-residue) plot of the QMEAN predicted errors for PopA. QMEAN scores for important functional sites (Phosphorilation- Activation- and Inhibitory- sites respectively) are depicted as arrows, indicating that the local environment of these regions is not located in problematic segments of the predicted structure.
3.2 PMP
To illustrate how to access functional and structural information for a given protein using the Protein Model Portal, we will use the example of the human Myeloid cell nuclear differentiation antigen protein (MNDA, UniProt accession code P41218). The MNDA protein is suggested to play a role in the granulocyte/monocyte cell-specific response to interferon (94–96).
3.2.1 Search options
PMP can be queried by submitting the entire amino acid sequence of a protein or a fragment of it. UniProt (80) proteins with identical or very similar sequences, will be identified and listed.
The portal can be also searched by database identifiers (e.g. UniProt, RefSeq (97), IPI (98), gi (99), Entrez (100)) or by keyword suggestions (e.g. “kinase”).
Models built based on a specific template structure can be also retrieved by entering either PDB accession codes (54) or structural genomics targets identifiers (101).
3.2.2 Results of the PMP query
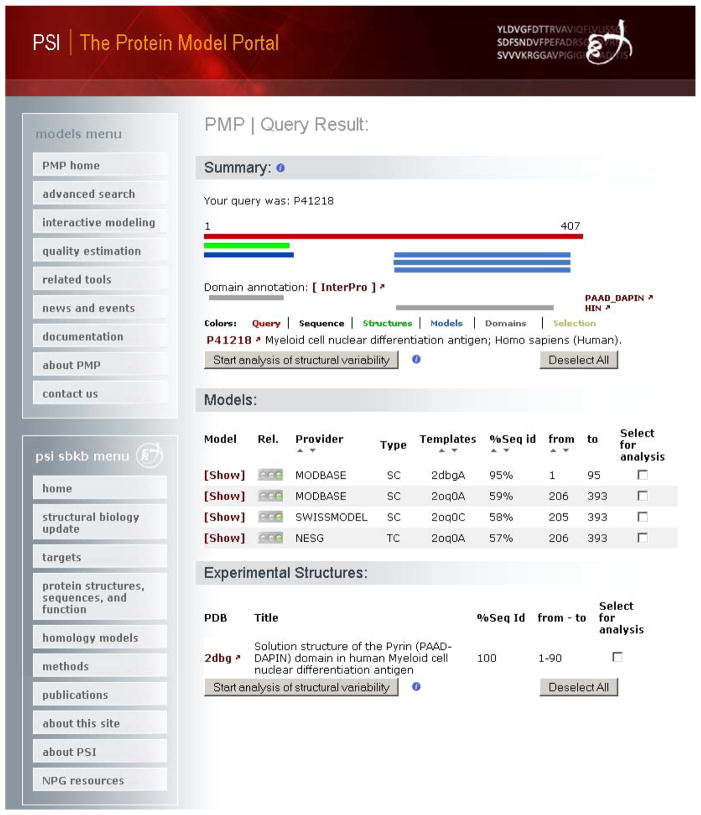
The results of the query are presented in a summary page (Fig. 5) with a graphical representation of the regions of the protein where structural information is available. Additionally functional annotation derived from UniProt and InterPro (81) (see Note 1) is provided. For the MNDA protein an experimental protein structure exists for the N-terminal Pyrin domain (PDB ID 2DBG (102)), a putative protein-protein interaction domain (103). Whereas for the C-terminal domain of unknown function, three protein structure models have been pre-computed by model resources accessible via PMP.
The graphical illustration of the matches is followed by a detailed list of the obtainable structural models for the protein of interest. Experimental protein structures in the PDB with more than 90% sequence identity to the target protein, are reported, if available.
Three models have been built for the MNDA protein by three resources accessible trough the portal: ModBase (55), Swiss-Model Repository (36), and NESG (104). Each single model is tagged with a color coded (“traffic lights”) as first indication about its reliability. In this example the models are based on a target-template alignment of about 60% sequence identity. Typically, models based on a target-template sequence alignment of this degree of similarity, are largely correct (7, 105, 106). Search results can be sorted based on different attributes, e.g. models provider, template identifier, target-template percentage of sequence identity and region of the target covered.
