Abstract
Background
IRX3 was recently reported as the effector of the FTO variants. We aimed to test IRX3's roles in the browning program and to evaluate the association between the genetic variants in IRX3 and human obesity.
Methods
IRX3 expression was examined in beige adipocytes in human and mouse models, and further validated in induced beige adipocytes. The browning capacity of primary preadipocytes was assessed with IRX3 knockdown. Luciferase reporter analysis and ChIP assay were applied to investigate IRX3's effects on UCP1 transcriptional activity. Moreover, genetic analysis of IRX3 was performed in 861 young obese subjects and 916 controls.
Results
IRX3 expression was induced in the browning process and was positively correlated with the browning markers. IRX3 knockdown remarkably inhibited UCP1 expression in induced mouse and human beige adipocytes, and also repressed the uncoupled oxygen consumption rate. Further, IRX3 directly bound to UCP1 promoter and increased its transcriptional activity. Moreover, 17 rare heterozygous missense/frameshift IRX3 variants were identified, with a significant enrichment in obese subjects (P = 0.038, OR = 2.27; 95% CI, 1.02–5.05).
Conclusions
IRX3 deficiency repressed the browning program of white adipocytes partially by regulating UCP1 transcriptional activity. Rare variants of IRX3 were associated with human obesity.
Keywords: IRX3, Beige adipocytes, Browning program, Genetic variants, Obesity
Highlights
-
•
IRX3 expression was positively correlated with the browning markers in both mice and humans.
-
•
IRX3 deficiency repressed the browning program in human and mouse cell models by regulating UCP1 transcriptional activity.
-
•
Rare variants of IRX3 were associated with human obesity.
While FTO is the first risk gene for obesity identified via genome-wide association studies, its exact mechanism remains unknown until two recent studies reported that IRX3, as FTO's target, probably inhibited the browning of white fat. However, whether IRX3 gene per se predisposes to obesity in humans and how IRX3 regulates the browning process remains unclear. Herein we combined cellular and genetic evidence to support that IRX3 promotes but not inhibit the browning process in both human and mouse cell models. Mechanistically, IRX3 increased the transcriptional activity of UCP1 through directly binding to its promoter. Thus, our findings provided an intact picture of IRX3's function in browning program and human obesity.
1. Introduction
Obesity is a complex disease determined by genetic and environmental factors. The heritability of obesity is as high as 40–70% (Stunkard et al., 1990; Allison et al., 1996), which makes decipherment of obese-related genes extremely important for obesity treatment. The strongest obesity-associated loci were consistently identified as located in the first and second introns of FTO (Scuteri et al., 2007; Dina et al., 2007; Frayling et al., 2007). However, the direct causality between FTO and obesity has not been clearly validated. Recently, two studies made a breakthrough in identifying IRX3 as the target of the risk loci in the FTO gene for human obesity (Smemo et al., 2014; Claussnitzer et al., 2015). One study showed the regulatory role of IRX3 in the browning of white adipose tissue (WAT) and obesity development through hypothalamus-specific (Insulin2-cre) Irx3 knockout mouse models (Smemo et al., 2014), while the other indicated that the FTO obesity-linked loci functioned as a body weight regulator by controlling IRX3 expression in the peripheral adipose tissue and IRX3 modulated the browning progress of WAT, which was further supported by adipose-specific (Ap2-cre) Irx3 knockout mice (Claussnitzer et al., 2015). However, the detailed effects of IRX3 on browning shift of white adipocytes remains undetermined.
Generation of inducible browning adipocytes (also called brown-like/beige adipocytes) confers beneficial effects against adiposity (Virtanen et al., 2009; Wu et al., 2012; Cypess et al., 2009). Beige adipocytes specialize in heat production by consuming a large amount of fatty acids and glucose, thus promoting energy expenditure. As a growing body of studies has demonstrated the existence of beige adipocytes in infants and adults (Cypess et al., 2009, 2013; Lidell et al., 2013) and its close association with reduced body weight has been discovered (Guerra et al., 1998), the browning program of WAT becomes an intriguing target to combat obesity and its metabolic consequences.
IRX3, a transcription factor belonging to the Iroquois family of homeobox genes, participates in the pattern and development of several tissues, including the central nervous system (Bellefroid et al., 1998), microvascular system (Scarlett et al., 2015), kidney (Reggiani et al., 2007), and heart (Zhang et al., 2011). Two heterozygous IRX3 mutations have been linked to idiopathic ventricular fibrillation in humans (Koizumi et al., 2015), suggesting that nonsynonymous variant in IRX3 could be functional and lead to certain diseases. With regard to the close association of IRX3 with the FTO obesity-linked loci, revealing IRX3's genetic profile can be of great importance to explain how genetic factors manipulate body weight alteration. However, whether the genetic variants of IRX3 are associated with human obesity and how IRX3 regulates the browning process remain unclear.
In the present study, we aimed to clarify the direct role and possible mechanism of IRX3 in the browning program of white adipocytes by using diverse browning models and in vitro experiments. We found that Irx3 disruption dramatically inhibited the browning program of preadipocytes, showing reduced Ucp1 expression and uncoupled oxygen consumption, and the effects might be achieved at least partially through inhibiting Ucp1 transcriptional activity. Targeted exons sequencing methods have been widely and successfully used to explore functional rare variants in relation to obesity and type 2 diabetes (Pearce et al., 2013; Ramachandrappa et al., 2013; Flannick et al., 2014; Bonnefond et al., 2012). Therefore, we further conducted targeted sequencing of IRX3 coding sequence in the lean and young obese subjects and found that rare IRX3 variants were significantly associated with human obesity. These findings provide more extensive information regarding the regulatory role of IRX3 in the browning process and human obesity.
2. Methods
2.1. Mouse Adipose Tissues
Male C57BL/6J mice at eight weeks old were subjected to cold room (4 °C) stress for one week (Wang et al., 2013) or received daily intraperitoneal injections of CL-316,243 (1 mg/kg, Sigma-Aldrich, C5976) for 10 days as described previously (Dempersmier et al., 2015). The dissected tissues were rapidly frozen and stored in liquid nitrogen. All the animal experiments were approved by the Animal Care Committee of Shanghai Jiao Tong University School of Medicine.
2.2. RNA Extraction and Real-Time PCR
Samples were homogenized with TRIzol reagent (Thermo Fisher Scientific Inc., 15596018) and total RNA was extracted according to the manufacturer's instructions. 1 μg of RNA was reverse transcribed to cDNA with a reverse transcription kit (Promega, A3500). Real-time PCR was carried out on the QuantStudio Dx system (Thermo Fisher Scientific Inc.) using SYBR Green Supermix (Takara Bio, DRR041A). The primers used in this study are provided in Table S1. Quantitative amounts of gene expression were normalized to the housekeeping gene mouse 36b4 or human βactin and analyzed using the 2− ΔΔCT methods. The results are representative of at least three independent experiments.
2.3. Western Blotting
Western blotting (WB) was performed as previously described (Wang et al., 2013). Proteins were assessed with the following antibodies: anti-IRX3 antibody (Abcam, ab25703), anti-GFP antibody (Cell Signaling Technology, 2956s), anti-actin antibody (Santa Cruz Biotechnology, sc-8432), anti-UCP1 antibody (Alpha Diagnostic, ucp1-a), anti-HSP90 antibody (Cell Signaling Technology, 4877s), anti-PGC-1α antibody (Millipore, ab3242), anti-AP2 antibody (Cell Signaling Technology, 3544s), and horseradish peroxidase-conjugated (HRP)-linked secondary antibody (Cell Signaling Technology, 7076, 7074). The representative blotting bands were repeated in at least three mice. The results are representative of at least three independent experiments.
2.4. Morphological Studies
Adipose tissues were stained with hematoxylin and eosin (HE) according to the standard protocols. For immunohistochemistry, rabbit anti-UCP1 antibody (Alpha Diagnostic, ucp1-a, diluted 1:400) or rabbit anti-IRX3 (Abcam, ab25703, diluted 1:1000) was used to incubate the sections, followed by incubation with HRP-linked secondary antibody (Dako, k500711). Electron microscopy of browning adipose tissue was performed as previously described (Wang et al., 2013). To determine the localization of IRX3 in the browning adipocytes, beige cells after eight days induction were fixed and incubated with rabbit anti-IRX3 antibody (Abcam, ab25703) and mouse anti-UCP1 antibody (R&D, MAB6158), followed by incubation with Alexa Fluor 594-conjugated goat anti-rabbit secondary antibody (ThermoFisher, R37117), 488–conjugated goat anti-mouse secondary antibody (ThermoFisher, A32723), and DAPI (SouthernBiotech, 0110-20). Images were acquired with a confocal laser-scanning microscope (Zeiss). All the images were representative of three independent experiments.
