Abstract
Advances in developmental cardiology have increased our understanding of the early aspects of heart differentiation. However, understanding noncoding RNA (ncRNA) transcription and regulation during this process remains elusive. Here, we constructed transcriptomes for both long noncoding RNAs (lncRNAs) and circular RNAs (circRNAs) in four important developmental stages ranging from early embryonic to cardiomyocyte based on high-throughput sequencing datasets, which indicate the high stage-specific expression patterns of two ncRNA types. Additionally, higher similarities of samples within each stage were found, highlighting the divergence of samples collected from distinct cardiac developmental stages. Next, we developed a method to identify numerous lncRNA and circRNA regulators whose expression was significantly stage-specific and shifted gradually and continuously during heart differentiation. We inferred that these ncRNAs are important for the stages of cardiac differentiation. Moreover, transcriptional regulation analysis revealed that the expression of stage-specific lncRNAs is controlled by known key stage-specific transcription factors (TFs). In addition, circRNAs exhibited dynamic expression patterns independent from their host genes. Functional enrichment analysis revealed that lncRNAs and circRNAs play critical roles in pathways that are activated specifically during heart differentiation. We further identified candidate TF-ncRNA-gene network modules for each differentiation stage, suggesting the dynamic organization of lncRNAs and circRNAs collectively controlled cardiac differentiation, which may cause heart-related diseases when defective. Our study provides a foundation for understanding the dynamic regulation of ncRNA transcriptomes during heart differentiation and identifies the dynamic organization of novel key lncRNAs and circRNAs to collectively control cardiac differentiation.
Keywords: Cardiac differentiation, Dynamic transcriptome, lncRNA and circRNA, Regulatory network, Function
Graphical Abstract
Highlights
-
•
Dynamic lncRNA and circRNA transcriptomes of heart differentiation were constructed.
-
•
Numerous key lncRNA and circRNA regulators with stage-specific expression during heart differentiation were identified.
-
•
LncRNAs and circRNAs play critical roles in pathways that are activated specifically during heart differentiation.
-
•
Regulatory network analysis revealed dynamic organization of lncRNA and circRNA during cardiac differentiation.
Understanding noncoding RNA (ncRNA) transcription and regulation during cardiac differentiation remains elusive. This study performed transcriptome analysis for both long noncoding RNAs (lncRNAs) and circular RNAs (circRNAs) during four important development stages ranging from early embryonic to cardiomyocyte. A method was developed to identify numerous lncRNA and circRNA regulators whose expression was significantly stage-specific and shifted gradually and continuously during heart differentiation. These ncRNAs were regulated by stage-specific transcription factors and candidate TF-ncRNA-gene network modules were identified. These data suggest the dynamic organization of lncRNAs and circRNAs collectively control cardiac differentiation, which may cause heart-related diseases when defective.
1. Introduction
Pluripotent-to-cardiomyocyte strategies provide invaluable models for understanding the mechanisms of cell fate determination and offer considerable opportunities in cardiac regenerative medicine (Chong et al., 2014; Tompkins et al., 2016). Heart differentiation processes depend on the precise control of gene expression patterns and disruption of gene transcriptional networks may induce congenital heart disease (Tan et al., 2002; Liu et al., 2015). In addition, epigenetic mechanisms, such as noncoding RNA (ncRNA) regulators have recently been related to cardiac development and disease (Li et al., 2013; Di Salvo, 2015; Xu et al., 2016). However, we currently lack a detailed understanding of dynamic ncRNA expression patterns during developmental transitions in the cardiac lineage.
A large part of the mammalian genome has been demonstrated to transcribe into ncRNAs, and long noncoding RNAs (lncRNAs) have emerged as critical regulators of gene expression (Derrien et al., 2012; Ching et al., 2016; Tompkins et al., 2016). Although our understanding of lncRNA functions is still emerging, lines of evidence have demonstrated that lncRNAs are important regulators of heart development and disease. For example, the lncRNA Fendrr has been demonstrated to act as a modulator of gene activity in mouse hearts (Grote et al., 2013). In addition, the lncRNA Braveheart has also been found to interact with the PRC2 complex during cardiac commitment (Klattenhoff et al., 2013). Genome-wide RNA sequencing has also been employed as a method for defining the lncRNA expression signature of developing hearts, and > 100 annotated and newly described lncRNAs have been identified during mouse heart development (Devaux et al., 2015). The lncRNA Mhrt, which localize in nuclei of cardiomyocytes, was weakly expressed in fetal mouse hearts, but was abundant in adult hearts (Wu and Arora, 2015). Together, these findings support the important roles of lncRNAs in cardiac specification and differentiation. However, the exact expression patterns and functions of lncRNAs during human heart differentiation have not yet been systematically studied.
In addition to lncRNAs, circular RNAs (circRNAs) have recently emerged as another class of noncoding RNAs. CircRNAs are much more stable than linear RNAs and may play key roles in many biological processes, including heart development and heart-associated diseases (Salzman, 2016). Discovery of the human circRNA CDR1as acting as a negative regulator of the miRNA miR-7 demonstrated the regulatory potential of circRNAs (Memczak et al., 2013). The circRNA Foxo3 circular RNA (circ-Foxo3) was found to be highly expressed in heart samples from aged patients and mice (Du et al., 2016). This circRNA can interact with multiple factors (such as ID-1, E2F1 and FAK) associated with stress and senescence responses to promote cardiac senescence. Moreover, a heart-related circRNA (HRCR) has been demonstrated to act as a miR-223 sponge to inhibit cardiac hypertrophy and heart failure (Wang et al., 2016). Employing bioinformatics and sequencing technology, Jakobi et al. compiled a catalog of 575 candidate circRNAs in adult murine hearts (Jakobi et al., 2016). They found that many of those candidate circRNAs coincide with disease-associated gene loci, such as circRNAs, originating from Ryr2, Hectd1 and Ppp2r3a. These observations led us to systematically investigate the expression of circRNAs in human heart differentiation.
In this study, to acquire a complete map of lncRNA and circRNA expression and their potential functions during human heart differentiation, we integrated RNA sequencing data to determine changes in the expression of lncRNAs, circRNAs and protein coding genes at sequential stages of differentiation: undifferentiated (ESC), mesoderm (MES), cardiac progenitor (CP) and definitive cardiomyocyte (CM). Focusing on lncRNA and circRNA, we characterized the dynamic expression patterns of noncoding RNA regulators during cardiogenesis. Enrichment analysis revealed key transcription factors (TFs) during heart differentiation. Finally, we identified key functional modules that are formed by lncRNAs and protein coding genes in each differentiation stage by regulatory network analysis. Our analysis defines the repertoire of noncoding transcripts during cardiac differentiation, and enhances our understanding of the important roles of lncRNAs and circRNAs in heart differentiation.
2. Materials and Methods
2.1. LncRNA and Protein Coding Gene Expression During Human Heart Differentiation
Genome-wide expression datasets were obtained from the Gene Expression Omnibus (GEO) public database under accession number GSE64417 (Szabo et al., 2015). Specifically, this dataset included RNA-seq data for four heart differentiation stages: undifferentiated (day 0, ESC), mesoderm (day 2, MES), cardiac progenitor (day 5, CP) and definitive cardiomyocytes (day 14, CM). These data were sequenced by Szabo et al. on the Illumina HiSeq 2500 platform and each stage was performed in biological triplicate, with 3–8 technical replicates each. In total, 71 samples were included in our analysis.
For hESC data at sequential stages of differentiations, raw fastq files from Sequence Read Archive (SRA) were downloaded and processed using Tophat (2.1.0) (Trapnell et al., 2009) for alignment and Cufflinks (2.2.1) (Trapnell et al., 2010) for assembly. All default options for these tools were used. The human reference genome GRCh38 and the corresponding gtf annotation were downloaded from Ensembl (release 81). Then, we harvested the expression profile of 71 samples and kept the genes with fragments per kilobase of transcript per million mapped reads (FPKM) > 1 in at least one heart sample. Based on the biotype annotation in the gtf file, we divided the whole gene expression profile into protein coding and lncRNA parts. In addition, lncRNAs were classified as lincRNAs, antisense lncRNAs, 3′-overlapping ncRNAs, sense-overlapping RNAs, processed transcripts and sense intronic types based on their annotation.
