Abstract
Autism spectrum disorder (ASD) is one of the most prevalent neurodevelopmental conditions worldwide. There is growing awareness that ASD is highly comorbid with gastrointestinal distress and altered intestinal microbiome, and that host-microbiome interactions may contribute to the disease symptoms. However, the paucity of knowledge on gut-brain axis signaling in autism constitutes an obstacle to the development of precision microbiota-based therapeutics in ASD. To this end, we explored the interactions between intestinal microbiota, gut physiology and social behavior in a BTBR T+Itpr3tf/J mouse model of ASD. Here we show that a reduction in the relative abundance of very particular bacterial taxa in the BTBR gut – namely, bile-metabolizing Bifidobacterium and Blautia species, - is associated with deficient bile acid and tryptophan metabolism in the intestine, marked gastrointestinal dysfunction, as well as impaired social interactions in BTBR mice. Together these data support the concept of targeted manipulation of the gut microbiota for reversing gastrointestinal and behavioral symptomatology in ASD, and offer specific plausible targets in this endeavor.
Keywords: Autism, BTBR mouse, Gut microbiota, Intestinal permeability, Intestinal transit, Bile acids, Serotonin, Tryptophan
Highlights
-
•
BTBR mice display a reduced abundance of bile-metabolizing Bifidobacterium and Blautia bacterial species in the intestine.
-
•
BTBR mice demonstrate deficient bacterial metabolism of bile moieties in the gut.
-
•
Changes in the gut microbiota are associated with marked gastrointestinal distress and reduced sociability in BTBR mice.
There is growing awareness that the gut microbiome may contribute to gastrointestinal and behavioral symptomatology of autism spectrum disorder (ASD). However, the exact mechanisms by which intestinal bacteria can affect gut-brain axis signaling in autism are as yet poorly understood. Here we explore interactions between intestinal microbiota, gut physiology and behavior in a mouse model of ASD. We show that a reduction in the abundance of particular intestinal bacteria in “autistic” mice is associated with gastrointestinal distress and social behavior deficits. This work supports the concept of targeting the gut microbiota for reversing gastrointestinal symptoms in ASD, and identifies specific plausible targets for microbiota-based interventions.
1. Introduction
Autism spectrum disorder (ASD) is one of the most serious neurodevelopmental conditions worldwide, affecting on average 1 in 150 children (Lyall et al., 2017). The hallmarks of ASD include impaired social communication skills and repetitive behaviors, frequently co-occurring with intellectual disability (Association, 2013). There are limited treatment options for ASD symptoms, and thus there is a great need to develop strategies that would improve the quality of life in ASD subjects. More recently, the gut microbiome has emerged as a novel important player and therapeutic target in autism (Vuong and Hsiao, 2017). Intestinal bacteria can orchestrate a range of gastrointestinal (GI) functions (Sommer and Backhed, 2013), and therefore are poised to affect gut-brain axis signaling in ASD. Animal studies have demonstrated that the gut microbiota plays a crucial role in anxiety (Neufeld et al., 2011), cognitive performance (Desbonnet et al., 2015), repetitive behaviors (Desbonnet et al., 2014) and social communications (Buffington et al., 2016; Sherwin et al., 2017). There is a growing body of evidence that ASD subjects display substantial alterations in microbiota composition, along with marked symptoms of GI distress, including changes to bowel habits, increased intestinal permeability, bloating and abdominal pain (Angelis et al., 2013; D'Eufemia et al., 1996; Kang et al., 2013; Luna et al., 2017; McElhanon et al., 2014; Tomova et al., 2015). In a recent small open-label clinical study, transplantation of standardized human gut microbiota to ASD-diagnosed children induced an improvement of GI function, as well as a reduction of behavioral ASD scoring (Kang et al., 2017). However, we do not yet know whether alterations in the gut microbiota in autism can in itself affect either behavioral or GI symptoms of ASD.
Identification of specific bacterial taxa and associated bacterial metabolites that can trigger changes in GI physiology and modulate the onset of autistic behaviors is thus an important quest in the search for precision microbiota-based therapeutics in ASD (Gilbert et al., 2013). Preclinical research is crucial in this endeavor (Hsiao et al., 2013; Kumar and Sharma, 2016). The BTBR T+ Itpr3tf/J mouse strain is one of the most widely used models of ASD. Similar to humans, these animals display robust deficits in social interactions and enhanced engagement in repetitive behaviors (Meyza et al., 2013; Moy et al., 2007). Moreover, the autistic-like behavior of BTBR mice is largely driven by multiple genetic alterations (Meyza and Blanchard, 2017), but is also sensitive to environmental influences, such as dietary intervention (Ruskin et al., 2013). Changes to a few bacterial taxa have also been recently reported in the BTBR intestine (Coretti et al., 2017; Klein et al., 2016; Newell et al., 2016). Together, these findings strengthen the utility of the BTBR strain in understanding dysfunctional microbiota-gut-brain signaling in ASD. To this end, we characterized GI physiology in BTBR mice and found substantial abnormalities in both intestinal barrier function and intestinal transit. We identified changes in bile acid and tryptophan metabolism as putative mechanisms underlying GI dysfunction in these animals. We further associated the observed changes in GI physiology with a reduction in the abundance of certain bacterial taxa in the BTBR microbiota.
2. Materials and Methods
2.1. Animals
C57BL/6 J (Harlan, UK) and BTBR T+ Itpr3tf/J (The Jackson Laboratory, US) adult male mice were group housed under a 12 h light-dark cycle; standard rodent chow and water were available ad libitum. All experiments were performed in accordance with European guidelines following approval by University College Cork Animal Ethics Experimentation Committee.
2.2. Study Design
Three independent cohorts participated in the study (Fig. S1). Detailed information on procedures with references is provided in Supplemental Methods.
2.3. Behavioral Assessment
Social behavior was analyzed in the three-chamber, social transmission of food preference and resident-intruder tests; compulsive behavior in the marble burying and self-grooming tests; anxiety-like behavior in the open field and elevated plus maze tests; depressive-like behavior in the forced swim test; cognitive performance in the novel object recognition test.
2.4. Plasma Corticosterone in Response to an Acute Stressor
Corticosterone levels were measured in plasma prior to (baseline) and after the onset of the forced swim test.
2.5. Fecal Water Content
Water content in feces was calculated as the difference in weight after desiccation of fecal pellets.
2.6. Whole Intestinal Transit
A test mouse was administered with carmine dye by oral gavage; the latency for the excretion of the first red-colored fecal pellet was recorded.
2.7. Colonic Transit
An artificial pellet was inserted into the proximal end of the isolated colon, and the spontaneous propagation of the pellet in anterograde direction was video recorded. The latency to initiate the propagation and the velocity of propagation were scored.
2.8. Intestinal Permeability
Intestinal tissue samples were mounted into the Ussing chambers. Short-circuit current (Isc) was recorded in a zero voltage clamp mode; transepithelial electrical resistance (TEER) was measured by discharging a 2 mV pulse. 4 kDa FITC-dextran was added to the mucosal chamber at a final concentration of 2.5 mg/mL; 200 μL samples were collected from the serosal chamber every 30 min for the following 3 h.
2.9. Myenteric Plexus Morphology
Myenteric plexus (MP) of distal colon was subjected to immunoflourescent staining against choline acetyltransferase (ChAT), HuC/HuD and nNOS. To analyze the morphology of MP, the following parameters were calculated in anti-ChAT stained tissues: the area occupied by MP; the mean thickness and the contact ratio of MP; the mean Feret diameter of interganglionic space. The numbers of HuC/HuD- and nNOS-positive myenteric neurons were quantified per visual field.
2.10. Tryptophan, Kynurenine and Serotonin Levels
Serotonin was measured in intestinal tissue samples by HPLC coupled to electrochemical detection. Kynurenine and tryptophan concentrations were determined in plasma by HPLC coupled to ultraviolet (230 nm) and fluorescent (254 nm excitation and 404 nm emission wavelengths) detection, accordingly.
2.11. Gene Expression Analysis
Total RNA was extracted from the intestinal tissue and the front lobe of the liver with the Qiagen RNeasy Plus Universal Mini kit, and from the right hippocampus with the Ambion mirVana™ miRNA Isolation Kit. Genes of interest were amplified with real-time RT-PCR using TaqMan probes (Table S1). Data were analyzed with the comparative cycle threshold method (2−ΔΔCt) and presented as a fold change vs. C57BL/6 group.
2.12. Bile Acid Levels in Plasma and Fecal Material
Fecal and plasma bile acids and salts were extracted following addition of deuterated internal standards (IS). Extracted acids were resuspended in 50% methanol, injected in triplicate and assessed in negative electrospray mode through a C18 Acquity column using a LCT Premier mass spectrometer (Waters Corp). Assessment of extraction efficiency was performed using IS, and samples were quantified by standard curve construction for individual bile moieties using targetlynx software (Waters).
2.13. Microbiota Composition and Short Chain Fatty Acids Analysis in the Cecal Content
DNA was extracted using the Qiagen QIAmp Fast DNA Stool Mini Kit coupled with an initial bead-beating step. The V3-V4 hypervariable region of the 16S rRNA gene was amplified and prepared for sequencing as outlined in the Illumina 16S Metagenomic Sequencing Library Protocol. Samples were sequenced at Clinical-Microbiomics (Copenhagen, Denmark) on the Illumina MiSeq platform using a 2 × 300 bp kit. Reads were assembled, processed and analyzed following the pipeline, described in Supplemental Methods. Short chain fatty acids (SCFAs) were measured by gas chromatography, using a Varian 3500 GC flame-ionization system fitted with a ZB-FFAP column.
2.14. Statistical Analysis
Normally distributed data are presented as mean ± SEM; unpaired two-tailed t-test and mixed-design ANOVA were applied to analyze the differences between groups. Non-parametric datasets, as well as discrete data, are presented as median (IQR). Mann-Whitney test was used for between-group comparisons. Spearman's rank correlation coefficient was employed for the correlation analysis in the pooled datasets. A p value < 0.05 was deemed significant in all cases.
