Abstract
Therapeutic approaches to fight Alzheimer's disease include anti-Amyloidβ (Aβ) antibodies and secretase inhibitors. However, the blood-brain barrier (BBB) limits the brain exposure of biologics and the chemical space for small molecules to be BBB permeable. The Brain Shuttle (BS) technology is capable of shuttling large molecules into the brain. This allows for new types of therapeutic modalities engineered for optimal efficacy on the molecular target in the brain independent of brain penetrating properties. To this end, we designed BACE1 peptide inhibitors with varying lipid modifications with single-digit picomolar cellular potency. Secondly, we generated active-exosite peptides with structurally confirmed dual binding mode and improved potency. When fused to the BS via sortase coupling, these BACE1 inhibitors significantly reduced brain Aβ levels in mice after intravenous administration. In plasma, both BS and non-BS BACE1 inhibitor peptides induced a significant time- and dose-dependent decrease of Aβ. Our results demonstrate that the BS is essential for BACE1 peptide inhibitors to be efficacious in the brain and active-exosite design of BACE1 peptide inhibitors together with lipid modification may be of therapeutic relevance.
Keywords: BACE1, Alzheimer's disease, Blood brain barrier, CNS delivery
Highlights
-
•
Dual active and exosite designed BACE1 inhibitors with cell and in vivo activity.
-
•
X-ray structures explain the potency and selectivity of the BACE1 inhibitors.
-
•
The BACE1 peptide inhibitors become active within the brain mediated by Brain Shuttle transport across the blood brain barrier.
Alzheimer's disease (AD) is a devastating brain disorder that affects many people. This work describe how specifically designed inhibitors against the enzyme BACE1, which is known to produce toxic molecules that is claimed to be involved in causing AD. The BACE1 inhibitors are very effective and selective as they bind to two sites at the same time to this enzyme and accumulate at the cell membrane where the enzyme is located. As these inhibitors are too big to penetrate into the brain we use the Brain Shuttle technology. Potentially this approach could be used to treat people with the risk of getting AD.
1. Introduction
The neurotoxic polypeptide β-Amyloid (Aβ) is likely to play a central role in the pathogenesis of Alzheimer's disease (AD) as described in the amyloid cascade hypothesis (Hardy and Higgins, 1992). AD is pathologically characterized by extracellular aggregation of Aβ and intracellular accumulation of hyperphosphorylated tau protein into neurofibrillary tangles (Braak and Braak, 1991). The predominant Aβ species is Aβ40, a peptide of 40 amino acids length, and the more amyloidogenic species Aβ42. Prevention of amyloid formation or clearance of existing Aβ early in AD progression is considered as important strategy against AD. Beta-secretase 1 (BACE1) cleaves the type-I transmembrane protein APP to form the N-terminus of Aβ. Due to catalyzing the initial step in Aβ production, BACE1 has become a key therapeutic target for preventing its subsequent aggregation into toxic aggregates (Cai et al., 2001). The first generation of BACE1 inhibitors consisted of large peptidomimetic transition state analogs, which showed poor in vivo pharmacological properties (Ghosh et al., 2012). Subsequently, issues such as serum-half life and blood brain barrier (BBB) permeability have been addressed by designing non-peptidic, lipophilic, small molecule BACE1 inhibitors. Orally bioavailable small molecule BACE1 inhibitors of the third generation show brain Aβ reduction in animal models and are currently being investigated in human clinical trials (Yan, 2016). Data from several clinical trials show that orally dosing once a day reduces CSF Aβ levels, but non-target related side effects like liver toxicity and skin depigmentation have been observed (Vassar, 2014), and are potentially mechanism-based side effects of BACE2 inhibition. Small molecules are currently the main drug modality for brain disorders as large molecules such as antibodies are in most case excluded as they are too big to cross the BBB into the brain.
A promising approach to facilitate delivery of large molecules across the BBB is to take advantage of receptors that mediate transcytosis, an endogenous process in which larger ligands are transported through the endothelial cell barrier (Fishman et al., 1987; Roberts et al., 1993; Friden et al., 1991; Freskgard and Urich, 2016). We recently developed the Brain Shuttle (BS) technology and showed enhanced brain exposure of antibodies (Niewoehner et al., 2014). Consequently, this technology allows us to develop and utilize more potent and selective BACE1 inhibitors as we are not restricted to stringent physicochemical properties, such as small in size, lipid soluble molecules and escape high level efflux transporters for sufficient BBB penetration and entering the CNS.
Without limiting the chemical space to obtain BBB penetrant molecules, we now describe our initial attempts towards optimization of BACE peptide inhibitor sequences and subsequent generation of BACE1 selective molecules with high potency. Also, we provide evidence in support of dual active-exosite peptide inhibitors with unique properties. We show that optimization of binding to the BACE1 active site increases selectivity of the inhibitory peptides. Further we show that exosite binding distal to the active site is critical for achieving cellular potency. Finally, we show that significant Aβ level reduction in wild-type mice after a single intravenous dose of potent BACE1 peptide inhibitors can be accomplished. However, we only block significantly Aβ production in the brain when conjugating the BACE1 peptide inhibitors to the BS for active transport across the BBB by targeting the endogenous Transferrin Receptor (TfR).
2. Material and Methods
2.1. BACE-1 Inhibitor Peptide Synthesis
The general procedures for the CEM Liberty Microwave Peptide Synthesizer (0.1 or 0.25 mMol scale) include treating the washed and pre-swelled resin (435 mg or 1.09 g, 0.1 or 0.25 mMol, TentaGel S RAM (Load: 0.23 mMol/g), (Rapp Polymere, Cat: S30023) with a solution of piperidine 20% in dimethylformamide (DMF) (7.0 or 10 mL) under microwave condition at 50 °C for 3 min for initial deprotection of Fmoc. The resin was washed with DMF and treated with a solution of piperidine 20% in DMF (7.0 or 10 mL) under microwave condition at 75 °C for 5 min for deprotection. To the washed and preswelled resin was added a solution of amino acids, 0.2 M in DMF (2.5 or 5.0 mL, 5.0 or 4.0 eq.) followed by a solution of COMU® 0.5 M in DMF (1.0 or 2.0 mL, 5.0 or 4.0 eq.), (CAS: 1075198-30-9, Iris Biotech, Cat: RL-1175.1000) followed by a solution of diisopropylethylamine (DIPEA) 2 M in N-Methyl-2-pyrrolidone (NMP) (0.5 or 1.0 mL, 10.0 or 8.0 eq.). To couple the amino acids this reaction mixture was treated under microwave condition at 75 °C for 5 min.
2.2. C-Terminal Modification of BACE-1 Inhibitor Peptides
MMT cleavage of Lys(ɛMMT) (0.1 mMol scale): The MMT protected peptide on the resin, prepared by incorporating Fmoc-Lys(MMT)-OH in the first coupling cycle, was washed with CH2Cl2 and then treated with a solution of CH2Cl2:TFA:triisopropylsilane 93:1:6 (5 mL) for 1 h at room temperature on the shaker. The resin was washed with DMF for further coupling.
General procedure for coupling of “Chol” (1.0 mMol scale): The deprotected, with DMF washed and pre-swelled resin was treated with a solution of Cholesteryl hydrogen succinate (2.43 g, 5.0 eq.), (CAS: 1510-21-0, Sigma-Aldrich, Cat: C6512) and COMU (2.14 g, 5.0 eq.), (CAS: 1075198-30-9, Iris Biotech, Cat: RL-1175.1000) and DIPEA (2.04 mL, 12.0 eq.) in 50 mL DMF for 1 h at room temperature on the shaker.
General procedure for coupling of “Chol'ether’” (0.1 mMol scale): The deprotected, with DMF washed and pre-swelled resin was treated with a solution of compound 8 (163 mg, 3.0 eq., see Supplement Fig. 3) and COMU (128 mg, 3.0 eq.), (CAS: 1075198-30-9, Iris Biotech, Cat: RL-1175.1000) and DIPEA (102 μL, 6.0 eq.) in 5.0 mL DMF for 1 h at room temperature on the shaker.
General procedure for final cleavage (0.1 mMol scale): The resin was washed with CH2Cl2 and then treated with a solution of TFA: TIS:water 95:2.5:2.5 (5 mL) for 30 min at room temperature on the shaker. The resin was filtered. The crude peptide was precipitated with diethyl ether (35 mL). The suspension was centrifuged and the solvent was decanted. The solid was dissolved in acetonitrile and water and freeze-dried to get the crude peptide.
C-terminal Lys-palmitoyl containing peptides were prepared by incorporating Fmoc-Lys(ε-palmitoyl)-OH (Bachem B-2530) in solid phase peptide synthesis.
General procedure for purification and MS characterization: The crude product was dissolved in acetonitrile and water (containing 0.1% TFA) and then purified by preparative HPLC. For the lipidated peptides, column YMC-Actus Pro C8, 5 μm, 75 × 30 mm with a gradient of water (containing 0.1% TFA): acetonitrile 70: 30 to 10: 90 and with a flow of 40 mL/min for 10 min was used. For the non-lipidated peptides, column YMC-Actus Pro ODS-AQ C18, 5 μm, 75 × 30 mm with a gradient of water (containing 0.1% TFA): acetonitrile 95: 5 to 50: 50 and with a flow of 40 mL/min for 10 min was used. Peptide homogeneity was confirmed by analytical reversed-phase HPLC in 0.1% TFA with an ACN gradient using a X Bridge C8 (5 μm, 0.46 × 5 cm) for lipidated peptides or a YMC Triart C18 column (5 μm, 0.46 × 5 cm) for non-lipidated peptides. Peptide molecular weights were confirmed by LC–ESI-MS. Sample separation was performed on an Agilent 1200 HPLC system by using a Waters Acquity BEH 130 C18 column (150 × 2.1 mm, pore size 1.7 μm). A column temperature of 60 °C and a flow rate of 0.35 ml/min were used throughout the gradient with Eluent A (water + 5% Acetonitrile + 0.05% TFA) and Eluent B (Acetonitrile + 0.05% TFA). Separation gradient was from 0% to 95% Eluent B in 14 min. Total run time was 18 min. Mass spectrometry was performed on a Thermo LTQ FT Ultra mass spectrometer in positive ion mode with electrospray ionization (ESI). For data analysis Xcalibur software (Ver. 2.1) was used.
2.3. Thermodynamic Solubility Measurements
Equilibrium solubilities are summarized in Table 1 and were measured in phosphate buffer, 0.05 M at pH 6.5 as described by Ottaviani et al. (2015).
Table 1.
Peptide structure-activity-relationship and solubility.
| Peptide | Sequence | Cell (Aβ lowering) IC50 (nM) |
Solubility pH 6.5 (μg/mL) |
|---|---|---|---|
| Pep#1 | Glu-Val-Asn-Sta-Val-Ala-Glu-DPro-NH2 | inactive (> 40 μM) | – |
| Pep#2 | Glu-Val-Asn-Sta-Val-Ala-Glu-DPro-Lys-NH2 | inactive (> 40 μM) | – |
| Pep#3 | Glu-Val-Asn-Sta-Val-Ala-Glu-DPro-Lys(ε-Chol'ester’)-NH2 | 0.40 | > 3000 |
| Pep#4 | Glu-Val-Asn-Sta-Val-Ala-Glu-DPro-Lys(ε-Palmitoyl)-NH2 | 6.0 | < 1.0 |
| Pep#5 | Ac-Tyr-Pro-Tyr-Phe-Ile-Pro-Leu-NH2 | Inactive (> 40 μM) | – |
| Pep#6 | Ac-Tyr-Pro-Tyr-Phe-Leu-Pro-Ile-Ser-Ala-Lys-NH2 | inactive (> 40 μM) | – |
| Pep#6a | Ac-Ala-Leu-Tyr-Pro-Tyr-Phe-Leu-Pro-Ile-Ser-Ala-Lys-NH2 | a | |
| Pep#7 | Tyr-Pro-Tyr-Phe-Ile-Pro-Leu-Gly-Gly-Gly-Glu-Val-Asn-Sta-Val-Ala-Glu-DPro-Lys(Chol'ester’)-NH2 | 0.027 | < 1.0 |
| Pep#8 | Tyr-Pro-Tyr-Phe-Ile-Pro-Leu-NH(CH2)5CO-Glu-Val-Asn-Sta-Val-Ala-Glu-DPro-Lys(Chol'ester’)-NH2 | 0.075 | – |
| Pep#9 | Tyr-Pro-Tyr-Phe-Ile-Pro-DLys-Gly-DLys-Gly-Glu-Val-Asn-Sta-Val-Ala-Glu-DPro-Lys(Chol'ester’)-NH2 | 0.006 | > 718 |
| Pep#10 | Tyr-Pro-Tyr-Phe-Ile-Pro-Ala-Gly-DLys-Gly-Glu-Val-Asn-Sta-Val-Ala-Glu-DPro-Lys(Chol'ester’)-NH2 | 0.004 | > 2450 |
| Pep#11 | Tyr-Pro-Tyr-Phe-Ile-Pro-DLys-Gly-DLys-Gly-Glu-Val-Asn-Sta-Val-Ala-Glu-DPro-Lys(Palmitoyl)-NH2 | 0.20 | > 3000 |
| Pep#12 | Tyr-Pro-Tyr-Phe-Ile-Pro-DLys-Gly-DLys-Gly-Glu-Val-Asn-Sta-Val-Ala-Glu-DPro-NH2 | 19 | – |
| Pep#13 | H-Tyr-Pro-Tyr-Phe-Ile-Pro-DLys-PEG(4)-Glu-Val-Asn-Sta-Val-Ala-Glu-DPro-NH2 | 45 | – |
| Pep#14 | Gly-Gly-Gly-Tyr-Pro-Tyr-Phe-Ile-Pro-DLys-Gly-DLys-Gly-Glu-Val-Asn-Sta-Val-Ala-Glu-DPro-Lys(Chol'ester’)-NH2 | 0.57 | – |
| Pep#15 | Gly-Gly-Gly-Tyr-Pro-Lys-Phe-Ile-Pro-Leu-Gly-DLys-Gly-Glu-Val-Asn-Sta-Val-Ala-Glu-DPro-Lys(Palmitoyl)-NH2 | 0.34 | – |
| Pep#16 | Gly-Gly-Gly-Tyr-Pro-Tyr-Phe-Ile-Pro-DLys-Gly-DLys-Gly-Glu-Val-Asn-Sta-Val-Ala-Glu-DPro-NH2 | 6.4 | – |
Compound (peptide 2 in Kornacker et al., 2005) used in the ternary complex BACE1 ectodomain structure (accession code 5MCO). A KD of 0.63 μM for Pep#6a has been reported determined using a biochemical approach isothermal calorimetry (ICT).