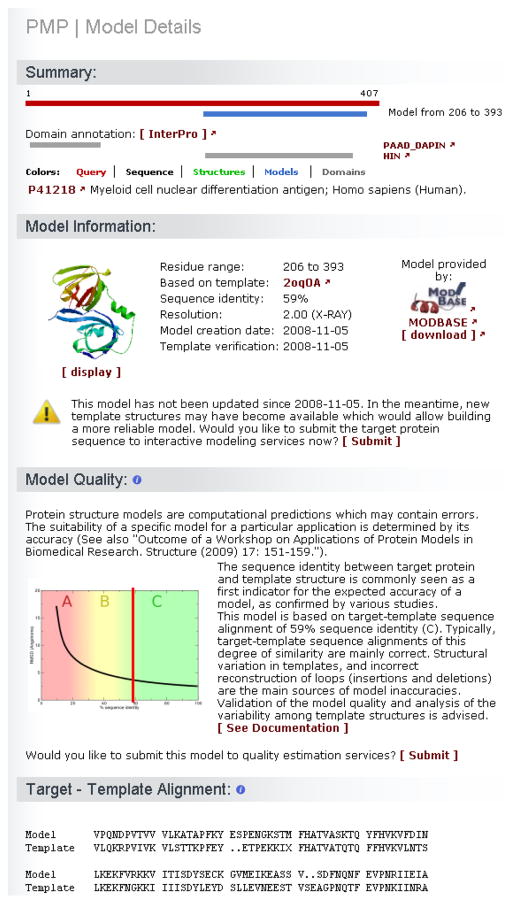
For each model the “Model Details” page provides further information (Fig. 6) about (1) the range of the modeled region, (2) the template used, (3) the target-template alignment the model was based on, (4) when the model was first created and verified, (5) the expected quality of the model, (6) a link to submit the model to quality estimation services and (7) the URL to the model database to download the model coordinates file. The protein structure models can be also visualized using the web browser applet Jmol (70).
In case the model has not been updated for a while a sign warns that new structures may have become available which would allow building a more reliable model. The target protein can be submitted directly to the interactive modeling services to compute models based on the most recent templates library (Fig. 6). In our example, some models have not been updated for a while and some regions exist for which structural information is not available, it is worthwhile triggering a new round of calculations. As of 11-11-2010 the results of interactive modeling show that there are no new templates that could be used instead of 2OQ0 (107) to model the C-terminal domain.
Fig. 5.
Protein Model Portal (PMP) query results for the human myeloid cell nuclear differentiation antigen protein (UniProt P41218 (94, 95), represented as a red bar). For the first 90 residues of this protein an experimentally solved structure (green bar) is deposited in the PDB database (PDB ID 2dbg (102)). The protein structure corresponds to the PPAD_DAPIN N-terminal domain of the protein. For the C-terminal HIN domain, three homology models are obtainable from the PMP model providers ModBase, SWISS-MODEL and NESG. Below the graphical representation a list of models and information about the structure is available. Additional information is accessible by clicking the corresponding model or PDB ID links. A subset of models or structures can be selected for further structural comparison.
Fig. 6.
PMP model details. For each model, target-template sequence identity, experimental annotation regarding the template, and cross-references to the model provider is available. A link allows users to automatically submit the protein sequence to interactive modeling servers for generating an updated prediction. The sequence alignment between the target and the template sequences is indicated, and a plot of the evolutionary distance between target and template gives an estimate about the expected accuracy of the model. Specialized model quality estimation tools can be automatically invoked for the model at hand to provide a more in depth assessment.
3.2.3 Protein model and structure comparison
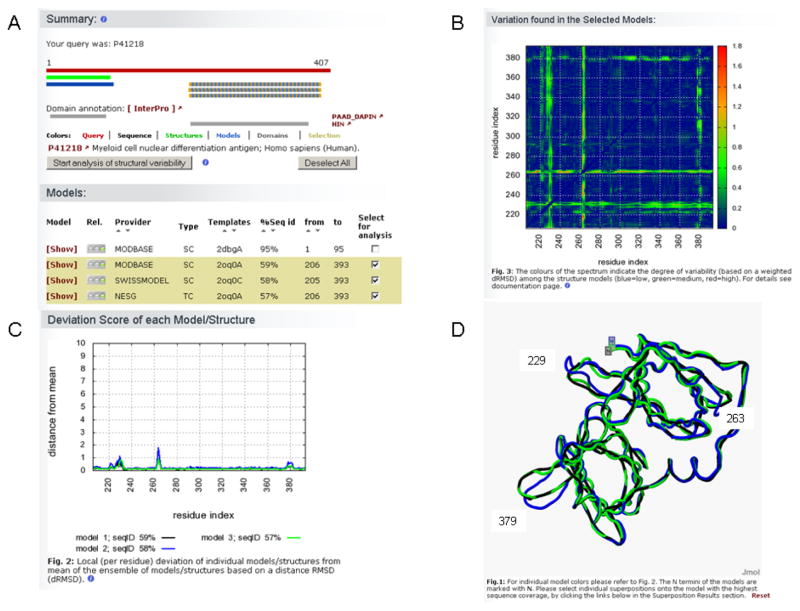
Models submitted by the different participating sites have been generated using various algorithmic approaches with different strengths and weaknesses. Also the quality of individual models highly depends on the evolutionary proximity to the selected structural templates. Finally, experimental structures may show structural variation due to domain motions, mobile loops, induced fit, etc. For these reasons, in the results page models and experimental structures spanning a common range can be selected to analyze their structural variability (Fig. 7A).
