Abstract
Transcripts of key enzymes in the Leloir pathway of galactose metabolism in mouse livers are significantly increased after chronic high-fat/high-sucrose feeding. UDP-galactose-4-epimerase (GALE) is the last enzyme in this pathway that converts UDP-galactose to UDP-glucose and was previously identified as a downstream target of the endoplasmic reticulum (ER) stress effector spliced X-box binding protein 1, suggesting an interesting cross talk between galactose and glucose metabolism in the context of hepatic ER stress and whole-body metabolic fitness. However, its specific role in glucose metabolism is not established. Using an inducible and tissue-specific mouse model, we report that hepatic overexpression of Gale increases gluconeogenesis from pyruvate and impairs glucose tolerance. Conversely, genetic reduction of Gale in liver improves glucose tolerance. Transcriptional profiling identifies trefoil factor 3 (Tff3) as one of the downstream targets of GALE. Restoration of Tff3 expression corrects glucose intolerance in Gale-overexpressing mice. These studies reveal a new link between hepatic GALE activity and whole-body glucose homeostasis via regulation of hepatic Tff3 expression.
Introduction
Hepatic endoplasmic reticulum (ER) stress is enhanced as a result of insulin resistance and diabetes (1–4). ER stress is also physiologically modulated by the availability of nutrients (5). One of the most widely studied ER stress effectors is spliced X-box binding protein 1 (Xbp1s), which we have previously demonstrated to regulate glucose metabolism, potentially via modulating UDP-galactose-4-epimerase (Gale) expression in the liver (5).
GALE is a bifunctional enzyme, catalyzing the reversible conversion of UDP-galactose (UDP-Gal) to UDP-glucose (UDP-Glc) as part of the Leloir pathway of galactose metabolism (6), and the formation of UDP-N-acetylgalactosamine (UDP-GalNAc) from UDP-N-acetylglucosamine (UDP-GlcNAc) in the presence of NAD+, an essential step in generating sugar moieties for glycoprotein synthesis (7).
Gale is dynamically regulated in various metabolic settings (8–10). We have shown that hepatic Gale expression is suppressed during fasting and gradually increased after refeeding, similar to Xbp1s (5,10). In this study, we found that GALE protein was upregulated after prolonged high-fat/high-sucrose (HFHS) diet exposure, so we set out to investigate the role of GALE in glucose homeostasis and to determine the downstream mediators of its metabolic impact. Hepatic Gale overexpression was achieved by engineering a transgenic (TG) mouse model utilizing the tetracycline-responsive element (TRE) transcriptional activation system, and Gale knocking down was achieved via a short hairpin RNA (shRNA) targeting Gale mRNA delivered specifically to the liver by engineered adeno-associated viruses (AAVs). Gale overexpression leads to impaired glucose homeostasis and insulin sensitivity, whereas a Gale knockdown ameliorates glucose intolerance and reduces serum cholesterol levels.
Our transcriptional profiling efforts identified trefoil factor 3 (Tff3) to be coordinately regulated by GALE. TFF3, also known as intestinal trefoil factor (ITF), is a stable secretory protein highly expressed in gastrointestinal mucosa (11–14). Its function is not well defined, but it is thought to protect the mucosa from insults by stabilizing the mucus layer and healing of the intestinal epithelium (13). Beyond acting locally, significant amounts of TFF3 protein are present in circulation. Recently, hepatic TFF3 protein has been suggested to be a mediator of glucose homeostasis in diabetic or obese mice (15,16).
Whereas we find Tff3 expression is acutely downregulated by galactose, it is not affected by lipids. This suggests that galactose-derived products of the GALE enzymatic reaction lead to inhibition of Tff3 expression. Overexpressing Tff3 in the liver using adeno-associated viral transduction (AAV) restores the glucose intolerance observed in Gale TG mice. Based on these observations, we hypothesize that the GALE-mediated galactose metabolic pathway regulates hepatic Tff3 expression, which in turn modulates whole-body glucose homeostasis.
Research Design and Methods
Animals
Animal care and experimental protocols were approved by the Institutional Animal Care and Use Committee of the UT Southwestern (UTSW) Medical Center. Liver-specific albumin-Cre TG (stock no. 003574) and Rosa26-loxP-STOP-loxP-rtTA TG (stock no. 005572) mice were purchased from The Jackson Laboratory. TRE-Gale mice were generated by subcloning the mouse Gale gene into the pTRE vector (Clontech Laboratories) with a rabbit β-globin 3′ untranslated region. Mice were randomly allocated to experimental groups, and weight matching was ensured at the beginning of experimental protocols. Investigators were not blinded to treatment groups during studies.
Metabolic Phenotyping
Glucose tolerance test (GTT), pyruvate tolerance test, insulin tolerance test (ITT), and triglyceride clearance test were performed as previously described (17–19). Liver triglyceride extraction and quantification were performed at the UTSW Metabolic Phenotyping Core using a previously published protocol (17). Liver glycogen was extracted in water and assayed with a protocol provided in the Glycogen Colorimetric/Fluorometric Assay Kit from BioVision (catalog no. K646-100). Serum parameters (albumin, bilirubin, aspartate transaminase [AST], alanine transaminase [ALT], cholesterol, triglyceride, VLDL, LDL, and direct HDL) were measured and calculated with a VITROS analyzer (Ortho Clinical Diagnostics).
RT-qPCR, RNA Sequencing, and Analysis
RNA was isolated from frozen tissues by homogenization in TRIzol Reagent (Invitrogen) with the manufacturer-provided protocol (20). RNA (1 µg) was used to transcribe cDNA using a reverse transcription kit (Bio-Rad). RT-qPCR primers were obtained from Harvard PrimerBank (https://pga.mgh.harvard.edu/primerbank/) (21). The mRNA levels were calculated using the comparative threshold cycle method, normalized to gene Rps16. Strand-specific mRNA sequencing and analysis were performed at Sequencing and Bioinformatics Core at the McDermott Center for Human Growth and Development with the standardized protocol (22).
