Abstract
To characterize type 2 diabetes (T2D)-associated variation across the allele frequency spectrum, we conducted a meta-analysis of genome-wide association data from 26,676 T2D case and 132,532 control subjects of European ancestry after imputation using the 1000 Genomes multiethnic reference panel. Promising association signals were followed up in additional data sets (of 14,545 or 7,397 T2D case and 38,994 or 71,604 control subjects). We identified 13 novel T2D-associated loci (P < 5 × 10−8), including variants near the GLP2R, GIP, and HLA-DQA1 genes. Our analysis brought the total number of independent T2D associations to 128 distinct signals at 113 loci. Despite substantially increased sample size and more complete coverage of low-frequency variation, all novel associations were driven by common single nucleotide variants. Credible sets of potentially causal variants were generally larger than those based on imputation with earlier reference panels, consistent with resolution of causal signals to common risk haplotypes. Stratification of T2D-associated loci based on T2D-related quantitative trait associations revealed tissue-specific enrichment of regulatory annotations in pancreatic islet enhancers for loci influencing insulin secretion and in adipocytes, monocytes, and hepatocytes for insulin action–associated loci. These findings highlight the predominant role played by common variants of modest effect and the diversity of biological mechanisms influencing T2D pathophysiology.
Introduction
Type 2 diabetes (T2D) has rapidly increased in prevalence in recent years and represents a major component of the global disease burden (1). Previous efforts to use genome-wide association studies (GWAS) to characterize the genetic component of T2D risk have largely focused on common variants (minor allele frequency [MAF] >5%). These studies have identified close to 100 loci, almost all of them currently defined by common alleles associated with modest (typically 5–20%) increases in T2D risk (2–6). Direct sequencing of whole genomes or exomes offers the most comprehensive approach for extending discovery efforts to the detection of low-frequency (0.5% < MAF < 5%) and rare (MAF <0.5%) risk and protective alleles, some of which might have greater impact on individual predisposition. However, extensive sequencing has thus far been limited to relatively small sample sizes (at most, a few thousand cases), restricting power to detect rarer risk alleles even if they are of large effect (7–9). Although evidence of rare variant associations has been detected in some candidate gene studies (10,11), the largest study to date, involving exome sequencing in ∼13,000 subjects, found little trace of rare variant association effects (9).
Here, we implement a complementary strategy that makes use of imputation into existing GWAS samples from the DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium with sequence-based reference panels (12). This strategy allows the detection of common and low-frequency (but not rare) variant associations in extremely large samples (13) and facilitates the fine-mapping of causal variants. We performed a European ancestry meta-analysis of GWAS with 26,676 T2D case and 132,532 control subjects, and we followed up our findings in additional independent European ancestry studies of 14,545 T2D case and 38,994 control subjects genotyped using the Metabochip (4). All contributing studies were imputed against the March 2012 multiethnic 1000 Genomes Project (1000G) reference panel of 1,092 whole-genome–sequenced individuals (12). Our study provides near-complete evaluation of common variants with much improved coverage of low-frequency variants, and the combined sample size considerably exceeds that of the largest previous T2D GWAS meta-analyses in individuals of European ancestry (4). In addition to genetic discovery, we fine-mapped novel and established T2D-associated loci to identify regulatory motifs and cell types enriched for potential causal variants, as well as pathways through which T2D-associated loci increase disease susceptibility.
Research Design and Methods
Research Participants
The DIAGRAM stage 1 meta-analyses comprises 26,676 T2D case and 132,532 control subjects (effective sample size Neff = 72,143 individuals, defined as 4/[(1/Ncases) + (1/Ncontrols)]) from 18 studies genotyped using commercial genome-wide single nucleotide variant (SNV) arrays (Supplementary Table 1). The Metabochip stage 2 follow-up comprises 14,545 T2D case and 38,994 control subjects (Neff = 38,645) from 16 nonoverlapping stage 1 studies (4,14). We performed additional follow-up in 2,796 T2D case and 4,601 control subjects from the European Prospective Investigation into Cancer and Nutrition-InterAct (EPIC-InterAct) study (15) and in 9,747 T2D case and 61,857 control subjects from the Resource for Genetic Epidemiology on Adult Health and Aging (GERA) study (16) (Supplementary Material).
Statistical Analyses
We imputed autosomal and X chromosome SNVs using the all-ancestries 1000G reference panel (1,092 individuals from Africa, Asia, Europe, and the Americas [March 2012 release]) using minimac (17) or IMPUTE2 (18). After imputation, from each study we removed monomorphic variants or those with imputation quality r2-hat < 0.3 (minimac) or proper-info <0.4 (IMPUTE2, SNPTEST). Each study performed T2D association analysis using logistic regression, adjusting for age, sex, and principal components for ancestry, under an additive genetic model. We performed inverse-variance weighted fixed-effect meta-analyses of the 18 stage 1 GWAS (Supplementary Table 1). Fifteen of the 18 studies repeated analyses also adjusting for BMI. SNVs reaching suggestive significance P < 10−5 in the stage 1 meta-analysis were followed up. Novel loci were selected using the threshold for genome-wide significance (P < 5 × 10−8) in the combined stage 1 and stage 2 meta-analysis. For the 23 variants with no proxy (r2 ≥ 0.6) available in Metabochip with 1000G imputation in the fine-mapping regions, the stage 1 result was followed up in EPIC-InterAct and GERA (Neff = 40,637), both imputed to 1000G variant density (Supplementary Material). Summary-level statistics from the stage 1 GWAS meta-analysis are available online at http://diagram-consortium.org/downloads.html.
Approximate Conditional Analysis With GCTA
We performed approximate conditional analysis in the stage 1 sample using GCTA v1.24 (19,20). We analyzed SNVs in the 1-Mb window around each lead variant, conditioning on the lead SNV at each locus (Supplementary Material) (21). We considered loci to contain multiple distinct signals if multiple SNVs reached locus-wide significance (P < 10−5), accounting for the approximate number of variants in each 1-Mb window (14).
Fine-Mapping Analyses Using Credible Set Mapping
To identify 99% credible sets of causal variants for each distinct association signal, we performed fine-mapping for loci at which the lead independent SNV reached P < 5 × 10−4 in the stage 1 meta-analysis. We performed credible set mapping using the T2D stage 1 meta-analysis results to obtain the minimal set of SNVs with cumulative posterior probability >0.99 (Supplementary Material).
Type 1 Diabetes/T2D Discrimination Analysis
Given the overlap between loci previously associated with type 1 diabetes (T1D) and the associated T2D loci, we used an inverse-variance weighted Mendelian randomization approach (22) to test whether this was likely to reflect misclassification of T1D case subjects as individuals with T2D in the current study (Supplementary Material).
Expression Quantitative Trait Locus Analysis
To look for potential biological overlap of T2D lead variants and expression quantitative trait locus (eQTL) variants, we extracted the lead (most significantly associated) eQTL for each tested gene from existing data sets for a range of tissues (Supplementary Material). We concluded that a lead T2D SNV showed evidence of association with gene expression if it was in high linkage disequilibrium (LD) (r2 > 0.8) with the lead eQTL SNV (P < 5 × 10−6).
Hierarchical Clustering of T2D-Related Metabolic Phenotypes
Starting with the T2D-associated SNVs, we obtained T2D-related quantitative trait z scores from published HapMap-based GWAS meta-analysis for the following: fasting glucose, fasting insulin adjusted for BMI, HOMA for β-cell function, and HOMA for insulin resistance (23); 2-h glucose adjusted for BMI (24); proinsulin (25); corrected insulin response (CIR) (26); BMI (27); and HDL cholesterol, LDL cholesterol, total cholesterol, and triglycerides (28). When an association result for an SNV was not available, we used the results for the variant in highest LD and only for variants with r2 > 0.6. We performed clustering of phenotypic effects using z scores for association with T2D risk alleles and standard methods (Supplementary Material) (29).
Functional Annotation and Enrichment Analysis
We tested for enrichment of genomic and epigenomic annotations using chromatin states for 93 cell types (after excluding cancer cell lines) from the National Institutes of Health (NIH) Roadmap Epigenomics Project, as well as binding sites for 165 transcription factors from the Encyclopedia of DNA Elements (ENCODE) project (30) and Pasquali et al. (31). Using fractional logistic regression, we then tested for the effect of variants with each cell type and transcription factor annotation on the variant posterior probabilities (πc) using all variants within 1 Mb of the lead SNV for each distinct association signal from the fine-mapping analyses (Supplementary Material). In each analysis, we considered an annotation significant if it reached a Bonferroni-corrected P < 1.9 × 10−4 (i.e., 0.05/258 annotations).
Pathway Analyses With DEPICT
We used the Data-driven Expression Prioritized Integration for Complex Traits (DEPICT) tool (32) to 1) prioritize genes that may represent promising candidates for T2D pathophysiology and 2) identify reconstituted gene sets that are enriched in genes from associated regions and might be related to T2D biological pathways. As input, we used independent SNVs from the stage 1 meta-analysis SNVs with P < 10−5 and lead variants at established loci (Supplementary Material). For the calculation of empirical enrichment P values, we used 200 sets of SNVs randomly drawn from entire genome within regions matching by gene density; we performed 20 replications for false discovery rate (FDR) estimation. Supplementary tables, supplementary material, and DEPICT analyses are available online at http://diagram-consortium.org/2017_Scott_DIAGRAM_1000G/.
