Abstract
Introduction:
Trypanosomes are protozoan parasites of vertebrates transmitted by blood-sucking tsetse fly. Trypanosomes remain a constant threat to the lives of humans and animals throughout large regions of Africa.
Aims and Objectives:
This study investigated the presence, prevalence, and species of trypanosome parasite in tsetse flies caught in two areas of no previous documented history of trypanosome infection.
Materials and Methods:
For this purpose, 63 and 77 nonterenal tsetse flies were collected from Oji River and Emene areas of Enugu State Nigeria, respectively. Genomic DNA was isolated from the whole tsetse fly using genomic DNA extraction kit. Identification and characterization of trypanosome were done using two approaches: the amplification of internal transcribed spacer 1 of ribosomal DNA and the use of primers specific to Trypanozoon.
Results:
In Oji River, of 63 tsetse flies collected, the identification of trypanosome parasite was done on 57 flies and 6 (10.71%) tsetse flies were infected with trypanosome parasite. Six flies were infected with Trypanosoma Congolense, 2 with Trypanosoma Vivax, and 1 with Trypanosoma brucei. Two mixed infections of T. vivax and T. congolense and 1 mixed infection of T. brucei and T. congolense was also identified. In Emene, of 77 tsetse flies collected, the identification of trypanosome parasite was done on 66 flies and 11 (16.6%) tsetse flies were infected with trypanosome parasite. Nine flies were infected with T. congolense, 2 with T. vivax, and 3 with T. brucei. Mixed infections identified include 2 mixed infections of T. brucei and T. congolense and 1 mixed infections of T. vivax and T. brucei. None of the subspecies of T. brucei were detected using species specific primers.
Discussion:
This study shows the parasitological evidence on the occurrence of animal African trypanosomiasis and also demonstrated that there is likely no active transmission of human African trypanosomiasis in the study areas.
Conclusion:
This study shows that there is likely no active transmission of human African trypanosomiasis going on in these localities since no human infective form of the parasite was detected.
Keywords: Trypanosoma brucei, Trypanosoma congolense, Trypanosoma vivax, trypanosome, tsetse fly
INTRODUCTION
Tsetse fly is a vector of trypanosome parasite that feed on vertebrate blood and transmits trypanosomiasis, causing sleeping sickness in humans and nagana in animals.[1] Despite all the control measures put in place for the control and eradication of tsetse fly in Nigeria, the vectors have continued to spread to new areas. Trypanosomiasis, both of humans and animals, is one of the most important factors restricting economic development in Africa.[2] Tsetse flies through the cyclical transmission of trypanosomiasis to both humans and animals have greatly influenced food production, natural-resource utilization, and the pattern of human settlement in Sub-Saharan Africa. It is estimated that the annual direct production losses in cattle alone amount to between US$6000 and $12,000 million, while animal deaths may reach 3 million.[3]
Animals tend to be more infected than humans since there are more species of trypanosomes that can infect them and the fly preferentially feeds on nonhuman animal hosts.[4] Nigeria is faced with problems of social unrest, poverty, malnutrition, disease, food insecurity among others, and trypanosomiasis has forced many farmers away from the more productive lands and grazing sites to subsist in marginal agricultural areas which have negative impact on their livelihood and agricultural production.[5] Tryanosomiasis has direct impact on livestock productivity, reducing meat and milk production. It also has direct impacts on human settlement[6] and reduces productivity of the people with the disease, family members who care for the ill, and the rural residents who fear that they might contract the disease thereby making individual become burden to the family and the resources.[7] Certain degree of success had been achieved in the control of trypanosomiasis in humans and animals by the use of bush clearing, trypanocidal drugs, trapping, and sterile insect technique; it was realized that the only permanent solution to the problem lies in the eradication of the fly.[8] Little information exists about trypanosome prevalence hence the need to update information on the status of trypanosomiasis in these areas. The purpose of this study was to identify the species of tsetse flies circulating in the study locations and to isolate and characterize the species of trypanosomes from tsetse flies.
MATERIALS AND METHODS
Study location
This study was carried out in Oji River (6°16'N, 7°16'E) and Emene (6°29'N, 7°34'E) areas of Enugu State, Nigeria [Figure 1]. These areas were selected based on reported outbreak of tsetse fly in these locations.
Figure 1.