Fig. 7.
PMP structure comparison results. Structural differences can be analyzed in case several structures or models are available for the same region of a protein. (A) The comparative models available for the C-terminal domain of the myeloid cell nuclear differentiation antigen protein were compared. A subset of models or structures can be selected either by clicking the corresponding bars in the graphical synopsis or by checking the boxes of the lists. (B) A two-dimensional matrix indicates which regions of the analyzed structures deviate most among each others (blue=low, green=medium and red=high variability). For the comparative models of the antigen protein these regions are located around residues 230, 260 and 380. (C) The plot shows the magnitude of the deviation (residue based) of individual models (or structures) from the mean of the ensemble of the analyzed macromolecules. (D) The variability among models or structures can be visualized as structural superposition. In plots (C) and (D) each comparative model is represented by a different color (black=ModBase, blue=Swiss-Model, green=NESG models). As expected, regions of the models showing small differences around residues 230, 260 and 380 of the antigen protein are located in loops region on the surface of the protein, which were reconstructed differently by the various modeling methods.
Differences within the ensemble of models and experimental structures can be identified using a matrix that shows the deviations of Cα distances of the collection of models (Fig. 7B).
In particular for each model or structure, regions of the protein that deviate more from the ensemble are shown in a plot (Fig. 7C).
The details of the superposed structures can be also visualized in page using Jmol (70) (Fig. 7D).
Whereas for the N-terminal domain of MNDA an experimental structure has been solved, for the C-terminal domain three structural models are available. As mentioned before the accuracy for these models are expected to be high and since all resources used the same template, the structural variations among them is expected to be low (Fig. 7). Some minor deviations are in fact observed around residues 230, 260 and 280 corresponding to loops region of the protein (Fig. 7D) which have been modeled differently by the various modeling servers.
3.2.4 Interactive modeling
Model accuracy crucially depends on the availability of suitable template structures. Model repositories contain precompiled models based on the best available templates at the time of modeling. However, in the meantime better templates might have been released, which would allow for producing a higher quality model. Therefore, PMP provides a service interface (called “Interactive Modeling”) where to submit target protein sequences to several established modeling services (29, 47, 55, 56, 108) and initiate a new template selection and modeling process for the protein of interest. Depending on the type of resource, protein structure models coordinate files are either sent as attachment to an e-mail or can be retrieved via the corresponding service website.
For the region of MNDA spanning residues ~ 90 to 200, there is no pre-computed structural information available trough PMP, however when submitting the target sequence to the interactive modeling services, ModWeb server calculate a new model structure based on template 3na7 (109) spanning residues 62–157. The sequence identity of the alignment used to build the model is relatively low (27%) and the results should be taken with caution and further analyzed by quality estimation tools.
3.2.5 Quality estimation resources
Various model quality estimation tools have been developed by the community to analyze different structural features of protein models in order to judge the correctness of structural predictions.
The accuracy of a pre-computed model can be estimated using state of the art model quality estimation tools (43, 51, 58), directly from the “Model Details” page.
Alternatively, any coordinate file (PDB format; see Note 11) can be submitted to the “Quality estimation” interface of the portal.
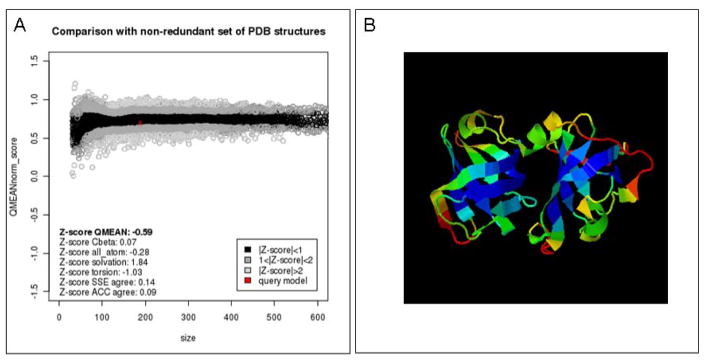
The three models generated for the C-terminal domains of the MNDA protein are estimated to be mainly correct with a medium to high quality scores especially for the beta barrels core parts of the structure (Fig. 8). On the contrary the model for the region spanning residues ~ 90 to 200 belongs to the low to bad quality range as expected for target-template sequence alignments below 30% sequence identity.
Fig. 8.
Model quality estimation. The quality of the model of the C-terminal domain of the myeloid cell nuclear differentiation antigen protein was analyzed using one of the tools accessible from the PMP portal, the QMEAN scoring function. (A) The global estimated energy of the antigen protein (red cross) is compared to the QMEAN energy estimates (51, 92) for a non-redundant set of high-quality experimental protein crystal structures of similar length, and their deviation from the expected distributions is represented as Z-scores. The QMEAN quality estimate for a C-terminal model (Fig. 6) lies within 0 to 1 standard deviations from the mean values, suggesting overall a very good expected quality for this model, comparable to experimental structures (B) The QMEAN method also allows predicting expected errors on a per residue basis. The model is colored according to the QMEAN score where blue regions represent regions predicted as reliable and red as potentially unreliable, respectively.