Results
Novel Loci Detected in T2D GWAS and Metabochip-Based Follow-up
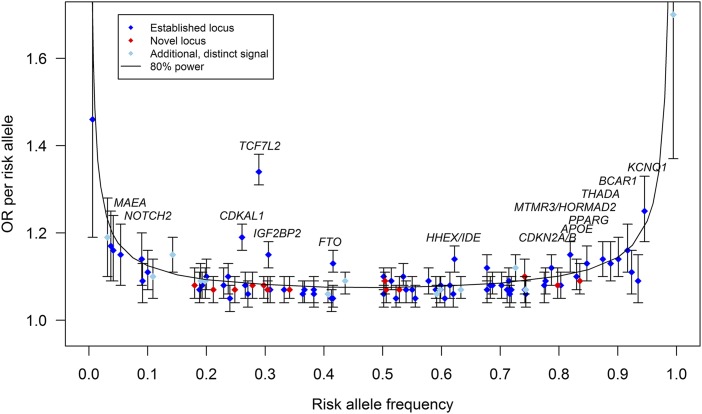
The stage 1 GWAS meta-analysis included 26,676 T2D case and 132,532 control subjects and evaluated 12.1 million SNVs, of which 11.8 million were autosomal and 260,000 mapped to the X chromosome. Of these, 3.9 million variants had MAF between 0.5 and 5%, a near fifteen-fold increase in the number of low-frequency variants tested for association compared with previous array-based T2D GWAS meta-analyses (2,4) (Supplementary Table 2). Of the 52 signals showing promising evidence of association (P < 10−5) in stage 1, 29 could be followed up in the stage 2 Metabochip data. In combined stage 1 and stage 2 data, 13 novel loci were detected at genome-wide significance (Table 1, Fig. 1, Supplementary Fig. 1A–D, and Supplementary Table 3).
Table 1.
Novel loci associated with T2D from the combination of 1000G-imputed GWAS meta-analysis (stage 1) and Metabochip follow-up (stage 2)
| Locus name* | Stage 1 |
Stage 2 |
Stage 1 + stage 2 |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr:position | SNV† | EA/ NEA | EAF | OR (95% CI) | P value | Chr:position | SNV‡ | r2 with lead SNV | EA/ NEA | EAF | OR (95% CI) | P value | OR (95% CI)¢ | P value | |
| ACSL1 | 4:185708807 | rs60780116 | T/C | 0.84 | 1.09 (1.06–1.13) | 7.38 × 10−8 | 4:185714289 | rs1996546 | 0.62 | G/T | 0.86 | 1.08 (1.03–1.13) | 5.60 × 10−4 | 1.09 (1.06–1.12) | 1.98 × 10−10 |
| HLA-DQA1 | 6:32594309 | rs9271774 | C/A | 0.74 | 1.10 (1.06–1.14) | 3.30 × 10−7 | 6:32594328 | rs9271775 | 0.91 | T/C | 0.80 | 1.08 (1.03–1.13) | 7.59 × 10−4 | 1.09 (1.06–1.12) | 1.11 × 10−9 |
| SLC35D3 | 6:137287702 | rs6918311 | A/G | 0.53 | 1.07 (1.04–1.10) | 6.67 × 10−7 | 6:137299152 | rs4407733 | 0.92 | A/G | 0.52 | 1.05 (1.02–1.08) | 1.63 × 10−3 | 1.06 (1.04–1.08) | 6.78 × 10−9 |
| MNX1 | 7:157027753 | rs1182436 | C/T | 0.80 | 1.08 (1.05–1.12) | 8.30 × 10−7 | 7:157031407 | rs1182397 | 0.92 | G/T | 0.85 | 1.06 (1.02–1.11) | 4.38 × 10−3 | 1.08 (1.05–1.10) | 1.71 × 10−8 |
| ABO | 9:136155000 | rs635634 | T/C | 0.18 | 1.08 (1.05–1.12) | 3.59 × 10−7 | 9:136154867 | rs495828 | 0.83 | T/G | 0.20 | 1.06 (1.01–1.10) | 1.23 × 10−2 | 1.08 (1.05–1.10) | 2.30 × 10−8 |
| PLEKHA1 | 10:124186714 | rs2292626 | C/T | 0.50 | 1.09 (1.06–1.11) | 1.75 × 10−12 | 10:124167512 | rs2421016 | 0.99 | C/T | 0.50 | 1.05 (1.02–1.08) | 2.30 × 10−3 | 1.07 (1.05–1.09) | 1.51 × 10−13 |
| HSD17B12 | 11:43877934 | rs1061810 | A/C | 0.28 | 1.08 (1.05–1.11) | 5.29 × 10−9 | 11:43876435 | rs3736505 | 0.92 | G/A | 0.30 | 1.05 (1.01–1.08) | 4.82 × 10−3 | 1.07 (1.05–1.09) | 3.95 × 10−10 |
| MAP3K11 | 11:65364385 | rs111669836 | A/T | 0.25 | 1.07 (1.04–1.10) | 7.43 × 10−7 | 11:65365171 | rs11227234 | 1.00 | T/G | 0.24 | 1.05 (1.01–1.08) | 8.77 × 10−3 | 1.06 (1.04–1.09) | 4.12 × 10−8 |
| NRXN3 | 14:79945162 | rs10146997 | G/A | 0.21 | 1.07 (1.04–1.10) | 4.59 × 10−6 | 14:79939993 | rs17109256 | 0.98 | A/G | 0.21 | 1.07 (1.03–1.11) | 1.27 × 10−4 | 1.07 (1.05–1.09) | 2.27 × 10−9 |
| CMIP | 16:81534790 | rs2925979 | T/C | 0.30 | 1.08 (1.05–1.10) | 2.72 × 10−8 | 16:81534790 | rs2925979 | 1.00 | T/C | 0.31 | 1.05 (1.02–1.08) | 3.06 × 10−3 | 1.07 (1.04–1.09) | 2.27 × 10−9 |
| ZZEF1 | 17:4014384 | rs7224685 | T/G | 0.30 | 1.07 (1.04–1.10) | 2.00 × 10−7 | 17:3985864 | rs8068804 | 0.95 | A/G | 0.31 | 1.07 (1.03–1.11) | 4.11 × 10−4 | 1.07 (1.05–1.09) | 3.23 × 10−10 |
| GLP2R | 17:9780387 | rs78761021 | G/A | 0.34 | 1.07 (1.05–1.10) | 5.49 × 10−8 | 17:9791375 | rs17676067 | 0.87 | C/T | 0.31 | 1.03 (1.00–1.07) | 3.54 × 10−2 | 1.06 (1.04–1.08) | 3.04 × 10−8 |
| GIP | 17:46967038 | rs79349575 | A/T | 0.51 | 1.07 (1.04–1.09) | 2.61 × 10−7 | 17:47005193 | rs15563 | 0.78 | G/A | 0.54 | 1.04 (1.01–1.07) | 2.09 × 10−2 | 1.06 (1.03–1.08) | 4.43 × 10−8 |
*The nearest gene is listed; this does not imply this is the biologically relevant gene. †Lead SNV types: all map outside transcripts except rs429358 (missense variant) and rs1061810 (3′ untranslated region). ‡Stage 2: proxy SNV (r2 > 0.6 with stage 1 lead SNV) was used when no stage 1 SNV was available. ¢The meta-analysis OR is aligned to the stage 1 SNV risk allele. Chr, chromosome; EA, effect allele; EAF, effect allele frequency; NEA, noneffect allele.
Figure 1.
The effect sizes of the established (blue diamonds, N = 69, P < 5 × 10−4) (Supplementary Material), novel (red diamonds, N = 13), and additional distinct (sky blue diamonds, N = 13) (Supplementary Table 7) signals according to their risk allele frequency (Supplementary Table 3). The additional distinct signals are based on approximate conditional analyses. The distinct signal at TP53INP1 led by rs11786613 (Supplementary Table 7) is plotted (sky blue diamond). This signal did not reach locus-wide significance but was selected for follow-up because of its low frequency and absence of LD with previously reported signal at this locus. The power curve shows the estimated effect size for which we had 80% power to detect associations. Established common variants with OR >1.12 are annotated.
Lead SNVs at all 13 novel loci were common. Although detected here using 1000G imputed data, all 13 were well captured by variants in the HapMap CEU (Central EUrope) reference panel (two directly, 10 via proxies with r2 > 0.8, and one via proxy with r2 = 0.62) (Supplementary Material). At all 13, lead variants defined through 1000G and those seen when the SNP density was restricted to HapMap content had broadly similar evidence of association and were of similar frequency (Supplementary Fig. 2 and Supplementary Table 3). Throughout this article, loci are named for the gene nearest to the lead SNV, unless otherwise specified (Table 1 and Supplementary Material).
Adjustment for BMI revealed no additional genome-wide significant associations for T2D and, at most known and novel loci, there were only minimal differences in statistical significance and estimated T2D effect size between BMI-adjusted and unadjusted models. The four signals at which we observed a significant effect of BMI adjustment (Pheterogeneity <4.4 × 10−4; based on 0.05/113 variants currently or previously reported to be associated with T2D at genome-wide significance) were FTO and MC4R (at which the T2D association is known to reflect a primary effect on BMI) and TCF7L2 and SLC30A8 (at which T2D associations were strengthened after BMI-adjustment) (Supplementary Fig. 3 and Supplementary Table 4).