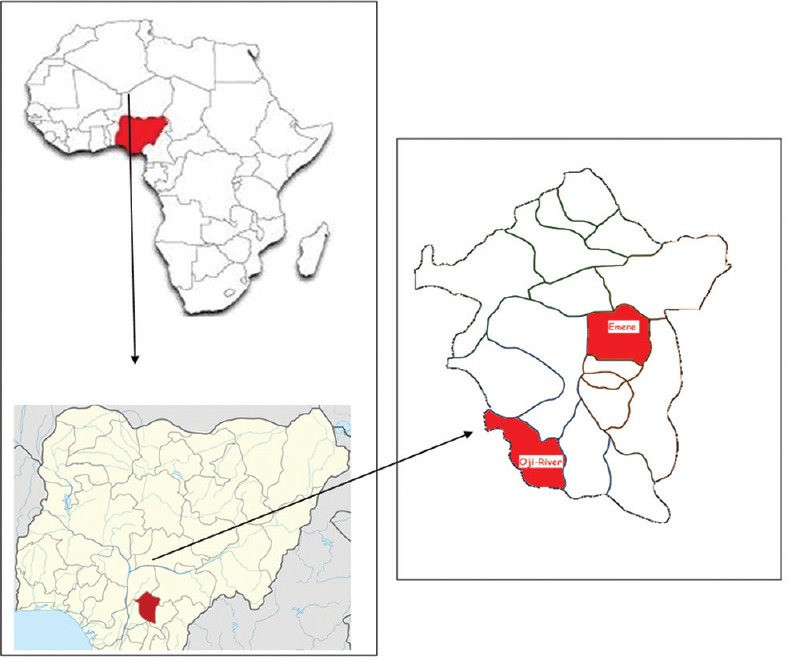
Sampled sites in Oji River and Emene
Entomological survey
Using pyramidal traps, a total of 63 and 77 nonterenal tsetse flies were collected in each location, respectively. The flies were identified using keys for identification of glossina species and stored in absolute ethanol until DNA extraction.
Identification of species of tsetse flies
Tsetse fly species identification was done based on give characteristics for each species by means of identification key. Each tsetse fly species has its own identification key. Such keys work by having a list of questions, usually in pairs, each of which normally asks you to look at a given feature of the insect and to decide whether it is similar to the first description or not. The answer you give will then lead you to the next question, giving you another choice. At the end of the sequence of questions and answers, the key will tell you which species the tsetse fly is.[1]
Genomic DNA extraction
In the laboratory, the flies were removed from the ethanol and allowed to stand at room temperature for the ethanol to evaporate followed by crushing of the whole fly with mortar and pestle. DNA was extracted using Bioneer AccuPrep Genomic DNA Extraction Kit according to the manufactures instruction. The purity of the extracted DNA samples was checked by measuring the ratio of A260/A280 and samples with values below 1.6 and above 1.9 were discarded.
Polymerase chain reaction amplification
Trypanosome infection and trypanosome species identification were done using polymerase chain reaction (PCR) based on the amplification of the internal transcribed spacer 1 (ITS) of the ribosomal DNA.[9] An 11 μl reaction mixture containing 3.5 μl of PCR Master Mix (promega), 0.5 μl of each primer (ITS1 CF: 5'-CCGGAAGTTCACCGATATTG-3'; ITS1 BR: 5'-TTGCTGCGTTCTTCAACGAA-3'),[10] 3.5 μl of autoclaved water, and 3.0 μl of sample DNA was subjected to PCR conditions which involved an initial denaturation step at 95°C for 2 min, followed by 30 cycles of 95°C for 30 s, 58°C for 30 s, 72°C for 1 min with a final extension step at 72°C for 5 min. A 5 μl of amplified PCR product was electrophoresed in 1.5% agarose stained with ethidium bromide (0.5 μg/ml) and the product visualized under ultraviolet illumination. The size of the electrophoresed amplified PCR products was used in determining the species of the trypanosome infection as shown in Table 1.
Table 1.