2.2. Identification of circRNAs During Heart Differentiation
To identify circRNAs, rRNA-depleted RNA-seq data were analyzed using the circBase pipeline with default settings (Memczak et al., 2013; Glazar et al., 2014). Briefly, the find_circ pipeline was used to find potential circRNAs from the unaligned reads. The alignment was implemented by Bowtie2 (2.2.5) and the genome build GRCh38 was used in our analysis. Here, we filtered the results for circRNAs that had at least two unique reads and calculated the reads per million (RPM) as the expression of circular RNAs. CircRNAs expressed in at least two samples were retained for further analysis. To identify the host genes of the identified circRNAs, we intersected the circRNA and GRCh38 gtf files using bedtools (2.25.0) (Quinlan and Hall, 2010). Moreover, we used bedtools to map the circRNAs to coding sequences (CDS), 3′ untranslated regions (UTRs), 5′ UTR, antisense transcripts, intronic regions, intergenic regions and ncRNA genes.
2.3. Principal Component Analysis and Hierarchical Analysis
To explore the underlying structure of gene, lncRNA and circRNA expression during heart differentiation, principal component analysis (PCA) was performed on the entire expression profile of all the differentiation stages using the R programming language (Team, 2015). In addition, hierarchical clustering analysis using the R package ‘pheatmap’ (Kolde, 2015) was applied to expression of the top 500 genes, lncRNAs or circRNAs with high expression variations across all samples. The dendrogram was constructed based on the average distance algorithm.
2.4. Stage-Specific lncRNAs, circRNAs and Genes
To identify differentiation stage-specific protein coding genes, lncRNAs and circRNAs, we used an analysis of variance (ANOVA) model with BH-corrected p < 0.05 to select RNAs that were differentially expressed among the differentiation stages. Next, these genes, lncRNAs or circRNAs were filtered based on fold change (FC) between two adjacent differentiation stages. The previous differentiation stage was set as the denominator (MES vs ESC, CP vs MES, CM vs CP). Genes, lncRNAs and circRNAs with FC > 2 were considered to be up-regulated during differentiation and grouped into the ‘Up’ pattern. Genes, lncRNAs and circRNAs with FC < 0.5 were grouped into the ‘Down’ pattern, and the remaining genes were considered non-differentially expressed and grouped into ‘Maintain’. Thus, all genes, lncRNAs and circRNAs were grouped into one of 27 possible patterns (including UUU, UUM, UUD, UMU, UMM, UMD, UDU, UDM, UDD, MUU, MUM, MUD, MMU, MMM, MMD, MDU, MDM, MDD, DUU, DUM, DUD, DMU, DMM, DMD, DDU, DDM and DDD).
2.5. Regulation and Functional Analysis of lncRNAs, circRNAs and Genes
LncRNAs and genes were further clustered according to their expression patterns and we focused on groups with consistent trends during differentiation. Homer (Heinz et al., 2010) was used to identify motifs in the promoters of lncRNAs and genes (encompassing regions 1500 bp upstream and 500 bp downstream of the translation start site) from each cluster. The software arguments ‘-noweight’ and ‘-nlen 0’ were adopted and all of the lncRNAs or genes served as the background set in the motif analysis.
To reveal the functions of lncRNAs and circRNAs with different expression patterns, the co-expressed genes of lncRNAs in each cluster were identified using the Pearson correlation analysis (PCC). For each pair of lncRNA (x) and protein coding gene (y), we calculated the PCC as follows:
where RX was the expression for lncRNA x in samples and RY was the expression for gene y in samples. μRX and μRY were the mean expression of lncRNA x and gene y. σRX and σRY were the standard deviation of RX and RY. E was the expectation. Only pairs with FDR < 0.01 were included for functional enrichment analysis. We made a functional enrichment analysis for each cluster using Bioconductor package ‘TCGAbiolinks’ based on hypergeometric test (Colaprico et al., 2016). Analyses were not performed for clusters with too few co-expressed genes.
2.6. Regulatory Network Analysis of lncRNAs, circRNAs and Genes
The expression profiles of stage-specific genes, lncRNAs and circRNAs were integrated to serve as inputs for the weighted gene co-expression network. Weighted correlation network analysis (WGCNA) (Langfelder and Horvath, 2008) was used for the construction of co-expression networks and identification of the stage-specific modules. Specifically, the pair-wise co-expression matrix Aij was generated, which is a symmetric n ∗ n matrix. Co-expression similarity between gene i and j was defined as the absolute value of the correlation:
The topological overlap matrix (TOM) was then determined based on the co-expression similarity matrix T = [tij]. The TOM similarity between two genes was defined as follows:
where the index u ran across all genes of the network and ki = ∑uaiu. Basically, TOM is an indicator of the agreement between sets of neighboring genes of i and j. The TOM was clustered, and gene modules were identified. Module characterizations were depicted using the expression profiles of eigen-genes.
In addition, we collected 49 heart differentiation-related genes from the literature. To identify heart disease-related genes and lncRNAs, we downloaded the heart disease-related single-nucleotide polymorphisms (SNPs) from GWAS catalog (Welter et al., 2014). Here, we focused on genes and lncRNAs in proximity (within 100 kb) to disease-related SNPs (Wagschal et al., 2015). This process was achieved using bedtools. In total, we identified 70 genes and 94 lncRNAs for further analysis. Moreover, we also added transcription factors into these functional modules. The sequences of lncRNAs were downloaded from UCSC, and ‘match’ was used to predict whether the TF motifs were presented in the promoters of lncRNAs. To identify context-specific TF-lncRNA regulation, we used co-expression analysis to filter sequence-based regulation. Here, we focused on stage-specific expressed lncRNAs, which were those with more than two-fold expression in specific differentiation stages. Genes co-expressed with lncRNAs or circRNAs in the top 1% and with correlation coefficient (absolute value) > 0.9 were retrieved for visualization of the modules. For gene-circRNA pairs, we required the correlation coefficients (absolute value) to be > 0.45.
2.7. Literature Curation of Heart-Related Genes
To investigate whether the genes in the network modules were associated with heart-related functions, we searched PubMed and explore the co-occurrence of genes and heart-related keywords (‘heart’, ‘differentiation’ and ‘cardiac’). This process was performed by R package ‘RISmed’ (https://cran.r-project.org/web/packages/RISmed/index.html).
3. Results
3.1. Global Gene Expression Patterns During Heart Differentiation
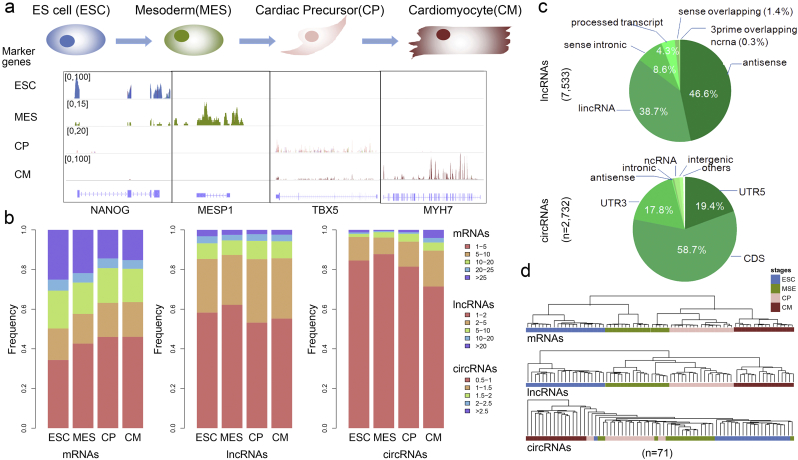
To investigate the temporal expression transcriptome profiles of human heart maturation, we collected RNA-Seq-based datasets at defined stages of cardiogenesis (Szabo et al., 2015), including ESC, MES, CP and CM. After read mapping and transcriptome assembly, 15,574 mRNAs, 7533 lncRNAs and 1702 circRNAs were expressed during heart maturation (Fig. S1). Specifically, some marker genes were specifically expressed at corresponding stages, such as the pluripotency genes OCT4 and NANOG that were expressed at higher levels in the ESC stage, and mesodermal markers (BRACHYURY and MESP1) and cardiac transcription factors (ISL1 and TBX5) that were expressed in the MES and CP stages. In addition, MYH6 and MYH7 were specifically expressed in the CM stage (Fig. 1a and Fig. S2).
