Abstract
The acute phase protein orosomucoid-1 (Orm1) is mainly expressed by hepatocytes (HPCs) under stress conditions. However, its specific function is not fully understood. Here, we report a role of Orm1 as an executer of HPC proliferation. Increases in serum levels of Orm1 were observed in patients after surgical resection for liver cancer and in mice undergone partial hepatectomy (PH). Transcriptome study showed that Orm1 became the most abundant in HPCs isolated from regenerating mouse liver tissues after PH. Both in vitro and in vivo siRNA-induced knockdown of Orm1 suppressed proliferation of mouse regenerating HPCs and human hepatic cells. Microarray analysis in regenerating mouse livers revealed that the signaling pathways controlling chromatin replication, especially the minichromosome maintenance protein complex genes were uniformly down-regulated following Orm1 knockdown. These data suggest that Orm1 is induced in response to hepatic injury and executes liver regeneration by activating cell cycle progression in HPCs.
Keywords: Orosomucoid-1, Hepatocyte proliferation, Lipocalin, Cell cycle, Bioinformatics
Highlights
-
•
Serum Orm1 levels increased approximately 1.3- to 2.5-folds in both humans and mice after partial hepatectomy.
-
•
Transcriptome analysis revealed that Orm1 mostly induced in hepatocytes as a regulator of mouse liver regeneration.
-
•
Orm1 knockdown in mice impaired liver regeneration with poor hepatocyte growth and suppressed cell cycle signaling.
Orosomucoid-1 (Orm1) is an acute phase protein mainly expressed by hepatocytes under stress conditions. Beginning from the finding that Orm1 was induced after partial hepatectomy in humans and mice, we showed enrichment of Orm1 in regenerating hepatocytes of hepatectomized mice by transcriptome analysis and following culture and animal experiments. Knockdown of Orm1 in mice resulted in decreases in hepatocyte growth accompanying suppressed signaling in controlling chromatin replication. Therefore, Orm1 would be a potential therapeutic and prognostic biomarker for liver diseases, especially after surgical resection of cancer-bearing liver, through its newly found ability to stimulate the cell cycle in regenerating hepatocytes.
1. Introduction
Orosomucoid-1 (Orm1), also known as alpha-1-acid glycoprotein 1 (AGP1), is a member of lipocalin protein family, which acts as the carrier of basic and neutrally charged lipophilic compounds under normal physiological conditions. In addition, it is known as an acute phase protein, which is expressed in response to stressful conditions such as tissue injury, inflammation, or infection (Lee et al., 2010). There are emerging evidences from proteomic studies that both the urinary and serum Orm1 may serve as predictors of therapeutic response and diagnostic and prognostic biomarkers for inflammatory diseases such as chronic heart failure (Agra et al., 2017) and cancers such as bladder cancer (Li et al., 2016), lung squamous cell carcinoma (Ayyub et al., 2016), breast cancer (Alexander et al., 2004) as well as hepatocellular carcinoma (HCC) (Falleti et al., 1993). However, the specific function of Orm1 under stresses has not yet been fully elucidated.
Liver regeneration is a complicated but coordinated multistep process mediated by integration of multiple signals involved in cytokines, growth factors and metabolic networks (Michalopoulos, 1990). Normal liver regeneration requires spatially and temporally precise interactions between different populations of liver-composing cells, including liver sinusoidal endothelial cells (LSECs) and hepatocytes (HPCs), to reconstitute liver structure and function (Ding et al., 2014, Hu et al., 2014). However, its executers have not been fully identified. Loss of liver function caused by viral hepatitis, cirrhosis, and liver damage from alcohol or drugs is a life-threatening condition. Increased understanding of the regenerative process should also shed light on clinical applications such as the treatment of acute liver failure (ALF), which is a highly lethal disorder with abrupt loss of hepatic metabolic and immunological function (Bernal et al., 2010, Mao et al., 2014). It would also have clinical implication for the development of predictive biomarkers for the prognosis of portal vein embolization (PVE) before surgical resection for liver cancer. PVE is a technique used to increase future remnant liver volume. However, a few patients fail to achieve sufficient growth of the liver tissue or suffer from tumor progression following PVE (de Graaf et al., 2009, Treska et al., 2011).
Here, we addressed a role of Orm1 as an executer promoting cell cycle of HPCs during liver regeneration. We observed increases in serum levels of Orm1 in patients after surgical resection for HCC and in mice undergone partial hepatectomy (PH), implying that Orm1 might be induced in order to promote liver regeneration. Using Cap analysis of gene expression (CAGE)-based transcriptome analysis (Carninci et al., 2005), we showed that lipocalin family genes were mostly enriched in mouse primary HPCs during liver regeneration and among these genes Orm1 was characterized as the most major biomarker of liver regeneration by Bayesian network analysis. Consistently, knockdown of Orm1 in mice resulted in decreases in HPC growth accompanying suppressed signaling in controlling chromatin replication. These results highlight that the HPC-derived lipocalin protein, Orm1 has potential to be a prognostic biomarker and potential therapeutic target for impaired regeneration in liver.
2. Materials and Methods
2.1. Clinical Samples
The study was performed in accordance with the Declaration of Helsinki and was approved by the Ethics Committee for Biomedical Research of the Jikei University School of Medicine, the Hospital Ethics Committee of Tokyo University, and the RIKEN Institute Research Ethics Committee. The patients had signed a written informed consent prior to study. Serum samples were collected from 10 patients who had undergone liver resection for HCC. Formalin-fixed paraffin-embedded liver tumor tissues and normal adjacent tissues obtained from HCC patients were purchased from Proteogenex (Culver City, CA, USA).
2.2. Experimental Animals
All experiments were performed in accordance with protocols approved by the RIKEN Institutional Animal Use and the Care Administrative Advisory Committees and adhered to the guidelines in the Institutional Regulation for Animal Experiments and Fundamental Guidelines for Proper Conduct of Animal Experiment and Related Activities in Academic Research Institutions under the jurisdiction of the Ministry of Education, Culture, Sports, Science and Technology, Japan. Male C57BL6/J mice (age, 6–14 weeks) were housed under constant temperature (22 °C ± 1 °C) with free access to food and water.
2.3. PH Experiment
The mice were randomly divided into PH or control sham groups with no blinding. For transcriptome analysis, three independent biological replicates at each time point were analyzed. For in vivo RNA interference experiment, four independent biological replicates at each time point were analyzed. After anesthetized with isoflurane gas and disinfection of the skin with 70% ethanol, the abdomen is incised in the median line (15–20 mm) and opened with clip forceps. The beginning of the median and left lateral lobes is ligated with 5-0 silk thread, and the lobes are hepatectomized. After spraying antibiotics in the cavity, the abdominal wall is sutured with 6-0 nylon thread. The animals are kept warm while recovering from anesthesia (Akita et al., 2002).
2.4. Primary Cell Isolation and RNA Extraction
Primary mouse LSECs and HPCs were isolated from mouse livers at 2 h, 30 h, 48 h and 1 week after PH and at 2 h after the sham operation. The liver was perfused with collagenase solution and HPCs were collected by centrifugation at 50 × g for 2 min at 4 °C for 3 times. The pelleted HPCs were then cultured in William's Medium E (Sigma Chemical Company, St. Louis, MO, USA). LSECs were isolated using purified anti-mouse CD146 (RRID: AB_1731991, ME-9F1, BioLegend, San Diego, CA, USA) and dynabeads labelled with M-450 sheep anti-rat IgG (Life Technologies, Gaithersburg, MD, USA) (Akita et al., 2002). The isolated LSECs were cultured in DMEM/F-12 medium (Gibco, Invitrogen, Grand Island, NY, USA) supplemented with 10% FBS, 1% penicillin/streptomycin and 50 mg/mL endothelial mitogen. Total RNA samples were then extracted from the cells using an RNeasy Kit (Qiagen, Valencia, CA, USA), and the amount and purity of the isolated RNA was evaluated using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
2.5. In Vivo RNA Interference
A Stealth RNAi™ Pre-Designed siRNA targeting mouse Orm1 (siOrm1), UUGAGACUCCCGAAGCUCUAUUGUG, and a paired control siRNA (siCtl), CACAAUAGAGCUUCGGGAGUCUCAA, were purchased from Life Technologies (Gaithersburg, MD, USA). Transfection was performed via a single tail vein injection of siRNA (1 mg/kg) using a next generation lipid based carrier Invivofectamine 3.0 reagent (Life Technologies) (Eguchi et al., 2016). Knockdown efficiency was verified 3 days post-siRNA injection in serum protein levels and in transcript levels in whole liver tissues.