The expected band sizes on amplification of different species of trypanosomes

Those that tested positive to Trypanosoma brucei were further amplified using primers specific to T. brucei rhodesiense (SRA For: 5'-TACTGTTGTTGTACCGCCGC-3'; SRA Rev: 5'-GACAACAAGTACCTTGGCGC-3')[11] and T. brucei gambiense (TgsGP For: 5'-GCTGCTGTGTTCGGAGAGC-3'; TgsGP Rev: 5'-GCCATCGTGCTTGCCTCTC-3').[12] All amplifications were performed using 25 μl reaction mixtures containing 2.5 μl of Super Taq 10x buffer, 0.5 μl of SRA For or TgsGP For primer, 0.5 μl of SRA Rev or TgsGP Rev primer, 0.2 μl of 100 mM dNTP, 1 μl of DNA polymerase, 14.55 μl of distilled water, 0.75 μl of MgCl2, and 5 μl of sample DNA. For TgsGp primers, the PCR condition involved an initial denaturation step at 95°C for 2 min, followed by 37 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 30 s with a final extension step at 72°C for 5 min. For SRA primers, the PCR condition involved an initial denaturation step at 95°C for 2 min, followed by 37 cycles of 94°C for 1 min, 68°C for 1 min, 72°C for 1 min with a final extension step at 72°C for 5 min. 5 μl of PCR product was mixed with standard loading dye and electrophoresed alongside 100 bp DNA ladder and negative control in 1.5% agarose stained with ethidium bromide (0.5 μg/ml) and the product visualized and photographed under ultraviolet illumination. The size of the electrophoresed amplified PCR products [Table 1] was used in determining the subspecies of T. brucei.
RESULTS
All the trapped tsetse flies were identified as Glossina palpalis palpalis. In Oji River, 63 tsetse flies were collected, the DNA sample of each fly was extracted and the identification of trypanosome infection was performed on 56 tsetse fly DNA samples. Of the 56 tsetse fly DNA samples analyzed, the amplification of ITS 1 of the ribosomal DNA identified 6 (10.71%) tsetse fly DNA samples infected by at least one trypanosome species [Figure 2]. Among these 6 infections, 3 mixed infections involving different trypanosome species were identified: 2 mixed infections of Trypanosoma vivax and Trypanosoma congolense and 1 mixed infection of T. brucei and T. congolense. Six flies were infected with T. congolense, 2 with T. vivax, and 1 with T. brucei [Figure 3].
Figure 2.
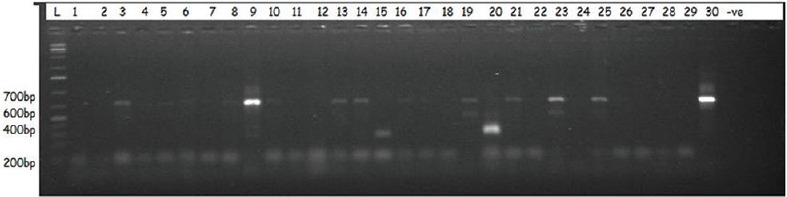
A representative agarose gel picture showing the resolution of the amplified products of samples using internal transcribed spacer 1 primers in tsetse flies collected in Oji River and Emene
Figure 3.
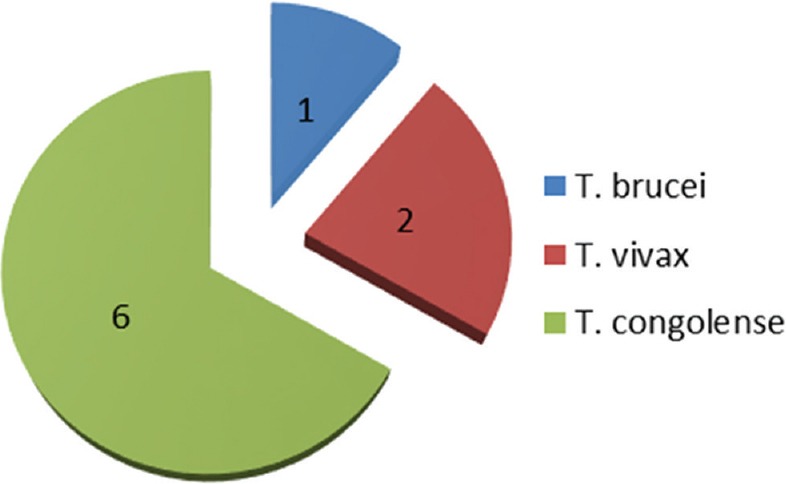
Trypanosome species infection in tsetse flies collected in Oji River: 6 flies were infected with Trypanosoma congolense; 2 with Trypanosoma vivax, and 1 with Trypanosoma brucei
In Emene, 77 tsetse flies were collected, the DNA sample of each fly was extracted, and the identification of trypanosomes was performed on 66 tsetse fly DNA samples. Of the 66 DNA samples analyzed, the amplification of the ITS 1 of the ribosomal DNA identified 11 (16.6%) DNA samples infected by at least one trypanosome species [Figure 2]. Among these 11 infections, three mixed infections involving different trypanosome species were identified: two mixed infections of T. brucei and T. congolense and one mixed infection of T. vivax and T. brucei. Nine flies were infected with T. congolense, 2 with T. vivax, and 3 with T. brucei [Figure 4].