Fig. 1.
Global transcriptome expression patterns during heart differentiation. (a) Read coverage visualization of stage-specific marker genes using the Integrative Genomics Viewer. The peak heights indicate the number of reads mapped to the corresponding genomic region. (b) Distribution of different abundances of genes, lncRNAs and circRNAs within each stage. Compared with mRNAs, the majority of lncRNAs as well as circRNAs were enriched at a lower expression level. (c) Genome annotation of lncRNAs (n = 7533) and circRNAs (n = 2732) showing that the majority of lncRNAs were mapped to antisense (46.6%) or lincRNA (38.7%), and the majority of circRNAs were derived from a CDS (58.7%). (d) Hierarchical clustering analysis suggested that samples (n = 71) within the same stage could be clustered together according to the top 500 variant genes, lncRNAs and circRNAs.
Analysis of the global expression patterns of mRNAs, lncRNAs and circRNAs during cardiac differentiation showed that the expression of lncRNAs was lower than that of mRNAs (Fig. 1b and Fig. S3). Approximately 60% of the lncRNAs had FPKM values between 1 and 2 in the four differentiation stages, whereas only approximately 4% had FPKM values > 20. Next, we mapped lncRNAs to genomic regions and found that 38.7% of the lncRNAs were derived from intergenic regions (Fig. 1c), suggesting that high-throughput sequencing allows the investigation of more noncoding RNAs that play key roles in cardiogenesis. In addition, evidence has demonstrated that neural tissues are highly enriched in circRNAs compared to other tissues (Veno et al., 2015). Although the expression of circRNAs was even lower than that of lncRNAs (Fig. 1b), several circRNAs (such as, circ-SLC8A1) were supported by about 100 junction-spanning reads (Fig. S4). Consistent with previous studies in other tissues (Rybak-Wolf et al., 2015), these heart-expressed circRNAs were usually derived from coding sequences (CDS, 58.7%) and 5′ UTR exons (19.4%, Fig. 1c).
Next, we performed principal component analysis (PCA) to reveal the underlying structure of the whole transcriptome from distinct differentiation stages. PCA analysis showed that samples from each stage were well aligned by the mRNA and lncRNA expression profiles (Fig. S5a). In addition, all the samples were clustered by the top 500 genes, lncRNAs and circRNAs with higher expression variation. Clustering analysis showed that samples of the same stages were clustered together well (Fig. 1d and Fig. S5b). These analyses showed a higher similarity of samples within each stage and highlighted the divergence of samples collected from distinct cardiac developmental stages, suggesting that overall gene, lncRNA and circRNA expression patterns change gradually over the course of heart maturation.
3.2. Dynamic Transcriptional Profiles Diverge at Distinct Differentiation Stages
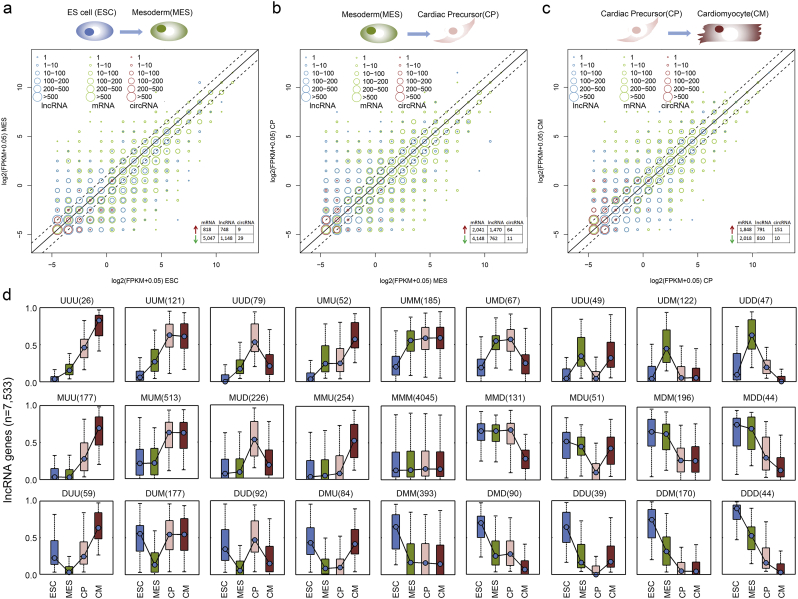
Based on the patterns revealed by PCA and clustering analysis, RNA expression changes gradually during cardiogenesis. We therefore identified temporally differentially expressed genes, lncRNAs and circRNAs using the ANOVA approach. Based on a false discovery rate (FDR) < 0.05, 3488 lncRNAs, 10,115 mRNAs and 198 circRNAs with temporal expression were identified (Fig. 2a-c and Supplementary Table S1, Supplementary Table S2, Supplementary Table S3). Unexpectedly, mRNAs and lncRNAs were changed mostly during the first three stages. Specifically, 1470 lncRNAs were up-regulated during MES differentiation (Fig. 2b), suggesting that lncRNAs are likely important for cell commitment. However, most circRNAs (161) were differently expressed during the differentiation from CP to CM. Of these circRNAs, most were up-regulated in CM (Fig. 2c), indicating that circRNAs might play key roles in heart cell specification.
Fig. 2.
Dynamic transcriptional profiles diverge at distinct differentiation stages. (a) Bubble charts of differentially expressed genes, lncRNAs and circRNAs during ESC to MES transformation. The x-axis indicates log2-transformed expression levels at lower stages and the sizes of the bubbles represent the frequency of genes (green), lncRNAs (blue) and circRNAs (red) at the corresponding expression levels. The dummy line indicates a two-fold change between the sequential stages. The number of up-regulated and down-regulated mRNAs, lncRNAs and circRNAs are shown in the right-bottom table. (b) Bubble charts of differentially expressed genes, lncRNAs and circRNAs during the MES to CP transformation. (c) Bubble charts of differentially expressed genes, lncRNAs and circRNAs during the CP to CM transformation. (d) The 27 expression patterns of lncRNAs. LncRNAs were grouped into the U (upregulated), D (downregulated) or M (maintain) based on two-fold change of expression between adjacent differentiation stages. Each pattern contains four boxplots corresponding to four sequential stages. The y-axis represents normalized expression values, and the poly lines indicate expression variance trends during stage progressions. The number of lncRNAs within the pattern is displayed in the bracket. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
By comparing any two adjacent differentiation stages, using the younger stage as the denominator (see Methods), 27 possible patterns of lncRNA and mRNA expression were identified (Fig. 2d and Fig. S6). LncRNAs and mRNAs were non-randomly represented across all these patterns. Relatively few lncRNAs and mRNAs exhibited continuously increased (UUU, 26 lncRNAs and 23 protein coding genes) or decreased (DDD, 44 lncRNAs and 239 protein coding genes) expression during heart differentiation, as most lncRNAs (53.70% or 4045/7533) and mRNAs (35.05% or 5459/15,574) remained unchanged during cardiogenesis. However, the MES to CP differentiation triggers significant changes in lncRNAs (MUM or MDM). This process also revealed some known lncRNAs that were demonstrated to play critical roles in heart, including LNC-ROR and KCNQ1OT1 (Fig. S7).