2.5. Human Adipose Tissue Samples
Human browning omental adipose tissue for immunohistochemistry was obtained from clinically and pathologically diagnosed patients with pheochromocytoma and sex-, age-, and BMI-matched control subjects. Primary human preadipocytes in stromal vascular fraction (SVF) from subcutaneous WAT (sWAT) for the induction to beige adipocytes were obtained from an underaged subjects. The biopsy samples of abdominal sWAT and omental WAT (oWAT) for the comparison of IRX3 expression were obtained from 21 severely obese subjects and 9 lean subjects. Obese patients with secondary or syndromic obesity-related diseases were excluded. Detailed characteristics of the subjects are presented in Table S2. The adipose tissue pieces were obtained during surgical procedures followed by immediate storage in liquid nitrogen and fixation in formalin. The human study was approved by the Institutional Review Board of Ruijin Hospital, Shanghai Jiao Tong University School of Medicine. Written informed consent was provided from each participant prior to inclusion in the study.
2.6. Stromal Vascular Fraction Isolation, Beige/Brown and White Adipocyte Differentiation
SVFs were isolated from the inguinal WAT (IWAT), epididymal WAT (EWAT) or brown adipose tissue (BAT) of male mice with C57BL/6J or 129/Sv background and induced into fully differentiated beige/brown adipocytes or white adipocytes as previously described (Wang et al., 2013). Briefly, the adipose tissue was minced and then digested with type II collagenase (Sigma) in 10 mM HEPES (Sigma) for 30 min at 37 °C. Digested adipose tissue was filtered through a 40 μm cell strainer to remove large undigested pieces. After centrifuge for 10 min, SVF cells were then resuspended with DMEM/F12 medium. For the induction of beige/brown adipocytes, SVF cells were plated onto 48-well plates to reach confluence and incubated with medium supplemented with 5 μg/ml insulin (Eli Lilly, HI0240), 0.5 mM isobutylmethylxanthine (Sigma-Aldrich, I7018), 1 μM dexamethasone (Sigma-Aldrich, D4902), 1 nM T3 (Sigma-Aldrich, T2877), and 1 μM rosiglitazone (Sigma-Aldrich, R2408) for four days, and subsequently in medium with insulin, T3, and for another four days. For the induction to white adipocytes, cells were incubated with medium supplemented with 5 μg/ml insulin, 0.5 mM isobutylmethylxanthine, and 1 μM dexamethasone for two days, and subsequently in medium with insulin 5 μg/ml insulin for six days.
2.7. Cell Culture
SVFs from murine or human were cultured in DMEM/F12 supplemented with 10% FBS, 1% P/S, 1 mM L-glutamine, and 10 ng/ml murine basic fibroblast growth factor (R&D Systems, 3139-FB-025). HEK293T cells were purchased from ATCC.
2.8. Lentiviral shRNA Generation
Lentiviral mouse Irx3 shRNA constructs and lentiviral human IRX3 constructs were made using pLKO-PGK-RFP and pLKO-CMV-RFP lentivector cloning, respectively. We verified all constructs using DNA sequencing; the sequences of mouse Irx3 shRNA were ccctatccaatgtgctttcat for Irx3 shRNA-1 and tctgtgtatgattaccttaaa for Irx3 shRNA-2. The sequences for human IRX3 shRNA were ttgtaagcatgtccgtgtata for hIRX3 shRNA-1 and gtttgtttgtccggttgattt for hIRX3 shRNA-2. The cells were infected with the lentivirus to knockdown Irx3/IRX3 24 h after seeding.
2.9. Oil Red O Staining
After induction to mature adipocytes for eight days, mature adipocytes underwent Oil Red O staining as previously described (Wang et al., 2013). In brief, the cells were fixed with 4% paraformaldehyde for 30 min, fully rinsed, air dried, and incubated with Oil Red O (Sigma-Aldrich) for 30 min. The images were observed directly with microscope (Olympus).
2.10. Triglyceride Measurement
After the induction for 8 days, mature beige adipocytes were collected for triglyceride (TG) measurement guided by the manufacturer's protocols (BioVision, K622-100).
2.11. Oxygen Consumption Rate Measurements
SVFs were seeded in an XF24 V28 microplate (Seahorse Bioscience) coated with poly-L-lysine. After 24 h, the cells were infected with lentivirus and subsequently differentiated into beige adipocytes for five days. Oxygen Consumption Rate (OCR) was measured as previously described (Wang et al., 2013) using an XF24 analyzer (Seahorse Bioscience) in accordance with the manufacturer's instructions. In brief, the cells were washed with Seahorse XF base medium with 25 mM D-glucose, 2 mM sodium pyruvate, and 2 mM LG, followed by incubation with 525 μl assay medium with or without 2% fatty acid-free BSA at 37 °C in a non-CO2 incubator (Seahorse Bioscience) for 1 h. 75 μl respiratory inhibitors (Seahorse Bioscience, 103015-100) was loaded into the injection port to reach the final concentration of 1 mg/ml oligomycin, 1.5 mM FCCP, 0.5 mM antimycin A and 0.5 mg/ml rotenone to detect the uncoupled respiration, maximal respiration, and non-mitochondrial respiration, respectively. The final OCR results were standardized to the protein concentration. The results are representative of at least three independent experiments.
2.12. Plasmid Construction
The coding region of IRX3 PCR amplified from a human cDNA clone (NM_0243362) was sub-cloned into the pEGFP-C1 vector (N-terminal GFP-tag) between the HindIII and BamHI restriction sites. Plasmids of murine Ucp1 promoter and human UCP1 promoter, which was 4 kb proximal to the transcriptional start site (TSS), was constructed into the pGL-4.12 vector (Promega) as previously described (Zhang et al., 2014). Mutagenesis was generated using a site-directed mutagenesis kit (Agilent Technologies). Full-length coding sequences for all the plasmids were verified by DNA sequencing.
2.13. Luciferase Reporter Assay
Luciferase reporter assay was conducted in HEK293T cells. After 48 h transfection with 800 ng pEGFP-C1 vector or pEGFP-C1-IRX3 plasmids, along with 1 ng pRL-TK (or alternatively 1 ng pRL-SV40) and 200 ng UCP1/Ucp1 promoter reporter construct, HEK293T cells were harvested for luciferase activity assessment using a dual-luciferase reporter assay system (Promega). The final results were normalized to Renilla luciferase activity. The results are representative of at least three independent experiments.
2.14. Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation (ChIP) assay was carried out to investigate the interaction between IRX3 and the Ucp1 promoter using a commercial kit (Millipore, 17–371) in accordance with the manufacturer's instructions. Nuclei were extracted from the mature beige adipocytes induced from preadipocytes within IWAT of C57BL/6J mice. For immunoprecipitation, equal aliquots of cell lysates were incubated with anti-IRX3 antibody or IgG overnight at 4 °C. The precipitated DNA fragments were analyzed by real-time PCR with the following mouse Ucp1 primers: forward primer 5′-GAGTAGCCGAAGGGTTCAGG-3′; reverse primer 5′-TCCCATAGGAAGCGTCATGC-3′′.
2.15. Participants in Case-Control Genetic Study
In stage one, we used an in-home whole-exome sequencing database that included 227 young obese subjects and 219 lean controls from eastern China as described in our recent study (Liu et al., 2017; Zou et al., 2017). It has been established that the population shows high genetic homogeneity to minimize any effects of population stratification (Liu et al., 2017). All the patients were continuously recruited from 2007 to 2011 from the specialized obesity outpatient clinic of Ruijin Hospital, Shanghai Jiao Tong University School of Medicine. Participants were excluded if they had secondary or syndromatic obesity; previous pharmacological, diet, or exercise intervention; infectious disease, hepatic or renal dysfunction, or malignant tumors. Trained physicians used height-weight scales to take measurements of the height and weight three times for each subject; mean values of each variable were used for further analysis. Genetic and basic characteristic information were obtained from the Genetics of Obesity in Chinese Youngs (GOCY) study (Wang et al., 2013; Liu et al., 2017; Zou et al., 2017; Hong et al., 2013) that was previously established by Ruijin Hospital and registered in ClinicalTrials.gov. In stage two, 634 young obese patients and 697 controls were additionally recruited into the study according to the same procedure. This study was also approved by the Institutional Review Board of Ruijin Hospital, Shanghai Jiao Tong University School of Medicine. Written informed consent was obtained from each participant.
2.16. Genotyping
Genomic DNA was isolated from peripheral blood using a commercial blood genomic DNA extraction kit (Qiagen). Sanger sequencing was performed to identify nonsynonymous variants, including frameshift, nonsense, deletion, or missense mutations in IRX3 (Gene ID: 79,191) exons with minor allele frequency (MAF) < 1%. Primers used to amplify the IRX3 exons are shown in Table S3.
2.17. Sequence Alignment of IRX3 Orthologs Related to the Studied Variants
Sequence alignment of IRX3 across different species related to the variants identified in obese subjects and controls was conducted using a ClustalW2 alignment program (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The comparison included the IRX3 amino acid sequence of human (Homo sapiens, NP_077312.2), mouse (Mus musculus, NP_032419.2), rat (Rattus norvegicus, NP_001100883.1), chimpanzee (Pan troglodytes, XP_510969.2), and dog (Canis lupus familiaris, XP_544403.2).