Figure 4.
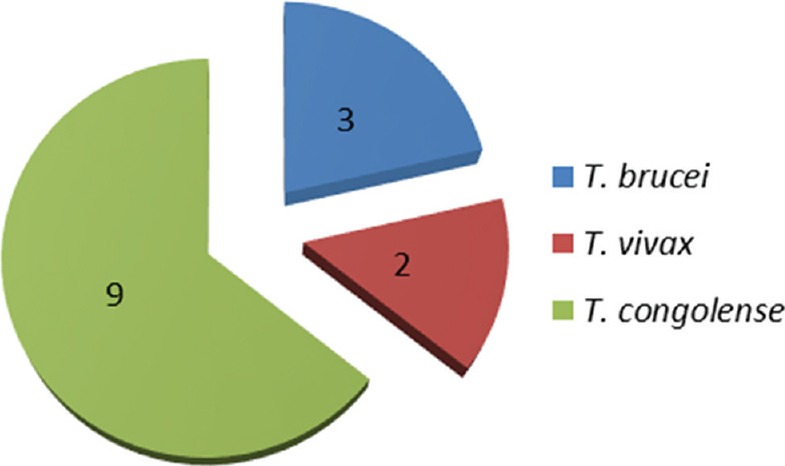
Trypanosome species infection in tsetse flies collected in Emene: 9 flies were infected with Trypanosoma congolense; 2 with Trypanosoma vivax, and 3 with Trypanosoma brucei
DISCUSSION
This investigation shows the presence of tsetse flies in the studied locations with G. palpalis palpalis being the only species of tsetse fly captured in the two locations and this could be attributed to the riverine nature of the study sites. This species normally inhabits vegetations associated with surface waters such as riverine and swampy of mangrove forest.[13] The same species of tsetse fly was recorded in the study done at northern Nigeria,[14] Malanga sleeping sickness focus of Democratic Republic of Congo,[15] Fontem sleeping sickness focus of Cameroon,[16] and Jomoro district of the western region of Ghana.[17]
Molecular tools have been developed for the identification and characterization of different species of trypanosomes found in tsetse fly.[18,19] Studies[20,21] have shown that the use of DNA-based methods for identification of trypanosomes is being favored over microscopy. Using molecular method, our investigation shows the presence of T. brucei, T. congolense, and T. vivax trypanosome parasites in the tsetse flies collected from the two different locations indicating that there is an ongoing transmission of trypanosome parasite. The different trypanosome species identified in this study are in agreement with results reported in tsetse flies of other regions of Africa.[15] Our results suggest that trypanosome infection probably remains a serious threat to animals in these areas. The result of our study revealed that 10.7% and 16.6% of the tsetse flies in Oji River and Emene, respectively, were infected by at least one trypanosome species. If we take into consideration the fact that these investigations were performed using whole tsetse fly, we can therefore say that the prevalence reported here was not underestimated since the trypanosomes affecting both the mid-gut (procyclic trypomasigotes) and salivary gland (metacyclic trypomasigotes) were detected in this study. Numerous single and mixed infections were identified in this study and these observations corroborate the earlier report in Fontem sleeping sickness focus of Cameroon,[16] Simanjiro, Northern Tanzania,[22] Malanga sleeping sickness focus of Democratic Republic of Congo,[15] and Kaura Local Government area of Kaduna State, Nigeria.[23] These results show that whatever the region, single and mixed trypanosome infections of different species are frequent in tsetse flies. The simplest route through which a tsetse fly can acquire mixed infection is by feeding on a host that is infected with different species or subspecies (concurrent infection). Another way is when the fly successively feeds on two or more infected hosts each of which is infected with a different species of trypanosome (sequential infection).[24]
The percentage of mixed infections in tsetse flies can vary between countries, and even within different regions of the same country. For example, 5.35% (Oji River) and 4.54% (Emene) of mixed infections were observed in this study, whereas 7.02% were reported in a study carried out at Malanga (Kimpese) sleeping sickness focus of the Democratic Republic of Congo.[15] In Tanzania and Côte d'Ivoire, mixed infections were reported in 13.5%[11] and 25%,[12] respectively, in tsetse flies. Among the trypanosome species identified, T. congolense was the most common species found. This is in line with the results obtained in pigs[25] and in tsetse flies.[26] The zero infection of T. brucei rhodesiense and T. brucei gambiense obtained in this study is similar to those obtained in tsetse flies.[22] Considering the impacts of trypanosome infections in animals, it is obvious that the high infection rate of these parasites recorded in this study will impact negatively on the economy of the study areas by affecting animal breeding in terms of animal health, productivity, quality, and quantity of meat thereby contributing to poverty, malnutrition, and hunger.
