Abstract
The selective translation of maternal mRNAs encoding cell-fate determinants drives the earliest decisions of embryogenesis that establish the vertebrate body plan. This chapter will discuss studies in Xenopus laevis that provide insights into mechanisms underlying this translational control. Xenopus has been a powerful model organism for many discoveries relevant to the translational control of maternal mRNAs because of the large size of its oocytes and eggs that allow for microinjection of molecules and the relative ease of manipulating the oocyte to egg transition (maturation) and fertilization in culture. Consequently, many key studies have focused on the expression of maternal mRNAs during the oocyte to egg transition (the meiotic cell cycle) and the rapid cell divisions immediately following fertilization. This research has made seminal contributions to our understanding of translational regulatory mechanisms, but while some of the mRNAs under consideration at these stages encode cell-fate determinants, many encode cell cycle regulatory proteins that drive these early cell cycles. In contrast, while maternal mRNAs encoding key developmental (i.e., cell-fate) regulators that function after the first cleavage stages may exploit aspects of these foundational mechanisms, studies reveal that these mRNAs must also rely on distinct and, as of yet, incompletely understood mechanisms. These findings are logical because the functions of such developmental regulatory proteins have requirements distinct from cell cycle regulators, including becoming relevant only after fertilization and then only in specific cells of the embryo. Indeed, key maternal cell-fate determinants must be made available in exquisitely precise amounts (usually low), only at specific times and in specific cells during embryogenesis. To provide an appreciation for the regulation of maternal cell-fate determinant expression, an overview of the maternal phase of Xenopus embryogenesis will be presented. This section will be followed by a review of translational mechanisms operating in oocytes, eggs, and early cleavage-stage embryos and conclude with a discussion of how the regulation of key maternal cell-fate determinants at the level of translation functions in Xenopus embryogenesis. A key theme is that the molecular asymmetries critical for forming the body axes are established and further elaborated upon by the selective temporal and spatial regulation of maternal mRNA translation.
Keywords: Xenopus, Maternal mRNA, Regulated translation, Embryonic asymmetry
2.1 Introduction
2.1.1 Oogenesis and Oocyte Maturation: Maternal mRNAs Set the Stage
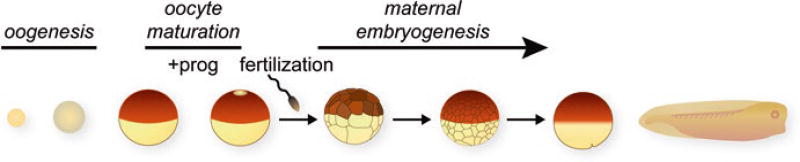
A significant amount of the early development of Xenopus is deterministic, meaning that embryonic cells destined to specific cell fates accumulate distinct proteins called maternal determinants (Heasman 2006a; White and Heasman 2008). Maternal determinants, regulatory proteins that function in cell-cell signaling, translational control, mRNA processing, chromatin remodeling, and other processes that can alter cell fates are translated from stored maternal mRNAs that are accumulated in eggs prior to fertilization during oogenesis (Fig. 2.1).
Fig. 2.1.
Maternal stages of Xenopus development. Summary diagram of oogenesis, oocyte maturation, fertilization, and cleavage stages of embryogenesis. The maternal period of embryonic development begins at fertilization and continues until the maternal mRNAs and proteins are eliminated and replaced by zygotic products during the maternal to zygotic transition
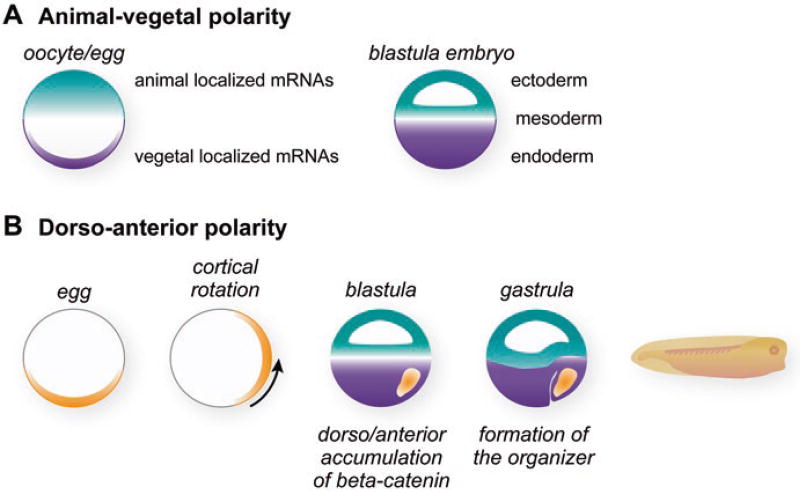
Oocytes form and grow over a period of months to develop into fully mature stage 6 oocytes with a distinctive darkly pigmented animal hemisphere and a lightly pigmented vegetal hemisphere (Fig. 2.1). These pigmentation differences mark the animal-vegetal axis of the oocyte and are carried over into eggs during oocyte maturation and then passed on to embryos after fertilization. The animal and vegetal hemispheres are a striking visual manifestation of the elaborate molecular events that create animal-vegetal distributions of macromolecules that function in cell-fate decisions, particularly localized mRNAs that encode cell-fate determinants (Medioni et al. 2012; King et al. 2005; Houston 2013) (Fig. 2.2a).
Fig. 2.2.
Formation of asymmetries and specific cell types during Xenopus early development. (a) Animal-vegetal polarity is established during oogenesis by the partitioning of molecules to animal and vegetal hemispheres. These partitioned molecules are exemplified by specific mRNAs that become localized during oogenesis to vegetal cortex and the animal hemisphere (Medioni et al. 2012; King et al. 2005; Houston 2013). After fertilization the cells of the embryo inherit these molecular asymmetries when they contribute to the formation of the ectoderm, the mesoderm, and the endoderm germ layers. (b) After fertilization asymmetry in the dorso/anterior to ventral/posterior dimension is established as a result of cortical rotation (Gerhart et al. 1989; Houston 2012). Wnt11 is translocated from the vegetal pole of the egg during cortical rotation to create the embryonic asymmetry in the dorso/anterior to ventral/posterior dimension (Schroeder et al. 1999). Wnt11 activates Wnt signaling to direct the accumulation of beta-catenin protein. The high levels of beta-catenin cause the dorso/anterior cells to induce and give rise to the organizer (Heasman et al. 1994). At the gastrula stage, the organizer produces extracellular signals that influence and pattern the adjacent cells of each germ layer (Gerhart et al. 1991; Harland and Gerhart 1997)
Growing Xenopus oocytes are transcriptionally active and accumulate mRNAs as they develop (oogenesis stages 1–6). Oocytes are thus accumulating the maternal mRNA population required to drive the later stages of oogenesis, the initial cell divisions following fertilization, and the subsequent maternal stages of embryonic development. Some maternal mRNAs translated in oocytes generate proteins used by oocytes and eggs, while others are stored in translationally inactive states until later in maternal embryogenesis when their encoded proteins are needed. These stored mRNAs include a specialized cohort that are transported and anchored to the vegetal cortex of the oocyte (Medioni et al. 2012; King et al. 2005; Houston 2013) and another group that are transported and concentrated in the animal hemisphere (Fig. 2.2a). These localized mRNAs encode proteins that help establish animal and vegetal cell identity that defines the animal-vegetal axis of the oocyte. This animal-vegetal axis is carried over into the fertilized egg and early embryo. Thus the complex control of maternal mRNA expression in both space and time that governs embryogenesis and the formation of distinct cell types begins in the oocyte.
At the completion of oogenesis, fully grown stage 6 oocytes arrest in meiosis (Fig. 2.1). In response to the hormone progesterone, they are released from meiotic arrest (oocyte maturation), complete meiosis, and become fertilizable eggs released by the mother. During oocyte maturation some localized mRNAs are released from their storage forms and are translated into proteins (Cragle and MacNicol 2014a; Standart and Minshall 2008). This translation contributes to the conversion of the mRNA asymmetries formed in the oocyte to protein asymmetries present in the egg that will be inherited by cells of the embryo. Some maternal mRNAs encode cell cycle proteins and fundamental cell structural proteins needed to drive the first rapid cell divisions in the fertilized embryo. Thus the regulated translation of maternal mRNAs prepares the embryo for the rapid cell divisions that immediately follow fertilization and generates proteins that guide formation of the vertebrate body plan (Heasman 2006a; Cragle and MacNicol 2014a; Gray and Wickens 1998; Richter and Lasko 2011).
2.1.2 Embryonic Development: Cortical Rotation Establishes Embryonic Asymmetries
At fertilization, animal-vegetal asymmetries that began during oogenesis are further elaborated and additional embryonic asymmetries are established (Fig. 2.2). In particular, the first cell division after fertilization is long (90 min) compared to the subsequent divisions (20–30 min). During this elongated first cell cycle, the Xenopus egg undergoes a cytoplasmic rearrangement in which the outer cortex rotates with respect to the heavy inner yolk mass. This movement occurs directionally along a longitudinal line centered on the sperm entry point in the animal hemisphere. The result is that the vegetal cortex (probably associated with localized mRNAs and/or proteins) moves upwards 30° toward the animal hemisphere. This process is referred to as cortical rotation (Gerhart et al. 1989; Houston 2012; Vincent and Gerhart 1987) (Fig. 2.2b). Cortical rotation creates new molecular asymmetries in the embryo in the horizontal dimension, the so-called dorsal/ventral axis of the embryo that is perpendicular to the animal-vegetal axis. Cells that arise in the path of the upward displacement will form the Nieuwkoop inducing center, the organizer, and anterior structures of the embryo, while cells along the path of the downward displacement will form posterior structures (Gerhart et al. 1989, 1991; Houston 2012; Heasman 2006b). Thus additional critical decisions about the body plan have already been made during this first cell cycle and involve the asymmetric re-localization of cell-fate regulators.
The identities of the mRNAs and proteins directly transported and/or activated by cortical rotation remain incompletely described (Houston 2012; Heasman 2006b). However, it is clear that the Wnt signaling pathway is activated in blastula cells along the pathway of cortical rotation (Heasman et al. 1994; Schohl and Fagotto 2002). Wnt signaling stabilizes the beta-catenin protein. Beta-catenin’s translocation to the nucleus will activate the transcription of genes that establish the Spemann organizer, a critical-inducing center that forms and functions after zygotic transcription begins (Hikasa and Sokol 2013) (Fig. 2.2b). The organizer in turn emits signals to nearby tissues, driving the cell movements of gastrulation and patterning the adjacent germ layers (Gerhart et al. 1991; De Robertis 2006; Harland and Gerhart 1997). Thus, the organizer has its roots in the earliest events of embryogenesis following fertilization that include maternal mRNA localization events.
2.1.3 Later Embryonic Development: The Organizer and Patterning of Germ Layers
During the blastula and gastrula stages, the embryo in the animal-vegetal dimension is partitioned into groups of cells that will form the three germ layers: the ectoderm, the mesoderm, and the endoderm (Fig. 2.2a) (Gerhart and Keller 1986). Each layer represents a group of progenitor cells whose fate is restricted to derivatives characteristic of that layer. For example, cells of the ectoderm germ layer will give rise to ectoderm and neuroectoderm cell types. Organizer signals promote cells to differentiate as anterior derivatives of each germ layer (Gerhart et al. 1991; De Robertis 2006; Harland and Gerhart 1997). The particular germ layer derivatives that form are a function of proximity to the organizer and its signals. For example, ectodermal cells exposed to organizer signals form anterior neural structures—the anterior parts of the nervous system that include the brain and the anterior spinal cord. In contrast, cells of the ectodermal germ layer that do not receive organizer signals form primitive ectoderm and posterior neural derivatives such as the posterior spinal cord.
2.2 Examining the Biological Functions of Maternal mRNAs in Xenopus Embryogenesis
The above sections make clear that the early stages of Xenopus embryogenesis rely on the expression of maternal mRNAs. In this section a key method that has allowed distinct functions to be assigned to many maternal mRNAs in Xenopus will be discussed. Over the last decade, this method has allowed the list of functional maternal mRNAs in Xenopus to grow substantially to the point where knowledge about functional maternal mRNAs in Xenopus is fairly equivalent to that in a powerful genetic vertebrate model, zebrafish (Houston 2013).
The challenge of demonstrating that a particular Xenopus maternal mRNA encodes a cell-fate regulator has been addressed with the development of methods that eliminate specific maternal mRNAs from eggs prior to their fertilization (Torpey et al. 1992; Heasman et al. 1991; Olson et al. 2012; Schneider et al. 2010; Hulstrand et al. 2010). In this method, modified antisense oligonucleotides complimentary to the maternal mRNA of interest are microinjected into stage 6 oocytes causing the degradation of the target maternal mRNA. The injected oocytes are marked with a colored dye and then reinserted into an ovulating female frog. The oocytes mature in the female and she then lays eggs that can be fertilized. The eggs containing the depleted mRNA are easily identified and distinguished from the control eggs by the marker dye. Monitoring the embryonic phenotypes of the experimental and control embryos, by classic morphological and molecular techniques, allows the biological function of the maternal mRNA to be determined. For example, the maternal VegT mRNA encodes a T-box transcription factor. Embryos depleted of VegT mRNA lack endoderm, indicating that the VegT functions to regulate processes required for endoderm formation (Zhang et al. 1998). In another example, oligonucleotide-directed depletion of the maternal mRNA encoding the Wnt11 ligand leads to mutant embryos lacking dorso/anterior structures that also fail to express zygotic organizer genes such as Xnr3 (Tao et al. 2005). Thus, Wnt11 signaling is required for organizer formation and function. This depletion method, combined with some knowledge of how the maternal Wnt11 mRNA is controlled, connects the cellular phenomenon of cortical rotation to the molecular localization of a specific maternal mRNA and ultimately to the function of a key regulatory tissue, the organizer, that performs its role later in development after the onset of zygotic transcription (Gerhart et al. 1991; Harland and Gerhart 1997).
2.3 Localized mRNAs in Formation of the Germ Layers and Embryonic Asymmetries
Localized maternal mRNAs and the proteins they encode participate in at least three important processes in Xenopus embryos: (1) primordial germ cell formation, (2) germ layer formation, and (3) formation of embryonic asymmetries and polarities. Specific vegetally localized mRNAs, such as the mRNA encoding the Nanos protein, reside in the germplasm that ultimately gives rise to the primordial germ cells (PGCs). The control of germ cell formation and its regulation by maternal mRNAs have been discussed recently (see Chap. 8) (Lai et al. 2012; Lai and King 2013; King 2014).