2.18. Statistical Analyses
Spearman's correlation analysis was performed to examine the associations between the expression of Irx3 and brown adipocyte-related genes. Differences of the frequency distribution of rare IRX3 variants between case patients and control subjects were calculated using the χ2 test. Unless otherwise stated, data was shown as mean ± S.E.M, and the results were compared by 2-tailed t-tests with α level of 0.05. P values < 0.05 were considered to be significantly different. As to basic experiments, data were generated from three independent experiments. Analyses were carried out with SAS version 9.3 (SAS Institute, New York, NY, USA).
3. Result
3.1. IRX3 was Induced in the Browning Program of WATs in Both Mice and Humans
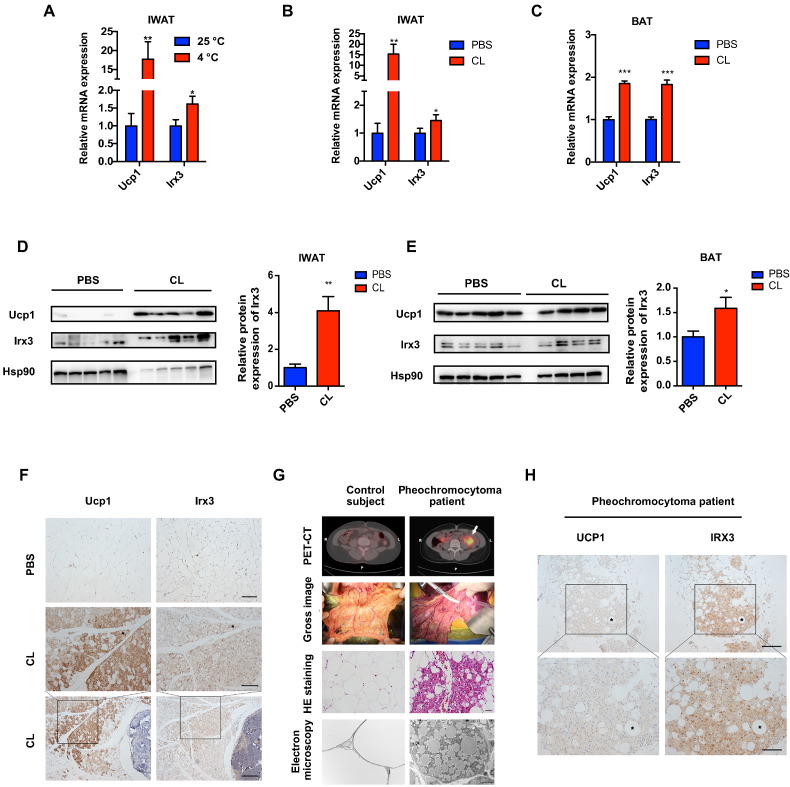
To identify the involvement of IRX3 in the generation of beige adipocytes within WAT, we first detected IRX3 expression during the induction of the browning program. In rodents, the browning program of white fat depots can be activated by prolonged cold stress or β3-adrenergic agonist CL-316,243 (Rosen and Spiegelman, 2014; Himms-Hagen et al., 1994). We found that cold stress (4 °C) increased the mRNA levels of Irx3 in IWAT coincident with the induction of brown adipocyte marker, Ucp1 (Fig. 1A). We did not observe an increase in IRX3 protein levels under cold stress (Supplementary Fig. 1). Interestingly, a significantly higher mRNA and protein level of Irx3 was also observed in the IWAT and classical BAT of the mice injected with CL-316,243 than the control group, in parallel with the induction of Ucp1 (Fig. 1B–F). Pheochromocytoma patients, characterized by excess β3-adrenergic activation, have abundant beige adipocytes (Puar et al., 2016). We therefore further validated IRX3 expression in the beige adipocytes from pheochromocytoma patients. The browning fat of pheochromocytoma patients were confirmed by the enhanced 18F-2-deoxy-2-fluoro-d-glucose uptake by PET/CT scan, tissue morphology, increased mitochondria under electron microscopy, and positive UCP1 protein staining (Fig. 1G–H). Consistently, we found a positive staining of IRX3 protein expression in the beige adipocytes (Fig. 1H). These results together suggested that IRX3 expression is induced in the browning process of WATs in both mice and humans, especially under β3-adrenergic stimulation.
Fig. 1.
IRX3/Irx3 expression is induced in browning adipose tissues.
(A) Relative mRNA levels of Ucp1 and Irx3 in IWAT from mice under cold stress (4 °C) for one week (n = 6) and at 25 °C (n = 8). (B–C) Relative mRNA levels of Ucp1 and Irx3 in IWAT (B) and BAT (C) from mice subjected to PBS or CL-316,243 for 10 days. For A–C, gene expression was normalized to 36b4. (D–E) Protein levels of Ucp1 and Irx3 (left) and the quantification value of Irx3 relative to Hsp90 (right) in IWAT (D) and BAT (E) in the mice subjected to PBS or CL-316,243 treatment. (F) Representative images of immunohistochemical staining in IWAT from mice subjected to PBS or CL-316,243 treatment (scale bar, 50 μm for the upper and middle panels, and 100 μm for the bottom panel). (G) PET-CT, gross image, HE staining (scale bar, 20 μm), and electron microscopy (scale bar, 5 μm) of the browning WAT from pheochromocytoma patients. (H) Representative images of immunohistochemical staining in oWAT from pheochromocytoma patients (scale bar, 100 μm for the upper panel, and 50 μm for the bottom panel). Data were presented as mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001. The results are representative of at least three independent experiments. IWAT, inguinal white adipose tissue. BAT, brown adipose tissue.
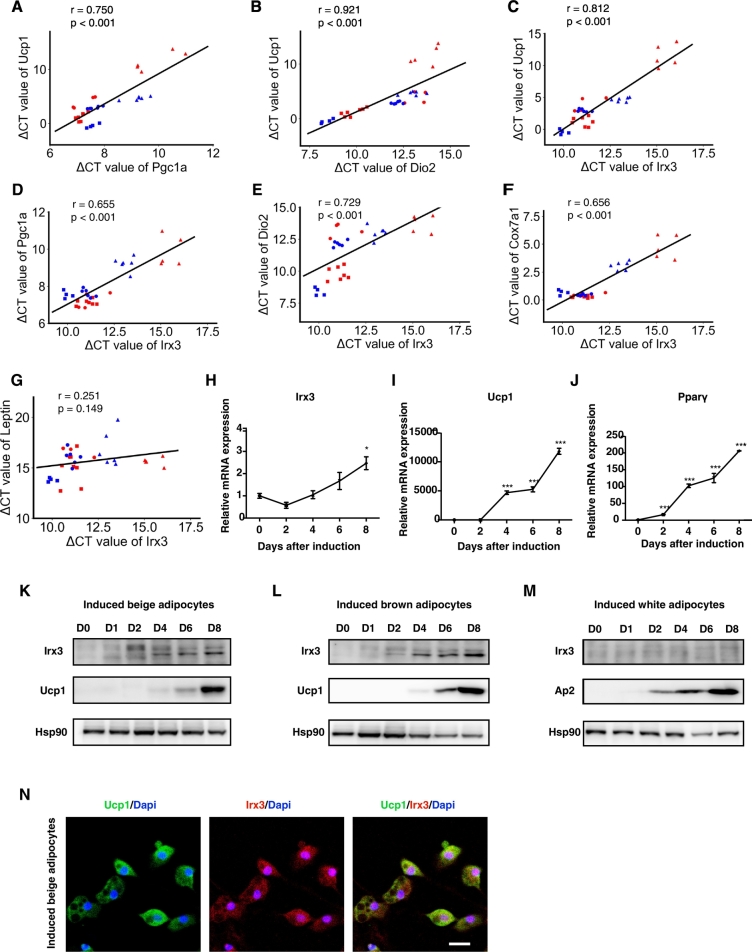
3.2. IRX3 mRNA Levels were Positively Associated with Beige Adipocyte Markers
Primary preadipocytes residing in SVFs from EWAT, IWAT, and BAT, have the distinct capacity to differentiate into brown-like adipocytes (Macotela et al., 2012). We next isolated and differentiated SVFs from the three types of adipose tissues of two mouse strains with 129/Sv and C57BL/6J genetic backgrounds, respectively, to examine the correlation between the expression of Irx3 and beige adipocyte markers. After an 8-day induction with beige/brown adipogenic procedures, Ucp1 expression level was positively correlated with Pgc-1α and Dio2 mRNA levels among different beige adipocytes (Fig. 2A-B). Concomitantly, Irx3 mRNA levels presented a positive correlation with the expression of beige adipocyte-associated genes such as Ucp1, Pgc-1α, Dio2, and Cox7a1 (Fig. 2C–F). No correlation between Irx3 and white adipocyte marker Leptin was observed (Fig. 2G). We further detected Irx3 expression during the time course of beige/brown adipocyte differentiation. We found that Irx3 expression was increased during beige/brown adipocyte differentiation, in parallel with the induction of Ucp1 and Pparγ (Fig. 2H–L), while Irx3 expression was not induced during white adipocyte differentiation, as compared with the increase in Ap2 gene expression (Fig. 2M). Besides, Irx3 and Ucp1 were co-expressed in mature beige adipocytes (Fig. 2N). These findings further supported the possible promoting effects of Irx3 on the browning program of white adipocytes.