2.4. Crystallization of BACE1 in Complex with Peptide Inhibitors
Co-crystallization of BACE1 in complex with Pep#16: BACE1 ectodomain protein (A14-T454) at a concentration of 0.9 mg/ml in 20 mM Tris buffer (pH 7.2), 150 mM sodium chloride was mixed with a 4-fold molar excess of ligand Pep#16 and incubated overnight at 4 °C. Prior to crystallization experiments, the protein was concentrated to 20 mg/ml and centrifuged at 20,000 × g. The crystallization droplets were set up at 22 °C by mixing 0.16 μL of protein solution with 0.04 μL reservoir solution (Procomplex Screen, Qiagen) in vapour diffusion sitting drop experiments. Crystals appeared within 10 days out of 0.1 M magnesium acetate, 0.1 M sodium acetate (pH 4.5) and 8% (w/v) PEG8000 as precipitant. Seeding with BACE1 crystals was used to improve the hit rate in the initial screening experiments.
Co-crystallization of ternary BACE1 in complex with GRL-8234(Chang et al., 2011) and Pep#6: BACE1 ectodomain mutant protein (A14-T454, K307A) at a concentration of 0.1 mg/ml in 20 mM Tris buffer (pH 7.2), 150 mM sodium chloride was mixed with active-site ligand GRL-8234, in a 8-fold molar excess for 2 days at 4 °C. Subsequently, Pep#6 was added in a 30-fold molar excess and after incubation for 2 h at 4 °C the protein was concentrated to 20.5 mg/ml. Crystallization droplets were set up at 22 °C by mixing 0.14 μL of protein solution with 0.06 μL reservoir solution (Procomplex Screen, Qiagen) in vapour diffusion sitting drop experiments. Small needle shaped crystals appeared within 2 days out of various conditions containing PEG as precipitant. Crystals were improved with respect to size through seeding experiments and the final crystals used for data collection were obtained out of 0.1 M ammonium acetate, 0.1 M sodium acetate (pH 4.0) and 15% (w/v) PEG4000.
Co-crystallization of BACE1 with Pep#3: BACE1 ectodomain mutant protein (A14-T454, K307A) at a concentration of 0.1 mg/ml in 20 mM Tris buffer (pH 7.2), 150 mM sodium chloride was mixed with a 15-fold molar excess of compound Pep#3 and after incubation overnight at 4 °C the protein was concentrated to 16.6 mg/ml. Initial crystals were obtained out of 0.1 M sodium chloride, 0.1 M HEPES (pH 7.5), 1.6 M ammonium sulfate in vapour diffusion sitting drop experiments by mixing 0.14 μL of protein solution with 0.06 μL reservoir solution (Crystal Screen HT, Hampton Research). Crystals were optimized by detergent screening (Detergent Screen HT, Hampton Research). Addition of 98 mM of HEGA-9 yielded large triangular shaped crystals which were used for data collection.
3. Molecular Modeling
Since the electron density of the cholesterol conjugate in Pep#3 was not resolved, a conformational ensemble was generated with Omega (OMEGA 2.5.1.4: OpenEye Scientific Software, Santa Fe, NM., http://www.eyesopen.com.). The peptide portion was kept fixed and conformations clashing with the protein were removed afterwards. From the remaining conformer ensemble a few representative members were manually selected.
The model of the Brain Shuttle peptide conjugate binding to BACE1 together with the cholesterol lipid anchor in a phospholipid bilayer was manually constructed from various components. The Computational Molecular Biophysics Group at the Georg-August-Universität Göttingen provides the coordinates of membrane patches which had been equilibrated via Molecular Dynamics simulations (http://cmb.bio.uni-goettingen.de/downloads.html). From three DOPC patches with 20% cholesterol a membrane model was built into which the X-ray structure of BACE1. A cholesterol anchor was attached and all lipid components within van-der-Waals radius of the embedded BACE-1 complex were removed. The model of a Brain Shuttle antibody was attached via the sortase recognition motif to the N-terminus of the peptide with one glycine being removed. The C-terminal transmembrane helix was modelled and manually attached to BACE1. No further energy minimization of the resulting multi-component structure was performed.
3.1. N-terminal Sortase Coupling of BACE-1 Inhibitor Peptides to the Brain Shuttle Construct
The mouse transferrin receptor (TfR) brain shuttle antibody (BS antibody) was engineered in a one-armed (sFab) IgG format as described (Niewoehner et al., 2014). Knobs-into-holes technology was used to favor heterodimeric pairing of one heavy chain carrying the TfR antibody fragment to a free heavy chain. In order to enable sortase coupling of BACE-1 inhibitor peptides to the BS antibody and subsequent purification of the conjugates, the free heavy chain was extended by a sortase recognition sequence (Popp et al., 2009), a small linker and a 6xHis tag, namely LPETGGSGSHHHHHH.
Expression and purification of one-armed Fc-derivatives: For transient expression of the recombinant antibody Fc-derivatives, expression plasmids encoding light and heavy chain were co-transfected into HEK293 suspension cells which were subsequently cultivated in serum free medium at 37 °C at 8% CO2. Seven days after transfection, the Fc containing cell culture supernatants were clarified by centrifugation at 14′000 × g for 30 min and filtration through a sterile filter (0.22 μm). Cell culture supernatants were stored at − 20 °C until purification. The recombinant Fc-fragments were purified from the supernatant by protein-A chromatography. After filtration over a 0.2 μm MF75TM (Fisher Scientific), clarified cell culture supernatants were applied on a PBS buffer equilibrated MAbSelectSure Protein A (5–50 ml) column (GE Healthcare). After washing with equilibration buffer, one-armed Fc-fragments were eluted with 25 mM citrate buffer, pH 3.0. Desired fractions were neutralized with Tris buffer, pooled, concentrated (Amicon 15 K Ultra centrifugal filter), and subjected to SEC (Superdex200 HiLoad 26/60, GE Healthcare) equilibrated with 50 mM histidin pH 7.5, 150 mM NaCl, 5 mM CaCl2. The protein concentration of purified protein was determined by measuring the optical density (OD) at 280 nm, with background correction at OD 320 nm, using the molar extinction coefficient calculated on the basis of the amino acid sequence (Pace and Betowski, 1995). The homogeneity of the BS antibody was confirmed by Coomassie brilliant blue staining after SDS-PAGE on 4–20% Tris-Glycine gels (NuPAGE® Pre-Cast gel system, Invitrogen), in the presence or absence of 5 mM 1,4-dithiotreitol. In addition, purity of the samples was quantified by integration of peaks in the CE-SDS chromatograms run on a LabChip GXII Touch HT (Perkin Elmer) for reduced and non-reduced samples with a non-covalent procedure for detection. Sample preparation was done by heating at 95 °C for 5 min in a SDS containing sample buffer before application to the Protein Express Chip (Perkin Elmer).
Sortase-A conjugation of the brain shuttle antibodies: The enzyme used in this study was a soluble construct of S. aureus sortase A lacking the N-terminal transmembrane region (delta24 aa) and carrying an engineered C-terminal 6xHis-tag. Reactions with 100 μM peptide, 25 μM BS antibody and 50 μM sortase A were incubated 5 h at RT and analyzed qualitatively by SDS-PAGE under reducing conditions. Basic composition of the conjugation buffer was 50 mM Tris/HCl pH 8.2, 300 mM NaCl, 5 mM CaCl2, and 10% glycerol (Buffer A).
Preparation of BS-BACE1 conjugate: Buffer A was used for the conjugation of the non-lipidated peptide. For purification of the conjugate the reaction mixture was then applied to a 1 ml His-Trap column (GE Healthcare), equilibrated in buffer A to remove unconjugated BS antibody and His-tagged sortase A. The flow through was collected, then concentrated and further applied to a Superdex S200 (16/60) column (GE Healthcare) equilibrated in 20 mM histidine pH 6.0, 140 mM NaCl to remove unconjugated peptide. Conjugate containing fractions were pooled and concentrated to ca. 3–4 mg/ml.
Preparation of BS-BACE1 (palm) conjugate: 1.3 mM (2-hydroxypropyl)-β-cyclodextrin was added to buffer A for the conjugation and purification (IMAC and SEC chromatography) using palmitylated BACE-1 inhibitor peptide.
Preparation of the BS-BACE1 (chol) conjugate: Buffer A was supplemented with 1.3 mM (2-hydroxypropyl)-β-cyclodextrin for the conjugation of the cholesteroylated BACE1 inhibitor peptide as well as for two steps of the purification process. Between IMAC and the final SEC in the cyclodextrin containing buffer A, an additional SEC without cyclodextrin was performed to separate conjugated and non-conjugated BS antibody.
Routinely, the quality of the conjugates was assessed in-house by capillary SDS-PAGE (as described above), HPLC, mass spectrometry and analytical ultracentrifugation.
Purity of the conjugates was checked by using an Agilent 1200 series HPLC system equipped with a C8 Poroshell 300SB column (5 μm, 1 × 75 mm microbore (Agilent)). The column temperature was adjusted to 70 °C and the flow rate set to 0.68 ml/min. Eluent A was 0.1% TFA in water and eluent B 0.08% TFA in acetonitrile. A graduated separation gradient was run from 20 to 75% eluent B in 2.85 min. The conjugates were analyzed in the oxidized or in the reduced form after treatment with high excess of DTT.
For characterization of the proteins LC ESI-MS was used. To the sample aliquots (approx. 2 μg each) in 100 mM Tris buffer pH 7.8, 2 μl 500 mM DTT was added and incubated for 15 min at 40 °C. Sample separation was performed by using a capillary UPLC system (Waters nanoAcquity) on an Agilent Poroshell C8 reversed-phase column (0.5 × 75 mm, pore size 5 μm). A column temperature of 65 °C and a flow rate of 70 μl/min was used throughout the gradient with eluent A (0.1% formic acid in water) and eluent B (0.1% formic acid in acetonitrile). Separation gradient was from 20 to 60% eluent B in either 50 min or 4 min, respectively. Mass spectrometry was performed on a Waters Premier XE Mass Spectrometer (Waters) with electrospray ionization source. For data analysis the Waters MassLynx software (Ver. 4.1) was used, and deconvolution of spectra was done with the MaxEnt1 software module.
3.2. Measuring Association States of Peptides or Brain-Shuttle Peptide Conjugates via Analytical Ultracentrifugation (AUC)
The brain shuttle conjugates were prepared at approximately 30 μM and centrifuged at 42,000 rpm and 20 °C in 3 mm SedVel60K centerpieces (Spin Analytical) on a Proteome Lab XLI analytical ultracentrifuge equipped with an AnTi60 rotor (Beckman Coulter). Control runs with samples of 30 μM solutions of the free peptides were centrifuged at 60,000 rpm and 20 °C in 12 mm SedVel60K centerpieces. Sedimentation velocity data (radial absorbance scans at 280 nm for conjugates, 275 nm and 235 nm for peptides) were analyzed with Sedfit (Schuck, 2000) and sedimentation coefficient distributions with s20,w values corrected for buffer viscosity and density were plotted with GUSSI (Brautigam, 2015). The AUC buffer used for BS-BACE1 (and corresponding un-conjugated peptide) was 20 mM histidine pH 6, 140 mM NaCl. This buffer was supplemented with 1.3 mM hydroxypropyl-β-cyclodextrin for the acylated derivatives BS-BACE1 (palm) and BS-BACE1 (chol) (and corresponding un-conjugated peptides).
4. IgG ELISA
After treatment left brain hemispheres were homogenized in 4 volumes of RIPA buffer (Pierce; 89,901) for 55 s at 6500 rpm in Green Bead Tubes (Roche; 03358941001) using a MagNa Lyser (Roche, 40400559). The brain homogenates were incubated for 2 h at 4 °C and subsequently centrifuged at 14000 rpm for 15 min. The brain supernatants were 1:200 diluted in 1 × PBS, 0.5% BSA and underwent together with the 1:10,000 diluted plasma samples a human IgG1/IgG4 sandwich ELISA. The assay was developed in-house to capture the anti-kappa domain and to detect anti-human Fcg. In short, capture antibody was added to streptavidin-coated 96-well plates and incubated for 1 h at room temperature. After removing excess antibody, the brain and plasma samples were added and incubated for 1 h. Following a washing step the human anti-TfR antibody was detected with a Digoxigenin-labeled detection antibody (1 h incubation). After washing, anti-Digoxigenin-POD (Roche; 11633716001) was added for 1h followed by a washing step and adding of TMB substrate solution (Roche; 11484281001). Absorbance was read at 450 nm after stopping colour development with 1 M H2SO4.
4.1. Immobilization, Binding Assay for BACE1 and BACE2 Proteins and SPR Data Analysis
C-terminal His-tagged full lengths BACE-1 and N-terminal T7-tagged BACE2 (amino acids: 20-465) proteins were immobilized via standard amino coupling on SPR sensor CM5 on Biacore instruments T200 or 3000 (GE Healthcare, Uppsala, Sweden).
4.1.1. Protein Immobilization
Immobilization of BACE1 and BACE2 proteins was performed in the running buffer containing 25 mM Na acetate, 150 mM NaCl, 0.01% P20, 3 mM EDTA, pH 4.5 at 25 °C and 35 °C, respectively. In the first step the carboxyl groups of CM5 sensor surface were transformed to the reactive succinimide esters by contacting the sensor surface with a solution of 0.2 M N-ethyl-N-dimethylaminopolycarboodiimide (EDC) and 0.05 M N-hydroxysuccinimide (NHS) for 7 min. After the activation step the sensor surfaces in parallel channels were contacted with BACE1 or BACE2 (~ 5 μg/ml in 10 mM sodium acetate, pH 4.5) protein solution to achieve surface densities of ~ 8000 RUs, respectively. Finally, the excess of activated carboxylic groups on the sensor surface was deactivated with ethanolamine solution (1 M, pH 8.5, 7 min).
4.1.2. Binding Assay
Binding assay was performed at 18 °C, at the same buffer conditions as proteins' immobilization. Peptides were analyzed in titration experiments (single cycle kinetic with 5 concentrations or multiple cycle kinetic with 10 concentrations, both with dilution factor of 2) on two protein surfaces and reference surface (activated and deactivated CM5 sensor surface) in parallel on the same sensor. Two exo-site peptides and an active-site small molecule ligand were used as reference molecules to monitor binding activity of BACE1 and BACE2 during the SPR run.