3. Results
3.1. BTBR Mice Display Marked Changes in the Intestinal Microbiota
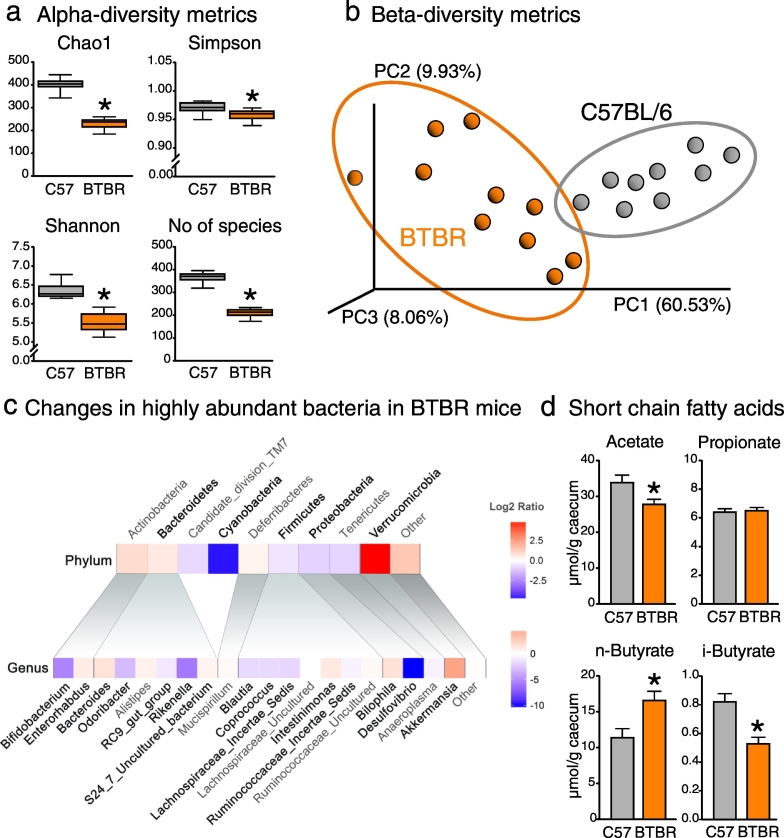
The 16S sequencing revealed a substantial reduction in the bacterial species diversity in the BTBR cecum (Fig. 1a). Principal coordinate analysis showed a clear separation of BTBR and C57BL/6 microbiotas (Fig. 1b). We detected 9 phyla, 23 families and 41 bacterial genera in C57BL/6 and BTBR mice (Table S2). At the phylum level, BTBR mice displayed a distinct shift towards an increase in Bacteroidetes and a decrease in Firmicutes; these are two major bacterial phyla altogether comprising > 90% of murine intestinal microbiota (Fig. 1c). The bloom in Bacteroidetes was mainly driven by an increase in S24–7 uncultured bacterium, a dominant genus of S24–7 family. Interestingly, this seems to have happened at the expense of minor Bacteroidetes genera such as Odoribacter, Parabacteroides, RC9 gut group and Rikenella, which all showed a reduction in relative abundance. A decrease in Firmicutes was largely driven by Lachnospiraceae family, in particular Blautia bacteria, but also Lachnospiraceae_Incertae Sedis and Coprococcus. BTBR mice also displayed an increase in Akkermansia, Bacteroides and Bilophila genera, as well as a reduction in Bifidobacterium and Desulfovibrio (Table S2). With such a prominent shift in the microbiota composition, it was perhaps not surprising that BTBR mice also displayed significant changes in the SCFAs levels in the gut lumen (Fig. 1d). Overall, BTBR mice seem to have a very robust phenotype of the gut microbiota. For example, a similar increase in Akkermansia and a reduction in Bifidobacterium numbers in the BTBR cecum were shown in previous reports (Klein et al., 2016; Newell et al., 2016). In the fecal material of BTBR mice, Coretti et al. found a different pattern of changes in the microbiota composition (Coretti et al., 2017); however, care should be taken when comparing cecal and fecal bacterial communities.
Fig. 1.
Altered microbiota composition and short chain fatty acid levels in BTBR mice (a–b) BTBR cecal microbiota showed lower diversity of bacterial species, and was clearly separated from the microbiota of C57BL/6 mice. (c) Heatmap of log2 fold change ratio showing increases (red) or decreases (blue) in most abundant bacteria in BTBR compared to C57BL/6 mice. Significant changes are bolded (for details, see Table S2). (d) Short chain fatty acid cecal levels were differentially changed in BTBR mice. *p < 0.05 for BTBR vs. C57BL/6 group. Statistical analysis: data are presented as median with IQR and min/max values as error bars on (a), and as mean + SEM on (d). N = 9 for C57BL/6 and N = 10 for BTBR group. U(17) = 0.000, p < 0.0005 for Chao1 index; U(17) = 15.5, p = 0.013 for Simpson index; U(17) = 0.000, p < 0.0005 for Shannon index; U(17) = 0.000, p < 0.0005 for No of species, Mann-Whitney test. t(17) = 2.424, p = 0.027 for acetate; t(17) = − 0.265, p = 0.794 for propionate; t(17) = − 2.887, p = 0.010 for n-butyrate; t(17) = 4.023, p = 0.001 for i-butyrate, unpaired t-test.
3.2. BTBR Mice Show Delayed Intestinal Transit
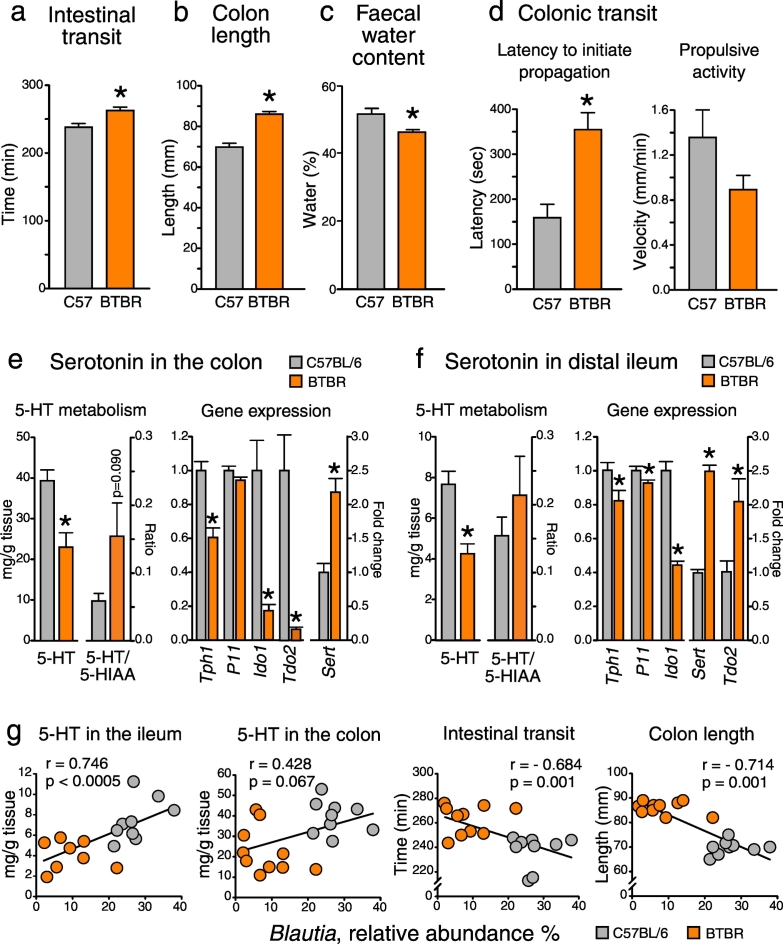
Among GI comorbidities associated with ASD, changes in bowel habits, in particular constipation, are the most prevalent (McElhanon et al., 2014). Hence, we examined intestinal motility, and found that whole intestinal transit was significantly delayed in BTBR mice (Fig. 2a). Further, BTBR mice displayed an elongated colon and decreased fecal water contents (Fig. 2b–c), both observations suggesting that the propagation of fecal matter along the colon was substantially slowed. The follow-up ex vivo analysis of colonic motility provided clear evidence of impaired transit in the BTBR large intestine. Indeed, it took almost twice as long to initiate spontaneous propagation of the pellet along the colon, and there was a trend towards a reduction in the velocity of oral-to-anal propulsion in BTBR mice (Fig. 2d). Measuring small intestinal transit in BTBR mice would be of interest for the future studies, particularly bearing in mind that impaired small intestinal motility was previously reported in a rat model of ASD (Kim et al., 2013).
Fig. 2.
Delayed intestinal transit and impaired serotonin metabolism in BTBR mice: implication of microbiota (a–c) BTBR mice showed delayed whole intestinal transit, as well as an elongated colon and decreased fecal water contents. (d) Colonic transit was impaired in BTBR mice. (e–f) Serotonin (5-HT) tissue levels were reduced in both small and large intestine of BTBR mice. This was coupled with alterations in the expression of genes involved in tryptophan metabolism. (g) Intestinal transit rate and 5-HT levels were strongly associated with the relative abundance of Blautia bacteria (see also Table S3). *p < 0.05 for BTBR vs. C57BL/6 group. Statistical analysis: data are presented as mean + SEM; n = 8 for C57BL/6 and n = 6 for BTBR group for (d), n = 9–10 in either group for the rest. t(18) = − 4.524, p < 0.0005 for the intestinal transit time; t(18) = − 14.053, p < 0.0005 for the colon length; t(12) = − 4.107, p = 0.001 for the latency to initiate propagation; t(11) = 1.599, p = 0.138 for the propulsive activity; t(17) = 3.597, p = 0.002 for 5-HT levels and t(15) = − 1.814, p = 0.09 for 5-HT/5-HIAA ratio in the colon; t(17) = 5.069, p < 0.0005 for Tph1, t(18) = 1.928, p = 0.070 for P11, t(16) = 4.578, p = 0.003 for Ido1, t(18) = 3.835, p = 0.006 for Tdo2 and t(18) = − 4.851, p < 0.0005 for Sert gene expression in the colon; t(17) = 4.164, p = 0.001 for 5-HT levels and t(8) = − 0.930, p = 0.380 for 5-HT/5-HIAA ratio in the ileum; t(17) = 2.311, p = 0.034 for Tph1, t(17) = 2.533, p = 0.021 for P11, t(16) = 9.546, p < 0.0005 for Ido1, t(18) = − 14.655, p < 0.0005 for Sert and t(18) = − 2.739, p = 0.013 for Tdo2 gene expression in the ileum, unpaired t-test. Spearman's rho was employed for the correlation analysis in the pooled datasets.
3.3. Impaired Serotonin Production in the BTBR Intestine
The release of mucosal serotonin (5-HT) triggers intestinal peristalsis (Heredia et al., 2013). To address the potential contribution of 5-HT metabolism to the delayed transit in the BTBR intestine, we assessed 5-HT availability in colonic and ileal tissue samples. We observed a 50% reduction in 5-HT tissue levels in both small and large intestine of BTBR mice, which was coupled with a down-regulation of Tph1 and an up-regulation of Sert gene expression (Fig. 2e–f). Tph activity determines the amount of 5-HT which is produced from dietary tryptophan and released from enterochromaffin cells, while Sert controls the rate of 5-HT re-uptake and consequent breakdown in enterocytes (Mawe and Hoffman, 2013). In line with the gene expression data, the ratio between 5-HIAA (the major metabolite of 5-HT) and 5-HT tended to be increased in the BTBR intestine (Fig. 2e–f). Together, these findings suggest reduced bioavailability of 5-HT in the BTBR gut due to a decrease in 5-HT synthesis and an increase in 5-HT breakdown. Alternatively to 5-HT, dietary tryptophan in the intestine can be metabolized along the kynurenine route. Ido1 and Tdo2 are two rate-limiting enzymes in this metabolic pathway. We found that the expression levels of both Ido1 and Tdo2 genes were markedly down-regulated in the BTBR colon. Interestingly, opposite changes in Ido1 and Tod2 genes were observed in the small intestine of BTBR mice (Fig. 2e–f).
3.4. Intestinal Transit and Serotonin Levels Correlate with the Abundance of Certain Bacterial Taxa
Gut microbes, in particular spore-forming clostridia, have been shown to regulate 5-HT metabolism in the GI tract (Yano et al., 2015). To this end, we investigated whether observed changes in the intestinal transit and 5-HT levels in the gut were associated with alterations in the abundance of any particular bacterial taxa. We found that both 5-HT tissue levels and intestinal transit rate were strongly correlated with the relative abundance of Firmicutes bacteria; this correlation was mainly driven by the Blautia genus of the Clostridiales order (Fig. 2g). There were also strong correlations between the abundance of Blautia and Tph1, Sert and Ido1 gene expression levels (Table S3). Similar, but weaker associations were observed with the Lachnospiraceae Incertae Sedis genus of the same order. These findings confirm the importance of clostridia bacteria in the maintenance of normal peristalsis and 5-HT synthesis in the gut.