2.6. CAGE Transcriptome Analysis
A simplified version of the CAGE protocol, deepCAGE, using a single-molecule sequencer HeliScope, which can avoid linker ligation, PCR, and enzymatic cleavage, was applied to generate transcriptional profiling of LSECs and HPCs during liver regeneration (Kanamori-Katayama et al., 2011). Detailed methods for the CAGE data generation and normalization are provided in the FANTOM5 main paper (Forrest et al., 2014). Data downloads and freely available genomic tools are summarized in the FANTOM database (RRID: SCR_000788, http://fantom.gsc.riken.jp/5/) (Severin et al., 2014) and are available in Table S1.
2.7. IHC Staining and Immunofluorescence Staining of Orm1 in Liver Sections
Tissue staining was performed as previously described (Hara et al., 2014). Liver sections were deparaffinized and heated to 98 °C in Target Retrieval Solution (DAKO Corporation, Carpinteria, CA, USA) in a microwave for 10 min for antigen retrieval. For histology, the sections were stained with Myer's hematoxylin solution and 1% Eosin Y solution (H&E) (Muto Pure Chemicals, Tokyo, Japan). For mouse liver sections, after blocking with 5% normal goat serum in PBS containing 0.1% Tween-20 (PBST) for 30 min at room temperature, the sections were incubated with rabbit anti-mouse Orm1 (2 μg/mL; PAA816Mu01; Cloud-Clone Corp) or control rabbit IgG overnight at 4 °C. For human liver sections, the sections were blocked in PBS containing 10% FBS for 1 h at room temperature and incubated with goat anti-human Orm1 (RRID: AB_2158195, 0.5 μg/mL; sc-51018; Santa Cruz Biotechnology; Santa Cruz, CA, USA) and goat IgG overnight at 4 °C. For blocking/competition, Orm1 antibody was treated with a five-fold (by weight) excess of blocking peptide (sc-51018 P; Santa Cruz Biotechnology) in 500 μL PBS overnight at 4 °C. EnVision + System-HRP (DAKO Corporation) was used as the second antibody. The sections were counterstained with hematoxylin and quantified by ImageJ software 1.6.0 (National Institutes of Health, Bethesda, MD, USA) and the IHC profiler plugin (Varghese et al., 2014). For double immunofluorescence staining of Orm1 and Ki-67 in human liver sections, the sections were blocked in Mouse on Mouse (M.O.M.™) Blocking Reagent (MKB-2213; Vector Laboratories; Burlingame, CA, USA) for 1 h and in PBS containing 10% FBS for another 1 h at room temperature. The sections were then incubated with goat anti-human Orm1 antibody (0.5 μg/mL; sc-51018; Santa Cruz Biotechnology) and goat IgG overnight at 4 °C. On the next day, the sections were stained with donkey anti-goat IgG alexa555 second antibody (RRID: AB_141788, A-21432; Invitrogen). After wash, the sections were incubated with mouse anti-human Ki-67 antibody (RRID: AB_393778, 1.25 μg/mL; 550609; BD Biosciences; San Jose, CA, USA) and mouse IgG overnight at 4 °C. The sections were then stained with donkey anti-mouse IgG alexa488 second antibody (RRID: AB_141607, A-21202; Invitrogen) and the nuclei were visualized by DAPI (Wako Industries, Osaka, Japan). Immunofluorescence staining signals were detected with a Zeiss LSM 700 laser scanning confocal microscope (Carl Zeiss Inc., Jena, Germany).
2.8. Immunofluorescence Staining of Orm1 in Primary Mouse HPCs
Primary mouse HPCs were isolated as described above and seeded in Greiner 96-well microtiter plates (Greiner Bio-One, Monroe, NC, USA) at a concentration of 1.5 × 105 cells/mL. The cells were treated with PDGF-BB (PeproTech, Rocky Hill, NJ, USA, #100-14B, 20 ng/mL) in DMEM containing 2% FBS for 48 h. Then, the cells were fixed in 4% paraformaldehyde (PFA) for 10 min and incubated with 0.1% Triton X-100 in PBS for 5 min at room temperature. After blocking with 5% FBS-PBS for 1 h at room temperature, cells were incubated with rabbit anti-mouse Orm1 antibody (1 μg/mL; PAA816Mu01; Cloud-Clone Corp) or control rabbit IgG overnight at 4 °C. The cells were then washed and stained with donkey anti-rabbit Cy5-conjugated secondary antibody (1:500, Invitrogen), while nuclei was visualized by Hoechst staining (Wako Industries). Immunofluorescence staining signals were detected with a Zeiss LSM 700 laser scanning confocal microscope (Carl Zeiss Inc.) or an ImageXpressMICRO High Content Screening System (Molecular Devices, Sunnyvale, CA, USA) and morphological analysis was performed using MetaXpress Image Analysis software (Molecular Devices).
2.9. Cell Culture
The human liver cancer cell lines FLC4 (RRID:CVCL_D204), FLC7 (RRID: CVCL_2805), and HepG2 (RRID: CVCL_0027), and the immortalized human liver endothelial cell line M1 were kindly supplied by Prof. Tomokazu Matsuura (The Jikei University School of Medicine, Tokyo, Japan) (Fujise et al., 1990, Matsuura et al., 1998). Human hepatic stellate cells (LX2; RRID: CVCL_5792) were kindly provided by Prof. Scott Friedman (Mount Sinai Hospital, New York) (Xu et al., 2005). All the cell lines have been tested as negative for mycoplasma contamination. The cells were maintained in DMEM (Wako Industries) containing 10% FBS, 100 U/mL penicillin/streptomycin and 2 mM l-glutamine and grown at 37 °C in a humidified 5% CO2 incubator.
2.10. Enzyme-Linked Immunosorbent Assay (ELISA)
Serum Orm1 concentrations were measured using ELISA kits (SEA816HU and SEA816MU for human and mouse serum samples, respectively; Cloud-Clone Corp., Houston, TX, USA). Serum samples were diluted appropriately in PBS and assayed and determined in a plate reader (ARVO MX, Perkin Elmer Inc., Waltham, MA, USA) at 450 nm according to the manufacturer's instructions.
2.11. Real-Time RT-PCR
PCR reactions were performed using the Roche LightCycler® 96 Real-Time PCR System (Roche Diagnostic Co., Ltd., Tokyo, Japan) with SsoAdvanced™ SYBR® Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA). Detailed methods for PCR analyses were previously described (Qin et al., 2013). The sequences of the primers are summarized in Table S2.
2.12. In Vitro RNA Interference
A siRNA targeting human Orm1 (sc-60133) (siOrm1) and a control siRNA (sc-37007) (siCtl) were purchased from Santa Cruz Biotechnology. FLC4 cells were plated in 96-well plates (1 × 104 cells/well) for cell viability analysis and 24-well plates (5 × 104 cells/well) for RNA isolation and transfected with 100 nM siRNAs using Lipofectamine 2000 (Life Technologies).
2.13. Cell Proliferation Assay
Cell viability was determined using the Cell Counting Kit-8 (Dojindo Molecular Technologies, Tokyo, Japan) in a plate reader (ARVO MX, Perkin Elmer Inc.) at 450 nm as previously described (Tatsukawa et al., 2011). For gain-of-function analysis, 24 h after siRNA transfection, FLC4 cells were treated with recombinant human Orm1 (CLPRO889, Cedarlane, Burlington, NC, USA) in DMEM containing 2% FBS for 24 h before cell viability determination.
2.14. Knowledge-Based Pathway Analysis
To explore the biological interpretation of the transcriptome data, the canonical pathway was identified using knowledge-based functional analysis software Ingenuity Pathways Analysis (IPA; RRID: SCR_008653, Ingenuity Systems, Mountain View, CA, USA) as previously described (Qin et al., 2012). Moreover, the causal analytics tool “upstream regulator analysis” was used to identify upstream regulatory molecules and associated mechanisms of the observed expression changes. Upstream regulator analysis is one of the causal analytics algorithms in IPA that was developed to identify the upstream molecules in the data set that can explain the observed expression changes. One of the statistical measures of the upstream regulator analysis is the activation z-score, which can be used to find likely regulating molecules based on a statistically significant pattern match of up- and down-regulation, and also to predict the activation state (either activated or inhibited) of a putative regulator (Kramer et al., 2014). An absolute z-score > 2 was considered as significant. In addition, functional annotation analysis was applied in SEA of GeneSpring GX13.0 (RRID: SCR_010972, Agilent Technologies, Palo Alto, CA, USA) or in the freely available tool DAVID (RRID: SCR_003033, http://david.abcc.ncifcrf.gov/) (Huang da et al., 2009).