Fig. 2.
The expression of Irx3 was correlated with the beige/brown adipocyte associated genes in vitro.
(A–G) In fully differentiated beige/brown adipocytes from preadipocytes isolated from IWAT, EWAT, and BAT of C57BL/6J and 129/Sv mice, Ucp1 mRNA expression was correlated with Pgc-1α (A) and Dio2 (B). Irx3 mRNA expression showed positive correlation with mRNA levels of (C) Ucp1, (D) Pgc-1α, (E) Dio2, and (F) Cox7a1. Irx3 mRNA expression showed no correlation with Leptin (G). Cells from C57BL/6J were shown in red, and from 129/Sv were in blue. Cells from IWAT, EWAT, and BAT were shown as dot, triangle, and square, respectively. The correlation was analyzed using ΔCT. (H-J) (H) Irx3 (I) Ucp1 and (J) Pparγ mRNA expression during the time course of beige adipocyte differentiation cultures (n = 3). Quantitative amounts of gene expression were normalized to the housekeeping gene 36b4. (K–L) Irx3 and Ucp1 protein levels at different time points during beige adipocyte differentiation from IWAT SVFs (K), during brown adipocyte differentiation from BAT SVFs (L). (M) Protein expression of Irx3 and Ap2 at different time points during white adipocyte differentiation from IWAT SVFs. (N) Intracellular location of Ucp1 and Irx3 protein in the fully differentiated beige adipocytes. Ucp1 protein, Irx3 protein and nucleus were indicated in green, red, and blue, respectively. Scar bar, 20 μm. Data were presented as mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001. The results are representative of at least three independent experiments.
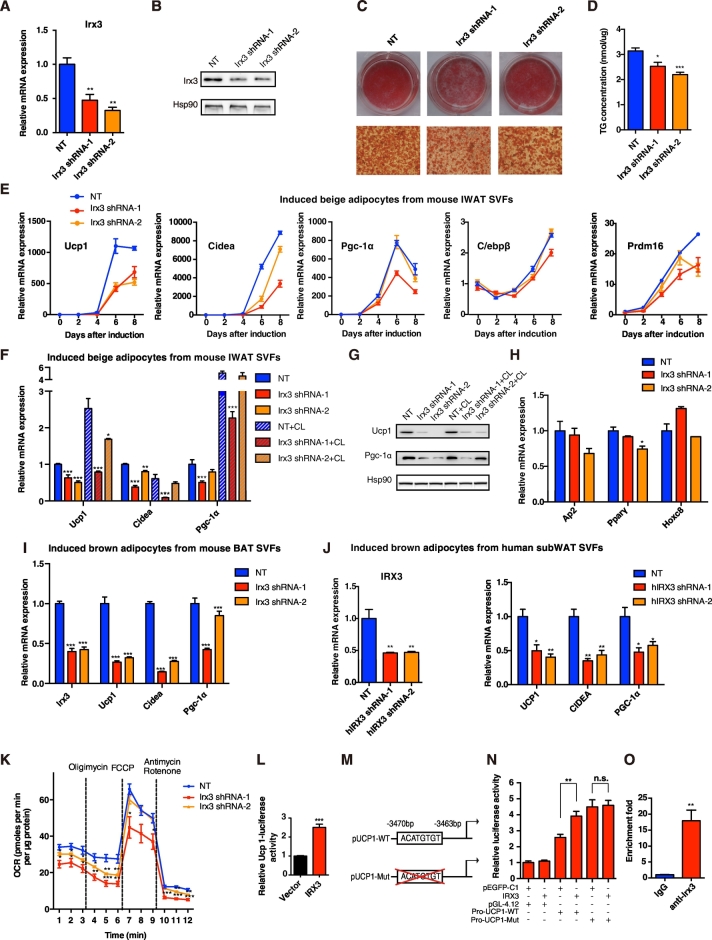
3.3. Irx3 Deficiency Repressed Beige Adipocyte Program and Biological Function in vitro
To avoid the non-specific or compensatory influence of other tissues in vivo, we next examined the cell-autonomous effects of Irx3 on the differentiation of beige/brown adipocytes from preadipocytes in SVFs of IWAT and BAT in vitro, respectively. We efficiently knocked down Irx3 expression in SVFs with lentiviral Irx3 shRNA (Fig. 3A-B) and found that Irx3 reduction led to less lipid accumulation after beige adipocyte differentiation as determined by Oil Red O staining (Fig. 3C) and triglyceride quantification (Fig. 3D). Furthermore, Irx3 knockdown repressed the expression of Prdm16 at early stage of differentiation and markedly inhibited terminal brown adipocyte markers Ucp1, Cidea and Pgc-1α at late stages (Fig. 3E). Of note, Ucp1, which is the specific marker for brown adipocytes and functions in uncoupling oxidative metabolism from ATP production (Shabalina et al., 2013), was robustly reduced in beige adipocytes with Irx3 knockdown, which was more obvious under CL-316,243 treatment (Fig. 3F). Pgc-1α, a transcriptional co-regulator of mitochondrial biogenesis (Liang and Ward, 2006), and Cidea, which promoted lipolysis and thermogenesis, were also significantly decreased in beige adipocytes with Irx3 knockdown compared to the controls (Fig. 3F). Besides, protein levels of Ucp1 and Pgc-1α showed similar changes upon Irx3 knockdown (Fig. 3G). Adipocyte differentiation-related genes, Ap2 and Pparγ, and white adipocyte-specific marker Hoxc8 showed no or mild changes with Irx3 knockdown (Fig. 3H). Other white adipocyte makers such as Glut4, Serpina3k and Wdnm1-like, were not significantly inhibited by Irx3 knockdown, and the latter two were rather increased (Supplementary Fig. 2). Furthermore, Irx3 knockdown also inhibited Ucp1, Cidea and Pgc-1α expression in differentiated brown adipocytes from SVFs in classical BAT (Fig. 3I). More importantly, IRX3 repression significantly inhibited human beige adipocyte program, reflected by reduced UCP1, CIDEA and PGC-1α expression in differentiated human SVFs (Fig. 3J). These results together suggested that IRX3 knockdown inhibited the browning program from preadipocytes in WATs of both mice and humans. Beige adipocyte functions in dissipating heat by maintaining a high mitochondrial OCR. Accordingly, measurement of the oxygen respiration ability of the fully differentiated beige adipocytes showed that both basal and uncoupled respiration rates were significantly decreased with or without normalization when Irx3 was reduced (Fig. 3K and Supplementary Fig. 3A). In addition, OCR measurement using 2% fatty acid-free BSA obtained similar results (Supplementary Fig. 3B). When normalized all OCR data to basal OCR, the relative uncoupled OCR to basal respiration also showed decreased trend by Irx3 knockdown, although not reaching statistical significance (Supplementary Fig. 3C). Collectively, our findings suggested that IRX3 deficiency repressed the beige/brown adipocytes induction from preadipocytes, indicating the promoting effects of Irx3/IRX3 on the browning program.
Fig. 3.
Knockdown of Irx3 repressed the differentiation of SVFs toward beige adipocytes in vitro.