CONCLUSION
It is clearly shown in this study that there is likely no active transmission of human African trypanosomiasis going on in these localities since no human infective form of the parasite was detected. This study has shown the parasitological evidence on the occurrence of animal African trypanosomiasis as three animal infective trypanosomes (T. congolense, T. vivax and T. b brucei) are widely distributed in the areas which indicates that the areas are endemic for trypanosomiasis.
We therefore recommend that HAT and AAT surveillance as well as vector control program should be put in place in these areas to reduce the incidence of trypanosomiasis.
Financial support and sponsorship
We thank African Research League (ARL) for their financial support.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
We appreciate the management and entire staff of Nigerian Institute for Trypanosomiasis Research (NITR), South-East Zonal Office, Enugu, for the provision of tsetse fly traps and logistics support.
REFERENCES
- 1.Food and agricultural organization of the united nations programme against African trypanosomosis. Tsetse and Tryponosomosis Information. 2014;37:2. [Google Scholar]
- 2.Wilson SG, Morris KR, Lewis IJ, Krog E. The effects of trypanosomiasis on rural economy with special reference to the Sudan, Bechuanaland and West Africa. Bull World Health Organ. 1963;28:595–613. [PMC free article] [PubMed] [Google Scholar]
- 3.Hursey BS, Slingenbergh J. The tsetse fly and its effects on Agriculture in Sub-Saharan Africa. World Anim Rev. 1995;84:67–73. [Google Scholar]
- 4.Owaga ML. Observations on the efficacy of Buffalo urine as a potent olfactory attractant for Glossina pallidipes Austen. Int J Trop Insect Sci. 1985;6:561–6. [Google Scholar]
- 5.Malele I, Nyingilili H, Msangi A. Factors defining the distribution limit of tsetse infection and the implication for livestock sector in Tanzania. Afr J Agric Res. 2011;6:2341–7. [Google Scholar]
- 6.Takele A, Abebe G. A survey of trypanosomiasis in Gamu Gofa region (Ethiopia) Rev Elev Med Vet Pays Trop. 1988;41:271–6. [PubMed] [Google Scholar]
- 7.World Health Organisation Control and Surveillance of African Trypanosomiasis. WHO Expert Committee on the Control and Surveillance of Human African Trypanosomiasis 2013. WHO Technical Report Series, No. 984. 2013 [PubMed] [Google Scholar]
- 8.Putt SN, Shaw AP. The Socio-Economic Effects of the Control of Tsetse Transmitted Trypanosomiasis in Nigeria, Proceedings of the 3rd International Symposium on Veterinary Epidemiology and Economics. 1982 [Google Scholar]
- 9.Masiga DK, Smyth AJ, Hayes P, Bromidge TJ, Gibson WC. Sensitive detection of trypanosomes in tsetse flies by DNA amplification. Int J Parasitol. 1992;22:909–18. doi: 10.1016/0020-7519(92)90047-o. [DOI] [PubMed] [Google Scholar]
- 10.Njiru ZK, Constantine CC, Guya S, Crowther J, Kiragu JM, Thompson RC, et al. The use of ITS1 rDNA PCR in detecting pathogenic African trypanosomes. Parasitol Res. 2005;95:186–92. doi: 10.1007/s00436-004-1267-5. [DOI] [PubMed] [Google Scholar]
- 11.Adams ER, Hamilton PB, Rodrigues AC, Malele II, Delespaux V, Teixeira MM, et al. New Trypanosoma (Duttonella) vivax genotypes from tsetse flies in East Africa. Parasitology. 2010;137:641–50. doi: 10.1017/S0031182009991508. [DOI] [PubMed] [Google Scholar]