4.1.3. Data Analysis
SPR data processing and analysis was performed using BiaEvaluation Software (version 2.0 (Biacore T200) and 4.1 (Biacore 3000)), and GraphPadPrism (version 6.04). All SPR binding curves monitored on the binding active channels were subtracted by signals from the reference channel (activated and deactivated sensor surface) and by buffer signals (double referencing). To extract kinetic parameters the binding data were globally fit to the mathematical binding model describing a one-to-one interaction. Equilibrium analysis was performed for peptide showing fast dissociation. Here, the amplitude of SPR signals at equilibrium (measured in the association phase at time of 100 s) was plotted against peptide concentration and fit to the one-to-one interaction model with four parameters with fixed maximal signal derived from normalized binding data monitored for a positive control (GraphPadPrism software).
4.2. Cell-Based IC50 Assay
The Aβ-lowering potency of BS coupled and non-coupled BACE-1 inhibitors was determined in vitro using HEK293 cells stably transfected with wild-type human APP (APP695) cDNA. Cells were seeded in 96-well Microtiter plates (Falcon, #353075) in Iscoveˈs cell culture medium (Gibco, #21980-032; with 10% (v/v) fetal bovine serum (Gibco, #26140-079), penicillin/streptomycin (Gibco, #15140-122)) to about 80% confluency. The BACE1 inhibitors were added in serial dilutions with the respective construct dilution buffers as control. After overnight incubation at 37 °C and 5% CO2 in a humidified incubator, the culture supernatants were harvested and the Aβ40 concentration subsequently analyzed using the human amyloid-β 1–40 AlphaLISA kit (PerkinElmer, #AL275C). Following manufacturersˈ instructions, 6 μL of culture supernatants were added to 2 μL of AlphaLISA anti-human Aβ acceptor beads mix (50 μg/mL beads with 5 nM biotinylated anti-Aβ 1–40 antibody in 1 × AlphaLISA HiBlock buffer) in a PerkinElmer OptiPlate-384 (#6007290) and incubated for 1 h at room temperature. To this mixture 16 μL of streptavidin donor beads were added (250 ng/mL in 1 × AlphaLISA buffer). Following an incubation of 30 min at room temperature the light emission was recorded at 615 nm on an EnVision-2104 multilabel reader (PerkinElmer, 1040564). Levels of Aβ40 in the culture supernatants were calculated as percentage of maximum signal. The IC50 values were calculated using the Excel XLfit (Microsoft Excel 2010) software.
4.3. Uptake of BS-BACE1 Constructs in Brain endothelial Cells In Vitro
Uptake and localization of the Brain Shuttle at different time points after a single dose of 100 nM was assessed in bEnd.3 (ATCC CRL2299) immortalized mouse brain endothelial cells. Cells were seeded onto 4-chamber slides (MilliCell EZ, PEZCS0416) coated with attachment factor solution (Cell applications Inc., 123-100) until 80% confluency. BS-coupled BACE1 peptide inhibitors were added to serum-free cell culture medium (Gibco, 31331) for 30 min and 240 min, with the respective construct dilution buffers as negative control and non-coupled BS as positive control. After incubation at 37 °C and 5% CO2 in a humidified incubator, excess construct was removed and cells subsequently fixed in 4% formaldehyde (AlfaAesa, 43,368) for 10 min at 4 °C. For fluorescence immunolabeling, fixed cells were incubated with fluorescently-labeled anti-human IgG (1:1000; Invitrogen, A11013) for 1 h at room temperature in HBSS buffer (Gibco, 14,175) containing 1% normal goat serum (Sigma, G9023) and 0.05% saponin (Sigma, 47036). After removing excess secondary antibody, nuclear counterstaining with DAPI (2 μg/mL; Roche, 10236276001) was performed. Stained cells were viewed with a Leica TCS SP5 confocal laser scanning microscope. Secondary antibodies alone did not produce significant staining (data not shown). Imaging and analysis parameters are described below in immunohistochemistry section.
4.4. Animal care and handling
All animal procedures were performed in strict adherence to the Swiss federal regulations on animal protection and approved by the appropriate governmental authorities, to the rules of the AAALAC and with the explicit approval of the local veterinary authority (permission number 1902). Mice had access to prefiltered sterile water and standard mouse chow (Kliba Nafag, #3436) ad libitum and were housed under a reversed day–night cycle.
4.5. Treatment
Nine-weeks-old female C57BL/6 J mice were intravenously (i.v.) single injected with BACE-1 inhibitory peptides with varying terminal moieties in doses ranging from 0.003 to 3 mg/kg. BS conjugates of the same peptides were i.v. injected at 10 mg/kg and compared to the equimolar concentration of unconjugated peptides 5, 24, and 48 h after administration. A single injection of the respective buffers was used as vehicle control. Three animals were included per group. At the indicated time points, animals were anesthetized, brain and plasma were collected and immediately frozen on powdered dry ice for further analyzes.
4.6. Uptake of BS-BACE1 Constructs in Brain Endothelial Cells In Vivo
After the brain was removed from the cranium, left and right hemispheres were separated with a cut in the sagittal plane. Localization of the BS at different time points after single dose of 10 mg/kg was assessed in the left hemisphere by confocal microscopy. For fluorescence immunolabeling, 10-μm-thick tissue sections were prepared on HistoBond slides (Marienfeld 0800001) using a cryotome (Leica Biosystems CM3050S). Sections were fixed in ice-cold acetone for 3 min, blocked for 20 min in 1% ovalbumin (Sigma, A5503), 1% BSA (Sigma, A9647) and 1% goat serum (Sigma, G9023), and incubated with directly labeled endothelial specific anti-CD31 antibody (1:100; Serotec, MCA2388A647) and anti-human IgG antibody (1:1000; Invitrogen, A21433) for 1 h at room temperature. Nuclear counterstaining with DAPI (2 μg/mL; Roche, 10236276001) was performed. Stained tissue sections were viewed with a Leica TCS SP5 confocal laser scanning microscope using a 63 × objective lens (numerical aperture of 1.4, pinhole set to 1.0 airy unit) with different zoom factors (up to 5). A sequential scan procedure was applied during image acquisition and parameters were adjusted to use the full dynamic range of the photomultipliers. Typically, stacks of 4 to 8 images (512 × 512 pixels) were taken and analyzed at 122-nm intervals through the z axis of the section. Representative images from the examined samples were chosen for figure editing. Digital images were processed using the software Imaris x64 7.7.2 (Bitplane).
4.7. Beta Amyloid (40) ELISA
After treatment the brain was removed from the cranium and left and right hemispheres were separated with a cut in the sagittal plane. Frozen right hemispheres were homogenized in 4 volumes of 2% diethanolamine (Sigma; D8885) for 20 s at 4000 rpm in Green Bead Tubes (Roche; 03358941001) using a MagNa Lyser (Roche, 40400559). The brain homogenates were incubated overnight on ice and subsequently centrifuged at 4 °C for 60 min at 50,000 rpm. The supernatants were 1:10 diluted in 50 mM NaCl and underwent a solid phase extraction (SPE). Blood samples were collected at the indicated time points in tubes containing EDTA (Milian; TOM-14), followed by centrifugation for 8 min at 1600 × g, and 1:10 dilution of the plasma in 0.2% diethanolamine. The processed brain homogenates and plasma samples were loaded onto 60-μm Oasis SPE plates (Waters; 186000679) and washed with increasing percentages of methanol. Samples were eluted from the SPE plates in 2% ammonium hydroxide diluted in 90% methanol and dried under nitrogen at 55 °C in a TurboVap®96 (Caliper LifeSciences). After, samples were reconstituted 1:1 in standard diluent provided in the β-amyloid (40) ELISA kit (Wako, 294-64701). The ELISA was performed according to the manufacturer protocol, using the provided human β-amyloid (1–40) as standard. Statistical analysis was done by two-way ANOVA followed by Dunnettˈs multiple-comparison test using GraphPad Prism 6. p values < 0.05 were considered significant: ****p < 0.0001, ***p = 0.0001 to 0.001, **p = 0.001 to 0.01, *p = 0.01 to 0.05, ns = non-significant (p > 0.05).
4.8. Human Neural Stem Cell-Based IC50 Assay
Human neurons were differentiated from SB-AD3 induced pluripotent stem cells (iPSCs) obtained through the StemBANCC consortium (Innovative Medicine Initiative; http://stembancc.org/). iPSCs were reprogrammed from fibroblasts of an healthy adult donor. Briefly, neural induction was obtained by embryoid body formation and dual SMAD inhibition (Chambers et al., 2009; Boissart et al., 2013), obtaining neural progenitor cells that were expanded and patterned for one week. Subsequently, the cells were re-plated in presence of BDNF (20 ng/ml; Peprotech, 450-02), GDNF (10 ng/ml; Peprotech, 450-10), cAMP (0.5 mM; BIOLOG Life Science, D009), and ascorbic acid phosphate (100 μM; Sigma, A8960), and kept in these conditions for a total of six weeks (Costa et al., 2016). The neuronal identity of the culture was verified by immunostaining for GFAP (Dako, Z033401), HuC/D (Invitrogen, A21271), and anti-Map2 (Neuromics, CH22103). Fully differentiated cells were treated for 48 h with a serial dilution of BACE1palm, BACE1chol, BACE1, or small molecule β-secretase (Jacobsen et al., 2014) and γ-secretase (Tolcher et al., 2012) inhibitors; vehicle controls were run in parallel (20 mM histidine pH 6, 140 mM NaCl, 1.3 mM 2-hydroxypropyl-beta-cyclodextrin (for the peptides), or DMSO (Sigma) (for the small molecules)). Experimental setup included cells from three independent differentiations and two technical replicates each dose (0.128–10,000 nM), and cells from one differentiation and two technical replicates each dose (0.001024–80 nM). The culture supernatants were collected and the Aβ40 concentration subsequently analyzed using a custom-conjugated AlphaLISA kit. Following manufacturersˈ instructions, 10 μL of culture supernatants were added to 5 μL anti-human Aβ acceptor beads mix (2.5 μg/mL Bap24-labeled (Brockhaus et al., 1998, Hock et al., 2003) beads with 0.25 nM biotinylated anti-Aβ 1–40 antibody 4G8 (BioLegend, 800704) and incubated for 3 h at room temperature. To this mixture 30 μg/mL streptavidin donor beads were added (Perkin Elmer, 6760002S). Following an incubation of 30 min at room temperature the light emission was recorded at 615 nm. Statistical analysis of Aβ40 levels was done by comparing treated with vehicle-treated cells using GraphPad Prism 6. Values plotted are means ± SEM.
4.9. FACS Analysis
Specific binding of conjugates to mouse BA/F3 pro-B cells expressing mouse transferrin receptor or unspecific attachment to HEK293 cells was tested by FACS analysis. Cells were harvested by centrifugation, washed once with PBS and 5 × 104 cells incubated with a 100 pM to 200 nM dilution series of the conjugates in 100 μL RPMI/10% FCS for 1.5 h on ice. After 2 washes with RPMI/10% FCS, cells were incubated with goat-anti-human IgG coupled to Phycoerythrin (Jackson Immunoresearch) at a dilution of 1:100 in RPMI/10% FCS for 1.5 h on ice. Cells were again washed, resuspended in RPMI/10% FCS and Phycoerythrin fluorescence measured on a FACS-Canto instrument (Becton-Dickinson).
4.10. Western Blot Analysis
The left hemisphere of BS-BACE1 treated animals was divided by a midline coronal cut and the forebrain was prepared for TfR expression analysis. The brains were homogenized in RIPA buffer (Thermo Scientific, 89901) with added protease inhibitors (Roche, 05056489001) using MagNA Lyser Green Beads (Roche, 03358941001) for 20 s at speed 4000. 30 μg of total protein lysates were diluted in LDS sample buffer (Invitrogen, NP0007) with added reducing agent (Invitrogen, NP0004). Samples underwent SDS-PAGE (4–12% gel, Invitrogen WG1402) and were transferred with the Trans-Blot® TurboTM Transfer System (BIO-RAD) onto nitrocellulose membranes (Trans-Blot® TurboTM Transfer Pack, BIO-RAD 170-4159). For western blot analysis, anti-TfR antibody (AbD Serotec, MCA1033G) was used, followed by labeling with anti-rat IgG-HRP (Amersham, NA935V). Chemiluminescence was detected with SuperSignal® West Dura Extended Duration Substrate (Thermo Scientific, 34076) using a Fusion FX7 system (Vilber Lourmat). The same membranes used to detect TfR were stripped, blocked, and re-probed with anti-β-actin antibody (Abcam, ab49900).
4.11. Immunocytochemistry
Uptake and localization of the BS at different time points after a single dose of 100 nM was assessed in bEnd.3 (ATCC CRL2299) immortalized mouse brain endothelial cells. Cells were seeded onto 4-chamber slides (MilliCell EZ, PEZCS0416) coated with attachment factor solution (Cell applications Inc., 123-100) until 80% confluency. BS-coupled BACE-1 peptide inhibitors were added to serum-free cell culture medium (Gibco, 31,331) for 10 min, 30 min and 240 min, with the respective construct dilution buffers as negative control and uncoupled BS as positive control. After incubation at 37 °C and 5% CO2 in a humidified incubator, excess construct was removed and cells subsequently fixed in 4% formaldehyde (AlfaAesa, 43368) for 10 min at 4 °C. For fluorescence immunolabeling, fixed cells were incubated with fluorescently-labeled anti-human IgG (1:1000; Invitrogen, A11013) for 1 h at room temperature in HBSS buffer (Gibco, 14,175) containing 1% normal goat serum (Sigma, G9023) and 0.05% saponin (Sigma, 47036). After removing excess secondary antibody, nuclear counterstaining with DAPI (2 μg/mL; Roche, 10236276001) was performed. Stained cells were viewed with a Leica TCS SP5 confocal laser scanning microscope. Secondary antibodies alone did not produce significant staining (data not shown). Imaging and analysis parameters are described above in “immunohistochemistry” section.