Acknowledgments
The authors would like to thank Konstantin Arnold for his dedicated support of the SWISS-MODEL service, Jürgen Haas for his commitment to new developments in PMP, and all members of the group for fruitful discussions.
Funding: The development and operation of SWISS-MODEL was supported by the SIB Swiss Institute of Bioinformatics; The Protein Model Portal of the Nature PSI Structural Biology Knowledgebase was supported by the National Institutes of Health NIH as a sub-grant with Rutgers University, under Prime Agreement Award Numbers: 3U54GM074958-04S2 and 1U01 GM093324-01.
Footnotes
InterPro is a collection of protein “signatures” used for the classification and automatic annotation of proteins. InterPro classifies sequences at super-family, family and subfamily levels and predicts the occurrence of functional domains, repeats and functional sites.
Intrinsically disordered regions in proteins have been associated with important biological functions involved for instance in cellular signaling and transcription regulation (110). Disordered regions often interfere with crystallization, and are therefore typically missing in experimental structures (unless in complex with other partners). Attempts to model intrinsically disordered regions using comparative techniques are therefore in most cases not such a good idea.
In case no evolutionary related template(s) for a given target protein can be found, it is not possible to reliably build a 3D structure model of this protein based on comparative/homology modeling techniques. De novo approaches (i.e. without using information from homologous templates) may be applied instead. However, it should be noted that despite advances in the field, de novo (or ab initio) techniques are restricted to relatively small proteins.
The “substitution matrix” is one of the important parameters of Blast/Profile Blast algorithms. The matrix allows evaluating and calculating the score of two aligned protein (or DNA) sequences. Different substitution matrixes have been specifically designed to change the scope and tune sequence database search. In particular, the choice of the substitution matrix influences the sensitivity vs. the selectivity of the search. The sensitivity of a query is defined as the ability of detecting remote homologs, but possibly including false matches. On the other side, selectivity ensures a more stringent search minimizing the number of false positives, at the cost of missing some true homologs. In particular, for the BLOSUM type of substitution matrices, a higher index (e.g. BLOSUM 80) indicates a more selective type of search, whereas a lower index (e.g. BLOSUM 45) will results in a more sensitive query. For more information please see the BLAST documentation at the NCBI server (111).
Profile Blast consist of two main steps, in the first one a profile is constructed from closely related sequences detected by a standard Blast search against a non-redundant protein sequence database. The profile is a representation of the group of aligned homologous sequences. This step can be iterated to extend the profile with new, more distantly related sequences. In the second step, the profile is used to perform a Blast search of the SMTL sequence library to look for related proteins with known structure. The parameters of both steps can be adjusted to shift the balance between selectivity and sensitivity of the search (see Note 4).
In HMM-HMM based alignment tools, both the query sequence and the sequences in the library are represented as HMM-based profiles. Therefore, the search is usually done against a culled version of the PDB database library, i.e. structures with similar sequences (e.g. 70% sequence identity) are clustered together.
In sequence database searches the E- (or expected) value associated with the results indicates the statistical significance of a given match (or hit). Each match is associated with a score (S), with higher scores indicating better results. The E-value estimates the probability of obtaining by chance a number of matches with this score (S) in a database of a particular size. In other words, the closer the E-value is towards 0, the more significant the alignment (between the query and the sequence found in the database) is. Similarly, the P- (or probability) value describes the probability that an alignment with this score (S) occurs by chance in a database of this size. The closer the P-value is towards 0, the better the alignment is.
In the best-case-scenario one would detect a statistical significant template covering the entire length of the protein of interest. Very often however, templates spanning only part of the query protein are detected. In this case, it is advisable to try to increase the sensitivity of the template detection methods, by additionally searching only those regions of the protein for which no templates were detected. Often, several non-continuous structural templates are detected which allow to model the target protein in separate fragments. Prediction of the relative orientation of isolated domains with comparative modeling methods is only feasible if a) one of the templates contains significant overlap with both domains, and b) their relative orientation is structurally well conserved.
The selection of the most suitable template should take into account not only the sequence similarity to the target protein, but also consider the quality of the experimental structure (e.g. resolution of the experimental technique), ligand molecules which may influence the local conformation of biding sites, or alternative conformations indicating structural variability observed within the protein family.
The development of sequence alignment algorithms is an active field of research in bioinformatics. For a (non-exhaustive) list of alignment tools employed in the filed of protein structure prediction please see (86).
A simple PDB-like file containing the coordinates of the template structure. For more information about PDB file format please refer to the corresponding documentation on the wwPDB website (112).