Western Blotting
Protein extractions were performed as previously described (18,23). Primary antibodies phospho-Akt (Ser473) (catalog no. 4060; Cell Signaling Technology) and total Akt (catalog no. 2920; Cell Signaling Technology), GALE (catalog no. ab155997; Abcam), tubulin (catalog no. sc-53030; Santa Cruz Biotechnology), O-linked glycosylation (catalog no. ab2739; Abcam), and actin (catalog no. A4700; Sigma-Aldrich) were used at 1:1,000 dilutions and detected using a secondary immunoglobulin G labeled with infrared dyes emitting at 680 nm (catalog no. 926-68070 and 926-68076; LI-COR Bioscience) or 800 nm (catalog no. 926-32211; LI-COR Bioscience) (both at 1:10,000 dilutions) and then visualized on a LI-COR Odyssey infrared scanner (LI-COR Bioscience). The scanned data were analyzed using Odyssey version 3.0 software (LI-COR Bioscience).
TFF3 ELISA
Mouse serum TFF3 levels were measured using an ELISA kit from LSBio (catalog no. LS-F24055) according to the manufacturer-provided protocol.
Histology
Livers were excised and fixed overnight in 10% PBS-buffered formalin and were thereafter stored in 50% ethanol. Tissues were sectioned (5 μm), rehydrated, and stained at the UTSW Molecular Pathology Core.
AAV Treatment
AAVs (serotype 8) were custom packed at SignaGen Laboratories. The Gale shRNA sequence for AAV preparation is AAGAACTTGGACTTGCCGTAG. Gale shRNA was delivered by tail vein injection either at 5 E+10 or 1.0 E+11 genomic copies per mouse, and Tff3, Escherichia coli GALE (eGale), and Plesiomonas shigelloides UDP-N-acetylglucosamine 4-epimerase (WBgU) AAVs were delivered by tail vein injection at 1.0 E+11 genomic copies per mouse with proper controls. Mice were studied at least 4 weeks after AAV injection to achieve maximal expression.
Metabolite Measurement by Mass Spectrometry
UDP-Glc/UDP-Gal and UDP-GlcNAc/UDP-GalNAc ratio in frozen liver tissues were determined at the UTSW Metabolic Phenotyping Core using a Nexera X2 UHPLC system coupled to a Shimadzu LCMS-8060 triple quadrupole mass spectrometer operating the dual ion source in electrospray (negative mode).
Statistical Analysis
All values are expressed as the mean ± SEM. The significance between the mean values for each study was evaluated by Student t test for comparisons of two groups. ANOVA was used for comparisons of more than two groups. P ≤ 0.05 is regarded as statistically significant.
Results
Generation of Hepatocyte-Specific Gale TG Mice
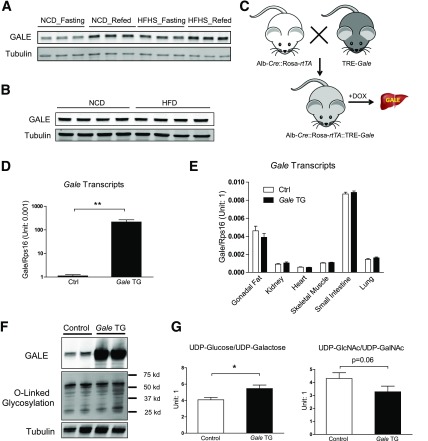
Gale expression is upregulated by refeeding (10) and prolonged HFHS diet feeding (Supplementary Fig. 1A), with short-term HFHS diet feeding trended to increase hepatic Gale levels (Supplementary Fig. 1B). GALE protein levels showed the same pattern with refeeding and prolonged HFHS feeding (Fig. 1A). In contrast, high-fat diet alone did not increase GALE protein levels (Fig. 1B), suggesting that the dietary carbohydrate component may be the dominant regulator for hepatic GALE induction.
Figure 1.
Generation of hepatocyte-specific Gale TG mice. A: GALE protein levels in livers from mice on NCD or HFHS diet with overnight fasting or 6-h refeeding after overnight fasting. B: GALE protein levels in livers from mice on NCD or HFD. C: Breeding schematic for Gale TG mice. D: Gale mRNA levels in Gale TG liver (n = 4 for each group). E: Gale mRNA levels in other tissues (n = 6 for each group). F: GALE protein levels and protein O-linked glycosylation levels in Gale TG liver. G: UDP-Glc/UDP-Gal and UDP-GlcNAc/UDP-GalNAc ratios in control and Gale TG livers 8 days after transgene induction (n = 9 and 8 for control and Gale TG livers, respectively). *P < 0.05; **P < 0.01, comparison by Student t test. All data are mean ± SEM. Ctrl, control; DOX, doxycycline.
In order to investigate whether hepatic GALE impacts glucose homeostasis, we generated TRE-Gale mice, which were bred with mice homozygous for Albumin-Cre and Rosa-flox-stop-flox-rtTA transgenes to achieve hepatocyte-specific Gale overexpression. In the resulting triple TG mice, liver-specific Gale transgene expression can be achieved by supplementation of doxycycline in the diet (600 mg ⋅ kg−1) (Fig. 1C–E). GALE protein levels were significantly increased under these conditions, whereas pan-O-linked protein glycosylation levels remained the same (Fig. 1F and Supplementary Fig. 1C and D). The ratio of hepatic UDP-Glc to UDP-Gal levels was increased in Gale TG livers, and the ratio of UDP-GlcNAc to UDP-GalNAc levels had a strong trend of being suppressed by Gale overexpression (Fig. 1G), suggesting that GALE is enzymatically active in our mouse model. Similar trends were observed in HFHS-induced obese mice (Supplementary Fig. 1E), in agreement with the observation of increased hepatic GALE levels in those mice. Importantly, hepatic Gale overexpression had no effect on weight gain of the mice on HFHS diet (Supplementary Fig. 1F).