5. Measuring Brain Shuttle levels via huIgG ELISA
After treatment left brain hemispheres were homogenized in 4 volumes of RIPA buffer (Thermo Scientific; 89,901) for 55 s at 6500 rpm in Green Bead Tubes (Roche; 03358941001) using a MagNa Lyser (Roche, 40400559). The brain homogenates were incubated for 2 h at 4 °C and subsequently centrifuged at 14000 rpm for 15 min. The brain supernatants were 1:200 diluted in 1 × PBS, 0.5% BSA and underwent together with the 1:10,000 diluted plasma samples a human IgG1/IgG4 sandwich ELISA. The assay was developed in-house to capture the anti-kappa domain and to detect anti-human Fcg. In short, capture antibody was added to streptavidin-coated 96-well plates and incubated for 1 h at room temperature. After removing excess antibody, the brain and plasma samples were added and incubated for 1 h. Following a washing step the human anti-TfR antibody (the BS) was detected with a Digoxigenin-labeled detection antibody (1 h incubation). After washing, anti-Digoxigenin-POD (Roche; 11633716001) was added for 1 h followed by a washing step and addition of TMB substrate solution (Roche; 11484281001). Absorbance was read at 450 nm after stopping colour development with 1 M H2SO4. Statistical analysis was done by t-test comparing BS-BACE1 treated animals with control IgG dosed animals at respective time points using GraphPad Prism 6. Values plotted are means ± SEM (triplicates each). p values < 0.05 were considered significant: *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ns = non-significant (p > 0.05).
5.1. Newly Described Data Sets Availability
Coordinates and structure factors described in this work have been deposited in Protein Data Bank under accession codes 5MBW (Pep#3), 5MCO (GRL-8234/Pep#6) and 5MCQ (Pep#16).
6. Results
6.1. Design of Lipidated Active Site BACE1 Peptide Inhibitors
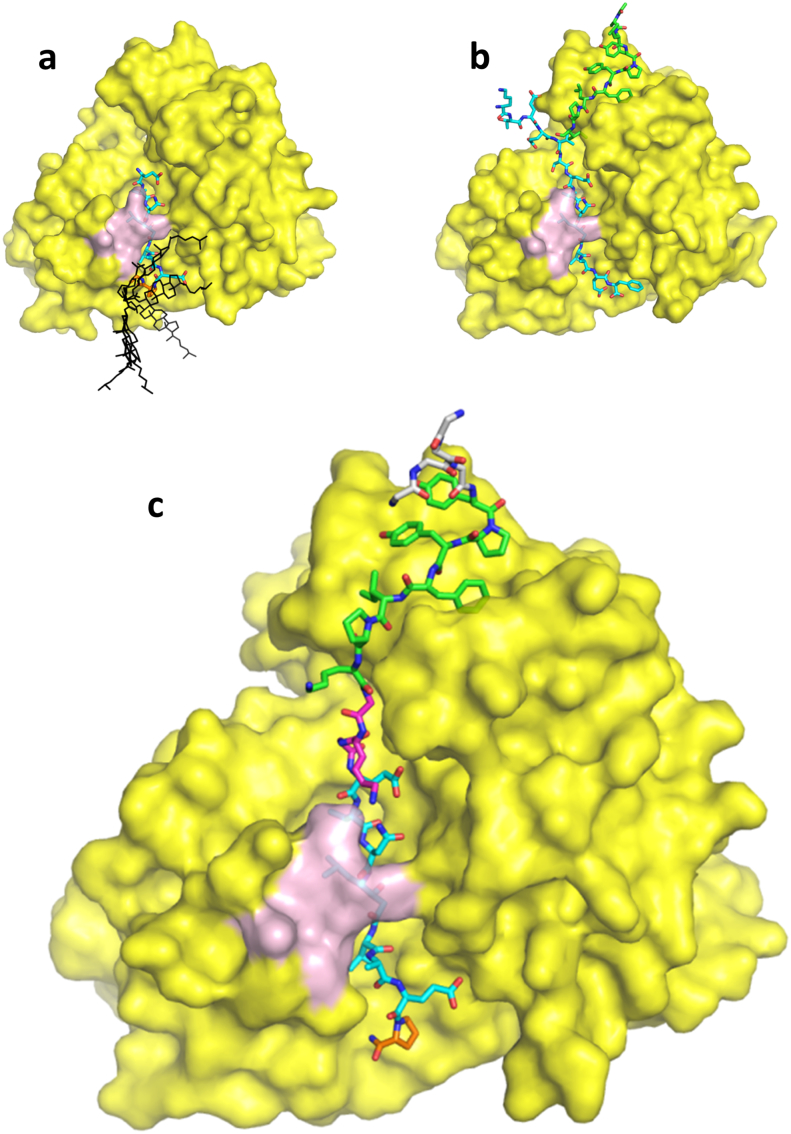
For effective and specific inhibition of BACE1, knowledge regarding the structure and function of BACE1 is essential. BACE1 inhibitory peptides based on the statine transition state mimetic (Gruninger-Leitch et al., 2002; Tung et al., 2002) were C-terminally modified with lysine. The Lys-amino side chain was acylated with palmitoyl (palm) or coupled to cholesterol (chol) via a succinate linker, as an ester bond or via an ether bond. Highly potent peptides were identified when screening in a cellular Aβ lowering assay using a cell-line transfected with human wild-type APP cDNA (Table 1 and Supplement Table 1–5). Without the lipid moiety all active site inhibitors were inactive in the cell assay. The X-ray crystal structure of BACE1 ectodomain was determined in complex with C-terminally modified Pep#3 at 2.95 Å (Fig. 1a). The peptide is spanning the BACE1 active site as expected. The D-Pro residue is inducing a turn in the inhibitor peptide structure so that its C-terminal lysine residue is positioned on the backside of the flexible BACE1 flap region (amino acids 131-161). The lysine side chain, is only poorly resolved and the conjugated chol molecule is not visible in the experimental electron density maps. Therefore, it can be assumed that the conjugated chol does not contribute to the binding affinity by direct interactions with the BACE1 ectodomain but instead promote the local concentration at the cell surface in favor of BACE1 inhibition. Similar findings have previously been described by linking a BACE1 inhibitor peptide to a sterol moiety through a long polyglycol linker (Rajendran et al., 2008). The C-terminal lysine residue of Pep#3 is positioned adjacent to the BACE1 ectodomain C-terminus, from where the full-length protein continues with a membrane anchoring α-helix. (Fig. 1a). This explain why a long linker is not necessary as the chol structure is presented and inserted into the plasma membrane by the already lipid surface-positioned BACE1 ectodomain (see also Fig. 3b).
Fig. 1.
Crystal structures of BACE1 ectodomain in complex with peptide inhibitors. The molecular surface of the BACE1 ectodomain is displayed in yellow, its flap region (amino acids 131–136) indicated pink. Peptide inhibitors are shown in sticks representation. (a) BACE1 in complex with the cholesterol conjugated Pep#3 (cyan). The cholesterol is not resolved in the electron density map. In order to visualize the mobility of the cholesterol lipid anchor several representative conformers are depicted (black). (b) BACE1 in complex with the exosite Pep#6 (green) and the active site inhibitor GRL-8234 (omitted for clarity). Superimposed in cyan is the APP derived active site inhibitor P10-P4′ StatVal (Turner et al., 2005). (c) BACE1 in complex with the dual, active and exosite binding peptide inhibitor Pep#16. The functionalities of the peptide from N- to C-terminus are sortase recognition motif (grey), exosite binding (green), linker (magenta), active site binding (cyan) and lipid anchor attachment (orange).
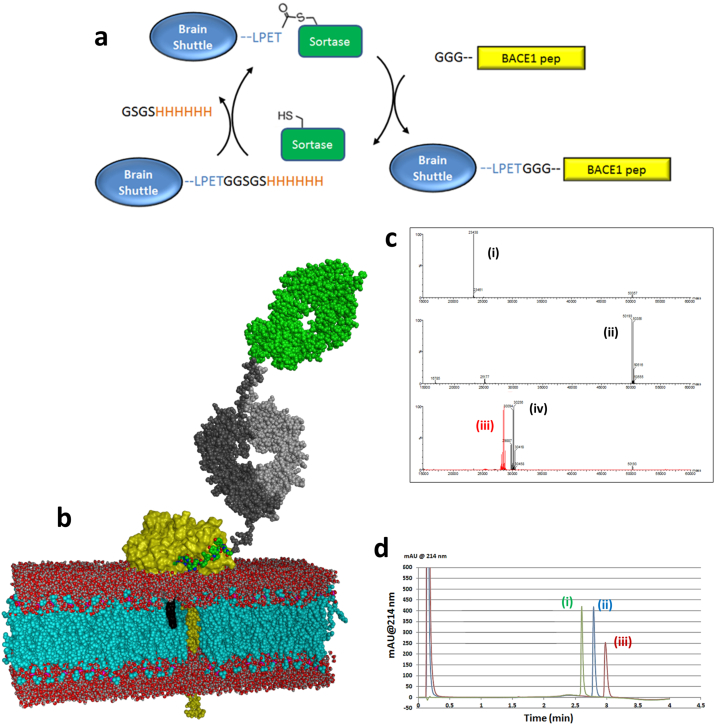
Fig. 3.
Conjugation of BACE1 peptide inhibitors to the Brain Shuttle. (a) Schematic overview of the conjugation of BACE1 peptide inhibitors to the Brain Shuttle using sortase coupling. (b) Model of the Brain Shuttle peptide conjugate binding to BACE1 at the plasma membrane. Brain shuttle module (Fab) in green, Fc fragment in grey, BACE1 in yellow, BACE1 peptide inhibitor in green and the phospholipid bilayer in red/blue. The cholesterol anchor is shown with a black surface, the modelled BACE1 transmembrane helix is depicted with a yellow space-filling atom representation. A part of the membrane has been removed to display the transmembrane portions. (c) Spectra from mass spectrometry. Peak: (i) CL/VL, (calc. mass 23,438 Da); (ii) CH3/CH2/CL1/CL/VL (calc. mass 50,191 Da for G0F-glycosylated chain, 50,353 Da for G1F), (iii) CH3/CH2/CL1 (calc. mass 28,447 Da for G0F-glycosylated chain, 28,609 Da for G1F) and (iv) CH3/CH2/CL1 – BACE1chol (calc. mass 30,095 Da for G0F-glycosylated chain, 50,257 Da for G1F). (d) RP-HPLC profiles showing exemplary the purity of the BACE1chol. Peak; (i) Brain Shuttle, (ii) Brain Shuttle-BACE1chol and (iii) BACE1chol.
6.2. BACE1 Exosite Peptide Structure and Optimization of Dual-Site Inhibitors
The X-ray crystal structure of a ternary complex between BACE1 ectodomain and the previously described exosite binding peptide Pep#6 (Kornacker et al., 2005) and an active site inhibitor GRL-8234(Chang et al., 2011) was determined at 2.5 Å resolution (Supplement Fig. 4 and Table 3). The central Tyr-Pro-Tyr-Phe-Leu-Pro-Ile sequence of exosite Pep#6 binds around the beta strand forming BACE1 residues 327-332. The N-terminal Ala-Leu of Pep#6 is solvent exposed suggesting that no additional spacer is required for N-terminal fusion. The C-terminal Ser-Ala-Lys sequence of Pep#6 is not resolved in the ternary complex crystal structure. The identified exosite is distinct from the pocket occupied by an APP fragment (P10-P4′StatVal) (Turner et al., 2005) representing the natural substrate (Fig. 1b). Superposition of the BACE1 crystal structures with the active site Pep#3 (chol-active site) (Fig. 1a) and the exosite peptide Pep#6 (Fig. 1b) shows a 12 Å distance between the Cα atoms of the C-terminal Ile of Pep#6 and the N-terminal Glu of Pep#3. Consequently, active-site/exosite fusion peptides where designed with 2–5 amino acid linkers, as well as n-alkyl and PEG linkers of equivalent length. First, linker length was investigated and a minimal length was required for optimal activity. In the glycine (Gly) series a triple glycine was optimal and no further improvement was evident by extending the oligo Gly chain (Table 1 and Supplement Tables 1–5). In the alkyl series a minimum C5 spacer gave the strongest inhibition. Cell potency in the sub-nanomolar range was also found for the oligoPEG series with both PEG(3) and PEG(4) spacers (Supplement Tables 1–5). Poor aqueous solubility of the first set of dual-site inhibitors led to significant aggregation as determined by analytical ultra-centrifugation (AUC) experiments. In the lipid modified series, a d-lysine scan was performed along with the oligo Gly linker to identify better soluble but still potent inhibitors such as Pep#9, Pep#10 and Pep#11 (Table 1). The d-lysine isomer was chosen for potential protease stability. Cooperative binding at both active- and exosites also translated to good cellular activity in the absence of lipid functionality. Cellular activity was much dependent on an optimized sequence, dual-site inhibitor. For the oligo Gly linker, the sequence specific incorporation of two DLys resulting in peptide Pep#12 was tolerated, whereas using DLys in combination with an oligo PEG linker as in Pep#13 led to loss of potency in the Aβ lowering cell assay (Table 1).
Table 3.
Statistics for X-ray data processing and model refinement.
| Lipidated peptide complex (Pep#3) | Ternary complex (GRL-8234, Pep#6a) | Fusion peptide complex (Pep#16) | |
|---|---|---|---|
| PDB accession number | 5MBW | 5MCO | 5MCQ |
| Data processinga | |||
| Space group | F23 | P3221 | P3221 |
| Unit cell axes [Å] | 206.8/206.8/206.8 | 101.5/101.5/117.2 | 101.9/101.9/116.4 |
| Resolution limits [Å] | 47.45–2.95 (3.13–2.95) | 48.74–2.49 (2.62–2.49) | 48.58–1.82 (1.91–1.82) |
| Completeness [%] | 100.0 (100.0) | 100.0 (100.0) | 99.9 (99.9) |
| I/σ(I) | 10.2 (1.4) | 9.9 (1.9) | 16.7 (1.3) |
| Multiplicity | 20.2 (21.0) | 9.6 (8.9) | 10.2 (10.1) |
| Refinement | |||
| R/Rfree [%] | 16.3/21.6 | 17.5/22.9 | 17.8/22.4 |
| Rmsd bond length [Å] | 0.013 | 0.019 | 0.020 |
| Rmsd bond angles [°] | 1.7 | 2.1 | 1.9 |
Number in parenthesis are values for the highest of ten resolution shells.