The establishment of the primary germ layers, the ectoderm, the mesoderm, and the endoderm, is one of the critical processes controlled by localized mRNAs (Medioni et al. 2012; King et al. 2005; Houston 2013) (Fig. 2.2a). The partitioning of different mRNAs to distinct regions of the oocyte and egg establishes polarities and the conditions for creating cell-fate differences during embryogenesis (Heasman 2006a; White and Heasman 2008; Houston 2012). mRNAs localized during oogenesis are released from their cytoskeletal anchors either at later stages of oogenesis or during the completion of meiosis (oocyte maturation to form an egg). Importantly, the released mRNAs and proteins do not diffuse freely through the cytoplasm but instead remain concentrated near their original sites of localization. After fertilization the new cells that form during the initial cell divisions in cleavage-stage embryos capture these mRNAs and their surrounding cytoplasm. As a consequence, vegetally localized mRNAs become concentrated in vegetal cells that will give rise to endoderm, while animally localized mRNAs become concentrated in animal cells that will give rise to ectoderm.
The importance of localized maternal mRNAs for the formation of the germ layers is supported by loss-of-function experiments that deplete specific maternal mRNAs from developing embryos, as discussed above (see Sect. 2.2). A role for the vegetally localized maternal VegT mRNA was discussed above. Another vegetally localized mRNA, the maternal Vg1 mRNA, encodes a secreted ligand that can activate TGFβ signaling. Vg1-depleted embryos exhibit defects that indicate that Vg1 is required for endoderm and mesoderm formation and anterior cell types (Birsoy et al. 2006). In contrast, the Foxi2 maternal mRNA encodes a transcription factor and is localized to animal cells. Depletion of Foxi2 causes defects in ectoderm formation by disrupting the normal activation of zygotic genes important for the ectoderm (Cha et al. 2012). Thus, specific localized mRNAs provide important links between the asymmetries formed in the oocyte and the establishment of germ layers during embryogenesis.
Finally, localized mRNAs provide the molecular basis for establishing the asymmetries that form during the initial stages of embryogenesis and organize the vertebrate body plan (Houston 2012, 2013). During the earliest steps of Xenopus development, there are at least two types of embryonic asymmetries that must form animal-vegetal and dorso/anterior. mRNA localization in the oocyte creates an unequal distribution of molecules in the animal-vegetal dimension that persist after fertilization to contribute to the animal-vegetal axis of the embryo (Fig. 2.2a). The second asymmetries form as a result of cortical rotation and are keys for establishing the early signaling centers of the embryo: the Nieuwkoop center and the Spemann organizer that are located to one side of the embryo (Gerhart et al. 1989, 1991; Houston 2012) (Fig. 2.2b). While the importance of cortical rotation is unequivocal, our understanding in molecular terms of exactly what it accomplishes remains incomplete. The challenge is to explain, at the level of molecular mechanism, how both type asymmetries influence development.
2.4 Signaling Pathways and Their Activation During Maternal Xenopus Development
Multiple signal transduction pathways are present in Xenopus embryos, and their regulated activation is necessary to guide the normal events of development. These pathways include the fibroblast growth factor (FGF), Wnt, bone morphogenetic protein (BMP), and Vg1/Nodal pathways. Each of these pathways functions in different regions of the embryo to direct the formation and patterning of the embryonic germ layers (Heasman 2006b; Schohl and Fagotto 2002; Harland and Gerhart 1997). Each pathway includes a similar set of components: extracellular ligands, transmembrane receptor(s), and intracellular signaling proteins that transduce the signal from the cell surface to the nucleus. Most of these components are encoded by maternal mRNAs whose regulated translation helps control pathway function and localization.
The FGF pathway relies upon specific cell surface tyrosine kinase receptors (FGFRs) that are activated upon binding by an FGF ligand (Dorey and Amaya 2010; Goetz and Mohammadi 2013). The activated receptor transduces signals by phosphorylation and activation of cytoplasmic MAPK, PI3K, and PLCγ pathways. Depleting the maternal FGFR1 RNA gives rise to embryos with defects in gastrulation as well as defects in genes associated with the mesoderm (Yokota et al. 2003).
Binding of Wnt ligands to the Frizzled and LRP5/6 receptor proteins activates the Wnt pathway (Hikasa and Sokol 2013). This triggers a series of events that culminate in the stabilization of beta-catenin. Once beta-catenin accumulates, it translocates to the nucleus where, in concert with Tcf transcription factors, it regulates the expression of specific genes, most notably the genes of Spemann’s organizer. The Wnt pathway is elaborate, consisting of both activators and inhibitors of signaling to allow for the integration of molecular cues from other signaling pathways. Loss-of-function experiments have provided conclusive evidence for the importance of maternal Wnt signaling. For example, embryos depleted of the mRNAs encoding components needed for pathway activation, such as the Wnt11 ligand, lack or have reduced amounts of organizer cells and exhibit defects in dorso/anterior axis formation (Tao et al. 2005). In contrast, embryos depleted of the mRNAs for pathway inhibitors, such as Axin, have enlarged organizers, ectopically express organizer genes, and give rise to embryos with enlarged head and anterior structures (Kofron et al. 2001).
BMPs and the Vg1/Nodal proteins are members of the transforming growth factor beta (TGFβ) superfamily of ligands (Wu and Hill 2009; Moustakas and Heldin 2009; Ramel and Hill 2012). The different ligands activate specific versions of a core pathway. Each group of ligands binds and activates heteromeric cell surface receptors. The activated serine/threonine kinase of the ligand-bound receptor phosphorylates cytoplasmic Smad proteins, and the modified proteins translocate to the nucleus where, in conjunction with other transcription factors, they active specific genes. This general scheme applies to each type of TGFβ ligand. Distinct signaling outcomes result from the use of different components, usually the receptors and Smad transcription factors used for each type of ligand (Wu and Hill 2009; Ramel and Hill 2012). There are shared components used by multiple pathways, such as the common Smad4 factor. In addition, some receptors may mediate signaling by multiple ligands, and such results have led to some confusing receptor nomenclature.
BMP signaling in Xenopus embryos is first activated coincident with the onset of zygotic transcription; however, this activation requires maternal signaling proteins (Faure et al. 2000). Thus controlling the synthesis of maternal BMP pathway components is necessary to create a functional pathway. The BMP pathway functions in the posterior cells and is activated when BMP ligand binds the type I and type II serine/threonine kinase receptors. Activation of the kinase domains results in phosphorylation of the receptor Smads 1, 5, and/or 8. The phosphorylated Smads function in complex with Smad4 and other transcription factors to activate the transcription of BMP-responsive genes.
The Vg1 mRNA encodes a growth factor ligand of the TGFβ family and is localized to the vegetal cortex of Xenopus oocytes (Weeks and Melton 1987; Melton 1987). Loss-of-function experiments demonstrate a critical role for Vg1 in formation of the germ layers; embryos depleted of Vg1 mRNA lack endoderm and have reduced amounts of mesoderm (Birsoy et al. 2006). The Vg1 ligand signals through the same pathway as the Nodal TGFβ ligands. The receptors for these ligands are referred to as activin receptors, and upon ligand binding they phosphorylate the Smad2 protein. While loss-of-function analysis demonstrates a clear maternal requirement for the Vg1 ligand and hence a requirement for a pathway to transduce Vg1 signals, biochemical experiments monitoring the timing of signaling indicate that the active pathway cannot be detected until the blastula stages, coincident with the activation of zygotic transcription (Schohl and Fagotto 2002; Faure et al. 2000; Lee et al. 2001).
In summary, ligands, receptors, signaling proteins, and transcription factors of key signaling pathways are critical for transducing signals that guide the initial steps of development. Yet despite the importance of these proteins, we are only beginning to understand the processes that control their synthesis. Much remains to be learned about how regulated translation of maternal mRNAs impinges on the assembly and function of signaling pathways that guide development.
2.5 Translational Control Mechanisms Operating During Xenopus Oocyte Maturation and Early Cleavage Stages of Embryogenesis
While we now know the identity of several maternal mRNAs that encode key cell-fate determinants that drive development, we have relatively little direct knowledge about the specific translational mechanisms that may control their translation. In fact, most of our knowledge about molecular mechanisms that control maternal mRNA translation comes from studies of maternal mRNAs that drive oocyte maturation, the second phase of the meiotic cell cycle (Groppo and Richter 2009; Richter 2007; MacNicol and MacNicol 2010; Weill et al. 2012). While these mRNAs do not encode cell-fate determinants, examination of their regulation has provided important insights into mRNA regulatory mechanisms that serve as a useful foundation for examining maternal mRNAs encoding cell-fate determinants and their possible modes of regulation. In this section, the translational regulation of maternal mRNAs during oocyte maturation will be discussed to serve as context for the subsequent discussions about mRNAs encoding developmentally relevant cell-fate regulators.
The translational state of an mRNA depends upon the sequence elements it contains and the proteins that bind these elements and influence the mRNA’s interaction with ribosomes. These complexes of proteins bound to an mRNA (mRNA ribonucleoprotein particles (mRNPs)) are the determining factors of whether an mRNA engages ribosomes and is actively translated into protein or is stored in a translationally repressed state. Therefore, a key to understanding the behaviors of specific mRNAs with respect to translation is to define their mRNP composition and its dynamics during changes in its translational status. For example, stored Xenopus maternal mRNAs are translationally repressed due to their association with general repressor proteins such as FRGY2, XP54 (DDX6), and RAP55 (Colegrove-Otero et al. 2005a; Minshall et al. 2001, 2007; Tanaka et al. 2006, 2014; Tafuri and Wolffe 1993; Ranjan et al. 1993; Deschamps et al. 1992). A comprehensive discussion of the RNA-binding proteins that mediate general translational repression in Xenopus oocytes and embryos is beyond the scope of this review (see Cragle and MacNichols for a thorough discussion (Cragle and MacNicol 2014a)). However, the following section will present a brief overview of the mechanisms of translation control that operate during Xenopus oocyte maturation and early cleavage stages with an emphasis on what is known about the functional sequence elements and their cognate binding proteins.
2.5.1 Control of Translation During Xenopus Oocyte Maturation: Regulated mRNA Polyadenylation
The regulated addition of adenylates to the 3′ end of maternal mRNAs, referred to as poly(A) tail lengthening or polyadenylation, is a mechanism used to control the translational activation of specific mRNAs during oocyte maturation (Cragle and MacNicol 2014a; Standart and Minshall 2008; Richter and Lasko 2011). The majority of eukaryotic mRNAs are cleaved and polyadenylated in the nucleus in two coupled reactions that recognize the conserved 5′-AAUAAA-3′ present in their 3′ untranslated regions (UTRs). Once mRNAs enter the cytoplasm, the poly(A) tails of many mRNAs are further subjected to both poly(A) tail lengthening and shortening (deadenylation) (Moore 2005). Depending on cell type, these changes can affect mRNA stability, translational activity, or both. During Xenopus oocyte maturation, the vast majority of maternal mRNAs are stable, regardless of poly(A) tail length. However, some mRNAs that have poly(A) tails and are translationally active in oocytes lose these structures during maturation, in a process called deadenylation, and become translationally inactive (Fox and Wickens 1990; Hyman and Wormington 1988). In contrast, other mRNAs with very short poly(A) tails that are translationally inactive in oocytes undergo poly(A) tail lengthening and translational activation during oocyte maturation (Weill et al. 2012; Sheets et al. 1994; Ivshina et al. 2014). In the following sections, two different mechanisms of translational activation of mRNAs coupled to polyadenylation will be discussed.
2.5.2 CPE/CPEB-Dependent Polyadenylation and Translation
Many Xenopus mRNAs that are polyadenylated during oocyte maturation contain cytoplasmic polyadenylation elements (CPE) in their 3′UTRs that serve as binding sites for the CPE-binding protein-1 (CPEB1) (Weill et al. 2012; Ivshina et al. 2014; Pique et al. 2008; Fox et al. 1989; McGrew et al. 1989; Hake and Richter 1994). These elements are distinct from the nuclear recognized AAUAAA element and are generally comprised of U-rich elements. CPE-containing mRNAs are repressed in stage 6 oocytes, and this repression is mediated by CPEB via CPEB-interacting proteins, such as Maskin and 4ET (Minshall et al. 2007; Stebbins-Boaz et al. 1999). The Maskin and 4ET repressor proteins are tethered to CPE-containing mRNAs via their interactions with CPEB, and they block translation of their bound mRNAs by preventing the productive assembly of a translation initiation complex on the 5′ cap structure of the mRNA, a necessary step in translational activation. In response to progesterone, the hormone that directs oocyte maturation, CPEB is phosphorylated (Mendez et al. 2000). Phosphorylation of CPEB triggers several important events including the dissociation of the Maskin and 4ET repressor proteins from the CPEB-mRNA complex. In addition, the CPEB-bound Gld2 poly(A) polymerase becomes activated and thus adds a poly(A) tail to CPE-containing mRNAs (Kwak et al. 2004; Barnard et al. 2004). The elongated poly(A) tail recruits poly(A)-binding protein (PABP), and PABP stimulates translational initiation through mechanisms that are not fully understood but probably involve its interactions with the 5′ cap structure (Gray et al. 2000). Thus, CPE-containing mRNAs are translationally activated during maturation by a dual mechanism: the relief from Maskin and 4ET repression combined with the stimulation provided by an elongated poly(A) tail bound by PABP. Other studies suggest that, the mRNA encoding another specificity factor for polyadenylation, CPEB4 is a substrate for CPEB1 polyadenylation (Novoa et al. 2010; Igea and Mendez 2010). Results suggest that these two CPEB proteins function sequentially to mediate temporally distinct polyadenylation events.
2.5.3 Musashi-Dependent Polyadenylation
Some Xenopus mRNAs that are polyadenylated during maturation do not contain CPEs but instead contain binding elements for the Musashi protein, referred to as Musashi binding elements or MBEs (Charlesworth et al. 2006; Arumugam et al. 2010). mRNAs containing MBEs are bound by the Musashi protein, contain short poly(A) tails, and are translationally repressed in oocytes by an unknown mechanism. In response to progesterone and oocyte maturation, Musashi is phosphorylated, and the protein directs the polyadenylation of MBE-containing mRNAs, through the recruitment and/or activation of a poly(A) polymerase (Cragle and MacNicol 2014b; Arumugam et al. 2012).