3.5. BTBR Mice Display Deficits in the Development of the Enteric Nervous System
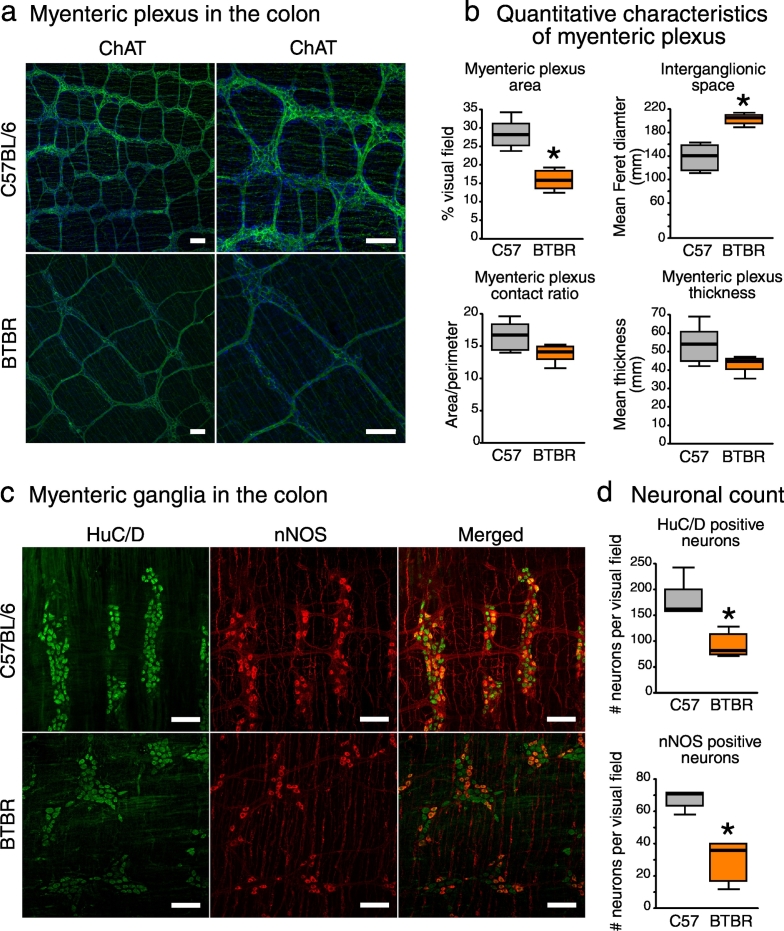
Healthy motility of the GI tract relies heavily on the enteric nervous system (ENS). We thus aimed to investigate whether delayed colonic transit in BTBR mice could have also been attributed to abnormal development of motor circuits in the ENS. For this, we stained the myenteric plexus (MP) in distal colon against choline acetyltransferase (ChAT), which is expressed in a vast majority of myenteric neurons including excitatory motor neurons, and against neuronal NO synthase (nNOS), which is expressed in inhibitory motor neurons of MP (Furness, 2000). ChAT staining showed the fine structure of MP with ganglia and interganglionic fibers (Fig. 3a). The morphometric analysis revealed a substantial reduction in the spatial density of MP in BTBR mice: the area occupied by MP was decreased, while the interganglionic space was increased (Fig. 3b). Furthermore, we found a 50% reduction in the numbers of inhibitory motor neurons (nNOS) and total myenteric neurons (pan-neuronal HuC/HuD marker) (Fig. 3c–d). These data suggest an abnormal development of both excitatory and inhibitory motor neurons in BTBR mice. These are interesting data in the context of recent findings indicating that the gut microbiota is essential for the development of the ENS (Collins et al., 2014; Obata and Pachnis, 2016).
Fig. 3.
BTBR mice display deficits in the development of the enteric nervous system (a) Anti-ChAT staining (green) in whole mount preparations of distal colon revealed the fine structure of myenteric plexus (MP), with ganglia and interconnecting fibers. Cellular nuclei were stained with DAPI (blue). (b) The spatial density of MP was decreased in BTBR mice. (c) Myenteric ganglia were co-stained against pan-neuronal marker HuC/HuD (green) and nNOS (red). (d) BTBR mice showed a reduction in total and nNOS-positive neuronal numbers. Scale bar represents 100 μm *p < 0.05 for BTBR vs. C57BL/6 group. Statistical analysis: data are presented as median with IQR and min/max values as error bars; n = 5 in either group. U(9) = 0.000, p = 0.004 for MP area; U(9) = 30.000, p = 0.004 for interganglionic space; U(9) = 6.000, p = 0.126 for MP contact ratio; U(9) = 5.000, p = 0.082 for MP thickness; U(8) = 0.000, p = 0.008 for HuC/HuD neurons; U(8) = 0.000, p = 0.008 for nNOS neurons, Mann-Whitney test.
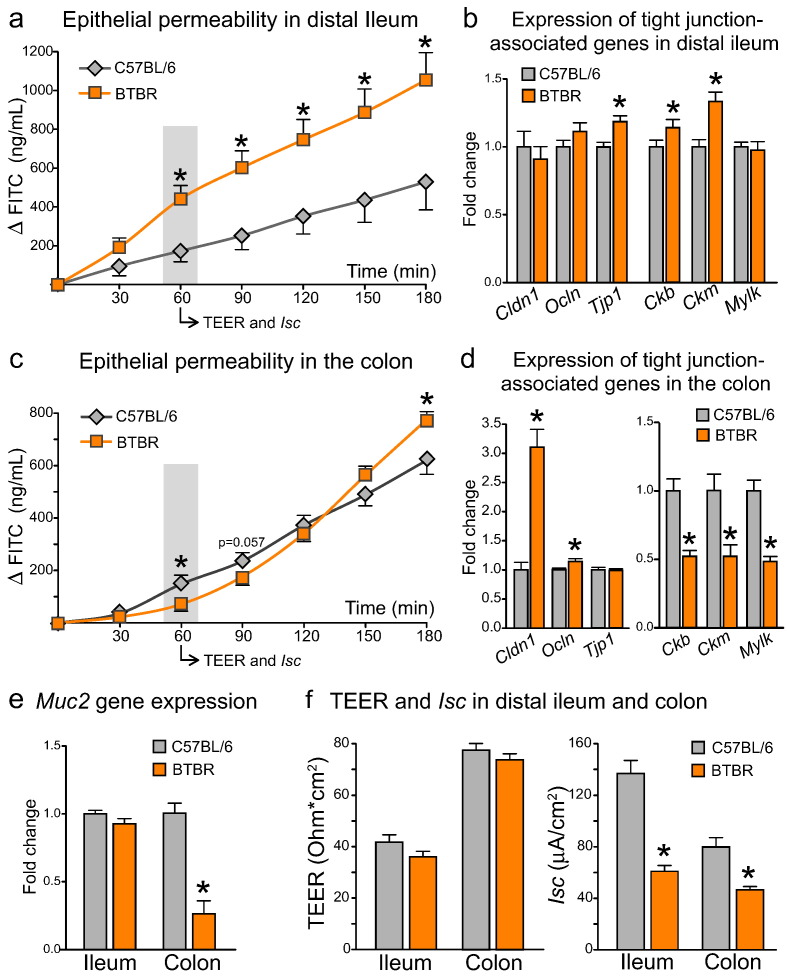
3.6. BTBR Mice Have Differential Changes in Epithelial Permeability in Small and Large Intestine
Along with altered bowel habits, ASD has been associated with impaired intestinal barrier function (D'Eufemia et al., 1996; de Magistris et al., 2010; Fiorentino et al., 2016). To characterize epithelial permeability in the BTBR intestine, we measured the efficacy of macromolecular diffusion across the epithelium ex vivo (4 kDa FITC flux). In the distal ileum, we observed a dramatic increase in epithelial permeability in BTBR mice (Fig. 4a). FITC flux was increased approximately two-fold at the 60 min time point and remained consistently elevated over the next two hours of the assay (p = 0.019 for the effect of genotype, F(1;14) = 7.090). Macromolecular permeability of intestinal epithelium is largely dependent on the proper function of tight junctions (TJs) (Zihni et al., 2016). Despite an increase in FITC flux, we failed to find substantial alterations in the gene expression levels of either TJ proteins (Cldn1, Ocln and Tjp1) or proteins regulating the assembly and energy supply to TJs (Ckb, Ckm, Mylk) (Glover et al., 2013; Shen et al., 2006) (Fig. 4b).
Fig. 4.
Differential changes to epithelial permeability and tight junction-associated gene expression in small and large intestine of BTBR mice (a) In distal ileum, BTBR mice showed a marked increase in FITC transepithelial permeability, with no substantial changes to either tight junction-associated genes (b) or mucin2 gene expression (e). (c) In the colon of BTBR mice, FITC permeability was increased in the end, but decreased in the beginning of the incubation period. The biphasic response of FITC flux was coupled with an up-regulation of tight junction protein genes, but a down-regulation of creatine kinase and mucin2 genes (d–e). (f) TEER was not changed, while Isc was substantially decreased in the BTBR intestine. *p < 0.05 for BTBR vs. C57BL/6 group. Statistical analysis: data are presented as mean + SEM, n = 10 in either group. FITC permeability in distal ileum: F(6;84) = 63.339, p < 0.0005 for the effect of Time, F(1;14) = 7.090, p = 0.019 for the effect of Genotype, F(6;84) = 7.177, p = 0.015 for the Time × Genotype interaction. FITC permeability in the colon: F(6;102) = 383.858, p < 0.0005 for the effect of Time, F(1;17) = 0.061, p = 0.808 for the effect of Genotype, F(6;102) = 9.198, p = 0.002 for the Time × Genotype interaction. Datasets were analyzed with mixed-design ANOVA with Genotype as an independent factor and Time as a repeated measure. Mean values in each time point were further compared between groups with unpaired t-test. t(14) = 1.582, p = 0.136 for TEER; t(14) = 6.862, p < 0.0005 for Isc; t(18) = 0.624, p = 0.541 for Cldn1, t(18) = − 1.429, p = 0.170 for Ocln, t(18) = − 2.768, p = 0.013 for Tjp1, t(15) = − 3.292, p = 0.005 for Ckb, t(16) = − 3.731, p = 0.002 for Ckm, t(15) = 0.217, p = 0.831 for Mylk, t(17) = 1.278, p = 0.218 for Muc2 gene expression in distal ileum, unpaired t-test. t(18) = 1.101, p = 0.285 for TEER; t(18) = 4.407, p = 0.001 for Isc; t(18) = − 6.319, p < 0.0005 for Cldn1, t(18) = − 2.273, p = 0.035 for Ocln, t(18) = 0.104, p = 0.918 for Tjp1, t(17) = 4.791, p < 0.0005 for Ckb, t(17) = 3.255, p = 0.005 for Ckm, t(17) = 5.751, p < 0.0005 for Mylk, t(17) = 6.261, p < 0.0005 for Muc2 gene expression in the colon, unpaired t-test.
Colonic tissue showed a very distinct profile of macromolecular permeability in BTBR mice (Fig. 4c). During the first 90 min of the assay, FITC flux was slightly decreased (p = 0.002 for the time × genotype interaction, F(6;102) = 9.198; p = 0.003 for 60 min and p = 0.057 for 90 min). However, during the following 90 min, FITC flux in BTBR samples substantially accelerated, overtaking the control group and reaching significantly higher levels by the end of the assay (p = 0.029 for 180 min). Gene expression analysis shed some light on the biphasic response of FITC permeability (Fig. 4d). On one hand, a marked increase in Cldn1 gene expression can explain a functionally “tighter” epithelium in the BTBR colon at baseline. On the other hand, a 50% down-regulation of creatine kinase Ckb and Ckm genes suggests a reduced efficacy of phosphocreatine metabolic shuttle turnover, which is critically important for the maintenance of energy supply to TJs (Glover et al., 2013). This would favor a rapid increase in macromolecular permeability in BTBR samples in ex vivo conditions, when tissue specimens are exposed to limited oxygen and substrate supply. Furthermore, these findings suggest that the BTBR colon might be less resilient to any metabolic stress in vivo, such as intestinal inflammation or hypoxia.