Gale Overexpression Acutely Impairs Glucose Tolerance
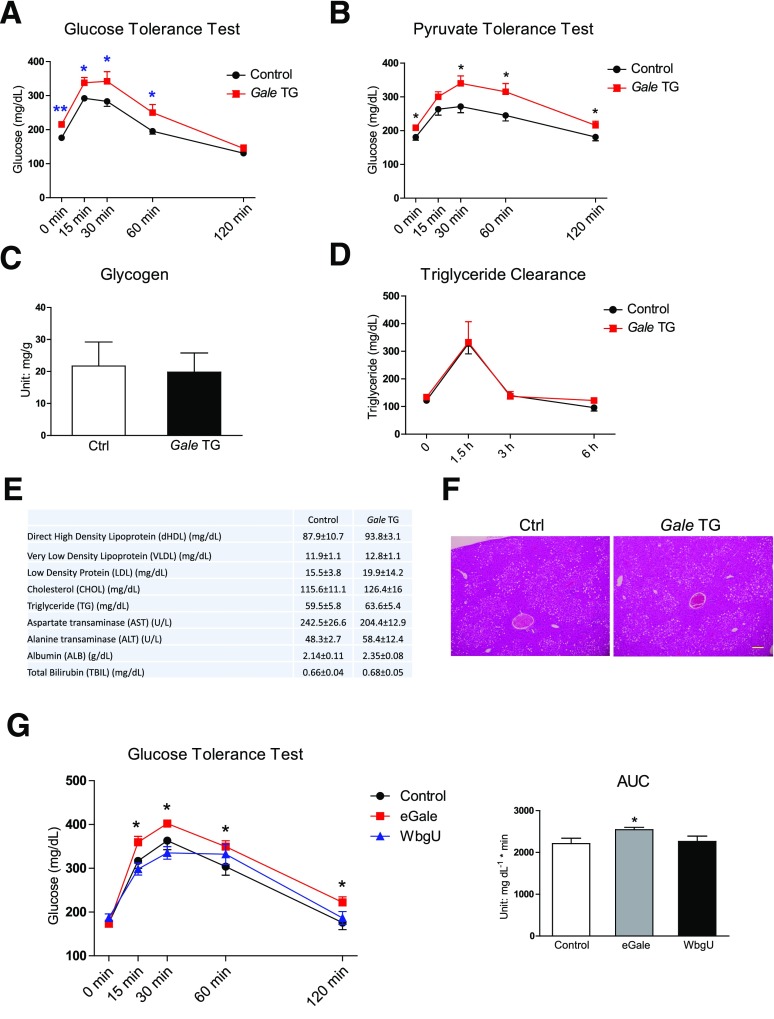
As short as 8 days after doxycycline supplementation, Gale TG mice maintained on normal chow diet (NCD) displayed a higher basal fasting glucose and an impaired glucose tolerance (Fig. 2A). This impairment was at least partially due to an increase in gluconeogenesis, evidenced by further elevated serum glucose levels after a pyruvate challenge (Fig. 2B). Similar impairments in glucose tolerance and pyruvate tolerance were observed in separate cohorts of mice that were fed HFHS diet for 8–9 weeks prior to Gale transgene induction for 8 days (Supplementary Fig. 2A and B), suggesting that hepatic Gale overexpression further worsened glycemic control in mice with preexisting obesity and insulin resistance. Gale TG mice had fasting insulin levels similar to control mice, pointing to the liver as the primary culprit for the impaired glucose homeostasis, independent of any β-cell dysfunction (Supplementary Fig. 2C). Gale TG livers had similar levels of gluconeogenic gene expression, except a slight increase in G6pc expression (Supplementary Fig. 2D). Short-term hepatic overexpression of Gale also had no effect on glycogen synthesis during refeeding after overnight fasting (Fig. 2C) or triglyceride clearance (Fig. 2D), nor any effects on serum lipid levels after overnight fasting (Fig. 2E and Supplementary Fig. 2E). Histological analysis also showed similar levels of lipid accumulation in Gale TG livers compared with control livers 3 weeks after transgene induction (Fig. 2F).
Figure 2.
Gale overexpression acutely impairs glucose tolerance. A: GTT of control and Gale TG mice 8 days after transgene induction on NCD (n = 9 and 8 for control and Gale TG groups, respectively). Glucose (1.25 g ⋅ kg−1) was intraperitoneally injected. B: Pyruvate tolerance test of overnight-fasted control and Gale TG mice 8 days after transgene induction on NCD (n = 10 and 8 for control and Gale TG groups, respectively). Sodium pyruvate (1 g ⋅ kg−1) was intraperitoneally injected. C: Liver glycogen content measured in control (Ctrl) and Gale TG mice 8 days after transgene induction on NCD. Mice were fasted overnight and then refed for 4 h (n = 7 and 5 for control and Gale TG groups, respectively). D: Serum triglycerides during an oral triglyceride tolerance test in control and Gale TG mice 3 weeks after transgene induction on HFHS diet (n = 13 and 6 for control and Gale TG mice, respectively). Oral gavage was used to administer 15 µL ⋅ g−1 body weight Intralipid (20%). E: Serum lipids and serum liver enzyme levels 8 days after transgene induction on NCD (n = 8 for both control and Gale TG groups). F: Representative hematoxylin-eosin staining of liver tissue, 3 weeks after transgene induction on HFHS diet. Scale bar = 50 µm. G: GTT of mice 4 weeks after eGale or WbgU AAV infection (n = 10 for each group). The area under the curve (AUC) is shown on the right. *P < 0.05; **P < 0.01, comparison by Student t test. All data are mean ± SEM.
GALE activities toward UDP-Gal and UDP-GalNAc have been shown to play different roles in Drosophila development (24). In fact, in bacteria, the two enzymatic activities are independently carried out by two separate enzymes: eGale that catalyzes the UDP-Gal and UDP-Glc interconversion and WbgU that catalyzes the UDP-GlcNAc and UDP-GalNAc interconversion (24). To dissect the contribution of each enzymatic activity of GALE to the changes in glucose metabolism observed in our mouse model, gene sequences for eGale and WbgU with optimized mammalian codon usage were each packaged into an AAV to achieve expression in liver. Similar to hepatic Gale overexpression, ectopic expression of eGale impaired glucose tolerance. In contrast, WbgU overexpression had no effect on glucose tolerance (Fig. 2G). This suggests that the impairment of glucose tolerance by hepatic overexpression of Gale is mediated by the portion of the enzymatic activity that is involved in the interconversion of UDP-Gal and UDP-Glc. The AAV-mediated expression was highly enriched in liver, with no detectable mRNA in white adipose tissue and very low levels in heart and skeletal muscle (Supplementary Fig. 2F). Overexpression of eGale increased the UDP-Glc/UDP-Gal ratio and had no effect on the UDP-GlcNAc/UDP-GalNAc ratio (Supplementary Fig. 2G); in contrast, overexpression of WbgU decreased the UDP-GlcNAc/UDP-GalNAc ratio but had no effect on the UDP-Glc/UDP-Gal ratio (Supplementary Fig. 2H). Altogether this suggests that eGale and WbgU were enzymatically active and functional in vivo.