2.15. Microarray Analysis
Total RNA was isolated from mouse liver tissues at 48 h after PH treated with siControl or siOrm as described above. After fragmentation of complementary RNA, microarray studies were performed by the RIKEN Research Resource Center using Affymetrix GeneChip Mouse Genome 430A 2.0 Array. The arrays were scanned using a GenePix4000B Microarray Scanner (Axon Instruments, Foster City, CA, USA). Data analysis was performed with GeneSpring GX13.0 (Agilent Technologies). Signal intensities for each probe were normalized to the 75th percentile without baseline transformation. Genes that were differentially expressed following Orm1 knockdown were identified by a fold change of > 2 and selected for pathway analysis. All data are MIAME compliant, and the raw data have been deposited in the Gene Expression Omnibus (RRID: SCR_007303, www.ncbi.nlm.nih.gov/geo, accession no. GSE83733).
2.16. Data Mining
Orm1 expression in human tissues was searched in the freely available FANTOM5 database SSTAR (Semantic catalog of Samples, Transcription Initiation and Regulator; http://fantom.gsc.riken.jp/5/sstar/). To understand the role of Orm1 in injured human liver regeneration, a clinical data set containing gene expression profiling of 10 normal livers and 4 HBV-associated ALF livers was downloaded from the Gene Expression Omnibus (accession no. GSE38941) (Nissim et al., 2012).
2.17. Multivariate Analyses
Unsupervised principle component analysis (PCA), partial least squares-discriminant analysis (PLS-DA) and orthogonal partial least squares-discriminant analysis (OPLS-DA) were run using the free-web software MetaboAnalyst 2.5 (RRID: SCR_015539, www.metaboanalyst.ca). PLS-DA is a partial least squares regression of a set Y of binary variables describing the categories of a categorical variable on a set X of predictor variables (Perez-Enciso and Tenenhaus, 2003). In this study, the class categories (cell types or time points) of each sample were set as the dependent categorical variable Y, and the expression level of genes was set as the independent predictor variables X. In the PLS-DA model, variable importance in the projection (VIP) (Wold et al., 2001) is a weighted sum of squares of the PLS loadings taking into account the amount of explained Y-variation in each dimension. The sum of squares of all VIP's is equal to 1, so the higher the VIP value, the more relevant for explaining Y and the larger contribution to the establishment of the PLS-DA model. Variables with VIP larger than 1 are the most relevant for explaining Y (Wold et al., 2001). VIP values > 1 were considered to be important in the present study. Correlations of gene expression between LSECs and HPCs were calculated according to the Pearson's correlation coefficients method in R Bioconductor (RRID: SCR_001905). A high correlation coefficient (| r | > 0.9) was collected for further network analysis. A multivariate statistical analysis technique Bayesian network analysis was applied to explore the regulatory networks of the causal complex using the web-based RX-Taogen software (http://extaogen.nies.go.jp/) (Yamanaka et al., 2004). The replicate exchange time was set as 20,000. A core concept of Bayesian modeling is to represent causal dependencies and express a conditional dependence structure among variables by computing the prior distribution over all variables (Friston and Kiebel, 2009). Therefore, elements that exert more effects should be located higher in the hierarchy of a calculated network model (He et al., 2012).
2.18. Statistical Analyses
Quantitative data are expressed as the means plus the standard deviation of three replicates from at least two independent experiments. The statistical significance of differences was assessed using Student's t-test, Fisher's exact test, Mann-Whitney U test or ANOVA with post-hoc Tukey HSD Calculator for multiple comparison. Values of P < 0.05 were considered to indicate statistical significance.
3. Results
3.1. Orm1 is Induced in Humans and Mice After PH
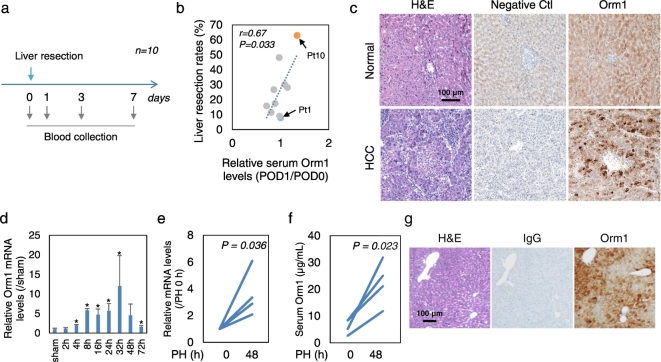
Postoperative serum Orm1 levels were examined in 10 patients who had undergone surgical resection for HCC (Fig. 1a). Changes in serum levels of Orm1 at 24 h post-liver resection were significantly positively correlated with the ratio of the resected liver volume to total liver volume (Fig. 1b). In a patient undergoing nearly 70% liver resection, serum Orm1 levels increased to 1.3-fold at 24 h post-liver resection and then declined to levels almost the same as that before liver resection at 3 and 7 days post-liver resection (Fig. S1a). In contrast, no increase in serum Orm1 levels was observed in a patient undergoing 8% liver resection (Fig. S1a). Immunohistochemistry (IHC) staining of Orm1 in human liver sections showed a strong Orm1 expression in HCC tissues than that in the normal adjacent tissues (Fig. 1c and Fig. S1b). This is in accordance with the elevated levels of serum Orm1 observed in HCC patients from previous reports (Falleti et al., 1993). Furthermore, double immunofluorescence staining revealed the existence of Orm1+ Ki67+ cells in HCC tissues (Fig. S1c). In addition, microarray data mining of a human ALF data set (GSE38941) revealed that Orm1 gene expression was significantly decreased in hepatitis B virus (HBV)-associated ALF livers compared to normal livers (Fig. S1d). These data suggested that Orm1 might be induced in order to promote liver regeneration.
Fig. 1.
Orm1 is induced in humans and mics after PH. (a)–(c) Human data. (a) Postoperative serum Orm1 levels were examined in 10 patients who had undergone liver resection for HCC. (b) Correlation between changes in serum Orm1 at 24 h post-liver resection with the rates of liver resection (the resection volume counts per total liver volume counts). POD, postoperative days. (c) Representative photographs of IHC staining of Orm1 in commercially obtained liver sections of HCC and normal adjacent tissues in HCC patients. As a negative control (Ctl), human Orm1 antibody that had been incubated with a 5-fold (by weight) excess of blocking peptide. (d)–(g) Mouse data. (d) Time course of Orm1 gene expression in whole-liver tissues after PH (n = 3). (e) Orm1 transcript levels in whole-liver tissues and (f) serum protein levels at 48 h after PH (n = 4). (g) IHC staining of Orm1 in mouse liver tissues at 48 h after PH. P-value was assessed using Student's t-test or Mann-Whitney U test. *P < 0.05 indicates statistical significance. Scale bars, 100 μm.
We further undertook a clinical investigation of the correlation between Orm1 and biomarkers of liver functions, including serum alanine transaminase (ALT), aspartate transaminase (AST) and bilirubin (Bil) levels and international normalized ratio of prothrombin time (PT.INR). Of interest, hierarchical cluster analysis based on the Pearson correlation coefficients between postoperative serum changes of Orm1 and liver function biomarkers divided the patients into two groups: the “Correlated” (n = 4) and “Non-correlated” groups (n = 6) (Fig. S2a and b). Moreover, significantly different postoperative serum levels of Orm1 were observed between these two groups, suggesting the existence of differential responses in Orm1 expression (Fig. S2c). No significant difference was observed in liver resection rates between these two groups (P = 0.42 in two-tailed Student's t-test), while risk-factor analysis demonstrated that HCC patients suffering from chronic hepatitis and/or cirrhosis (Fig. S2d), who probably carrying immunodeficiency, showed poor correlation between Orm1 and liver function biomarkers (Hochepied et al., 2003).
Our data imply that Orm1 might be induced in order to promote liver regeneration. To further explore this hypothesis, we observed Orm1 expression during hepatic regeneration in a mouse model of PH. The time course analysis demonstrated Orm1 gene expression in liver tissues was increased as early as 4 h after PH (Fig. 1d). Furthermore, individual paired increase in Orm1 transcript levels (Fig. 1e) in whole-liver tissues and serum protein levels (Fig. 1f) was observed before and 48 h after PH. IHC staining of Orm1 in liver tissues of partially hepatectomized mice showed a strong expression of Orm1 at 48 h after PH in HPCs positioned in the central vein and portal vein areas (Fig. 1g), suggesting a potent interaction between Orm1 induction and angiocrine signals.
