Abstract
To elucidate mechanisms that regulate Vβ rearrangement, we generated and analyzed mice with a V(D)J recombination reporter cassette of germline Dβ-Jβ segments inserted into the endogenous Vβ14 locus (Vβ14Rep). As a control, we first generated and analyzed mice with the same Dβ-Jβ cassette targeted into the generally expressed c-myc locus (c-mycRep). Substantial c-mycRep recombination occurred in both T and B cells and initiated concurrently with endogenous Dβ to Jβ rearrangements in thymocytes. In contrast, Vβ14Rep recombination was restricted to T cells and initiated after endogenous Dβ to Jβ rearrangements, but concurrently with endogenous Vβ14 rearrangements. Thus, the local chromatin environment imparts lineage and developmental stage-specific accessibility upon the inserted reporter. Although Vβ14 rearrangements occur on only 5% of endogenous TCRβ alleles, the Vβ14Rep cassette underwent rearrangement on 80–90% of alleles, supporting the suggestion that productive coupling of accessible Vβ14 segments and DJβ complexes influence the frequency of Vβ14 rearrangements. Strikingly, Vβ14Rep recombination also occurs on TCRβ alleles lacking endogenous Vβ to DJβ rearrangements, indicating that Vβ14 accessibility per se is not subject to allelic exclusion.
During lymphocyte development, TCR and Ig V region exons are assembled from germline V, D, and J segments. V(D)J recombination is initiated by the lymphocyte-specific RAG 1 and 2 proteins, which introduce DNA double strand breaks between a pair of participating gene segments and their flanking recombination signal sequences (RSs),4 with the RAG-generated ends then being joined by the nonhomologous end joining pathways to complete the process (1). Chromosomal V(D)J recombination is regulated within the contexts of lineage specificity, developmental stage specificity, and allelic exclusion via modulation of accessibility of participating V, D, and J gene segments to the RAG endonuclease (2, 3). However, despite intense efforts, much remains to be learned about the molecular mechanisms that determine V(D)J recombinational accessibility and the factors that influence the choice of particular gene segments for recombination (4, 5).
TCR V region exons are assembled in a highly regulated fashion during αβ T cell development (3, 6). In CD4−/CD8−“double-negative” (DN) thymocytes, TCRβ V region exons are assembled in an ordered fashion with Dβ to Jβ joining initiating in CD44+/CD25+ stage II DN thymocytes before Vβ rearrangement to a preassembled DJβ complex in CD44−/CD25+ stage III DN thymocytes (7). TCRβ locus Dβ to Jβ rearrangements occur on both alleles, while the Vβ to DJβ rearrangement step is thought to occur on one allele at a time (3, 8, 9). Following the assembly and expression of in-frame (productive) VβDJβ rearrangements on the first allele, further Vβ to DJβ rearrangements on the second allele are prevented via feedback regulation to enforce TCRβ locus allelic exclusion (3, 8, 9). However, Vβ to DJβ rearrangements can occur on the second allele following assembly of out-of-frame (nonproductive) VβDJβ rearrangements on the first allele (3, 8, 9). Expression of productive VβDJβ rearrangements in DN thymocytes also signals differentiation into CD4+/CD8+“double-positive” (DP) thymocytes and initiation of Vα to Jα rearrangements (6). The assembly and expression of productive VαJα rearrangements leads to cell surface expression of αβ TCRs that signal differentiation into CD4+ or CD8+“single-positive” thymocytes, which exit the thymus as αβ T cells.
The molecular mechanisms that direct the assembly of endogenous TCRβ V region exons have not been elucidated. Ordered TCRβ rearrangement is likely mediated by developmental stage-specific modulation of Vβ, Dβ, and Jβ recombinational accessibility, intrinsic properties of the participating RSs, and other chromosomal factors such as distance (10–12). In addition, Vβ rearrangement likely involves factors that actively promote the physical juxtaposition of RAG-accessible Vβ/Dβ RSs separated across large chromosomal distances (4, 5). Specific replacement of the endogenous Vβ14 RS with the 3′Dβ1 RS, which possesses a 5- to 10-fold higher intrinsic ability to recombine with 5′Dβ RSs (13), led to a corresponding increase in the frequency of primary Vβ14 to DJβ rearrangements (11). These findings led to the suggestion that this particular RS replacement enhances the likelihood that juxtaposed Vβ14 segments and DJβ complexes generate productive synaptic complexes by increasing RAG binding to the Vβ14 RS and/or by increasing RAG-mediated cleavage. In the context of this interpretation, it was suggested that endogenous Vβ14 segments may actually be juxtaposed with DJβ complexes much more frequently than they rearrange (11), further implying that Vβ14 segments also may be recombinationally accessible in a much higher percentage of developing αβ T cells than the frequency with which they rearrange to DJβ complexes.
To test the notion that Vβ14 segments become recombinationally accessible much more frequently than they rearrange, we wished to directly monitor RAG access to the endogenous Vβ14 locus, rather than use correlative measures of V(D)J recombinational accessibility, such as active germline transcription, nuclease sensitivity, open chromatin structure, or expression of an inserted reporter gene (14–17). For this purpose, we have developed a V(D)J recombination reporter cassette and assessed its ability to rearrange when inserted in place of the endogenous Vβ14 RS.
Materials and Methods
Generation of targeting constructs and probes
The Vβ14Rep targeting vector was constructed in pLNTK using a 2.3-kb NdeI fragment for the 5′ homology arm and a 2.4-kb NdeI-SphI fragment containing Vβ14Rep for the 3′ homology arm. Vβ14Rep was created by first replacing the 236-bp NdeI-BglI fragment containing the Vβ14 RS with an PCR product that amplified genomic sequence between the NdeI site and the Vβ14 RS and also introduced EcoRI and BglII sites just inside the NdeI site and a ClaI site just inside of the BglI site. The 582-bp AccI-EcoRV fragment spanning the 5′Dβ1 RS and just 3′ of Jβ1.1 was blunt-end ligated into this ClaI site. The 5′Vβ14 probe is a 1.4-kb PstI-NdeI fragment. The 3′Vβ14 probe is a 0.7-kb SphI-HindIII fragment. The CWΔP probe is a 1.5-kb HindIII fragment. The c-mycRep targeting vector was constructed in pLNTK using a 4.5-kb SphI genomic fragment for the 5′ homology arm and a 3.0-kb SphI genomic fragment for the 3′ homology arm. The 762-bp AccI-EcoRV fragment containing Dβ1 and Jβ1.1 was inserted at the unique DraIII site in the 5′ homology arm using blunt-end ligation. The c-mycA probe is a 1.5-kb XbaI fragment. The c-mycB probe is a 1.6-kb XhoI-KpnI fragment; the c-mycD probe is a 700-bp XhoI-BamH1 fragment.
Gene targeting and generation of embryonic stem (ES) cells
The Vβ14Rep targeting vector was electroporated into Jβ1ω/ωES cells (18), while the c-mycRep targeting vector was electroporated into TC1 ES cells as described (19) to generate c-mycRepNeo and Vβ14RepNeo ES cells, respectively. c-mycRepNeo clones were identified by Southern blotting using the 5′ probe on EcoR1-digested DNA, and confirmed with the 3′ probe (c-myc+, 20 kb; c-mycRepNeo, 12 kb). Vβ14RepNeo clones were identified by Southern blot analysis with the Vβ14 5′ probe on BamHI-digested DNA (Vβ14ω, 18 kb; Vβ14RepNeo, 8 kb) and confirmed with the Vβ14 3′ probe on EcoRI-digested DNA (Vβ14ω, 5 kb; Vβ14RepNeo, 4.24 kb) The pgk-Neor gene was removed from independently targeted clones by infection with AdenoCre- and Cre-deleted clones identified by Southern blot analysis of BglII-digested DNA with the CWΔP probe (Vβ14ω, 3.6 kb; Vβ14RepNeo, 4 kb; Vβ14Rep, 2 kb).