6.3. Crystal Structure of BACE1 with Dual Site Lipid-Free Inhibitory Peptide
The X-ray crystal structure of BACE1 ectodomain in complex with the active and exosite binding Pep#16 (Fig. 1c) was determined. The active site binding part of the peptide inhibitor superimposes very well with the active site only binder Pep#3. Equally, the exosite binding Tyr-Pro-Tyr-Phe-Leu-Pro sequence superimposes well with the exosite only binder Pep#6. The N-terminal Gly-Gly-Gly sortase recognition motif of Pep#16 is solvent exposed and at least two conformations coexist. The exosite/linker sequence DLys(1)-Gly-DLys(2)-Gly is well resolved with the exception of the solvent exposed side chain of the exosite DLys(1). The linker DLys(2) side chain is forming a water-bridged H-bond with the side chain of the BACE1 flap residue Gln134. The largely extended conformation of the linker region of Pep#16 provides a rationale for the observed minimal linker requirement of three amino acids. Reduction of the linker lengths to two amino acids is no longer sufficient to bridge the two BACE1 binding sites resulting in loss of exosite binding. New syntheses incorporating N-terminal triple Gly were performed and three representative candidates Pep#14, Pep#15 and Pep#16 were prepared with good cellular activity (Table 1). Having identified three sub-classes of dual site inhibitors i) chol-, ii) palmitoyl, iii) lipid-free, the next steps towards BS ligation were undertaken.
6.4. BACE1 Selectivity and High potency is Achievable with Dual-Site Inhibitory Peptides
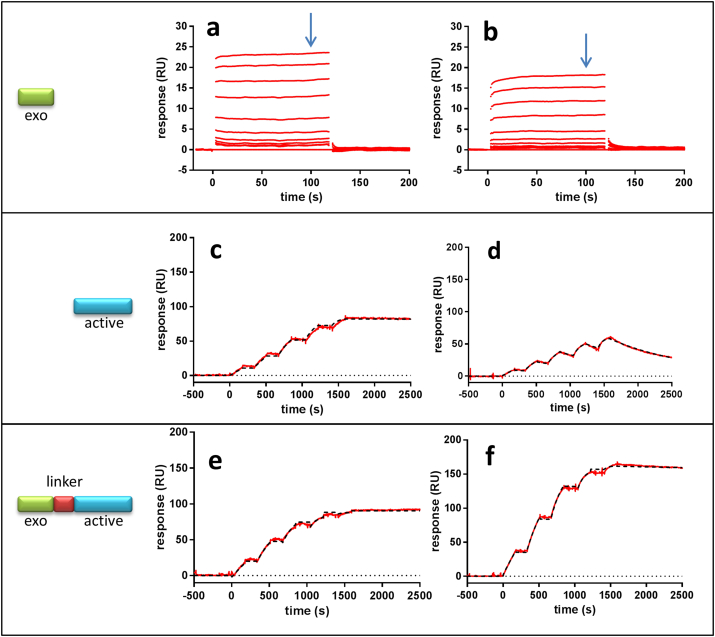
The crystal structure of BACE1 indicates that the proteolytic site is largely similar to most human aspartic proteases (Hong et al., 2000). These findings suggest that potential cross-inhibition of BACE1 inhibitors with other crucial aspartic proteases need to be considered. Among these, cathepsin D and E and especially BACE2 have received the most attention. In order to investigate possibilities for selectivity in different regions of the BACE1 structure we compared different parts of a dual BACE1 inhibitor with the most closely and important secretase, BACE2. BACE2 shares 45% amino acid identity with BACE1 (Farzan et al., 2000). Since all BACE1 small molecule inhibitors currently under clinical evaluation also block BACE2, it is relevant to investigate if our dual site peptide inhibitors possess a selectivity profile between BACE1 and BACE2. Surface plasmon resonance (SPR) based assay was used where either BACE1 or BACE2 was the immobilized target. The exosite peptide (Pep#6) used in this study does not discriminate between the two secretases (Fig. 2a and b). However, the SPR results clearly showed that high selectivity towards BACE1 in the active site is achievable (Fig. 2c and d) with Pep#1. When the exosite peptide is fused to the selective active site peptide (Pep#1) the potency is enhanced for both secretases (Fig. 2e and f), but the selectivity is likely maintained even though the precise binding constant was not possible to establish due to their high potency driven by slow dissociation rate (Pep#12). The binding constants from the SPR experiments are shown in Table 2. Taken together, these data illustrate the possibility to generate selective BACE1 versus BACE2 peptide inhibitors, at least within the active site but potentially also by exploring new chemistry for the exosite.
Fig. 2.
SPR binding data demonstrate the influence of a dual site inhibitor and BACE1 selectivity. Binding curves (a, c and e) on the BACE1 protein surface and (b, d and f) on BACE2 protein. Binding curves for exosite Pep#6 (a and b) monitored in a multiple cycle kinetic experiment up to 50 μM. Binding curves for active-site Pep#1 (c and d) monitored in a single cycle kinetic titration experiment up to 200 nM with dilution factor of 2 (red). Binding curves for dual-site inhibitor Pep#12 (e and f) monitored in a single cycle kinetic titration experiment up to 1000 nM with dilution factor of 2 (red). Binding data (c-f) were superimposed with mathematically calculated binding curves describing one-to-one interaction (black). The exosite peptide (a and b) binds equally well to BACE1 and BACE2. The affinity of exosite peptide was calculated in equilibrium analysis (Suppl. Fig. 2). The active site peptide (c and d) binds stronger to BACE1 as do the dual site peptide inhibitor (e and f). The binding data and kinetic parameters are shown in Table 2.
Table 2.
SPR binding data and kinetic parameters for peptides monitored over BACE1 and BACE2 protein surfaces.
| Peptide | Protein | kon (1/Ms) | koff (1/s) | KD (M) | Selectivity for BACE1b |
|---|---|---|---|---|---|
| Exo-Pep#6 | BACE1 | 2.5·10− 5 | |||
| BACE2 | 6.6·10− 5 | ||||
| Active-Pep#1 | BACE1 | 2.5–4.0·103 | < 1·10− 6 | < 1·10− 9 | |
| BACE2 | 2.7–3.1·103 | 7.8–8.3·10− 4 | 2.5–3.1·10− 7 | > 280 | |
| Active-exo-Pep#12 | BACE1 | 1.1–1.3·105 | < 1·10− 6 | < 1·10− 11 | |
| BACE1 | 1.1–1-3·105 | 1.2–2.5·10− 5 | 1.1–2.0·10− 10 | > 15a |
As slow dissociation rate constant (koff) monitored for this interaction is on the limit of the instrument resolution the effective affinity and thus selectivity might be significantly higher.
Selectivity for BACE1 calculated as ratio KD(BACE2)/KD(BACE1).
6.5. Ligation of BACE1 Peptide Inhibitors to the Brain Shuttle
As the BBB blocks most molecules from entering the brain including these types of BACE1 peptide inhibitors, fusions between the BS and the BACE1 peptide inhibitors was investigated to allow BBB crossing and substantial brain exposure. For this we used an enzyme-based conjugation technique which relies on Staphylococcus aureus Sortase A (SrtA) (Rashidian et al., 2013). In order to enable SrtA coupling of N-terminal triple Gly modified BACE-1 inhibitor peptides to the BS antibody and subsequent purification of the conjugates, the short heavy chain was extended by a small linker, a SrtA recognition sequence, and a 6xHis-tag, LPETGGSGSHHHHHH. Triple Gly modified peptides pep#14-16 were prepared and determined to be potent BACE1 inhibitors in the cellular Aβ lowering assay (Table 1). The schematic SrtA reaction is illustrated in Fig. 3a, where SrtA mediate the site-specific C-terminal BS labeling by first cleaving the threonine-glycine bond followed by the N-terminally engineered series of Gly residues resolving the intermediate and thus regenerating the active site cysteine on the SrtA and conjugating the peptide bond to the N-terminus of the BACE1 peptide inhibitors. The 6xHis-tag was used to remove any unreacted BS construct as this is still attached to the BS construct. Based on the X-ray structures (Fig. 1) we hypothesized that the complete BS-BACE1 constructs would then be free to engage with the BACE1 ectodomain on the cell surface (Fig. 3b) by inserting the dual inhibitor within the BACE1 structure and positioning the lipid moiety into the lipid layer. The completion and specificity of the SrtA reaction was confirmed by LC/MS traces (Fig. 3c). Since the reaction was carried out with an excess of SrtA we further purified the products obtained from the reaction by RP-HPLC (Fig. 3d). Homogenity and monodispersity of the BS-BACE-1 preparations were verified by analytical ultracentrifugation. Successful conjugation of the BACE1 peptide inhibitors to the BS allowed us to explore their properties both in vitro in cells and in vivo in the periphery and within the CNS compartment.
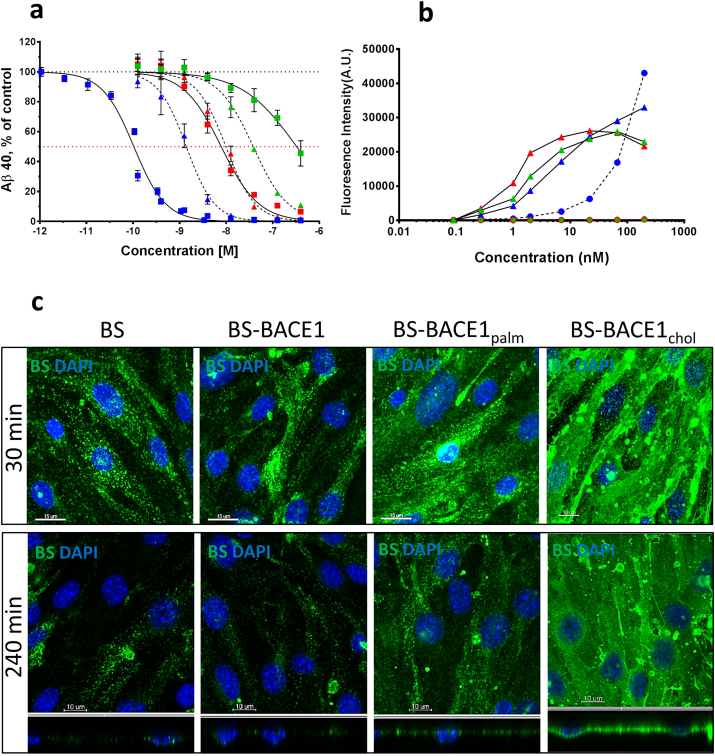
6.6. In Vitro Cellular Potency of BS-BACE1 Peptide Constructs and the Importance of Lipidation
The six compounds, free peptides or BS-conjugated pep#14-16, taken further into in vivo testing were investigated in a cellular system for BACE1 inhibition. Again, we used the HEK293 heterologous system where the cells are transfected with wild-type human APP for measuring Aβ inhibition. All free BACE1 inhibitory peptides (Pep#14, Pep#15 and Pep#16) and corresponding BS conjugated constructs blocked production of Aβ40 (Fig. 4a). The lipidated BACE1 peptides (Pep#14 and Pep#15) were more potent in the cellular system illustrating the role the lipid play in membrane anchoring as suggested by the X-ray structure (Fig. 1a). The potency of BACE1 (Pep#16) and BACE1chol peptide (Pep#14) was decreased and increased, respectively, when linked to the BS. This substantial improvement in BACE1chol peptide potency when fused to the BS is unclear, but could be explained by improved solubility when attached to the large BS protein. To investigate the lipid effect in binding to the cells we measured binding of the BS-BACE1 constructs to cells with and without TfR expression (Fig. 4b). As expected, all the constructs bound to cells which express TfR via the BS but only the BS-BACE1chol construct showed substantial interaction with cells lacking TfR expression. This is likely due to the interaction of the cholesterol moiety with the cellular lipid layer and in particular lipid rafts, as it has been shown that cholesterol is a critical structural component of lipid rafts (Brown and London, 1998). To further analyze these findings we performed immunocytochemistry on the three BS-BACE1 constructs and compared these to the non-conjugated BS. We could confirm that the BS-BACE1chol construct has the highest tendency to interact with the cells (Fig. 4c). In contrary, BS-BACE1palm (Pep#15) and the non-conjugated BS were taken up into intracellular vesicles in a time dependent fashion.
Fig. 4.
Aβ inhibition, TfR binding, and cell uptake of BACE1 peptide inhibitors and BS-BACE1 peptide conjugates in vitro. (a) Aβ production inhibition in HEK293 cells stably transfected with wild-type human APP (APP695) cDNA. Cells were incubated with the different compounds for 16 h (n = 3). Both free and BS-conjugated BACE1 peptide inhibitors blocked Aβ production. Pep#16 BACE1 (green triangle) Pep#15 BACE1palm (red triangle) Pep#14 BACE1chol (blue triangle) BS-BACE1 (green square) BS-BACE1palm (red square) BS-BACE1chol (blue square). The BS-BACE1chol peptide was most potent and is enhanced by the attached to the BS. (b) Cell binding measured by FACS on cells with and without TfR expression. BA/F3 cells (triangles) expressing TfR showed binding of BS-BACE1 (green), BS-BACE1palm (red) and BS-BACE1chol (blue). HEK cells (circle) with no TfR expression showed no binding of BS-BACE1 (green) and BS-BACE1palm (red), but binding of BS-BACE1chol (blue). (c) Uptake and localization of the BS 30 and 240 min after a single dose of 100 nM was assessed in bEnd.3 mouse brain endothelial cells. Unconjugated BS showed time dependent cell uptake, as did BS-BACE1 and BS-BACE1palm. Strong cell surface binding was seen for the BS-BACE1chol construct. X–Y axis (square) and X–Z axis (rectangle).