3.2. Transcriptional Network Analysis Identifies Orm1 as a Regulator in Liver Regeneration
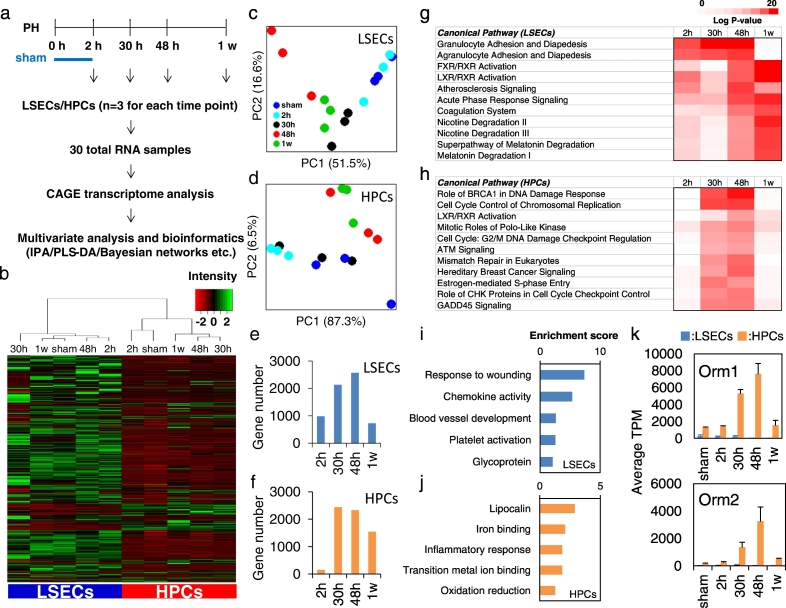
To further understand the role of Orm1 in the coordinated signal transduction controlling liver regeneration, transcriptional profiling of LSECs and HPCs isolated at 5 different time points during liver regeneration, at 2 h, 30 h, 48 h or 1 w after 70% PH and at 2 h after the sham operation, was measured using high-throughput CAGE technology (Fig. 2a and Table S1). Hierarchical clustering demonstrated diverse expression profiles between LSECs and HPCs (Fig. 2b). Cell-type specificity of several previously reported LSEC and HPC markers were confirmed at their transcription levels (Fig. S3a) and in orthogonal partial least squares-discriminant analysis (OPLS-DA) modeling (Fig. S3b) and upstream regulator analysis of the Ingenuity Pathways Analysis (IPA) program (Fig. S3c and Table S3). Principle component analysis (PCA) showed strong and reproducible time effects in both LSEC and HPC datasets (Fig. 2c and d). Compared with the sham control, the most significant differentially expressed genes were observed at 30 h and 48 h in LSECs (Fig. 2e) and HPCs (Fig. 2f). A large number of significantly and differentially expressed genes were observed as early as 2 h after PH in LSECs than in HPCs. This is in accord with the previous reports that LSECs contribute to liver regeneration in the early phases to stimulate HPC proliferation (Ding et al., 2010). Canonical pathways analysis of the IPA program revealed that enrichment in LSECs for genes encoding molecules regulating inflammation reaction pathways occurred as early as 2 h after PH (Fig. 2g and Table S4), while in HPCs, pathways related to cell cycle control were abundantly enriched at 30 h and 48 h after PH (Fig. 2h), suggesting a potential interaction between early injury response in LSECs and later cell cycle regulation in HPCs.
Fig. 2.
Transcriptional profiling of LSECs and HPCs during liver regeneration. (a) Schematic overview of transcriptome experimental procedures (n = 3). (b) Hierarchical clustering with Ward's method of 16,499 genes measured by CAGE technology in LSECs and HPCs. The mean average from three biological replicates at each time point was shown. Low expression tags were deleted and the tags that were expressed at least at one time point in either LSECs or HPCs were kept with the expression cutoff at 1 tags per million (TPM). PCA score plots of differentially expressed genes in the process of liver regeneration for (c) LSECs and (d) HPCs. Numbers of significantly differentially expressed genes compared with the sham control at each time point in (e) LSECs and (f) HPCs. Top canonical pathways associated with significantly differentially expressed genes during liver regeneration in (g) LSECs and (h) HPCs in IPA program. Functional annotations identified using DAVID software with top (i) LSEC and (j) HPC genes selected by evaluating the relative contribution of each gene to differential gene expression during the process of liver regeneration in PLS-DA modeling. (k) Expression of Orm1 and Orm2 in LSECs and HPCs during liver regeneration measured by CAGE analysis.
Accordingly, genes that regulate mouse liver regeneration in LSECs and HPCs were identified in the PLS-DA model according to variable importance in the projection (VIP) scores (Fig. S4a and b and Table S5). Of interest, further functional annotation analysis in DAVID bioinformatics resources uncovered that the most associated functional annotations in LSECs included wound healing functions such as response to wounding, blood vessel development, platelet activation, and glycoprotein (Fig. 2i), while those in HPCs were associated with lipocalin including Orm1, iron binding, and oxidation reduction (Fig. 2j). Then, Pearson's correlation coefficients were calculated between the regulatory genes in LSECs and HPCs (Fig. S4c) and Bayesian network analysis was performed on the highly correlated genes using the TAO-Gen algorithm. Notably, the node for Orm1 in HPCs was located at the top of the predicted network hierarchy and a high correlation was observed between Orm1 and Orm2, two Orm isoforms that cooperatively interact with each other (Han et al., 2010), implicating the role of ORM gene family especially Orm1 as an upstream regulator in liver regeneration (Fig. S5). The expression of Orm1 and Orm2 significantly increased in HPCs at 30 h and peaked at 48 h after PH, while low expression of Orm1 was observed in LSECs (Fig. 2k).
3.3. Regulatory Role of Orm1 on the Proliferation of Mouse Regenerating HPCs and Human Hepatic Cells
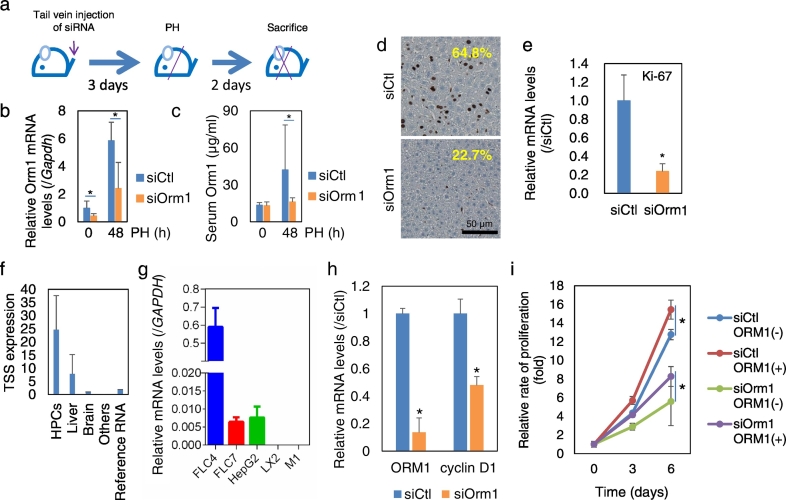
Next, the role of Orm1 on mouse liver regeneration was examined by siRNA-induced loss-of-function analysis (Fig. 3a). In vivo knockdown of Orm1 with its siRNA administered to mice via the tail vein significantly suppressed the Orm1 expression by 60% at 48 h after PH at both transcript levels in whole-liver tissue (Fig. 3b) and serum protein levels (Fig. 3c). In addition, inhibited expression of the proliferating marker Ki-67 levels was observed in regenerating livers at 48 h after PH, suggesting that Orm1 facilitates the proliferation of HPCs in regenerating livers (Fig. 3d and e).
Fig. 3.
Regulatory role of Orm1 on the proliferation of mouse regenerating HPCs and human hepatic cells. (a) Schematic overview of loss-of-function experimental procedures. Orm1 expression at transcript levels in (b) whole-liver tissue and (c) serum protein levels. (d) IHC staining and (e) gene expression of cell proliferation marker Ki-67 in whole-liver tissue at 48 h post-PH. The average percentage of Ki-67 positive cells presented in (d) was quantified from five randomly selected areas in three slides from each mouse (n = 2–4). Scale bar, 50 μm. siCtl, control siRNA; siOrm1, Orm1 siRNA. The quantitative data were presented as the mean plus standard error (n = 4). (f) Data mining of Orm1 expression in human tissues obtained from the freely available FANTOM5 database. Reference RNA, commercially obtained universal human reference total RNA. Others, other human hepatic non-parenchymal cells, primary cells, tissues and cancer cell lines. (g) The gene expression of Orm1 was examined in human liver cancer cell lines FLC4, FLC7, and HepG2, human hepatic stellate cell line LX2, and immortalized human liver endothelial cell line M1 using real-time RT-PCR. Gene expression was normalized to that of GAPDH. Effect of Orm1 knockdown on (h) gene expression of Orm1 and cyclin D1 and (i) cell proliferation of human hepatic cell line FLC4 in the in the absence [ORM1(−)] and presence [ORM1(+)] of 25 ng/mL recombinant human Orm1. The quantitative data were presented as the mean plus the standard deviation of three replicates. *P < 0.05 assessed using two-tailed Student's t-test, Mann-Whitney U test or ANOVA with post-hoc Tukey HSD Calculator for multiple comparison.