Generation of mice and lymphocytes
Vβ14Rep/ω lymphocytes were generated through RAG2-deficient blastocyst complementation (RDBC) as described (20). Germline Vβ14Rep/WT and Vβ14Rep/Rep mice obtained from breeding RDBC-derived chimeric mice with 129SvEv mice. c-mycRepNeo ES cells were injected into C57BL6 blastocyts to generate chimeras for germline transmission of the c-mycRepNeo allele. Chimeras were bred to 129SvEv mice to generate c-mycRepNeo mice. c-mycRepNeo mice were bred to E2A-Cre-transgenic mice (21) to remove the pgk-Neor gene. c-mycRep mice were identified by Southern blot analysis of XhoI-digested DNA and hybridization to the c-mycD probe (5.5-kb c-mycRep; 7.5-kb c-mycRepNeo; 4.7-kb c-myc+). Resulting c-mycRep mice were further bred to 129SvEv mice to outcross the E2A-Cre transgene, which was verified by PCR using primers 5′-CCTGGAAAATGCTTCTGTC CG-3′ and 5′-CAGGGTGTTATAAGCAATCCC-3′ specific for the Cre gene. These studies have been reviewed and approved by the Institutional Animal Care and Use Committees of Boston Children’s Hospital and the Children’s Hospital of Philadelphia.
Flow cytometry analysis and cell sorting
Thymocytes and splenocytes were stained with PE-conjugated anti-CD4 and FITC-conjugated anti-CD8 or PE-conjugated anti-TCRβ and FITC-conjugated anti-Vβ14 (BD Pharmingen). FACS data acquisition and analysis was performed on a FACSCalibur flow cytometer using CellQuest software (BD Biosciences). Cell sorting of DNII and DNIII thymocytes was performed using a MoFlo cell sorter (DakoCytomation) following staining of CD4-depleted thymocytes (Miltenyi Biotec) with FITC-conjugated anti-CD8α, FITC-conjugated anti-CD4, FITC-conjugated anti-TCRβ, FITC-conjugated anti-B220, FITC-conjugated anti-TCRγδ, PE-conjugated anti-CD25, and CYC-conjugated anti-CD44.
PCR analysis of rearrangements
Vβ14Rep Dβ to Jβ rearrangements were detected by PCR using the 5′Vβ14 5′-TGTCTTTGGTGACTTCTGACTTG-3′ and Jβ1.1 5′-CAACGTG AGTCTGGTTCCTTTACC-3′ primers and probed with the P1 primer 5′-CCTACAACTGTGAGTCTGGTTCCTTTACC-3′. Endogenous TCRβ locus Dβ to Jβ rearrangements were detected by PCR as previously described (18). c-mycRep Dβ to Jβ rearrangements were detected by PCR using the 5′c-myc 5′-GAAGACTGCGGTGAGTCGTGATCT-3′ and Jβ1.1 primers and probed with the P1 primer. For sequence analysis, PCR products representing Vβ14Rep or endogenous TCRβ locus Dβ to Jβ1 rearrangements were cloned into the pGEM T-Easy vector and then sequenced with either the T7 or Sp6 primers. For seminested PCR analysis of Vβ14Rep or endogenous TCRβ locus Dβ to Jβ rearrangements, the second PCR were conducted with the 5′Vβ14a 5′-AAATCAAGCCCTAAC CTCTAC-3′ and the Jβ1.1 primer (Vβ14Rep) or with the 5′ primer 5′-TGTCTTTGGTGACTTCTGACTTG-3′ and the P2 primer 5′-CCT GACTTCCACCCGAGGTT-3′ (endogenous), using the first PCR product as template and 32 cycles of amplification.
Southern analysis of rearrangements in hybridomas
T cell hybridoma clones were produced by fusion of Con A and IL-2-stimulated T cells with the thymoma cell line BW-1100.129.237 as described (19). B cell hybridoma clones were produced by fusion of LPS stimulated splenocytes with the myeloma cell line NS1 as described (22). TCR αβ+ and Vβ14+ T cell hybridomas were selected by flow cytometry and IgM+ B cell hybridomas were selected by ELISA for further analysis. Genomic DNA was isolated, digested with EcoRI, and analyzed by PCR or Southern blotting and hybridization with the following probes: the 3′-JH4 probe—a 1.5-kb HindIII-EcoR1 fragment, the 3′-Jβ1 probe—a 0.777-kb Drd1 fragment, the VDJβ probe—a 0.743-kb AflIII-HaeIII fragment, the 5′-Vβ14 probe, the 3′-Vβ14 probe, the CWΔP probe, and the DJβ1 probe amplified with primers 5′-AATCTTAAGGGGTGAAGAGAGG-3′ and 5′-ATTCTGTCTGTCCCAAGGCCC-3′.
Results
The Dβ-Jβ cassette functions as reporter of V(D)J recombinational accessibility
Within the context of a TCRβ minilocus (TCRβPF), Dβ1 to Jβ1.1 rearrangement in developing lymphocytes is dependent upon the presence of transcriptional elements, but occurs independent of TCRβPF chromosomal integration site (10, 23–25); while, despite the presence of a TATA box in the 5′Dβ1 RS, germline Dβ1/Jβ1 transcription and Dβ1 to Jβ1 rearrangement are both absolutely dependent upon the upstream pDβ1 promoter (25–30). Thus, we reasoned that insertion of a germline Dβ1 and Jβ1.1 genomic fragment lacking the pDβ1 promoter (the Dβ-Jβ cassette) into particular genomic loci could be used as a reporter to directly monitor V(D)J recombinational accessibility of the local chromatin environment by assaying Dβ-Jβ cassette rearrangements throughout lymphocyte development. If so, recombination of the Dβ-Jβ cassette inserted into a generally transcribed locus should occur in both B and T lymphocytes and initiate concurrently with endogenous Dβ to Jβ rearrangements in DNII thymocytes when Rag1/Rag2 are first expressed. The c-myc locus is transcribed throughout B and T cell development (31, 32). Thus, to test this notion, we used Cre-loxP-mediated gene targeting to generate mice with germline Dβ1 and Jβ1.1 segments and their RSs inserted into the first intron of the c-myc locus (the c-mycRep mutation) (Fig. 1A) on a single allele (c-mycRep/WT mice). As expected, lymphocyte development was indistinguishable between c-mycRep/WT and wild-type (WT) mice (data not shown).
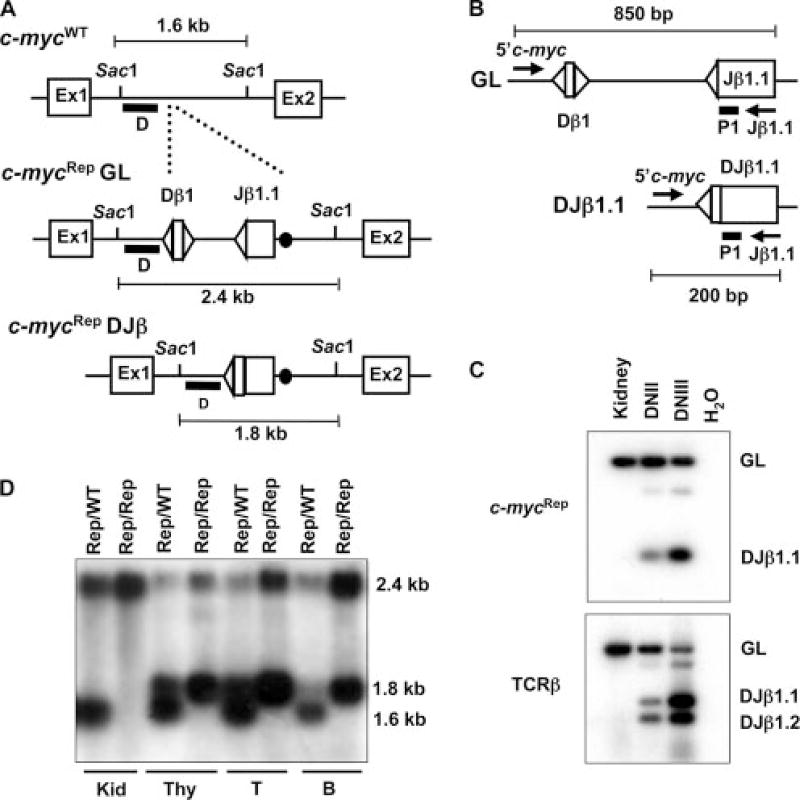
FIGURE 1.