Insights Into Genetic Architecture of T2D
In this meta-analysis, we tested 3.9 million low-frequency variants (r2 ≥ 0.3 or proper-info ≥0.4; minor allele present in ≥3 studies) for T2D association, constituting 96.7% of the low-frequency variants ascertained by the 1000G European panel (March 2012) (Supplementary Table 2). For variants with risk allele frequencies (RAF) of 0.5%, 1%, or 5%, we had 80% power to detect association (P < 5 × 10−8) for allelic odds ratios (ORs) of 1.80, 1.48, and 1.16, respectively, after accounting for imputation quality (Fig. 1 and Supplementary Table 5). Despite the increased coverage and sample size, we identified no novel low-frequency variants at genome-wide significance (Fig. 1).
Since we had only been able to test 29 of the 52 promising stage 1 signals on the Metabochip, we investigated whether this failure to detect low-frequency variant associations with T2D could be a consequence of selective variant inclusion on the Metabochip. Among the remaining 23 variants, none reached genome-wide significance after aggregating with GWAS data available from EPIC-InterAct. Six of these 23 SNVs had MAF <5%, and for these we performed additional follow-up in the GERA study. However, none reached genome-wide significance in a combined analysis of stage 1, EPIC-InterAct, and GERA (a total of 39,219 case and 198,990 control subjects) (Supplementary Table 6). Therefore, despite substantially enlarged sample sizes that would have allowed us to detect low-frequency risk alleles with modest effect sizes, the overwhelming majority of variants for which T2D association can be detected with these sample sizes are themselves common.
To identify loci containing multiple distinct signals, we performed approximate conditional analysis within the established and novel GWAS loci and detected two such novel common variant signals (Supplementary Table 7) (19,20). At the ANKRD55 locus, we identified a previously unreported distinct (Pconditional < 10−5) association signal led by rs173964 (Pconditional = 3.54 × 10−7, MAF = 26%) (Supplementary Table 7 and Supplementary Fig. 4). We also observed multiple signals of association at loci with previous reports of such signals (4,14), including CDKN2A/B (three signals in total), DGKB and KCNQ1 (six signals), and HNF4A and CCND2 (three signals) (Supplementary Table 7 and Supplementary Fig. 4). At CCND2, in addition to the main signal with lead SNV rs4238013, we detected 1) a novel distinct signal led by a common variant, rs11063018 (Pconditional = 2.70 × 10−7, MAF = 19%) and 2) a third distinct signal led by a low-frequency protective allele (rs188827514, MAF = 0.6%; ORconditional = 0.60, Pconditional = 1.24 × 10−6) (Supplementary Fig. 5A and Supplementary Table 7), which represents the same distinct signal as that at rs76895963 (Pconditional = 1.0) reported in the Icelandic population (Supplementary Fig. 5B) (7). At HNF4A, we confirmed recent analyses (obtained in partially overlapping data) (14) that a low-frequency missense variant (rs1800961, p.Thr139Ile, MAF = 3.7%) is associated with T2D and is distinct from the known common variant GWAS signal (which we mapped here to rs12625671).
We evaluated the trans-ethnic heterogeneity of allelic effects (i.e., discordance in the direction and/or magnitude of estimated ORs) at novel loci on the basis of Cochran's Q statistics from the largest T2D trans-ancestry GWAS meta-analysis to date (2). Using reported summary statistics from that study, we observed no significant evidence of heterogeneity of effect size (Bonferroni correction PCochran's Q < 0.05/13 = 0.0038) between major ancestral groups at any of the 13 loci (Supplementary Table 8). These results are consistent with these loci being driven by common causal variants that are widely distributed across populations.
1000G Variant Density for Identification of Potentially Causal Genetic Variants
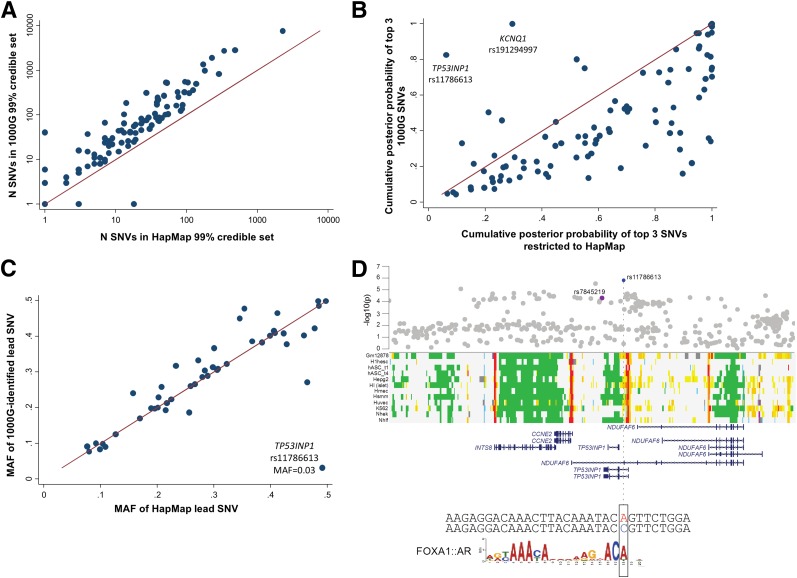
We used credible set fine-mapping (33) to investigate whether 1000G imputation allowed us to better resolve the specific variants driving 95 distinct T2D association signals at 82 loci (Supplementary Material). The 99% credible sets included between 1 and 7,636 SNVs; 25 included fewer than 20 SNVs, 16 fewer than 10 (Supplementary Tables 9 and 10). We compared 1000G-based credible sets with those constructed from HapMap SNVs alone (Fig. 2B and Supplementary Table 9). At all but three of the association signals (two at KCNQ1 and rs1800961 at HNF4A), 1000G imputation resulted in larger credible sets (median increase of 34 variants) spanning wider genomic intervals (median interval size increase of 5 kb) (Fig. 2B and Supplementary Table 9). The 1000G-defined credible sets included >85% of the SNVs in the corresponding HapMap sets (Supplementary Table 9). Despite the overall larger credible sets, we asked whether 1000G imputation enabled an increase in the posterior probability afforded to the lead SNVs, but we found no evidence to this effect (Fig. 2C).
Figure 2.
A: The number (N) of SNVs included in 99% credible sets when performed on all SNVs compared with when analyses were restricted to those SNVs present in HapMap. B: The cumulative πc of the top three SNVs among all 1000G SNVs and after restriction to HapMap SNVs is shown. While the low-frequency SNV at TP53INP1 (rs11786613) did not reach the threshold for a distinct signal in approximate conditional analyses, we fine-mapped both this variant and the previous common signal separately after reciprocal conditioning, which suggested they were independent. C: The MAF of the lead SNV identified in current analyses compared with that identified among SNVs present in HapMap. D: The association of the low-frequency variant rs11786613 (blue) and that of the previous lead variant at this locus, rs7845219 (purple). The low-frequency variant overlaps regulatory annotations active in pancreatic islets, among other tissues, and the sequence surrounding the A allele of this variant has an in silico recognition motif for a FOXA1:AR (androgen receptor) protein complex.
Within the 50 loci previously associated with T2D in Europeans (4), which had at least modest evidence of association in the current analyses (P < 5 × 10−4), we asked whether the lead SNV in 1000G-imputed analysis was of similar frequency to that observed in HapMap analyses. Only at TP53INP1 was the most strongly associated 1000G-imputed SNV (rs11786613, OR = 1.21, P = 1.6 × 10−6, MAF = 3.2%) of substantially lower frequency than the lead HapMap-imputed SNV (3) (rs7845219, MAF = 47.7%) (Fig. 2A). rs11786613 was neither present in HapMap nor on the Metabochip (Supplementary Fig. 6). Reciprocal conditioning of this low-frequency SNV and the previously identified common lead SNV (rs7845219, OR = 1.05, P = 5.0 × 10−5, MAF = 47.5%) indicated that the two signals were likely to be distinct but the signal at rs11786613 did not meet our threshold (Pconditional < 10−5) for locus-wide significance (Supplementary Fig. 4).
Pathophysiological Insights From Novel T2D Associations
Among the 13 novel T2D-associated loci, many (such as those near HLA-DQA1, NRXN3, GIP, ABO, and CMIP) included variants previously implicated in predisposition to other diseases and traits (r2 > 0.6 with the lead SNV) (Supplementary Table 3 and Supplementary Material). For example, the novel association at SNV rs1182436 lies ∼120 kb upstream of MNX1, a gene implicated in pancreatic hypoplasia and neonatal diabetes (34–36).
The lead SNV rs78761021 at the GLP2R locus, encoding the receptor for glucagon-like peptide 2, is in strong LD (r2 = 0.87) with a common missense variant in GLP2R (rs17681684, D470N, P = 3 × 10−7). These signals were strongly dependent and mutually extinguished in reciprocal conditional analyses, consistent with the coding variant being causal and implicating GLP2R as the putative causal gene (Supplementary Fig. 7). While previously suggested to regulate energy balance and glucose tolerance (37), GLP2R has primarily been implicated in gastrointestinal function (38,39). In contrast, GLP1R, encoding the glucagon-like peptide 1 receptor (the target for a major class of T2D therapies [40]), is more directly implicated in pancreatic islet function, and variation at this gene has been associated with glucose levels and T2D risk (41).