- 12.Djohan V, Kaba D, Rayaissé JB, Dayo GK, Coulibaly B, Salou E, et al. Detection and identification of pathogenic trypanosome species in tsetse flies along the Comoé River in Côte d'Ivoire. Parasite. 2015;22:18. doi: 10.1051/parasite/2015018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.World Health Organisation. WHO Expert Committee the Control and Survelance of African Trypanosomiasis. Geneva, Switzerland: WHO Technical Report Series; 1995. p. 881. [Google Scholar]
- 14.Isaac C, Ciosi M, Hamilton A, Scullion KM, Dede P, Igbinosa IB, et al. Molecular identification of different trypanosome species and subspecies in tsetse flies of Northern Nigeria. Parasit Vectors. 2016;9:301. doi: 10.1186/s13071-016-1585-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simo G, Silatsa B, Flobert N, Lutumba P, Mansinsa P, Madinga J, et al. Identification of different trypanosome species in the mid-guts of tsetse flies of the Malanga (Kimpese) sleeping sickness focus of the Democratic Republic of Congo. Parasit Vectors. 2012;5:201. doi: 10.1186/1756-3305-5-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simo G, Njitchouang GR, Melachio TT, Njiokou F, Cuny G, Tazoacha A. Population genetics of Trypanosoma brucei circulating in Glossina palpalis palpalis and domestic animals of the Fontem sleeping sickness focus of Cameroon. Parasit Vectors. 2014;7:156. doi: 10.1186/1756-3305-7-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Apaatah F. Trypanosome Prevalence in Pigs and Tsetse Flies from Jomoro District in the Western Region of Ghana, Thesis (MPhil)-University of Ghana. 2014 doi: 10.1016/j.vprsr.2020.100444. [DOI] [PubMed] [Google Scholar]
- 18.Enyaru JC, Ouma JO, Malele II, Matovu E, Masiga DK. Landmarks in the evolution of technologies for identifying trypanosomes in tsetse flies. Trends Parasitol. 2010;26:388–94. doi: 10.1016/j.pt.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 19.Wastling SL, Welburn SC. Diagnosis of human sleeping sickness: Sense and sensitivity. Trends Parasitol. 2011;27:394–402. doi: 10.1016/j.pt.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 20.Lehane MJ, Msangi AR, Whitaker CJ, Lehane SM. Grouping of trypanosome species in mixed infections in Glossina pallidipes. Parasitology. 2000;120(Pt 6):583–92. doi: 10.1017/s0031182099005983. [DOI] [PubMed] [Google Scholar]
- 21.Njiru ZK, Makumi JN, Okoth S, Ndungu JM, Gibson WC. Identification of trypanosomes in Glossina pallidipes and G. longipennis in Kenya. Infect Genet Evol. 2004;4:29–35. doi: 10.1016/j.meegid.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Salekwa LP, Nnko HJ, Ngonyoka A, Estes AB, Agaba M, Gwakisa PS. Relative abundance of tsetse fly species and their infection rates in Simanjiro, Northern Tanzania. Livest Res Rural Dev. 2014;26:213. [Google Scholar]
- 23.Ahmed AB. High trypanosome infections in Glossina palpalis palpalis robineau-desvoidy 1830 in Southern Kaduna state, Nigeria. Sci World J. 2007;2:1–7. [Google Scholar]
- 24.Morlais I, Grebaut P, Bodo JM, Djoha S, Cuny G, Herder S. Detection and identification of trypanosomes by polymerase chain reaction in wild tsetse flies in Cameroon. Acta Trop. 1998;70:109–17. doi: 10.1016/s0001-706x(98)00014-x. [DOI] [PubMed] [Google Scholar]
- 25.Simo G, Asonganyi T, Nkinin SW, Njiokou F, Herder S. High prevalence of Trypanosoma brucei gambiense group 1 in pigs from the Fontem sleeping sickness focus in Cameroon. Vet Parasitol. 2006;139:57–66. doi: 10.1016/j.vetpar.2006.02.026. [DOI] [PubMed] [Google Scholar]
- 26.Kubi C, Billiouw M, Van Den Bossche P. Age prevalence of trypanosomal infections in female Glossina morsitans morsitans (Diptera: Glossinidae) on the plateau area of Eastern Zambia. Onderstepoort J Vet Res. 2007;74:223–9. doi: 10.4102/ojvr.v74i3.125. [DOI] [PubMed] [Google Scholar]