We further investigated whether Orm1 might play a regulatory role in the proliferation of human hepatic cells. Data mining of the functional annotation of the mammalian genome (FANTOM) database, consisted of CAGE expression profiling of 1000 human RNA samples from primary cell samples, human tissues and cancer cell lines (Arner et al., 2015), suggested that Orm1 was predominantly expressed by HPCs (Fig. 3f). Gene expression analysis of a panel of human liver cell lines using PCR also showed that Orm1 was strongly expressed by HPCs but did not express in hepatic stellate cell line LX2 and LSEC cell line M1 (Fig. 3g). In a human HCC functional liver cell-4 cell line (FLC4), which has a similar gene expression profile as human liver (Laurent et al., 2012), silencing Orm1 expression by siRNA inhibited cell proliferation, accompanying the lowered expression of cell cycle-associated gene cyclin D1 (Fig. 3h and i). In contrast, treatment with recombinant human Orm1 protein significantly promoted cell growth in both control and Orm1 siRNAs-transfected cells (Fig. 3i).
3.4. Molecular Targets of Orm1 in Regenerating Mouse Livers
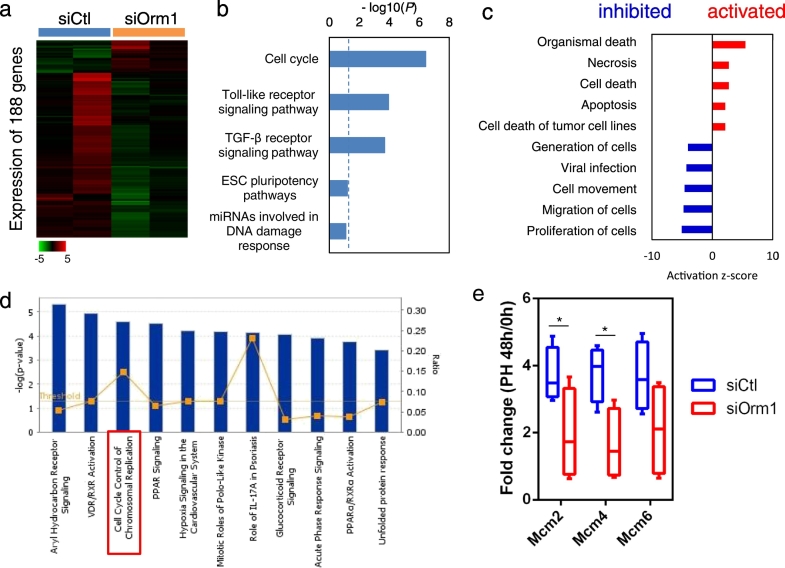
Finally, microarray analysis was performed to identify the molecular targets of Orm1 in regenerating mouse liver at 48 h after PH and a total of 188 differentially expressed genes were identified with a fold change > 2 (Fig. 4a and Table S6). Functional annotation analysis using Single Experiment Analysis (SEA) suggested that cell cycle signaling pathways were under the control by Orm1 in regenerating mouse livers (Fig. 4b). Diseases or Functions Annotation in IPA also suggested that “organismal death” and “necrosis” were activated, while “proliferation of cells” and “migration of cells” were inhibited in the regenerating livers treated with siOrm1 compared with the control livers (Fig. 4c). Notably, IPA pathway analysis indicated that the DNA helicase complex MCM genes including MCM2 and MCM4, were involved in the enriched signaling pathway of “cell cycle control of chromosomal replication” (Fig. 4d), which was also enriched in HPCs during liver regeneration (Fig. 2h). RT-PCR analysis verified that siRNA-mediated knockdown of Orm1 alone did not inhibit the expression of MCM genes in mouse livers but significantly suppressed their induction at 48 h after PH (Fig. 4e). Further pathway analysis revealed that cytokines such as interleukin-6 (IL-6) and tumor necrosis factor (TNF)-dependent signaling pathways were involved in the regulatory effect of Orm1 on MCM gene expression (Fig. S6). It was reported that Orm1 could specifically bind to the C-C chemokine receptor type 5 (CCR5) and regulated the production of inflammatory chemokines (Atemezem et al., 2001, Lei et al., 2016). Of interest, upstream regulator analysis in IPA suggested that the activation z-score of platelet-derived growth factor-BB (PDGF-BB) is − 2.5 (Fig. S7a), supporting the parallel change in PDGF-BB and Orm1 observed in mouse regenerating HPCs using CAGE analysis (Fig. S7b). In addition, an inductive effect of PDGF-BB on Orm1 expression was also observed in freshly isolated primary mouse HPCs (Fig. S7c). The mitogenic effect of PDGFs, mostly due to activation of cell cycle pathways, has been suggested in rat and mouse HPCs (Bowen et al., 2014, Limaye et al., 2008). Together, these data suggest that Orm1 plays important roles in liver regeneration, especially in controlling HPC's cell cycle.
Fig. 4.
Molecular targets of Orm1 in mouse regenerating livers. Microarray analysis (GSE83733) was performed in control siRNA (siCtl) and Orm1 siRNA (siOrm1)-injected mouse livers at 48 h post-PH (n = 2). (a) Heatmap visualization of 188 differentially expressed genes with a fold change of > 2 in the livers between groups of mice receiving siOrm1 or siCtl. (b) Top five associated signaling pathways performed using SEA analysis in GeneSpring GX13. (c) The top diseases or functions annotation and (d) top canonical pathway analysis performed in IPA platform. The pathways were ranked according to their-log10 of P values. The ratio indicates the number of enriched genes of interest relative to the total number of genes associated with that pathway in the IPA database. (e) Gene expression levels of MCM2, MCM4 and MCM6 involved in the enriched “cell cycle control of chromosomal replication” singling pathway in whole-liver tissues before (PH 0 h) and at 48 h post-PH (PH 48 h) were verified using RT-PCR and presented as fold change compared to PH 0 h (n = 4). The data were presented in a Box-and-Whisker plot. *P < 0.05 assessed using the Mann-Whitney U test or two-tailed Student's t-test.
4. Discussion
Regulation of liver cell proliferation is a key event to control organ size during hepatic development and regeneration (Paneda et al., 2002). There are increasing evidences that liver regeneration is initiated by reentry of quiescent hepatocytes into cell cycle and proliferation (Schaub et al., 2014, Yanger et al., 2014). Therefore, overcoming growth arrest of adult differentiated HPCs plays a crucial role in liver regeneration. The mechanism involved in liver regeneration is complex and mediated by the integration of multiple signals. As HGF and epidermal growth factor (EGF) are the only well-established direct mitogens for HPCs, liver regeneration is completely abolished when their signals are deleted (Paranjpe et al., 2016). Besides, non-mitogenic cytokines as well as paracrine mediators are also important in orchestratedly controlling HPC proliferation and paracrine cell interactions (Michalopoulos, 2017). Our data suggested Orm1 acts as an additional regulator of regenerating HPC proliferation.
The well-known lipocalin family protein Lcn2 has been suggested to be a promising biomarker for the early diagnosis of renal injury (Mishra et al., 2005). Importantly, it was recently reported that Lcn2 plays an important role in regulating bacterial infection and liver regeneration. The underlying mechanism is related in part to the deregulating cell cycle regulators such as cyclin-dependent kinases of HPCs (Xu et al., 2015). This was in accordance with our results that Orm1 knockdown suppressed the expression of genes controlling the cell cycle and chromosomal replication in proliferating HPCs. These findings highlight the importance of the HPCs-derived lipocalin protein family in regulating liver regeneration. Microarray analysis in Orm1 knockdown mouse regenerating livers pointed out the signaling pathways that control chromatin replication, especially the MCM genes, as the candidate targets involved in the cell cycle control by Orm1. It is well-known that the MCM complex plays essential roles throughout DNA replication, which promotes recovery from arrest and resumes mitotic growth through direct interactions with checkpoint and recombination proteins (Bailis et al., 2008). Further study to evaluate the predictive value of serum levels of lipocalin family proteins for liver regeneration, such as in the prognosis of PVE, should have clinical significance. In addition, future study in the development of approaches to regulate the expression of lipocalin family genes should have clinical implications in the treatment of liver failure.