2.6 Xenopus Embryo-Specific Translational Control Mechanisms: Poly(A) Removal and Addition
While most studies of translational control in Xenopus have focused on oocyte maturation, some have focused on translational control mechanisms in cleavage-stage embryos. For example, the Eg1, Eg2, Eg5, and c-mos mRNAs are all deadenylated and translationally repressed after fertilization (Sheets et al. 1994; Le Guellec et al. 1991). These processes require specific sequence elements in the target mRNAs that recruit the CELF1 protein (also called EDEN and CUGBP1) (Paillard et al. 1998). Conversely, translation of the maternals Cl1, Cl2, and activin receptor mRNAs in Xenopus embryos is activated following fertilization, coincident with their polyadenylation (Paris and Philippe 1990; Paris et al. 1988; Simon et al. 1992, 1996; Simon and Richter 1994). The embryo-specific polyadenylation of these mRNAs requires specific 3′UTR sequence elements, termed embryonic CPEs (eCPEs) that are distinct from CPEs that direct mRNA polyadenylation during maturation (Charlesworth et al. 2013). Consistent with this distinction between stage-specific regulatory mechanisms, the Cl1, and Cl2 eCPEs are bound by ElrA RNA-binding protein (Good 1995). Embryonic specific polyadenylation also occurs on the maternal mRNA encoding the nuclear lamin B1 protein, coincident with its translation in embryos (Ralle et al. 1999). However, the lamin B1 mRNA does not contain eCPE sequences with obvious similarity to the Cl1 and Cl2 mRNAs, suggesting that translational activation during Xenopus embryogenesis may occur by multiple parallel pathways.
The poly(rC)-binding protein αCP2 can recruit cytoplasmic poly(A) polymerase activity to mRNAs in Xenopus embryos, and this recruitment relies on C-rich sequences recognized by αCP2 (Vishnu et al. 2011). This mechanism is specific for embryos and is not active in oocytes. The αCP2 protein polyadenylates mRNAs that contain C-rich CPEs in their 3′UTRs in close proximity to the conserved hexa-nucleotide signal AAUAAA. While it is clear that αCP2 can function in embryos as a specificity factor for a unique form of cytoplasmic polyadenylation, it is unclear what endogenous mRNAs are normally substrates for this protein.
2.7 Translational Control of Cell-Fate Determinants During Maternally Controlled Embryogenesis
Translational control of mRNAs encoding cell-fate determinants involves distinct mechanisms that elaborate on the basic mechanisms discussed above and/or employ unique mechanisms such as mRNA localization and cell-type-specific repression. These specialized mechanisms are probably critical for the precise control required by cell-fate regulators to work properly within the Xenopus embryo. This section will discuss specific examples and mechanisms relevant to translational control of mRNAs that encode critical cell-fate determinants.
2.7.1 Translational Control Mechanisms Related to Localized mRNAs
As discussed earlier, the localization of specific RNAs to subcellular domains of oocytes and eggs restricts the ultimate cellular destination of these mRNAs and their encoded proteins in the developing embryo (Medioni et al. 2012; King et al. 2005; Houston 2012). However, this mechanism only works if the localized mRNAs are translationally inactive during transport to prevent spatially inappropriate expression of protein. Specific examples will be discussed here.
2.7.1.1 Vg1 mRNA
The Vg1 mRNA encodes a growth factor ligand of the TGFβ family and is localized to the vegetal cortex of Xenopus oocytes (Weeks and Melton 1987; Melton 1987). Embryos depleted of Vg1 mRNA lack endoderm and have reduced amounts of mesoderm (Birsoy et al. 2006). Translation of the Vg1 mRNA is repressed until it is localized to the vegetal cortex in stage 6 oocytes. This repression in growing oocytes is mediated through a translational control element (TCE) contained within the 3′UTR of the Vg1 mRNA, located adjacent to the sequences for localization (Otero et al. 2001; Wilhelm et al. 2000). The translational repression operates independently of polyadenylation. The ElrA RNA-binding protein, a member of the ELAV family, has been implicated in mediating this repression, but the precise mechanisms remain under investigation (Colegrove-Otero et al. 2005b).
2.7.1.2 VegT mRNA
The VegT mRNA encodes a T-box transcription factor, and the mRNA is localized to the vegetal cortex of fully grown stage 6 oocytes (Zhang and King 1996; Lustig et al. 1996; Stennard et al. 1996). Embryos depleted of VegT maternal mRNA do not form endoderm, and they exhibit defects in the production of signals needed to induce mesoderm (Zhang et al. 1998). As discussed above, localized mRNAs are subject to specific mechanisms of repression while they are transported during oogenesis (Medioni et al. 2012; King et al. 2005). At subsequent stages of development, the translation of localized mRNAs must be activated, but in general these later activation steps are poorly understood. An exception is the XSeb4R protein that acts as a positive regulator of VegT mRNA translation in embryos (Souopgui et al. 2008). XSeb4R is an RRM-containing RNA-binding protein that exerts its effects on the VegT mRNA by directly binding to sequences within the 3′UTR of the mRNA. The mechanism by which this binding enhances translation or stability is unknown.
2.7.1.3 Wnt11b mRNA
Maternal mRNA depletion also reveals a role for vegetally localized Wnt11 (also called Wnt11b) in Xenopus axis formation (Hikasa and Sokol 2013) (see Sect. 2.4). Wnt11 mRNA translation regulation is connected to cortical rotation. In oocytes and eggs, Wnt11 mRNA is closely associated with the vegetal cortex. After fertilization Wnt11 mRNA in embryos is uniformly distributed between dorsal and ventral blastomeres of cleaving embryos, but the Wnt11 mRNA in dorsal cells is polyadenylated more extensively than in ventral cells (Schroeder et al. 1999; Flachsova et al. 2013). This differential polyadenylation is sensitive to treatments that disrupt cortical rotation, such as UV light treatment. In addition, Wnt11 mRNA in dorsal cells is preferentially associated with polyribosomes compared to the mRNA in ventral cells, indicating that it is being actively translated in dorsal cells. These observations suggest a connection between cortical rotation, dorsal cell polyadenylation, and translational activation of Wnt11 mRNA. However, it is worth noting that other studies raise questions about the polyadenylation status of Wnt11 mRNA (Tao et al. 2005). The results of these studies suggest that a significant fraction of the Wnt11 mRNA itself is translocated to the dorsal cells during cortical rotation. This movement of the Wnt11 mRNA followed by its translational activation is sufficient to explain differences in Wnt11 protein expression.
2.7.2 Translational Regulation of the FGFR Signaling Pathway
Xenopus embryos provide many advantages for analyzing signaling mechanisms in a developmental context, and the fibroblast growth factor (FGF) pathway was one of the first investigated (Amaya et al. 1991). This pathway relies upon specific cell surface receptors (FGFRs) that possess cytoplasmic tyrosine kinase domains (Dorey and Amaya 2010; Lea et al. 2009). These receptors are activated to initiate signaling when an FGF ligand binds to the FGF receptor causing it to multimerize and activate its tyrosine kinase. The activated kinase phosphorylates specific cytoplasmic proteins to transduce the signal (Goetz and Mohammadi 2013).
In Xenopus, mRNAs encoding FGFRs and several different FGF ligands are present maternally (Dorey and Amaya 2010; Lea et al. 2009). Depletion of the maternal FGFR1 mRNA or expression of a dominant negative FGFR causes specific defects in gastrulation and gene expression (Yokota et al. 2003; Amaya et al. 1991). Antibody staining experiments reveal that translation of the FGFR1 maternal mRNA is highly regulated, with RNA translation repressed in oocytes and only activated during oocyte maturation (Amaya et al. 1991; Musci et al. 1990). This regulation relies upon a sequence element in the 3′UTR of the FGFR1 mRNA, the translational inhibitory element (TIE) that efficiently represses translation in oocytes (Robbie et al. 1995). The proteins that repress by binding the TIE have not been identified, but relief of TIE repression requires specific signaling events that are activated during oocyte maturation, and these mechanisms function independent of polyadenylation. In embryos the FGFR1 mRNA is also translationally activated, but only in cells of the animal hemisphere. The mechanistic basis of FGFR1 spatially restricted expression is unknown (Cornell et al. 1995).
2.7.3 Translational Regulation of the Bone Morphogenetic Protein (BMP) Signaling Pathway
In vertebrate organisms the BMP signaling pathway is important for multiple aspects of embryonic development and adult organ homeostasis (Moustakas and Heldin 2009; Ramel and Hill 2012; Plouhinec et al. 2011). This pathway consists of a related family of extracellular ligands that signal through heteromeric cell surface receptors. Signaling commences when a BMP ligand binds to the receptor complex and activates its cytoplasmic serine/threonine kinase. The activated kinase phosphorylates the cytoplasmic Smad1/5/8 proteins, and the modified Smads translocate to the nucleus where they guide the transcription of specific genes (Heasman 2006a; Smith 2009).
Interference with BMP signaling during maternally controlled stages of Xenopus embryogenesis, by overexpression of wild-type or mutant BMP ligands, receptors, or intracellular Smad effector proteins, severely disrupts the formation of mesoderm and ectoderm/neuroectoderm derivatives and the associated patterns of gene expression (Heasman 2006a; White and Heasman 2008; Kimelman 2006; Kimelman and Pyati 2005). For example, BMP receptor proteins lacking their cytoplasmic domains retain the ability to interact with BMP ligands. But these truncated receptors cannot transduce signals, and they act as “ligand sinks” that reduce normal ligand-dependent signaling. Xenopus embryos expressing BMP receptors lacking their cytoplasmic domains develop secondary axes that contain ectopic neural and mesodermal cell types (Graff et al. 1994; Mishina et al. 1995; Suzuki et al. 1994; Maeno et al. 1994; New et al. 1997; Frisch and Wright 1998). These examples, along with other functional studies, demonstrate the importance of BMP signaling for Xenopus embryogenesis.
mRNAs encoding the proteins of the BMP pathway are present maternally, but for many components it is not known whether or not the corresponding proteins are expressed (Graff et al. 1994, 1996; Nishimatsu et al. 1992a, b; Hawley et al. 1995; Suzuki et al. 1997). While BMP signaling in embryos is first activated coincident with the onset of zygotic transcription (Faure et al. 2000), this activation requires the maternal signaling proteins. Thus controlling the synthesis of maternal BMP pathway could provide a way to regulate pathway signaling.
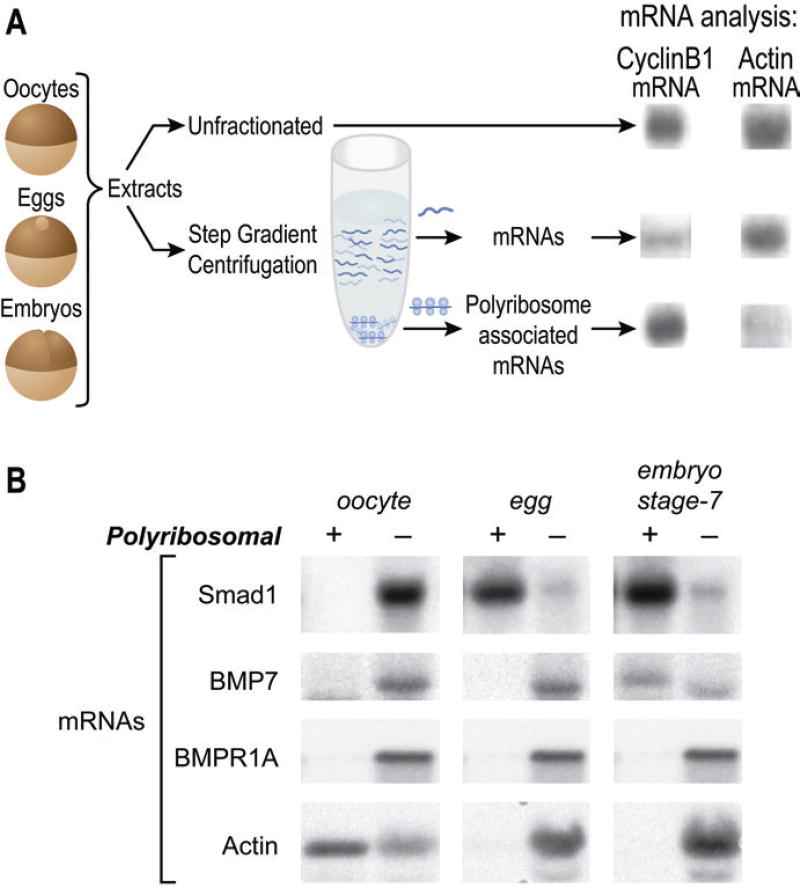
Polyribosome association assays reveal that the mRNAs encoding different signaling proteins of the BMP pathway exhibit a diverse set of regulatory behaviors (Fig. 2.3) (Fritz and Sheets 2001). While each of the mRNAs in the BMP pathway is inefficiently associated with polyribosomes in immature oocytes, indicating their translational inactivity, each is recruited to polyribosomes at a distinct subsequent developmental stage (Fig. 2.3). Specifically, the mRNA encoding the Smad1 transcription factor and the mRNA encoding the BMP receptor, referred to as the activin A receptor, type I (ACVR1a, also called ALK2), are recruited to polyribosomes during oocyte maturation, whereas the mRNAs encoding the BMP7 ligand and the BMP receptor referred to as the activin A receptor, type IIA (ACVR2a, also called XSTK9), are recruited during the early stages of embryogenesis. The BMP receptor1a mRNA (BMPR1a, also referred to as ALK3) is not associated with polyribosomes until after the onset of zygotic transcription. Thus, the translation of the maternal mRNAs of the BMP pathway is highly regulated with different mRNAs exhibiting a distinct pattern of temporal control (Fritz and Sheets 2001) (Fig. 2.3). These distinctions may help coordinate the proper assembly of the entire pathway in space and time in the embryo or may indicate that some pathway components have independent functions.
Fig. 2.3.
Regulated translation of the mRNAs encoding proteins of the BMP pathway. (a) The isolation of polyribosomes from Xenopus oocytes, eggs, and embryos (Sheets et al. 2010). At the right, total (unfractionated) RNA from eggs, non-polyribosomal RNA from eggs, and polyribosomal RNA from eggs were analyzed with blot hybridization using probes to the cyclin B1 and cytoskeletal actin mRNAs. (b) Polyribosome and non-polyribosome fractions were prepared from Xenopus laevis oocytes, eggs, and stage 7 blastula embryos (Fritz and Sheets 2001). Total RNAs isolated from the fractions were analyzed by blot hybridization. Filters were hybridized with probes to detect Xenopus mRNAs encoding the Smad1 transcription factor, the BMP7 ligand, BMPR1a receptor, and cytoskeletal actin proteins. A representative RNA blot analyzing each mRNA is shown. Recreated from Fritz, B.R. and M.D. Sheets Developmental Biology, 2001. 236(1): p. 230–243, with permission from Elsevier
As predicted from prior work in oocytes, fertilized eggs, and early cleavage-stage embryos, the polyadenylation state of each mRNA coincided with their state of polyribosome recruitment (Fritz and Sheets 2001). For example, the poly(A) tail of Smad1 mRNA is lengthened during oocyte maturation when this message becomes efficiently recruited to polyribosomes. In contrast, the BMP7 mRNA becomes associated with polyribosomes and polyadenylated during embryogenesis. Thus, while the timing of polyribosome association differs for each mRNA, in each case, polyribosome loading is coincident with poly(A) tail elongation. This suggests that the temporal control of polyadenylation governs when and potentially how efficiently each mRNA is translationally activated.