The epithelial mucus layer also helps to maintain the physical barrier between the gut and the lumen (Johansson et al., 2011). Interestingly, the expression of Muc2, a gene encoding mucin 2, which is the major constituent of mucus layer, was down-regulated in the large intestine of BTBR mice, but not in distal small intestine (Fig. 4e). Despite marked alterations in FITC permeability, only marginal changes in ion conductance (TEER) were observed in either ileum or colon samples of BTBR mice (Fig. 4f, left panel). Although at first glance counterintuitive, these findings are supported by the current model of junctional gate (Zihni et al., 2016), whereby ions and molecules use different pathways to diffuse through the tight junction. TEER and macromolecular permeability of the intestinal epithelium can thus be differentially regulated and even functionally decoupled (Balda et al., 1996).
3.7. Reduced Activity of Electrolyte Transport in the BTBR Intestine
We analyzed Isc values in BTBR and C57BL/6 intestine as a measure of electrolyte transport across the epithelium. In both small and large intestine, Isc was substantially decreased in BTBR mice (Fig. 4f, right panel). This suggests either a reduction in Na+ absorption from the gut lumen and/or a decrease in Cl− secretion into the lumen. Interestingly, we observed a down-regulation of Slc5a1 gene in the BTBR ileum (Fig. S2a). Slc5a1 encodes apical Na+/glucose co-transporter, which is known to enhance intestinal Isc in the presence of luminal glucose (Berglund et al., 2001; Turner et al., 1997). Activation of Cl− secretion in the colon is associated with water retention in the lumen and, when reaching pathological levels, diarrhea (Thiagarajah and Verkman, 2012). In this regard, a reduction in Cl− secretion in the BTBR colon would be consistent with decreased fecal water contents and delayed colonic transit (Fig. 2b–d).
3.8. Intestinal Permeability Correlates With the Abundance of Certain Bacterial Taxa
Gut bacteria and their metabolites can modulate intestinal barrier function (Kelly et al., 2015); therefore, we examined the associations between microbiota composition and intestinal macromolecular permeability (Table S3). Interestingly, here we observed a different correlation profile in comparison with the one found for intestinal transit and 5-HT metabolism. From the Firmicutes phylum, correlations with Blautia genus were largely lost. Instead, a strong negative association between the abundance of Ruminococcaceae Incertae Sedis bacteria and FITC permeability was observed. In Bacteroidetes, a strong positive correlation between FITC flux and S24–7 uncultured bacterium abundance was present. Many of these differences can be driven by a unique metabolic profile of each bacterial genus, including the production of different biologically active compounds, such as SCFAs or indoles (Kennedy et al., 2017; Sandhu et al., 2017).
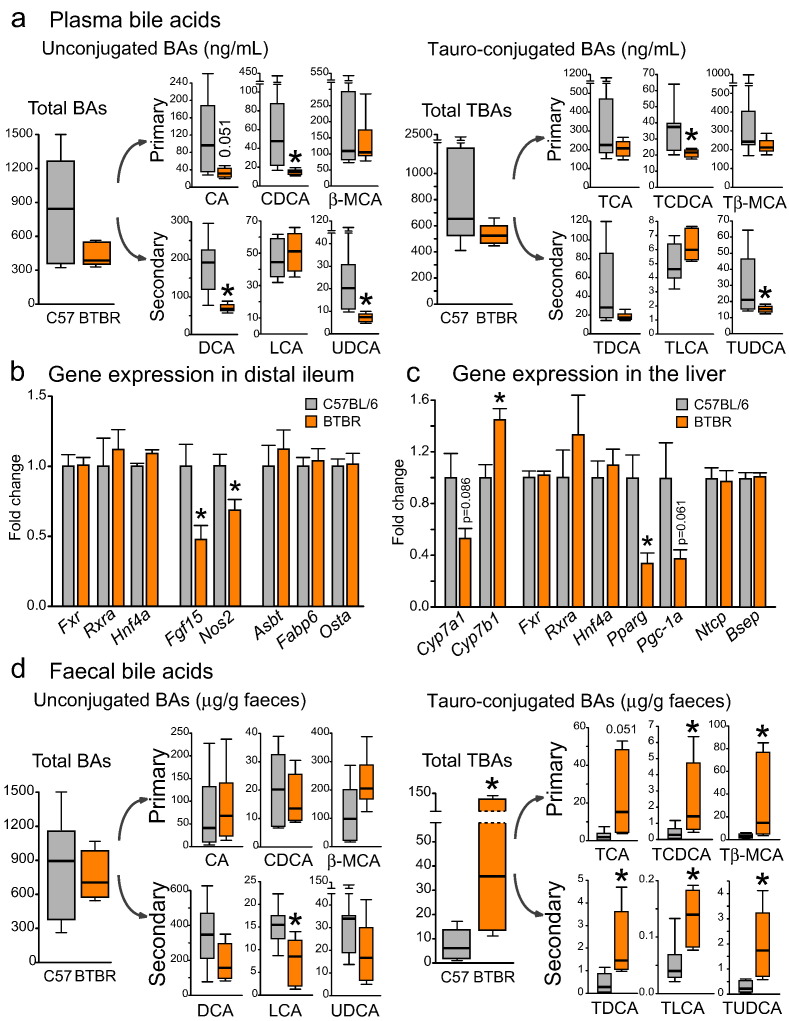
3.9. BTBR Mice Display Reduced Plasma Bile Acid Levels and Deficient Bile Acid Signaling in the Ileum
Epithelial permeability data (Fig. 4) clearly demonstrated functional differences between the colon and the ileum in BTBR mice. One of the unique features of the terminal small intestine is the active uptake of the bile from the intestinal lumen (de Aguiar Vallim et al., 2013). Primarily, bile acids (BAs) assist in the digestion of dietary lipids and regulate lipid metabolism. In the small intestine, they are also very important for the maintenance of epithelial barrier function (Gadaleta et al., 2011; Inagaki et al., 2006). Hence, we measured BAs in plasma and fecal matter of BTBR and C57BL/6 mice (Fig. 5 and Table S4). In plasma, the levels of tauro-conjugated and unconjugated BAs were overall decreased in BTBR mice (Fig. 5a). About 95% of the bile released into the gut is actively re-absorbed in distal ileum to ensure the complete enterohepatic circulation. Thus, a decrease in plasma BAs in BTBR mice could be attributed to a deficit in either BA synthesis or BA re-uptake. We did not find any changes in the gene expression of intestinal BA transporters (Asbt, Fabp6, Ostα) or liver BA transporters (Ntcp, Bsep) across the groups (Fig. 5b–c). These findings rule out the BA malabsorption in BTBR mice. Instead, there was a trend towards a reduction in the expression of Cyp7a1 gene, which encodes a rate-limiting enzyme in the classical pathway of BA synthesis in the liver (Fig. 5c). Interestingly, the expression of Cyp7b1 gene, which encodes a rate-limiting enzyme in the alternative pathway of BA synthesis, was up-regulated in BTBR mice. When absorbed in distal ileum, BAs activate the farnesoid X receptor (FXR) in enterocytes. The expression levels of Fxr gene, as well as its partners Rxrα and Hnf4α, were unchanged in the BTBR ileum (Fig. 5b). However, the downstream targets of this receptor, in particular Fgf15 and Nos2, were substantially down-regulated. Intestinal Fgf15 is a potent inhibitor of Cyp7a1 expression and BA synthesis (Kong et al., 2012). Hence, it is surprising that the suppression of the intestinal Fxr/Fgf15 signaling pathway failed to up-regulate Cyp7a1 gene expression in the liver of BTBR mice. On the other hand, Cyp7a1 expression can be activated by the nuclear receptors Pgc-1α and Ppar-γ (Duan et al., 2012; Shin et al., 2003). Similar to Cyp7a1, the expression levels of both Pgc-1α and Pparγ genes were decreased in the BTBR liver (Fig. 5c), suggesting that multiple nuclear receptors are implicated in the decreased BA synthesis observed in BTBR mice.
Fig. 5.
Altered bile acid signatures and deficient bacterial metabolism of bile moieties in BTBR mice (a) In plasma, a reduction in unconjugated and tauro-conjugated BA levels was seen in BTBR mice. (b–c) The gene expression levels of BA transporters were not altered in the distal ileum or liver of BTBR mice. In contrast, Cyp7a1 and Pparg genes in the liver, as well as Fgf15 and Nos2 genes in the ileum were down-regulated in BTBR mice. (d) In the fecal matter of BTBR mice, a dramatic increase in all tauro-conjugated BAs and a trend towards a reduction in unconjugated secondary BAs were observed. *p < 0.05 for BTBR vs. C57BL/6 group. Statistical analysis: data are presented as median with IQR and min/max values as error bars on (a) and (d), and as mean + SEM on (b) and (c). U(11) = 11.000, p = 0.181 for total BAs, U(11) = 7.000, p = 0.051 for CA, U(11) = 2.000, p = 0.005 for CDCA, U(11) = 19.000, p = 0.836 for β-MCA, U(10) = 1.500, p = 0.005 for DCA, U(11) = 25.000, p = 0.628 for LCA, U(11) = 0.000, p = 0.001 for UDCA, U(10) = 10.000, p = 0.268 for total TBAs, U(10) = 13.000, p = 0.530 for TCA, U(10) = 5.000, p = 0.048 for TCDCA, U(10) = 8.000, p = 0.149 for Tβ-MCA, U(10) = 10.000, p = 0.268 for TDCA, U(10) = 27.000, p = 0.149 for TLCA, U(10) = 2.000, p = 0.010 for TUDCA levels in plasma, n = 7 in C57BL/6 and n = 6 in BTBR group, Mann-Whitney test. U(11) = 16.000, p = 0.534 for total BAs, U(11) = 25.000, p = 0.628 for CA, U(11) = 17.000, p = 0.628 for CDCA, U(11) = 34.000, p = 0.073 for β-MCA, U(11) = 10.000, p = 0.138 for DCA, U(11) = 6.000, p = 0.035 for LCA, U(11) = 10.000, p = 0.138 for UDCA, U(11) = 39.000, p = 0.008 for total TBAs, U(11) = 35.000, p = 0.051 for TCA, U(11) = 37.000, p = 0.022 for TCDCA, U(11) = 38.000, p = 0.014 for Tβ-MCA, U(11) = 40.000, p = 0.005 for TDCA, U(11) = 39.000, p = 0.008 for TLCA, U(11) = 42.000, p = 0.001 for TUDCA levels in the fecal matter, n = 7 in C57BL/6 and n = 6 in BTBR group, Mann-Whitney test. t(18) = − 0.081, p = 0.936 for Fxr, t(18) = − 0.488, p = 0.632 for Rxra, t(18) = − 0.726, p = 0.477 for Hnf4a, t(17) = 2.776, p = 0.013 for Fgf15, t(18) = 2.807, p = 0.012 for Nos2, t(18) = − 0.102, p = 0.920 for Asbt, t(18) = − 0.373, p = 0.714 for Fabp6, t(18) = − 0.118, p = 0.907 for Osta gene expression in distal ileum, n = 10 in either group, unpaired t-test. t(11) = 1.887, p = 0.086 for Cyp7a1, t(12) = − 3.157, p = 0.008 for Cyp7b1, t(12) = − 0.274, p = 0.788 for Fxr, t(12) = − 0.905, p = 0.383 for Rxra, t(12) = − 0.525, p = 0.609 for Hnf4a, t(11) = 3.189, p = 0.009 for Pparg, t(12) = 2.190, p = 0.061 for Pgc-1a, t(12) = 0.167, p = 0.870 for Ntcp, t(12) = − 0.274, p = 0.788 for Bsep gene expression in the liver, n = 8 in C57BL/6 and n = 6 in BTBR group, unpaired t-test.