Generation and analysis of c-mycRep mice. A, Schematic diagrams of the WT c-myc locus (WT), the c-myc locus with the inserted germline Dβ1 and Jβ1.1 segments (c-mycRep). The RSs are depicted as triangles and the loxP site as a black circle. Open boxes depict the relative locations of the three c-myc exons. The relative location of the SacI sites and the sizes of the SacI-digested genomic fragments for WT, c-mycRep germline (GL), and c-mycRep DJβ alleles are indicated. ■, The location of the 5′Vβ14, DJβ1, and 3′Vβ14 probes. B, Schematic diagrams of the GL and Dβ to Jβ rearranged (DJβ) c-mycRep alleles. The relative location of the 5′c-myc and Jβ1.1 primers are indicated with arrows. ■, The location of the P1 probe. The sizes of the PCR products for c-mycRep GL and c-mycRep DJβ alleles are indicated. C, PCR analysis of c-mycRep Dβ to Jβ rearrangements, endogenous Dβ to Jβ rearrangements, and endogenous Vβ14 to DJβ rearrangements using the 5′c-myc/Jβ1.1. 5′Dβ1/P2, and Vβ14/P2 primer sets on genomic DNA isolated from Vβ14Rep/ω kidneys and sort-purified DNII or DNIII thymocytes. Products corresponding GL and rearranged (DJβ1.1) c-mycRep alleles, as well as endogenous TCRβ rearrangements, are indicated. D, Southern blot analysis of c-mycRep Dβ to Jβ rearrangements using probe D on SacI digested genomic DNA isolated from c-mycRep/WT or c-mycRep/Rep kidney, thymus, T cells, or B cells. Restriction fragments representing GL and rearranged (DJβ1.1) c-mycRep alleles, as well as endogenous c-myc alleles, as indicated.
To characterize developmental stage specificity of c-mycRep recombination, we analyzed c-mycRep Dβ to Jβ rearrangements and endogenous TCRβ locus Dβ to Jβ rearrangements by PCR in sort-purified DNII and DNIII thymocytes. PCR with a primer that hybridizes to c-myc sequences just upstream of the inserted Dβ-Jβ cassette (the 5′c-myc primer) and a primer that hybridizes to sequences within the Jβ1.1 segment (the Jβ1.1 primer) (Fig. 1B) should specifically amplify an 850-bp product from the germline c-mycRep allele and a 200-bp product from potential Dβ to Jβ rearranged c-mycRep alleles (Fig. 1B). Endogenous TCRβ locus Dβ1 to Jβ1.1 and Dβ1 to Jβ1.2 rearrangements can be detected by PCR using a primer that hybridize 5′ of Dβ1 and 3′ of Jβ1.2(18). PCR products corresponding to c-mycRep Dβ1 to Jβ1.1 rearrangements and endogenous TCRβ locus Dβ1 to Jβ1.1 rearrangements were detectable using genomic DNA isolated from DNII and DNIII thymocytes (Fig. 1C). Notably, the ratio of the level of c-mycRep Dβ1 to Jβ1.1 rearrangements in DNII cells compared with DNIII cells (0.4) was similar to the ratio of the level of endogenous Dβ1 to Jβ1.1 rearrangements in DNII cells compared with DNIII cells (0.35). We cloned and sequenced 20 of these 200-bp PCR products and found that they were c-mycRep DJβ1.1 joins indistinguishable from similarly obtained endogenous TCRβ locus DJβ1.1 joins (data not shown), demonstrating that c-mycRep undergoes bona fide V(D)J recombination. Thus, as expected, c-mycRep recombination initiates concurrently with Rag1/Rag2 expression in developing thymocytes.
To evaluate whether c-mycRep recombination occurs at a substantial level in B and T lineage cells, we conducted Southern blot analysis with the myc D probe on SacI-digested genomic DNA isolated from the kidney, thymus, ConA/IL-2-stimulated T cells, and LPS-stimulated B cells of c-mycRep/WT and c-mycRep/Rep mice. The myc D probe hybridizes to a 1.6-kb SacI fragment on the c-mycWT allele, a 2.4-kb SacI fragment on the unrearranged c-mycRep allele, and a 1.8 kb SacI fragment on potential Dβ1 to Jβ1.1 rearranged c-mycRep alleles due to the deletion of sequences between Dβ1 and Jβ1.1 (Fig. 1A). We observed no rearrangement in c-mycRep/WT and c-mycRep/Rep kidneys, as indicated by the absence of the 1.8-kb band (Fig. 1D). However, we found c-mycRep Dβ to Jβ rearrangement in c-mycRep/WT and c-mycRep/Rep thymocytes, T cells, and B cells, as indicated by the presence of the 1.8-kb SacI fragment and a lessened intensity of the 2.4-kb SacI fragment. The ratios of the 1.8-kb rearranged to 2.4-kb rearranged band in c-mycRep/WT and c-mycRep/Rep tissues suggest that ~50% of c-mycRep alleles rearrange in B and T cells (Fig. 1D).
To precisely quantify the level of c-mycRep recombination that occurs in developing B and T cells, we generated clonal αβ T cell and B cell hybridomas and assayed for c-mycRep Dβ to Jβ rearrangements by conducting PCR on their genomic DNA using the 5′myc and Jβ1.1 primers. In c-mycRep/WT αβ T cell hybridomas, c-mycRep Dβ to Jβ rearrangements occurred on 76 of 113 (67%) alleles, while 37 of 113 (33%) contained unrearranged c-mycRep alleles (Table I). The analysis of c-mycRep/Rep αβ T cell hybridomas revealed that 33 of 104 (32%) contained c-mycRep rearrangements on both alleles, 57 of 104 (54%) contained c-mycRep rearrangements on a single allele, and 14 of 104 (14%) contained two unrearranged c-mycRep alleles (Table II). The same analysis of c-mycRep/WT B cell hybridomas revealed that 26 of 60 (43%) alleles contained rearranged c-mycRep, while 34 of 60 (56.6%) alleles remained unrearranged (Table I). In addition, the analysis of homozygous c-mycRep/Rep B cell hybridomas demonstrated that 9 of 92 (10%) contained c-mycRep rearrangements on both alleles, 49 of 92 (53%) contained c-mycRep rearrangements on one allele, and 34 of 92 (37%) harbored no rearrangements (Table II). Although we find a lower level of rearrangement in B cell hybridomas as compared with T cell hybridomas, these data indicate that the c-mycRep cassette is RAG accessible in the majority of developing B and αβ T cells. Notably, the rearrangement levels observed make the Dβ-Jβ cassette a readily discernable marker of a chromosomal locus that is accessible for V(D)J recombination and allow an estimate of the minimal level of recombinational accessibility.
Table I.
c-mycRep rearrangements in c-mycRep/WT hybridomas
| Cell Type | Total # Cells | c-mycRep Status | # Cells (%) |
|---|---|---|---|
| αβ T cells | 113 | Germline | 37 (33) |
| DJβ | 76 (67) | ||
| IgM+ B cells | 60 | Germline | 34 (57) |
| DJβ | 26 (43) |
Table II.