Although the underlying molecular mechanism has not yet been elucidated, there are emerging evidences from proteomic studies that both the urinary and serum Orm1 may serve as predictors of therapeutic response and diagnostic and prognostic biomarkers for inflammatory diseases such as chronic heart failure (Agra et al., 2017, Yin et al., 2004) and cancers such as bladder cancer (Li et al., 2016), lung squamous cell carcinoma (Ayyub et al., 2016), breast cancer (Alexander et al., 2004) as well as HCC (Falleti et al., 1993). There is a well-recognized link between tissue regeneration and tumorigenesis, which share common molecular pathways in controlling cell growth (Charni et al., 2017). During the repeated liver damage and compensatory regeneration, aberrant stabilization and activation of pro-regenerative regulators such as c-Myc has contributed to the development of liver cancers (Dauch et al., 2016). In addition, liver regeneration after surgical resection or chemical damage may also facilitate tumor growth and recurrence (Shi and Line, 2014). Therefore, it is reasonable that signaling pathways related to carcinogenesis such as “Role of BRCA1 in DNA Damage Response” and “Hereditary Breast Cancer Signaling” were also enriched in HPCs during liver regeneration. Interestingly, there are growing evidences that Lcn2 is a promising target and a diagnostic and prognostic marker for cholangiocarcinoma (Chiang et al., 2016) and breast cancer (Leng et al., 2009). In this study, suppressed cell proliferation of HCC cells following Orm1 knockdown suggest that Orm1, as an important regulator for normal HPC proliferation during liver regeneration, may serve as a potential therapeutic target in HCC.
Endothelial cells have well-established functions in regulating organ morphogenesis, maintenance and regeneration (Rafii et al., 2016). Spatially and temporally release of paracrine trophogenes, known as angiocrine factors, by endothelial cells after organ injury sustains the homeostasis of resident stem cell and guides the regeneration and repair of organs (Ding et al., 2010). In contrast, deletion of angiocrine factors such as PDGF-BB in endothelial cells disrupts organogenesis and the repair of adult organs (Bjarnegard et al., 2004, Ding et al., 2011). The inductive effect of PDGF-BB on Orm1 expression was observed in primary mouse HPCs, suggesting that the increase in Orm1 expression by HPCs during liver regeneration might reflect endothelial cell-derived inductive signals.
In summary, based on the combination of genome-wide transcriptome analysis and statistical bioinformatics, in vitro and in vivo functional analyses, and clinical observations, we found and characterized Orm1, which was predominantly induced in HPCs in response to hepatic injury, as a promising executer promoting cell cycle of HPCs during liver regeneration (Fig. 5). We propose that the lipocalin family proteins including Orm1 will serve as prognostic biomarkers and potential therapeutic targets for liver diseases.
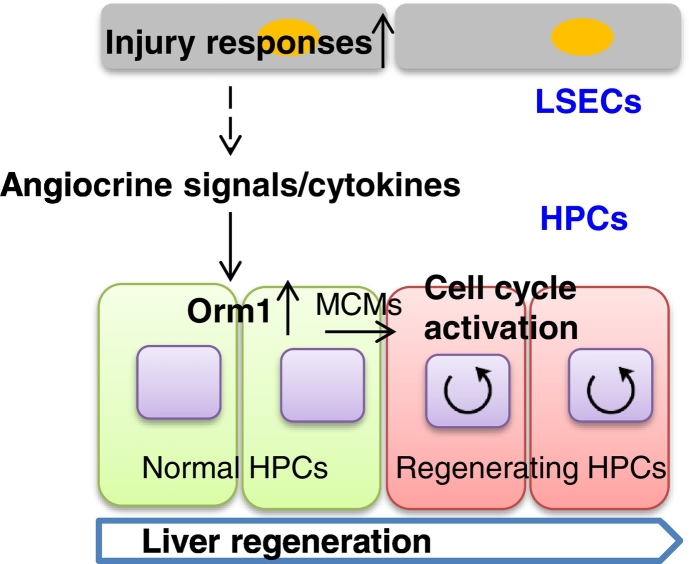
Fig. 5.
Schematic diagram of a regulatory role of Orm1 on regenerating HPC proliferation. In this study, beginning with the transcriptome profiling of LSECs and HPCs during mouse liver regeneration, early transcriptional changes in injury response pathways in LSECs was observed, followed by activation of cell cycle control pathways in HPCs, and Orm1, predominantly expressed in HPCs, was found and characterized as a promising factor responsible for that by regulating HPC proliferation during liver regeneration.
The following are the supplementary data related to this article.
Raw datasets of CAGE transcriptome analysis. Total RNA samples of LSECs and HPCs were extracted at 5 different time points (2 h, 30 h, 48 h or 1 week after PH and at 2 h from sham control) during liver regeneration. Then, transcriptional profiling of 16,499 genes was measured in LSECs and HPCs using the CAGE technology.
The primers used in this study.
List of upstream regulators of differentially expressed genes between LSECs and HPCs. Upstream regulator analysis was performed on a dataset of 9885 differentially expressed genes between LSECs and HPCs using the IPA program.
List of upstream regulators governing LSECs and HPCs during liver regeneration. Upstream regulator analysis was performed on the dataset of significantly differentially expressed genes by the comparison with the sham control at each time point in LSECs and HPCs using the IPA program.
List of VIP scores of regulatory genes of LSECs and HPCs. Regulatory genes governing LSECs and HPCs during liver regeneration were identified in the PLS-DA model with VIP scores > 1.
Raw datasets of microarray analysis. A 40% PH was performed 3 days after in vivo knockdown of Orm1 by a single tail vein injection of its siRNA using Invivofectamine 3.0. Then, gene expression profiles of regenerating mouse livers at 48 h after PH were measured using the Affymetrix GeneChip Mouse Genome 430A 2.0 Array. Data normalization and analysis were performed with GeneSpring GX13.0. The signal intensities for each probe were normalized to the 75th percentile without baseline transformation and filtered by percentile (1 out of 4 samples have values between 20 and 100 percentiles).
Supplementary figures
Funding Sources
This work was supported by a Grant-in-Aid for Young Scientists (B) (JP16K19378) from the Ministry of Education, Culture, Sports, Science and Technology of Japan to X.-Y.Q. and the Research on the Innovative Development and the Practical Application of New Drugs for Hepatitis B (H24-B Drug Discovery-Hepatitis-General-003) from the Japan Agency for Medical Research and Development to S.K. FANTOM5 was made possible by research grants for the RIKEN Omics Science Center and the Innovative Cell Biology by Innovative Technology (Cell Innovation Program) from the MEXT to Y.H. It was also supported by research grants for the RIKEN Preventive Medicine and Diagnosis Innovation Program (RIKEN PMI) to Y.H. and the RIKEN Centre for Life Science Technologies, Division of Genomic Technologies (RIKEN CLST (DGT)) from the MEXT, Japan. The funder had no role in study design, data collection, data analysis, interpretation and writing of the report.
Conflicts of Interest
The authors have no conflicts of interest to declare.
Author Contributions
X.-Y.Q., P.C., Y.H., A.R.R.F. and S.K. designed the study. X.-Y.Q., M.H., E.A., I.I., H.T. Y.F., C.D., H.K., T.L., M.I., H. Suzuki and A.R.R.F. performed the experiments. Y. K., K. N., T.M. and N. K. prepared the clinical samples. F.W., J.K. and H. Sone assisted in the data analysis. X.-Y.Q., M.H., E.A., A.R.R.F. and S.K. wrote the manuscript.
Acknowledgments
Acknowledgments
We thank Mrs. Misako Shirai (The Jikei University School of Medicine, Japan) for assistance with preparation of clinical samples.