Regarding circRNAs, only one circRNAs exhibited continuously increased expression (circ-PCMTD1), and one circRNA exhibited continuously decreased expression (circ-TUBA1B, Fig. S8). Expression of most circRNAs (103) was triggered by CP differentiation (MMU, (Table S3). Specifically, two circRNAs circ-TTN were up-regulated from the MES to CM stage (MUU), suggesting their critical roles in cardiogenesis (Tan et al., 2017). However, we observed that the expression patterns of its host gene (TTN) was distinct from this circRNA, suggesting that they function independently from the linear genes. Taken together, these results suggested a dynamic transcriptome during heart differentiation, and our analyses identified several potential non-coding regulators and will facilitate additional studies of lncRNAs and circRNAs in cardiogenesis.
3.3. Stage-Specific Regulation and Cellular Functions of lncRNAs During Cardiac Differentiation
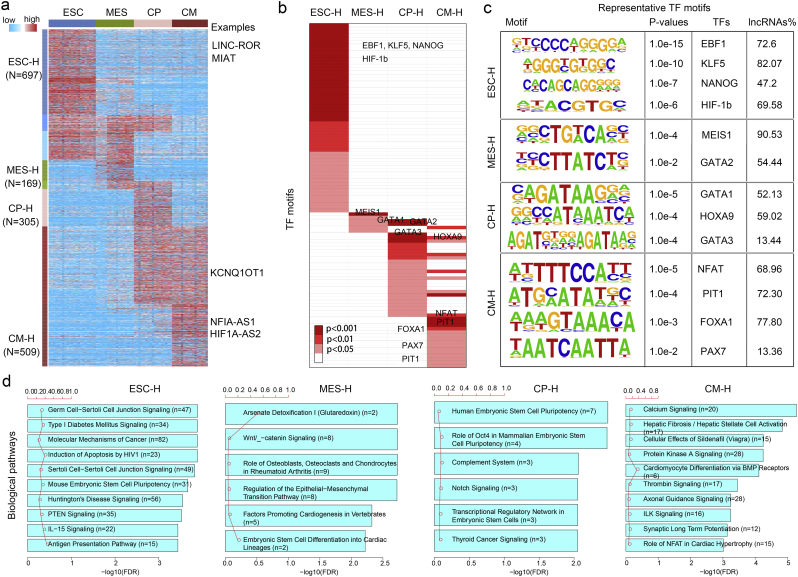
The above analyses indicated that lncRNAs are expressed in a stage-specific manner during cardiac differentiation, which raised the question of which factors regulate stage-specific expression patterns. To address this central question, we focused on eight groups of lncRNAs without inverse expression during differentiation (Fig. 3a). Among these lncRNAs, we identified 697, 169, 305 and 509 lncRNAs that were highly expressed at the ESC, MES, CP and CM stages, respectively. Several representative lncRNAs were also identified, including LNC-ROR, KCNQ1OT1 and NFIA-AS1. LNC-ROR has been shown to contribute to the maintenance of embryonic stem cells. In this study, this lncRNA was highly expressed in ESC, and its expression was decreased during cardiac differentiation (Fig. 3a and Fig. S7). In addition, KCNQ1OT1 has been shown to effect transcription via chromatin flexibility, and access to enhancers in the developing heart (Korostowski et al., 2012; Devaux et al., 2015) and to be highly expressed in cardiac progenitors and cardiomyocytes (Fig. 3a and Fig. S7). Notably, NFIA-AS1, which was shown to act as a regulator to control NFIA-regulated miR-382-5p expression in atherosclerosis (Hu et al., 2015; Ballantyne et al., 2016), showed a stage-specific expression pattern in cardiomyocytes (Fig. S7).
Fig. 3.
Stage-specific regulation and cellular functions of lncRNAs during cardiac differentiation. (a) Dynamic expression profile of lncRNAs. The expression patterns of lncRNAs were further clustered and stage-specific highly expressed clusters are labeled with ‘-H’ with the corresponding color. Some examples are listed beside the cluster. (b) Stage-specific enrichment of TF motifs. Each cell represents a TF motif that enriched in the promoters of stage-specific lncRNAs and the color indicates enrichment significance (p values). (c) Representative TF motifs in the different stages that were labeled in (b). (d) Biological processes enriched by stage-specific genes co-expressed with lncRNAs. The bars represent the statistical significance of functional enrichment analysis with the numbers of genes. The red line indicates the ratio of listed genes found in each pathway over the total number of genes in that pathway. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Given that transcription factors (TFs) can act as master regulators of gene expression programs (Budden et al., 2015; Uosaki et al., 2015), we hypothesized that motifs for TFs that drive cardiac differentiation would be enriched in the promoters of lncRNAs. We found that TF motifs showed stage-specific enrichment patterns (Fig. 3b). Some TF motifs were over-represented at each differentiation stage, including those that regulate the ESC state (NANOG) and cardiac development (Fig. 3c, such as MEIS, GATA, NFAT, etc.). MEIS1 has been implicated in heart development (Wamstad et al., 2012), and our analysis suggested that approximately 90.53% of all lncRNAs are enriched for the MEIS1 motif. Moreover, NFAT coupled with calcineurin regulates cardiomyocyte maturation (Graef et al., 2001; Hogan et al., 2003; Schulz and Yutzey, 2004), but abnormal signaling will result in pathological cardiac hypertrophy (Molkentin, 2004; Wilkins et al., 2004) and heart failure (Diedrichs et al., 2004). We observed that approximately 68.96% of the lncRNAs that were highly expressed at the CM stage could bind NFAT (Fig. 3c). These results indicated that the stage-specific expression of lncRNAs are strictly regulated by these TFs.
The regulation and function of stage-specific protein-coding genes also revealed that they are regulated by specific TFs and enriched for heart-associated functions (Fig. S9). We thus employed ‘guilt by association’ to identify the corresponding stage-specific protein-coding genes that are co-expressed with specifically expressed lncRNAs at each stage (Pearson's correlation FDR < 0.01) and performed functional enrichment analysis. Our analysis revealed that lncRNAs were indeed involved in cardiac specification and differentiation. For instance, CP-specific lncRNAs were enriched for ‘notch signaling pathway’ (Fig. 3d), which is a crucial cell-fate regulator in the developing heart (Niessen and Karsan, 2008; de la Pompa, 2009; Kwon et al., 2009; de la Pompa and Epstein, 2012). Mutations in this pathway have been associated with human congenital heart defects, such as aortic valve disease (Garg et al., 2005) and ventricular septal defects. In addition, we observed that several familiar signals including calcium, PKA and ILK signaling were enriched at the CM stage (Fig. 3d). Lines of evidence have suggested that these signals play essential roles in cardiomyocytes. For example, protein kinase A/C can regulate cardiac L-type calcium channels, and abnormal calcium remodelling may cause heart disease (Kamp and Hell, 2000). ILK signaling can promote cardiac cell migration, survival and repair (Bock-Marquette et al., 2004; Hannigan et al., 2007). Consequently, these results indicate that lncRNAs exhibit spatially and temporally dynamic expression under the control of specific TFs and could serve as important regulators during cardiac differentiation.