c-mycRep rearrangements in c-mycRep/Rep hybridomas
| Cell Type | Total # Cells | c-mycRep Status | # Cells (%) |
|---|---|---|---|
| αβ T | 104 | Germline/Germline | 14 (14) |
| Germline/DJβ | 57 (54) | ||
| DJβ/DJβ | 33 (32) | ||
| IgM+ B | 92 | Germline/Germline | 34 (37) |
| Germline/DJβ | 49 (53) | ||
| DJβ/DJβ | 9 (10) |
Vβ14Rep recombination mirrors endogenous Vβ14 rearrangement
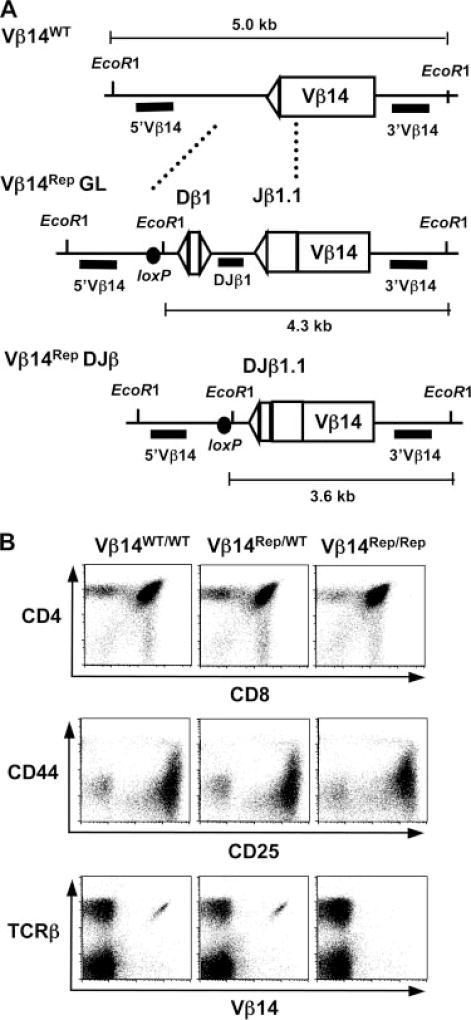
After validating that the Dβ-Jβ cassette can serve as a marker of V(D)J recombinational accessibility, we proceeded to test whether Vβ14 segments become recombinationally accessible in developing αβ T cells at a similar frequency to which they rearrange. For this purpose, we used Cre-loxP-mediated gene targeting to replace the endogenous Vβ14 RS with the Dβ-Jβ cassette (Fig. 2A) on a single TCRβ allele of Jβ1ω/ω ES cells to generate Vβ14Rep/ω ES cells. The Jβ1ω locus lacks the endogenous Dβ2-Jβ2 locus, so that all TCRβ rearrangements involve Dβ1-Jβ1 segments, but otherwise its rearrangement is indistinguishable from the WT TCRβ locus and it supports normal αβ T cell development (18). The gene targeting also introduced a single loxP site and a unique EcoRI site to distinguish between Vβ14Rep and endogenous Vβ14 rearrangements (Fig. 2A). Due to deletion of the endogenous Vβ14 RS and the orientation of the inserted Dβ1 and Jβ1.1 RSs, Vβ14Rep alleles are not capable of assembling productive Vβ14DJβ1 rearrangements. We used Vβ14Rep/ω ES cells and RAG2-deficient blastocyst complementation (RDBC) (20) to generate chimeric mice with Vβ14Rep/ω lymphocytes and bred these mice with 129SvEv (WT) mice to establish germline Vβ14Rep/WT and Vβ14Rep/Rep mice. Flow cytometric analysis of thymocytes and peripheral lymphocytes isolated from WT, Jβ1ω/ω, Vβ14Rep/ω, Vβ14Rep/WT, and Vβ14Rep/Rep mice demonstrated that the Vβ14Rep allele had no discernable effect on gross αβ T cell development (Fig. 2B). However, as expected, thymocytes and splenocytes isolated from Vβ14Rep/Rep mice completely lack cell surface expression of Vβ14 (Fig. 2B).
FIGURE 2.
Generation and characterization of Vβ14Rep/ω, Vβ14Rep/WT, and Vβ14Rep/Rep mice. A, Schematic diagrams of the WT Vβ14 locus (WT), the Vβ14 locus with the inserted germline Dβ1 and Jβ1.1 segments (Vβ14Rep), and the Dβ to Jβ rearranged Vβ14Rep locus (DJβ). The RSs are depicted as triangles and the loxP site as a black circle. The relative location of the EcoRI sites and the sizes of the EcoRI-digested genomic fragments for WT, Vβ14Rep, and DJβ alleles are indicated. Solid black bar shows the location of the 5′Vβ14, DJβ1, and 3′Vβ14 probes. B, Flow cytometric analysis of thymocytes and splenocytes from 4- to 6-wk-old WT, Vβ14Rep/WT, and Vβ14Rep/Rep mice. Shown are representative anti- CD4-PE/-CD8-FITC and anti-CD44-PE/-CD25-FITC stains of thymocytes and anti-TCRβ-PE/-Vβ14-FITC stains of splenocytes.
To evaluate whether Vβ14Rep recombination occurs in Vβ14Rep/ω lymphocytes, we first conducted PCR on genomic DNA isolated from Vβ14Rep/ω ES cells and Vβ14Rep/ω thymocytes using a primer that hybridizes to TCRβ locus sequences just 5′ of the inserted Dβ-Jβ cassette (the 5′Vβ14 primer) and the Jβ1.1 primer (Fig. 3A). This primer pair should specifically amplify an 850-bp product from the germline Vβ14Rep allele and a 200-bp product from potential Dβ to Jβ rearranged Vβ14Rep alleles (Fig. 3A). PCR products the expected sizes for both germline and Dβ to Jβ rearranged Vβ14Rep alleles were amplified from Vβ14Rep/ω thymocytes (Fig. 3B), while only PCR products corresponding to germline Vβ14Rep alleles were amplified from Vβ14Rep/ω ES cell genomic DNA (Fig. 3B). We cloned and sequenced 18 of these 200-bp PCR products and found that they were Vβ14Rep DJβ1.1 joins indistinguishable from similarly obtained endogenous TCRβ locus DJβ1.1 joins (data not shown), demonstrating that Vβ14Rep also undergoes bona fide V(D)J recombination.
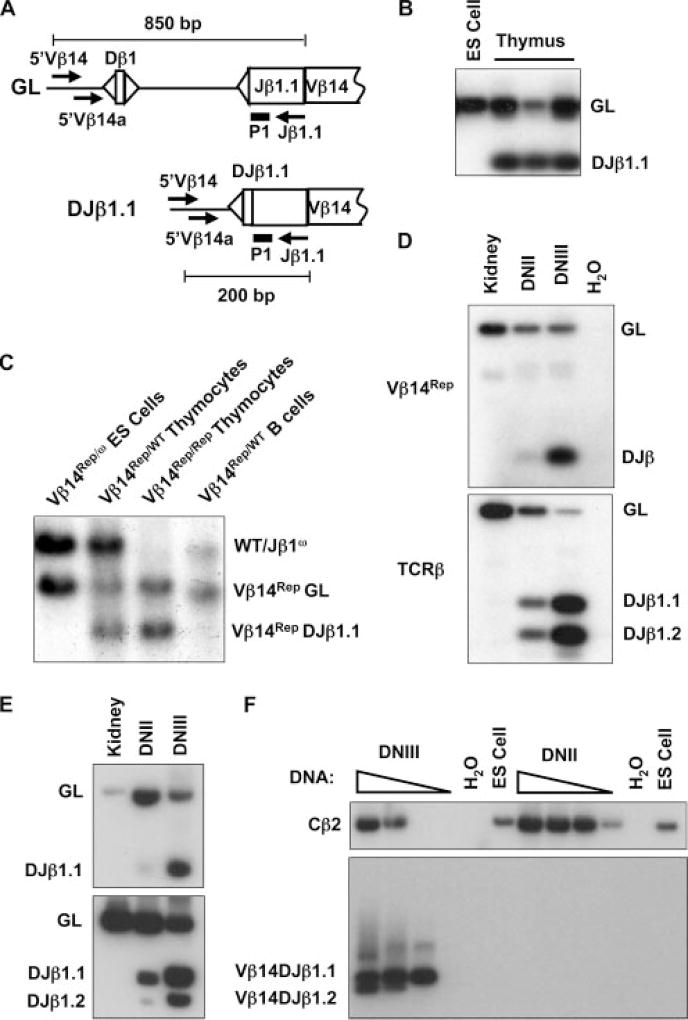
FIGURE 3.