3.10. BTBR Mice Display a Deficit in the Microbial Conversion of Bile
Bile that is not re-absorbed in terminal small intestine undergoes further transformation by the intestinal microbiota (Joyce and Gahan, 2016). First, gut bacteria de-conjugate BAs from taurine and glycine. Second, unconjugated primary BAs (cholic acid (CA), chenodeoxycholic acid (CDCA) and muricholic acid (MCA) in mice) are processed into secondary BAs, such as deoxycholic acid (DCA), lithocholic acid (LCA) and ursodeoxycholic acid (UDCA). In the fecal matter of BTBR mice, we observed a remarkable increase in all conjugated BAs, for both primary and secondary conjugates (Fig. 5d, right panel). Further, we saw a significant reduction in secondary bile acid LCA, with a similar trend in DCA and UDCA (Fig. 5d, left panel). Moreover, the ratio between secondary and primary unconjugated BAs was significantly decreased in BTBR mice (Fig. S3). These data indicate that the BTBR microbiota has a reduced capacity to metabolize host bile.
3.11. Changes in the Gut Microbiota are Associated with a Robust Autistic-like Behavioral Phenotype
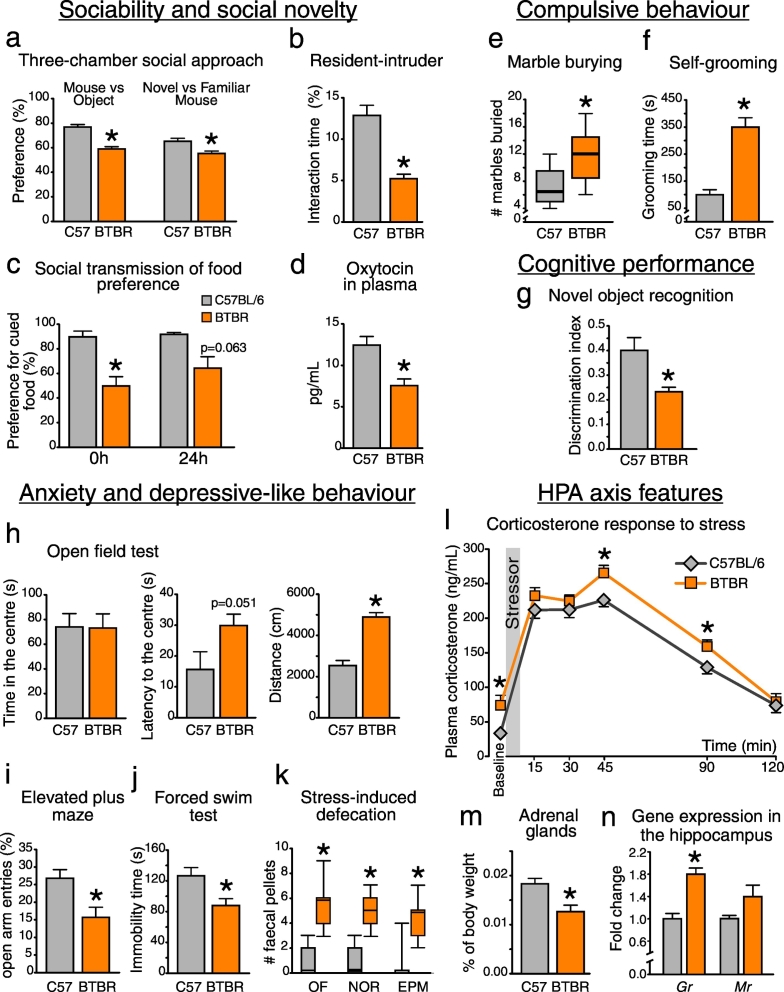
We characterized the ASD-relevant behavioral phenotype in BTBR mice (Fig. 6). In agreement with previous reports (Meyza et al., 2013), BTBR mice demonstrated impaired social interactions (a–c) and enhanced engagement in repetitive behaviors (e–f), as well as increased anxiety levels (h–k). Here we found decreased levels of social behavior-related hormone oxytocin in BTBR plasma (d). Further, BTBR mice displayed higher baseline levels of the stress hormone corticosterone, as well as elevated corticosterone values at the peak of the acute stress response and during the subsequent recovery phase (Fig. 6l, p < 0.05 for the effect of genotype, F(1;16) = 8.193). Despite this hyper-corticosterone phenotype, BTBR mice had smaller adrenal glands, and increased expression of the glucocorticoid receptor gene in the hippocampus (Fig. 6m–n). Interestingly, the expression of Cyp11a1, a gene encoding the first rate-limiting enzyme in the production of corticosterone, was substantially up-regulated in the distal ileum of BTBR mice (Fig. S2b). Taken on its own, this transcriptional shift does not necessarily implicate an increase in intestinal hormonal levels. However, if local production of corticosterone was elevated in the BTBR ileum, this could explain increased levels of circulating hormone (Fig. 6l) (Mukherji et al., 2013). This could also affect the GI physiology in BTBR mice including an up-regulation of Tdo2 gene in distal ileum (Fig. 2f, see also Discussion).
Fig. 6.
Autistic-like behavioral phenotype in BTBR mice (a–d) BTBR mice displayed impaired social interactions, as well as decreased plasma levels of oxytocin. (e–f) BTBR mice were largely engaged in compulsive behaviors. They showed impaired working memory performance (g), enhanced locomotor activity and anxiety-like behavior (h, i and k). (j) No evidence of depressive-like behavior was observed. (l) BTBR mice displayed elevated plasma corticosterone levels at baseline and when being exposed to an acute stressor (forced swim test). (m–n) BTBR mice had smaller adrenal glands and increased expression of glucocorticoid receptor gene in the hippocampus. *p < 0.05 for BTBR vs. C57BL/6 group. Statistical analysis: data are presented as median with IQR and min/max values as error bars on (e) and (k), and as mean + SEM on the rest, n = 10 in either group. t(18) = 8.153, p < 0.0005 for Mouse vs Object and t(17) = 3.707, p = 0.002 for Novel vs Familiar Mouse preference; t(18) = 5.678, p < 0.0005 for the RI interaction; t(13) = 3.485, p = 0.004 and t(13) = 2.029, p = 0.063 for the cued food preference at 0 h and 24 h time points, respectively; t(17) = 3.623, p = 0.002 for oxytocin levels; t(18) = − 6.303, p < 0.0005 for self-grooming time; t(18) = 3.040, p = 0.011 for the NOR discrimination index; t(18) = 0.059, p = 0.953 for the time in the center, t(18) = − 2.090, p = 0.051 for the latency to enter the center and t(18) = − 7.099, p < 0.0005 for the distance travelled in the OF test; t(17) = 2.947, p = 0.009 for open arm entries in the EPM test; t(18) = 2.785, p = 0.012 for the immobility time in the FST test; t(18) = 3.229, p = 0.005 for the adrenal glands weight; t(17) = − 5.480, p < 0.0005 for the Gr and t(17) = − 1.845, p = 0.094 for the Mr. gene expression, unpaired t-test. U(18) = 83.000, p = 0.011 for # of buried marbles; U(18) = 99.000, p < 0.0005, U(18) = 98.000, p < 0.0005 and U(18) = 83.000, p = 0.001 for # of fecal pellets in the OF, NOR and EPM tests, respectively, Mann-Whitney test. Plasma Corticosterone: F(5;80) = 209.091, p < 0.0005 for the effect of Time, F(1;16) = 8.193, p = 0.011 for the effect of Genotype, F(5;80) = 1.534, p = 0.221 for the Time × Genotype interaction. Datasets were analyzed with mixed-design ANOVA with Genotype as an independent factor and Time as a repeated measure. Mean values in each time point were further compared between groups with unpaired t-test.
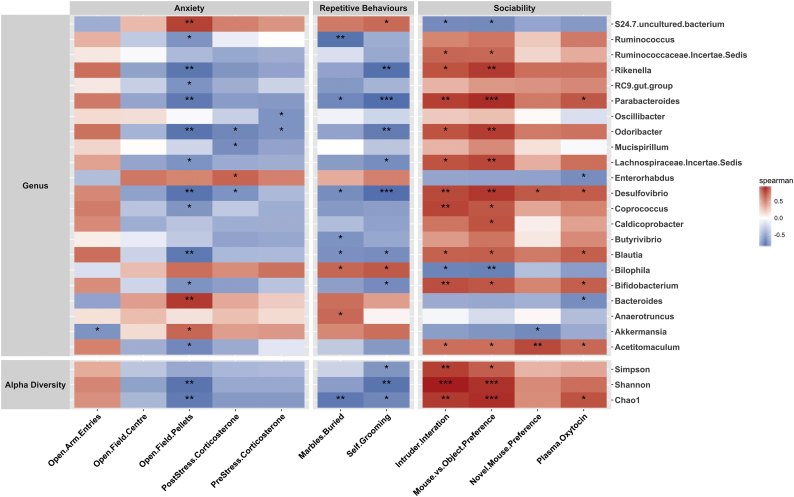
Correlation analysis between the relative abundance of bacterial taxa and behavioral scores showed an impressively strong association between the gut microbiota and the behavioral phenotype (Fig. 7). Rikenella, Parabacteroides, Odoribacter, Desulfovibrio, Blautia and Bifidobacterium species (all reduced in abundance in BTBR mice) were positively correlated with the social behavior scores and negatively correlated with the repetitive behaviors and anxiety phenotype. For the S24–7 family, Bilophila and Bacteroides species (all increased in abundance in BTBR mice), a negative association with sociability levels and a positive association with compulsive and anxiety-like behaviors was observed (Fig. 7). Given the tight interactions between the gut microbiota and GI functions (Table S3), it is perhaps not surprising that behavioral scores were also associated with changes in the intestinal serotonin levels and other GI parameters assessed (Fig. S4).
Fig. 7.
A strong association between behavioral scores and gut microbiota composition A correlation matrix showing positive (red) and negative (blue) associations between behavioral scores and the relative abundance of bacterial genera in BTBR and C57BL/6 mice. Significant correlations are marked with asterisks. *p < 0.05, **p < 0.01 and ***p < 0.001, Spearman's rank correlation analysis, Benjamini-Hochberg adjusted p values with a FDR of 0.05.