The observation that the timing of polyadenylation differs for each mRNA suggests the involvement of specific regulatory mechanisms. For example, the poly(A) tails of the Smad1 and ACVR1a mRNAs are elongated during oocyte maturation (Fritz and Sheets 2001). This suggests that polyadenylation of these mRNAs is controlled by the CPE/CPEB-dependent mechanism that functions on mRNAs during maturation (Ivshina et al. 2014; Fernandez-Miranda and Mendez 2012). In support of this idea, both the Smad1 and ACVR1a mRNAs contain putative CPE sequence elements for maturation-specific polyadenylation in their 3′UTRs. The Xenopus BMP7 mRNA is polyadenylated during the initial cleavage stages that follow fertilization (Fritz and Sheets 2001). Other mRNAs polyadenylated at fertilization contain eCPE sequence elements (see Sect. 2.6) in their 3′UTRs (Charlesworth et al. 2013). However, BMP7 mRNA lacks obvious eCPEs, suggesting that different RNA recognition factors or even distinct mechanisms operate during the blastula stages of developmental.
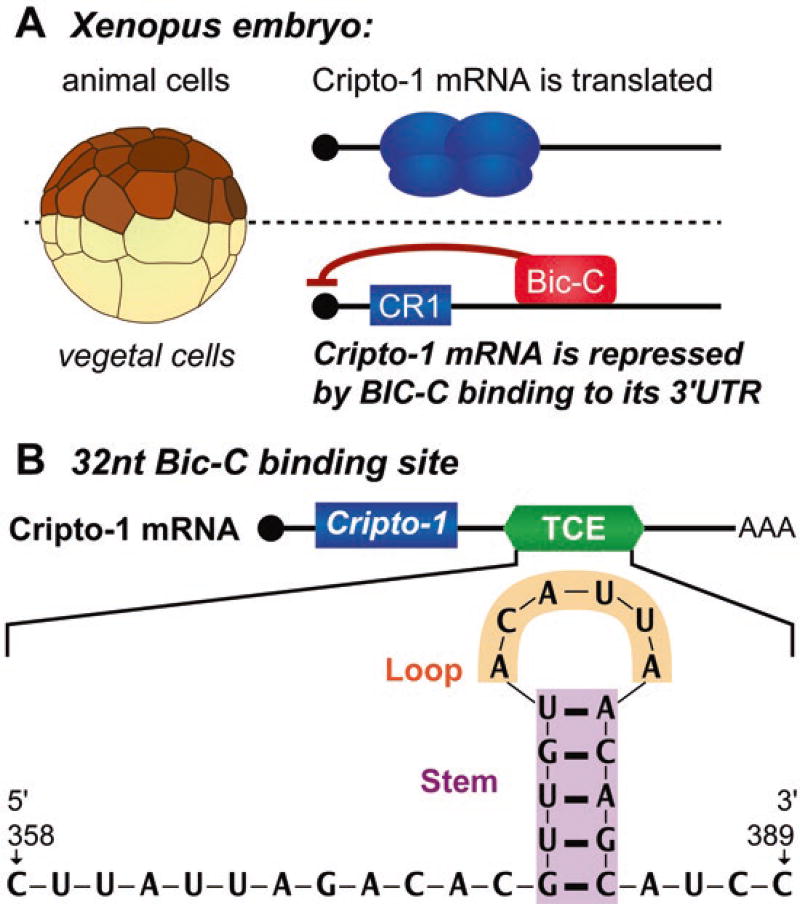
2.7.4 Translational Control of Cripto-1 mRNA: Cell-Specific Repression
The above examples suggest that differential timing of polyadenylation reflects and/or causes temporal differences in translational activation during embryogenesis. Studies of Cripto-1 mRNA translation reveal that cell-type-specific translational repression mechanisms can add an additional layer of spatial control on top of this temporal control. Significantly, the capacity for this spatial control of translation later in embryogenesis, mediated by a cell-type-specific repressor named Bicaudal-C (Bic-C), is actually established by early mechanisms that cause the vegetal localization of maternal Bic-C mRNA (Zhang et al. 2013). Thus, asymmetries established early in development are used to establish more refined asymmetries at later stages, as described further in the following sections.
Cripto proteins are secreted co-receptors of the Nodal signaling pathway, a pathway that is critical for normal vertebrate development (Klauzinska et al. 2014). Mutant alleles of Cripto genes cause severe embryonic defects in mouse and zebrafish (Gritsman et al. 1999; Ding et al. 1998). The Xenopus Cripto-1 protein (also called xCR1 or FRL1) was discovered as an interaction partner of the fibroblast growth factor receptor (FGFR1) (Kinoshita et al. 1995). Subsequent experiments indicated that Cripto-1 could also bind Wnt ligands and affect their ability to initiate signaling (Tao et al. 2005). These data reveal that Cripto-1’s effect on signaling in Xenopus could involve other crucial signaling pathways in addition to the Nodal pathway. Regardless, depletion of the maternal Cripto-1 mRNA in Xenopus embryos alters cell-fate decisions and severely disrupts axis formation (Tao et al. 2005). These phenotypes are similar to those observed with embryos depleted of the maternal Wnt11 mRNA (Tao et al. 2005), suggesting that Wnt11 and Cripto-1 impact the same developmental events. Thus, while the precise pathways affected by Cripto-1 in Xenopus embryos remain to be determined, it is clear that Cripto-1 and the mechanisms that control its maternal expression are critical for Xenopus development.
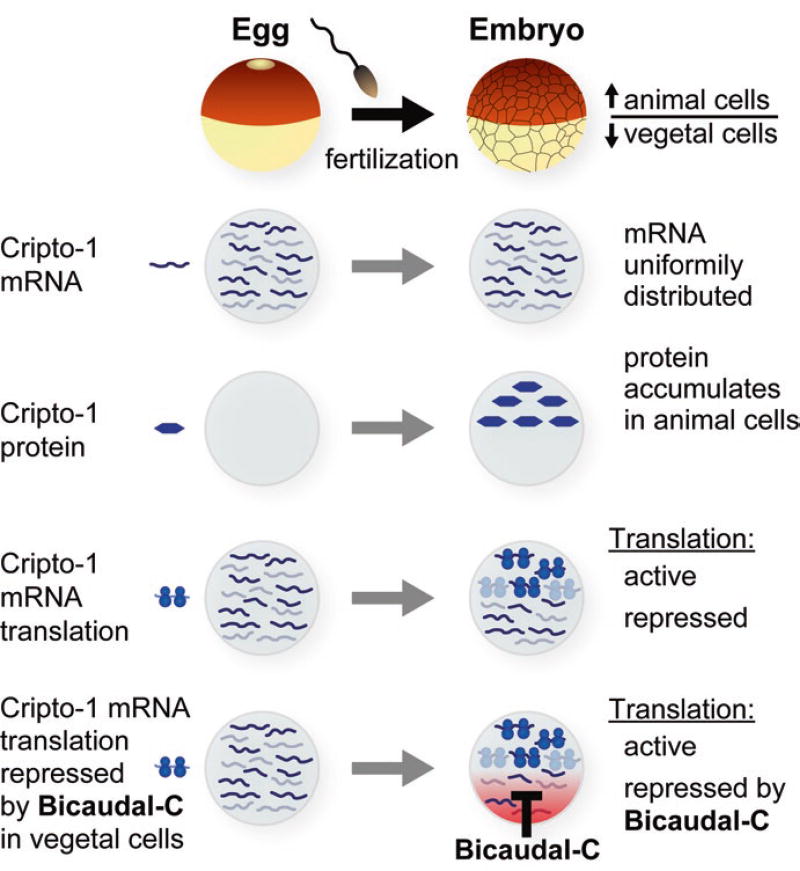
In contrast to some of the cell-fate determinant-encoding mRNAs discussed above, the maternal Cripto-1 mRNA is uniformly distributed throughout the Xenopus embryo (Dorey and Hill 2006) (Fig. 2.4). That is, there is no spatial control of Cripto-1 mRNA per se. However, importantly, the Cripto-1 protein accumulates only within the cells of the marginal zone and animal hemisphere (Dorey and Hill 2006) (Fig. 2.4). This effect could be accomplished by differential translation of Cripto-1 mRNA or differential stability of Cripto-1 protein or both mechanisms. However, polyribosome association experiments provide strong evidence for differential translational activity of the Cripto-1 mRNA in animal versus vegetal cells, with translation being more efficient in animal cells (Zhang et al. 2009). Indeed, additional experiments reveal that Cripto-1 mRNA translation is regulated both temporally and spatially in the embryo. In oocytes and eggs, Cripto-1 is translationally repressed, but upon fertilization it is translationally active, but only within the cells of the animal hemisphere (Fig. 2.4). When the Cripto-1 mRNA becomes translationally active, it is polyadenylated throughout all cells of the embryo, even though translational activation of Cripto-1 mRNA is confined to animal cells (Zhang et al. 2009).
Fig. 2.4.
Spatial accumulation of the Cripto-1 protein in Xenopus embryos is controlled by regulated translation of the Cripto-1 mRNA. In Xenopus, the Cripto-1 mRNA is uniformly distributed throughout the egg and all embryonic cells. The Cripto-1 protein is only produced in animal cells of embryos after fertilization, but not vegetal cells (Cragle and MacNicol 2014a). Differential accumulation of the Cripto-1 protein in Xenopus embryos is due to the spatially regulated translation of the Cripto-1 mRNA. Translation is activated in animal cells and repressed in vegetal cells (Heasman et al. 1994; Zhang et al. 2009). The Bicaudal-C (Bic-C) protein is responsible for repression functions in the vegetal cells (Heasman 2006a; Houston 2013; Zhang et al. 2013, 2014). The Bic-C mRNA is localized to the vegetal cortex of developing oocytes, and as a consequence after fertilization, Bic-C is restricted to vegetal cells (Schohl and Fagotto 2002)
A luciferase reporter assay designed to measure translational efficiency quantitatively within embryos was used to define the translational control mechanisms responsible for the cell-type-specific expression of Cripto-1 mRNA. Briefly, building on the foundational knowledge from studies in oocytes, eggs, and cleavage-stage embryos, the luciferase-coding region was engineered as a fusion to the 3′UTR of Cripto-1 mRNA. A second luciferase fusion gene was generated fused instead to the 3′UTR cyclin B1 mRNA. Cyclin B1, in contrast to Cripto-1 mRNA, is equally associated with polyribosomes in animal and vegetal cells. These reporter mRNAs were independently injected into either the animal or vegetal cells of 4–8 cell embryos that were allowed to develop to the late blastula stage at which point they were harvested and assayed for luciferase activity. The translation of the luciferase reporter containing the 3′UTR of Cripto-1 mRNA but not the control gene fusion containing the 3′UTR of cyclin B1 mRNA is repressed in the vegetal cell-injected embryos (Zhang et al. 2009). These experiments led to three important conclusions: (1) spatial translational differences in Cripto-1 mRNA translation were caused by a vegetal cell-specific translational repression; (2) repression could be directed by sequences present in the 3′UTR of the Cripto-1 mRNA, nicely following the paradigm established for translational control in Xenopus oocytes and in other systems; and (3) spatially controlled translational regulation could be recapitulated in living embryos with a sensitive luciferase assay that would facilitate dissection of the relevant repressive elements and their cognate proteins. Indeed, subsequent deletion-mapping experiments identified a subregion of the Cripto-1 3′UTR that was sufficient for efficient repression, a region referred to as the translational control element (TCE) (Zhang et al. 2009).
2.7.5 Bicaudal-C Is a Vegetal Cell Translational Repressor
The simplest hypothesis for the observations concerning Cripto-1 mRNA regulation is that vegetal cells contain a cell-specific repressor protein(s) that animal cells lack and that this protein(s) binds an element(s) within the TCE identified in the mapping experiments outlined above. The TCE contains binding sites for the repressor proteins pumilio and CUGBP1 (also called CELF1), but while mutational analysis revealed that these binding sites contribute to repression, pumilio and CUGBP1 proteins are uniformly distributed in embryonic cells, suggesting that they are unlikely to be responsible for the cell-type specificity of repression (Zhang et al. 2009). Therefore a targeted functional approach was taken, exploiting what is known about localized mRNAs. This approach identified Bicaudal-C (Bic-C) as the RNA-binding protein responsible for vegetal cell specificity of Cripto-1 translational repression (Zhang et al. 2013) (Figs. 2.4 and 2.5). Bic-C protein is enriched in vegetal cells, or predicted to be, because the Bic-C mRNA is localized to these cells (Wessely and De Robertis 2000). A key experiment involved injecting the mRNA encoding Bic-C into animal cells (where Bic-C is normally not present). It was observed that ectopic expression of Bic-C was sufficient to mediate vegetal cell-specific repression of relevant luciferase reporter mRNAs. Significantly, when Bic-C is expressed in animal cells, it binds to the native Cripto-1 mRNA and to reporter RNAs when they contain the TCE (Zhang et al. 2013). These results strongly support the idea that Bic-C is responsible for the specificity of vegetal cell repression and is largely sufficient for this repression. Importantly, a tethered translation assay also supports a direct and robust role for Bic-C in mRNA translational repression (Zhang et al. 2013) (Figs. 2.4 and 2.5). A reasonable hypothesis for the role CUGBP1 and pumilio proteins in Cripto-1 mRNA repression is that these proteins simply help stabilize Bic-C binding to target mRNAs, though direct biochemical experiments to test this idea remain to be done. Regardless, the ectopic animal cell injection assay provides a powerful tool to further investigate Bic-C protein’s structure and function and offers a platform to advance both biological and medically useful knowledge concerning this important RNA-binding protein.
Fig. 2.5.
Spatially regulated translation of the Cripto-1 mRNA by the Bic-C repressor. (a) Bic-C represses the Cripto-1 mRNA in vegetal cells of Xenopus embryos. The Cripto-1 mRNA is translated in animal cells but repressed in vegetal cells. Vegetal cell repression requires Bic-C binding to its recognition element, a sequence within the Cripto-1 mRNA’s 3′UTR (Heasman 2006a; Houston 2013; Zhang et al. 2013, 2014). (b) The 32nt Bic-C binding site and its predicted stem-loop structure identified from the translational control element (TCE) of the Cripto-1 mRNA 3′UTR (Heasman 2006a; Zhang et al. 2014)
2.7.6 The Bic-C RNA-Binding Element
The results described above were exploited to define a Bic-C RNA binding site, the first defined for any Bic-C ortholog (Zhang et al. 2014). Bic-C proteins contain multiple heteronucleoprotein K homology (KH) domains that are known to function as sequence-specific RNA-binding modules (Gamberi and Lasko 2012; Hollingworth et al. 2012; Teplova et al. 2011; Nakel et al. 2010; Valverde et al. 2008). The N-terminal region of Xenopus Bic-C containing these domains was sufficient for specific binding to the TCE, indicating, as predicted, that the TCE contains a Bic-C binding element(s) (Zhang et al. 2009, 2013). In vitro RNA-binding and RNA protection experiments using purified Bic-C were used to define a minimal site within TCE that bound Bic-C. The Bic-C target site in the Cripto-1 mRNA’s 3′UTR is a 32-nucleotide element that mutational analyses revealed contains a stem-loop structure (Zhang et al. 2014) (Fig. 2.5). While the sequence of the stem is not critical, its structure is. In contrast, the sequence of the loop region is important. In addition, this Bic-C binding element contains a 10-nucleotide region 5′ to the stem loop that may also provide sequence specificity to binding (Fig. 2.5). This 32-nucleotide binding site is necessary and sufficient for translational repression in the ectopic animal cap assay (Zhang et al. 2014). Together these results support a hypothesis in which Bic-C’s role in Xenopus maternal development is executed by its direct translational repression of select target mRNAs within the vegetal cells of embryos. The identification of a Bic-C binding element will guide further detailed analysis of the Bic-C RNA interface and ultimately may facilitate the identification of additional Bic-C target mRNAs in embryos and relevant adult tissues.