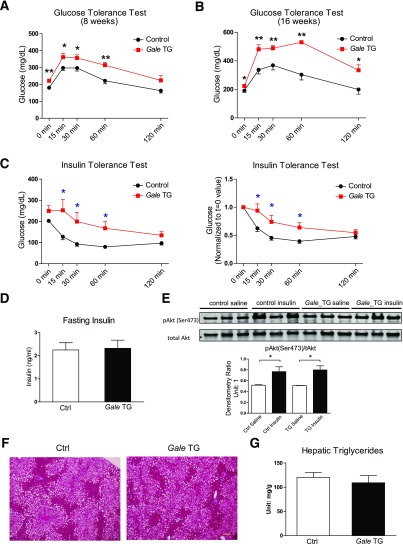
Chronic Gale Overexpression Impairs Glucose Tolerance and Results in Insulin Resistance
Sustained Gale overexpression induced by chronic doxycycline supplementation in HFHS-fed mice further impaired glucose tolerance (Fig. 3A and B) and whole-body insulin sensitivity (Fig. 3C), whereas fasting insulin was not affected (Fig. 3D). The same dose of insulin that was used in ITTs increased Akt Ser473 phosphorylation in control livers and Gale TG livers to a similar extent (Fig. 3E). Gale TG livers displayed similar histomorphology and triglyceride content compared with control livers (Fig. 3F and G). Interestingly, total liver wet weight normalized by body weight was increased after long-term Gale induction by 22%, and dry liver weight showed a similar trend of increase (Supplementary Fig. 3A and B).
Figure 3.
Chronic Gale overexpression impairs glucose tolerance and results in insulin resistance. A: GTT of control and Gale TG mice with transgene expression and HFHS diet challenge concurrently for 8 weeks (n = 9 and 8 for control and Gale TG groups, respectively). Glucose (0.75 g ⋅ kg−1) was intraperitoneally injected. B: GTT of mice previously fed with HFHS diet for 8 weeks and then 16 weeks of transgene induction with continued HFHS diet feeding (n = 5 for each group). Glucose (0.75 g ⋅ kg−1) was intraperitoneally injected. C: ITT in 4 h–fasted control and Gale TG mice after 16 weeks of HFHS diet feeding and concurrently 16 weeks of transgene induction (n = 9 and 8 for control and Gale TG groups, respectively). Insulin was intraperitoneally injected (2 units ⋅ kg−1 body weight). Left, original glucose reading; right, glucose readings were normalized to starting reading. D: Fasting insulin in control (Ctrl) and Gale TG mice after 16 weeks of HFHS diet feeding and 16 weeks of transgene induction (n = 4 for each group). E: Western blotting of insulin-stimulated phospho-Akt [pAkt(Ser473)] levels in livers from mice with 17 weeks HFHS diet feeding and transgene overexpression. Insulin was intraperitoneally injected (2 units ⋅ kg−1 body weight) before tissue harvest 15 min later. Quantification is shown below the blot (n = 3 for each group). F: Representative trichrome staining of liver tissues from mice after 22 weeks HFHS diet feeding and transgene induction. Scale bar = 50 µm. G: Hepatic triglyceride contents from mice with 22 weeks HFHS diet feeding and transgene induction (n = 8 for each group). *P < 0.05; **P < 0.01, comparison by Student t test. All data are mean ± SEM.
Knocking Down Hepatic Gale Expression Improves Glucose Homeostasis
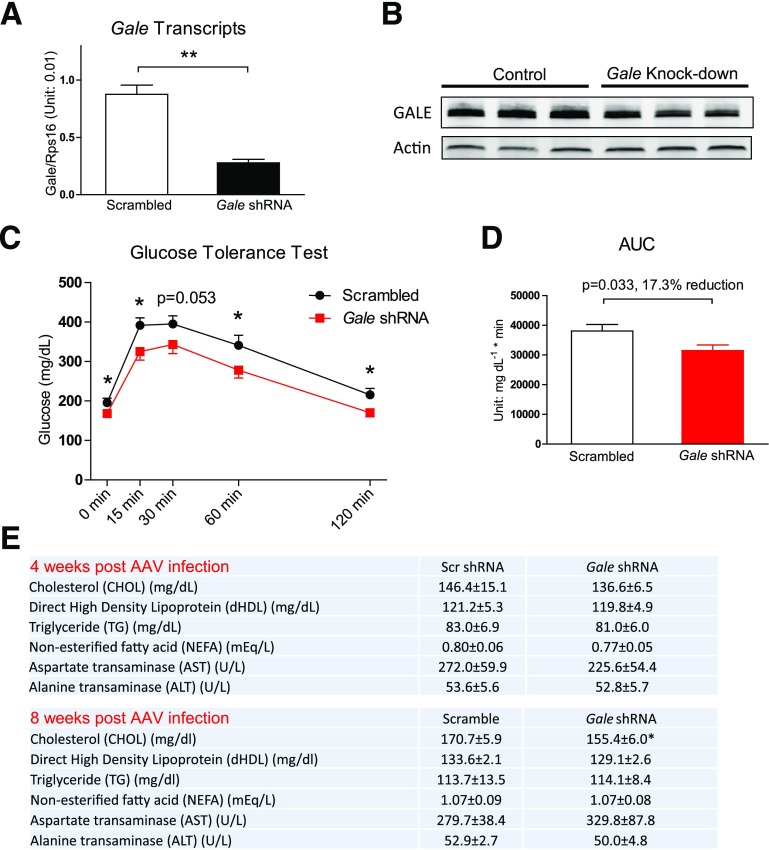
After establishing that hepatic overexpression of Gale impairs glucose metabolism, we sought to test whether genetic knocking down of Gale would protect or improve whole-body glucose homeostasis during nutrient overload.