(A–B) (A) Relative mRNA and (B) protein levels of Irx3 in SVFs from IWAT were efficiently knocked down after a 3-day infection of mouse Irx3 lentiviral shRNA (n = 3 for each group). (C–D) (C) Oil Red O staining and (D) TG concentration of the mature beige adipocytes under eight-day differentiation (n = 6 for each group). (E) Relative mRNA expression of Ucp1, Cidea, Pgc-1α, C/ebpβ, and Prdm16 at different time points during beige adipocyte differentiation from IWAT SVFs (n = 3–4 for different groups). (F–G) (F) Relative mRNA (n = 4) and (G) protein expression of brown adipocyte marker genes of the induced beige adipocytes from mouse IWAT SVTs for eight days with the infection of mouse Irx3 lentiviral shRNA, with or without CL-316,243 activation. (H) Relative mRNA expression of adipocyte differentiation-related genes in beige adipocytes (n = 4). (I) Relative mRNA expression of the indicated genes in the induced brown adipocytes from mouse BAT SVFs for eight days, with the infection of mouse Irx3 lentiviral shRNA (n = 4). (J) Relative mRNA levels of IRX3 in SVFs from human sWAT after a 3-day infection of human IRX3 lentiviral shRNA (left), and the mRNA levels of brown adipocyte marker genes in the SVFs under 2-day differentiation (n = 4). (K) OCR measurement of the beige adipocytes under 5-day differentiation, with the infection of mouse Irx3 lentiviral shRNA (n = 5). (L–N) (L) Ucp1 promoter-luciferase reporter activity was measured in HEK293T cells transfected with pEGFP-C1 vector or IRX3 for 48 h. (M) Schematic diagram of the mutant mouse Ucp1 promoter deleting ACATGTGT among the − 3470 to − 3463 bp region proximal to the TSS of the Ucp1 gene. (N) Transcriptional activity of wild-type or mutant Ucp1 promoter was measured with IRX3 overexpression. (O) qPCR of Ucp1 promoter binding region which was recruited by Irx3 antibody in 8-day induced beige adipocytes, as analyzed by ChIP. Fold enrichment of Ucp1 promoter was given (n = 3). For the measurement of luciferase activity, cells were seeded on 24-well plate and transfected with 800 ng pEGFP-C1 vector or IRX3, 200 ng Ucp1 promoter construct, and 1 ng pRL-SV40, followed by the harvest for luciferase activity assessment using a dual-luciferase reporter assay system (Promega). Luciferase activity was corrected for Renilla luciferase activity (n = 3–4 for each group). For qPCR data, mRNA expression was normalized to 36b4 for mouse genes and βACTIN for human genes. SVF cells were isolated from IWAT and BAT of C57BL/6J mice or human sWAT wherever mentioned, and IRX3 shRNA were introduced to cells 24 h after seeding. Data were presented as mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001. The results are representative of at least three independent experiments. NT, no targeting.
3.4. IRX3 Overexpression Increased UCP1 Transcriptional Activity
IRX3 is a transcription factor and we found that IRX3 knockdown significantly repressed UCP1 expression in the beige adipocytes, based on which we further explored whether IRX3 could directly regulate the transcriptional activity of UCP1 promoter. Previous studies have demonstrated that IRX3 exerts its transcriptional activity by specifically binding to the motif of ACAnnTGT (Bilioni et al., 2005), which is also presented as ACATGTGT among the − 3470 to − 3463 bp region proximal to the TSS of the mouse Ucp1 gene. Significantly, IRX3 overexpression increased the transcriptional activity of Ucp1 promoter containing this motif in a luciferase reporter assay in HEK293T cells (Fig. 3L). Further, we constructed the mutant Ucp1 promoter plasmids with IRX3 binding motif deletion as indicated in Fig. 3M, which abolished IRX3-enhanced Ucp1 promoter activity (Fig. 3N), while point mutations (ACATGTGT replaced with ATATGTAT) (Bilioni et al., 2005) in IRX3 binding motif of Ucp1 promoter did not abolish IRX3’s activating effects on Ucp1 promoter activity (Supplementary Fig. 4A). In addition, we also constructed human UCP1 promoter containing five putative IRX3 binding regions (ACAnnTGT) and five corresponding point mutations with the sequence from ACAnnTGT to ATAnnTAT. Consistently, we found that IRX3 overexpression also promoted human UCP1 promoter activity while point mutations did not abolish IRX3’s activating effects on UCP1 promoter (Supplementary Fig. 4B). To further examine the direct binding effects of Irx3 on Ucp1 promoter, we conducted ChIP assay and found that Irx3 bound to the putative binding motif on the Ucp1 promoter in the induced mature beige cells (Fig. 3O), strengthening the result that the transcriptional activity of UCP1 promoter was enhanced by IRX3. These results suggested that IRX3 promoted UCP1 transcriptional activity at least in part through the ACATGTGT binding site.
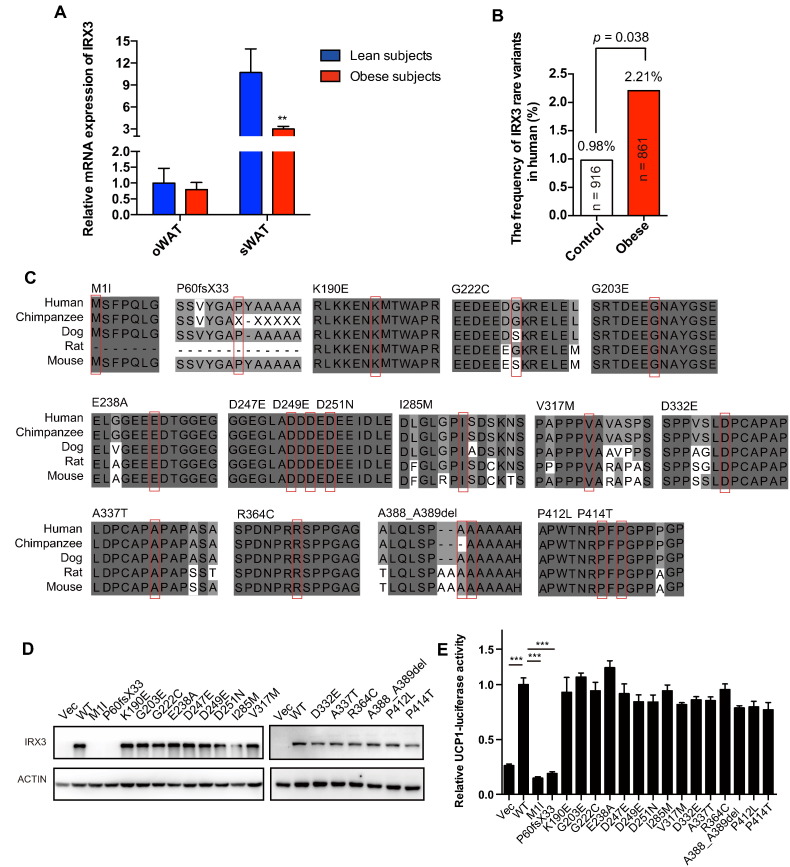
3.5. The Association Between IRX3 and Human Obesity
Taken together, we have identified the promoting effects of IRX3 on the browning program. To further explore the association of IRX3 with human obesity, we examined IRX3 expression in human adipose tissues and detected significantly reduced IRX3 mRNA levels in the sWAT of obese subjects compared to those of non-obese individuals (Fig. 4A). Moreover, we evaluated the effects of IRX3 variants on human obesity in a two-stage case-control study including 227 young obese subjects (BMI, 35.1–61.7 kg/m2) and 219 lean controls (BMI, 17.5–23.0 kg/m2) in the first stage; and 634 young obese subjects (BMI, 30.0–63.3 kg/m2) and 697 lean control subjects (BMI, 17.5–23.0 kg/m2) in the second stage (Table 1). Stage one identified 5 cases carrying rare nonsynonymous IRX3 variants with a MAF < 1% but none in the controls; and stage two screened 11 variants from 14 obese subjects and 9 variants from lean subjects. In total, 19 obese patients and 9 lean subjects (2.21% vs. 0.98%) had rare nonsynonymous IRX3 variants (P = 0.038; OR = 2.27; 95% CI, 1.02–5.05) (Fig. 4B), indicating that rare nonsynonymous variants of IRX3 were associated with the development of human obesity.
Fig. 4.
The association of IRX3 and human obesity.
(A) Relative mRNA levels of IRX3 in oWAT or sWAT from normal weight (n = 9) and obese subjects (n = 21) measured by qPCR. Gene expression was normalized to βACTIN. (B) Comparison of the frequency of the IRX3 rare variants in normal weight subjects and obese patients. (C) Sequence conservation analysis of the IRX3 orthologs related to variants identified in obese subjects and controls. Mutant sites are shown with a red box. Dark gray represents residues that are completely conserved, mild gray partially conserved, and light gray shows similar residues. (D) Protein expression levels of WT and mutant IRX3 in HEK293T cells. GFP antibody was used to indicate IRX3, and actin antibody served as a loading control. (E) Ucp1 promoter-luciferase reporter activity was measured in HEK293T cells transfected with pEGFP-C1 vector, WT or mutant IRX3 for 48 h (n = 3). Normalized luciferase activities are shown as fold change and compared to WT IRX3 (n = 4). oWAT, omental white adipose tissue. sWAT, subcutaneous white adipose tissue. Data were presented as mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001. The results are representative of at least three independent experiments.
Table 1.