We also observed associations with T2D centered on rs9271774 near HLA-DQA1 (Table 1), a region showing a particularly strong association with T1D (42). There is considerable heterogeneity within, and overlap between, the clinical presentations of T1D and T2D, but these can be partially resolved through measurement of islet cell autoantibodies (43). Such measures were not uniformly available across studies contributing to our meta-analysis (Supplementary Table 1). We therefore considered whether the adjacency between T1D and T2D risk loci was likely to reflect misclassification of individuals with autoimmune diabetes as case subjects in the current study.
Three lines of evidence make this unlikely. First, the lead T1D-associated SNV in the HLA region (rs6916742) was only weakly associated with T2D in the current study (P = 0.01), and conditioning on this variant had only modest impact on the T2D association signal at rs9271774 (Punconditional = 3.3 × 10−7; Pconditional = 9.1 × 10−6). Second, of 52 published genome-wide significant T1D association GWAS signals, 50 were included in the current analysis: only six of these reached even nominal association with T2D (P < 0.05; Supplementary Fig. 8), and at one of these six (BCAR1), the T1D risk allele was protective for T2D. Third, in genetic risk score analyses, the combined effect of these 50 T1D signals on T2D risk was of only nominal significance (OR = 1.02 [95% CI 1.00–1.03], P = 0.026), and significance was eliminated when the six overlapping loci were excluded (OR = 1.00 [95% CI 0.98–1.02], P = 0.73). In combination, these findings argue against substantial misclassification and indicate that the signal at HLA-DQA1 is likely to be a genuine T2D signal.
Potential Genes and Pathways Underlying the T2D Loci: eQTL and Pathway Analysis
cis-eQTLs analyses highlighted four genes as possible effector transcripts: ABO (pancreatic islets), PLEKHA1 (whole blood), and HSD17B12 (adipose, liver, muscle, whole blood) at the respective loci and HLA-DRB5 expression (adipose, pancreatic islets, whole blood) at the HLA-DQA1 locus (Supplementary Table 11).
We next asked whether large-scale gene expression data, mouse phenotypes, and protein–protein interaction networks could implicate specific gene candidates and gene sets in the etiology of T2D. Using DEPICT (32), 29 genes were prioritized as driving observed associations (FDR <0.05), including ACSL1 and CMIP among the genes mapping to the novel loci (Supplementary Table 12). These analyses also identified 20 enriched reconstituted gene sets (FDR <5%) falling into four groups (Supplementary Fig. 9) (complete results, including gene prioritization, can be downloaded from http://diagram-consortium.org/2017_Scott_DIAGRAM_1000G/). These included pathways related to mammalian target of rapamycin (mTOR) based on coregulation of the IDE, TLE1, SPRY2, CMIP, and MTMR3 genes (44).
Overlap of Associated Variants With Regulatory Annotations
We observed significant enrichment for T2D-associated credible set variants in pancreatic islet active enhancers and/or promoters (log odds [β] = 0.74, P = 4.2 × 10−8) and FOXA2 binding sites (β = 1.40, P = 4.1 × 10−7), as previously reported (Supplementary Table 13) (14). We also observed enrichment for T2D-associated variants in coding exons (β = 1.56, P = 7.9 × 10−5), in EZH2-binding sites across many tissues (β = 1.35, P = 5.3 × 10−6), and in binding sites for NKX2.2 (β = 1.73, P = 4.1 × 10−8) and PDX1 (β = 1.46, P = 7.4 × 10−6) in pancreatic islets (Supplementary Fig. 10).
Even though credible sets were generally larger, analyses performed on the 1000G imputed results produced stronger evidence of enrichment than equivalent analyses restricted to SNVs present in HapMap. This was most notably the case for variants within coding exons (β = 1.56, P = 7.9 × 10−5 in 1000G compared with β = 0.68, P = 0.62 in HapMap) and likely reflects more complete capture of the true causal variants in the more densely imputed credible sets. Single lead SNVs overlapping an enriched annotation accounted for the majority of the total posterior probability (πc > 0.5) at seven loci. For example, the lead SNV (rs8056814) at BCAR1 (πc = 0.57) overlaps an islet enhancer (Supplementary Fig. 11A), while the newly identified low-frequency signal at TP53INP1 overlaps an islet promoter element (rs117866713, πc = 0.53) (Fig. 2D) (31).
We applied hierarchical clustering to the results of diabetes-related quantitative trait associations for the set of T2D-associated loci from the current study, identifying three main clusters of association signals with differing impact on quantitative traits (Supplementary Table 9). The first, including GIPR, C2CDC4A, CDKAL1, GCK, TCF7L2, GLIS3, THADA, IGF2BP2, and DGKB, involved loci with a primary impact on insulin secretion and processing (26,29). The second cluster captured loci (including PPARG, KLF14, and IRS1) disrupting insulin action. The third cluster, showing marked associations with BMI and lipid levels, included NRXN3, CMIP, APOE, and MC4R but not FTO, which clustered alone.
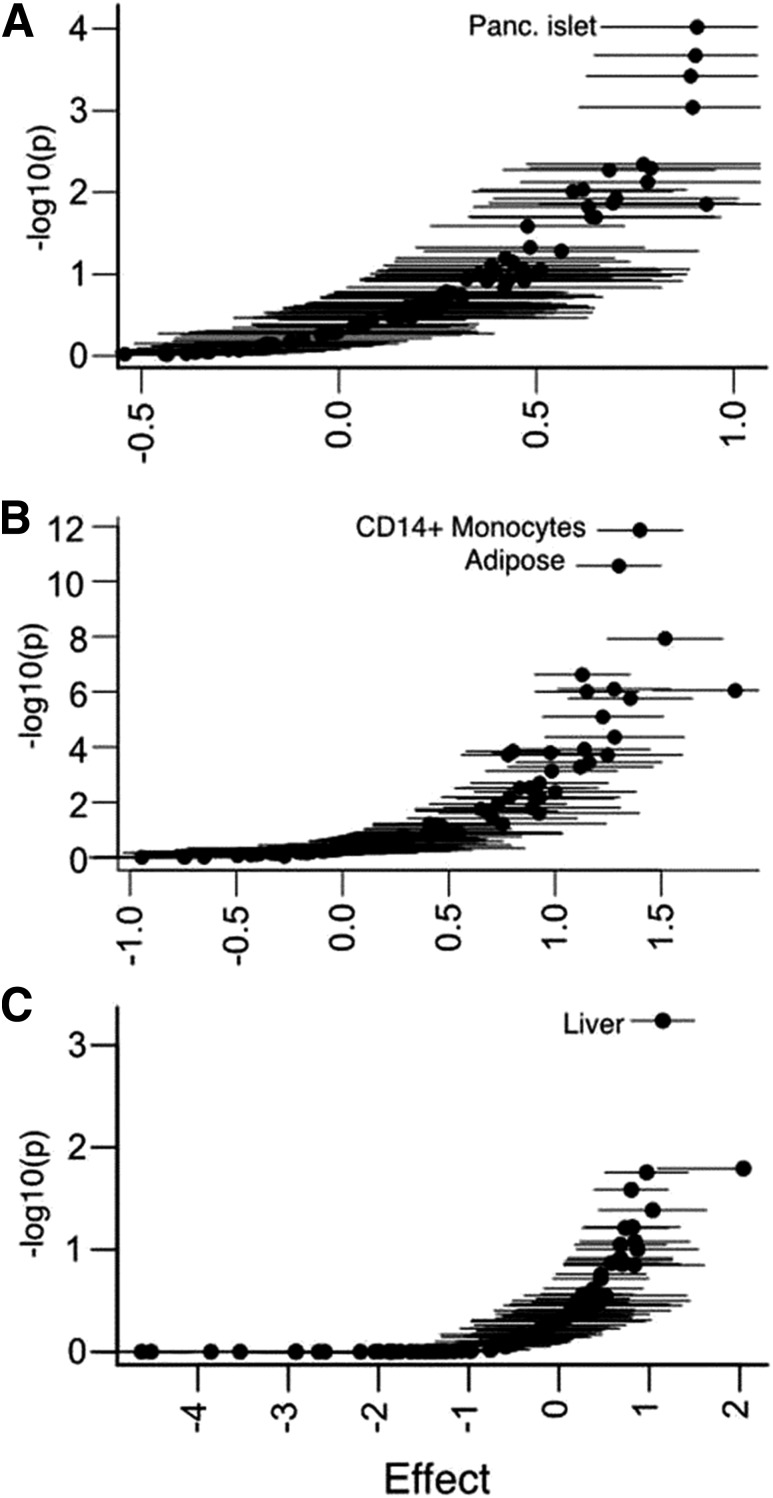
In regulatory enhancement analyses, we observed strong tissue-specific enrichment patterns broadly consistent with the phenotypic characteristics of the physiologically stratified locus subsets. The cluster of loci disrupting insulin secretion showed the most marked enrichment for pancreatic islet regulatory elements (β = 0.91, P = 9.5 × 10−5). In contrast, the cluster of loci implicated in insulin action was enriched for annotations from adipocytes (β = 1.3, P = 2.7 × 10−11) and monocytes (β = 1.4, P = 1.4 × 10−12), and that characterized by associations with BMI and lipids showed preferential enrichment for hepatic annotations (β = 1.15, P = 5.8 × 10−4) (Fig. 3A–C). For example, at the novel T2D-associated CMIP locus, previously associated with adiposity and lipid levels (28,45), the lead SNV (rs2925979, πc = 0.91) overlaps an active enhancer element in both liver and adipose tissue, among others (Supplementary Fig. 11B).