4. Discussion
The BTBR mouse strain is a very popular and useful animal model of ASD, with a robust autistic-like behavioral phenotype (Meyza et al., 2013). Here we show that social behavior deficits in BTBR mice are associated with marked GI distress, evidenced by changes in intestinal permeability, transit and the ENS morphology, as well as with altered microbiota composition. BTBR mice displayed profound changes in the cecal microbiota, with 18 out of 44 identified bacterial genera being differentially altered (Table S2). These included bacterial taxa that have previously been shown to be important for host health, and some of them associated with ASD: Akkermansia, Bifidobacterium, Blautia, Desulfovibrio, Bilofila, etc. (Kang et al., 2017; Vuong and Hsiao, 2017). A significant reduction in the relative abundance of Blautia and Bifidobacterium genera is of particular interest with respect to GI physiology in BTBR mice. Indeed, Blautia bacteria belong to the Clostridiales order, representatives of which have been shown to modulate 5-HT metabolism in the host by increasing the expression of Tph1 in the gut, activating 5-HT synthesis and accelerating GI motility (Yano et al., 2015). Furthermore, Blautia and Bifidobacterium species are involved in the transformation of host bile. Bifidobacterium, along with Lactobacillus and a few other taxa, express bile salt hydrolase enzymes, which carry out de-conjugation of BAs from taurine or glycine (Begley et al., 2006); this is the first step in bacterial conversion of bile (Joyce and Gahan, 2016). Of note, lactobacilli also tended to be decreased in the BTBR gut (Table S2). Unconjugated primary BAs are further transformed into secondary BAs as a result of 7α-dehydroxylation activity, which is present in some members of Clostridium cluster XIVa including Blautia species (Ridlon et al., 2013). Therefore, a substantial loss in Blautia and Bifidobacterium species in the BTBR microbiota strongly suggests that the efficacy of bacteria-mediated bile transformation, as well as the effect of bacteria on 5-HT production are impaired in the BTBR intestine.
The follow-up analysis of bile moieties and tryptophan metabolites in the BTBR gut confirmed the sequencing-based functional predictions. First, we observed a dramatic increase in all conjugated BAs, as well as a reduction in secondary BA pool in BTBR fecal matter (Fig. 5d and Fig. S3). These data indicate that both bile salt hydrolase and 7α-dehydroxylation activities are compromised in the BTBR microbiota. Secondly, we found a marked reduction in 5-HT bioavailability in BTBR small and large intestine due to a decrease in 5-HT synthesis and an increase in 5-HT breakdown (Fig. 2e–f). Strikingly, intestinal 5-HT levels and Tph1 gene expression were strongly and positively correlated with the relative abundance of Blautia genus (Fig. 2g, Table S3). Hence, the observed reduction in the abundance of Blautia species could underlie the deficit in 5-HT production in the BTBR gut.
Changes in either tryptophan metabolism or the microbial conversion of bile can have a tremendous impact on GI physiology. Secondary BAs, produced by gut bacteria, have a unique signaling profile in the GI tract as compared with primary bile moieties. For example, secondary BAs were shown to activate colonic motility by stimulating the release of intestinal 5-HT and CGRP (Alemi et al., 2013). Therefore, a reduction in secondary BA levels could contribute to the delayed colonic transit in BTBR mice (Fig. 2b–d). Further, tauro-conjugated β-MCA has recently been shown to be a potent natural antagonist of Fxr receptor in mice and to inhibit its downstream signaling (Sayin et al., 2013). Hence, an increase in Tβ-MCA intestinal levels can explain the observed down-regulation of Fgf15 gene expression in the BTBR ileum (Fig. 5b and d). In addition to the deficits in bacteria-mediated bile conversion, BTBR mice displayed a reduction in plasma BA levels, most likely due to a decrease in BA synthesis along the classical Cyp7a1-mediated pathway (Fig. 5a and c). This can have deleterious consequences for the intestine as well. BAs are toxic for many bacterial species; they also induce the expression of anti-microbial defense genes in the host (including iNOS, RegIIIγ) (Inagaki et al., 2006; Joyce et al., 2014). In fact, a reduction in the luminal BA levels has been associated with small intestine bacterial overgrowth, activation of inflammation, and subsequent damage to the epithelium (Inagaki et al., 2006; Lorenzo-Zúñiga et al., 2003). A down-regulation of iNos (Nos2) gene expression in the ileum of BTBR mice (Fig. 5b) suggests that anti-microbial defense can be compromised in the BTBR intestine, which in turn could contribute to the impairment of small intestine barrier function (Fig. 4a).
Similar to bile moieties, tryptophan metabolites are involved in the regulation of essential GI functions. 5-HT activates intestinal peristalsis and electrolyte secretion, supports the development of the ENS and amplifies inflammatory responses in the gut (Ghia et al., 2009; Heredia et al., 2013; Liu et al., 2009; Mawe and Hoffman, 2013). Hence, a decrease in 5-HT bioavailability can explain delayed intestinal transit and reduced activity of electrolyte transport seen in BTBR mice (Figs. 2a–d, 4f). Interestingly, metabolism of tryptophan along the kynurenine route also seems to be affected in BTBR mice. The expression levels of Ido1 and Tdo2 genes, which encode alternative rate-limiting enzymes in this pathway, were markedly altered in the BTBR intestine (Fig. 2e–f). Tdo2 gene expression is responsive to stress and is under a tight control of glucocorticoids (Green and Curzon, 1975). Strikingly, we found an increased expression of Cyp11a1 gene in the BTBR ileum (Fig. S2b), which could have resulted in elevated production of corticosterone and an up-regulation of Tdo2 locally in the BTBR ileum (Fig. 2f). The observed changes in intestinal Ido1 and Tdo2 gene expression are very unlikely to have a direct effect on the brain in BTBR mice. Indeed, > 90% of tryptophan is catabolized down to kynurenine in the liver, with liver Tdo2 activity determining the systemic levels of kynurenine, and the availability of tryptophan for the brain (Kennedy et al., 2017). To this end, neither the expression of liver Tdo2 nor the plasma levels of tryptophan was altered in BTBR mice (Fig. S5a–b). However, this will certainly affect the intestinal levels of kynurenine and its metabolites (kynurenic and quinolinic acids), and local effects on the BTBR gut physiology are thus very plausible. For instance, kynurenic acid is known to slow the intestinal transit by inhibiting the enteric NMDA receptors, as well as to dampen the inflammatory response in the gut (Érces et al., 2012; Kaszaki et al., 2008; Keszthelyi et al., 2009). Moreover, both 5-HT and kynurenine metabolites can alter the activity of intestinal vagal afferents, and hence, might directly affect vagal signaling to the brain (Slattery et al., 2006). Of note, recent evidence suggests altered 5-HT and kynurenine metabolism in ASD (Boccuto et al., 2013; Lim et al., 2015; Muller et al., 2016).
Altered tryptophan metabolism and inadequate bacterial conversion of bile are thus very likely to contribute to both intestinal dysmotility and impaired gut barrier function in BTBR mice. Maintenance of epithelial integrity is critically important not only for gut health, but also for brain health. The phenomenon of “leaky” gut has recently been extended beyond GI disorders, such as irritable bowel syndrome, to psychiatric diseases, such as depression (Kelly et al., 2015). Dysfunctional intestinal barrier in BTBR mice can permit the microbiota to induce low-grade inflammation in the gut. This shift towards a pro-inflammatory state, particularly in combination with an anxious phenotype (Fig. 6h–l), can in turn trigger the activation of neuroinflammatory responses in the brain (Rea et al., 2016; Wohleb et al., 2015) and exaggerate autistic-like behavioral deficits in BTBR mice. Indeed, the BTBR mouse has been shown to display a pro-inflammatory phenotype that is thought to contribute to the development of ASD-like behavior (Careaga et al., 2015; Heo et al., 2011; Onore et al., 2013). Interestingly, we found that deficits in social behavior in BTBR mice were accompanied by a decrease in plasma oxytocin levels (Fig. 6d). Oxytocin is one of the key hormones involved in supporting the salience of social interactions, and is known to be modulated by intestinal microbiota (Buffington et al., 2016; Varian et al., 2017).
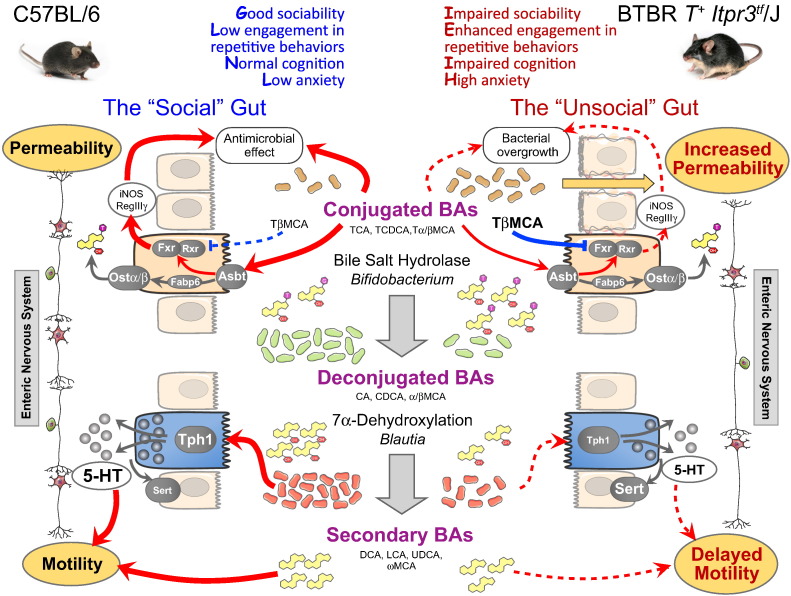
In summary, here we show that the BTBR mouse displays marked GI dysfunction (Fig. 8). Similar to human ASD, BTBR mice have “leaky” intestinal epithelium and delayed intestinal transit. We further identify deficient bacterial transformation of bile moieties and impaired tryptophan metabolism in the BTBR intestine as potential mechanisms underlying GI symptoms in these animals. Taking advantage of microbiota sequencing and physiological data, we associate these functional changes with a reduction in the relative abundance of Bifidobacterium and Blautia bacteria in the BTBR microbiota. These data offer specific targets in the gut microbiota for improving gastrointestinal and, perhaps, behavioral symptomatology in the BTBR mouse model of autism, and support the concept of microbiota-based interventions in ASD.
Fig. 8.
Putative mechanisms linking changes in the gut microbiota with GI malfunction in the BTBR mouse model of human ASD. In BTBR mice, autistic-like behavioral phenotype is associated with altered microbiota and marked GI distress: increased epithelial permeability (1) and delayed intestinal motility (2). (1) Reduced abundance of bile-metabolizing Bifidobacterium species increases the fraction of tauro-conjugated BAs (e.g. tauromuricholic acid (TβMCA)) in the GI tract. TβMCA inhibits Fxr signaling and thus supresses anti-microbial defense of the host (iNOS, RegIIIγ). We hypothesise that loss of Fxr signaling in BTBR mice causes small intestinal bacterial overgrowth and, as a result, an increase in epithelial permeability. This effect is further exaggerated by deficient synthesis of BAs, which normally prevent bacterial overgrowth. (2) Healthy intestinal transit relies on many factors, including the ENS, secondary BAs and 5-HT (all deficient in BTBR mice). Secondary BA levels are reduced; 5-HT bioavailability is decreased due to a lower production (down-regulation of Tph1) and a higher breakdown (up-regulation of Sert) rates. Lower abundance of clostridia bacteria, including Blautia species, potentially contributes to these changes. Further, the BTBR myeneric plexus shows reduced numbers of excitatory and inhibitory neurons. All this can cause delayed intestinal motility in BTBR mice.
Funding Sources & Conflicts of Interest
The APC Microbiome Institute is a research institute funded by Science Foundation Ireland (SFI) through the Irish Government's National Development Plan. J.F·C, T.G.D, C.S., S.A.J. and C.G.M.G. are supported by SFI (Grant Nos. SFI/12/RC/2273). S.A.J is also funded by SFI-EU 16/ERA-HDHL/3358. J.F·C, C.S. and T.G.D have research support from Mead Johnson, Cremo, 4D Pharma, Suntory Wellness, and Nutricia. J.F.C, C.S., T.G.D and G.C. have spoken at meetings sponsored by food and pharmaceutical companies. All other authors report no biomedical financial interests or potential conflicts of interest.
