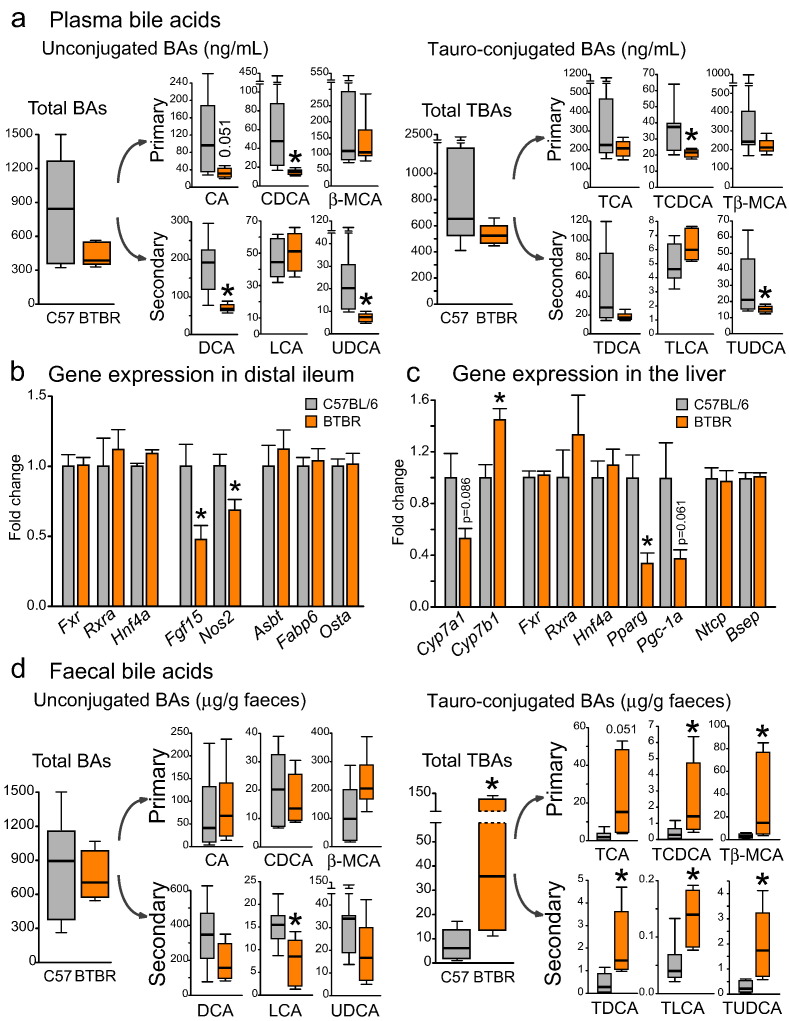
Fig. 5.
Altered bile acid signatures and deficient bacterial metabolism of bile moieties in BTBR mice (a) In plasma, a reduction in unconjugated and tauro-conjugated BA levels was seen in BTBR mice. (b–c) The gene expression levels of BA transporters were not altered in the distal ileum or liver of BTBR mice. In contrast, Cyp7a1 and Pparg genes in the liver, as well as Fgf15 and Nos2 genes in the ileum were down-regulated in BTBR mice. (d) In the fecal matter of BTBR mice, a dramatic increase in all tauro-conjugated BAs and a trend towards a reduction in unconjugated secondary BAs were observed. *p < 0.05 for BTBR vs. C57BL/6 group. Statistical analysis: data are presented as median with IQR and min/max values as error bars on (a) and (d), and as mean + SEM on (b) and (c). U(11) = 11.000, p = 0.181 for total BAs, U(11) = 7.000, p = 0.051 for CA, U(11) = 2.000, p = 0.005 for CDCA, U(11) = 19.000, p = 0.836 for β-MCA, U(10) = 1.500, p = 0.005 for DCA, U(11) = 25.000, p = 0.628 for LCA, U(11) = 0.000, p = 0.001 for UDCA, U(10) = 10.000, p = 0.268 for total TBAs, U(10) = 13.000, p = 0.530 for TCA, U(10) = 5.000, p = 0.048 for TCDCA, U(10) = 8.000, p = 0.149 for Tβ-MCA, U(10) = 10.000, p = 0.268 for TDCA, U(10) = 27.000, p = 0.149 for TLCA, U(10) = 2.000, p = 0.010 for TUDCA levels in plasma, n = 7 in C57BL/6 and n = 6 in BTBR group, Mann-Whitney test. U(11) = 16.000, p = 0.534 for total BAs, U(11) = 25.000, p = 0.628 for CA, U(11) = 17.000, p = 0.628 for CDCA, U(11) = 34.000, p = 0.073 for β-MCA, U(11) = 10.000, p = 0.138 for DCA, U(11) = 6.000, p = 0.035 for LCA, U(11) = 10.000, p = 0.138 for UDCA, U(11) = 39.000, p = 0.008 for total TBAs, U(11) = 35.000, p = 0.051 for TCA, U(11) = 37.000, p = 0.022 for TCDCA, U(11) = 38.000, p = 0.014 for Tβ-MCA, U(11) = 40.000, p = 0.005 for TDCA, U(11) = 39.000, p = 0.008 for TLCA, U(11) = 42.000, p = 0.001 for TUDCA levels in the fecal matter, n = 7 in C57BL/6 and n = 6 in BTBR group, Mann-Whitney test. t(18) = − 0.081, p = 0.936 for Fxr, t(18) = − 0.488, p = 0.632 for Rxra, t(18) = − 0.726, p = 0.477 for Hnf4a, t(17) = 2.776, p = 0.013 for Fgf15, t(18) = 2.807, p = 0.012 for Nos2, t(18) = − 0.102, p = 0.920 for Asbt, t(18) = − 0.373, p = 0.714 for Fabp6, t(18) = − 0.118, p = 0.907 for Osta gene expression in distal ileum, n = 10 in either group, unpaired t-test. t(11) = 1.887, p = 0.086 for Cyp7a1, t(12) = − 3.157, p = 0.008 for Cyp7b1, t(12) = − 0.274, p = 0.788 for Fxr, t(12) = − 0.905, p = 0.383 for Rxra, t(12) = − 0.525, p = 0.609 for Hnf4a, t(11) = 3.189, p = 0.009 for Pparg, t(12) = 2.190, p = 0.061 for Pgc-1a, t(12) = 0.167, p = 0.870 for Ntcp, t(12) = − 0.274, p = 0.788 for Bsep gene expression in the liver, n = 8 in C57BL/6 and n = 6 in BTBR group, unpaired t-test.