2.7.7 Branching Out: Identification of Bic-C Target mRNAs Reveals a Bic-C Regulatory Network that Transforms Bic-C-dependent Repression into Distinct Cell-Fate Programs
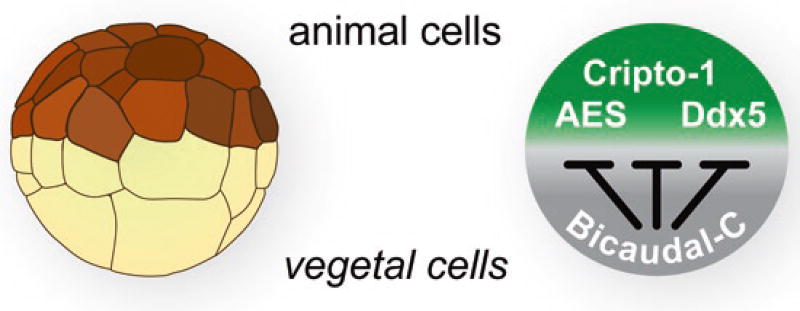
RNA-binding proteins regulate multiple mRNAs to mediate their biological functions. To identify additional mRNA targets, Bic-C was immunoprecipitated from Xenopus embryos, and the associated RNAs were analyzed with RNA-Seq. This approach identified 62 new putative Bic-C targets from Xenopus embryos (Zhang et al. 2013). Of course, this approach identifies RNAs based on their association with Bic-C in living embryos but does not reveal whether they are indeed functionally repressed by Bic-C. Importantly, several of the 62 mRNA were validated as bona fide Bic-C repression targets using the translational assays initially developed for the study of Cripto-1 mRNA (Zhang et al. 2013).
A critical issue is to determine whether the proteins encoded by these Bic-C target mRNAs do indeed function in embryogenesis and how their repression by Bic-C contributes to their function. Notably, many of the putative Bic-C target mRNAs encode proteins known to function in developmentally relevant pathways. For example, the Dpy30 mRNA encodes a histone methyltransferase important for cell-fate decisions in ES cells (Jiang et al. 2011), while the BCCIP mRNA encodes a protein that guides progenitor cells in neural development (Huang et al. 2012). In addition to Cripto-1, other Bic-C target mRNAs encode proteins implicated in Nodal/TGFβ signaling, including the Smad4b (Chang et al. 2006) and Oct25 transcription factors (Cao et al. 2008), and Coco (referred to as Dand5 in mammals), a secreted signaling antagonist (Bell et al. 2003; Vonica and Brivanlou 2007; Bates et al. 2013). Cripto-1 has also been implicated as a regulator of Wnt signaling (Kraus et al. 2012; Maisonneuve et al. 2009), and therefore it is interesting that some Bic-C target mRNAs, such as the AES/GRG5 transcription factor (Costa et al. 2013) and the Ddx5 RNA helicase (Guturi et al. 2014), encode proteins known to affect Wnt signaling.
These observations lead to a model in which Bic-C controls a network of mRNA and many mRNAs in the network encode embryonic cell-fate regulators (Fig. 2.6). This suggests that Bic-C acts as the control point of a posttranscriptional regulatory network that establishes the proper balance of maternal proteins in the embryo essential for normal development. Precedence for this kind of control by RNA-binding proteins has been documented in other systems (Hogan et al. 2008; Gerber et al. 2006; Ule and Darnell 2006; Licatalosi and Darnell 2010; Kershner and Kimble 2010). Future experiments will require a systematic examination of the functions of the Bic-C targets in the embryo with the goal of understanding how different amounts of repression establish the proper balance of cell-fate determinants throughout the embryo to drive normal development.
Fig. 2.6.
The Bic-C posttranscriptional network (Zhang et al. 2013). The vegetal cells of developing Xenopus embryos contain a high concentration of Bic-C that represses the translation of specific targets such as the AES, Cripto-1, and Ddx5 mRNAs. The protein products of Bic-C target mRNAs are potentially concentrated in animal cells due to their repression by Bic-C in vegetal cells
2.8 Summary: Maternal mRNA Regulation Establishes and Elaborates Molecular Asymmetries in the Embryo
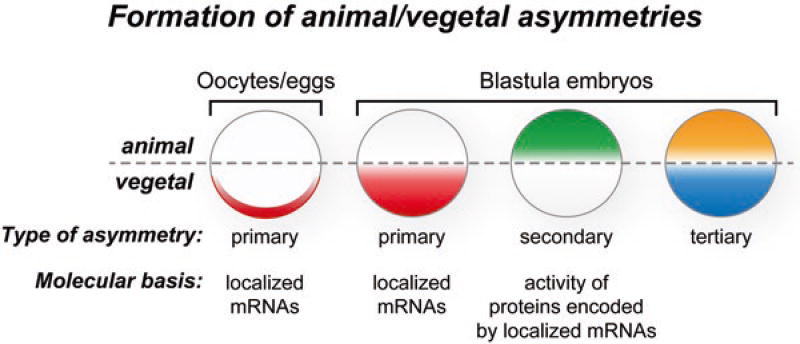
As discussed earlier, the primary animal-vegetal asymmetry in Xenopus is established early in embryogenesis by localizing mRNAs and proteins in oogenesis, oocyte maturation, and the early cleavage stages of embryos (Medioni et al. 2012; King et al. 2005; Houston 2012) (Fig. 2.7). The localization of these molecules may establish immediate molecular asymmetries, for example, if the molecule is an active protein or an mRNA that is translated immediately after localization. In addition, this early localization can serve as preparation for a molecular asymmetry that will not become evident until later in development, if, for instance, it involves an mRNA that is translationally activated only during later stages of development. In particular, localization of Bic-C mRNA to vegetal cells is predicted to lead to the accumulation of Bic-C protein in this region of the embryo that in turn represses mRNAs, such as Cripto-1, later in embryogenesis (Figs. 2.6 and 2.7). While the Bic-C protein asymmetry is predicted to run from the vegetal to animal hemisphere, the Cripto-1 protein asymmetry runs in the opposite direction (Dorey and Hill 2006) (Figs. 2.6 and 2.7). Given that Bic-C has a number of targets, it is plausible that Bic-C-dependent repression helps establish a number of distinct protein gradients that each differ based on intrinsic translational activation capacities, affinity for Bic-C, and other factors but share with the Cripto-1 protein the same basic directionality.
Fig. 2.7.
Model for the formation of asymmetries during maternal Xenopus development. During oogenesis specific mRNAs are localized to the vegetal cortex, while others are concentrated in the animal hemisphere. These mRNAs and potentially other components (e.g., proteins and metabolites) form the molecular basis of the animal-vegetal polarity of the fully grown stage 6 oocyte. The translation of some localized mRNAs into proteins during oocyte maturation creates additional animal-vegetal asymmetries present in unfertilized eggs. After fertilization the asymmetries of the egg are inherited by specific blastomeres to generate the initial animal-vegetal polarity of the embryo. During embryogenesis the activities of the animal and vegetal localized mRNAs and proteins create additional asymmetries. For example, the Bic-C repressor blocks translation of the Cripto-1 mRNA in vegetal cells, and as a result the Cripto-1 protein accumulated in animal cells. This is an example of a secondary asymmetry that results from the activity of the molecules localized in oocytes and eggs. In this way mRNA localization differences in the animal-vegetal hemispheres that begin in oocytes are carried into the embryo and elaborated upon both temporally and spatially into secondary and tertiary gradients
In this way, mRNA localization differences in the animal-vegetal hemispheres that begin in oocytes are carried into the embryo and reinforced and elaborated upon both temporally and spatially into secondary and tertiary gradients by combining translational repression and temporal modes of translation activation (Fig. 2.7). The animal-vegetal molecular differences are accompanied by molecular asymmetries that run perpendicular to this axis. In particular, during the first cell cycle following fertilization, cortical rotation serves to localize mRNAs and proteins toward one side of the embryo, establishing molecular asymmetries that produce the dorso/anterior-ventral/posterior axis (Gerhart et al. 1989; Houston 2012). The molecular differences formed by the animal-vegetal and dorso/anterior-ventral/posterior axes ultimately impinge upon the formation of Spemann’s organizer that in turn establishes new postzygotic molecular asymmetries (Gerhart et al. 1989, 1991; Harland and Gerhart 1997). While postzygotic embryonic development is complex and highly orchestrated and balanced, it all starts with direct and precise forms of molecular regulation involving the translational control of maternal mRNAs that are beginning to be understood.
2.9 Conclusion
Xenopus laevis has served as an important model for biological research for over 50 years, contributing to fundamental knowledge of cell cycle processes and vertebrate development (Gerhart and Keller 1986; Gurdon 1964, 1977, 1988, 2013; Brown 1967; Dawid 1965; Wu and Gerhart 1980; Scharf and Gerhart 1980; Gurdon et al. 1958; Elsdale et al. 1958). A major advantage of this model was and remains this organism’s ability to produce large number of eggs in response to a simple injection of hormone. Moreover, these eggs could be fertilized in petri dishes, producing populations of synchronously developing embryos for observation and experimentation. In addition the relatively large eggs and oocytes were ideally suited for molecular and biochemical analysis, including microinjection of defined molecules and the preparation of cellular extracts that could function in DNA replication, mRNA processing, chromosome segregation, and cell cycle oscillations (Murray et al. 1989; Murray and Kirschner 1989). These advantages have inspired generations of scientists to establish their own colonies of African clawed frogs and embrace the joy and frustrations of amphibian husbandry. While the quest to understand maternally controlled development and particularly the role of maternal mRNAs was and is not confined to Xenopus, the success of antisense approaches to target maternal mRNAs in this organism and subsequently follow development of the resulting “mutant” embryos and their control siblings suggests that Xenopus can continue to provide critical insights and indeed even lead the way toward a deeper molecular understanding of the maternal stages of animal development and the role of mRNA translational regulation (Olson et al. 2012; Hulstrand et al. 2010). Thus, from the intellectual perspective, it is a truly unique and exciting time for studying Xenopus maternal mRNAs and defining their roles in embryonic development. The future of maternal mRNA research in Xenopus is no longer limited by major technical challenges. As in many fascinating and important biological fields today, the major limitation this field faces is the availability of sufficient support and resources.
Acknowledgments
We thank Laura Vanderploeg for her graphic insights and preparing figures. Work in the Sheets lab is supported by NSF grant 1050395 and NIH grants (R21HD069345 and R21HD076828). Megan Dowdle is supported by a SciMed GRS Advanced Opportunity Fellowship through University of Wisconsin-Madison Graduate School and Biotechnology Training Program through the National Institute of General Medical Sciences of the National Institutes of Health (T32GM008349). Work in the Fox lab is supported by NIH grant (NIGMS R01GM56890).