3.4. Functional Analysis of circRNAs Associated With Cardiogenesis
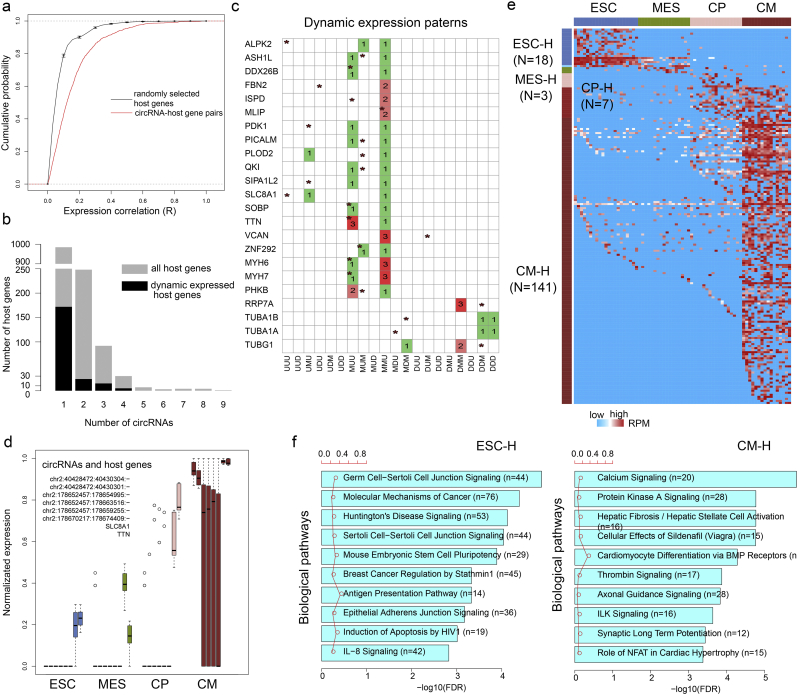
Recent studies have suggested that circRNA expression does not always correlate with expression of the linear transcript from which it is derived (Memczak et al., 2013; Salzman et al., 2013). This could be due to differences in their biogenesis. Although most circRNAs were not co-expressed with their host genes, we observed an obvious co-expression relationship between circRNAs and their corresponding mRNAs compared with random mRNAs (Fig. 4a, p < 2.2e-16). In addition, we observed that > 400 host genes from which circRNAs were derived could process more than one circRNAs (Fig. 4b) but their expression patterns could be different. In total, we identified 23 differentially expressed genes that were capable of generating more than one circRNA (Fig. 4c). All of these host genes could generate at least one circRNA with a different expression pattern. For example, SLC8A1 possess two identical circRNAs but have different expression pattern within two of the circRNAs (Fig. 4d). SLC8A1 showed continually up-regulation during heart differentiation (UUU), but one circRNA had the UMU pattern, while the other one had the MMU pattern. There is one circRNA derived from TTN is also with distinct pattern from its host gene (Fig. 4d).
Fig. 4.
Functional analysis of circRNAs. (a) Cumulative distribution of the expression correlation between circRNAs and their host genes. The bar plot represents of coefficients variances in random conditions. (b) Barplot of the counts of host genes, which transcribed various numbers of circRNAs. (c) Lattice graph showing the dynamic expression patterns of circRNAs and their host genes. Host genes with more than one circRNAs are shown, and the dynamic expression patterns without ‘MMM’ are shown. The asterisk indicates host gene, and the colored cells represent one or more circRNAs filled with digits. (d) Boxplots of the expression of SLC8A1 and TTN together with circRNAs during cardiac differentiation. (e) Dynamic expression profile of circRNAs (n = 169) in cardiogenesis. (f) Biological pathways of stage-specific circRNAs in the stages ESC and CM.
CircRNAs are widely expressed in cell-type or tissue-specific patterns, and a subset of circRNAs appears to be conserved across species (Jeck and Sharpless, 2014; Qu et al., 2015). In addition, many studies suggest that circRNAs display dynamic expression patterns during development (Memczak et al., 2013; Rybak-Wolf et al., 2015), which is what we observed during cardiogenesis (Fig. 4e). The dynamic changes in abundance suggested that circRNAs may play a key role in cardiogenesis. We therefore performed functional enrichment analysis using ‘guilt by association’ and found that the pathways in which circRNAs participated were basically consistent with specification and differentiation (Fig. 4f), including mouse ESC pluripotency being enriched at the ESC stage and calcium signaling and cardiomyocyte differentiation via BMP receptors being enriched at the CM stage.
3.5. Network Analysis Reveals That lncRNAs and circRNAs are Associated With Cardiac Differentiation
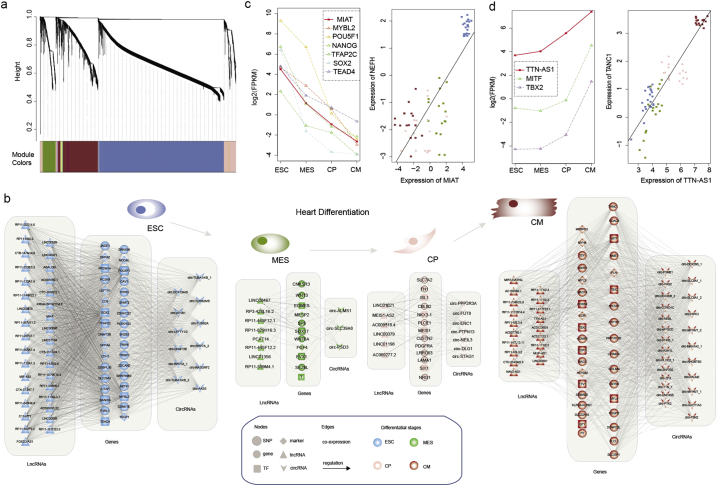
A usual notion suggests that molecular networks are central to biological function and networks describing a certain biological process may depend on biological contexts such as cell type. We hypothesized that interactions among genes, lncRNAs and circRNAs partly expose their biological processes in cardiogenesis. We then performed weighted gene co-expression network analysis for stage-specific lncRNAs, circRNAs and genes (including 4876 genes, 1680 lncRNAs and 169 circRNAs). Based on the expression profiles of eigen-genes, we identified six stage-specific modules (Fig. 5a). Next, we focused on four modules that showed strong stage-specific expression. Then, we extracted the top 1% of genes that were co-expressed with lncRNAs or circRNAs from the co-expressed modules and identified some key lncRNAs that may play roles in heart-related diseases. We observed that the genes and lncRNAs in this core sub-network show dynamic expression during cardiac differentiation (Fig. S10–S13).
Fig. 5.
Network view of heart differentiation related modules. (a) Network heat map plot of the lncRNA-gene-circRNA co-expression network. Branches in the hierarchical clustering dendrogram correspond to modules. Color-coded module membership is displayed in the colored bars below the heat map. (b) Network view of lncRNA, circRNA, gene and TF modules. From left to right, the lncRNAs, genes and circRNAs are shown in specific heart differentiation stages. The gray lines indicate co-expression relationships, and the arrow lines indicate transcriptional regulation. Nodes with black edges indicate that the presence of heart disease-related SNPs around the genes or lncRNAs. (c)–(d) Expression of representative lncRNAs, TFs and regulated protein coding genes during heart differentiation.
In addition, our results indicated that the dynamic expression of lncRNAs and genes was strictly regulated by TFs. We thus included TF regulation into these modules (Fig. 5b and Table S4). Our network analysis revealed some lncRNAs, as well as the regulators mediated their dynamic expression that play critical roles in heart differentiation (Table S5). MIAT has been shown to exhibit significantly increased expression in Ang II-induced cardiac hypertrophy and to contribute to pathological development by suppressing miR-150 expression in cardiomyocytes (Zhu et al., 2016). We observed that this lncRNA was highly expressed at the ESC stage and co-expressed with NEFH (Fig. 5c). Specifically, the promoter region of this lncRNA can be bound by ES-specific TFs, such as POU5F1, NANOG and SOX2. These TFs were co-expressed with MIAT (Fig. 5c). These results show that with the strict regulation imposed by these ES-specific TFs, the lncRNA MIAT might play critical roles in ESC by targeting NEFH coding genes. In addition, our analyses revealed another lncRNA (TTN-AS1) that might play important roles in heart differentiation. This lncRNA exhibited increased expression during heart differentiation (Fig. 5d), and was regulated by MIFT and TBX2. MITF has been demonstrated to play an essential role in β-adrenergic–induced cardiac hypertrophy (Tshori et al., 2006), and Tbx2 is a determinant in the local repression of chamber-specific gene expression and chamber differentiation (Christoffels et al., 2004). Moreover, we found that this lncRNA was co-expressed with TANC1, which has been shown to be highly expressed in in human hearts (Arking et al., 2011). All these results point to the important roles of TTN-AS1 in heart differentiation.