Rare IRX3 variants were enriched in young obese subjects in comparison to lean controls.
| Ntotal (male) |
Age (yrs) |
BMI (kg/m2) |
RareIRX3variant |
|||||
|---|---|---|---|---|---|---|---|---|
| Median (inter-quartile range) | Range | Median(inter-quartile range) | Range | N(%) | OR (95% CI) | Pvalue | ||
| Stage 1 | ||||||||
| Cases | 227 (98) | 21 (18, 25) | 13.0–30.0 | 38.4 (36.8, 41.2) | 35.0–61.7 | 5 (2.20) | – | 0.061 |
| Controls | 219 (92) | 38.4 (36.8, 41.2) | 13.0–30.0 | 20.4 (19.5, 21.6) | 17–23.0 | 0 (0) | ||
| Stage 2 | ||||||||
| Cases | 634 (262) | 22 (18, 26) | 12.0–30.0 | 34.6 (32.9, 37.3) | 30.0–63.3 | 14 (2.21) | 1.73 (0.74, 4.02) | 0.20 |
| Controls | 697 (244) | 25 (22, 28) | 14.0–58.0 | 19.5 (18.3, 21.0) | 17.5–23.0 | 9 (1.29) | ||
| Combined | ||||||||
| Cases | 861 (360) | 22 (18, 25) | 12.0–30.0 | 35.9 (33.6, 38.9) | 30.0–63.3 | 19 (2.21) | 2.27 (1.02, 5.05) | 0.038 |
| Controls | 916 (336) | 24 (21, 27) | 13.0–58.0 | 19.8 (18.6, 1.1) | 17.5–23.0 | 9 (0.98) | ||
N, sample size; Yrs, years; BMI, body mass index; OR, odds ratio; CI, confidence interval.
P values were calculated using χ2 tests.
A total of 17 rare nonsynonymous variants in heterozygous form were identified in the studied cohort (Table 2). Most of these rare IRX3 variants were highly conserved among various species, implying the functional importance for the IRX3 protein (Fig. 4C). Some of the variants were predicted to be damaged or possibly damaged by MutationTaster, PolyPhen-2, and SIFT (Table 2). To better validate the function of these variants, we further conducted functional examination of the nonsynonymous variants. We found that two variants, M1I and P60fsX33, markedly changed the protein expression, while no obvious alteration in expression of other variants was observed (Fig. 4D and Supplementary Fig. 5). More importantly, M1I and P60fsX33 mutants markedly eliminated wild-type IRX3-induced Ucp1 transcriptional activity (Fig. 4E), confirming their functional damage. This result suggested that the genetic defects of IRX3 could possibly repress the browning program in humans at least in part by regulating UCP1 transcriptional activity to increase the risk of obesity. These findings together suggested the involvement of IRX3 deficiency in human obesity.
Table 2.
17 rare IRX3 variants identified in young obese subjects and lean controls.
| Positiona | Nucleotide changeb | Amino acid changeb | State | Cases | Controls | BMI of cases (kg/m2) | BMI of controls (kg/m2) | Mutation taster# | SIFT§ | PolyPhen2‡ |
|---|---|---|---|---|---|---|---|---|---|---|
| chr16:54319960 | c.G3 > A | p.M1I | Het | 1 | 0 | 41.7 | – | Deleterious | Deleterious | Possibly damaging |
| chr16:54319787-54319788 | c.175_176delGC | p.P60fsX33 | Het | 0 | 1 | – | 21.6 | – | – | – |
| chr16:54319225 | c.568A > G | p.K190E | Het | 0 | 1 | – | 20.7 | Damaging | Damaging | Probably damaging |
| chr16:54319185 | c.608G > A | p.G203E | Het | 1 | 0 | 36.3 | – | Damaging | Damaging | Probably damaging |
| chr16:54319129 | c.664G > T | p.G222C | Het | 1 | 0 | 38.7 | – | Damaging | Damaging | Benign |
| chr16:54319080 | c.713A > C | p.E238A | Het | 4 | 1 | 34.2, 36.9, 37.0, 45.3 | 18.2 | Tolerated | Tolerated | Benign |
| chr16:54319052 | c.741C > G | p.D247E | Het | 2 | 1 | 30.8, 32.7 | 21.2 | Tolerated | Tolerated | Benign |
| chr16:54319046 | c.747C > G | p.D249E | Het | 2 | 0 | 37.3, 39.4 | – | Tolerated | Tolerated | Benign |
| chr16:54319042 | c.751G > A | p.D251N | Het | 1 | 1 | 41.1 | 20.3 | Damaging | Damaging | Possibly damaging |
| chr16:54318938 | c.855T > G | p.I285M | Het | 1 | 0 | 42.6 | – | Tolerated | Tolerated | Benign |
| chr16:54318844 | c.949G > A | p.V317 M | Het | 0 | 1 | – | 21.1 | Tolerated | Tolerated | Benign |
| chr16:54318797 | c.996C > A | p.D332E | Het | 0 | 1 | – | 21.6 | Tolerated | Tolerated | Benign |
| chr16:54318784 | c.1009G > A | p.A337T | Het | 0 | 1 | – | 19.9 | Tolerated | Tolerated | Benign |
| chr16:54318703 | c.1090C > T | p.R364C | Het | 1 | 0 | 40.6 | – | Damaging | Damaging | Probably damaging |
| chr16:54318631-54318626 | c.1162_1167delGCCGCC | p.A388_A389del | Het | 0 | 1 | – | 21.8 | – | – | – |
| chr16:54318558 | c.1235C > T | p.P412L | Het | 3 | 0 | 34.2, 37.7, 38.9 | – | Damaging | Damaging | Possibly damaging |
| chr16:54318553 | c.1240C > A | p.P414T | Het | 2 | 0 | 33.8, 39.1 | – | Tolerated | Tolerated | Benign |
Het, heterozygous; BMI, body mass index. Functional prediction was conducted by Mutation taster# (http://mutationtaster.org), SIFT§ (http://sift.jcvi.org), and PolyPhen2‡ (http://genetics.bwh.harvard.edu/pph2/).
NCBI Build 37.
Variations are based on RefSeq records NM_024336.2 and NP_077312.2.
4. Discussion
In this study, we showed that IRX3/Irx3 expression was induced in the browning process of WATs in both humans and mice, and its knockdown led to noticeably impaired ability of beige/brown adipocytes program in vitro, possibly by inhibiting Ucp1 promoter transcriptional activity. We further identified the association between rare nonsynonymous IRX3 variants with human obesity risk. Taken together, we demonstrated that IRX3 played a promoting role in the browning of WAT and its rare variants are associated with human obesity.
Weight loss by increasing energy expenditure is an attractive therapeutic approach for obesity and its complications, especially since the identification of heat-dissipating brown adipocytes in human adults (Cypess et al., 2009, 2013; Lidell et al., 2013). The present study demonstrated the effects of IRX3 on the browning process by the following evidence: (1) IRX3/Irx3 reduction by shRNA in vitro dramatically inhibited the browning program and energy expenditure of beige adipocytes, as reflected by the reduced UCP1 levels and oxygen consumption ability, which is a key mechanism for the development of obesity (Wang et al., 2013; Zhang et al., 2014; Lee et al., 2014); (2) IRX3 significantly activated UCP1 transcriptional activity; (3) two loss-of-function IRX3 mutants identified in human largely abolished this activating effect. This evidence well documented the direct promoting effects of IRX3 on the browning program. Consistently, IRX3/Irx3 expression was induced during the browning process of WAT in humans (pheochromocytoma patients) and β3-adrenergic agonist-stimulated mice, supporting the physiological role of IRX3 in browning. Irx3 expression was also increased during the time course of beige/brown adipocyte differentiation, supporting the promoting effects of IRX3/Irx3 on browning program. Obviously, this conclusion was opposite to previous reports regarding IRX3’s inhibiting effects on browning in mouse models (Smemo et al., 2014). By using global Irx3-knockout mice and hypothalamus-conditional dominant negative Irx3 transgenic mice with non-C57BL/6J background strains, Smemo et al. found that both mouse strains showed increased Ucp1 expression in perigonadal WAT, indicating its inhibiting role in the browning process of WAT (Smemo et al., 2014). However, another study by Claussnitzer et al. reported that IRX3 knockdown increased UCP1 expression only in human preadipocytes with FTO risk allele but did not show any effect on UCP1 expression in human cells with FTO wild-type allele (Claussnitzer et al., 2015), indicating that IRX3 may inhibit browning dependent on certain genetic background. Besides, we showed that IRX3 knockdown significantly inhibited human beige adipocyte program. Whether the discrepancy between our and Claussnitzer's study is due to the genetic background difference like Caucasian versus Chinese, or due to preadipocyte characterization, for instance, the SVFs from adipose tissues in underaged in our study versus SVFs from adults in Claussnitzer's study, could not be excluded. The detailed underlying mechanism is largely unknown and certainly will be worth exploring in future. Investigation of the effect of Irx3 on the browning program in vivo using the adipose tissue specific knockout mouse model which is different from the dominant negative Irx3 overexpressed model, as used in previous studies Smemo et al., 2014, and with different genetic background such as C57BL/6J strains might give more information regarding the discrepancy. Taken together, these findings may suggest that IRX3 plays a crucial regulatory role in the browning program possibly based on the specific genetic background, and genetic-based stratification may be needed to determine the effectivity of anti-obesity intervention by targeting IRX3.