References
- Amaya E, Musci TJ, Kirschner MW. Expression of a dominant negative mutant of the FGF receptor disrupts mesoderm formation in Xenopus embryos. Cell. 1991;66(2):257–270. doi: 10.1016/0092-8674(91)90616-7. http://www.ncbi.nlm.nih.gov/pubmed/1649700. [DOI] [PubMed] [Google Scholar]
- Arumugam K, Wang Y, Hardy LL, MacNicol MC, MacNicol AM. Enforcing temporal control of maternal mRNA translation during oocyte cell-cycle progression. EMBO J. 2010;29(2):387–397. doi: 10.1038/emboj.2009.337. http://www.ncbi.nlm.nih.gov/pubmed/19959990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam K, MacNicol MC, Wang Y, Cragle CE, Tackett AJ, Hardy LL, MacNicol AM. Ringo/cyclin-dependent kinase and mitogen-activated protein kinase signaling pathways regulate the activity of the cell fate determinant Musashi to promote cell cycle re-entry in Xenopus oocytes. J Biol Chem. 2012;287(13):10639–10649. doi: 10.1074/jbc.M111.300681. http://www.ncbi.nlm.nih.gov/pubmed/22215682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119(5):641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Bates TJ, Vonica A, Heasman J, Brivanlou AH, Bell E. Coco regulates dorsoventral specification of germ layers via inhibition of TGFbeta signalling. Development. 2013;140(20):4177–4181. doi: 10.1242/dev.095521. http://www.ncbi.nlm.nih.gov/pubmed/24026124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell E, Munoz-Sanjuan I, Altmann CR, Vonica A, Brivanlou AH. Cell fate specification and competence by Coco, a maternal BMP, TGFbeta and Wnt inhibitor. Development. 2003;130(7):1381–1389. doi: 10.1242/dev.00344. http://www.ncbi.nlm.nih.gov/pubmed/12588853. [DOI] [PubMed] [Google Scholar]
- Birsoy B, Kofron M, Schaible K, Wylie C, Heasman J. Vg 1 is an essential signaling molecule in Xenopus development. Development. 2006;133(1):15–20. doi: 10.1242/dev.02144. http://www.ncbi.nlm.nih.gov/pubmed/16308332. [DOI] [PubMed] [Google Scholar]
- Brown DD. The genes for ribosomal RNA and their transcription during amphibian development. Curr Top Dev Biol. 1967;2:47–73. doi: 10.1016/s0070-2153(08)60283-5. http://www.ncbi.nlm.nih.gov/pubmed/4943365. [DOI] [PubMed] [Google Scholar]
- Cao Y, Siegel D, Oswald F, Knochel W. Oct25 represses transcription of nodal/activin target genes by interaction with signal transducers during Xenopus gastrulation. J Biol Chem. 2008;283(49):34168–34177. doi: 10.1074/jbc.M803532200. http://www.ncbi.nlm.nih.gov/pubmed/18922797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha SW, McAdams M, Kormish J, Wylie C, Kofron M. Foxi2 is an animally localized maternal mRNA in Xenopus, and an activator of the zygotic ectoderm activator Foxi1e. PLoS One. 2012;7(7):e41782. doi: 10.1371/journal.pone.0041782. http://www.ncbi.nlm.nih.gov/pubmed/22848601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Brivanlou AH, Harland RM. Function of the two Xenopus smad4s in early frog development. J Biol Chem. 2006;281(41):30794–30803. doi: 10.1074/jbc.M607054200. http://www.ncbi.nlm.nih.gov/pubmed/16908518. [DOI] [PubMed] [Google Scholar]
- Charlesworth A, Wilczynska A, Thampi P, Cox LL, MacNicol AM. Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation. EMBO J. 2006;25(12):2792–2801. doi: 10.1038/sj.emboj.7601159. http://www.ncbi.nlm.nih.gov/pubmed/16763568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth A, Meijer HA, de Moor CH. Specificity factors in cytoplasmic polyadenylation. Wiley Interdiscip Rev RNA. 2013;4(4):437–461. doi: 10.1002/wrna.1171. http://www.ncbi.nlm.nih.gov/pubmed/23776146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colegrove-Otero LJ, Minshall N, Standart N. RNA-binding proteins in early development. Crit Rev Biochem Mol Biol. 2005a;40(1):21–73. doi: 10.1080/10409230590918612. [DOI] [PubMed] [Google Scholar]
- Colegrove-Otero LJ, Devaux A, Standart N. The Xenopus ELAV protein ElrB represses Vg1 mRNA translation during oogenesis. Mol Cell Biol. 2005b;25(20):9028–9039. doi: 10.1128/MCB.25.20.9028-9039.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell RA, Musci TJ, Kimelman D. FGF is a prospective competence factor for early activin-type signals in Xenopus mesoderm induction. Development. 1995;121(8):2429–2437. doi: 10.1242/dev.121.8.2429. [DOI] [PubMed] [Google Scholar]
- Costa AM, Pereira-Castro I, Ricardo E, Spencer F, Fisher S, da Costa LT. GRG5/AES interacts with T-cell factor 4 (TCF4) and downregulates Wnt signaling in human cells and zebrafish embryos. PLoS One. 2013;8(7):e67694. doi: 10.1371/journal.pone.0067694. http://www.ncbi.nlm.nih.gov/pubmed/23840876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cragle CE, MacNicol AM. In: From Oocyte to Fertilizable Egg, in Xenopus Development. Kloc M, ZKubiak J, editors. John Wiley & Sons, Inc; Oxford: 2014. [DOI] [Google Scholar]
- Cragle C, MacNicol AM. Musashi protein-directed translational activation of target mRNAs is mediated by the poly(A) polymerase, germ line development defective-2. J Biol Chem. 2014b;289(20):14239–14251. doi: 10.1074/jbc.M114.548271. http://www.ncbi.nlm.nih.gov/pubmed/24644291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid IB. Deoxyribonucleic acid in amphibian eggs. J Mol Biol. 1965;12(3):581–599. doi: 10.1016/s0022-2836(65)80313-8. http://www.ncbi.nlm.nih.gov/pubmed/5892910. [DOI] [PubMed] [Google Scholar]
- De Robertis EM. Spemann’s organizer and self-regulation in amphibian embryos. Nat Rev Mol Cell Biol. 2006;7(4):296–302. doi: 10.1038/nrm1855. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16482093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps S, Viel A, Garrigos M, Denis H, le Maire M. mRNP4, a major mRNA-binding protein from Xenopus oocytes is identical to transcription factor FRG Y2. J Biol Chem. 1992;267(20):13799–13802. http://www.ncbi.nlm.nih.gov/pubmed/1629179. [PubMed] [Google Scholar]
- Ding J, Yang L, Yan YT, Chen A, Desai N, Wynshaw-Boris A, Shen MM. Cripto is required for correct orientation of the anterior-posterior axis in the mouse embryo. Nature. 1998;395(6703):702–707. doi: 10.1038/27215. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9790191. [DOI] [PubMed] [Google Scholar]
- Dorey K, Amaya E. FGF signalling: diverse roles during early vertebrate embryogenesis. Development. 2010;137(22):3731–3742. doi: 10.1242/dev.037689. http://www.ncbi.nlm.nih.gov/pubmed/20978071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorey K, Hill CS. A novel Cripto-related protein reveals an essential role for EGF-CFCs in Nodal signalling in Xenopus embryos. Dev Biol. 2006;292(2):303–316. doi: 10.1016/j.ydbio.2006.01.006. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16497290. [DOI] [PubMed] [Google Scholar]
- Elsdale TR, Fischberg M, Smith S. A mutation that reduces nucleolar number in Xenopus laevis. Exp Cell Res. 1958;14(3):642–643. doi: 10.1016/0014-4827(58)90175-7. http://www.ncbi.nlm.nih.gov/pubmed/13562098. [DOI] [PubMed] [Google Scholar]
- Faure S, Lee MA, Keller T, ten Dijke P, Whitman M. Endogenous patterns of TGF beta superfamily signaling during early Xenopus development. Development. 2000;127(13):2917–2931. doi: 10.1242/dev.127.13.2917. [DOI] [PubMed] [Google Scholar]
- Fernandez-Miranda G, Mendez R. The CPEB-family of proteins, translational control in senescence and cancer. Ageing Res Rev. 2012;11(4):460–472. doi: 10.1016/j.arr.2012.03.004. http://www.ncbi.nlm.nih.gov/pubmed/22542725. [DOI] [PubMed] [Google Scholar]
- Flachsova M, Sindelka R, Kubista M. Single blastomere expression profiling of Xenopus laevis embryos of 8 to 32-cells reveals developmental asymmetry. Sci Rep. 2013;3:2278. doi: 10.1038/srep02278. http://www.ncbi.nlm.nih.gov/pubmed/23880666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox CA, Wickens M. Poly(A) removal during oocyte maturation: a default reaction selectively prevented by specific sequences in the 3′ UTR of certain maternal mRNAs. Genes Dev. 1990;4(12B):2287–2298. doi: 10.1101/gad.4.12b.2287. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=1980657. [DOI] [PubMed] [Google Scholar]
- Fox CA, Sheets MD, Wickens MP. Poly(A) addition during maturation of frog oocytes: distinct nuclear and cytoplasmic activities and regulation by the sequence UUUUUAU. Genes Dev. 1989;3(12B):2151–2162. doi: 10.1101/gad.3.12b.2151. [DOI] [PubMed] [Google Scholar]
- Frisch A, Wright CVE. Xbmprii, a novel xenopus type ii receptor mediating bmp signaling in embryonic tissues. Development. 1998;125(3):431–442. doi: 10.1242/dev.125.3.431. [DOI] [PubMed] [Google Scholar]
- Fritz BR, Sheets MD. Regulation of the mRNAs encoding proteins of the BMP signaling pathway during the maternal stages of Xenopus development. Dev Biol. 2001;236(1):230–243. doi: 10.1006/dbio.2001.0324. [DOI] [PubMed] [Google Scholar]
- Gamberi C, Lasko P. The bic-C family of developmental translational regulators. Comp Funct Genomics. 2012;2012:141386. doi: 10.1155/2012/141386. http://www.ncbi.nlm.nih.gov/pubmed/22611335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2006;103(12):4487–4492. doi: 10.1073/pnas.0509260103. http://www.ncbi.nlm.nih.gov/pubmed/16537387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart J, Keller R. Region-specific cell activities in amphibian gastrulation. Annu Rev Cell Biol. 1986;2:201–229. doi: 10.1146/annurev.cb.02.110186.001221. http://www.ncbi.nlm.nih.gov/pubmed/3548766. [DOI] [PubMed] [Google Scholar]
- Gerhart J, Danilchik M, Doniach T, Roberts S, Rowning B, Stewart R. Cortical rotation of the Xenopus egg: consequences for the anteroposterior pattern of embryonic dorsal development. Development. 1989;107(Suppl):37–51. doi: 10.1242/dev.107.Supplement.37. http://www.ncbi.nlm.nih.gov/pubmed/2699856. [DOI] [PubMed] [Google Scholar]
- Gerhart J, Doniach T, Stewart R. Organizing the Xenopus organizer. In: Keller RE, editor. Gastrulation. Plenum Press; New York: 1991. pp. 57–77. [Google Scholar]
- Goetz R, Mohammadi M. Exploring mechanisms of FGF signalling through the lens of structural biology. Nat Rev Mol Cell Biol. 2013;14(3):166–180. doi: 10.1038/nrm3528. http://www.ncbi.nlm.nih.gov/pubmed/23403721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good PJ. A conserved family of elav-like genes in vertebrates. Proc Natl Acad Sci U S A. 1995;92(10):4557–4561. doi: 10.1073/pnas.92.10.4557. http://www.ncbi.nlm.nih.gov/pubmed/7753842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff JM, Thies RS, Song JJ, Celeste AJ, Melton DA. Studies with a Xenopus BMP receptor suggest that ventral mesoderm-inducing signals override dorsal signals in vivo. Cell. 1994;79(1):169–179. doi: 10.1016/0092-8674(94)90409-x. [DOI] [PubMed] [Google Scholar]
- Graff JM, Bansal A, Melton DA. Xenopus Mad proteins transduce distinct subsets of signals for the TGF beta superfamily. Cell. 1996;85(4):479–487. doi: 10.1016/s0092-8674(00)81249-0. [DOI] [PubMed] [Google Scholar]
- Gray NK, Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- Gray NK, Coller JM, Dickson KS, Wickens M. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 2000;19(17):4723–4733. doi: 10.1093/emboj/19.17.4723. http://www.ncbi.nlm.nih.gov/pubmed/10970864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritsman K, Zhang J, Cheng S, Heckscher E, Talbot WS, Schier AF. The EGF-CFC protein one-eyed pinhead is essential for nodal signaling. Cell. 1999;97(1):121–132. doi: 10.1016/s0092-8674(00)80720-5. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10199408. [DOI] [PubMed] [Google Scholar]
- Groppo R, Richter JD. Translational control from head to tail. Curr Opin Cell Biol. 2009;21(3):444–451. doi: 10.1016/j.ceb.2009.01.011. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19285851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon JB. The transplantation of living cell nuclei. Adv Morphog. 1964;4:1–43. doi: 10.1016/b978-1-4831-9951-1.50004-8. http://www.ncbi.nlm.nih.gov/pubmed/5331922. [DOI] [PubMed] [Google Scholar]
- Gurdon JB. The croonian lecture, 1976. Egg cytoplasm and gene control in development. Proc R Soc Lond B Biol Sci. 1977;198(1132):211–247. doi: 10.1098/rspb.1977.0095. http://www.ncbi.nlm.nih.gov/pubmed/19752. [DOI] [PubMed] [Google Scholar]
- Gurdon JB. The origin of cell-type differences in early embryos. Cell Differ Dev. 1988;25(Suppl):1–6. doi: 10.1016/0922-3371(88)90092-5. http://www.ncbi.nlm.nih.gov/pubmed/3061583. [DOI] [PubMed] [Google Scholar]
- Gurdon JB. The egg and the nucleus: a battle for supremacy (Nobel Lecture) Angew Chem. 2013;52(52):13890–13899. doi: 10.1002/anie.201306722. http://www.ncbi.nlm.nih.gov/pubmed/24311340. [DOI] [PubMed] [Google Scholar]
- Gurdon JB, Elsdale TR, Fischberg M. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature. 1958;182(4627):64–65. doi: 10.1038/182064a0. http://www.ncbi.nlm.nih.gov/pubmed/13566187. [DOI] [PubMed] [Google Scholar]
- Guturi K, Sarkar M, Bhowmik A, Das N, Ghosh M. DEAD-box protein p68 is regulated by ss-catenin/transcription factor 4 to maintain a positive feedback loop in control of breast cancer progression. Breast Cancer Res. 2014;16(6):496. doi: 10.1186/s13058-014-0496-5. http://www.ncbi.nlm.nih.gov/pubmed/25499975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake LE, Richter JD. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell. 1994;79(4):617–627. doi: 10.1016/0092-8674(94)90547-9. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=7954828. [DOI] [PubMed] [Google Scholar]
- Harland R, Gerhart J. Formation and function of Spemann’s organizer. Annu Rev Cell Dev Biol. 1997;13:611–667. doi: 10.1146/annurev.cellbio.13.1.611. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9442883. [DOI] [PubMed] [Google Scholar]
- Hawley SH, Wunnenberg-Stapleton K, Hashimoto C, Laurent MN, Watabe T, Blumberg BW, Cho KW. Disruption of BMP signals in embryonic Xenopus ectoderm leads to direct neural induction. Genes Dev. 1995;9(23):2923–2935. doi: 10.1101/gad.9.23.2923. [DOI] [PubMed] [Google Scholar]