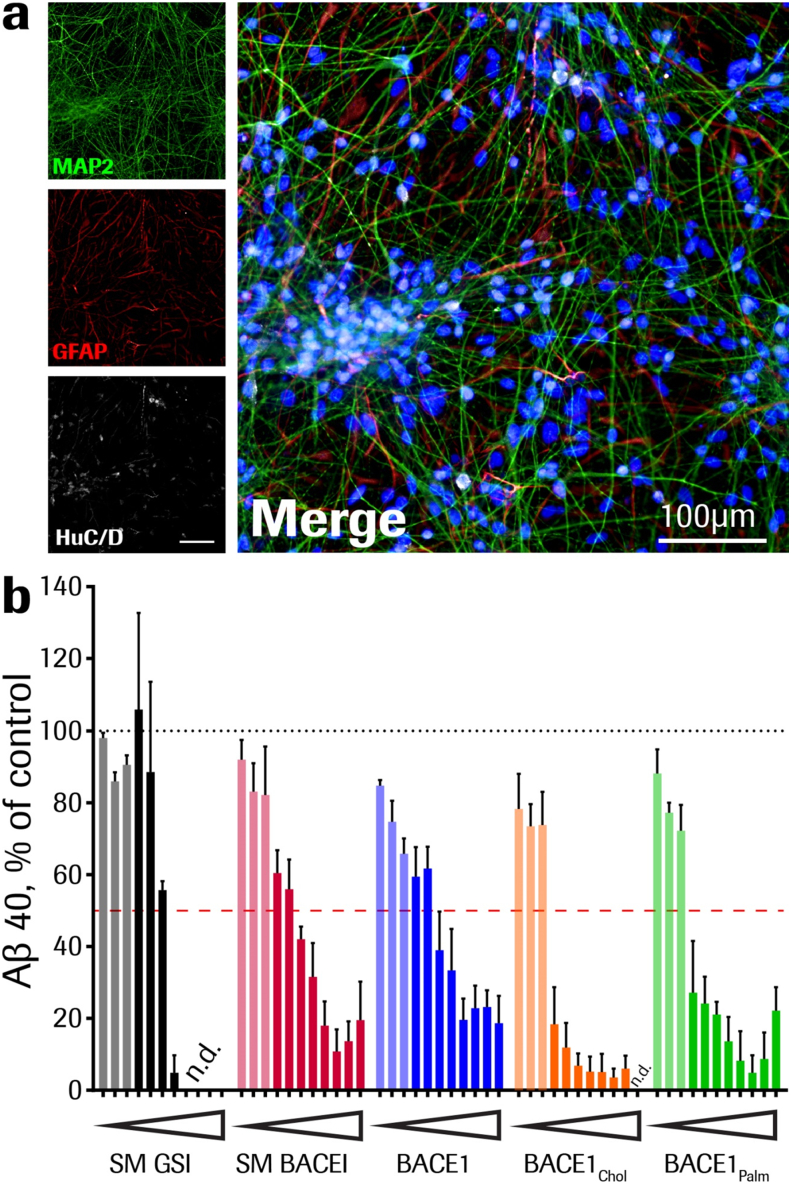
We also investigated the BACE1 inhibition profile for the free BACE1 inhibitory peptides (Pep#14, Pep#15 and Pep#16) in human stem-cell derived neurons. The neuronal identity was verified by staining for markers like Map2 and HuC/D (Fig. 5a). In this experiment we included a small molecule BACE1 inhibitor (SM BACE1) and a small molecule γ-secretase inhibitor (SM GSI) as reference compounds. All the BACE1 peptide inhibitors was able to reduce the Aβ production (Fig. 5b). As for the HEK293 heterologous system the lipidated BACE1 peptides (Pep#14 and Pep#15) were more potent in this more native cellular system illustrating the role the lipid play in membrane anchoring also on neurons. Notably, the SM GSI completely blocked Aβ production while the SM and peptide inhibitors BACE1 was unable to abolish the production completely.
Fig. 5.
Aβ inhibition by BACE1 peptide inhibitors and small molecule β- and γ-secretase inhibitors in vitro. (a) Human stem-cell derived neurons were cultured for six weeks and neuronal identity verified by staining for markers like Map2 (green) and HuC/D (white). The level of astrocytes in fully differentiated cells was verified with anti-GFAP (red) labeling. As reference, cell nuclei are stained with DAPI (blue). (b) Aβ production inhibition in human stem cell-derived neurons. Fully differentiated cells were incubated with the different compounds for 48 h at doses ranging from 10 to 0.001 μM, decreasing in half-logarithmic steps. Experimental setup included cells from three independent differentiations and two technical replicates each dose (0.128–10,000 nM; solid colors), and cells from one differentiation and two technical replicates each dose (0.001024–0.0256 nM; shaded colors). Both small molecule and BACE1 peptide inhibitors blocked Aβ production to a comparable extend, SM GSI (black bars), SM BACE1 (red bars), Pep#16 BACE1 (blue bars), Pep#15 BACE1palm (green bars) and Pep#14 BACE1chol (orange bars). All % values represent Aβ40 level changes in treated cells relative to respective vehicle-treated cells (means ± SEM). n.d. signal below detection limit.
6.7. In Vivo Dose-response Shows Maximum Achievable Aβ Reduction of Approximately 50% in Plasma
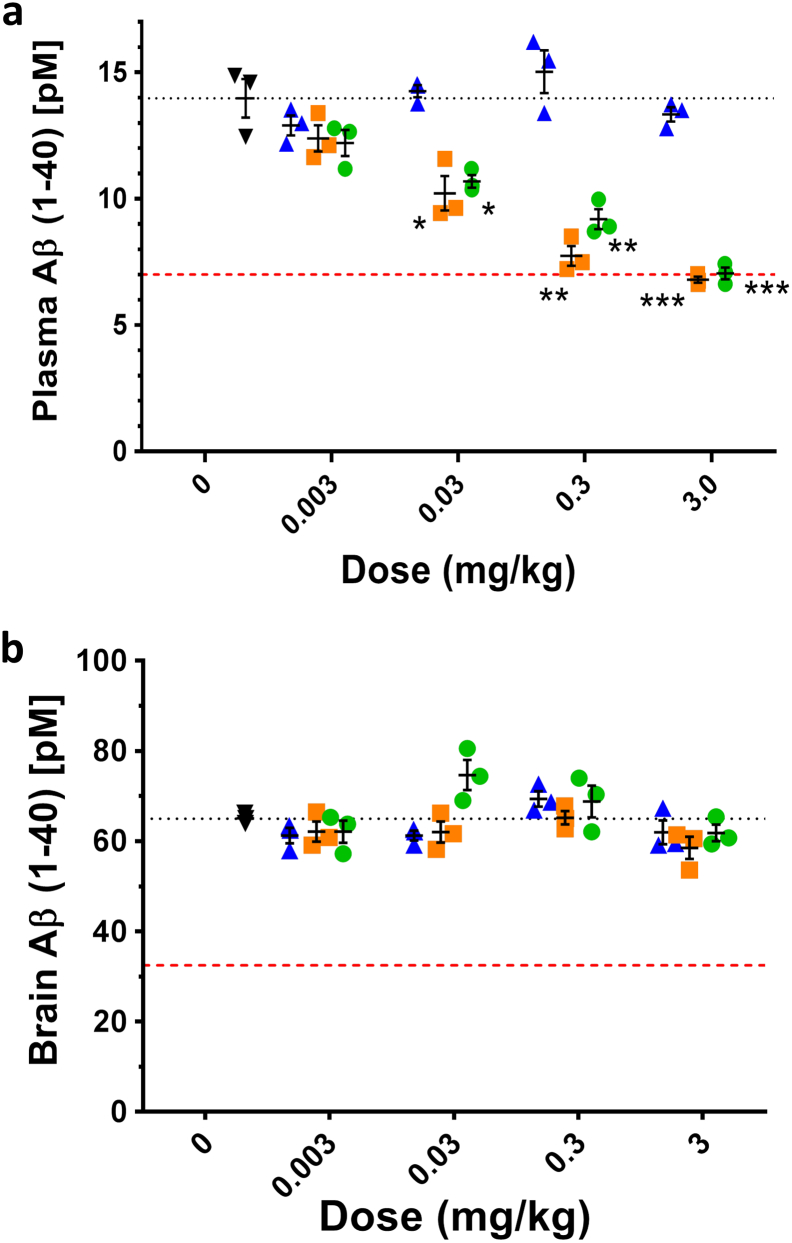
To investigate the degree of achievable Aβ reduction in vivo, uncoupled BACE1 peptide inhibitors were intravenously injected into wild type and plasma and brain samples collected 5 h post dose (Fig. 6). The dose response ranged from 0.003 mg/kg to 3 mg/kg. In plasma, injection of 3 mg/kg inhibitor solution resulted in the highest Aβ level reduction observed, namely 51% for BACE1chol (Pep#14) and 50% for BACE1palm (Pep#15) construct. This seems to be the maximum Aβ inhibition for the BACE1 specific peptide inhibitors used in this study as it reaches a plateau around 50% reduction in Aβ at the maximal concentration. Similar effects have been shown with a specific anti-BACE1 mAb and can be attributed to the finding that BACE1 only accounts for partial Aβ40 production in the periphery based on BACE1−/− mouse data (Atwal et al., 2011). This is different using the HEK293 heterologous cells system or the more physiological human stem-cell derived neuron cells where complete Aβ40 inhibition is possible (Figs. 4a and 5b). The non-lipidated BACE1 peptide possess no in vivo Aβ40 inhibition, likely due to its lower potency and/or faster plasma clearance (Fig. 6a). In contrast, the two lipidated BACE1 peptide inhibitors showed a dose-response in blocking Aβ production. Notably, there was no detectable BACE1 inhibition in brain (Fig. 6b), suggesting that the BACE1 peptides are not able to penetrate into the CNS compartment. The 50% Aβ40 inhibition data in wild type mice demonstrates the maximum Aβ inhibition achievable in vivo with these active-exosite BACE1 inhibitors. This finding is confirmed by an independent study conducted in wild-type rats, where again 50% reduction was accomplished this time with active site inhibitor peptides in rats at a dose of 5 mg/kg (Suppl. Fig. 1). The pharmacokinetic profiles illustrate the prolonged plasma half-life through the lipidation either by palm or chol. Comparable levels of Aβ reduction in wild type mice using an anti-BACE1 specific antibody have recently been described (Lu et al., 2016). It is not clear if this incomplete Aβ inhibition is due to excellent selectivity profile that these dual active/exo site inhibitors possess and if BACE1 accounts for only a fraction of the Aβ production, with the remainder coming from another secretase (Atwal et al., 2011), or if these peptide inhibitors only have access to certain tissues/cells/organelles leaving certain compartments free from BACE1 inhibition.
Fig. 6.
BACE1 peptide inhibitors inhibits peripheral but not central Aβ production after systemic dosing. (a) Dose-dependent inhibition in plasma for the two lipid conjugated compounds BACE1palm (green circle) and BACE1chol (orange square) but not for the non-lipidated BACE1 peptide inhibitor (blue triangle). Black dotted line indicates plasma Aβ (pM) of vehicle control (black tringle). (b) None of the peptide inhibitors blocked Aβ production in the CNS compartment. Colour-coding as in (a). Red dashed line represents 50% Aβ inhibition. Values plotted are means ± SEM (n = 3). *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 (t-test, compared to vehicle–dosed animals).
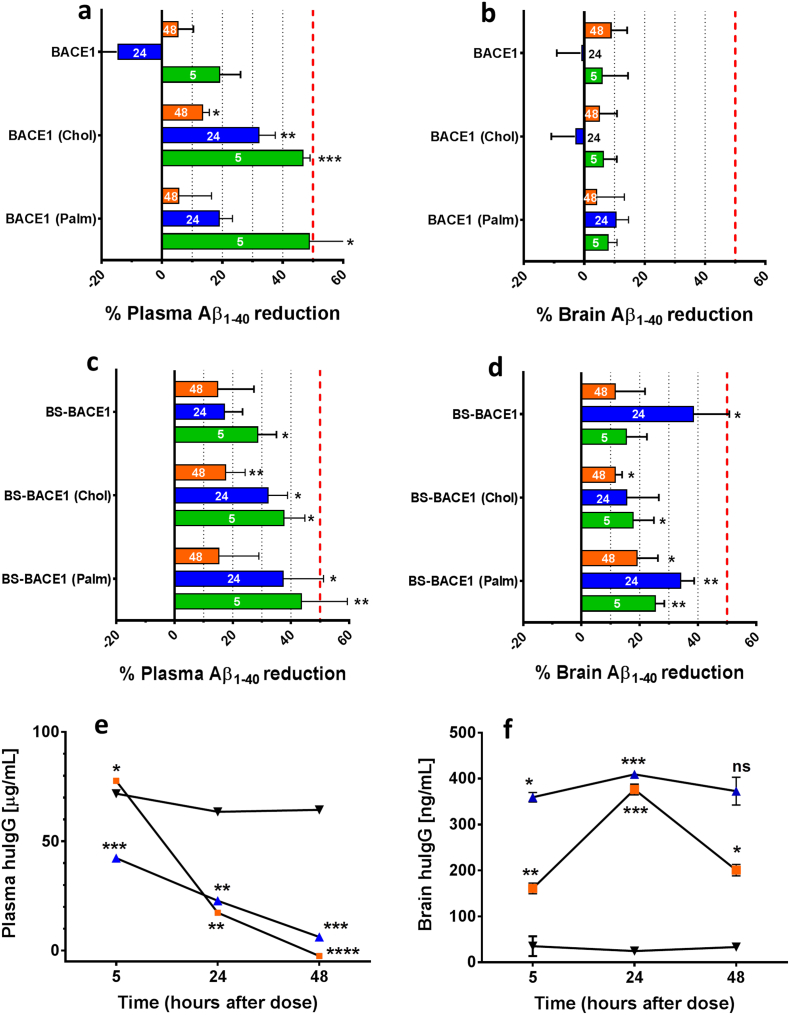
6.8. Brain-Shuttle Coupled BACE1 Inhibitors Decrease Aβ levels in Plasma and Brain
To determine how effective the BS-conjugated and non-conjugated BACE1 peptide inhibitors are at reducing Aβ production in plasma and CNS in vivo, we first conducted a single-dose experiment (0.3 mg/kg) in wild-type C57BL/6 mice evaluating plasma and brain Aβ reduction (Fig. 7a, b). Again, we see a clear Aβ reduction in plasma for the two lipidated non-conjugated BACE inhibitors, and this reduction was time-dependent. However, in the same animals no detectable brain Aβ reduction could be detected at any time point. Unfortunately we were not able to implement a PK assay to measure the free peptide concentration in plasma and brain. For comparison we tested BS-BACE1, BS-BACE1palm and BS-BACE1chol in vivo by intravenous application in mice. A single dose of 10 mg/kg of the BS-BACE1 constructs were applied and brain and plasma samples were collected at five, 24 and 48 h post injection (Fig. 7c and d). Conjugation of the BACE1 inhibitors to the BS resulted in comparable potency in plasma as the unconjugated counterparts, except for the non-lipidated peptide variant where the BS marginally improved the potency in plasma. Importantly, in the brain in contrast to plasma, Aβ reduction could only be achieved after conjugation of the BACE1 inhibitors to the BS (Fig. 7d). The level of Aβ reduction was both highly significant and substantial, as the maximal inhibition in vivo with this BACE1 peptide inhibitors is about 50% (Fig. 6a). An independent repetition of the 24 h time point confirmed the observed Aβ reduction in plasma and brain parenchyma after BS-BACEpalm systemic administration (n = 5; data not shown). Based on the positive CNS inhibition data with the BS-coupled constructs, we set out to examine the pharmacokinetic (PK) profiles in mice up to 48 h. When we dosed 10 mg/mg of the BS-BACE1, BS-BACE1chol, and as a comparison a control mAb with similar size as the BS, we could show that both BS-BACE1 constructs were cleared much faster than the control mAb due TfR target mediated drug deposition (TMDD) (Fig. 7e). BS-BACE1chol showed higher concentration initially at 5 h than BS-BACE1, possibly attributed to the chol moiety reducing plasma clearance at earlier time points. The plasma AUC5-48h was 2.4–2.8 fold higher for the control mAb compare to the two BS-BACE1 constructs. However, the brain PK profile was very different (Fig. 7f). Both BS-BACE1 constructs showed significantly higher brain exposure and improvements in AUC5–48h compared to the control mAb, between 9.5 and 13.2 fold. The brain/plasma AUC ratio is 1.7% (BS-BACE1), 1.0% (BS-BACE1chol) and 0.04% (control mAb) illustrating the BBB transport capacity of the BS. Notably, the BS-BACE1 construct possesses a more rapid and prolonged brain exposure profile compare to BS-BACE1chol, suggesting that the cholesterol moiety influence the overall brain exposure profile.