Moreover, we also revealed some important circRNA regulators, such as circ-TTN. Based on network analysis, these circRNAs were co-expressed with some critical genes, including MYL4 (Fig. 5b). Evidence has indicated that the mutation of MYL4 leads to the disruption of sarcomere structure, atrial enlargement and electrical abnormalities associated with human atrial fibrillation (Orr et al., 2016). Our analysis elucidated another regulatory layer for this gene by targeting circRNAs in heart differentiation. Taken together, integrated network analysis revealed not only several critical lncRNA and circRNA regulators in heart differentiation, but also their upstream regulators and downstream target genes.
4. Discussion
To advance our understanding of the molecular underpinnings of heart differentiation, we performed a global survey of ncRNA transcriptome profiles across discrete stages of heart differentiation. Our analyses showed dramatic differential expression changes of lncRNAs and circRNAs during cardiogenesis, revealing a previously unrecognized complexity of cardiac differentiation.
An increasing number of studies have demonstrated the critical roles of lncRNA in heart development and differentiation. Some lncRNAs, such as Braveheart and Mhrt have been shown to exhibit aberrant expression in heart-related diseases. However, most studies focus on defining the regulatory functions of lncRNAs, whereas few investigations focus on assessing how lncRNA themselves are transcriptionally regulated (Tompkins et al., 2016). Here, we found that the atlas of ncRNA temporal expression was well correlated with transcriptional regulation changes. Most TF regulators, such as NANOG, MEIS1, GATA1 and NFAT, were activated or inactivated at specific heart differentiation stages. In addition, these stage-specific ncRNAs were well correlated with the biological functions of each differentiation stage. For example, lncRNAs highly expressed in the ESC stage were related to the ESC pluripotency pathway and those highly expressed in the CM stage were enriched in calcium, cardiomyocyte differentiation via BMP receptors and ILK signaling pathways. In addition to TF regulation, lines of evidence have also indicated that epigenetic regulation, including DNA methylation and histone modification play critical roles in regulating the expression of lncRNAs (Serra-Juhe et al., 2015; Zhong et al., 2016). Integration of multiple epigenetic datasets may provide novel insights into the regulation of lncRNAs during heart differentiation.
Deep sequencing techniques and advanced bioinformatics analysis methods recently enabled the characterization of thousands of circRNAs in numerous tissues and organisms. Emerging evidence also suggests that some circRNAs may have important biological functions or serve as diagnostic biomarkers in disease conditions, including heart differentiation and heart related diseases. Here, we observed that most circRNAs were also dynamically expressed during cardiogenesis. Specifically, they show distinct expression patterns with their host genes (such as circ-SLC8A1), indicating their independent functions. Concerning their biological function, circRNAs are hypothesized to serve as epigenetic miRNA sponges (Taulli et al., 2013). However, available experimental data are not very comprehensive, especially for investigating the global regulation and function of circRNAs. In this study, we inferred the circRNA functions based on the genes with which they were co-expressed and the identified circRNAs showed stage-specific functions during heart differentiation.
In this current study, we used the commonly used method (ANOVA) to identify the differentially expressed mRNAs, lncRNAs and circRNAs during heart differentiation (Yu et al., 2014; Zhang et al., 2016). There are a number of statistical tests specifically designed for RNA-Seq data, such as DESeq (Love et al., 2014) and edgeR (Robinson et al., 2010). To identify the genes that are more likely to be important, we also used DESeq to identify the DEGs. We found that the overlap of DEGs is very high (92.01% for lncRNAs; 75% for mRNAs), suggesting that to some extent our results were robust to the statistical tests. In addition, several pipelines have been developed to specifically identify the nonlinear reads and consequently predict the landscape of circRNAs based on deep sequencing datasets. Integration of the results derived from different methods are likely to reduce the false positives. Next, we also used CIRI (Szabo et al., 2015) to identify circRNAs. We found that the majority (64.80%) of circRNAs were also identified by this method. These overlapping circRNAs might be better candidates for further experimental validation (Table S3).
The current study utilized bioinformatics to determine the dynamic transcriptome landscapes of lncRNA and circRNA regulators during heart differentiation. Thus, it will be important to investigate the expression dynamic of the key regulators and their target genes by biochemical methods in the future. Collectively, our study provides a foundation for understanding lncRNA and circRNA expression and regulation during heart differentiation.
The following are the supplementary data related to this article.
Supplementary figures
The gene expression patterns during heart differentiation.
The lncRNA expression patterns during heart differentiation.
The circRNA expression patterns during heart differentiation.
The network modules for the specific stage of heart differentiation.
Literature curation of the genes, lncRNAs and circRNAs in network modules.
Funding Sources
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31571331 and 61502126), the China Postdoctoral Science Foundation (Grant Nos. 2016T90309, 2015M571436 and LBH-Z14134), the Natural Science Foundation of Heilongjiang Province (Grant No. QC2015020), the Weihan Yu Youth Science Fund Project of Harbin Medical University, the Harbin Special Funds of Innovative Talents on Science and Technology Research Project (Grant No. 2015RAQXJ091). The funding sources had no role in the design or conduction of the study.
Conflicts of Interest
The authors declare no conflicts of interests.
Author Contributions
JX, YZ and BC designed the experiments. YL, BC and JX interpreted the data and wrote the manuscript with input from other co-authors. JZ, CH and ND performed the data analysis. JL, JX and XL performed the regulatory network analysis and figure designs.
Contributor Information
Benzhi Cai, Email: caibz@ems.hrbmu.edu.cn.
Yunpeng Zhang, Email: zyp19871208@126.com.
Juan Xu, Email: xujuanbiocc@ems.hrbmu.edu.cn.