An in vitro verified Gale shRNA sequence was placed under the control of the U6 promoter and packaged into AAV for the liver infection. Four weeks after AAV infection, there was a 68% reduction in Gale transcript levels (Fig. 4A) and a 56% reduction in GALE protein levels (Fig. 4B and Supplementary Fig. 4A). These viruses are specific to the liver, as AAV-infected mice did not show any reduction in Gale transcripts in six other tissues surveyed (Supplementary Fig. 4B). Four weeks after infection, GTTs were significantly improved in mice with hepatic Gale reduction (Fig. 4C and D). Serum lipid profiles were not altered 4 weeks after AAV infection, but 8 weeks after infection, total plasma cholesterol levels were significantly reduced in Gale shRNA AAV–infected mice (Fig. 4E). Serum AST or ALT levels were not affected by Gale knockdown, suggesting that the knockdown did not cause liver damage (Fig. 4E). Hepatic triglyceride levels were not different between control and Gale knockdown groups, although a large intragroup variability was noted (Supplementary Fig. 4C).
Figure 4.
Knocking down hepatic Gale improves glucose tolerance. A: Gale mRNA levels in the liver 4 weeks after Gale shRNA AAV infection (n = 4 for each group). B: Western blotting of liver samples 4 weeks after Gale shRNA AAV infection (n = 3 for each group). C: GTT of mice 4 weeks after HFHS diet feeding with scrambled shRNA or Gale shRNA AAV infection (n = 12 and 13 for scrambled and Gale shRNA groups, respectively). Glucose (1.25 g ⋅ kg−1) was intraperitoneally injected. D: The area under the curve (AUC) of GTT curves shown in C. E: Serum lipid profile and liver enzyme levels 4 weeks (n = 5 for both scrambled [scr] and Gale shRNA groups) or 8 weeks (n = 7 and 8 for scrambled and Gale shRNA groups, respectively) after scrambled or Gale shRNA AAV infection. *P < 0.05; **P < 0.01, comparison by Student t test. All data are mean ± SEM.
The same dose of AAV was used for a separate cohort of mice that were kept on HFHS for 8 weeks before the infection, and it only led to 25% reduction in Gale mRNA in the liver and had no effect on fasting glucose (Supplementary Fig. 4D). When the amount of AAV for infection was doubled, the knocking down efficiency increased to 52%, and it led to a 28.3 mg/dL reduction in fasting glucose 4 weeks after infection (Supplementary Fig. 4E).
Restoration of Hepatic Tff3 Expression Rescues Glucose Intolerance in Gale TG Mice
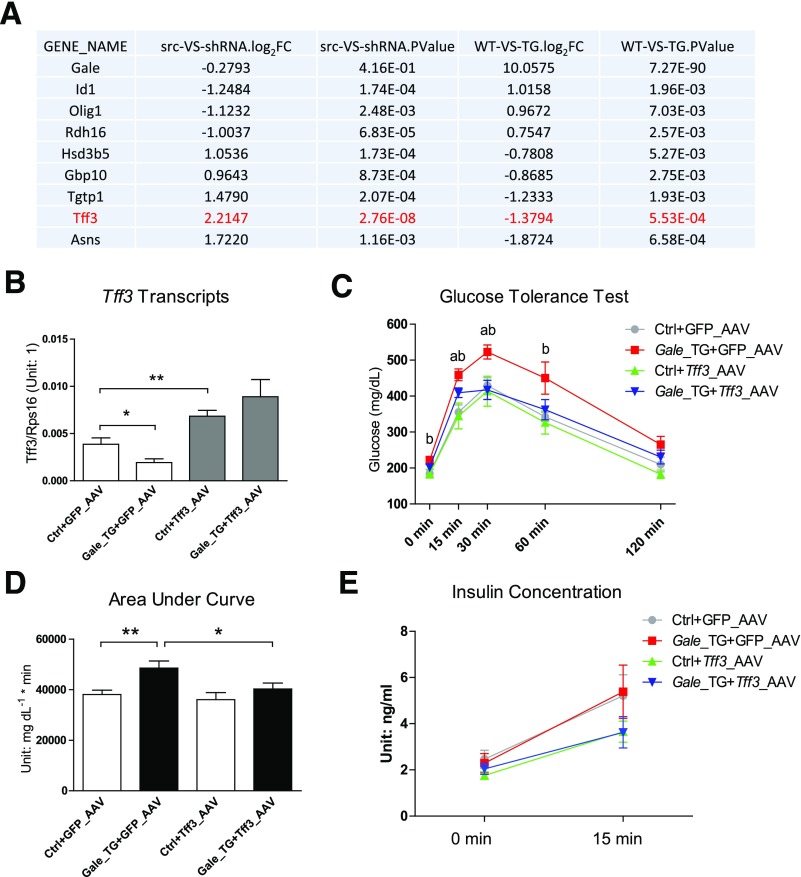
To determine the impact of GALE on gene expression and potential mechanisms by which GALE regulates glucose metabolism, we performed an RNA sequencing (RNA-Seq) experiment on livers from Gale TG overexpressing and Gale shRNA AAV–infected mice. Only eight genes showed significant changes in gene expression in opposite directions in both TG and knockdown livers (Fig. 5A). Among the genes coordinately regulated by GALE, Tff3 was suppressed in Gale-overexpressing livers and upregulated in Gale-knockdown livers. This Tff3 expression pattern was further supported by qPCR (Supplementary Fig. 5A). Given previous reports that hepatic Tff3 overexpression improves glucose metabolism in multiple mouse models of obesity or diabetes (15,16), we tested whether restoration of hepatic Tff3 expression could rescue glucose homeostasis in Gale TG mice. Mice infected with a Tff3-AAV showed a twofold increase in hepatic Tff3 mRNA levels compared with those infected with a control virus (GFP-AAV), and AAV-mediated Tff3 expression was not affected by the Gale transgene (Fig. 5B). Overexpression of Tff3 completely normalized glucose excursions in Gale TG mice, whereas it had no impact on glucose levels in control mice (Fig. 5C and D). This suggests that additional Tff3 transcripts above normal levels have no added benefits on global glucose metabolism. No impairment in basal or glucose-stimulated insulin secretion in Gale TG mice was observed (Fig. 5E). Basal and glucose-stimulated insulin levels in mice infected with Tff3 AAV showed a trend to be lower, independent of the Gale overexpression status of the mice (Fig. 5E). This is consistent with a previous report that showed that TFF3 could lower both basal and glucose-stimulated insulin levels (16). Serum TFF3 levels were in fact not altered by hepatic Gale overexpression or by Tff3 AAV infection (Supplementary Fig. 5B), suggesting that the rescue of glucose tolerance was mediated by a local effect of TFF3 in the liver. Tff3 overexpression did not affect serum triglyceride, nonesterified fatty acid, AST, or ALT levels (Supplementary Fig. 5C).