Figure 3.
T2D loci stratified by patterns of quantitative trait (e.g., glycemic, insulin, lipid, and anthropometric) effects show distinct cell-type annotation patterns. We hierarchically clustered loci based on endophenotype data and identified groups of T2D loci associated with measures of insulin secretion (A), insulin resistance (B), and BMI/lipids (C). We then tested the effect of variants in cell-type enhancer and promoter chromatin states on the posterior probabilities of credible sets for each group. We identified most significant effects among pancreatic (Panc.) islet chromatin for insulin secretion loci, CD14+ monocyte and adipose chromatin for insulin resistance loci, and liver chromatin for BMI/lipid loci.
Discussion
In this large-scale study of T2D genetics, in which individual variants were assayed in up to 238,209 subjects, we identified 13 novel T2D-associated loci at genome-wide significance and refined causal variant location for the 13 novel and 69 established T2D loci. We also found evidence for enrichment in regulatory elements at associated loci in tissues relevant for T2D and demonstrated tissue-specific enrichment in regulatory annotations when T2D loci were stratified according to inferred physiological mechanism.
We calculate that the present analysis, together with loci reported in other recent publications (9), brings the total number of independent T2D associations to 128 distinct signals at 113 loci (Supplementary Table 3). Lead SNVs at all 13 novel loci were common (MAF >15%) and of comparable effect size (1.07 ≤ OR ≤ 1.10) to previously identified common variant associations (2,4). Associations at the novel loci showed homogeneous effects across diverse ethnicities, supporting the evidence for coincident common risk alleles across ancestry groups (2). Moreover, we conclude that misclassification of diabetes subtype is not a major concern for these analyses and that the HLA-DQA1 signal represents genuine association with T2D, independent of nearby signals that influence T1D.
We observed a general increase in the size of credible sets with 1000G imputation compared with HapMap imputation. This is likely due to improved enumeration of potential causal common variants on known risk haplotypes rather than resolution toward low-frequency variants of larger effect driving common variant associations. These findings are consistent with the inference (arising also from the other analyses reported here) that the T2D risk signals identified by GWAS are overwhelmingly driven by common causal variants. In such a setting, imputation with denser reference panels, at least in ethnically restricted samples, provides more complete elaboration of the allelic content of common risk haplotypes. Finer resolution of those haplotypes that would provide greater confidence in the location of causal variants will likely require further expansion of trans-ethnic fine-mapping efforts (2). The distinct signals at the established CCND2 and TP53INP1 loci point to contributions of low-frequency variant associations of modest effect but indicate that even larger samples will be required to robustly detect association signals at low frequency. Such new large data sets might be used to expand the follow-up of suggestive signals from our analysis.
The discovery of novel genome-wide significant association signals in the current analysis is attributable primarily to increased sample size rather than improved genomic coverage. Although we queried a large proportion of the low-frequency variants present in the 1000G European reference haplotypes and had >80% power to detect genome-wide significant associations with OR >1.8 for the tested low-frequency risk variants, we found no such low-frequency variant associations in either established or novel loci. While low-frequency variant coverage in the current study was not complete, this observation adds to the growing evidence (2,4,9,46) that few low-frequency T2D risk variants with moderate to strong effect sizes exist in European ancestry samples and is consistent with a primary role for common variants of modest effect in T2D risk. The current study reinforces the conclusions from a recent study that imputed from whole-genome sequencing data—from 2,657 European T2D case and control subjects rather than 1000G—into a set of GWAS studies partially overlapping with the present meta-analysis. We demonstrated that the failure to detect low-frequency associations in that study is not overcome by a substantial increase in sample size (9). It is worth emphasizing that we did not, in this study, have sufficient imputation quality to test for T2D associations with rare variants and we cannot evaluate the collective contribution of variants with MAF <0.5% to T2D risk.
The development of T2D involves dysfunction of multiple mechanisms across several distinct tissues (9,29,31,47,48). When coupled with functional data, we saw larger effect estimates for enrichment of coding variants than observed with HapMap SNVs alone, consistent with more complete recovery of the causal variants through imputation using a denser reference panel. The functional annotation analyses also demonstrated that the stratification of T2D risk loci according to primary physiological mechanism resulted in evidence for consistent and appropriate tissue-specific effects on transcriptional regulation. These analyses exemplify the use of a combination of human physiology and genomic annotation to position T2D GWAS loci with respect to the cardinal mechanistic components of T2D development. Extension of this approach is likely to provide a valuable in silico strategy to aid prioritization of tissues for mechanistic characterization of genetic associations. Using the hypothesis-free pathway analysis of T2D associations with DEPICT (32), we highlighted a causal role of mTOR signaling pathway in the etiology of T2D not observed from individual loci associations. The mTOR pathway has previously been implicated in the link between obesity, insulin resistance, and T2D from cell and animal models (44,49).
The current results emphasize that progressively larger sample sizes, coupled with higher density sequence-based imputation (13), will continue to represent a powerful strategy for genetic discovery in T2D and in complex diseases and traits more generally. At known T2D-associated loci, identification of the most plausible T2D causal variants will likely require large-scale multiethnic analyses, where more diverse haplotypes, reflecting different patterns of LD, in combination with functional (31,50,51) data allow refinement of association signals to smaller numbers of variants (2).
Supplementary Material
Article Information
Funding.
ARIC. The Atherosclerosis Risk in Communities (ARIC) Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C; R01HL087641, R01HL59367, and R01HL086694; National Human Genome Research Institute contract U01HG004402; and NIH contract HHSN268200625226C. Infrastructure was partly supported by grant no. UL1RR025005, a component of the NIH and NIH Roadmap for Medical Research. The authors wish to acknowledge the many contributions of Dr. Linda Kao (Department of Epidemiology, Johns Hopkins School of Public Health), who helped direct the diabetes genetics working group in the ARIC Study until her passing in 2014. The authors thank the staff and participants of the ARIC study for their important contributions.
BioMe. This work is funded by the Icahn School of Medicine at Mount Sinai Institute for Personalized Medicine BioMe BioBank Program, which is supported by The Andrea & Charles Bronfman Philanthropies.
D2D2007. The FIN-D2D study has been financially supported by the hospital districts of Pirkanmaa, South Ostrobothnia, and Central Finland; the Finnish National Public Health Institute (National Institute for Health and Welfare); the Finnish Diabetes Association; the Ministry of Social Affairs and Health in Finland; the Academy of Finland (grant no. 129293), the European Commission (Directorate C-Public Health grant agreement no. 2004310); and Finland’s Slottery Machine Association.
DANISH. The study was funded by the Lundbeck Foundation and produced by the Lundbeck Foundation Centre for Applied Medical Genomics in Personalised Disease Prediction, Prevention and Care (LuCamp, www.lucamp.org) and Danish Council for Independent Research. The Novo Nordisk Foundation Center for Basic Metabolic Research is an independent research center at the University of Copenhagen, partially funded by an unrestricted donation from the Novo Nordisk Foundation (www.metabol.ku.dk).
DGI. Diabetes Genetics Initiative (DGI), this work was supported by a grant from Novartis. The Botnia Study was supported by grants from the Signe and Ane Gyllenberg Foundation, Swedish Cultural Foundation in Finland, Finnish Diabetes Research Society, the Sigrid Jusélius Foundation, Folkhälsan Research Foundation, Foundation for Life and Health in Finland, Jakobstad Hospital, Medical Society of Finland, Närpes Research Foundation and the Vasa and Närpes Health centers, the European Commission’s Seventh Framework Programme (FP7) (2007–2013), the European Network for Genetic and Genomic Epidemiology (ENGAGE), the Collaborative European Effort to Develop Diabetes Diagnostics (CEED3) (2008–2012), and the Swedish Research Council, including a Linné grant (no. 31475113580).
DGDG. Diabetes Gene Discovery Group (DGDG), this work was funded by Genome Canada, Génome Québec, and the Canada Foundation for Innovation. Cohort recruitment was supported by the Fédération Française des Diabetiques, INSERM, CNAMTS, Centre Hospitalier Universitaire Poitiers, La Fondation de France, and the Endocrinology-Diabetology department of the Corbeil-Essonnes Hospital. C. Petit, J.-P. Riveline, and S. Franc were instrumental in recruitment and S. Brunet, F. Bacot, R. Frechette, V. Catudal, M. Deweirder, F. Allegaert, P. Laflamme, P. Lepage, W. Astle, M. Leboeuf, and S. Leroux provided technical assistance. K. Shazand and N. Foisset provided organizational guidance. The authors thank all individuals who participated as case or control subjects in this study.
deCODE. The deCODE study was funded by deCODE Genetics/Amgen, Inc., and partly supported by ENGAGE HEALTH-F4-2007-201413. The authors thank the Icelandic study participants and the staff of deCODE Genetics core facilities and recruitment center for their contributions to this work.