Please make sure when submitting a multiple sequence alignment that the names of the proteins specified in the alignment contain only alphanumerical characters. Use short names for the proteins (e.g. “Q9A784”, “PopA_CAUCR”, 2wb4) and verify that the alignment contains the sequence of the structure template. The selected template should be part of the SMTL library (see: “Template library” Tools section of the server.)
A step by step tutorial how to use DeepView for oligomeric protein modeling is provided on the SWISS-MODEL server web site (http://swissmodel.expasy.org/) and (113).
References
- 1.Schwede T, Sali A, Eswar N, Peitsch MC. Protein Structure Modeling. In: Schwede T, Peitsch MC, editors. Computational Structural Biology. World Scientific; Singapore: 2008. pp. 3–35. [Google Scholar]
- 2.Baker D, Sali A. Protein structure prediction and structural genomics. Science. 2001;294:93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- 3.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 4.Sutcliffe MJ, Haneef I, Carney D, Blundell TL. Knowledge based modelling of homologous proteins, Part I: Three-dimensional frameworks derived from the simultaneous superposition of multiple structures. Protein Eng. 1987;1:377–384. doi: 10.1093/protein/1.5.377. [DOI] [PubMed] [Google Scholar]
- 5.Peitsch MC. ProMod and Swiss-Model: Internet-based tools for automated comparative protein modelling. Biochem Soc Trans. 1996;24:274–279. doi: 10.1042/bst0240274. [DOI] [PubMed] [Google Scholar]
- 6.Fiser A. Template-based protein structure modeling. Methods Mol Biol. 673:73–94. doi: 10.1007/978-1-60761-842-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moult J. A decade of CASP: progress, bottlenecks and prognosis in protein structure prediction. Curr Opin Struct Biol. 2005;15:285–289. doi: 10.1016/j.sbi.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 8.Arinaminpathy Y, Khurana E, Engelman DM, Gerstein MB. Computational analysis of membrane proteins: the largest class of drug targets. Drug Discov Today. 2009;14:1130–1135. doi: 10.1016/j.drudis.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwede T, Sali A, Honig B, Levitt M, et al. Outcome of a workshop on applications of protein models in biomedical research. Structure. 2009;17:151–159. doi: 10.1016/j.str.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peitsch MC. About the use of protein models. Bioinformatics. 2002;18:934–938. doi: 10.1093/bioinformatics/18.7.934. [DOI] [PubMed] [Google Scholar]
- 11.Tramontano A. The biological applications of protein models. In: Schwede T, Peitsch MC, editors. Computational Structural Biology. World Scientific Publishing; 2008. pp. 111–127. [Google Scholar]
- 12.Junne T, Schwede T, Goder V, Spiess M. The plug domain of yeast Sec61p is important for efficient protein translocation, but is not essential for cell viability. Mol Biol Cell. 2006;17:4063–4068. doi: 10.1091/mbc.E06-03-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant MA. Protein structure prediction in structure-based ligand design and virtual screening. Comb Chem High Throughput Screen. 2009;12:940–960. doi: 10.2174/138620709789824718. [DOI] [PubMed] [Google Scholar]
- 14.Takeda-Shitaka M, Takaya D, Chiba C, Tanaka H, et al. Protein structure prediction in structure based drug design. Curr Med Chem. 2004;11:551–558. doi: 10.2174/0929867043455837. [DOI] [PubMed] [Google Scholar]
- 15.Das R, Baker D. Prospects for de novo phasing with de novo protein models. Acta Crystallogr D Biol Crystallogr. 2009;65:169–175. doi: 10.1107/S0907444908020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giorgetti A, Raimondo D, Miele AE, Tramontano A. Evaluating the usefulness of protein structure models for molecular replacement. Bioinformatics. 2005;21(Suppl 2):ii72–76. doi: 10.1093/bioinformatics/bti1112. [DOI] [PubMed] [Google Scholar]
- 17.Topf M, Baker ML, Marti-Renom MA, Chiu W, et al. Refinement of protein structures by iterative comparative modeling and CryoEM density fitting. J Mol Biol. 2006;357:1655–1668. doi: 10.1016/j.jmb.2006.01.062. [DOI] [PubMed] [Google Scholar]