Author Contributions
Conceptualization, A.V.G, G.M., E.S., S.A.J., J.F.C.; Formal Analysis, A.V.G., A.B., S.A., D.K., V.P., K.M.; Investigation, A.V.G., S.A.J., G.M., A.B., E.S., S.A., I.F., D.K., A.M.P., V.P.; Writing – Original Draft, A.V.G.; Writing – Review & Editing, S.A.J., G.M., E.S., S.A., K.R., O.M., S.B., N.P.H., C.S., G.C., C.G.M.G., T.G.D., J.F.C.; Visualization, A.V.G., D.K., V.P.; Supervision, O.M., S.B., C.S., G.C., T.G.D., J.F.C.; Project Administration, A.V.G, G.M., K.R.; Funding Acquisition, C.S., S.A.J., C.G.M.G., T.G.D., J.F.C. S.A.J, G.M. and A.B. contributed equally to the study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2017.09.020.
Appendix A. Supplementary data
Supplementary material
References
- Alemi F., Poole D.P., Chiu J., Schoonjans K., Cattaruzza F., Grider J.R., Bunnett N.W., Corvera C.U. The receptor TGR5 mediates the prokinetic actions of intestinal bile acids and is required for normal defecation in mice. Gastroenterology. 2013;144:145–154. doi: 10.1053/j.gastro.2012.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelis M., Piccolo M., Vannini L., Siragusa S., Giacomo A., Serrazzanetti D.I., Cristofori F., Guerzoni M.E., Gobbetti M., Francavilla R. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS One. 2013:8. doi: 10.1371/journal.pone.0076993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Association, A.P . American Psychiatric Pub; 2013. Diagnostic and Statistical Manual of Mental Disorders (DSM-5®) [Google Scholar]
- Balda M.S., Whitney J.A., Flores C., González S., Cereijido M., Matter K. Functional dissociation of paracellular permeability and transepithelial electrical resistance and disruption of the apical-basolateral intramembrane diffusion barrier by expression of a mutant tight junction membrane protein. J. Cell Biol. 1996;134:1031–1049. doi: 10.1083/jcb.134.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley M., Hill C., Gahan C.G.M. Bile salt hydrolase activity in probiotics. Appl. Environ. Microbiol. 2006;72:1729–1738. doi: 10.1128/AEM.72.3.1729-1738.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund J.J., Riegler M., Zolotarevsky Y., Wenzl E., Turner J.R. Regulation of human jejunal transmucosal resistance and MLC phosphorylation by Na +-glucose cotransport. Am. J. Physiol. 2001;281:G1487–G1493. doi: 10.1152/ajpgi.2001.281.6.G1487. [DOI] [PubMed] [Google Scholar]
- Boccuto L., Chen C.-F., Pittman A.R., Skinner C.D., McCartney H.J., Jones K., Bochner B.R., Stevenson R.E., Schwartz C.E. Decreased tryptophan metabolism in patients with autism spectrum disorders. Mol. Autism. 2013;4:16. doi: 10.1186/2040-2392-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffington Shelly A., Di Prisco, Gonzalo V., Auchtung Thomas A., Ajami Nadim J., Petrosino Joseph F., Costa-Mattioli M. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell. 2016;165:1762–1775. doi: 10.1016/j.cell.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Careaga M., Schwartzer J., Ashwood P. Inflammatory profiles in the BTBR mouse: how relevant are they to autism spectrum disorders? Brain Behav. Immun. 2015;43:11–16. doi: 10.1016/j.bbi.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J., Borojevic R., Verdu E., Huizinga J., Ratcliffe E. Intestinal microbiota influence the early postnatal development of the enteric nervous system. Neurogastroenterol. Motil. 2014;26:98–107. doi: 10.1111/nmo.12236. [DOI] [PubMed] [Google Scholar]
- Coretti L., Cristiano C., Florio E., Scala G., Lama A., Keller S., Cuomo M., Russo R., Pero R., Paciello O. Sex-related alterations of gut microbiota composition in the BTBR mouse model of autism spectrum disorder. Sci Rep. 2017;7 doi: 10.1038/srep45356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Aguiar Vallim Thomas Q., Tarling Elizabeth J., Edwards Peter A. Pleiotropic roles of bile acids in metabolism. Cell Metab. 2013;17:657–669. doi: 10.1016/j.cmet.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magistris L., Familiari V., Pascotto A., Sapone A., Frolli A., Iardino P., Carteni M., De Rosa M., Francavilla R., Riegler G. Alterations of the intestinal barrier in patients with autism spectrum disorders and in their first-degree relatives. J. Pediatr. Gastroenterol. Nutr. 2010;51:418–424. doi: 10.1097/MPG.0b013e3181dcc4a5. [DOI] [PubMed] [Google Scholar]
- Desbonnet L., Clarke G., Shanahan F., Dinan T.G., Cryan J.F. Microbiota is essential for social development in the mouse. Mol. Psychiatry. 2014;19:146–148. doi: 10.1038/mp.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbonnet L., Clarke G., Traplin A., O'Sullivan O., Crispie F., Moloney R.D., Cotter P.D., Dinan T.G., Cryan J.F. Gut microbiota depletion from early adolescence in mice: implications for brain and behaviour. Brain Behav. Immun. 2015;48:165–173. doi: 10.1016/j.bbi.2015.04.004. [DOI] [PubMed] [Google Scholar]
- D'Eufemia P., Celli M., Finocchiaro R., Pacifico L., Viozzi L., Zaccagnini M., Cardi E., Giardini O. Abnormal intestinal permeability in children with autism. Acta Paediatr. 1996;85:1076–1079. doi: 10.1111/j.1651-2227.1996.tb14220.x. [DOI] [PubMed] [Google Scholar]
- Duan Y., Chen Y., Hu W., Li X., Yang X., Zhou X., Yin Z., Kong D., Yao Z., Hajjar D.P. Peroxisome proliferator-activated receptor γ activation by ligands and dephosphorylation induces proprotein convertase subtilisin kexin type 9 and low density lipoprotein receptor expression. J. Biol. Chem. 2012;287:23667–23677. doi: 10.1074/jbc.M112.350181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Érces D., Varga G., Fazekas B., Kovács T., Tőkés T., Tiszlavicz L., Fülöp F., Vécsei L., Boros M., Kaszaki J. N-methyl-d-aspartate receptor antagonist therapy suppresses colon motility and inflammatory activation six days after the onset of experimental colitis in rats. Eur. J. Pharmacol. 2012;691:225–234. doi: 10.1016/j.ejphar.2012.06.044. [DOI] [PubMed] [Google Scholar]
- Fiorentino M., Sapone A., Senger S., Camhi S.S., Kadzielski S.M., Buie T.M., Kelly D.L., Cascella N., Fasano A. Blood–brain barrier and intestinal epithelial barrier alterations in autism spectrum disorders. Mol. Autism. 2016;7:49. doi: 10.1186/s13229-016-0110-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furness J.B. Types of neurons in the enteric nervous system. J. Auton. Nerv. Syst. 2000;81:87–96. doi: 10.1016/s0165-1838(00)00127-2. [DOI] [PubMed] [Google Scholar]
- Gadaleta R.M., van Erpecum K.J., Oldenburg B., Willemsen E.C.L., Renooij W., Murzilli S., Klomp L.W.J., Siersema P.D., Schipper M.E.I., Danese S. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut. 2011;60(4):463–472. doi: 10.1136/gut.2010.212159. [DOI] [PubMed] [Google Scholar]
- Ghia J.E., Li N., Wang H., Collins M., Deng Y., El–Sharkawy R.T., Côté F., Mallet J., Khan W.I. Serotonin has a key role in pathogenesis of experimental colitis. Gastroenterology. 2009;137:1649–1660. doi: 10.1053/j.gastro.2009.08.041. [DOI] [PubMed] [Google Scholar]
- Gilbert J.A., Krajmalnik-Brown R., Porazinska D.L., Weiss S.J., Knight R. Toward effective probiotics for autism and other neurodevelopmental disorders. Cell. 2013;155:1446–1448. doi: 10.1016/j.cell.2013.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover L.E., Bowers B.E., Saeedi B., Ehrentraut S.F., Campbell E.L., Bayless A.J., Dobrinskikh E., Kendrick A.A., Kelly C.J., Burgess A. Control of creatine metabolism by HIF is an endogenous mechanism of barrier regulation in colitis. Proc. Natl. Acad. Sci. 2013;110:19820–19825. doi: 10.1073/pnas.1302840110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green A.R., Curzon G. Effects of hydrocortisone and immobilization on tryptophan metabolism in brain and liver of rats of different ages. Biochem. Pharmacol. 1975;24:713–716. doi: 10.1016/0006-2952(75)90248-8. [DOI] [PubMed] [Google Scholar]
- Heo Y., Zhang Y., Gao D., Miller V.M., Lawrence D.A. Aberrant immune responses in a mouse with behavioral disorders. PLoS One. 2011;6:e20912. doi: 10.1371/journal.pone.0020912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heredia D.J., Gershon M.D., Koh S.D., Corrigan R.D., Okamoto T., Smith T.K. Important role of mucosal serotonin in colonic propulsion and peristaltic reflexes: in vitro analyses in mice lacking tryptophan hydroxylase 1. J. Physiol. 2013;591:5939–5957. doi: 10.1113/jphysiol.2013.256230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao E.Y., McBride S.W., Hsien S., Sharon G., Hyde E.R., McCue T., Codelli J.A., Chow J., Reisman S.E., Petrosino J.F. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopment disorders. Cell. 2013:155. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki T., Moschetta A., Lee Y.-K., Peng L., Zhao G., Downes M., Yu R.T., Shelton J.M., Richardson J.A., Repa J.J. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc. Natl. Acad. Sci. U. S. A. 2006;103:3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M.E.V., Larsson J.M.H., Hansson G.C. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host–microbial interactions. Proc. Natl. Acad. Sci. 2011;108:4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce S.A., Gahan C.G.M. Bile acid modifications at the microbe-host interface: potential for nutraceutical and pharmaceutical interventions in host health. Annu. Rev. Food Sci. Technol. 2016;7:313–333. doi: 10.1146/annurev-food-041715-033159. [DOI] [PubMed] [Google Scholar]
- Joyce S.A., MacSharry J., Casey P.G., Kinsella M., Murphy E.F., Shanahan F., Hill C., Gahan C.G.M. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc. Natl. Acad. Sci. 2014;111:7421–7426. doi: 10.1073/pnas.1323599111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D.W., Park J.G., Ilhan Z.E., Wallstrom G., Labaer J., Adams J.B., Krajmalnik-Brown R. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS One. 2013:8. doi: 10.1371/journal.pone.0068322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D.-W., Adams J.B., Gregory A.C., Borody T., Chittick L., Fasano A., Khoruts A., Geis E., Maldonado J., McDonough-Means S. Microbiota Transfer Therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open-label study. Microbiome. 2017;5:10. doi: 10.1186/s40168-016-0225-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaszaki J., Palásthy Z., érczes D., Rácz A., Torday C., Varga G., Vécsei L., Boros M. Kynurenic acid inhibits intestinal hypermotility and xanthine oxidase activity during experimental colon obstruction in dogs. Neurogastroenterol. Motil. 2008;20:53–62. doi: 10.1111/j.1365-2982.2007.00989.x. [DOI] [PubMed] [Google Scholar]