References
- Arking D.E., Junttila M.J., Goyette P., Huertas-Vazquez A., Eijgelsheim M., Blom M.T., Newton-Cheh C., Reinier K., Teodorescu C., Uy-Evanado A., Carter-Monroe N., Kaikkonen K.S., Kortelainen M.L., Boucher G., Lagace C., Moes A., Zhao X., Kolodgie F., Rivadeneira F., Hofman A., Witteman J.C., Uitterlinden A.G., Marsman R.F., Pazoki R., Bardai A., Koster R.W., Dehghan A., Hwang S.J., Bhatnagar P., Post W., Hilton G., Prineas R.J., Li M., Kottgen A., Ehret G., Boerwinkle E., Coresh J., Kao W.H., Psaty B.M., Tomaselli G.F., Sotoodehnia N., Siscovick D.S., Burke G.L., Marban E., Spooner P.M., Cupples L.A., Jui J., Gunson K., Kesaniemi Y.A., Wilde A.A., Tardif J.C., O'Donnell C.J., Bezzina C.R., Virmani R., Stricker B.H., Tan H.L., Albert C.M., Chakravarti A., Rioux J.D., Huikuri H.V., Chugh S.S. Identification of a sudden cardiac death susceptibility locus at 2q24.2 through genome-wide association in European ancestry individuals. PLoS Genet. 2011;7:e1002158. doi: 10.1371/journal.pgen.1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballantyne M.D., McDonald R.A., Baker A.H. lncRNA/MicroRNA interactions in the vasculature. Clin. Pharmacol. Ther. 2016;99:494–501. doi: 10.1002/cpt.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock-Marquette I., Saxena A., White M.D., Dimaio J.M., Srivastava D. Thymosin beta4 activates integrin-linked kinase and promotes cardiac cell migration, survival and cardiac repair. Nature. 2004;432:466–472. doi: 10.1038/nature03000. [DOI] [PubMed] [Google Scholar]
- Budden D.M., Hurley D.G., Crampin E.J. Predictive modelling of gene expression from transcriptional regulatory elements. Brief. Bioinform. 2015;16:616–628. doi: 10.1093/bib/bbu034. [DOI] [PubMed] [Google Scholar]
- Ching T., Peplowska K., Huang S., Zhu X., Shen Y., Molnar J., Yu H., Tiirikainen M., Fogelgren B., Fan R., Garmire L.X. Pan-cancer analyses reveal long intergenic non-coding RNAs relevant to tumor diagnosis, subtyping and prognosis. EBioMedicine. 2016;7:62–72. doi: 10.1016/j.ebiom.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J.J., Yang X., Don C.W., Minami E., Liu Y.W., Weyers J.J., Mahoney W.M., Van Biber B., Cook S.M., Palpant N.J., Gantz J.A., Fugate J.A., Muskheli V., Gough G.M., Vogel K.W., Astley C.A., Hotchkiss C.E., Baldessari A., Pabon L., Reinecke H., Gill E.A., Nelson V., Kiem H.P., Laflamme M.A., Murry C.E. Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature. 2014;510:273–277. doi: 10.1038/nature13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels V.M., Hoogaars W.M., Tessari A., Clout D.E., Moorman A.F., Campione M. T-box transcription factor Tbx2 represses differentiation and formation of the cardiac chambers. Dev. Dyn. 2004;229:763–770. doi: 10.1002/dvdy.10487. [DOI] [PubMed] [Google Scholar]
- Colaprico A., Silva T.C., Olsen C., Garofano L., Cava C., Garolini D., Sabedot T.S., Malta T.M., Pagnotta S.M., Castiglioni I., Ceccarelli M., Bontempi G., Noushmehr H. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G., Lagarde J., Veeravalli L., Ruan X., Ruan Y., Lassmann T., Carninci P., Brown J.B., Lipovich L., Gonzalez J.M., Thomas M., Davis C.A., Shiekhattar R., Gingeras T.R., Hubbard T.J., Notredame C., Harrow J., Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux Y., Zangrando J., Schroen B., Creemers E.E., Pedrazzini T., Chang C.P., Dorn G.W., II, Thum T., Heymans S., Cardiolinc N. Long noncoding RNAs in cardiac development and ageing. Nat. Rev. Cardiol. 2015;12:415–425. doi: 10.1038/nrcardio.2015.55. [DOI] [PubMed] [Google Scholar]
- Di Salvo T.G. Epigenetic regulation in heart failure: part I RNA. Cardiol. Rev. 2015;23:213–228. doi: 10.1097/CRD.0000000000000071. [DOI] [PubMed] [Google Scholar]
- Diedrichs H., Chi M., Boelck B., Mehlhorn U., Schwinger R.H. Increased regulatory activity of the calcineurin/NFAT pathway in human heart failure. Eur. J. Heart Fail. 2004;6:3–9. doi: 10.1016/j.ejheart.2003.07.007. [DOI] [PubMed] [Google Scholar]
- Du W.W., Yang W., Chen Y., Wu Z.K., Foster F.S., Yang Z., Li X., Yang B.B. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2016 doi: 10.1093/eurheartj/ehw001. [DOI] [PubMed] [Google Scholar]
- Garg V., Muth A.N., Ransom J.F., Schluterman M.K., Barnes R., King I.N., Grossfeld P.D., Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- Glazar P., Papavasileiou P., Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef I.A., Chen F., Crabtree G.R. NFAT signaling in vertebrate development. Curr. Opin. Genet. Dev. 2001;11:505–512. doi: 10.1016/s0959-437x(00)00225-2. [DOI] [PubMed] [Google Scholar]
- Grote P., Wittler L., Hendrix D., Koch F., Wahrisch S., Beisaw A., Macura K., Blass G., Kellis M., Werber M., Herrmann B.G. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannigan G.E., Coles J.G., Dedhar S. Integrin-linked kinase at the heart of cardiac contractility, repair, and disease. Circ. Res. 2007;100:1408–1414. doi: 10.1161/01.RES.0000265233.40455.62. [DOI] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis‑regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan P.G., Chen L., Nardone J., Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- Hu Y.W., Zhao J.Y., Li S.F., Huang J.L., Qiu Y.R., Ma X., Wu S.G., Chen Z.P., Hu Y.R., Yang J.Y., Wang Y.C., Gao J.J., Sha Y.H., Zheng L., Wang Q. RP5-833A20.1/miR-382-5p/NFIA-dependent signal transduction pathway contributes to the regulation of cholesterol homeostasis and inflammatory reaction. Arterioscler. Thromb. Vasc. Biol. 2015;35:87–101. doi: 10.1161/ATVBAHA.114.304296. [DOI] [PubMed] [Google Scholar]
- Jakobi T., Czaja-Hasse L.F., Reinhardt R., Dieterich C. Profiling and validation of the circular RNA repertoire in adult murine hearts. Genomics Proteomics Bioinformatics. 2016;14:216–223. doi: 10.1016/j.gpb.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp T.J., Hell J.W. Regulation of cardiac L-type calcium channels by protein kinase A and protein kinase C. Circ. Res. 2000;87:1095–1102. doi: 10.1161/01.res.87.12.1095. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C.A., Scheuermann J.C., Surface L.E., Bradley R.K., Fields P.A., Steinhauser M.L., Ding H., Butty V.L., Torrey L., Haas S., Abo R., Tabebordbar M., Lee R.T., Burge C.B., Boyer L.A. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R. 2015. pheatmap: Pretty Heatmaps. [Google Scholar]
- Korostowski L., Sedlak N., Engel N. The Kcnq1ot1 long non-coding RNA affects chromatin conformation and expression of Kcnq1, but does not regulate its imprinting in the developing heart. PLoS Genet. 2012;8:e1002956. doi: 10.1371/journal.pgen.1002956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C., Qian L., Cheng P., Nigam V., Arnold J., Srivastava D. A regulatory pathway involving Notch1/beta-catenin/Isl1 determines cardiac progenitor cell fate. Nat. Cell Biol. 2009;11:951–957. doi: 10.1038/ncb1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Xu J., Chen H., Bai J., Li S., Zhao Z., Shao T., Jiang T., Ren H., Kang C., Li X. Comprehensive analysis of the functional microRNA-mRNA regulatory network identifies miRNA signatures associated with glioma malignant progression. Nucleic Acids Res. 2013;41 doi: 10.1093/nar/gkt1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Morley M., Brandimarto J., Hannenhalli S., Hu Y., Ashley E.A., Tang W.H., Moravec C.S., Margulies K.B., Cappola T.P., Li M., consortium, MA RNA-Seq identifies novel myocardial gene expression signatures of heart failure. Genomics. 2015;105:83–89. doi: 10.1016/j.ygeno.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., Loewer A., Ziebold U., Landthaler M., Kocks C., le Noble F., Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Molkentin J.D. Calcineurin-NFAT signaling regulates the cardiac hypertrophic response in coordination with the MAPKs. Cardiovasc. Res. 2004;63:467–475. doi: 10.1016/j.cardiores.2004.01.021. [DOI] [PubMed] [Google Scholar]