- 18.Topf M, Sali A. Combining electron microscopy and comparative protein structure modeling. Curr Opin Struct Biol. 2005;15:578–585. doi: 10.1016/j.sbi.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Zhu J, Cheng L, Fang Q, Zhou ZH, et al. Building and refining protein models within cryo-electron microscopy density maps based on homology modeling and multiscale structure refinement. J Mol Biol. 397:835–851. doi: 10.1016/j.jmb.2010.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guex N, Peitsch MC, Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis. 2009;30(Suppl 1):S162–173. doi: 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- 21.Brazas MD, Yamada JT, Ouellette BF. Providing web servers and training in Bioinformatics: 2010 update on the Bioinformatics Links Directory. Nucleic Acids Res. 2010;38(Suppl):W3–6. doi: 10.1093/nar/gkq553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Battey JN, Kopp J, Bordoli L, Read RJ, et al. Automated server predictions in CASP7. Proteins. 2007;69:68–82. doi: 10.1002/prot.21761. [DOI] [PubMed] [Google Scholar]
- 23.Pieper U, Webb BM, Barkan DT, Schneidman-Duhovny D, et al. ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 2011;39:D465–474. doi: 10.1093/nar/gkq1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chivian D, Baker D. Homology modeling using parametric alignment ensemble generation with consensus and energy-based model selection. Nucleic Acids Res. 2006;34:e112. doi: 10.1093/nar/gkl480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hildebrand A, Remmert M, Biegert A, Soding J. Fast and accurate automatic structure prediction with HHpred. Proteins. 2009;77(Suppl 9):128–132. doi: 10.1002/prot.22499. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larsson P, Skwark MJ, Wallner B, Elofsson A. Improved predictions by Pcons.net using multiple templates. Bioinformatics. 27:426–427. doi: 10.1093/bioinformatics/btq664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 29.Fernandez-Fuentes N, Madrid-Aliste CJ, Rai BK, Fajardo JE, et al. M4T: a comparative protein structure modeling server. Nucleic Acids Res. 2007;35:W363–368. doi: 10.1093/nar/gkm341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneidman-Duhovny D, Hammel M, Sali A. Macromolecular docking restrained by a small angle X-ray scattering profile. J Struct Biol. doi: 10.1016/j.jsb.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vroling B, Sanders M, Baakman C, Borrmann A, et al. GPCRDB: information system for G protein-coupled receptors. Nucleic Acids Res. 39:D309–319. doi: 10.1093/nar/gkq1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Devries ME, Skolnick J. Structure modeling of all identified G protein-coupled receptors in the human genome. PLoS Comput Biol. 2006;2:e13. doi: 10.1371/journal.pcbi.0020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marcatili P, Rosi A, Tramontano A. PIGS: automatic prediction of antibody structures. Bioinformatics. 2008;24:1953–1954. doi: 10.1093/bioinformatics/btn341. [DOI] [PubMed] [Google Scholar]
- 34.Sivasubramanian A, Sircar A, Chaudhury S, Gray JJ. Toward high-resolution homology modeling of antibody Fv regions and application to antibody-antigen docking. Proteins. 2009;74:497–514. doi: 10.1002/prot.22309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwede T, Diemand A, Guex N, Peitsch MC. Protein structure computing in the genomic era. Res Microbiol. 2000;151:107–112. doi: 10.1016/s0923-2508(00)00121-2. [DOI] [PubMed] [Google Scholar]
- 36.Kiefer F, Arnold K, Kunzli M, Bordoli L, et al. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37:D387–392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pieper U, Webb BM, Barkan DT, Schneidman-Duhovny D, et al. ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koh IY, V, Eyrich A, Marti-Renom MA, Przybylski D, et al. EVA: Evaluation of protein structure prediction servers. Nucleic Acids Res. 2003;31:3311–3315. doi: 10.1093/nar/gkg619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chothia C, Lesk AM. The relation between the divergence of sequence and structure in proteins. Embo J. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng J, Xu J. Low-homology protein threading. Bioinformatics. 2010;26:i294–300. doi: 10.1093/bioinformatics/btq192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benkert P, Tosatto SC, Schwede T. Global and local model quality estimation at CASP8 using the scoring functions QMEAN and QMEANclust. Proteins. 2009;77(Suppl 9):173–180. doi: 10.1002/prot.22532. [DOI] [PubMed] [Google Scholar]
- 42.McGuffin LJ, Roche DB. Rapid model quality assessment for protein structure predictions using the comparison of multiple models without structural alignments. Bioinformatics. 2010;26:182–188. doi: 10.1093/bioinformatics/btp629. [DOI] [PubMed] [Google Scholar]
- 43.Eramian D, Eswar N, Shen MY, Sali A. How well can the accuracy of comparative protein structure models be predicted? Protein Sci. 2008;17:1881–1893. doi: 10.1110/ps.036061.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melo F, Feytmans E. Scoring Functions for Protein Structure Prediction. In: Schwede T, Peitsch MC, editors. Computational Structural Biology. World Scientific Publishing; 2008. [Google Scholar]