Vβ14Rep recombination mirrors endogenous Vβ14 rearrangement. A, Schematic diagrams of the germline (GL) and Dβ to Jβ rearranged (DJβ1.1) Vβ14Rep alleles. The relative location of the 5′Vβ14, 5′Vβ14a, and Jβ1.1 primers are indicated with arrows. A solid black bar shows the location of the P1 probe. The sizes of the PCR products for Vβ14Rep GL and Vβ14Rep DJβ alleles are indicated. B, PCR analysis of Vβ14Rep recombination using the 5′Vβ14 and Jβ1.1 primers on genomic DNA isolated from Vβ14Rep/ω ES cells, thymocytes, and purified B cells. Products corresponding to GL and rearranged (DJβ1.1) Vβ14Rep alleles are indicated. C, Southern blot analysis of Vβ14Rep recombination using the 3′Vβ14 probe on EcoRI-digested genomic DNA isolated from Vβ14Rep/ω ES cells, Vβ14Rep/WT thymocytes, Vβ14Rep/Rep thymocytes, Vβ14Rep/WT purified B cells. Bands corresponding to WT, Jβ1ω (ω), GL Vβ14Rep (Vβ14Rep GL), and Dβ to Jβ rearranged Vβ14Rep (Vβ14Rep DJβ1.1) alleles are indicated. D–F, Analysis of Vβ14Rep Dβ to Jβ rearrangements, endogenous Dβ to Jβ rearrangements, and endogenous Vβ14 to DJβ rearrangements in developing thymocytes. Products corresponding GL and rearranged (DJβ1.1) Vβ14Rep alleles, as well as endogenous TCRβ rearrangements, are indicated. D, PCR using the 5′Vβ14/Jβ1.1 and 5′Dβ1/P2 primer sets on genomic DNA isolated from Vβ14Rep/WT kidneys and sort-purified DNII or DNIII Vβ14Rep/WT thymocytes. E, Seminested PCR analysis of Vβ14Rep and endogenous Dβ to Jβ rearrangements on genomic DNA isolated from Vβ14Rep/ω kidneys and sort-purified DNII or DNIII Vβ14Rep/ω thymocytes. F, Seminested PCR analysis of endogenous Vβ14 to DJβ rearrangements on genomic DNA isolated from Vβ14Rep/WT ES cells and sort-purified DNII or DNIII Vβ14Rep/WT thymocytes. Also shown are PCR amplifications of Cβ2 to demonstrate the presence of DNA in the DNII and ES cell reactions.
To determine whether Vβ14Rep recombination occurs at a substantial level in T and B lineage cells, we next conducted Southern blot analysis on EcoRI-digested genomic DNA isolated from Vβ14Rep/ω ES cells, Vβ14Rep/WT thymocytes, and Vβ14Rep/Rep thymocytes, and Vβ14Rep/WT B cells. The 3′Vβ14 probe hybridizes to a 5.0-kb EcoRI fragment from germline Jbω and WT alleles, a 4.3-kb fragment from germline Vβ14Rep alleles, and a 3.6-kb EcoRI fragment from Dβ to Jβ rearranged Vβ14Rep alleles due to deletion of the nucleotides between Vβ14Rep Dβ1 and Jβ1.1 segments (Fig. 2A). Southern blot analysis revealed the presence of 5.0-, 4.3-, and 3.6-kb bands in EcoRI-digested genomic DNA isolated from Vβ14Rep/WT and Vβ14Rep/Rep thymocytes, but only 5.0-and 3.6-kb bands in EcoRI-digested Vβ14Rep/WT B cell DNA (Fig. 3C). The ratios of the intensities of the 4.3- and 3.6-kb bands in Vβ14Rep/WT and Vβ14Rep/Rep thymocytes demonstrates that a substantial fraction of Vβ14Rep alleles recombined, indicating that the Vβ14 locus becomes recombinationally accessible in a higher percentage of thymocytes than Vβ14 rearrangement occurs. These data also reveal that Vβ14Rep recombination occurs in T, but not B, lymphocytes, mirroring the lineage-specific pattern of endogenous Vβ14 rearrangement. Critically, these data demonstrate that insertion of the Dβ-Jβ segments and their RSs into the Vβ14 locus does not promote recombinational accessibility in B cells. Thus, local chromatin environment imparts lineage-specific recombinational accessibility upon the inserted reporter.
To determine the developmental stage in which Vβ14Rep recombination initiates, we analyzed Vβ14Rep Dβ to Jβ rearrangements and endogenous TCRβ locus Dβ to Jβ rearrangements by PCR in sort-purified DNII and DNIII Vβ14Rep/WT thymocytes. PCR products corresponding to Vβ14Rep Dβ1 to Jβ1.1 rearrangements and endogenous TCRβ locus Dβ1 to Jβ1.1 rearrangements were both detectable using genomic DNA isolated from DNIII thymocytes (Fig. 3D). However, PCR products corresponding to Vβ14Rep Dβ1 to Jβ1.1 rearrangements were barely detectable using genomic DNA isolated from DNII cells (Fig. 3D), while PCR products corresponding to endogenous TCRβ locus Dβ1 to Jβ1.1 rearrangements were detectable at a substantial level using DNA isolated from DNII thymocytes (Fig. 3D), validating the presence of DNII cell genomic DNA. Notably, the ratio of the level of Vβ14Rep Dβ1 to Jβ1.1 rearrangements in DNII cells compared with DNIII cells (0.09) was significantly less than the ratio of the level of endogenous Dβ1 to Jβ1.1 rearrangements in DNII cells compared with DNIII cells (0.43). These experiments were conducted three times with similar results each time (data not shown). We also conducted seminested PCR analyses of Vβ14Rep Dβ to Jβ rearrangements, endogenous TCRβ locus Dβ to Jβ rearrangements, and endogenous Vβ14 to DJβ rearrangements in sort-purified DNII and DNIII Vβ14Rep/WT thymocytes. PCR products corresponding to Vβ14Rep Dβ1 to Jβ1.1 rearrangements were barely detectable using genomic DNA isolated from DNII cells (Fig. 3E); while, PCR products corresponding to endogenous TCRβ locus Dβ1 to Jβ1.1 rearrangements were detectable at a substantial level using DNA isolated from DNII thymocytes (Fig. 3E). Importantly, PCR products corresponding to Vβ14 to DJβ1.1 and DJβ1.2 rearrangements were detectable in DNIII, but not DNII, thymocytes (Fig. 3F), validating the purity of the sorted cells. In addition, a Cβ2 PCR product was detectable in both DNIII and DNIII thymocytes (Fig. 3F), demonstrating the presence of DNII cell genomic DNA. Thus, the developmental stage-specific initiation of Vβ14Rep recombinational accessibility largely mirrors that of endogenous Vβ14 rearrangements, which are readily detectable in DNIII, but not DNII, thymocytes (33).
Vβ14Rep recombination occurs in a much higher percentage of developing αβ T cells than Vβ14 rearrangement
To quantify the overall level of Vβ14 recombinational accessibility that occurs during αβ T cell development, we generated Vβ14Rep/ω αβ T cell hybridomas and analyzed Vβ14Rep and endogenous TCRβ rearrangements using a series of TCRβ locus probes on EcoRI-digested DNA (data not shown). Of the 76 clonal hybridomas analyzed, 47 (62%) contained Vβ14Rep Dβ to Jβ rearrangements, while only 11 (14%) contained germline Vβ14Rep alleles (Table III). In addition, we found that 18 (24%) contained Vβ14Rep alleles with endogenous Vβ, Dβ, or Jβ rearrangements to either Vβ14Rep Dβ segments or Vβ14Rep DJβ complexes (Table III), the identity of which were confirmed by sequence analysis of PCR-amplified joins (data not shown). Another three (4%) contained an aberrant Dβ1 rearrangement, most likely involving recombination between the endogenous 5′Dβ1 RS and one of several cryptic RSs located just 5′ of Vβ14Rep (GenBank AE000665; Table III; data not shown). Thus, although the majority of Vβ14Rep recombination events involve Vβ14Rep Dβ to Jβ rearrangements, Vβ14Rep also can target the rearrangement of endogenous Vβ, Dβ, and Jβ segments to Vβ14Rep Dβ segments or DJβ complexes. These data demonstrate that Vβ14Rep recombination occurs in a substantially higher percentage (at least 86%) of developing αβ T cells than the ~7% in which primary Vβ14 rearrangements occur (11).
Table III.