- Niessen K., Karsan A. Notch signaling in cardiac development. Circ. Res. 2008;102:1169–1181. doi: 10.1161/CIRCRESAHA.108.174318. [DOI] [PubMed] [Google Scholar]
- Orr N., Arnaout R., Gula L.J., Spears D.A., Leong-Sit P., Li Q., Tarhuni W., Reischauer S., Chauhan V.S., Borkovich M., Uppal S., Adler A., Coughlin S.R., Stainier D.Y., Gollob M.H. A mutation in the atrial-specific myosin light chain gene (MYL4) causes familial atrial fibrillation. Nat. Commun. 2016;7:11303. doi: 10.1038/ncomms11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Pompa J.L. Notch signaling in cardiac development and disease. Pediatr. Cardiol. 2009;30:643–650. doi: 10.1007/s00246-008-9368-z. [DOI] [PubMed] [Google Scholar]
- de la Pompa J.L., Epstein J.A. Coordinating tissue interactions: notch signaling in cardiac development and disease. Dev. Cell. 2012;22:244–254. doi: 10.1016/j.devcel.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu S., Yang X., Li X., Wang J., Gao Y., Shang R., Sun W., Dou K., Li H. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A., Stottmeister C., Glazar P., Jens M., Pino N., Giusti S., Hanan M., Behm M., Bartok O., Ashwal-Fluss R., Herzog M., Schreyer L., Papavasileiou P., Ivanov A., Ohman M., Refojo D., Kadener S., Rajewsky N. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32:309–316. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz R.A., Yutzey K.E. Calcineurin signaling and NFAT activation in cardiovascular and skeletal muscle development. Dev. Biol. 2004;266:1–16. doi: 10.1016/j.ydbio.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Serra-Juhe C., Cusco I., Homs A., Flores R., Toran N., Perez-Jurado L.A. DNA methylation abnormalities in congenital heart disease. Epigenetics. 2015;10:167–177. doi: 10.1080/15592294.2014.998536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo L., Morey R., Palpant N.J., Wang P.L., Afari N., Jiang C., Parast M.M., Murry C.E., Laurent L.C., Salzman J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015;16:126. doi: 10.1186/s13059-015-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan F.L., Moravec C.S., Li J., Apperson-Hansen C., McCarthy P.M., Young J.B., Bond M. The gene expression fingerprint of human heart failure. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11387–11392. doi: 10.1073/pnas.162370099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W.L., Lim B.T., Anene-Nzelu C.G., Ackers-Johnson M., Dashi A., See K., Tiang Z., Lee D.P., Chua W.W., Luu T.D., Li P.Y., Richards A.M., Foo R.S. A landscape of circular RNA expression in the human heart. Cardiovasc. Res. 2017;113:298–309. doi: 10.1093/cvr/cvw250. [DOI] [PubMed] [Google Scholar]
- Taulli R., Loretelli C., Pandolfi P.P. From pseudo-ceRNAs to circ-ceRNAs: a tale of cross-talk and competition. Nat. Struct. Mol. Biol. 2013;20:541–543. doi: 10.1038/nsmb.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team R.C. R Foundation for Statistical Computing; 2015. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Tompkins J.D., Jung M., Chen C.Y., Lin Z., Ye J., Godatha S., Lizhar E., Wu X., Hsu D., Couture L.A., Riggs A.D. Mapping human pluripotent-to-cardiomyocyte differentiation: methylomes, transcriptomes, and exon DNA methylation “memories”. EBioMedicine. 2016;4:74–85. doi: 10.1016/j.ebiom.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28(5):511. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tshori S., Gilon D., Beeri R., Nechushtan H., Kaluzhny D., Pikarsky E., Razin E. Transcription factor MITF regulates cardiac growth and hypertrophy. J. Clin. Invest. 2006;116:2673–2681. doi: 10.1172/JCI27643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uosaki H., Cahan P., Lee D.I., Wang S., Miyamoto M., Fernandez L., Kass D.A., Kwon C. Transcriptional landscape of cardiomyocyte maturation. Cell Rep. 2015;13:1705–1716. doi: 10.1016/j.celrep.2015.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veno M.T., Hansen T.B., Veno S.T., Clausen B.H., Grebing M., Finsen B., Holm I.E., Kjems J. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015;16:245. doi: 10.1186/s13059-015-0801-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagschal A., Najafi-Shoushtari S.H., Wang L., Goedeke L., Sinha S., deLemos A.S., Black J.C., Ramirez C.M., Li Y., Tewhey R., Hatoum I., Shah N., Lu Y., Kristo F., Psychogios N., Vrbanac V., Lu Y.C., Hla T., de Cabo R., Tsang J.S., Schadt E., Sabeti P.C., Kathiresan S., Cohen D.E., Whetstine J., Chung R.T., Fernandez-Hernando C., Kaplan L.M., Bernards A., Gerszten R.E., Naar A.M. Genome-wide identification of microRNAs regulating cholesterol and triglyceride homeostasis. Nat. Med. 2015;21:1290–1297. doi: 10.1038/nm.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamstad J.A., Alexander J.M., Truty R.M., Shrikumar A., Li F., Eilertson K.E., Ding H., Wylie J.N., Pico A.R., Capra J.A., Erwin G., Kattman S.J., Keller G.M., Srivastava D., Levine S.S., Pollard K.S., Holloway A.K., Boyer L.A., Bruneau B.G. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Long B., Liu F., Wang J.X., Liu C.Y., Zhao B., Zhou L.Y., Sun T., Wang M., Yu T., Gong Y., Liu J., Dong Y.H., Li N., Li P.F. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., Parkinson H. The NHGRI GWAS catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–6. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins B.J., Dai Y.S., Bueno O.F., Parsons S.A., Xu J., Plank D.M., Jones F., Kimball T.R., Molkentin J.D. Calcineurin/NFAT coupling participates in pathological, but not physiological, cardiac hypertrophy. Circ. Res. 2004;94:110–118. doi: 10.1161/01.RES.0000109415.17511.18. [DOI] [PubMed] [Google Scholar]
- Wu C., Arora P. Long noncoding Mhrt RNA: molecular crowbar unravel insights into heart failure treatment. Circ. Cardiovasc. Genet. 2015;8:213–215. doi: 10.1161/CIRCGENETICS.115.001019. [DOI] [PubMed] [Google Scholar]
- Xu J., Feng L., Han Z., Li Y., Wu A., Shao T., Ding N., Li L., Deng W., Di X., Wang J., Zhang L., Li X., Zhang K., Cheng S. Extensive ceRNA-ceRNA interaction networks mediated by miRNAs regulate development in multiple rhesus tissues. Nucleic Acids Res. 2016;44:9438–9451. doi: 10.1093/nar/gkw587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Fuscoe J.C., Zhao C., Guo C., Jia M., Qing T., Bannon D.I., Lancashire L., Bao W., Du T., Luo H., Su Z., Jones W.D., Moland C.L., Branham W.S., Qian F., Ning B., Li Y., Hong H., Guo L., Mei N., Shi T., Wang K.Y., Wolfinger R.D., Nikolsky Y., Walker S.J., Duerksen-Hughes P., Mason C.E., Tong W., Thierry-Mieg J., Thierry-Mieg D., Shi L., Wang C. A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat. Commun. 2014;5:3230. doi: 10.1038/ncomms4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Ma J., Long K., Jin L., Liu Y., Zhou C., Tian S., Chen L., Luo Z., Tang Q., Jiang A., Wang X., Wang D., Jiang Z., Wang J., Li X., Li M. Dynamic gene expression profiles during postnatal development of porcine subcutaneous adipose. PeerJ. 2016;4:e1768. doi: 10.7717/peerj.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J., Agha G., Baccarelli A.A. The role of DNA methylation in cardiovascular risk and disease: methodological aspects, study design, and data analysis for epidemiological studies. Circ. Res. 2016;118:119–131. doi: 10.1161/CIRCRESAHA.115.305206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.H., Yuan Y.X., Rao S.L., Wang P. LncRNA MIAT enhances cardiac hypertrophy partly through sponging miR-150. Eur. Rev. Med. Pharmacol. Sci. 2016;20:3653–3660. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures
The gene expression patterns during heart differentiation.
The lncRNA expression patterns during heart differentiation.
The circRNA expression patterns during heart differentiation.
The network modules for the specific stage of heart differentiation.
Literature curation of the genes, lncRNAs and circRNAs in network modules.