Fig. 7.
Brain-Shuttle mediated transport of BACE1 inhibitors necessary for Aβ production inhibition in the brain. (a and b) Relevant Aβ reduction 5–48 h after systemic application seen with the non-conjugated BACE1 peptide inhibitors in plasma (a) but not in brain (b). The two lipidated constructs BACE1palm and BACE1chol showed higher potency than BACE1. (c and d) Aβ reduction 5–48 h after systemic application of BS-conjugated BACE1 peptide inhibitors in plasma (c) as well as in the brain (d). Plasma reduction was similar between comparable BS-coupled and un-coupled BACE1 inhibitors and reached 50% over 48 h. In brain the highest reduction was seen 24 h after application. Bars are colour coded, 5 h (green), 24 h (blue) and 48 h (orange) post i.v. dosing. (e and f) Plasma pharmacokinetics of BS-BACE1 (blue), BS-BACE1chol (orange) and a control IgG (black) were analyzed 5–48 h after single intravenous dose of 10 mg/kg. The concentration of BS-BACE1 and BS-BACE1chol in plasma was lower and in brain higher than for the non-BBB permeable control IgG. AUC5–48h (μg h/ml) in plasma was 47.0 (BS-BACE1), 55.2 (BS-BACEchol) and 131.5 (control IgG). AUC5–48h (ng h/ml) in brain was 775.4 (BS-BACE1), 557.5 (BS-BACEchol) and 58.9 (control IgG). Red dashed line represents 50% Aβ inhibition. Values plotted are means ± SEM (n = 3). *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 (t-test, compared to control IgG–dosed animals).
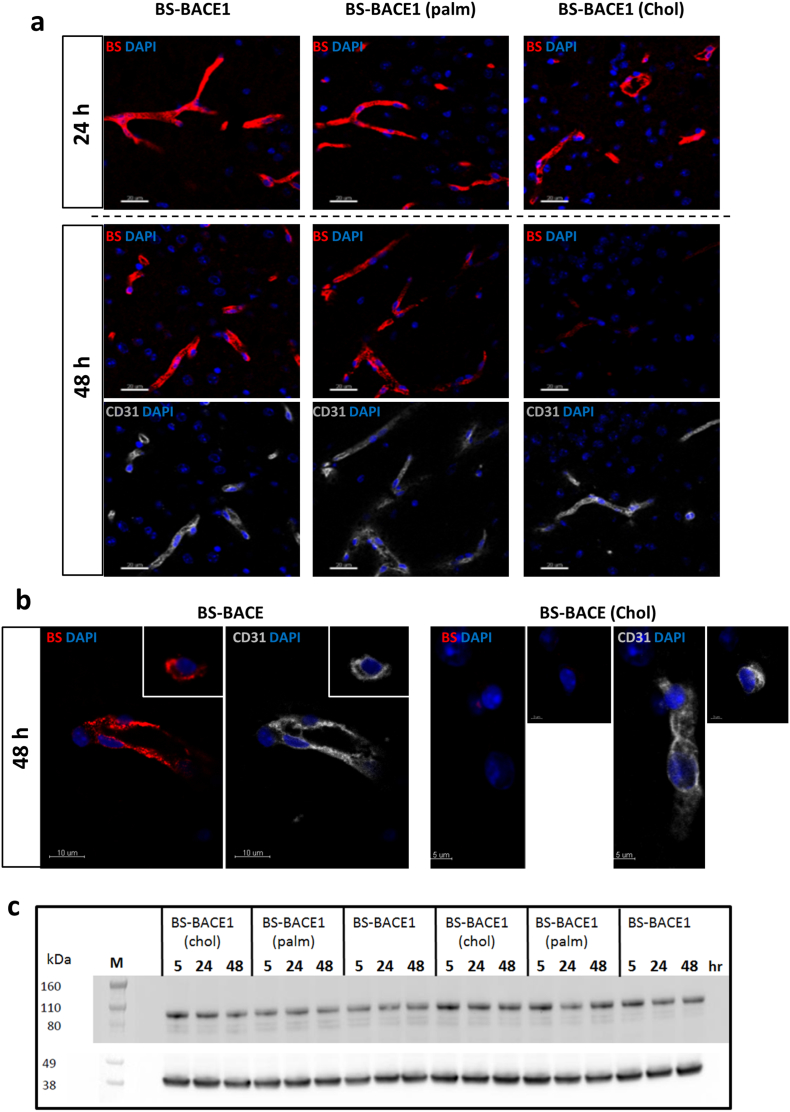
6.9. Lipidation with Cholesterol Change the Transport Kinetics Over the BBB Without Interfering with TfR Expression
In order to understand the difference in plasma and brain exposure for each BS-BACE1 construct we investigated the BS-mediated brain uptake and their distribution by immunohistochemistry. It is to be excpected that the different constructs should be detectable in brain capillaries if they enter the brain by crossing through the endothelial cells at the BBB. To address this, we compared the distribution of different BS-BACE1 constructs in brain tissue at different time points. Mice were dosed at 10 mg/kg and PBS-perfused after five, 24 and 48 h (Fig. 8a). Brain sections were stained with fluorescent labeled anti-human secondary IgG to visualize BS distribution. Notably, the staining profile for the BS-BACE1chol construct was in agreement with its brain Aβ reduction data and its brain PK profile. At 48 h post dose there was very low level of the BS-BACE1chol construct at the BBB. Representative high-magnification images further illustrate the different localization of the BS-BACE1chol construct compare to the non-lipidated BS-BACE1 construct (Fig. 8b). Furthermore, we also explored possible changes in the levels of TfR expression in the brain treated with the three constructs (Fig. 8c), but could not detect notable differences in TfR expression. Taken together these data suggests that certain lipids conjugated to a BS construct could influence uptake and also potentially the intracellular sorting within the brain endothelial cells at the BBB and thus influence the degree of CNS exposure.
Fig. 8.
Lipidation of the BACE1 inhibitor peptide impacts transport of the BS construct over the BBB (a and b) Localization of injected BS-BACE1, BS-BACE1palm, and BS-BACE1chol 24 and 48 h after single i.v. dose (each 10 mg/kg) relative to endothelial CD31 signal (grey). (a) BS-BACE1 and BACE-1palm were localized in mouse brain blood vessels 24 and 48 h post systemic application. Vascular localization of BS-BACE1chol was observed at 24 h but not after 48 h. (b) High-resolution images of the 48 h time point highlight differences in vascular (CD31) localization of BS-BACE1chol compared to BS-BACE1. (c) TfR expression in mouse brain 5–48 h after single intravenous dose (10 mg/kg) of BS-BACE1, BS-BACE1palm and BS-BACE1chol. TfR protein levels was stable (about 100 kDa) compared the control housekeeping gene β-actin (about 38 kDa).
7. Discussion
Over the past decade, substantial research efforts have been directed towards understanding BACE1 as a critical target for AD therapy. Our results illustrate how to avoid the restriction of BBB permeability in the BACE1 inhibitor design process, resulting in the discovery of cell and in vivo active lead series. Furthermore, using inhibitors with multiple mode of actions and interaction points with BACE1 opens up numerous options for designing BACE1 inhibitors with superior potency and selectivity. The lipidation strategy for membrane anchoring is an approach to enhance cellular activity, which we show using systemic administration translates into improved in vivo activity, both in plasma and in the CNS. We also provide structural data explaining how the lipid moiety attached to the BACE1 peptide inhibitor is positioned to interact favorable with the lipid cell membrane when the inhibitor is bound to the active site. Furthermore, although BACE1 peptide inhibitors have been developed which are active in a biochemical enzymatic assay, we show examples of peptides that are potent and very efficient in blocking Aβ production in a cellular assay. A key feature of these cellular active peptides is the dual active/exosite inhibition concept that leverages the simultaneous interaction with two different binding pockets on the BACE1 ectodomain. Interestingly, in the HEK293 heterologous cellular assay the BACE1 peptides completely inhibit the Aβ production while in vivo a maximum of 50% reduction was achieved. Similar data have been reported for a BACE1 bispecific antibody (Yu et al., 2011) and also for some non-peptidic BACE1 Inhibitors (May et al., 2011; Quartino et al., 2014). If this is due to BACE1 accounting for only partial Aβ production, with the remainder coming from another secretase or if these larger peptides and antibodies only have access to certain intracellular compartments where a portion of BACE1 is located (Ben Halima et al., 2016), remains to be investigated. It should be noted that complete inhibition of Aβ production was not achieved with a small molecule BACE1 inhibitor and the peptide inhibitors in the more physiological human stem-cell derived neurons, while the GSI complete block all Aβ production. BACE1 peptide inhibitors and the BS-conjugated constructs likely act in intracellular vesicles to inhibit BACE1-mediated processing of APP, an assumption based on previous studies that showed endocytosis as a necessary step in APP processing (Cirrito et al., 2008). Such a cellular compartmentalization of the BS-BACE1 constructs could potentially lead to improved APP substrate specificity. It has been shown that heterozygous BACE1 +/− mice, a model for 50% therapeutic inhibition of BACE1, exhibit only about 20% lowering of cerebral Aβ levels when crossed to APP transgenic mice (Laird et al., 2005, McConlogue et al., 2007). This suggests that likely > 50% BACE1 activity was inhibited by the BS-BACE1 peptide inhibitor to achieve the observed 50% reduction in Aβ levels. Nevertheless, a reduction of Aβ production by approximately 50% appears to be sufficient to rescue the cognitive decline in Tg2576 mice (Chang et al., 2011) and has been proposed as a good target for a therapeutic intervention strategy in humans (Vassar, 2014).
Most BBB penetrant small molecules inhibitors currently under development inhibit both BACE1 and BACE2. Therefore potential secondary effects of BACE2 inhibition will have to be taken into consideration (Evin, 2016). Our data illustrate the possibility to design highly potent BACE1 inhibitors with > 250-fold selectivity against BACE2. This is possible as the chemical space is not restrained to comply with the physicochemical boundary conditions for passive diffusion through the BBB. In this paper we have not focused on selectivity versus other proteases such as cathepsin D, but this also has to be addressed using dual site and lipid modified BACE1 peptide inhibitors linked to the BS.
One major obstacle for the treatment of neurodegenerative diseases like AD is that any effective therapeutic has to be able to cross the BBB. Therefore, BACE1 inhibitors traditionally have been designed as hydrophobic low molecular weight molecules with a reduced susceptibility to Pgp-mediated efflux at the BBB. To circumvent this, we conjugated our potent peptide inhibitors to the BS antibody fragment that targets endogenous TfR to cross the BBB. None of the BACE1 peptide inhibitors regardless of potency were active in CNS without linking them to the BS, thus suggest an important strategy to support and improve potency of CNS-targeting peptides. Furthermore, the duration of Aβ blockage was extended by the BS as the BS construct contains and IgG Fc portion which prolongs the plasma half-life through recycling by the neonatal FcRn. Consequently, administration BS-coupled therapeutics is predicted to be less frequent compared to a small molecule approach. Considering these factors, the advantage of combining drug modalities such as a BS antibody and peptides may become a promising treatment strategy.
These investigations successfully illustrate the concept of combining the brain transport technology BS with unique multifunctional BACE1 peptide inhibitors. We have shown that BACE1 can be targeted with highly potent and selective peptide inhibitors, both in the periphery and in the CNS, by reducing Aβ levels in vivo. New and alternative approaches to tackle promising targets like BACE1 are needed to further explore the therapeutic potential. Research must continue to improve understanding of the biology of BACE1 and the work presented here should be seen as a starting point for a concept that can be improved on many levels. Developing safe BACE1 inhibitory drugs remains challenging, despite the great promise shown in several ongoing clinical trials. In addition, our results illustrate that peptidomimetics may indeed be effective at targeting the CNS, and we therefore propose that promising CNS targets other than BACE1 may also be effectively targeted with peptides using the BS transport technology.
Acknowledgments
Acknowledgments
Authors thank Kersten Klar and Patrick Studer for peptide synthesis. Josiane Kohler for performing SPR binding experiment and Martine Stihle for crystallization. Michael Tischler for protein analytics, Johannes Erny and Siegfried Stolz for LC-MS analytics and Eric Kusznir for AUC measurements. Elvira da Silva for the derivation of neural progenitor cells from iPSCs. We would also like to thank our colleagues Alexander Bujotzek and Guy Georges from the Roche Innovation Center Munich for the model of the Brain Shuttle antibody. Part of the research leading to the iPSC-derived neurons' results has received support from the Innovative Medicines Initiative Joint Undertaking under grant agreement n° 115439, resources of which are composed of financial contribution from the European Union's Seventh Framework Programme (FP7/2007-2013) and EFPIA companies' in kind contribution. This publication reflects only the author's views and neither the IMI JU nor EFPIA nor the European Commission are liable for any use that may be made of the information contained therein.
Funding Sources
All funding was provided by the company F. Hoffmann-La Roche.
Conflict of Interest
All authors are under paid employment by the company F. Hoffmann-La Roche.
Author Contributions
E.K and P-O.F. planned the experiments. E.K. designed and synthesized compounds. N.R. performed all in vivo experiments. D.S. performed and coordinated all BS conjugation work. A.K. and J.B. performed protein crystallography work. W.G. performed molecular modeling, BS-BACE1pep/BACE1 structures and structural analysis. S.H. supervised and analyzed SPR binding data. J.N. performed the FACS experiments. A.R. performed analytical ultra-centrifugation (AUC) and MS experiments. C.C., C.P. and K.B. performed the human stem cell work. C.S. generated all cell inhibition data. D.S., A.G., J.E., J.H. and E.H. generated and analyzed the BS constructs. H. L. writing review and editing. E.K., N.R. and P-O.F. conceptualized and interpreted the data and drafted the manuscript.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2017.09.004.