Vβ14Rep rearrangements in Vβ14Rep/Rep hybridomas
| Cell Type | Total # | Vβ14Rep Status | # Cells (%) |
|---|---|---|---|
| Vβ14Rep germline | 11 (14) | ||
| Vβ14Rep DJβ | 47 (62) | ||
| αβ T cells | 76 | 5’Dβ1 RS to Vβ14Rep 3’Dβ RS | 3 (4) |
| DJβ1-Jβ1 RS to Vβ14Rep 3’Dβ RS | 3 (4) | ||
| Vβ to Vβ14Rep DJβ | 3 (4) | ||
| Vβ to Vβ14Rep Dβ | 2 (3) | ||
| VDJβ1-Jβ1 RS to Vβ14Rep 3’Dβ RS | 4 (5) | ||
| 5’Dβ1 to cRS | 3 (4) | ||
| IgM+ B cells | 37 | Vβ14Rep germline | 37 (100) |
In addition to their productive and selected VβDJβ rearrangements, ~60% of normal αβ T cell hybridomas contain DJβ rearrangements and ~40% contain out-of-frame VβDJβ rearrangements on their nonselected alleles (8, 18). Thus, to more rigorously address whether Vβ14Rep recombination occurs in a substantially higher percentage of developing αβ T cells than Vβ14 rearrangements occur, we also quantified Vβ14Rep and endogenous Vβ14 rearrangements on nonselected TCRβ alleles in αβ T cell hybridomas generated from Jβ1ω/ω, Vβ14Rep/ω, and Vβ14Rep/Rep mice. Southern analysis of TCRβ rearrangements in 92 Vβ14+ Jβ1ω/ω hybridomas demonstrated that only 4 (5%) contained endogenous Vβ14 to DJβ rearrangements on the nonselected allele. In contrast, Southern blot analysis of 43 Vβ14+ Vβ14Rep/ω αβ T cell hybridomas revealed that 36 (84%) contained Vβ14Rep recombination on the nonselected allele (Table IV). These recombination events included Vβ14Rep Dβ to Jβ rearrangements and endogenous Vβ, Dβ, or Jβ rearrangements to Vβ14Rep Dβ segments and DJβ complexes (data not shown). We next quantified Vβ14Rep recombination in a panel of 55 Vβ14Rep/Rep αβ T cell hybridomas. Of the 38 with endogenous DJβ rearrangement on the nonselected allele, 20 (52%) contained Vβ14Rep Dβ to Jβ rearrangements on both alleles (Table V). Together, these data demonstrate unequivocally that Vβ14Rep recombination occurs in a substantially higher percentage of developing αβ T cells than the percentage in which Vβ14 rearrangements occur. Thus, we conclude that endogenous Vβ14 segments are recombinationally accessible in a much higher percentage of thymocytes than they rearrange to DJβ complexes and Vβ14 accessibility per se is not subject to allelic exclusion.
Table IV.
Vβ14 locus rearrangements on non-selected alleles in Vβ14+ T cell hybridomas
| Genotype | Total # | Vβ14 or Vβ14Rep Status | # Cells (%) |
|---|---|---|---|
| Jβ1ω/ω | 92 | Vβ14 germline | 88 (95) |
| Vβ14 rearranged | 4 (5) | ||
| Vβ14Rep/ω | 43 | Vβ14Rep germline | 7 (16) |
| Vβ14Rep rearranged | 36 (84) |
Table V.
Vβ14Rep rearrangements in VβDJβ/DJβ Vβ14Rep/Rep T cell hybridomas
| Total # Cells | Allele 1 | Allele 2 | # Cells (%) |
|---|---|---|---|
| 38 | Germline | Germline | 9 (24) |
| Germline | DJβ | 9 (24) | |
| DJβ | DJβ | 20 (52) |
Discussion
We have shown here that local chromatin environment imparts lineage- and stage-specific accessibility upon an inserted Dβ-Jβ reporter cassette, allowing the cassette to function as a reporter of V(D)J recombinational accessibility of particular chromosomal loci. In developing thymocytes, the assembly of TCRβ V region exons is ordered with Dβ to Jβ rearrangements occurring before Vβ rearrangements (7, 34). Despite initiation of endogenous Dβ to Jβ rearrangement in DNII stage thymocytes, we found that Vβ14Rep Dβ to Jβ rearrangement predominantly tracks with endogenous Vβ14 to DJβ rearrangement in DNIII thymocytes. Thus, the endogenous Vβ14 segment largely becomes accessible for V(D)J recombination upon differentiation of thymocytes to the DNIII stage. These findings support the notion that ordered assembly of Vβ14DJβ complexes is mediated, at least in part, through the developmental stage-specific accessibility of the Dβ and Jβ portion of the TCRβ locus in DNII thymocytes and the Vβ14 segment in DNIII cells. In this regard, a small percentage (3%) of Jβ1Rep/ω αβ T cell hybridomas contained rearrangement of upstream Vβ segments directly to Vβ14Rep Dβ segments, but not directly to endogenous Dβ segments, suggesting that initiation of Dβ/Jβ accessibility in DNII thymocytes, before Vβ accessibility, may ensure formation of DJβ complexes before activation of Vβ rearrangement.
Endogenous Vβ to DJβ rearrangement must proceed through the physical juxtaposition of recombinationally accessible Vβ segments with DJβ complexes across large chromosomal distances (4, 5). Despite occurrence of Vβ14 to DJβ rearrangements in only 5% of Jβ1ω/ω αβ T cells, Vβ14Rep recombination occurred in 86% of Vβ14Rep/ω αβ T cells, on 84% of alleles in Vβ14 expressing Vβ14Rep/ω αβ T cells, and on both alleles in 42% of Vβ14Rep/Rep αβ T cells that contain endogenous Vβ to DJβ rearrangements on only one allele. Thus, endogenous Vβ14 segments are recombinationally accessible in a much higher percentage of developing thymocytes than that in which they actually undergo rearrangement. Previously, we demonstrated that specific replacement of the endogenous Vβ14 RS with the 3′Dβ1 RS resulted in an ~10-fold increase in the frequency of Vβ14 to DJβ rearrangements (11). This same RS replacement resulted in a corresponding increase in RAG-mediated cleavage of Vβ14 and Dβ segments in vitro (13). It seems unlikely that replacement of the endogenous Vβ14 RS with the 3′Dβ1 RS, either precisely or as part of the Dβ-Jβ cassette, would increase juxtaposition between Vβ14 segments and DJβ complexes. Moreover, the TATA box of the 5′Dβ1 RS is not sufficient to drive Dβ1-Jβ1 transcription or Dβ1 to Jβ1 rearrangement (25–30). Therefore, our current findings in combination with our earlier RS replacement study suggests that the frequency of Vβ14 rearrangements is determined by the productive coupling of recombinationally accessible Vβ14 segments and DJβ complexes. Finally, and most strikingly, our current observation that Vβ14Rep recombination occurs on TCRβ alleles lacking endogenous Vβ to DJβ rearrangements indicates that Vβ14 accessibility is not subject to allelic exclusion (discussed in detail below).
V(D)J recombination is thought to proceed via the initial assembly of the RAG proteins on one RS, followed by capture of the second RS to form a synaptic complex in which RAG-mediated cleavage occurs (35–37). RAG proteins most likely first assemble on RSs with 12-bp spacers 12-RSs) and capture RSs with 23-bp spacers 23-RSs) (36, 37). Thus, during Vβ14 to DJβ rearrangement, the RAG proteins may initially assemble on accessible 5′Dβ 12-RSs and capture Vβ14 23-RSs following structural changes in chromatin that bring Vβ14 segments in close proximity to RAG-bound DJβ complexes. In this context, Vβ14 segments could be rendered accessible for V(D)J recombination either before juxtaposition or, possibly, during synaptic complex formation through RAG2-mediated binding to, or RAG1-catalyzed ubiquitination of, histones within Vβ14 chromatin (5, 36–38). Alternatively, chromosomal factors may direct RAG assembly on accessible Vβ14 23-RSs, leading to the capture of 5′Dβ 12-RSs following juxtaposition of RAG-bound Vβ14 segments and DJβ complexes. In either scenario, the frequency of Vβ14 rearrangement would be determined either by the RAG-binding affinity or by the recombination potential of the RS sequence attached to Vβ14.