References
- Agra R.M., Varela-Roman A., Gonzalez-Ferreiro R., Vinuela J.E., Castro-Pais A., Fernandez-Trasancos A., Diaz-Rodriguez E., Alvarez E., Carreira M.C., Casanueva F.F. Orosomucoid as prognosis factor associated with inflammation in acute or nutritional status in chronic heart failure. Int. J. Cardiol. 2017;228:488–494. doi: 10.1016/j.ijcard.2016.11.134. [DOI] [PubMed] [Google Scholar]
- Akita K., Okuno M., Enya M., Imai S., Moriwaki H., Kawada N., Suzuki Y., Kojima S. Impaired liver regeneration in mice by lipopolysaccharide via TNF-alpha/kallikrein-mediated activation of latent TGF-beta. Gastroenterology. 2002;123:352–364. doi: 10.1053/gast.2002.34234. [DOI] [PubMed] [Google Scholar]
- Alexander H., Stegner A.L., Wagner-Mann C., Du Bois G.C., Alexander S., Sauter E.R. Proteomic analysis to identify breast cancer biomarkers in nipple aspirate fluid. Clin. Cancer Res. 2004;10:7500–7510. doi: 10.1158/1078-0432.CCR-04-1002. [DOI] [PubMed] [Google Scholar]
- Arner E., Daub C.O., Vitting-Seerup K., Andersson R., Lilje B., Drablos F., Lennartsson A., Ronnerblad M., Hrydziuszko O., Vitezic M. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science. 2015;347:1010–1014. doi: 10.1126/science.1259418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atemezem A., Mbemba E., Vassy R., Slimani H., Saffar L., Gattegno L. Human alpha1-acid glycoprotein binds to CCR5 expressed on the plasma membrane of human primary macrophages. Biochem. J. 2001;356:121–128. doi: 10.1042/0264-6021:3560121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyub A., Saleem M., Fatima I., Tariq A., Hashmi N., Musharraf S.G. Glycosylated Alpha-1-acid glycoprotein 1 as a potential lung cancer serum biomarker. Int. J. Biochem. Cell Biol. 2016;70:68–75. doi: 10.1016/j.biocel.2015.11.006. [DOI] [PubMed] [Google Scholar]
- Bailis J.M., Luche D.D., Hunter T., Forsburg S.L. Minichromosome maintenance proteins interact with checkpoint and recombination proteins to promote s-phase genome stability. Mol. Cell. Biol. 2008;28:1724–1738. doi: 10.1128/MCB.01717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal W., Auzinger G., Dhawan A., Wendon J. Acute liver failure. Lancet. 2010;376:190–201. doi: 10.1016/S0140-6736(10)60274-7. [DOI] [PubMed] [Google Scholar]
- Bjarnegard M., Enge M., Norlin J., Gustafsdottir S., Fredriksson S., Abramsson A., Takemoto M., Gustafsson E., Fassler R., Betsholtz C. Endothelium-specific ablation of PDGFB leads to pericyte loss and glomerular, cardiac and placental abnormalities. Development. 2004;131:1847–1857. doi: 10.1242/dev.01080. [DOI] [PubMed] [Google Scholar]
- Bowen W.C., Michalopoulos A.W., Orr A., Ding M.Q., Stolz D.B., Michalopoulos G.K. Development of a chemically defined medium and discovery of new mitogenic growth factors for mouse hepatocytes: mitogenic effects of FGF1/2 and PDGF. PLoS One. 2014;9 doi: 10.1371/journal.pone.0095487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Charni M., Aloni-Grinstein R., Molchadsky A., Rotter V. p53 on the crossroad between regeneration and cancer. Cell Death Differ. 2017;24:8–14. doi: 10.1038/cdd.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang K.C., Yeh T.S., Wu R.C., Pang J.S., Cheng C.T., Wang S.Y., Juang H.H., Yeh C.N. Lipocalin 2 (LCN2) is a promising target for cholangiocarcinoma treatment and bile LCN2 level is a potential cholangiocarcinoma diagnostic marker. Sci Rep. 2016;6 doi: 10.1038/srep36138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauch D., Rudalska R., Cossa G., Nault J.C., Kang T.W., Wuestefeld T., Hohmeyer A., Imbeaud S., Yevsa T., Hoenicke L. A MYC-aurora kinase A protein complex represents an actionable drug target in p53-altered liver cancer. Nat. Med. 2016;22:744–753. doi: 10.1038/nm.4107. [DOI] [PubMed] [Google Scholar]
- Ding B.S., Nolan D.J., Butler J.M., James D., Babazadeh A.O., Rosenwaks Z., Mittal V., Kobayashi H., Shido K., Lyden D. Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature. 2010;468:310–315. doi: 10.1038/nature09493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B.S., Nolan D.J., Guo P., Babazadeh A.O., Cao Z., Rosenwaks Z., Crystal R.G., Simons M., Sato T.N., Worgall S. Endothelial-derived angiocrine signals induce and sustain regenerative lung alveolarization. Cell. 2011;147:539–553. doi: 10.1016/j.cell.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B.S., Cao Z., Lis R., Nolan D.J., Guo P., Simons M., Penfold M.E., Shido K., Rabbany S.Y., Rafii S. Divergent angiocrine signals from vascular niche balance liver regeneration and fibrosis. Nature. 2014;505:97–102. doi: 10.1038/nature12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi A., De Mollerat Du, Jeu X., Johnson C.D., Nektaria A., Feldstein A.E. Liver Bid suppression for treatment of fibrosis associated with non-alcoholic steatohepatitis. J. Hepatol. 2016;64:699–707. doi: 10.1016/j.jhep.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falleti E., Pirisi M., Fabris C., Bortolotti N., Soardo G., Toniutto P., Gonano F., Bartoli E. Increase of serum alpha 1-acid glycoprotein despite the decline of liver synthetic function in cirrhotics with hepatocellular carcinoma. Eur. J. Clin. Chem. Clin. Biochem. 1993;31:407–411. doi: 10.1515/cclm.1993.31.7.407. [DOI] [PubMed] [Google Scholar]
- Forrest A.R., Kawaji H., Rehli M., Baillie J.K., de Hoon M.J., Haberle V., Lassmann T., Kulakovskiy I.V., Lizio M., Itoh M. A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston K., Kiebel S. Predictive coding under the free-energy principle. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2009;364:1211–1221. doi: 10.1098/rstb.2008.0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujise K., Nagamori S., Hasumura S., Homma S., Sujino H., Matsuura T., Shimizu K., Niiya M., Kameda H., Fujita K. Integration of hepatitis B virus DNA into cells of six established human hepatocellular carcinoma cell lines. Hepato-Gastroenterology. 1990;37:457–460. [PubMed] [Google Scholar]
- de Graaf W., van den Esschert J.W., van Lienden K.P., van Gulik T.M. Induction of tumor growth after preoperative portal vein embolization: is it a real problem? Ann. Surg. Oncol. 2009;16:423–430. doi: 10.1245/s10434-008-0222-6. [DOI] [PubMed] [Google Scholar]
- Han S., Lone M.A., Schneiter R., Chang A. Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc. Natl. Acad. Sci. U. S. A. 2010;107:5851–5856. doi: 10.1073/pnas.0911617107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara M., Kirita A., Kondo W., Matsuura T., Nagatsuma K., Dohmae N., Ogawa S., Imajoh-Ohmi S., Friedman S.L., Rifkin D.B. LAP degradation product reflects plasma kallikrein-dependent TGF-beta activation in patients with hepatic fibrosis. Spring. 2014;3:221. doi: 10.1186/2193-1801-3-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Imanishi S., Sone H., Nagano R., Qin X.Y., Yoshinaga J., Akanuma H., Yamane J., Fujibuchi W., Ohsako S. Effects of methylmercury exposure on neuronal differentiation of mouse and human embryonic stem cells. Toxicol. Lett. 2012;212:1–10. doi: 10.1016/j.toxlet.2012.04.011. [DOI] [PubMed] [Google Scholar]
- Hochepied T., Berger F.G., Baumann H., Libert C. Alpha(1)-acid glycoprotein: an acute phase protein with inflammatory and immunomodulating properties. Cytokine Growth Factor Rev. 2003;14:25–34. doi: 10.1016/s1359-6101(02)00054-0. [DOI] [PubMed] [Google Scholar]
- Hu J., Srivastava K., Wieland M., Runge A., Mogler C., Besemfelder E., Terhardt D., Vogel M.J., Cao L., Korn C. Endothelial cell-derived angiopoietin-2 controls liver regeneration as a spatiotemporal rheostat. Science. 2014;343:416–419. doi: 10.1126/science.1244880. [DOI] [PubMed] [Google Scholar]
- Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori-Katayama M., Itoh M., Kawaji H., Lassmann T., Katayama S., Kojima M., Bertin N., Kaiho A., Ninomiya N., Daub C.O. Unamplified cap analysis of gene expression on a single-molecule sequencer. Genome Res. 2011;21:1150–1159. doi: 10.1101/gr.115469.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent T., Murase D., Tsukioka S., Matsuura T., Nagamori S., Oda H. A novel human hepatoma cell line, FLC-4, exhibits highly enhanced liver differentiation functions through the three-dimensional cell shape. J. Cell. Physiol. 2012;227:2898–2906. doi: 10.1002/jcp.23033. [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Choi J.W., Hwang I., Lee J.W., Lee J.H., Kim A.Y., Huh J.Y., Koh Y.J., Koh G.Y., Son H.J. Adipocytokine orosomucoid integrates inflammatory and metabolic signals to preserve energy homeostasis by resolving immoderate inflammation. J. Biol. Chem. 2010;285:22174–22185. doi: 10.1074/jbc.M109.085464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H., Sun Y., Luo Z., Yourek G., Gui H., Yang Y., Su D.F., Liu X. Fatigue-induced Orosomucoid 1 acts on C-C chemokine receptor type 5 to enhance muscle endurance. Sci Rep. 2016;6 doi: 10.1038/srep18839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng X., Ding T., Lin H., Wang Y., Hu L., Hu J., Feig B., Zhang W., Pusztai L., Symmans W.F. Inhibition of lipocalin 2 impairs breast tumorigenesis and metastasis. Cancer Res. 2009;69:8579–8584. doi: 10.1158/0008-5472.CAN-09-1934. [DOI] [PubMed] [Google Scholar]