Appendix A. Supplementary data
Supplementary material
References
- Atwal J.K., Chen Y., Chiu C., Mortensen D.L., Meilandt W.J., Liu Y., Heise C.E., Hoyte K., Luk W., Lu Y., Peng K., Wu P., Rouge L., Zhang Y., Lazarus R.A., Scearce-Levie K., Wang W., Wu Y., Tessier-Lavigne M., Watts R.J. A therapeutic antibody targeting BACE1 inhibits amyloid-beta production in vivo. Sci. Transl. Med. 2011;3:84ra43. doi: 10.1126/scitranslmed.3002254. [DOI] [PubMed] [Google Scholar]
- Ben Halima S., Mishra S., Raja K.M., Willem M., Baici A., Simons K., Brustle O., Koch P., Haass C., Caflisch A., Rajendran L. Specific inhibition of beta-secretase processing of the Alzheimer disease amyloid precursor protein. Cell Rep. 2016;14:2127–2141. doi: 10.1016/j.celrep.2016.01.076. [DOI] [PubMed] [Google Scholar]
- Boissart C., Poulet A., Georges P., Darville H., Julita E., Delorme R., Bourgeron T., Peschanski M., Benchoua A. Differentiation from human pluripotent stem cells of cortical neurons of the superficial layers amenable to psychiatric disease modeling and high-throughput drug screening. Transl. Psychiatry. 2013;3 doi: 10.1038/tp.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H., Braak E. Neuropathological staging of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Brockhaus M., Grunberg J., Rohrig S., Loetscher H., Wittenburg N., Baumeister R., Jacobsen H., Haass C. Caspase-mediated cleavage is not required for the activity of presenilins in amyloidogenesis and NOTCH signaling. Neuroreport. 1998;9:1481–1486. doi: 10.1097/00001756-199805110-00043. [DOI] [PubMed] [Google Scholar]
- Brown D.A., London E. Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol. 1998;14:111–136. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- Cai H., Wang Y., Mccarthy D., Wen H., Borchelt D.R., Price D.L., Wong P.C. BACE1 is the major beta-secretase for generation of Abeta peptides by neurons. Nat. Neurosci. 2001;4:233–234. doi: 10.1038/85064. [DOI] [PubMed] [Google Scholar]
- Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W.P., Huang X., Downs D., Cirrito J.R., Koelsch G., Holtzman D.M., Ghosh A.K., Tang J. Beta-secretase inhibitor GRL-8234 rescues age-related cognitive decline in APP transgenic mice. FASEB J. 2011;25:775–784. doi: 10.1096/fj.10-167213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirrito J.R., Kang J.E., Lee J., Stewart F.R., Verges D.K., Silverio L.M., Bu G., Mennerick S., Holtzman D.M. Endocytosis is required for synaptic activity-dependent release of amyloid-beta in vivo. Neuron. 2008;58:42–51. doi: 10.1016/j.neuron.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa V., Aigner S., Vukcevic M., Sauter E., Behr K., Ebeling M., Dunkley T., Friedlein A., Zoffmann S., Meyer C.A., Knoflach F., Lugert S., Patsch C., Fjeldskaar F., Chicha-Gaudimier L., Kiialainen A., Piraino P., Bedoucha M., Graf M., Jessberger S., Ghosh A., Bischofberger J., Jagasia R. mTORC1 inhibition corrects neurodevelopmental and synaptic alterations in a human stem cell model of tuberous sclerosis. Cell Rep. 2016;15:86–95. doi: 10.1016/j.celrep.2016.02.090. [DOI] [PubMed] [Google Scholar]
- Evin G. Future therapeutics in Alzheimer's disease: development status of BACE inhibitors. BioDrugs. 2016;30:173–194. doi: 10.1007/s40259-016-0168-3. [DOI] [PubMed] [Google Scholar]
- Farzan M., Schnitzler C.E., Vasilieva N., Leung D., Choe H. BACE2, a beta -secretase homolog, cleaves at the beta site and within the amyloid-beta region of the amyloid-beta precursor protein. Proc. Natl. Acad. Sci. U. S. A. 2000;97:9712–9717. doi: 10.1073/pnas.160115697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman J.B., Rubin J.B., Handrahan J.V., Connor J.R., Fine R.E. Receptor-mediated transcytosis of transferrin across the blood-brain barrier. J. Neurosci. Res. 1987;18:299–304. doi: 10.1002/jnr.490180206. [DOI] [PubMed] [Google Scholar]
- Freskgard P.O., Urich E. Antibody therapies in CNS diseases. Neuropharmacology. 2017 Jul 1;120:38–55. doi: 10.1016/j.neuropharm.2016.03.014. [DOI] [PubMed] [Google Scholar]
- Friden P.M., Walus L.R., Musso G.F., Taylor M.A., Malfroy B., Starzyk R.M. Anti-transferrin receptor antibody and antibody-drug conjugates cross the blood-brain barrier. Proc. Natl. Acad. Sci. U. S. A. 1991;88:4771–4775. doi: 10.1073/pnas.88.11.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A.K., Brindisi M., Tang J. Developing beta-secretase inhibitors for treatment of Alzheimer's disease. J. Neurochem. 2012;120(Suppl. 1):71–83. doi: 10.1111/j.1471-4159.2011.07476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruninger-Leitch F., Schlatter D., Kung E., Nelbock P., Dobeli H. Substrate and inhibitor profile of BACE (beta-secretase) and comparison with other mammalian aspartic proteases. J. Biol. Chem. 2002;277:4687–4693. doi: 10.1074/jbc.M109266200. [DOI] [PubMed] [Google Scholar]
- Hardy J.A., Higgins G.A. Alzheimer's disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- Hock C., Konietzko U., Streffer J.R., Tracy J., Signorell A., Muller-Tillmanns B., Lemke U., Henke K., Moritz E., Garcia E., Wollmer M.A., Umbricht D., de Quervain D.J., Hofmann M., Maddalena A., Papassotiropoulos A., Nitsch R.M. Antibodies against beta-amyloid slow cognitive decline in Alzheimer's disease. Neuron. 2003;38:547–554. doi: 10.1016/s0896-6273(03)00294-0. [DOI] [PubMed] [Google Scholar]
- Hong L., Koelsch G., Lin X., Wu S., Terzyan S., Ghosh A.K., Zhang X.C., Tang J. Structure of the protease domain of memapsin 2 (beta-secretase) complexed with inhibitor. Science. 2000;290:150–153. doi: 10.1126/science.290.5489.150. [DOI] [PubMed] [Google Scholar]
- Jacobsen H., Ozmen L., Caruso A., Narquizian R., Hilpert H., Jacobsen B., Terwel D., Tanghe A., Bohrmann B. Combined treatment with a BACE inhibitor and anti-Abeta antibody gantenerumab enhances amyloid reduction in APPLondon mice. J. Neurosci. 2014;34:11621–11630. doi: 10.1523/JNEUROSCI.1405-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornacker M.G., Lai Z., Witmer M., Ma J., Hendrick J., Lee V.G., Riexinger D.J., Mapelli C., Metzler W., Copeland R.A. An inhibitor binding pocket distinct from the catalytic active site on human beta-APP cleaving enzyme. Biochemistry. 2005;44:11567–11573. doi: 10.1021/bi050932l. [DOI] [PubMed] [Google Scholar]
- Laird F.M., Cai H., Savonenko A.V., Farah M.H., He K., Melnikova T., Wen H., Chiang H.C., Xu G., Koliatsos V.E., Borchelt D.R., Price D.L., Lee H.K., Wong P.C. BACE1, a major determinant of selective vulnerability of the brain to amyloid-beta amyloidogenesis, is essential for cognitive, emotional, and synaptic functions. J. Neurosci. 2005;25:11693–11709. doi: 10.1523/JNEUROSCI.2766-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Hoyte K., Montgomery W.H., Luk W., He D., Meilandt W.J., Zuchero Y.J., Atwal J.K., Scearce-Levie K., Watts R.J., Deforge L.E. Characterization of a sensitive mouse Abeta40 PD biomarker assay for Alzheimer's disease drug development in wild-type mice. Bioanalysis. 2016;8:1067–1075. doi: 10.4155/bio-2016-0003. [DOI] [PubMed] [Google Scholar]
- May P.C., Dean R.A., Lowe S.L., Martenyi F., Sheehan S.M., Boggs L.N., monk S.A., Mathes B.M., Mergott D.J., Watson B.M., Stout S.L., Timm D.E., Smith Labell E., Gonzales C.R., Nakano M., Jhee S.S., Yen M., Ereshefsky L., Lindstrom T.D., Calligaro D.O., Cocke P.J., Greg Hall D., Friedrich S., Citron M., Audia J.E. Robust central reduction of amyloid-beta in humans with an orally available, non-peptidic beta-secretase inhibitor. J. Neurosci. 2011;31:16507–16516. doi: 10.1523/JNEUROSCI.3647-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcconlogue L., Buttini M., Anderson J.P., Brigham E.F., Chen K.S., Freedman S.B., Games D., Johnson-Wood K., Lee M., Zeller M., Liu W., Motter R., Sinha S. Partial reduction of BACE1 has dramatic effects on Alzheimer plaque and synaptic pathology in APP transgenic mice. J. Biol. Chem. 2007;282:26326–26334. doi: 10.1074/jbc.M611687200. [DOI] [PubMed] [Google Scholar]
- Niewoehner J., Bohrmann B., Collin L., Urich E., Sade H., Maier P., Rueger P., Stracke J.O., Lau W., Tissot A.C., Loetscher H., Ghosh A., Freskgard P.O. Increased brain penetration and potency of a therapeutic antibody using a monovalent molecular shuttle. Neuron. 2014;81:49–60. doi: 10.1016/j.neuron.2013.10.061. [DOI] [PubMed] [Google Scholar]
- Ottaviani G., Wendelspiess S., Alvarez-Sanchez R. Importance of critical micellar concentration for the prediction of solubility enhancement in biorelevant media. Mol. Pharm. 2015;12:1171–1179. doi: 10.1021/mp5006992. [DOI] [PubMed] [Google Scholar]
- Pace C.M., Betowski L.D. Measurement of high-molecular-weight polycyclic aromatic hydrocarbons in soils by particle beam high-performance liquid chromatography-mass spectrometry. J. Am. Soc. Mass Spectrom. 1995;6:597–607. doi: 10.1016/1044-0305(95)00249-D. [DOI] [PubMed] [Google Scholar]
- Popp M.W., Antos J.M., Ploegh H.L. Site-specific protein labeling via sortase-mediated transpeptidation. Curr. Protoc. Protein Sci. 2009 doi: 10.1002/0471140864.ps1503s56. (Chapter 15, Unit 15 3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quartino A., Huledal G., Sparve E., Luttgen M., Bueters T., Karlsson P., Olsson T., Paraskos J., Maltby J., Claeson-Bohnstedt K., Lee C.M., Alexander R., Falting J., Paulsson B. Population pharmacokinetic and pharmacodynamic analysis of plasma Abeta40 and Abeta42 following single oral doses of the BACE1 inhibitor AZD3839 to healthy volunteers. Clin. Pharmacol. Drug Dev. 2014;3:396–405. doi: 10.1002/cpdd.130. [DOI] [PubMed] [Google Scholar]
- Rajendran L., Schneider A., Schlechtingen G., Weidlich S., Ries J., Braxmeier T., Schwille P., Schulz J.B., Schroeder C., Simons M., Jennings G., Knolker H.J., Simons K. Efficient inhibition of the Alzheimer's disease beta-secretase by membrane targeting. Science. 2008;320:520–523. doi: 10.1126/science.1156609. [DOI] [PubMed] [Google Scholar]
- Rashidian M., Dozier J.K., Distefano M.D. Enzymatic labeling of proteins: techniques and approaches. Bioconjug. Chem. 2013;24:1277–1294. doi: 10.1021/bc400102w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R.L., Fine R.E., Sandra A. Receptor-mediated endocytosis of transferrin at the blood-brain barrier. J. Cell Sci. 1993;104(Pt 2):521–532. doi: 10.1242/jcs.104.2.521. [DOI] [PubMed] [Google Scholar]
- Tolcher A.W., Messersmith W.A., Mikulski S.M., Papadopoulos K.P., Kwak E.L., Gibbon D.G., Patnaik A., Falchook G.S., Dasari A., Shapiro G.I., Boylan J.F., Xu Z.X., Wang K., Koehler A., Song J., Middleton S.A., Deutsch J., Demario M., Kurzrock R., Wheler J.J. Phase I study of RO4929097, a gamma secretase inhibitor of Notch signaling, in patients with refractory metastatic or locally advanced solid tumors. J. Clin. Oncol. 2012;30:2348–2353. doi: 10.1200/JCO.2011.36.8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung J.S., Davis D.L., Anderson J.P., Walker D.E., Mamo S., Jewett N., Hom R.K., Sinha S., Thorsett E.D., John V. Design of substrate-based inhibitors of human beta-secretase. J. Med. Chem. 2002;45:259–262. doi: 10.1021/jm0155695. [DOI] [PubMed] [Google Scholar]
- Turner R.T., 3rd, Hong L., Koelsch G., Ghosh A.K., Tang J. Structural locations and functional roles of new subsites S5, S6, and S7 in memapsin 2 (beta-secretase) Biochemistry. 2005;44:105–112. doi: 10.1021/bi048106k. [DOI] [PubMed] [Google Scholar]
- Vassar R. BACE1 inhibitor drugs in clinical trials for Alzheimer's disease. Alzheimers Res. Ther. 2014;6:89. doi: 10.1186/s13195-014-0089-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R. Stepping closer to treating Alzheimer's disease patients with BACE1 inhibitor drugs. Transl. Neurodegener. 2016;5:13. doi: 10.1186/s40035-016-0061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.J., Zhang Y., Kenrick M., Hoyte K., Luk W., Lu Y., Atwal J., Elliott J.M., Prabhu S., Watts R.J., Dennis M.S. Boosting brain uptake of a therapeutic antibody by reducing its affinity for a transcytosis target. Sci. Transl. Med. 2011;3:84ra44. doi: 10.1126/scitranslmed.3002230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material