Our previous RS replacement study which suggested that Vβ14 segments may be recombinationally accessible in a much higher percentage of thymocytes than the frequency with which they rearrange to DJβ complexes was conducted on a single allele in cells that contained an inactivated TCRβ locus on the other allele (11). Thus, we were unable to ascertain whether the paucity of αβ T cells with Vβ14 to DJβ rearrangements on both alleles was determined by the distinct modulation of Vβ14 accessibility on each allele or by the differential coupling of recombinationally accessible Vβ14 segments and DJβ complexes on each allele. Our current observation that Vβ14Rep recombination occurred on approximately half of TCRβ alleles that lack edogenous Vβ to DJβ rearrangements in Vβ14Rep/ω and Vβ14Rep/Rep αβ T cell hybridomas indicates that Vβ14Rep recombination is not subject to allelic exclusion. This finding demonstrates unequivocally that the assembly of a nonproductive (out-of-frame) VβDJβ rearrangement on the first TCRβ allele is not necessary to activate Vβ14 accessibility on the second allele. By FACS, we did not detect surface expression of Vβ14, or any Vβs, other than Vβ8 on T lineage cells of mice expressing a Vβ8DJβ transgene (A. C. Carpenter and C. H. Bassing, unpublished observations,), suggesting that, similar to the other Vβs (39), Vβ14 is subject to transgene feedback regulation and potentially to allelic exclusion. If so, our finding that Vβ14Rep recombination is not subject to allelic exclusion suggests that feedback regulation of Vβ14 rearrangement may be enforced by preventing the productive coupling of recombinationally accessible Vβ14 segments and DJβ complexes, rather than through inhibition of Vβ14 accessibility. Consistent with this notion, although Vβ14 remains recombinationally accessible in DP thymocytes (40), Vβ14 is not expressed on the cell surface with other Vβs, indicating that Vβ14 allelic exclusion may be maintained through a unique mechanism, such as the induction of apoptosis in DP cells undergoing rare Vβ14 to DJβ rearrangements (40). In contrast, transgenic overexpression of TCRβ chains may inhibit endogenous rearrangements, at least to some degree, by accelerated development and not normal feedback mechanisms (41). In this context, sequence analyses of limited numbers of VβDJβ joins in WT αβ T cells and direct Vβ14 to Jβ rearrangements in αβ T cells with specific TCRβ RS replacements revealed two in-frame rearrangements in 5–10% of cells (33, 42, 43), indicating that normal TCRβ expression may not inhibit Vβ to DJβ rearrangement and lead to allelic exclusion in all developing αβ T cells. Accordingly, our observation that Vβ14Rep recombination occurs on both alleles in a substantial percentage of αβ T cells may simply reflect that endogenous Vβ14 to DJβ rearrangements are not completely inhibited by feedback regulation. However, our current data cannot exclude the possibility that Vβ14Rep rearranges efficiently and on both alleles in DNIII thymocytes before the assembly and expression of TCRβ chains that signal inhibition of Vβ14 RAG accessibility. Perhaps the analysis of Vβ14Rep recombination and Vβ14 expression in thymocytes expressing VβDJβ transgenes or preassembled endogenous VβDJβ rearrangements may distinguish among these possibilities.
The Vβ14 segment is unique among Vβ segments due to its proximity to Dβ/Jβ segments (44), its rearrangement through inversion (44), and its continued accessibility in DP thymocytes (45). Therefore, the generation and analysis of mice containing replacement of additional Vβ RSs with inserted Dβ-Jβ cassettes will be required to determine whether the rearrangement of other Vβ segments is directed by similar mechanisms to those we have uncovered for Vβ14 in this study.
Acknowledgments
We thank Tiffany Borjeson for blastocyst injections and Atilla Fabian for cell sorting.
Footnotes
This work was supported by National Institutes of Health Grant AI20047 (to F.W.A.) and the Department of Pathology of the Children’s Hospital of Philadelphia (to C.H.B.). S.R. was supported by a Genentech/IDEC Fellowship from the American Cancer Society and the Dana Farber Cancer Institute Postdoctoral Training Program in Cancer Immunology. A.C.C. was supported by the Training Program in Immune System Development and Regulation at the University of Pennsylvania. C.H.B. is a Pew Scholar in the Biomedical Sciences. F.W.A. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations used in this paper: RSs, recombination signal sequence; DN, double negative; ES, embryonic stem; WT, wild type; RDBC, RAG2-deficient blastocys complementation.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Jung D, Alt FW. Unraveling V(D)J recombination; insights into gene regulation. Cell. 2004;116:299–311. doi: 10.1016/s0092-8674(04)00039-x. [DOI] [PubMed] [Google Scholar]
- 2.Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu. Rev. Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- 3.Jackson AM, Krangel MS. Turning T-cell receptor β recombination on and off: more questions than answers. Immunol. Rev. 2006;209:129–141. doi: 10.1111/j.0105-2896.2006.00342.x. [DOI] [PubMed] [Google Scholar]
- 4.Hesslein DG, Schatz DG. Factors and forces controlling V(D)J recombination. Adv. Immunol. 2001;78:169–232. doi: 10.1016/s0065-2776(01)78004-2. [DOI] [PubMed] [Google Scholar]
- 5.Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109(Suppl):S45–S55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 6.Krangel MS, Carabana J, Abbarategui I, Schlimgen R, Hawwari A. Enforcing order within a complex locus: current perspectives on the control of V(D)J recombination at the murine T-cell receptor α/δ locus. Immunol. Rev. 2004;200:224–232. doi: 10.1111/j.0105-2896.2004.00155.x. [DOI] [PubMed] [Google Scholar]
- 7.Godfrey DI, Kennedy J, Mombaerts P, Tonegawa S, Zlotnik A. Onset of TCR-β gene rearrangement and role of TCR-β expression during CD3−CD4−CD8− thymocyte differentiation. J. Immunol. 1994;152:4783–4792. [PubMed] [Google Scholar]
- 8.Mostoslavsky R, Alt FW, Rajewsky K. The lingering enigma of the allelic exclusion mechanism. Cell. 2004;118:539–544. doi: 10.1016/j.cell.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 9.Khor B, Sleckman BP. Allelic exclusion at the TCRβ locus. Curr. Opin. Immunol. 2002;14:230–234. doi: 10.1016/s0952-7915(02)00326-6. [DOI] [PubMed] [Google Scholar]
- 10.Sleckman BP, Bassing CH, Hughes MM, Okada A, D’Auteuil M, Wehrly TD, Woodman BB, Davidson L, Chen J, Alt FW. Mechanisms that direct ordered assembly of T cell receptor β locus V, D, and J gene segments. Proc. Natl. Acad. Sci. USA. 2000;97:7975–7980. doi: 10.1073/pnas.130190597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu C, Bassing CH, Jung D, Woodman BB, Foy D, Alt FW. Dramatically increased rearrangement and peripheral representation of Vβ14 driven by the 3'Dβ1 recombination signal sequence. Immunity. 2003;18:75–85. doi: 10.1016/s1074-7613(02)00515-0. [DOI] [PubMed] [Google Scholar]
- 12.Tourigny MR, Mazel S, Burtrum DB, Petrie HT. T cell receptor (TCR)-β gene recombination: dissociation from cell cycle regulation and developmental progression during T cell ontogeny. J. Exp. Med. 1997;185:1549–1556. doi: 10.1084/jem.185.9.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung D, Bassing CH, Fugmann SD, Cheng HL, Schatz DG, Alt FW. Extrachromosomal recombination substrates recapitulate beyond 12/23 restricted VDJ recombination in nonlymphoid cells. Immunity. 2003;18:65–74. doi: 10.1016/s1074-7613(02)00507-1. [DOI] [PubMed] [Google Scholar]