It is widely known that the browning process is characterized by the induction of UCP1, the uncoupling protein specialized in thermogenesis of mitochondria. A few transcriptional factors such as PPARγ are required for the commitment and differentiation of brown-like adipocytes by targeting UCP1 promoters. Although early studies focused on the regulating region of the UCP1 promoter located ~ 2.5 kb proximal to the TSS, Kong et al found that the transcription factor IRF4 could mediate the transcriptional activity of the UCP1 promoter outside this region (Kong et al., 2014). In addition, studies from the ENCODE consortium indicated that the entire 10 kb sequence upstream to UCP1's TSS could also be of function (https://genome.ucsc.edu/ENCODE/dataSummaryMouse.html). Here, we demonstrated that the IRX3-specific binding site ACATGTGT was located in − 3470 ~ – 3463 bp of UCP1's TSS, and deletion of this sequence ablated transcriptional activation by IRX3. Besides, ChIP assay further identified that Irx3 directly bound to this motif of Ucp1 promoter, providing the evidence that Irx3 could interact with the Ucp1 promoter to increase its transcriptional activity and might regulate thermogenesis as a novel transcriptional factor. It is also worth mentioning whether IRX3 also recruited PGC-1α or other co-activators to promote UCP1 transcription is largely unknown and warranted further investigation. In addition, we also found potential IRX3 binding site on the promoter of Cidea, another affected gene by IRX3. The transcriptional regulation of Cidea by IRX3 remained to be determined in future study. Another point that we want to mention is that deletion of the whole IRX3-binding domain in UCP1 promoter showed increased luciferase activity than wild-type UCP1 promoter, while point mutations in this domain did not increase UCP1 promoter activity, either did not affect IRX3’s promoting effects on UCP1 promoter activity (Supplementary Fig. 4A). These data suggested that the whole IRX3 binding domain might possess an inhibitory effect on UCP1 transcription. We speculate that there might be an inhibitory factor to competitively bind to this IRX3 binding domain to play an inhibitory role on UCP1 transcription activity. Overexpression of IRX3 increased wild-type UCP1 promoter activity but not the binding motif-deleted UCP1 promoter. This result suggested that IRX3 might release the inhibitive effects of this domain on UCP1 transcriptional activity. Taken together, IRX3 might be one activating component in a transcription machinery complex for induction of browning. Detailed mechanism may need more investigations in future study.
FTO and IRX3 could influence obesity organically (Claussnitzer et al., 2015). Although the strong association between FTO variations and obesity has been continuously reported, the genetic defects of the IRX3 gene per se on obesity have not been clarified until now. As the “missing heritability” for complicated disease is partly attributed to low-frequency and rare variants in the candidate genes with a relatively moderate to large effect size (Liu and Song, 2010; Eichler et al., 2010), it seems that targeted resequencing to identify such variants followed by functional validation is an effective strategy to uncover the genetic basis for complex disease including human obesity. In line with this strategy, we have previously identified that a gain-of-function variant in LGR4 (Wang et al., 2013; Zou et al., 2017) and rare loss-of-function variants in NPC1 (Liu et al., 2017) predispose to obesity. In this study, we first revealed the genetic structure of rare variants of IRX3 in relation to obesity. By targeted sequencing of IRX3 coding regions in a relatively large number of young obese and lean subjects, we found that rare nonsynonymous IRX3 variants were enriched in the obese population. Moreover, two nonsense mutants significantly abolished the promoting effect of IRX3 on UCP1 transcriptional activity. These findings together may suggest that loss-of-function mutation in IRX3 may increase human obesity risk by inhibiting browning capacity. More human studies were warranted to confirm this point. We also found significantly lower expression of IRX3 in the subcutaneous fat of obese subjects, which was consistent with two previous studies (Dankel et al., 2010; Landgraf et al., 2016), further supporting the involvement of IRX3 in human obesity. In addition, we want to mention that the two mutations were respectively detected in an obese subject and a normal-weight subject, which suggested that genetic heterozygosity for a severely dysfunctional IRX3 allele did not determine body weight following a classic Mendelian model, as seen in monogenic genes for obesity such as POMC (Flickinger and Salz, 1994; Krude et al., 1998), MC4R (Vaisse et al., 1998; Yeo et al., 1998), and LEP (Montague et al., 1997; Echwald et al., 1997). As such, IRX3 might interact with other obesity-related genes or environmental factors in a complicated manner to determine the penetrance for obesity, although its nonsynonymous variants increased the risk of obesity in humans. A larger-scale case-control replication study or population-based study with a highly homogeneous environmental and genetic background is needed for further investigation.
In summary, our cellular and genetic evidence supported a directly promoting effect of IRX3 on the browning program of white adipocytes by regulating UCP1 transcription and provided a new view for the association of rare variants in IRX3, as one candidate target of FTO variants, with human obesity risk.
Funding Sources
This research was supported by grants from the National Natural Science Foundation of China (81522011, 81570757, 81570758, 81370963, 81370934 and 81370949), National Basic Research Program of China (2015CB553601), Foundation for Innovative Research Groups of the National Natural Science Foundation of China (81621061), National International Science Cooperation Foundation (2015DFA30560), Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (20161306, 20171903), Shanghai Rising-Star Program (17QA1403300), Shanghai Municipal Commission of Health and Family Planning (2017YQ002).
Conflict of Interest
No potential conflicts of interest relevant to this article were reported.
Author Contributions
J.W., W.W., G.N., and W.G. conceived the project and designed the experiments. Y.Z. and P.L. carried out most of the experiments. N.C. assisted in some experiments. Y. Z., J.W. and R.L. wrote the paper. J.S., W.L., M.Y., S.Z., M.C., Y.S., and T.N. recruited obese and control participants in stages one. A.G., Q.C., T.N., and Q.M. provided rodent biological samples for association analysis. X.Y., J.J., X.D., and B.S. provided white adipose samples of obese subjects who received sleeve gastrectomy surgery for weight loss. B.Y. provided subcutaneous adipose samples of subject. W.G., Y.F.Z., J.H., W.W., and G.N. designed and set up the GOCY study. Y.S. assisted with statistical analysis. S.L. and S·K contributed valuable comments and advice on the manuscript. G.N., W.W., W.G., and J.W. are the guarantors of this work and, as such, had full access to all the data in the study and took responsibility for the integrity of the data and the accuracy of the data analysis.
Acknowledgments
Acknowledgements
We thank all the staff and participants for their contributions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2017.09.010.
Contributor Information
Guang Ning, Email: gning@sibs.ac.cn.
Weiqing Wang, Email: wqingw@hotmail.com.
Weiqiong Gu, Email: weiqionggu@163.com.
Jiqiu Wang, Email: wangjq@shsmu.edu.cn.