DILGOM. The DIetary, Lifestyle, and Genetic determinants of Obesity and Metabolic syndrome (DILGOM) study was supported by the Academy of Finland (grant no. 118065). V.Sa. was supported by the Academy of Finland (grant no. 139635) and the Finnish Foundation for Cardiovascular Research. S.Mӓ. was supported by the Academy of Finland (grant nos. 136895 and 263836). S.R. was supported by the Academy of Finland Centre of Excellence in Complex Disease Genetics (grant nos. 213506 and 129680), the Academy of Finland (grant no. 251217), the Finnish Foundation for Cardiovascular Research, and the Sigrid Jusélius Foundation.
DR’s EXTRA. The Dose Responses to Exercise Training (DR's EXTRA) Study was supported by the Ministry of Education and Culture of Finland (627; 2004–2011), the Academy of Finland (grant nos. 102318 and 123885), Kuopio University Hospital, the Finnish Diabetes Association, the Finnish Heart Association, the Päivikki and Sakari Sohlberg Foundation, and by grants from European Commission’s FP6 Integrated Project (EXGENESIS, LSHM-CT-2004-005272), the City of Kuopio, and the Social Insurance Institution of Finland (4/26/2010).
EGCUT. Estonian Genome Center of the University of Tartu (EGCUT) was supported by European Commision grant through the European Regional Development Fund (project no. 2014-2020.4.01.15-0012); PerMedI (TerVE EstRC); European Commision Horizon 2020 grants 692145, 676550, and 654248; and Estonian Research Council grant IUT20-60.
EMIL-Ulm. The EMIL Study received support by the State of Baden-Württemberg, Germany, the City of Leutkirch, Germany, and the German Research Council to B.O.B. (GRK 1041). The Ulm Diabetes Study Group received support from the German Research Foundation (DFG-GRK 1041) and the State of Baden-Württemberg Centre of Excellence Metabolic Disorders to B.O.B.
EPIC-InterAct. This work was funded by the European Commission’s Sixth Framework Programme (grant no. LSHM_CT_2006_037197). The authors thank all EPIC participants and staff for their contribution to the EPIC-InterAct study. The authors thank the laboratory team at the MRC Epidemiology Unit for sample management. I.B. was supported by grant WT098051.
FHS. This research was conducted in part using data and resources from the Framingham Heart Study (FHS) of the National Heart, Lung, and Blood Institute of the NIH and Boston University School of Medicine. The analyses reflect intellectual input and resource development from the FHS investigators participating in the SNP Health Association Resource (SHARe) project. This work was partially supported by the National Heart, Lung, and Blood Institute's FHS (contract no. N01‐HC‐25195) and its contract with Affymetrix, Inc., for genotyping services (contract no. N02‐HL‐6‐4278). A portion of this research utilized the Linux Cluster for Genetic Analysis (LinGA‐II) funded by the Robert Dawson Evans endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. The work is also supported by National Institute for Diabetes and Digestive and Kidney Diseases (NIDDK) grants R01-DK078616 (to J.B.M., J.D., and J.C.F.), K24-DK080140 (to J.B.M.), U01-DK085526 (to H.Che., J.D., and J.B.M.), and a Massachusetts General Hospital Research Scholars Award (to J.C.F.).
FUSION. The Finland-United States Investigation of NIDDM Genetics (FUSION) study was funded by NIH grants U01-DK062370, R01-HG000376, and R01-DK072193 and NIH intramural project no. ZIA HG000024. Genome-wide genotyping was conducted by the Johns Hopkins University Genetic Resources Core. Facility SNP Center at the Center for Inherited Disease Research (CIDR), with support from CIDR NIH contract no. N01-HG-65403.
GERA. Data came from a grant, the Resource for Genetic Epidemiology Research in Adult Health and Aging (RC2 AG033067, C. Schaefer [Kaiser Permanente Northern California Division of Research] and N. Risch [Institute for Human Genetics, University of California], principal investigators) awarded to the Kaiser Permanente Research Program on Genes, Environment and Health (RPGEH) and the UCSF Institute for Human Genetics. The RPGEH was supported by grants from the Robert Wood Johnson Foundation, the Wayne and Gladys Valley Foundation, the Lawrence Ellison Medical Foundation, Kaiser Permanente Northern California, and the Kaiser Permanente National and Northern California Community Benefit Programs.
GoDARTS. The Genetics of Diabetes Audit and Research in Tayside Scotland (GoDARTS) study was funded by the Wellcome Trust (084727/Z/08/Z, 085475/Z/08/Z, 085475/B/08/Z) and as part of the European Commission IMI-SUMMIT program. The authors acknowledge the support of the Health Informatics Centre, University of Dundee, for managing and supplying the anonymized data and NHS Tayside, the original data owner. The authors are grateful to all the participants who took part in the GoDARTS study, to the general practitioners, to the Scottish School of Primary Care for their help in recruiting the participants, and to the whole team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, and nurses.
Heinz Nixdorf Recall. The authors thank the Heinz Nixdorf Foundation (Chairman: M. Nixdorf, Past Chairman: G. Schmidt [deceased]) and the German Federal Ministry of Education and Research (BMBF) for the generous support of this study. An additional research grant was received from Imatron, Inc., South San Francisco, CA, which produced the electron beam computerized tomography scanners, and GE-Imatron, South San Francisco, CA, after the acquisition of Imatron, Inc. The authors acknowledge the support of the Sarstedt AG & Co. (Nümbrecht, Germany) concerning laboratory equipment. The authors received support of the Ministry of Innovation, Science and Research, Nordrhine Westfalia for the genotyping of the Heinz Nixdorf Recall (HNR) study participants. Technical support for the imputation of the HNR study data on the supercomputer Cray XT6m was provided by the Center for Information and Media Services, University of Duisburg-Essen. The authors are indebted to all the study participants and to the dedicated personnel of both the study center of the HNR study and the electron beam computerized tomography scanner facilities, D. Grönemeyer, Bochum, and R. Seibel, Mülheim, as well as to the investigative group, in particular U. Roggenbuck, U. Slomiany, E.M. Beck, A. Öffner, S. Münkel, M. Bauer, S. Schrader, R. Peter, and H. Hirche.
HPFS. The Health Professionals Follow-up Study (HPFS) was funded by the NIH grants P30 DK46200, DK58845, U01HG004399, and UM1CA167552.
IMPROVE and SCARFSHEEP. The IMPROVE study was supported by the European Commission (LSHM-CT-2007-037273), the Swedish Heart-Lung Foundation, the Swedish Research Council (8691), the Knut and Alice Wallenberg Foundation, the Foundation for Strategic Research, the Torsten and Ragnar Söderberg Foundation, the Strategic Cardiovascular Programme of Karolinska Institutet, and the Stockholm County Council (560183). The SCARFSHEEP study was supported by the Swedish Heart-Lung Foundation, the Swedish Research Council, the Strategic Cardiovascular Programme of Karolinska Institutet, the Strategic Support for Epidemiological Research at Karolinska Institutet, and the Stockholm County Council. B.S. acknowledges funding from the Magnus Bergvall Foundation and the Foundation for Old Servants. M.F. acknowledges funding from the Swedish e-science Research Center (SeRC). R.J.S. is supported by the Swedish Heart-Lung Foundation, the Tore Nilsson Foundation, the Thuring Foundation, and the Foundation for Old Servants. S.E.H. is funded by the British Heart Foundation (PG08/008).
KORAgen. The KORA (Cooperative Health Research in the Region of Augsburg) research platform was initiated and financed by the Helmholtz Zentrum München, German Research Center for Environmental Health, which is funded by the German Federal Ministry of Education and Research (BMBF) and by the State of Bavaria. The KORA research was supported within the Munich Center of Health Sciences (MC Health), Ludwig-Maximilians-Universität, as part of LMUinnovativ. Part of this project was supported by the German Center for Diabetes Research (DZD).
METSIM. The METabolic Syndrome In Men (METSIM) study was funded by the Academy of Finland (grant nos. 77299 and 124243).
NHS. Nurses' Health Study (NHS), this work was funded by the NIH grants P30 DK46200, DK58845, U01HG004399, and UM1CA186107.
PPP-Malmo-Botnia (PMB). The Prevalence, Prediction and Prevention of Diabetes (PPP)-Botnia study has been financially supported by grants from the Sigrid Jusélius Foundation, the Folkhälsan Research Foundation, the Ministry of Education in Finland, the Nordic Center of Excellence in Disease Genetics, the European Commission (EXGENESIS), the Signe and Ane Gyllenberg Foundation, the Swedish Cultural Foundation in Finland, the Finnish Diabetes Research Foundation, the Foundation for Life and Health in Finland, the Finnish Medical Society, the Paavo Nurmi Foundation, the Helsinki University Central Hospital Research Foundation, the Perklén Foundation, the Ollqvist Foundation, and the Närpes Health Care Foundation. The study has also been supported by the Municipal Health Care Center and Hospital in Jakobstad and Health Care Centers in Vasa, Närpes, and Korsholm. Studies from Malmö were supported by grants from the Swedish Research Council (SFO EXODIAB 2009-1039; LUDC 349-2008-6589, 521-2010-3490, 521-2010-3490, 521-2010-3490, 521-2007-4037, and 521-2008-2974; ANDIS 825-2010-5983), the Knut and Alice Wallenberg Foundation (KAW 2009.0243), the Torsten and Ragnar Söderbergs Stiftelser (MT33/09), the IngaBritt and Arne Lundberg’s Research Foundation (grant no. 359), and the Swedish Heart-Lung Foundation.