- 45.Zhou H, Zhou Y. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002;11:2714–2726. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 47.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y, Skolnick J. The protein structure prediction problem could be solved using the current PDB library. Proc Natl Acad Sci U S A. 2005;102:1029–1034. doi: 10.1073/pnas.0407152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peitsch MC. Protein modelling by E-Mail. BioTechnology. 1995;13:658–660. [Google Scholar]
- 50.van Gunsteren WF, Billeter SR, Eising AA, Hünenberger PH, et al. Biomolecular Simulations: The GROMOS96 Manual and User Guide. Zürich: VdF Hochschulverlag ETHZ; 1996. [Google Scholar]
- 51.Benkert P, Kunzli M, Schwede T. QMEAN server for protein model quality estimation. Nucleic Acids Res. 2009;37:W510–514. doi: 10.1093/nar/gkp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arnold K, Kiefer F, Kopp J, Battey JN, et al. The Protein Model Portal. J Struct Funct Genomics. 2009;10:1–8. doi: 10.1007/s10969-008-9048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berman HM, Westbrook JD, Gabanyi MJ, Tao W, et al. The protein structure initiative structural genomics knowledgebase. Nucleic Acids Res. 2009;37:D365–368. doi: 10.1093/nar/gkn790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pieper U, Webb BM, Barkan DT, Schneidman-Duhovny D, et al. ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 2011:D465–474. doi: 10.1093/nar/gkq1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ginalski K, Elofsson A, Fischer D, Rychlewski L. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics. 2003;19:1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- 58.McGuffin LJ. The ModFOLD server for the quality assessment of protein structural models. Bioinformatics. 2008;24:586–587. doi: 10.1093/bioinformatics/btn014. [DOI] [PubMed] [Google Scholar]
- 59.Hartshorn MJ. AstexViewer: a visualisation aid for structure-based drug design. J Comput Aided Mol Des. 2002;16:871–881. doi: 10.1023/a:1023813504011. [DOI] [PubMed] [Google Scholar]
- 60.Mulder N, Apweiler R. InterPro and InterProScan: tools for protein sequence classification and comparison. Methods Mol Biol. 2007;396:59–70. doi: 10.1007/978-1-59745-515-2_5. [DOI] [PubMed] [Google Scholar]
- 61.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 62.Jones DT, Ward JJ. Prediction of disordered regions in proteins from position specific score matrices. Proteins. 2003;53(Suppl 6):573–578. doi: 10.1002/prot.10528. [DOI] [PubMed] [Google Scholar]
- 63.Jones DT. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics. 2007;23:538–544. doi: 10.1093/bioinformatics/btl677. [DOI] [PubMed] [Google Scholar]
- 64.Altschul SF, Madden TL, Schaffer AA, Zhang J, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 66.Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 67.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 68.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 69.Hutchinson EG, Thornton JM. PROMOTIF--a program to identify and analyze structural motifs in proteins. Protein Sci. 1996;5:212–220. doi: 10.1002/pro.5560050204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org/
- 71.Stroud RM, Choe S, Holton J, Kaback HR, et al. 2007 annual progress report synopsis of the Center for Structures of Membrane Proteins. J Struct Funct Genomics. 2009;10:193–208. doi: 10.1007/s10969-008-9058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Elsliger MA, Deacon AM, Godzik A, Lesley SA, et al. The JCSG high-throughput structural biology pipeline. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:1137–1142. doi: 10.1107/S1744309110038212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Watson JD, Sanderson S, Ezersky A, Savchenko A, et al. Towards fully automated structure-based function prediction in structural genomics: a case study. J Mol Biol. 2007;367:1511–1522. doi: 10.1016/j.jmb.2007.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xiao R, Anderson S, Aramini J, Belote R, et al. The high-throughput protein sample production platform of the Northeast Structural Genomics Consortium. J Struct Biol. 2010;172:21–33. doi: 10.1016/j.jsb.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bonanno JB, Almo SC, Bresnick A, Chance MR, et al. New York-Structural GenomiX Research Consortium (NYSGXRC): a large scale center for the protein structure initiative. J Struct Funct Genomics. 2005;6:225–232. doi: 10.1007/s10969-005-6827-0. [DOI] [PubMed] [Google Scholar]
- 76.http://jcmm.burnham.org/.
