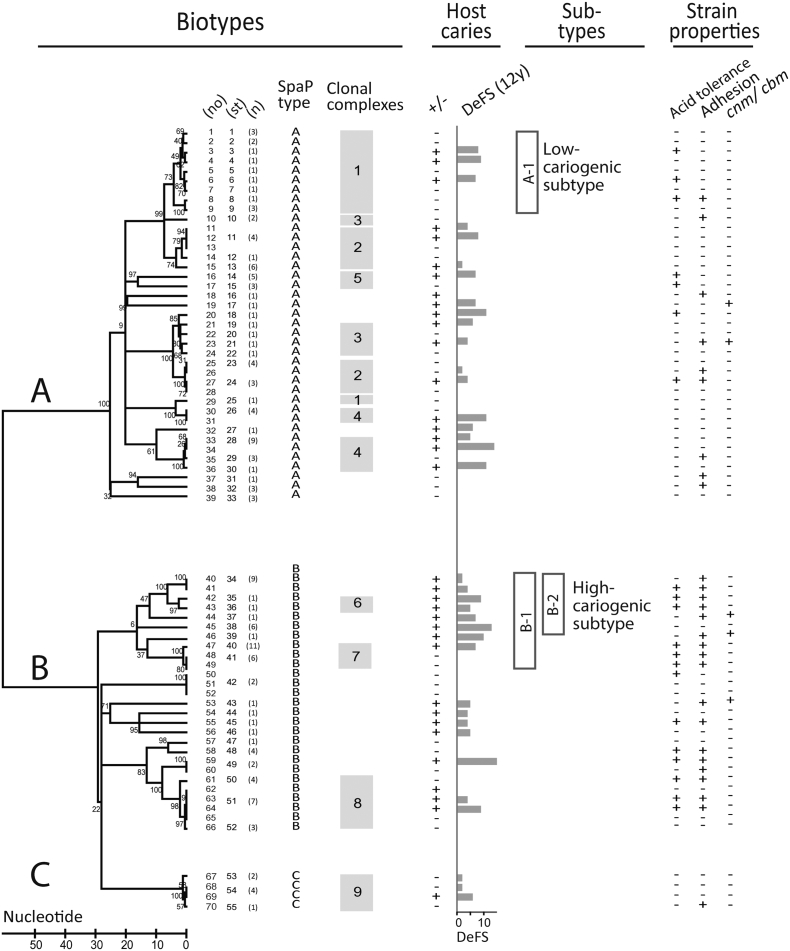
Fig. 2.
S. mutans biotypes A, B and C with corresponding SpaP adhesin subtypes of high (B-1) and low (A-1) cariogenicity. Clustering of S. mutans isolates from 35 caries (+) or 35 caries-free (−) extremes of infected children with caries status (+, − or DeFS at 12 years of age) based on neighbor joining and clonal complex analyses of spaP and housekeeping gene sequences. The isolates grouped both into biotypes A, B, and C with distinct SpaP A, B, and C adhesin types and clonal complexes C1–C9, and into genetically related subtypes that coincided with high caries (i.e. B-1, B-2 and C6, C7) or low caries (i.e. A-1 and C1) cases. The high- and low-cariogenic types also differed in acid tolerance and adhesion to DMBT1 and saliva. The clonal complexes share at least six identical alleles with at least one member in the group, all reflecting housekeeping alleles except for C8 which also shared the same spaP B allele. A total of 55 sequence types (ST) occurred among the 70 children. Numbers in parentheses mark STs shared in 15 children or identical STs in 25 children from which 74 additional strains were analyzed (mean 3, range 2 to 8 isolates/child).