PIVUS and ULSAM. Prospective Investigation of the Vasculature in Uppsala Seniors (PIVUS) and Uppsala Longitudinal Study of Adult Men (ULSAM), this work was funded by the Swedish Research Council, Swedish Heart-Lung Foundation, Knut and Alice Wallenberg Foundation, and Swedish Diabetes Foundation. Genome-wide genotyping was funded by the Wellcome Trust and performed by the SNP&SEQ Technology Platform in Uppsala (www.genotyping.se). The authors thank Tomas Axelsson, Ann-Christine Wiman, and Caisa Pöntinen for their assistance with genotyping. The SNP Technology Platform is supported by Uppsala University, Uppsala University Hospital, and the Swedish Research Council for Infrastructures.
Rotterdam Study. This work is funded by Erasmus Medical Center and Erasmus University, Rotterdam; Netherlands Organization for Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly (RIDE); the Ministry of Education, Culture and Science; the Ministry of Health, Welfare and Sport; the European Commission (DG XII); and the Municipality of Rotterdam. This study is also funded by the Research Institute for Diseases in the Elderly (014-93-015, RIDE2) and the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) project no. 050-060-810. The generation and management of GWAS genotype data for the Rotterdam Study is supported by NWO Investments (no. 175.010.2005.011, 911-03-012). The authors thank Pascal Arp, Mila Jhamai, Marijn Verkerk, Lizbeth Herrera, and Marjolein Peters for their help in creating the GWAS database. The authors thank the study participants, the staff from the Rotterdam Study, and the participating general practitioners and pharmacists.
STR. The Swedish Twin Registry (STR) was supported by grants from the U.S. NIH (AG028555, AG08724, AG04563, AG10175, and AG08861), the Swedish Research Council, the Swedish Heart-Lung Foundation, the Swedish Foundation for Strategic Research, the Royal Swedish Academy of Science, and ENGAGE (within the European Commission FP7 HEALTH-F4-2007-201413). Genotyping was performed by the SNP&SEQ Technology Platform in Uppsala (www.genotyping.se). The authors thank Tomas Axelsson, Ann-Christine Wiman, and Caisa Pöntinen for their excellent assistance with genotyping. The SNP&SEQ Technology Platform is supported by Uppsala University, Uppsala University Hospital, and the Swedish Research Council for Infrastructures.
WARREN 2/58BC and Wellcome Trust Case Control Consortium. Collection of the U.K. T2D cases was supported by Diabetes UK, BDA Research, and the UK Medical Research Council (Biomedical Collections Strategic Grant G0000649). The UK Type 2 Diabetes Genetics Consortium collection was supported by the Wellcome Trust (Biomedical Collections Grant GR072960). Metabochip genotyping was supported by the Wellcome Trust (Strategic Awards 076113, 083948, and 090367 and core support for the Wellcome Trust Centre for Human Genetics 090532) and analysis by the European Commission (ENGAGE HEALTH-F4-2007-201413), MRC (Project Grant G0601261), NIDDK (DK073490, DK085545, and DK098032), and Wellcome Trust (083270 and 098381). The Wellcome Trust Case Control Consortium is funded by Wellcome 076113 and 085475.
Institutional support for study design and analysis. This work was funded by MRC (G0601261), NIDDK (RC2-DK088389, U01-DK105535, U01-DK085545, and U01-DK105535), FP7 (ENGAGE HEALTH-F4-2007-201413), and the Wellcome Trust (090532, 098381, 106130, and 090367).
Individual funding for study design and analysis. J.T.-F. is a Marie-Curie Fellow (PIEF-GA-2012-329156). M.K. is supported by the European Commission under the Marie Curie Intra-European Fellowship (project MARVEL, PIEF-GA-2013-626461). C.L., R.A.S., and N.J.W. are funded by the Medical Research Council (MC_UU_12015/1). L.M. is partially supported by 2010–2011 PRIN funds of the University of Ferrara (holder: Guido Barbujani), in part sponsored by the European Foundation for the Study of Diabetes (EFSD) Albert Renold Travel Fellowships for Young Scientists, and sponsored by the fund promoting internationalization efforts of the University of Ferrara (holder: C.S.). A.P.M. is a Wellcome Trust Senior Fellow in Basic Biomedical Science (grant no. WT098017). M.I.M. is a Wellcome Trust Senior Investigator. J.R.B.P. is supported by the Wellcome Trust (WT092447MA). T.H.P. is supported by The Danish Council for Independent Research Medical Sciences (FSS), the Lundbeck Foundation, and the Alfred Benzon Foundation. I.P. was in part funded by the Elsie Widdowson Fellowship, the Wellcome Trust Seed Award in Science (205915/Z/17/Z), and the European Commission’s Horizon 2020 research and innovation programme (DYNAhealth, project no. 633595). B.F.V. is supported by the NIH/NIDDK (R01DK101478) and the American Heart Association (13SDG14330006). E.Z. is supported by the Wellcome Trust (098051). S.E.H. is funded by British Heart Foundation PG08/008 and University College London Biomedical Research Centre. V.Sa. was supported by the Academy of Finland (grant no. 139635) and by the Finnish Foundation for Cardiovascular Research.
Duality of Interest. I.B. owns stock in GlaxoSmithKline and Incyte. J.C.F. has received consulting honoraria from Pfizer and PanGenX. V.St., G.T., A.K., U.T., and K.Ste. are employed by deCODE Genetics/Amgen, Inc. E.I. is a scientific advisor for Precision Wellness, CELLINK, and Olink Proteomics for work unrelated to the present project. M.I.M. sits on advisory panels for Pfizer and Novo Nordisk; has received honoraria from Pfizer, Novo Nordisk, and Eli Lilly; and is a recipient of research funding from Pfizer, Novo Nordisk, Eli Lilly, Takeda, Sanofi, Merck, Boehringer Ingelheim, AstraZeneca, Janssen, Roche, Servier, and AbbVie. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. Writing and coordination group: R.A.S., L.J.S., R.M., L.M., K.J.G., M.K., J.D., A.P.M., M.Bo., M.I.M., I.P. Central analysis group: R.A.S., L.J.S., R.M., L.M., C.M., A.P.M., M.Bo., M.I.M., I.P. Additional lead analysts: L.M., K.J.G., M.K., N.P., T.H.P., A.D.J., J.D.E., T.F., Y.Le., J.R.B.P., L.J., A.U.J. GWAS cohort-level primary analysts: R.A.S., L.J.S., R.M., K.J.G., V.St., G.T., L.Q., N.R.V.Z., A.Ma., H.Che., P.A., B.F.V., H.G., M.M.-N., J.S.R., N.W.R., N.R., L.C.K., E.M.v.L., S.M.W., C.Fu., P.Kw., C.M., P.C., M.L., Y. Lu, C.D., D.T., L.Y., C.L., A.P.M., I.P. Metabochip cohort-level primary analysts: T.S., H.A.Ke., H.Chh., L.E., S.G., T.M.T., M.F., R.J.S. Cohort sample collection, phenotyping, genotyping, or additional analysis: R.A.S., H.G., R.B., A.B.H., A.K., G.Si., N.D.K., J.L., L.Lia., T.M., M.R., B.T., T.E., E.M., C.Fo., C.-T.L., D.Ry., B.I., V.L., T.T., D.J.C., J.S.P., N.G., C.T.H., M.E.J., T.J., A.L., M.C.C., R.M.v.D., D.J.H., P.Kr., Q.S., S.E., K.R.O., J.R.B.P., A.R.W., E.Z., J.T.-F., G.R.A., L.L.B., P.S.C., H.M.S., H.A.Ko., L.K., B.S., T.W.M., M.M.N., S.P., D.B., K.G., S.E.H., E.Tr., N.K., J.M., G.St., R.W., J.G.E., S.Mӓ., L.P., E.Ti., G.C., E.E., S.L., B.G., K.L., O.M., E.P.B., O.G., D.Ru., M.Bl., P.Ko., A.T., N.M.M., C.S., T.M.F., A.T.H., I.B., B.B., H.B., P.W.F., A.B.G., D.P., Y.T.v.d.S., C.L., N.J.W., K.Str., M.Bo., M.I.M. Metabochip cohort principal investigators: R.E., K.-H.J., S.Mo., U.d.F., A.H., M.S., P.D., P.J.D., T.M.F., A.T.H., S.R., V.Sa., N.L.P., B.O.B., R.N.B., F.S.C., K.L.M., J.T., T.H., O.P., I.B., C.L., N.J.W. GWAS cohort principal investigators: L.La., E.I., L.Lin., C.M.L., S.C., P.F., R.J.F.L., B.B., H.B., P.W.F., A.B.G., D.P., Y.T.v.d.S., D.A., L.C.G., C.L., N.J.W., E.S., C.M.v.D., J.C.F., J.B.M., E.B., C.G., K.Str., A.Me., A.D.M., C.N.A.P., F.B.H., U.T., K.Ste., J.D., M.Bo., M.I.M. I.P. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db16-1253/-/DC1.