- Heasman J. Maternal determinants of embryonic cell fate. Semin Cell Dev Biol. 2006a;17(1):93–98. doi: 10.1016/j.semcdb.2005.11.005. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16426874. [DOI] [PubMed] [Google Scholar]
- Heasman J. Patterning the early Xenopus embryo. Development. 2006b;133(7):1205–1217. doi: 10.1242/dev.02304. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16527985. [DOI] [PubMed] [Google Scholar]
- Heasman J, Holwill S, Wylie CC. Fertilization of cultured Xenopus oocytes and use in studies of maternally A inherited molecules. Methods Cell Biol. 1991;36:213–230. doi: 10.1016/s0091-679x(08)60279-4. [DOI] [PubMed] [Google Scholar]
- Heasman J, Crawford A, Goldstone K, Garner-Hamrick P, Gumbiner B, McCrea P, Kintner C, Noro CY, Wylie C. Overexpression of cadherins and underexpression of beta-catenin inhibit dorsal mesoderm induction in early Xenopus embryos. Cell. 1994;79(5):791–803. doi: 10.1016/0092-8674(94)90069-8. http://www.ncbi.nlm.nih.gov/pubmed/7528101. [DOI] [PubMed] [Google Scholar]
- Hikasa H, Sokol SY. Wnt signaling in vertebrate axis specification. Cold Spring Harb Perspect Biol. 2013;5(1):a007955. doi: 10.1101/cshperspect.a007955. http://www.ncbi.nlm.nih.gov/pubmed/22914799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6(10):e255. doi: 10.1371/journal.pbio.0060255. http://www.ncbi.nlm.nih.gov/pubmed/18959479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingworth D, Candel AM, Nicastro G, Martin SR, Briata P, Gherzi R, Ramos A. KH domains with impaired nucleic acid binding as a tool for functional analysis. Nucleic Acids Res. 2012;40(14):6873–6886. doi: 10.1093/nar/gks368. http://www.ncbi.nlm.nih.gov/pubmed/22547390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston DW. Cortical rotation and messenger RNA localization in Xenopus axis formation. Wiley interdisciplinary reviews. Dev Biol. 2012;1(3):371–388. doi: 10.1002/wdev.29. http://www.ncbi.nlm.nih.gov/pubmed/23801488. [DOI] [PubMed] [Google Scholar]
- Houston DW. Regulation of cell polarity and RNA localization in vertebrate oocytes. Int Rev Cell Mol Biol. 2013;306:127–185. doi: 10.1016/B978-0-12-407694-5.00004-3. http://www.ncbi.nlm.nih.gov/pubmed/24016525. [DOI] [PubMed] [Google Scholar]
- Huang YY, Lu H, Liu S, Droz-Rosario R, Shen Z. Requirement of mouse BCCIP for neural development and progenitor proliferation. PLoS One. 2012;7(1):e30638. doi: 10.1371/journal.pone.0030638. http://www.ncbi.nlm.nih.gov/pubmed/22292003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulstrand AM, Schneider PN, Houston DW. The use of antisense oligonucleotides in Xenopus oocytes. Methods. 2010;51(1):75–81. doi: 10.1016/j.ymeth.2009.12.015. http://www.ncbi.nlm.nih.gov/pubmed/20045732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman LE, Wormington WM. Translational inactivation of ribosomal protein mRNAs during Xenopus oocyte maturation. Genes Dev. 1988;2(5):598–605. doi: 10.1101/gad.2.5.598. http://www.ncbi.nlm.nih.gov/pubmed/2454870. [DOI] [PubMed] [Google Scholar]
- Igea A, Mendez R. Meiosis requires a translational positive loop where CPEB1 ensues its replacement by CPEB4. EMBO J. 2010;29(13):2182–2193. doi: 10.1038/emboj.2010.111. http://www.ncbi.nlm.nih.gov/pubmed/20531391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivshina M, Lasko P, Richter JD. Cytoplasmic polyadenylation element binding proteins in development, health, and disease. Annu Rev Cell Dev Biol. 2014;30:393–415. doi: 10.1146/annurev-cellbio-101011-155831. http://www.ncbi.nlm.nih.gov/pubmed/25068488. [DOI] [PubMed] [Google Scholar]
- Jiang H, Shukla A, Wang X, Chen WY, Bernstein BE, Roeder RG. Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell. 2011;144(4):513–525. doi: 10.1016/j.cell.2011.01.020. http://www.ncbi.nlm.nih.gov/pubmed/21335234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershner AM, Kimble J. Genome-wide analysis of mRNA targets for Caenorhabditis elegans FBF, a conserved stem cell regulator. Proc Natl Acad Sci U S A. 2010;107(8):3936–3941. doi: 10.1073/pnas.1000495107. http://www.ncbi.nlm.nih.gov/pubmed/20142496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimelman D. Mesoderm induction: from caps to chips. Nat Rev Genet. 2006;7(5):360–372. doi: 10.1038/nrg1837. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16619051. [DOI] [PubMed] [Google Scholar]
- Kimelman D, Pyati UJ. Bmp signaling: turning a half into a whole. Cell. 2005;123(6):982–984. doi: 10.1016/j.cell.2005.11.028. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16360027. [DOI] [PubMed] [Google Scholar]
- King ML. In: Germ-Cell Specification in Xenopus, in Xenopus Development. Kloc M, Kubiak JZ, editors. John Wiley & Sons, Inc; Oxford: 2014. [DOI] [Google Scholar]
- King ML, Messitt TJ, Mowry KL. Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol Cell. 2005;97(1):19–33. doi: 10.1042/BC20040067. http://www.ncbi.nlm.nih.gov/pubmed/15601255. [DOI] [PubMed] [Google Scholar]
- Kinoshita N, Minshull J, Kirschner MW. The identification of two novel ligands of the FGF receptor by a yeast screening method and their activity in Xenopus development. Cell. 1995;83(4):621–630. doi: 10.1016/0092-8674(95)90102-7. http://www.ncbi.nlm.nih.gov/pubmed/7585965. [DOI] [PubMed] [Google Scholar]
- Klauzinska M, Castro NP, Rangel MC, Spike BT, Gray PC, Bertolette D, Cuttitta F, Salomon D. The multifaceted role of the embryonic gene Cripto-1 in cancer, stem cells and epithelial-mesenchymal transition. Semin Cancer Biol. 2014;29:51–58. doi: 10.1016/j.semcancer.2014.08.003. http://www.ncbi.nlm.nih.gov/pubmed/25153355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofron M, Klein P, Zhang F, Houston DW, Schaible K, Wylie C, Heasman J. The role of maternal axin in patterning the Xenopus embryo. Dev Biol. 2001;237(1):183–201. doi: 10.1006/dbio.2001.0371. http://www.ncbi.nlm.nih.gov/pubmed/11518515. [DOI] [PubMed] [Google Scholar]
- Kraus MR, Clauin S, Pfister Y, Di Maio M, Ulinski T, Constam D, Bellanne-Chantelot C, Grapin-Botton A. Two mutations in human BICC1 resulting in Wnt pathway hyperactivity associated with cystic renal dysplasia. Hum Mutat. 2012;33(1):86–90. doi: 10.1002/humu.21610. http://www.ncbi.nlm.nih.gov/pubmed/21922595. [DOI] [PubMed] [Google Scholar]
- Kwak JE, Wang L, Ballantyne S, Kimble J, Wickens M. Mammalian GLD-2 homologs are poly(A) polymerases. Proc Natl Acad Sci U S A. 2004;101(13):4407–4412. doi: 10.1073/pnas.0400779101. http://www.ncbi.nlm.nih.gov/pubmed/15070731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, King ML. Repressive translational control in germ cells. Mol Reprod Dev. 2013;80(8):665–676. doi: 10.1002/mrd.22161. http://www.ncbi.nlm.nih.gov/pubmed/23408501. [DOI] [PubMed] [Google Scholar]
- Lai F, Singh A, King ML. Xenopus Nanos1 is required to prevent endoderm gene expression and apoptosis in primordial germ cells. Development. 2012;139(8):1476–1486. doi: 10.1242/dev.079608. http://www.ncbi.nlm.nih.gov/pubmed/22399685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guellec R, Paris J, Couturier A, Roghi C, Philippe M. Cloning by differential screening of a Xenopus cDNA that encodes a kinesin-related protein. Mol Cell Biol. 1991;11(6):3395–3398. doi: 10.1128/mcb.11.6.3395. http://www.ncbi.nlm.nih.gov/pubmed/1710028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea R, Papalopulu N, Amaya E, Dorey K. Temporal and spatial expression of FGF ligands and receptors during Xenopus development. Dev Dyn. 2009;238(6):1467–1479. doi: 10.1002/dvdy.21913. http://www.ncbi.nlm.nih.gov/pubmed/19322767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MA, Heasman J, Whitman M. Timing of endogenous activin-like signals and regional specification of the Xenopus embryo. Development. 2001;128(15):2939–2952. doi: 10.1242/dev.128.15.2939. http://www.ncbi.nlm.nih.gov/pubmed/11532917. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11(1):75–87. doi: 10.1038/nrg2673. http://www.ncbi.nlm.nih.gov/pubmed/20019688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig KD, Kroll KL, Sun EE, Kirschner MW. Expression cloning of a Xenopus T-related gene (Xombi) involved in mesodermal patterning and blastopore lip formation. Development. 1996;122(12):4001–4012. doi: 10.1242/dev.122.12.4001. http://www.ncbi.nlm.nih.gov/pubmed/9012520. [DOI] [PubMed] [Google Scholar]
- MacNicol MC, MacNicol AM. Developmental timing of mRNA translation—integration of distinct regulatory elements. Mol Reprod Dev. 2010;77(8):662–669. doi: 10.1002/mrd.21191. http://www.ncbi.nlm.nih.gov/pubmed/20652998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeno M, Ong RC, Suzuki A, Ueno N, Kung HF. A truncated bone morphogenetic protein 4 receptor alters the fate of ventral mesoderm to dorsal mesoderm: roles of animal pole tissue in the development of ventral mesoderm [see comments] Proc Natl Acad Sci U S A. 1994;91(22):10260–10264. doi: 10.1073/pnas.91.22.10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve C, Guilleret I, Vick P, Weber T, Andre P, Beyer T, Blum M, Constam DB. Bicaudal C, a novel regulator of Dvl signaling abutting RNA-processing bodies, controls cilia orientation and leftward flow. Development. 2009;136(17):3019–3030. doi: 10.1242/dev.038174. http://www.ncbi.nlm.nih.gov/pubmed/19666828. [DOI] [PubMed] [Google Scholar]
- McGrew LL, Dworkin-Rastl E, Dworkin MB, Richter JD. Poly(A) elongation during Xenopus oocyte maturation is required for translational recruitment and is mediated by a short sequence element. Genes Dev. 1989;3(6):803–815. doi: 10.1101/gad.3.6.803. [DOI] [PubMed] [Google Scholar]
- Medioni C, Mowry K, Besse F. Principles and roles of mRNA localization in animal development. Development. 2012;139(18):3263–3276. doi: 10.1242/dev.078626. http://www.ncbi.nlm.nih.gov/pubmed/22912410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton DA. Translocation of a localized maternal mRNA to the vegetal pole of Xenopus oocytes. Nature. 1987;328(6125):80–82. doi: 10.1038/328080a0. [DOI] [PubMed] [Google Scholar]
- Mendez R, Murthy KG, Ryan K, Manley JL, Richter JD. Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol Cell. 2000;6(5):1253–1259. doi: 10.1016/s1097-2765(00)00121-0. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11106762. [DOI] [PubMed] [Google Scholar]
- Minshall N, Thom G, Standart N. A conserved role of a DEAD box helicase in mRNA masking. RNA. 2001;7(12):1728–1742. doi: 10.1017/s135583820101158x. http://www.ncbi.nlm.nih.gov/pubmed/11780630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshall N, Reiter MH, Weil D, Standart N. CPEB interacts with an ovary-specific eIF4E and 4E-T in early Xenopus oocytes. J Biol Chem. 2007;282(52):37389–37401. doi: 10.1074/jbc.M704629200. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17942399. [DOI] [PubMed] [Google Scholar]
- Mishina Y, Suzuki A, Ueno N, Behringer RR. Bmpr encodes a type i bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes Dev. 1995;9(24):3027–3037. doi: 10.1101/gad.9.24.3027. [DOI] [PubMed] [Google Scholar]
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309(5740):1514–1518. doi: 10.1126/science.1111443. http://www.ncbi.nlm.nih.gov/pubmed/16141059. [DOI] [PubMed] [Google Scholar]
- Moustakas A, Heldin CH. The regulation of TGFbeta signal transduction. Development. 2009;136(22):3699–3714. doi: 10.1242/dev.030338. http://www.ncbi.nlm.nih.gov/pubmed/19855013. [DOI] [PubMed] [Google Scholar]
- Murray AW, Kirschner MW. Cyclin synthesis drives the early embryonic cell cycle. Nature. 1989;339(6222):275–280. doi: 10.1038/339275a0. http://www.ncbi.nlm.nih.gov/pubmed/2566917. [DOI] [PubMed] [Google Scholar]
- Murray AW, Solomon MJ, Kirschner MW. The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature. 1989;339(6222):280–286. doi: 10.1038/339280a0. http://www.ncbi.nlm.nih.gov/pubmed/2566918. [DOI] [PubMed] [Google Scholar]
- Musci TJ, Amaya E, Kirschner MW. Regulation of the fibroblast growth factor receptor in early Xenopus embryos. Proc Natl Acad Sci U S A. 1990;87(21):8365–8369. doi: 10.1073/pnas.87.21.8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakel K, Hartung SA, Bonneau F, Eckmann CR, Conti E. Four KH domains of the C. elegans Bicaudal-C ortholog GLD-3 form a globular structural platform. RNA. 2010;16(11):2058–2067. doi: 10.1261/rna.2315010. http://www.ncbi.nlm.nih.gov/pubmed/20823118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New HV, Kavka AI, Smith JC, Green JB. Differential effects on Xenopus development of interference with type IIA and type IIB activin receptors. Mech Dev. 1997;61(1–2):175–186. doi: 10.1016/s0925-4773(96)00639-9. [DOI] [PubMed] [Google Scholar]
- Nishimatsu S, Oda S, Murakami K, Ueno N. Multiple genes for Xenopus activin receptor expressed during early embryogenesis. FEBS Lett. 1992a;303(1):81–84. doi: 10.1016/0014-5793(92)80482-v. [DOI] [PubMed] [Google Scholar]
- Nishimatsu S, Iwao M, Nagai T, Oda S, Suzuki A, Asashima M, Murakami K, Ueno N. A carboxyl-terminal truncated version of the activin receptor mediates activin signals in early Xenopus embryos. FEBS Lett. 1992b;312(2–3):169–173. doi: 10.1016/0014-5793(92)80928-a. [DOI] [PubMed] [Google Scholar]
- Novoa I, Gallego J, Ferreira PG, Mendez R. Mitotic cell-cycle progression is regulated by CPEB1 and CPEB4-dependent translational control. Nat Cell Biol. 2010;12(5):447–456. doi: 10.1038/ncb2046. http://www.ncbi.nlm.nih.gov/pubmed/20364142. [DOI] [PubMed] [Google Scholar]
- Olson DJ, Hulstrand AM, Houston DW. Maternal mRNA knock-down studies: antisense experiments using the host-transfer technique in Xenopus laevis and Xenopus tropicalis. Methods Mol Biol. 2012;917:167–182. doi: 10.1007/978-1-61779-992-1_10. http://www.ncbi.nlm.nih.gov/pubmed/22956088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero LJ, Devaux A, Standart N. A 250-nucleotide UA-rich element in the 3′ untranslated region of Xenopus laevis Vg1 mRNA represses translation both in vivo and in vitro. RNA. 2001;7(12):1753–1767. http://www.ncbi.nlm.nih.gov/pubmed/11780632. [PMC free article] [PubMed] [Google Scholar]