Figure 5.
Restoration of hepatic Tff3 expression reuses glucose intolerance in Gale TG mice. A: List of genes that are coordinated regulated by Gale overexpression and knockdown. The list is generated by setting logCPM [log(count per million)] >0 (meaningfully detected genes) and P < 0.01 (significantly changed) for both comparisons. Only genes that are oppositely regulated by Gale overexpression and knockdown are shown. Gale gene was not significantly reduced in knockdown comparison and is placed in the list as a reference. Log2FC, log2(fold change). src-VS-shRNA, scrambled versus shRNA (comparison of CPM of shRNA group over src group); WT-VS-TG, wild type versus transgenic (comparison of CPM of TG group over WT group). B: Tff3 mRNA levels in control (Ctrl) and Gale TG livers 4 weeks after Tff3 AAV infection (n = 8 for each group). C: GTT of control and Gale TG mice 4 weeks after Tff3 AAV infection (n = 7, 7, 8, and 6 for Ctrl+GFP_AAV, Gale_TG+GFP_AAV, Ctrl+Tff3_AAV, and Gale_TG+Tff3_AAV groups, respectively). Glucose (1.25 g ⋅ kg−1) was intraperitoneally injected. a, Gale_TG+GFP_AAV vs. Gale_TG+Tff3_AAV, P < 0.05; b, Ctrl+GFP_AAV vs. Gale_TG+GFP_AAV, P < 0.05. D: The area under the curve for GTT curves in C. E: Fasting insulin (4 h) and glucose-stimulated insulin for control and Gale TG mice 5 weeks after Tff3 AAV infection (n = 7, 7, 8, and 6 for Ctrl+GFP_AAV, Gale_TG+GFP_AAV, Ctrl+Tff3_AAV, and Gale_TG+Tff3_AAV groups, respectively), 15 min after oral administration of 1.25 g ⋅ kg−1 glucose solution. *P < 0.05; **P < 0.01, comparison by Student t test. All data are mean ± SEM.
Tff3 May Be Directly Regulated by Metabolites Derived via GALE
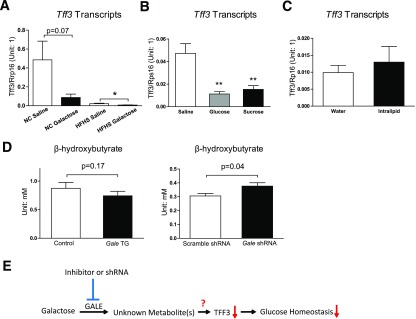
To test whether the hepatic Tff3 expression is acutely regulated by galactose, mice fed with NCD or HFHS diet for 12 weeks were challenged with 1 g ⋅ kg−1 body weight galactose or the same volume of saline by oral gavage, and their hepatic Tff3 expression levels were evaluated 1 h later. Galactose strongly reduced hepatic Tff3 expression in both NCD and HFHS mice, and HFHS mice had a much lower Tff3 expression level to begin with compared with NCD mice (Fig. 6A). However, this acute suppression of hepatic Tff3 was not specific to galactose; the same dose of glucose or sucrose suppressed Tff3 to a similar degree in mice fed on NCD (Fig. 6B). In contrast, a 15 µL ⋅ g−1 body weight oral Intralipid (20%) gavage had no effect on Tff3 expression (Fig. 6C). Importantly, none of these carbohydrate challenges altered hepatic Gale expression within this acute time scale, nor did Intralipid (Supplementary Fig. 6A and B). Altogether, these data suggest that dietary galactose and other carbohydrates, but not lipids, rapidly suppress hepatic Tff3 expression, presumably through metabolites derived from the GALE enzymatic activity. Serum β-hydroxybutyrate in Gale TG mice showed decreasing trends, indicating a decrease of fatty acid oxidation in the liver after Gale overexpression; in contrast, a significant increase in serum β-hydroxybutyrate was seen in Gale knockdown mice (Fig. 6E). Altogether these data suggest that galactose may enter glucose utilization pathways via GALE and generates some metabolites that subsequently inhibit fatty acid oxidation.
Figure 6.
Tff3 may be directly regulated by metabolites derived via GALE. A: Hepatic Tff3 mRNA levels in NCD and HFHS diet fed mice 1 h after 1 g ⋅ kg−1 galactose gavage (n = 4 for each group). B: Hepatic Tff3 mRNA levels in mice 1 h after 1 g ⋅ kg−1 body weight glucose or sucrose gavage (n = 8, 4, and 6 for saline, glucose, and sucrose groups, respectively). C: Hepatic Tff3 mRNA levels in mice 1 h after 15 μL ⋅ g−1 body weight Intralipid gavage (n = 8 for each group). D: Left, serum β-hydroxybutyrate levels in control and Gale TG mice (8 days after transgene induction); right, serum β-hydroxybutyrate levels in scrambled and Gale shRNA AAV–infected mice (4 weeks after AAV infection) (n = 8 and 7 for control and Gale TG mice and n = 7 and 8 for scramble shRNA and Gale shRNA groups, respectively). E: A model showing how GALE affects glucose homeostasis in the liver and how inhibition of GALE may improve glucose homeostasis. *P < 0.05; **P < 0.01, comparison by Student t test. All data are mean ± SEM.
Discussion
In the current study, a tissue-specific inducible system that allows bypassing the postnatal period (during which mouse pups greatly rely on lactose) was used to study the effects of the Leloir pathway enzyme GALE on glucose metabolism in the adult liver. We show that Gale is upregulated in the liver by overnutrition. Consistent with the notion that the elevation of endogenous hepatic GALE activity contributes to the diabetic phenotype, ectopic overexpression of Gale impairs glucose homeostasis, whereas genetic knocking down Gale protects mice from glucose intolerance. Although ectopic GALE protein level is supraphysiological, the magnitude of the changes in UDP-Glc/UDP-Gal and UDP-GlcNAC/UDP-GalNAC ratios after GALE overexpression is in line with what we observed in diet-induced obese mice, suggesting that our Gale TG mouse model in this study is still physiopathologically relevant. We then identify Tff3 as a downstream target of GALE, which is acutely regulated by galactose and other carbohydrates. Based on these observations, we propose that galactose-derived metabolites generated by GALE lead to an inhibition of hepatic Tff3 expression and thereby impair glucose homeostasis (Fig. 6E).