Deceased.
See accompanying article, p. 2741.
References
- 1.Global Burden of Disease Study 2013 Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 2015;386:743–800 [DOI] [PMC free article] [PubMed]
- 2.Mahajan A, Go MJ, Zhang W, et al.; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium; Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium; Mexican American Type 2 Diabetes (MAT2D) Consortium; Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples (T2D-GENES) Consortium . Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet 2014;46:234–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voight BF, Scott LJ, Steinthorsdottir V, et al.; MAGIC investigators; GIANT Consortium . Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 2010;42:579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris AP, Voight BF, Teslovich TM, et al.; Wellcome Trust Case Control Consortium; Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators; Genetic Investigation of ANthropometric Traits (GIANT) Consortium; Asian Genetic Epidemiology Network–Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium . Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 2012;44:981–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeggini E, Scott LJ, Saxena R, et al.; Wellcome Trust Case Control Consortium . Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 2008;40:638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dupuis J, Langenberg C, Prokopenko I, et al.; DIAGRAM Consortium; GIANT Consortium; Global BPgen Consortium; Anders Hamsten on behalf of Procardis Consortium; MAGIC investigators . New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 2010;42:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steinthorsdottir V, Thorleifsson G, Sulem P, et al. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat Genet 2014;46:294–298 [DOI] [PubMed] [Google Scholar]
- 8.Estrada K, Aukrust I, Bjørkhaug L, et al.; SIGMA Type 2 Diabetes Consortium . Association of a low-frequency variant in HNF1A with type 2 diabetes in a Latino population [published correction appears in JAMA 2014;312:1932]. JAMA 2014;311:2305–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuchsberger C, Flannick J, Teslovich TM, et al. The genetic architecture of type 2 diabetes. Nature 2016;536:41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Majithia AR, Flannick J, Shahinian P, et al.; GoT2D Consortium; NHGRI JHS/FHS Allelic Spectrum Project; SIGMA T2D Consortium; T2D-GENES Consortium . Rare variants in PPARG with decreased activity in adipocyte differentiation are associated with increased risk of type 2 diabetes [published correction appears in Proc Natl Acad Sci U S A 2014;111:16225]. Proc Natl Acad Sci U S A 2014;111:13127–13132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonnefond A, Clément N, Fawcett K, et al.; Meta-Analysis of Glucose and Insulin-Related Traits Consortium (MAGIC) . Rare MTNR1B variants impairing melatonin receptor 1B function contribute to type 2 diabetes. Nat Genet 2012;44:297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abecasis GR, Auton A, Brooks LD, et al.; 1000 Genomes Project Consortium . An integrated map of genetic variation from 1,092 human genomes. Nature 2012;491:56–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J, Bakshi A, Zhu Z, et al.; LifeLines Cohort Study . Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet 2015;47:1114–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaulton KJ, Ferreira T, Lee Y, et al.; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium . Genetic fine-mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat Genet 2015;47:1415–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langenberg C, Sharp S, Forouhi NG, et al.; InterAct Consortium . Design and cohort description of the InterAct Project: an examination of the interaction of genetic and lifestyle factors on the incidence of type 2 diabetes in the EPIC Study. Diabetologia 2011;54:2272–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cook JP, Morris AP. Multi-ethnic genome-wide association study identifies novel locus for type 2 diabetes susceptibility. Eur J Hum Genet 2016;24:1175–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 2012;44:955–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009;5:e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J, Ferreira T, Morris AP, et al.; Genetic Investigation of ANthropometric Traits (GIANT) Consortium; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium . Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet 2012;44:369–375, S1–S3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 2011;88:76–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.UK10K Consortium, Walter K, Min JL, et al. The UK10K project identifies rare variants in health and disease. Nature 2015;526:82–90 [DOI] [PMC free article] [PubMed]
- 22.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 2013;37:658–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manning AK, Hivert M-F, Scott RA, et al.; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium; Multiple Tissue Human Expression Resource (MUTHER) Consortium . A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet 2012;44:659–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saxena R, Hivert M-F, Langenberg C, et al.; GIANT consortium; MAGIC investigators . Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat Genet 2010;42:142–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strawbridge RJ, Dupuis J, Prokopenko I, et al.; DIAGRAM Consortium; GIANT Consortium; MuTHER Consortium; CARDIoGRAM Consortium; C4D Consortium . Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes 2011;60:2624–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prokopenko I, Poon W, Mägi R, et al. A central role for GRB10 in regulation of islet function in man. PLoS Genet 2014;10:e1004235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Speliotes EK, Willer CJ, Berndt SI, et al.; MAGIC; Procardis Consortium . Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet 2010;42:937–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Willer CJ, Schmidt EM, Sengupta S, et al.; Global Lipids Genetics Consortium . Discovery and refinement of loci associated with lipid levels. Nat Genet 2013;45:1274–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dimas AS, Lagou V, Barker A, et al.; MAGIC Investigators . Impact of type 2 diabetes susceptibility variants on quantitative glycemic traits reveals mechanistic heterogeneity. Diabetes 2014;63:2158–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunham I, Kundaje A, Aldred SF, et al.; ENCODE Project Consortium . An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pasquali L, Gaulton KJ, Rodríguez-Seguí SA, et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet 2014;46:136–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pers TH, Karjalainen JM, Chan Y, et al.; Genetic Investigation of ANthropometric Traits (GIANT) Consortium . Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun 2015;6:5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maller JB, McVean G, Byrnes J, et al.; Wellcome Trust Case Control Consortium . Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet 2012;44:1294–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flanagan SE, De Franco E, Lango Allen H, et al. Analysis of transcription factors key for mouse pancreatic development establishes NKX2-2 and MNX1 mutations as causes of neonatal diabetes in man. Cell Metab 2014;19:146–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melé M, Ferreira PG, Reverter F, et al.; GTEx Consortium . Human genomics. The human transcriptome across tissues and individuals. Science 2015;348:660–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonnefond A, Vaillant E, Philippe J, et al. Transcription factor gene MNX1 is a novel cause of permanent neonatal diabetes in a consanguineous family. Diabetes Metab 2013;39:276–280 [DOI] [PubMed] [Google Scholar]
- 37.Guan X. The CNS glucagon-like peptide-2 receptor in the control of energy balance and glucose homeostasis. Am J Physiol Regul Integr Comp Physiol 2014;307:R585–R596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy KG, Bloom SR. Gut hormones and the regulation of energy homeostasis. Nature 2006;444:854–859 [DOI] [PubMed] [Google Scholar]
- 39.GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015;348:648–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drucker DJ, Nauck MA. The incretin system: glucagon-like peptide-1 receptor agonists and dipeptidyl peptidase-4 inhibitors in type 2 diabetes. Lancet 2006;368:1696–1705 [DOI] [PubMed] [Google Scholar]
- 41.Wessel J, Chu AY, Willems SM, et al.; EPIC-InterAct Consortium . Low-frequency and rare exome chip variants associate with fasting glucose and type 2 diabetes susceptibility. Nat Commun 2015;6:5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bradfield JP, Qu H-Q, Wang K, et al. A genome-wide meta-analysis of six type 1 diabetes cohorts identifies multiple associated loci. PLoS Genet 2011;7:e1002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.National Institute for Health and Care Excellence. Type 1 diabetes in adults: diagnosis and management [article online]. 2015. Available from nice.org.uk/guidance/ng17. Accessed 16 March 2017 [PubMed]
- 44.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 2011;12:21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shungin D, Winkler TW, Croteau-Chonka DC, et al.; ADIPOGen Consortium; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GEFOS Consortium; GENIE Consortium; GLGC; ICBP; International Endogene Consortium; LifeLines Cohort Study; MAGIC Investigators; MuTHER Consortium; PAGE Consortium; ReproGen Consortium . New genetic loci link adipose and insulin biology to body fat distribution. Nature 2015;518:187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Agarwala V, Flannick J, Sunyaev S, Altshuler D; GoT2D Consortium . Evaluating empirical bounds on complex disease genetic architecture. Nat Genet 2013;45:1418–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stumvoll M, Goldstein BJ, van Haeften TW. Type 2 diabetes: principles of pathogenesis and therapy. Lancet 2005;365:1333–1346 [DOI] [PubMed] [Google Scholar]
- 48.Parker SCJ, Stitzel ML, Taylor DL, et al.; NISC Comparative Sequencing Program; National Institutes of Health Intramural Sequencing Center Comparative Sequencing Program Authors; NISC Comparative Sequencing Program Authors . Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci USA 2013;110:17921–17926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dann SG, Selvaraj A, Thomas G. mTOR Complex1-S6K1 signaling: at the crossroads of obesity, diabetes and cancer. Trends Mol Med 2007;13:252–259 [DOI] [PubMed] [Google Scholar]
- 50.Claussnitzer M, Dankel SN, Klocke B, et al.; DIAGRAM+Consortium . Leveraging cross-species transcription factor binding site patterns: from diabetes risk loci to disease mechanisms. Cell 2014;156:343–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farh KK, Marson A, Zhu J, et al. Genetic and epigenetic fine-mapping of causal autoimmune disease variants. Nature 2015;518:337–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.