- 14.Liang HE, Hsu LY, Cado D, Schlissel MS. Variegated transcriptional activation of the immunoglobulin κ locus in pre-b cells contributes to the allelic exclusion of light-chain expression. Cell. 2004;118:19–29. doi: 10.1016/j.cell.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 15.Blackwell TK, Moore MW, Yancopoulos GD, Suh H, Lutzker S, Selsing E, Alt FW. Recombination between immunoglobulin variable region gene segments is enhanced by transcription. Nature. 1986;324:585–589. doi: 10.1038/324585a0. [DOI] [PubMed] [Google Scholar]
- 16.Yancopoulos GD, Alt FW. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 1985;40:271–281. doi: 10.1016/0092-8674(85)90141-2. [DOI] [PubMed] [Google Scholar]
- 17.McMurry MT, Krangel MS. A role for histone acetylation in the developmental regulation of VDJ recombination. Science. 2000;287:495–498. doi: 10.1126/science.287.5452.495. [DOI] [PubMed] [Google Scholar]
- 18.Bassing CH, Alt FW, Hughes MM, D’Auteuil M, Wehrly TD, Woodman BB, Gartner F, White JM, Davidson L, Sleckman BP. Recombination signal sequences restrict chromosomal V(D)J recombination beyond the 12/23 rule. Nature. 2000;405:583–586. doi: 10.1038/35014635. [DOI] [PubMed] [Google Scholar]
- 19.Sleckman BP, Bardon CG, Ferrini R, Davidson L, Alt FW. Function of the TCR α enhancer in αβ and γδ T cells. Immunity. 1997;7:505–515. doi: 10.1016/s1074-7613(00)80372-6. [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Lansford R, Stewart V, Young F, Alt FW. RAG-2-deficient blastocyst complementation: an assay of gene function in lymphocyte development. Proc. Natl. Acad. Sci. USA. 1993;90:4528–4532. doi: 10.1073/pnas.90.10.4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. USA. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zarrin AA, Alt FW, Chaudhuri J, Stokes N, Kaushal D, Du Pasquier L, Tian M. An evolutionarily conserved target motif for immunoglobulin class-switch recombination. Nat. Immunol. 2004;5:1275–1281. doi: 10.1038/ni1137. [DOI] [PubMed] [Google Scholar]
- 23.Ferrier P, Krippl B, Blackwell TK, Furley AJ, Suh H, Winoto A, Cook WD, Hood L, Costantini F, Alt FW. Separate elements control DJ and VDJ rearrangement in a transgenic recombination substrate. EMBO J. 1990;9:117–125. doi: 10.1002/j.1460-2075.1990.tb08087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okada A, Mendelsohn M, Alt F. Differential activation of transcription versus recombination of transgenic T cell receptor β variable region gene segments in B and T lineage cells. J. Exp. Med. 1994;180:261–272. doi: 10.1084/jem.180.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sikes ML, Gomez RJ, Song J, Oltz EM. A developmental stage-specific promoter directs germline transcription of DβJβ gene segments in precursor T lymphocytes. J. Immunol. 1998;161:1399–1405. [PubMed] [Google Scholar]
- 26.Oestreich KJ, Cobb RM, Pierce S, Chen J, Ferrier P, Oltz EM. Regulation of TCRβ gene assembly by a promoter/enhancer holocomplex. Immunity. 2006;24:381–391. doi: 10.1016/j.immuni.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 27.Sikes ML, Suarez CC, Oltz EM. Regulation of V(D)J recombination by transcriptional promoters. Mol. Cell. Biol. 1999;19:2773–2781. doi: 10.1128/mcb.19.4.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sikes ML, Meade A, Tripathi R, Krangel MS, Oltz EM. Regulation of V(D)J recombination: a dominant role for promoter positioning in gene segment accessibility. Proc. Natl. Acad. Sci. USA. 2002;99:12309–12314. doi: 10.1073/pnas.182166699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitehurst CE, Schlissel MS, Chen J. Deletion of germline promoter PD β1 from the TCR β locus causes hypermethylation that impairs Dβ1 recombination by multiple mechanisms. Immunity. 2000;13:703–714. doi: 10.1016/s1074-7613(00)00069-8. [DOI] [PubMed] [Google Scholar]
- 30.Whitehurst CE, Chattopadhyay S, Chen J. Control of V(D)J recombinational accessibility of the Dβ1 gene segment at the TCR β locus by a germline promoter. Immunity. 1999;10:313–322. doi: 10.1016/s1074-7613(00)80031-x. [DOI] [PubMed] [Google Scholar]
- 31.Zimmerman KA, Yancopoulos GD, Collum RG, Smith RK, Kohl NE, Denis KA, Nau MM, Witte ON, Toran-Allerand D, Gee CE, et al. Differential expression of myc family genes during murine development. Nature. 1986;319:780–783. doi: 10.1038/319780a0. [DOI] [PubMed] [Google Scholar]
- 32.Douglas NC, Jacobs H, Bothwell AL, Hayday AC. Defining the specific physiological requirements for c-Myc in T cell development. Nat. Immunol. 2001;2:307–315. doi: 10.1038/86308. [DOI] [PubMed] [Google Scholar]
- 33.Wu C, Ranganath S, Gleason M, Woodman BB, Borjeson TM, Alt FW, Bassing CH. Restriction of endogenous TCRβ rearrangements to Vβ14 through selective recombination signal sequence modifications. Proc. Natl. Acad. Sci. USA. 2007;104:4002–4007. doi: 10.1073/pnas.0700081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Born W, Yague J, Palmer E, Kappler J, Marrack P. Rearrangement of T-cell receptor β-chain genes during T-cell development. Proc. Natl. Acad. Sci. USA. 1985;82:2925–2929. doi: 10.1073/pnas.82.9.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mundy CL, Patenge N, Matthews AG, Oettinger MA. Assembly of the RAG1/RAG2 synaptic complex. Mol. Cell. Biol. 2002;22:69–77. doi: 10.1128/MCB.22.1.69-77.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones JM, Gellert M. Ordered assembly of the V(D)J synaptic complex ensures accurate recombination. EMBO J. 2002;21:4162–4171. doi: 10.1093/emboj/cdf394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Curry JD, Geier JK, Schlissel MS. Single-strand recombination signal sequence nicks in vivo: evidence for a capture model of synapsis. Nat. Immunol. 2005;6:1272–1279. doi: 10.1038/ni1270. [DOI] [PubMed] [Google Scholar]
- 38.West KL, Singha NC, De Ioannes P, Lacomis L, Erdjument-Bromage H, Tempst P, Cortes P. A direct interaction between the RAG2 C terminus and the core histones is required for efficient V(D)J recombination. Immunity. 2005;23:203–212. doi: 10.1016/j.immuni.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 39.Uematsu Y, Ryser S, Dembic Z, Borgulya P, Krimpenfort P, Berns A, von Boehmer H, Steinmetz M. In transgenic mice the introduced functional T cell receptor β gene prevents expression of endogenous β genes. Cell. 1988;52:831–841. doi: 10.1016/0092-8674(88)90425-4. [DOI] [PubMed] [Google Scholar]
- 40.Mathieu N, Spicuglia S, Gorbatch S, Cabaud O, Fernex C, Verthuy C, Hempel WM, Hueber AO, Ferrier P. Assessing the role of the T cell receptor β gene enhancer in regulating coding joint formation during V(D)J recombination. J. Biol. Chem. 2003;278:18101–18109. doi: 10.1074/jbc.M212647200. [DOI] [PubMed] [Google Scholar]
- 41.Gartner F, Alt FW, Monroe R, Chu M, Sleckman BP, Davidson L, Swat W. Immature thymocytes employ distinct signaling pathways for allelic exclusion versus differentiation and expansion. Immunity. 1999;10:537–546. doi: 10.1016/s1074-7613(00)80053-9. [DOI] [PubMed] [Google Scholar]
- 42.Aifantis I, Buer J, von Boehmer H, Azogui O. Essential role of the pre-T cell receptor in allelic exclusion of the T cell receptor β locus. Immunity. 1997;7:601–607. doi: 10.1016/s1074-7613(00)80381-7. [DOI] [PubMed] [Google Scholar]
- 43.Smith CA, Graham CM, Thomas DB. Productive re-arrangement at both alleles of the T-cell receptor β-chain locus in CD4 T-cell clones specific for influenza haemagglutinin. Immunology. 1994;81:502–506. [PMC free article] [PubMed] [Google Scholar]
- 44.Glusman G, Rowen L, Lee I, Boysen C, Roach JC, Smit AF, Wang K, Koop BF, Hood L. Comparative genomics of the human and mouse T cell receptor loci. Immunity. 2001;15:337–349. doi: 10.1016/s1074-7613(01)00200-x. [DOI] [PubMed] [Google Scholar]
- 45.Senoo M, Shinkai Y. Regulation of Vβ germline transcription in RAG-deficient mice by the CD3epsilon-mediated signals: implication of Vβ transcriptional regulation in TCR β allelic exclusion. Int. Immunol. 1998;10:553–560. doi: 10.1093/intimm/10.5.553. [DOI] [PubMed] [Google Scholar]