Mammalian GALE catalyzes two reactions, one interconverting UDP-Gal and UDP-Glc and the other interconverting UDP-GlcNAc and UDP-GalNAc. Interconversion of UDP-GlcNAc and UDP-GalNAc can generate UDP-GalNAc from more abundant UDP-GlcNAc for protein glycosylation. This process is especially important during a shortage of galactose. In a previous study, we demonstrated enhanced insulin sensitivity in primary hepatocytes overexpressing Gale and impaired insulin-stimulated Akt phosphorylation when Gale was knocked down. We suggested that the underlying mechanism relied on the modulation of protein (likely insulin receptor) glycosylation via generation of UDP-GalNAc through GALE. However, whether this mechanism can translate in vivo remained unclear. In this study, we presented three pieces of evidence: first, infection with WbgU AAV did not improve GTT (Fig. 2G); second, insulin-stimulated Akt phosphorylation in Gale TG livers was the same as in control mice (Fig. 3E); and third, and more importantly, levels of pan-O-linked glycosylation of proteins were not different between control and Gale TG livers (Fig. 1F). These data argue against the idea that enhanced GALE activity in vivo can improve insulin sensitivity via enhanced protein glycosylation in the liver. Based on the comparable level of impaired glucose tolerance in mice infected with bacterial Gale (eGale) (which only interconverts UDP-Gal and UDP-Glc) and Gale TG mice, we believe it is likely one or more metabolites acutely generated via GALE that suppresses Tff3 expression to mediate the metabolic phenotype of Gale overexpression. Given that very few genes were changed in our RNA-Seq experiment, it is more likely that this suppression is via direct modulation of transcriptional regulatory elements of the Tff3 gene. Since glucose and sucrose suppress Tff3 expression to a similar degree as galactose does, the metabolite that modulates Tff3 expression should originate from a general carbohydrate metabolism pathway, such as glycolysis or the tricarboxylic acid cycle, rather than a specific galactose metabolic pathway. Future studies will be needed to identify the specific metabolite that mediates Tff3 expression.
Overexpression of Gale in the liver mainly affects glucose metabolism, without altering hepatic lipid accumulation (Fig. 2F and Fig. 3F and G) or serum lipid levels (Fig. 2E and Supplementary Fig. 2E). The decrease in serum β-hydroxybutyrate is likely secondary to increased metabolites from glucose that are derived from galactose via GALE. This type of interaction between glucose metabolism and lipid metabolism was first described more than a half century ago by Randle et al. (25). Interestingly, TFF3 also mainly affects glucose metabolism, whereas its effects on lipid metabolism are minimal (16), consistent with the notion that TFF3 is a major effector downstream of GALE.
TFF3 is secreted and present in circulation (26), but whether it acts as an endocrine hormone is unknown. A 50-kDa membrane glycoprotein has been identified to bind to TFF3 by in situ binding, but whether it is a cell surface receptor for TFF3 needs further investigation (27). Alternatively, it is possible that TFF3 may work cell autonomously, especially in the liver. This aspect of TFF3 in the regulation of hepatic metabolism remains not well characterized (15,16) and warrants further analysis.
Beyond GALE, other enzymes involved in galactose metabolism are also upregulated by HFHS diet feeding (Supplementary Fig. 6C). This may reflect an interesting interaction between galactose metabolism and glucose metabolism in the context of the metabolic syndrome; most of the systemic galactose may simply be converted to glucose used as a fuel source. Nevertheless, whether a generalized partial inhibition of galactose metabolism improves glucose homeostasis and whether all the underlying mechanisms converge on Tff3 deserve further investigation. One of the concerns for complete inhibition of the galactose metabolic pathway, including GALE, is the potential to cause galactosemia (28–30). However, all galactosemia patients are either homozygous or compound heterozygous for mutations in GALT, GALK, or GALE genes within the Leloir pathway, and carriers who are heterozygotes for any of these autosomal recessive mutations are not at risk for developing symptoms of generalized galactosemia. This suggests a high tolerance toward a partial inhibition of the Leloir pathway. Importantly, epimerase deficiency galactosemia caused by mutations in the GALE gene is usually a mild form of galactosemia. So we believe that GALE-specific inhibitors that lead to only a partial inhibition of GALE could be safely implemented clinically and hold the potential to be a new class of drugs for controlling glucose levels in states of metabolic dysregulation.
Supplementary Material
Article Information
Acknowledgments. The authors thank Chen Zhang and Steven Connell (Touchstone Diabetes Center, UTSW Medical Center) for technical assistance, the UTSW Medical Center Molecular Pathology Core for assistance in histology, the UTSW Metabolic Phenotyping Core facility for assistance in metabolic phenotyping, and the Genomics and Microarray Core at McDermott Center for Human Growth and Development for assistance in RNA-Seq.
Funding. This study was supported by a Lilly Innovation Fellowship Award to Y.Z., a Fonds de Recherche du Québec–Santé postdoctoral fellowship to S.Z., American Heart Association postdoctoral fellowship 16POST26420136 to M.S., National Institutes of Health (NIH) grant R01-DK104789 to R.K.G., and National Institute of Diabetes and Digestive and Kidney Diseases grants R01-DK55758, R01-DK099110, P01-DK088761, and P01-AG051459 as well as a grant from the Cancer Prevention and Research Institute of Texas (RP140412) to P.E.S. R.G. was partially supported by a Core C grant (7685) under P01-DK088761. C.X. was partially supported by NIH grant UL1TR001105.
Duality of Interest. Y.Z., P.X., Y.L., H.C., B.B.Z., and H.V.L. are employees and may hold stocks of Eli Lilly and Company. This does not alter the authors' adherence to Diabetes policies on sharing data and materials. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. Y.Z. designed the studies, performed the research, interpreted the results, and wrote the manuscript. S.Z., R.G., A.L.G., M.S., F.Z., P.X., Y.L., H.C., and O.Z. helped to perform the research and interpret the results. Y.D. generated and verified TRE-Gale mouse. D.A.S., R.K.G., and B.B.Z. provided reagents and consultation to the project and reviewed the manuscript. C.X. performed RNA-seq analysis. H.V.L. and P.E.S. designed and supervised the study and reviewed and revised the manuscript. All authors approved the final version of the manuscript. H.V.L. and P.E.S. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db17-0323/-/DC1.