Appendix A. Supplementary data
Supplementary material
References
- Allison D.B., Kaprio J., Korkeila M., Koskenvuo M., Neale M.C., Hayakawa K. The heritability of body mass index among an international sample of monozygotic twins reared apart. Int. J. Obes. Relat. Metab. Disord. 1996;20:501–506. [PubMed] [Google Scholar]
- Bellefroid E.J., Kobbe A., Gruss P., Pieler T., Gurdon J.B., Papalopulu N. Xiro3 encodes a Xenopus homolog of the Drosophila Iroquois genes and functions in neural specification. EMBO J. 1998;17:191–203. doi: 10.1093/emboj/17.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilioni A., Craig G., Hill C., McNeill H. Iroquois transcription factors recognize a unique motif to mediate transcriptional repression in vivo. Proc. Natl. Acad. Sci. U. S. A. 2005;102:14671–14676. doi: 10.1073/pnas.0502480102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefond A., Clément N., Fawcett K., Yengo L., Vaillant E., Guillaume J.L. Rare MTNR1B variants impairing melatonin receptor 1B function contribute to type 2 diabetes. Nat. Genet. 2012;44:297–301. doi: 10.1038/ng.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussnitzer M., Dankel S.N., Kim K.H., Quon G., Meuleman W., Haugen C. FTO obesity variant circuitry and adipocyte Browning in humans. N. Engl. J. Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cypess A.M., Lehman S., Williams G., Tal I., Rodman D., Goldfine A.B. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cypess A.M., White A.P., Vernochet C., Schulz T.J., Xue R., Sass C.A. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nat. Med. 2013;19:635–639. doi: 10.1038/nm.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankel S.N., Fadnes D.J., Stavrum A.K., Stansberg C., Holdhus R., Hoang T. Switch from stress response to homeobox transcription factors in adipose tissue after profound fat loss. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempersmier J., Sambeat A., Gulyaeva O., Paul S.M., Hudak C.S., Raposo H.F. Cold-inducible Zfp516 activates UCP1 transcription to promote browning of white fat and development of brown fat. Mol. Cell. 2015;57:235–246. doi: 10.1016/j.molcel.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dina C., Meyre D., Gallina S., Durand E., Körner A., Jacobson P. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- Echwald S.M., Rasmussen S.B., Sorensen T.I., Andersen T., Tybjaerg-Hansen A., Clausen J.O. Identification of two novel missense mutations in the human OB gene. Int. J. Obes. Relat. Metab. Disord. 1997;21:321–326. doi: 10.1038/sj.ijo.0800408. [DOI] [PubMed] [Google Scholar]
- Eichler E.E., Flint J., Gibson G., Kong A., Leal S.M., Moore J.H. Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannick J., Thorleifsson G., Beer N.L., Jacobs S.B., Grarup N., Burtt N.P. Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nat. Genet. 2014;46:357–363. doi: 10.1038/ng.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flickinger T.W., Salz H.K. The Drosophila sex determination gene snf encodes a nuclear protein with sequence and functional similarity to the mammalian U1A snRNP protein. Genes Dev. 1994;8:914–925. doi: 10.1101/gad.8.8.914. [DOI] [PubMed] [Google Scholar]
- Frayling T.M., Timpson N.J., Weedon M.N., Zeggini E., Freathy R.M., Lindgren C.M. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra C., Koza R.A., Yamashita H., Walsh K., Kozak L.P. Emergence of brown adipocytes in white fat in mice is under genetic control. Effects on body weight and adiposity. J. Clin. Invest. 1998;102:412–420. doi: 10.1172/JCI3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himms-Hagen J., Cui J., Danforth E.J., Taatjes D.J., Lang S.S., Waters B.L. Effect of CL-316,243, a thermogenic beta 3-agonist, on energy balance and brown and white adipose tissues in rats. Am. J. Phys. 1994;266:1371–1382. doi: 10.1152/ajpregu.1994.266.4.R1371. [DOI] [PubMed] [Google Scholar]
- Hong J., Shi J., Qi L., Cui B., Gu W., Zhang Y. Genetic susceptibility, birth weight and obesity risk in young Chinese. Int. J. Obes. 2013;37:673–677. doi: 10.1038/ijo.2012.87. [DOI] [PubMed] [Google Scholar]
- Koizumi A., Sasano T., Kimura W., Miyamoto Y., Aiba T., Ishikawa T. Genetic defects in a His-Purkinje system transcription factor, IRX3, cause lethal cardiac arrhythmias. Eur. Heart J. 2015;37(18):1469–1475. doi: 10.1093/eurheartj/ehv449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X., Banks A., Liu T., Kazak L., Rao R.R., Cohen P. IRF4 is a key thermogenic transcriptional partner of PGC-1alpha. Cell. 2014;158:69–83. doi: 10.1016/j.cell.2014.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krude H., Biebermann H., Luck W., Horn R., Brabant G., Gruters A. Severe early-onset obesity, adrenal insufficiency and red hair pigmentation caused by POMC mutations in humans. Nat. Genet. 1998;19:155–157. doi: 10.1038/509. [DOI] [PubMed] [Google Scholar]
- Landgraf K., Scholz M., Kovacs P., Kiess W., Körner A. FTO obesity risk variants are linked to adipocyte IRX3 expression and BMI of children - relevance of FTO variants to defend body weight in lean children. PLoS One. 2016;11 doi: 10.1371/journal.pone.0161739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P., Linderman J.D., Smith S., Brychta R.J., Wang J., Idelson C. Irisin and FGF21 are cold-induced endocrine activators of brown fat function in humans. Cell Metab. 2014;19:302–309. doi: 10.1016/j.cmet.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Ward W.F. PGC-1alpha: a key regulator of energy metabolism. Adv. Physiol. Educ. 2006;30:145–151. doi: 10.1152/advan.00052.2006. [DOI] [PubMed] [Google Scholar]
- Lidell M.E., Betz M.J., Dahlqvist Leinhard O., Heglind M., Elander L., Slawik M. Evidence for two types of brown adipose tissue in humans. Nat. Med. 2013;19:631–634. doi: 10.1038/nm.3017. [DOI] [PubMed] [Google Scholar]
- Liu S., Song Y. Building genetic scores to predict risk of complex diseases in humans: is it possible? Diabetes. 2010;59:2729–2731. doi: 10.2337/db10-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Zou Y., Hong J., Cao M., Cui B., Zhang H. Rare loss-of-function variants in NPC1 predispose to human obesity. Diabetes. 2017:db160877. doi: 10.2337/db16-0877. [DOI] [PubMed] [Google Scholar]
- Macotela Y., Emanuelli B., Mori M.A., Gesta S., Schulz T.J., Tseng Y.H. Intrinsic differences in adipocyte precursor cells from different white fat depots. Diabetes. 2012;61:1691–1699. doi: 10.2337/db11-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague C.T., Farooqi I.S., Whitehead J.P., Soos M.A., Rau H., Wareham N.J. Congenital leptin deficiency is associated with severe early-onset obesity in humans. Nature. 1997;387:903–908. doi: 10.1038/43185. [DOI] [PubMed] [Google Scholar]
- Pearce L.R., Atanassova N., Banton M.C., Bottomley B., Klaauw A.A., Revelli J.P. KSR2 mutations are associated with obesity, insulin resistance, and impaired cellular fuel oxidation. Cell. 2013;155:765–777. doi: 10.1016/j.cell.2013.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puar T., Berkel A., Gotthardt M., Havekes B., Hermus A.R., Lenders J.W. Genotype-dependent brown adipose tissue activation in patients with pheochromocytoma and paraganglioma. J. Clin. Endocrinol. Metab. 2016;101:224–232. doi: 10.1210/jc.2015-3205. [DOI] [PubMed] [Google Scholar]
- Ramachandrappa S., Raimondo A., Cali A.M., Keogh J.M., Henning E., Saeed S. Rare variants in single-minded 1 (SIM1) are associated with severe obesity. J. Clin. Invest. 2013;123:3042–3050. doi: 10.1172/JCI68016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiani L., Raciti D., Airik R., Kispert A., Brandli A.W. The prepattern transcription factor Irx3 directs nephron segment identity. Genes Dev. 2007;21:2358–2370. doi: 10.1101/gad.450707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen E.D., Spiegelman B.M. What we talk about when we talk about fat. Cell. 2014;156:20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarlett K., Pattabiraman V., Barnett P., Liu D., Anderson L.M. The proangiogenic effect of iroquois homeobox transcription factor Irx3 in human microvascular endothelial cells. J. Biol. Chem. 2015;290:6303–6315. doi: 10.1074/jbc.M114.601146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuteri A., Sanna S., Chen W.M., Uda M., Albai G., Strait J. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3 doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina I.P., Jong J.M., Kalinovich A.V., Cannon B., Nedergaard J. UCP1 in brite/beige adipose tissue mitochondria is functionally thermogenic. Cell Rep. 2013;5:1196–1203. doi: 10.1016/j.celrep.2013.10.044. [DOI] [PubMed] [Google Scholar]
- Smemo S., Tena J.J., Kim K.H., Gamazon E.R., Sakabe N.J., Gómez-Marín C. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stunkard A.J., Harris J.R., Pedersen N.L., McClearn G.E. The body-mass index of twins who have been reared apart. N. Engl. J. Med. 1990;322:1483–1487. doi: 10.1056/NEJM199005243222102. [DOI] [PubMed] [Google Scholar]
- Vaisse C., Clement K., Guy-Grand B., Froguel P. A frameshift mutation in human MC4R is associated with a dominant form of obesity. Nat. Genet. 1998;20:113–114. doi: 10.1038/2407. [DOI] [PubMed] [Google Scholar]
- Virtanen K.A., Lidell M.E., Orava J., Heglind M., Westergren R., Niemi T. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- Wang J., Liu R., Wang F., Hong J., Li X., Chen M. Ablation of LGR4 promotes energy expenditure by driving white-to-brown fat switch. Nat. Cell Biol. 2013;15:1455–1463. doi: 10.1038/ncb2867. [DOI] [PubMed] [Google Scholar]
- Wu J., Boström P., Sparks L.M., Ye L., Choi J.H., Giang A.H. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G.S., Farooqi I.S., Aminian S., Halsall D.J., Stanhope R.G., O'Rahilly S. A frameshift mutation in MC4R associated with dominantly inherited human obesity. Nat. Genet. 1998;20:111–112. doi: 10.1038/2404. [DOI] [PubMed] [Google Scholar]
- Zhang S.S., Kim K.H., Rosen A., Smyth J.W., Sakuma R., Delgado-Olguin P. Iroquois homeobox gene 3 establishes fast conduction in the cardiac His-Purkinje network. Proc. Natl. Acad. Sci. U. S. A. 2011;108:13576–13581. doi: 10.1073/pnas.1106911108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zhang H., Li B., Meng X., Wang J., Zhang Y. Berberine activates thermogenesis in white and brown adipose tissue. Nat. Commun. 2014;5:5493. doi: 10.1038/ncomms6493. [DOI] [PubMed] [Google Scholar]
- Zou Y., Ning T., Shi J., Chen M., Ding L., Huang Y. Association of a gain-of-function variant in LGR4 with central obesity. Obesity. 2017;25:252–260. doi: 10.1002/oby.21704. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material