- Paillard L, Omilli F, Legagneux V, Bassez T, Maniey D, Osborne HB. EDEN and EDEN-BP, a cis element and an associated factor that mediate sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J. 1998;17(1):278–287. doi: 10.1093/emboj/17.1.278. http://www.ncbi.nlm.nih.gov/pubmed/9427761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris J, Philippe M. Poly(A) metabolism and polysomal recruitment of maternal mRNAs during early Xenopus development. Dev Biol (Orlando) 1990;140(1):221–224. doi: 10.1016/0012-1606(90)90070-y. [DOI] [PubMed] [Google Scholar]
- Paris J, Osborne HB, Couturier A, Le Guellec R, Philippe M. Changes in the polyadenylation of specific stable RNA during the early development of Xenopus laevis. Gene. 1988;72(1– 2):169–176. doi: 10.1016/0378-1119(88)90139-4. http://www.ncbi.nlm.nih.gov/pubmed/2468559. [DOI] [PubMed] [Google Scholar]
- Pique M, Lopez JM, Foissac S, Guigo R, Mendez R. A combinatorial code for CPE-mediated translational control. Cell. 2008;132(3):434–448. doi: 10.1016/j.cell.2007.12.038. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18267074. [DOI] [PubMed] [Google Scholar]
- Plouhinec JL, Zakin L, De Robertis EM. Systems control of BMP morphogen flow in vertebrate embryos. Curr Opin Genet Dev. 2011;21(6):696–703. doi: 10.1016/j.gde.2011.09.001. http://www.ncbi.nlm.nih.gov/pubmed/21937218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralle T, Gremmels D, Stick R. Translational control of nuclear lamin B1 mRNA during oogenesis and early development of Xenopus. Mech Dev. 1999;84(1–2):89–101. doi: 10.1016/s0925-4773(99)00078-7. http://www.ncbi.nlm.nih.gov/pubmed/10473123. [DOI] [PubMed] [Google Scholar]
- Ramel MC, Hill CS. Spatial regulation of BMP activity. FEBS Lett. 2012;586(14):1929–1941. doi: 10.1016/j.febslet.2012.02.035. http://www.ncbi.nlm.nih.gov/pubmed/22710177. [DOI] [PubMed] [Google Scholar]
- Ranjan M, Tafuri SR, Wolffe AP. Masking mRNA from translation in somatic cells. Genes Dev. 1993;7(9):1725–1736. doi: 10.1101/gad.7.9.1725. http://www.ncbi.nlm.nih.gov/pubmed/8370522. [DOI] [PubMed] [Google Scholar]
- Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007;32(6):279–285. doi: 10.1016/j.tibs.2007.04.004. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17481902. [DOI] [PubMed] [Google Scholar]
- Richter JD, Lasko P. Translational control in oocyte development. Cold Spring Harb Perspect Biol. 2011;3(9):a002758. doi: 10.1101/cshperspect.a002758. http://www.ncbi.nlm.nih.gov/pubmed/21690213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbie EP, Peterson M, Amaya E, Musci TJ. Temporal regulation of the Xenopus FGF receptor in development: a translation inhibitory element in the 3′ untranslated region. Development. 1995;121(6):1775–1785. doi: 10.1242/dev.121.6.1775. http://www.ncbi.nlm.nih.gov/pubmed/7600993. [DOI] [PubMed] [Google Scholar]
- Scharf SR, Gerhart JC. Determination of the dorsal-ventral axis in eggs of Xenopus laevis: complete rescue of uv-impaired eggs by oblique orientation before first cleavage. Dev Biol. 1980;79(1):181–198. doi: 10.1016/0012-1606(80)90082-2. http://www.ncbi.nlm.nih.gov/pubmed/7409319. [DOI] [PubMed] [Google Scholar]
- Schneider PN, Hulstrand AM, Houston DW. Fertilization of Xenopus oocytes using the host transfer method. J Vis Exp. 2010;(45) doi: 10.3791/1864. http://www.ncbi.nlm.nih.gov/pubmed/21085101. [DOI] [PMC free article] [PubMed]
- Schohl A, Fagotto F. Beta-catenin, MAPK and Smad signaling during early Xenopus development. Development. 2002;129(1):37–52. doi: 10.1242/dev.129.1.37. [DOI] [PubMed] [Google Scholar]
- Schroeder KE, Condic NL, Eisenberg LM, Yost HJ. Spatially regulated translation in embryos: asymmetric expression of maternal Wnt-11 along the dorsal-ventral axis in Xenopus. Dev Biol. 1999;214(2):288–297. doi: 10.1006/dbio.1999.9426. [DOI] [PubMed] [Google Scholar]
- Sheets MD, Fox CA, Hunt T, Vande Woude G, Wickens M. The 3′-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes Dev. 1994;8(8):926–938. doi: 10.1101/gad.8.8.926. [DOI] [PubMed] [Google Scholar]
- Sheets MD, Fritz B, Hartley RS, Zhang Y. Polyribosome analysis for investigating mRNA translation in Xenopus oocytes, eggs and embryos. Methods. 2010;51(1):152–156. doi: 10.1016/j.ymeth.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Richter JD. Further analysis of cytoplasmic polyadenylation in Xenopus embryos and identification of embryonic cytoplasmic polyadenylation element-binding proteins. Mol Cell Biol. 1994;14(12):7867–7875. doi: 10.1128/mcb.14.12.7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Tassan JP, Richter JD. Translational control by poly(A) elongation during Xenopus development: differential repression and enhancement by a novel cytoplasmic polyadenylation element. Genes Dev. 1992;6(12B):2580–2591. doi: 10.1101/gad.6.12b.2580. [DOI] [PubMed] [Google Scholar]
- Simon R, Wu L, Richter JD. Cytoplasmic polyadenylation of activin receptor mRNA and the control of pattern formation in Xenopus development. Dev Biol. 1996;179(1):239–250. doi: 10.1006/dbio.1996.0254. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8873767. [DOI] [PubMed] [Google Scholar]
- Smith JC. Forming and interpreting gradients in the early Xenopus embryo. Cold Spring Harb Perspect Biol. 2009;1(1):a002477. doi: 10.1101/cshperspect.a002477. http://www.ncbi.nlm.nih.gov/pubmed/20066079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souopgui J, Rust B, Vanhomwegen J, Heasman J, Henningfeld KA, Bellefroid E, Pieler T. The RNA-binding protein XSeb4R: a positive regulator of VegT mRNA stability and translation that is required for germ layer formation in Xenopus. Genes Dev. 2008;22(17):2347–2352. doi: 10.1101/gad.479808. http://www.ncbi.nlm.nih.gov/pubmed/18765788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standart N, Minshall N. Translational control in early development: CPEB, P-bodies and germinal granules. Biochem Soc Trans. 2008;36(Pt 4):671–676. doi: 10.1042/BST0360671. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18631138. [DOI] [PubMed] [Google Scholar]
- Stebbins-Boaz B, Cao Q, de Moor CH, Mendez R, Richter JD. Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol Cell. 1999;4(6):1017–1027. doi: 10.1016/s1097-2765(00)80230-0. http://www.ncbi.nlm.nih.gov/pubmed/10635326. [DOI] [PubMed] [Google Scholar]
- Stennard F, Carnac G, Gurdon JB. The Xenopus T-box gene, Antipodean, encodes a vegetally localised maternal mRNA and can trigger mesoderm formation. Development. 1996;122(12):4179–4188. doi: 10.1242/dev.122.12.4179. http://www.ncbi.nlm.nih.gov/pubmed/9012537. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Thies RS, Yamaji N, Song JJ, Wozney JM, Murakami K, Ueno N. A truncated bone morphogenetic protein receptor affects dorsal-ventral patterning in the early Xenopus embryo [see comments] Proc Natl Acad Sci U S A. 1994;91(22):10255–10259. doi: 10.1073/pnas.91.22.10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Kaneko E, Ueno N, Hemmati-Brivanlou A. Regulation of epidermal induction by BMP2 and BMP7 signaling. Dev Biol (Orlando) 1997;189(1):112–122. doi: 10.1006/dbio.1997.8652. [DOI] [PubMed] [Google Scholar]
- Tafuri SR, Wolffe AP. Selective recruitment of masked maternal mRNA from messenger ribonucleoprotein particles containing FRGY2 (mRNP4) J Biol Chem. 1993;268(32):24255–24261. http://www.ncbi.nlm.nih.gov/pubmed/8226972. [PubMed] [Google Scholar]
- Tanaka KJ, Ogawa K, Takagi M, Imamoto N, Matsumoto K, Tsujimoto M. RAP55, a cytoplasmic mRNP component, represses translation in Xenopus oocytes. J Biol Chem. 2006;281(52):40096–40106. doi: 10.1074/jbc.M609059200. http://www.ncbi.nlm.nih.gov/pubmed/17074753. [DOI] [PubMed] [Google Scholar]
- Tanaka KJ, Ogawa K, Takagi M, Imamoto N, Matsumoto K, Tsujimoto M. RAP55, a cytoplasmic mRNP component, represses translation in Xenopus oocytes. J Biol Chem. 2014;289(30):20490. doi: 10.1074/jbc.A114.060905. http://www.ncbi.nlm.nih.gov/pubmed/25063841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Q, Yokota C, Puck H, Kofron M, Birsoy B, Yan D, Asashima M, Wylie CC, Lin X, Heasman J. Maternal wnt11 activates the canonical wnt signaling pathway required for axis formation in Xenopus embryos. Cell. 2005;120(6):857–871. doi: 10.1016/j.cell.2005.01.013. http://www.ncbi.nlm.nih.gov/pubmed/15797385. [DOI] [PubMed] [Google Scholar]
- Teplova M, Malinina L, Darnell JC, Song J, Lu M, Abagyan R, Musunuru K, Teplov A, Burley SK, Darnell RB, Patel DJ. Protein-RNA and protein-protein recognition by dual KH1/2 domains of the neuronal splicing factor Nova-1. Structure. 2011;19(7):930–944. doi: 10.1016/j.str.2011.05.002. http://www.ncbi.nlm.nih.gov/pubmed/21742260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torpey N, Wylie CC, Heasman J. Function of maternal cytokeratin in Xenopus development. Nature. 1992;357(6377):413–415. doi: 10.1038/357413a0. [DOI] [PubMed] [Google Scholar]
- Ule J, Darnell RB. RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr Opin Neurobiol. 2006;16(1):102–110. doi: 10.1016/j.conb.2006.01.003. http://www.ncbi.nlm.nih.gov/pubmed/16418001. [DOI] [PubMed] [Google Scholar]
- Valverde R, Edwards L, Regan L. Structure and function of KH domains. FEBS J. 2008;275(11):2712–2726. doi: 10.1111/j.1742-4658.2008.06411.x. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18422648. [DOI] [PubMed] [Google Scholar]
- Vincent JP, Gerhart JC. Subcortical rotation in Xenopus eggs: an early step in embryonic axis specification. Dev Biol. 1987;123(2):526–539. doi: 10.1016/0012-1606(87)90411-8. [DOI] [PubMed] [Google Scholar]
- Vishnu MR, Sumaroka M, Klein PS, Liebhaber SA. The poly(rC)-binding protein alphaCP2 is a noncanonical factor in X. laevis cytoplasmic polyadenylation. RNA. 2011;17(5):944–956. doi: 10.1261/rna.2587411. http://www.ncbi.nlm.nih.gov/pubmed/21444632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonica A, Brivanlou AH. The left-right axis is regulated by the interplay of Coco, Xnr1 and derriere in Xenopus embryos. Dev Biol. 2007;303(1):281–294. doi: 10.1016/j.ydbio.2006.09.039. http://www.ncbi.nlm.nih.gov/pubmed/17239842. [DOI] [PubMed] [Google Scholar]
- Weeks DL, Melton DA. A maternal mRNA localized to the vegetal hemisphere in Xenopus eggs codes for a growth factor related to TGF-beta. Cell. 1987;51(5):861–867. doi: 10.1016/0092-8674(87)90109-7. [DOI] [PubMed] [Google Scholar]
- Weill L, Belloc E, Bava FA, Mendez R. Translational control by changes in poly(A) tail length: recycling mRNAs. Nat Struct Mol Biol. 2012;19(6):577–585. doi: 10.1038/nsmb.2311. http://www.ncbi.nlm.nih.gov/pubmed/22664985. [DOI] [PubMed] [Google Scholar]
- Wessely O, De Robertis EM. The Xenopus homologue of Bicaudal-C is a localized maternal mRNA that can induce endoderm formation. Development. 2000;127(10):2053–2062. doi: 10.1242/dev.127.10.2053. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10769230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JA, Heasman J. Maternal control of pattern formation in Xenopus laevis. J Exp Zool B Mol Dev Evol. 2008;310(1):73–84. doi: 10.1002/jez.b.21153. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17219372. [DOI] [PubMed] [Google Scholar]
- Wilhelm JE, Vale RD, Hegde RS. Coordinate control of translation and localization of Vg1 mRNA in Xenopus oocytes. Proc Natl Acad Sci U S A. 2000;97(24):13132–13137. doi: 10.1073/pnas.97.24.13132. http://www.ncbi.nlm.nih.gov/pubmed/11087864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Gerhart JC. Partial purification and characterization of the maturation-promoting factor from eggs of Xenopus laevis. Dev Biol. 1980;79(2):465–477. doi: 10.1016/0012-1606(80)90131-1. http://www.ncbi.nlm.nih.gov/pubmed/7000582. [DOI] [PubMed] [Google Scholar]
- Wu MY, Hill CS. Tgf-beta superfamily signaling in embryonic development and homeostasis. Dev Cell. 2009;16(3):329–343. doi: 10.1016/j.devcel.2009.02.012. http://www.ncbi.nlm.nih.gov/pubmed/19289080. [DOI] [PubMed] [Google Scholar]
- Yokota C, Kofron M, Zuck M, Houston DW, Isaacs H, Asashima M, Wylie CC, Heasman J. A novel role for a nodal-related protein; Xnr3 regulates convergent extension movements via the FGF receptor. Development. 2003;130(10):2199–2212. doi: 10.1242/dev.00434. http://www.ncbi.nlm.nih.gov/pubmed/12668633. [DOI] [PubMed] [Google Scholar]
- Zhang J, King ML. Xenopus VegT RNA is localized to the vegetal cortex during oogenesis and encodes a novel T-box transcription factor involved in mesodermal patterning. Development. 1996;122(12):4119–4129. doi: 10.1242/dev.122.12.4119. http://www.cob.org.uk/Development/122/12/dev3554.html. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9012531. [DOI] [PubMed] [Google Scholar]
- Zhang J, Houston DW, King ML, Payne C, Wylie C, Heasman J. The role of maternal VegT in establishing the primary germ layers in Xenopus embryos. Cell. 1998;94(4):515–524. doi: 10.1016/s0092-8674(00)81592-5. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9727494. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Forinash KD, McGivern J, Fritz B, Dorey K, Sheets MD. Spatially restricted translation of the xCR1 mRNA in Xenopus embryos. Mol Cell Biol. 2009;29(13):3791–3802. doi: 10.1128/MCB.01865-08. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19364820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cooke A, Park S, Dewey CN, Wickens M, Sheets MD. Bicaudal-C spatially controls translation of vertebrate maternal mRNAs. RNA. 2013;19(11) doi: 10.1261/rna.041665.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Park S, Blaser S, Sheets MD. Determinants of RNA binding and translational repression by the Bicaudal-C regulatory protein. J Biol Chem. 2014;289(11):7497–7504. doi: 10.1074/jbc.M113.526426. http://www.ncbi.nlm.nih.gov/pubmed/24478311. [DOI] [PMC free article] [PubMed] [Google Scholar]