- 77.Nierman WC, Feldblyum TV, Laub MT, Paulsen IT, et al. Complete genome sequence of Caulobacter crescentus. Proc Natl Acad Sci U S A. 2001;98:4136–4141. doi: 10.1073/pnas.061029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aldridge P, Paul R, Goymer P, Rainey P, et al. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol Microbiol. 2003;47:1695–1708. doi: 10.1046/j.1365-2958.2003.03401.x. [DOI] [PubMed] [Google Scholar]
- 79.Jenal U, Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- 80.Wu CH, Apweiler R, Bairoch A, Natale DA, et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hunter S, Apweiler R, Attwood TK, Bairoch A, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chan C, Paul R, Samoray D, Amiot NC, et al. Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci U S A. 2004;101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wassmann P, Chan C, Paul R, Beck A, et al. Structure of BeF3- -modified response regulator PleD: implications for diguanylate cyclase activation, catalysis, and feedback inhibition. Structure. 2007;15:915–927. doi: 10.1016/j.str.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 84.De N, Pirruccello M, Krasteva PV, Bae N, et al. Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS Biol. 2008;6:e67. doi: 10.1371/journal.pbio.0060067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sigrist CJ, Cerutti L, de Castro E, Langendijk-Genevaux PS, et al. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010;38:D161–166. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dunbrack RL., Jr Sequence comparison and protein structure prediction. Curr Opin Struct Biol. 2006;16:374–384. doi: 10.1016/j.sbi.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 87.Waterhouse AM, Procter JB, Martin DM, Clamp M, et al. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- 89.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 90.Paul R, Abel S, Wassmann P, Beck A, et al. Activation of the diguanylate cyclase PleD by phosphorylation-mediated dimerization. J Biol Chem. 2007;282:29170–29177. doi: 10.1074/jbc.M704702200. [DOI] [PubMed] [Google Scholar]
- 91.Paul R, Abel S, Wassmann P, Beck A, et al. Activation of the diguanylate cyclase PleD by phosphorylation-mediated dimerization. J Biol Chem. 2007;282:29170–29177. doi: 10.1074/jbc.M704702200. [DOI] [PubMed] [Google Scholar]
- 92.Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ramachandran GN, Ramakrishnan C, Sasisekharan V. Stereochemistry of polypeptide chain configurations. J Mol Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 94.Briggs R, Dworkin L, Briggs J, Dessypris E, et al. Interferon alpha selectively affects expression of the human myeloid cell nuclear differentiation antigen in late stage cells in the monocytic but not the granulocytic lineage. J Cell Biochem. 1994;54:198–206. doi: 10.1002/jcb.240540208. [DOI] [PubMed] [Google Scholar]
- 95.Briggs RC, Briggs JA, Ozer J, Sealy L, et al. The human myeloid cell nuclear differentiation antigen gene is one of at least two related interferon-inducible genes located on chromosome 1q that are expressed specifically in hematopoietic cells. Blood. 1994;83:2153–2162. [PubMed] [Google Scholar]
- 96.Dawson MJ, Trapani JA, Briggs RC, Nicholl JK, et al. The closely linked genes encoding the myeloid nuclear differentiation antigen (MNDA) and IFI16 exhibit contrasting haemopoietic expression. Immunogenetics. 1995;41:40–43. doi: 10.1007/BF00188431. [DOI] [PubMed] [Google Scholar]
- 97.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, et al. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 99.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, et al. GenBank. Nucleic Acids Res. 2011;39:D32–37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Baxevanis AD. Searching NCBI databases using Entrez. Curr Protoc Bioinformatics Chapter. 2008;1(Unit 1):3. doi: 10.1002/0471250953.bi0103s24. [DOI] [PubMed] [Google Scholar]
- 101.Chen L, Oughtred R, Berman HM, Westbrook J. TargetDB: a target registration database for structural genomics projects. Bioinformatics. 2004;20:2860–2862. doi: 10.1093/bioinformatics/bth300. [DOI] [PubMed] [Google Scholar]
- 102.Saito K, Inoue M, Koshiba S, Kigawa T, et al. 2006 doi: 10.2210/pdb2dbg/pdb. [DOI] [Google Scholar]
- 103.Fairbrother WJ, Gordon NC, Humke EW, O’Rourke KM, et al. The PYRIN domain: a member of the death domain-fold superfamily. Protein Sci. 2001;10:1911–1918. doi: 10.1110/ps.13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.http://www.nesg.org/.
- 105.Koh IY, V, Eyrich A, Marti-Renom MA, Przybylski D, et al. EVA: Evaluation of protein structure prediction servers. Nucleic Acids Res. 2003;31:3311–3315. doi: 10.1093/nar/gkg619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kopp J, Bordoli L, Battey JND, Kiefer F, et al. Assessment of CASP7 Predictions for Template-Based Modeling Targets. Proteins: Structure, Function, and Bioinformatics. 2007;69:38–56. doi: 10.1002/prot.21753. [DOI] [PubMed] [Google Scholar]
- 107.Liao JCC, Lam R, Ravichandran M, Ma J, et al. 2007 doi: 10.2210/pdb2oq0/pdb. [DOI] [Google Scholar]
- 108.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Caly DL, O’Toole PW, Moore SA. The 2.2-Å structure of the HP0958 protein from Helicobacter pylori reveals a kinked anti-parallel coiled-coil hairpin domain and a highly conserved ZN-ribbon domain. J Mol Biol. 2010;403:405–419. doi: 10.1016/j.jmb.2010.08.051. [DOI] [PubMed] [Google Scholar]
- 110.Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, et al. Intrinsic disorder and functional proteomics. Biophys J. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.http://blast.ncbi.nlm.nih.gov/
- 112.http://www.wwpdb.org/docs.html.
- 113.Bordoli L, Kiefer F, Arnold K, Benkert P, et al. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc. 2009;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]