- Kelly J.R., Kennedy P.J., Cryan J.F., Dinan T.G., Clarke G., Hyland N.P. Breaking down the barriers: the gut microbiome, intestinal permeability and stress-related psychiatric disorders. Front. Cell. Neurosci. 2015;9:392. doi: 10.3389/fncel.2015.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy P.J., Cryan J.F., Dinan T.G., Clarke G. Kynurenine pathway metabolism and the microbiota-gut-brain axis. Neuropharmacology. 2017;112(Part B):399–412. doi: 10.1016/j.neuropharm.2016.07.002. [DOI] [PubMed] [Google Scholar]
- Keszthelyi D., Troost F.J., Masclee A.A.M. Understanding the role of tryptophan and serotonin metabolism in gastrointestinal function. Neurogastroenterol. Motil. 2009;21:1239–1249. doi: 10.1111/j.1365-2982.2009.01370.x. [DOI] [PubMed] [Google Scholar]
- Kim J.-W., Choi C.S., Kim K.C., Park J.H., Seung H., Joo S.H., Yang S.M., Shin C.Y., Park S.H. Gastrointestinal tract abnormalities induced by prenatal valproic acid exposure in rat offspring. Toxicol. Res. 2013;29:173–179. doi: 10.5487/TR.2013.29.3.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein M.S., Newell C., Bomhof M.R., Reimer R.A., Hittel D.S., Rho J.M., Vogel H.J., Shearer J. Metabolomic modeling to monitor host responsiveness to gut microbiota manipulation in the BTBRT + tf/j mouse. J. Proteome Res. 2016;15:1143–1150. doi: 10.1021/acs.jproteome.5b01025. [DOI] [PubMed] [Google Scholar]
- Kong B., Wang L., Chiang J.Y.L., Zhang Y., Klaassen C.D., Guo G.L. Mechanism of tissue-specific farnesoid X receptor in suppressing the expression of genes in bile-acid synthesis in mice. Hepatology. 2012;56:1034–1043. doi: 10.1002/hep.25740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H., Sharma B. Minocycline ameliorates prenatal valproic acid induced autistic behaviour, biochemistry and blood brain barrier impairments in rats. Brain Res. 2016;1630:83–97. doi: 10.1016/j.brainres.2015.10.052. [DOI] [PubMed] [Google Scholar]
- Lim C.K., Essa M.M., de Paula Martins R., Lovejoy D.B., Bilgin A.A., Waly M.I., Al-Farsi Y.M., Al-Sharbati M., Al-Shaffae M.A., Guillemin G.J. Altered kynurenine pathway metabolism in autism: implication for immune-induced glutamatergic activity. Autism Res. 2015 doi: 10.1002/aur.1565. [DOI] [PubMed] [Google Scholar]
- Liu M.-T., Kuan Y.-H., Wang J., Hen R., Gershon M.D. 5-HT4 receptor-mediated neuroprotection and neurogenesis in the enteric nervous system of adult mice. J. Neurosci. 2009;29:9683–9699. doi: 10.1523/JNEUROSCI.1145-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo-Zúñiga V., Bartolí R., Planas R., Hofmann A.F., Viñado B., Hagey L.R., Hernández J.M., Mañé J., Alvarez M.A., Ausina V. Oral bile acids reduce bacterial overgrowth, bacterial translocation, and endotoxemia in cirrhotic rats. Hepatology. 2003;37:551–557. doi: 10.1053/jhep.2003.50116. [DOI] [PubMed] [Google Scholar]
- Luna R.A., Oezguen N., Balderas M., Venkatachalam A., Runge J.K., Versalovic J., Veenstra-VanderWeele J., Anderson G.M., Savidge T., Williams K.C. Distinct microbiome-neuroimmune signatures correlate with functional abdominal pain in children with autism spectrum disorder. Cell. Mol. Gastroenterol. Hepatol. 2017;3(2):218–230. doi: 10.1016/j.jcmgh.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyall K., Croen L., Daniels J., Fallin M.D., Ladd-Acosta C., Lee B.K., Park B.Y., Snyder N.W., Schendel D., Volk H. The Changing epidemiology of autism spectrum disorders. Annu. Rev. Public Health. 2017;38 doi: 10.1146/annurev-publhealth-031816-044318. (null) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawe G.M., Hoffman J.M. Serotonin signalling in the gut—functions, dysfunctions and therapeutic targets. Nat. Rev. Gastroenterol. Hepatol. 2013;10:473–486. doi: 10.1038/nrgastro.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElhanon B.O., McCracken C., Karpen S., Sharp W.G. Gastrointestinal symptoms in autism spectrum disorder: a meta-analysis. Pediatrics. 2014;133:872–883. doi: 10.1542/peds.2013-3995. [DOI] [PubMed] [Google Scholar]
- Meyza K.Z., Blanchard D.C. The BTBR mouse model of idiopathic autism – Current view on mechanisms. Neurosci. Biobehav. Rev. 2017;76:99–110. doi: 10.1016/j.neubiorev.2016.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyza K.Z., Defensor E.B., Jensen A.L., Corley M.J., Pearson B.L., Pobbe R.L.H., Bolivar V.J., Blanchard D.C., Blanchard R.J. The BTBR T + tf/J mouse model for autism spectrum disorders–in search of biomarkers. Behav. Brain Res. 2013;251:25–34. doi: 10.1016/j.bbr.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy S.S., Nadler J.J., Young N.B., Perez A., Holloway L.P., Barbaro R.P., Barbaro J.R., Wilson L.M., Threadgill D.W., Lauder J.M. Mouse behavioral tasks relevant to autism: phenotypes of 10 inbred strains. Behav. Brain Res. 2007;176:4–20. doi: 10.1016/j.bbr.2006.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji A., Kobiita A., Ye T., Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153:812–827. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- Muller C.L., Anacker A.M.J., Veenstra-VanderWeele J. The serotonin system in autism spectrum disorder: from biomarker to animal models. Neuroscience. 2016;321:24–41. doi: 10.1016/j.neuroscience.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld K.M., Kang N., Bienenstock J., Foster J.A. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol. Motil. 2011;23:255. doi: 10.1111/j.1365-2982.2010.01620.x. (e119) [DOI] [PubMed] [Google Scholar]
- Newell C., Bomhof M.R., Reimer R.A., Hittel D.S., Rho J.M., Shearer J. Ketogenic diet modifies the gut microbiota in a murine model of autism spectrum disorder. Mol. Autism. 2016;7:37. doi: 10.1186/s13229-016-0099-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata Y., Pachnis V. The effect of microbiota and the immune system on the development and organization of the enteric nervous system. Gastroenterology. 2016;151:836–844. doi: 10.1053/j.gastro.2016.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onore C.E., Careaga M., Babineau B.A., Schwartzer J.J., Berman R.F., Ashwood P. Inflammatory macrophage phenotype in BTBR T + tf/J mice. Front. Neurosci. 2013;7:158. doi: 10.3389/fnins.2013.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea K., Dinan T.G., Cryan J.F. The microbiome: A key regulator of stress and neuroinflammation. Neurobiol. Stress. 2016;4:23–33. doi: 10.1016/j.ynstr.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon J.M., Alves J.M., Hylemon P.B., Bajaj J.S. Cirrhosis, bile acids and gut microbiota. Gut Microbes. 2013;4:382–387. doi: 10.4161/gmic.25723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskin D.N., Svedova J., Cote J.L., Sandau U., Rho J.M., Kawamura M., Jr., Boison D., Masino S.A. Ketogenic diet improves core symptoms of autism in BTBR mice. PLoS One. 2013;8:e65021. doi: 10.1371/journal.pone.0065021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu K.V., Sherwin E., Schellekens H., Stanton C., Dinan T.G., Cryan J.F. Feeding the microbiota-gut-brain axis: diet, microbiome, and neuropsychiatry. Transl. Res. 2017;179:223–244. doi: 10.1016/j.trsl.2016.10.002. [DOI] [PubMed] [Google Scholar]
- Sayin Sama I., Wahlström A., Felin J., Jäntti S., Marschall H.-U., Bamberg K., Angelin B., Hyötyläinen T., Orešič M., Bäckhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Shen L., Black E.D., Witkowski E.D., Lencer W.I., Guerriero V., Schneeberger E.E., Turner J.R. Myosin light chain phosphorylation regulates barrier function by remodeling tight junction structure. J. Cell Sci. 2006;119:2095–2106. doi: 10.1242/jcs.02915. [DOI] [PubMed] [Google Scholar]
- Sherwin E., Dinan T.G., Cryan J.F. Recent developments in understanding the role of the gut microbiota in brain health and disease. Ann. N. Y. Acad. Sci. 2017:1–21. doi: 10.1111/nyas.13416. [DOI] [PubMed] [Google Scholar]
- Shin D.-J., Campos J.A., Gil G., Osborne T.F. PGC-1α activates CYP7A1 and bile acid biosynthesis. J. Biol. Chem. 2003;278:50047–50052. doi: 10.1074/jbc.M309736200. [DOI] [PubMed] [Google Scholar]
- Slattery J.A., Page A.J., Dorian C.L., Brierley S.M., Blackshaw L.A. Potentiation of mouse vagal afferent mechanosensitivity by ionotropic and metabotropic glutamate receptors. J. Physiol. 2006;577:295–306. doi: 10.1113/jphysiol.2006.117762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F., Backhed F. The gut microbiota - masters of host development and physiology. Nat. Rev. Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- Thiagarajah J.R., Verkman A.S. CFTR inhibitors for treating diarrheal disease. Clin. Pharmacol. Ther. 2012;92:287–290. doi: 10.1038/clpt.2012.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomova A., Husarova V., Lakatosova S., Bakos J., Vlkova B., Babinska K., Ostatnikova D. Gastrointestinal microbiota in children with autism in Slovakia. Physiol. Behav. 2015;138 doi: 10.1016/j.physbeh.2014.10.033. [DOI] [PubMed] [Google Scholar]
- Turner J.R., Rill B.K., Carlson S.L., Carnes D., Kerner R., Mrsny R.J., Madara J.L. Physiological regulation of epithelial tight junctions is associated with myosin light-chain phosphorylation. Am. J. Phys. Cell Physiol. 1997;273:C1378–C1385. doi: 10.1152/ajpcell.1997.273.4.C1378. [DOI] [PubMed] [Google Scholar]
- Varian B.J., Poutahidis T., DiBenedictis B.T., Levkovich T., Ibrahim Y., Didyk E., Shikhman L., Cheung H.K., Hardas A., Ricciardi C.E. Microbial lysate upregulates host oxytocin. Brain Behav. Immun. 2017;61:36–49. doi: 10.1016/j.bbi.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong H.E., Hsiao E.Y. Emerging roles for the gut microbiome in autism spectrum disorder. Biol. Psychiatry. 2017;81:411–423. doi: 10.1016/j.biopsych.2016.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohleb E.S., McKim D.B., Sheridan J.F., Godbout J.P. Monocyte trafficking to the brain with stress and inflammation: a novel axis of immune-to-brain communication that influences mood and behavior. Neuroinflam. Behav. 2015:34. doi: 10.3389/fnins.2014.00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano Jessica M., Yu K., Donaldson Gregory P., Shastri Gauri G., Ann P., Ma L., Nagler Cathryn R., Ismagilov Rustem F., Mazmanian Sarkis K., Hsiao Elaine Y. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zihni C., Mills C., Matter K., Balda M.S. Tight junctions: from simple barriers to multifunctional molecular gates. Nat. Rev. Mol. Cell Biol. 2016;17:564–580. doi: 10.1038/nrm.2016.80. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material