- Li F., Yu Z., Chen P., Lin G., Li T., Hou L., Du Y., Tan W. The increased excretion of urinary orosomucoid 1 as a useful biomarker for bladder cancer. Am. J. Cancer Res. 2016;6:331–340. [PMC free article] [PubMed] [Google Scholar]
- Limaye P.B., Bowen W.C., Orr A.V., Luo J., Tseng G.C., Michalopoulos G.K. Mechanisms of hepatocyte growth factor-mediated and epidermal growth factor-mediated signaling in transdifferentiation of rat hepatocytes to biliary epithelium. Hepatology. 2008;47:1702–1713. doi: 10.1002/hep.22221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao S.A., Glorioso J.M., Nyberg S.L. Liver regeneration. Transl. Res. 2014;163:352–362. doi: 10.1016/j.trsl.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura T., Kawada M., Hasumura S., Nagamori S., Obata T., Yamaguchi M., Hataba Y., Tanaka H., Shimizu H., Unemura Y. High density culture of immortalized liver endothelial cells in the radial-flow bioreactor in the development of an artificial liver. Int. J. Artif. Organs. 1998;21:229–234. [PubMed] [Google Scholar]
- Michalopoulos G.K. Liver regeneration: molecular mechanisms of growth control. FASEB J. 1990;4:176–187. [PubMed] [Google Scholar]
- Michalopoulos G.K. Hepatostat: liver regeneration and normal liver tissue maintenance. Hepatology. 2017;65:1384–1392. doi: 10.1002/hep.28988. [DOI] [PubMed] [Google Scholar]
- Mishra J., Dent C., Tarabishi R., Mitsnefes M.M., Ma Q., Kelly C., Ruff S.M., Zahedi K., Shao M., Bean J. Neutrophil gelatinase-associated lipocalin (NGAL) as a biomarker for acute renal injury after cardiac surgery. Lancet. 2005;365:1231–1238. doi: 10.1016/S0140-6736(05)74811-X. [DOI] [PubMed] [Google Scholar]
- Nissim O., Melis M., Diaz G., Kleiner D.E., Tice A., Fantola G., Zamboni F., Mishra L., Farci P. Liver regeneration signature in hepatitis B virus (HBV)-associated acute liver failure identified by gene expression profiling. PLoS One. 2012;7:e49611. doi: 10.1371/journal.pone.0049611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paneda C., Gorospe I., Herrera B., Nakamura T., Fabregat I., Varela-Nieto I. Liver cell proliferation requires methionine adenosyltransferase 2A mRNA up-regulation. Hepatology. 2002;35:1381–1391. doi: 10.1053/jhep.2002.32538. [DOI] [PubMed] [Google Scholar]
- Paranjpe S., Bowen W.C., Mars W.M., Orr A., Haynes M.M., DeFrances M.C., Liu S., Tseng G.C., Tsagianni A., Michalopoulos G.K. Combined systemic elimination of MET and epidermal growth factor receptor signaling completely abolishes liver regeneration and leads to liver decompensation. Hepatology. 2016;64:1711–1724. doi: 10.1002/hep.28721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Enciso M., Tenenhaus M. Prediction of clinical outcome with microarray data: a partial least squares discriminant analysis (PLS-DA) approach. Hum. Genet. 2003;112:581–592. doi: 10.1007/s00439-003-0921-9. [DOI] [PubMed] [Google Scholar]
- Qin X.Y., Kojima Y., Mizuno K., Ueoka K., Muroya K., Miyado M., Zaha H., Akanuma H., Zeng Q., Fukuda T. Identification of novel low-dose bisphenol a targets in human foreskin fibroblast cells derived from hypospadias patients. PLoS One. 2012;7:e36711. doi: 10.1371/journal.pone.0036711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X.Y., Wei F., Tanokura M., Ishibashi N., Shimizu M., Moriwaki H., Kojima S. The effect of acyclic retinoid on the metabolomic profiles of hepatocytes and hepatocellular carcinoma cells. PLoS One. 2013;8 doi: 10.1371/journal.pone.0082860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafii S., Butler J.M., Ding B.S. Angiocrine functions of organ-specific endothelial cells. Nature. 2016;529:316–325. doi: 10.1038/nature17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub J.R., Malato Y., Gormond C., Willenbring H. Evidence against a stem cell origin of new hepatocytes in a common mouse model of chronic liver injury. Cell Rep. 2014;8:933–939. doi: 10.1016/j.celrep.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin J., Lizio M., Harshbarger J., Kawaji H., Daub C.O., Hayashizaki Y., Consortium F., Bertin N., Forrest A.R. Interactive visualization and analysis of large-scale sequencing datasets using ZENBU. Nat. Biotechnol. 2014;32:217–219. doi: 10.1038/nbt.2840. [DOI] [PubMed] [Google Scholar]
- Shi J.H., Line P.D. Effect of liver regeneration on malignant hepatic tumors. World J. Gastroenterol. 2014;20:16167–16177. doi: 10.3748/wjg.v20.i43.16167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsukawa H., Sano T., Fukaya Y., Ishibashi N., Watanabe M., Okuno M., Moriwaki H., Kojima S. Dual induction of caspase 3- and transglutaminase-dependent apoptosis by acyclic retinoid in hepatocellular carcinoma cells. Mol. Cancer. 2011;10:4. doi: 10.1186/1476-4598-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treska V., Topolcan O., Vrzalova J., Skalicky T., Sutnar A., Liska V., Fichtl J., Narsanska A., Ferda J., Treskova I. Predictive value of serum biomarkers in patients after portal vein embolization (PVE): a pilot study. Anticancer Res. 2011;31:339–344. [PubMed] [Google Scholar]
- Varghese F., Bukhari A.B., Malhotra R., De A. IHC profiler: an open source plugin for the quantitative evaluation and automated scoring of immunohistochemistry images of human tissue samples. PLoS One. 2014;9 doi: 10.1371/journal.pone.0096801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold S., Sjostrom M., Eriksson L. PLS-regression: a basic tool of chemometrics. Chemom. Intell. Lab. Syst. 2001;58:109–130. [Google Scholar]
- Xu L., Hui A.Y., Albanis E., Arthur M.J., O'Byrne S.M., Blaner W.S., Mukherjee P., Friedman S.L., Eng F.J. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut. 2005;54:142–151. doi: 10.1136/gut.2004.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M.J., Feng D., Wu H., Wang H., Chan Y., Kolls J., Borregaard N., Porse B., Berger T., Mak T.W. Liver is the major source of elevated serum lipocalin-2 levels after bacterial infection or partial hepatectomy: a critical role for IL-6/STAT3. Hepatology. 2015;61:692–702. doi: 10.1002/hep.27447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka T., Toyoshiba H., Sone H., Parham F.M., Portier C.J. The TAO-Gen algorithm for identifying gene interaction networks with application to SOS repair in E. coli. Environ. Health Perspect. 2004;112:1614–1621. doi: 10.1289/txg.7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanger K., Knigin D., Zong Y., Maggs L., Gu G., Akiyama H., Pikarsky E., Stanger B.Z. Adult hepatocytes are generated by self-duplication rather than stem cell differentiation. Cell Stem Cell. 2014;15:340–349. doi: 10.1016/j.stem.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W.H., Chen J.W., Jen H.L., Chiang M.C., Huang W.P., Feng A.N., Young M.S., Lin S.J. Independent prognostic value of elevated high-sensitivity C-reactive protein in chronic heart failure. Am. Heart J. 2004;147:931–938. doi: 10.1016/j.ahj.2003.11.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Raw datasets of CAGE transcriptome analysis. Total RNA samples of LSECs and HPCs were extracted at 5 different time points (2 h, 30 h, 48 h or 1 week after PH and at 2 h from sham control) during liver regeneration. Then, transcriptional profiling of 16,499 genes was measured in LSECs and HPCs using the CAGE technology.
The primers used in this study.
List of upstream regulators of differentially expressed genes between LSECs and HPCs. Upstream regulator analysis was performed on a dataset of 9885 differentially expressed genes between LSECs and HPCs using the IPA program.
List of upstream regulators governing LSECs and HPCs during liver regeneration. Upstream regulator analysis was performed on the dataset of significantly differentially expressed genes by the comparison with the sham control at each time point in LSECs and HPCs using the IPA program.
List of VIP scores of regulatory genes of LSECs and HPCs. Regulatory genes governing LSECs and HPCs during liver regeneration were identified in the PLS-DA model with VIP scores > 1.
Raw datasets of microarray analysis. A 40% PH was performed 3 days after in vivo knockdown of Orm1 by a single tail vein injection of its siRNA using Invivofectamine 3.0. Then, gene expression profiles of regenerating mouse livers at 48 h after PH were measured using the Affymetrix GeneChip Mouse Genome 430A 2.0 Array. Data normalization and analysis were performed with GeneSpring GX13.0. The signal intensities for each probe were normalized to the 75th percentile without baseline transformation and filtered by percentile (1 out of 4 samples have values between 20 and 100 percentiles).
Supplementary figures