References
- 1.Boden G. Endoplasmic reticulum stress: another link between obesity and insulin resistance/inflammation? Diabetes 2009;58:518–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flamment M, Hajduch E, Ferré P, Foufelle F. New insights into ER stress-induced insulin resistance. Trends Endocrinol Metab 2012;23:381–390 [DOI] [PubMed] [Google Scholar]
- 3.Salvadó L, Palomer X, Barroso E, Vázquez-Carrera M. Targeting endoplasmic reticulum stress in insulin resistance. Trends Endocrinol Metab 2015;26:438–448 [DOI] [PubMed] [Google Scholar]
- 4.Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell 2010;140:900–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng Y, Wang ZV, Tao C, et al. The Xbp1s/GalE axis links ER stress to postprandial hepatic metabolism. J Clin Invest 2013;123:455–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holden HM, Rayment I, Thoden JB. Structure and function of enzymes of the Leloir pathway for galactose metabolism. J Biol Chem 2003;278:43885–43888 [DOI] [PubMed] [Google Scholar]
- 7.Thoden JB, Wohlers TM, Fridovich-Keil JL, Holden HM. Human UDP-galactose 4-epimerase. Accommodation of UDP-N-acetylglucosamine within the active site. J Biol Chem 2001;276:15131–15136 [DOI] [PubMed] [Google Scholar]
- 8.Renaud HJ, Cui JY, Lu H, Klaassen CD. Effect of diet on expression of genes involved in lipid metabolism, oxidative stress, and inflammation in mouse liver-insights into mechanisms of hepatic steatosis. PLoS One 2014;9:e88584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Somel M, Creely H, Franz H, et al. Human and chimpanzee gene expression differences replicated in mice fed different diets. PLoS One 2008;3:e1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang F, Xu X, Zhou B, He Z, Zhai Q. Gene expression profile change and associated physiological and pathological effects in mouse liver induced by fasting and refeeding. PLoS One 2011;6:e27553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitt H, Wundrack I, Beck S, et al. A third P-domain peptide gene (TFF3), human intestinal trefoil factor, maps to 21q22.3. Cytogenet Cell Genet 1996;72:299–302 [DOI] [PubMed] [Google Scholar]
- 12.Chinery R, Williamson J, Poulsom R. The gene encoding human intestinal trefoil factor (TFF3) is located on chromosome 21q22.3 clustered with other members of the trefoil peptide family. Genomics 1996;32:281–284 [DOI] [PubMed] [Google Scholar]
- 13.Podolsky DK. Mechanisms of regulatory peptide action in the gastrointestinal tract: trefoil peptides. J Gastroenterol 2000;35(Suppl. 12):69–74 [PubMed] [Google Scholar]
- 14.Wong WM, Poulsom R, Wright NA. Trefoil peptides. Gut 1999;44:890–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xue Y, Shen L, Cui Y, et al. Tff3, as a novel peptide, regulates hepatic glucose metabolism. PLoS One 2013;8:e75240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ge H, Gardner J, Wu X, et al. Trefoil factor 3 (TFF3) is regulated by food intake, improves glucose tolerance and induces mucinous metaplasia. PLoS One 2015;10:e0126924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Y, Gao Y, Tao C, et al. Connexin 43 mediates white adipose tissue beiging by facilitating the propagation of sympathetic neuronal signals. Cell Metab 2016;24:420–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morley TS, Xia JY, Scherer PE. Selective enhancement of insulin sensitivity in the mature adipocyte is sufficient for systemic metabolic improvements. Nat Commun 2015;6:7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang QA, Tao C, Jiang L, et al. Distinct regulatory mechanisms governing embryonic versus adult adipocyte maturation. Nat Cell Biol 2015;17:1099–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Y, Pereira RO, O’Neill BT, et al. Cardiac PI3K-Akt impairs insulin-stimulated glucose uptake independent of mTORC1 and GLUT4 translocation. Mol Endocrinol 2013;27:172–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Spandidos A, Wang H, Seed B. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res 2012;40:D1144–D1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park J, Kim M, Sun K, An YA, Gu X, Scherer PE. VEGF-A-expressing adipose tissue shows rapid beiging and enhanced survival after transplantation and confers IL4-independent metabolic improvements. Diabetes 2017;66:1479–1490 [DOI] [PMC free article] [PubMed]
- 24.Daenzer JM, Sanders RD, Hang D, Fridovich-Keil JL. UDP-galactose 4′-epimerase activities toward UDP-Gal and UDP-GalNAc play different roles in the development of Drosophila melanogaster. PLoS Genet 2012;8:e1002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Randle PJ, Garland PB, Hales CN, Newsholme EA. The glucose fatty-acid cycle. Its role in insulin sensitivity and the metabolic disturbances of diabetes mellitus. Lancet 1963;1:785–789 [DOI] [PubMed] [Google Scholar]
- 26.Liu SQ, Roberts D, Zhang B, Ren Y, Zhang LQ, Wu YH. Trefoil factor 3 as an endocrine neuroprotective factor from the liver in experimental cerebral ischemia/reperfusion injury. PLoS One 2013;8:e77732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan XD, Hsueh W, Chang H, Wei KR, Gonzalez-Crussi F. Characterization of a putative receptor for intestinal trefoil factor in rat small intestine: identification by in situ binding and ligand blotting. Biochem Biophys Res Commun 1997;237:673–677 [DOI] [PubMed] [Google Scholar]
- 28.Fridovich-Keil J, Bean L, He M, Schroer R. Epimerase deficiency galactosemia. In GeneReviews. Pagon RA, Adam MP, Ardinger HH, et al., Eds. Seattle, WA, University of Washington, 2011
- 29.Maceratesi P, Daude N, Dallapiccola B, et al. Human UDP-galactose 4′ epimerase (GALE) gene and identification of five missense mutations in patients with epimerase-deficiency galactosemia. Mol Genet Metab 1998;63:26–30 [DOI] [PubMed] [Google Scholar]
- 30.Pey AL, Padín-Gonzalez E, Mesa-Torres N, Timson DJ. The metastability of human UDP-galactose 4′-epimerase (GALE) is increased by variants associated with type III galactosemia but decreased by substrate and cofactor binding. Arch Biochem Biophys 2014;562:103–114 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.