Abstract
Type 2 diabetes (T2D) affects more than 415 million people worldwide, and its costs to the health care system continue to rise. To identify common or rare genetic variation with potential therapeutic implications for T2D, we analyzed and replicated genome-wide protein coding variation in a total of 8,227 individuals with T2D and 12,966 individuals without T2D of Latino descent. We identified a novel genetic variant in the IGF2 gene associated with ∼20% reduced risk for T2D. This variant, which has an allele frequency of 17% in the Mexican population but is rare in Europe, prevents splicing between IGF2 exons 1 and 2. We show in vitro and in human liver and adipose tissue that the variant is associated with a specific, allele-dosage–dependent reduction in the expression of IGF2 isoform 2. In individuals who do not carry the protective allele, expression of IGF2 isoform 2 in adipose is positively correlated with both incidence of T2D and increased plasma glycated hemoglobin in individuals without T2D, providing support that the protective effects are mediated by reductions in IGF2 isoform 2. Broad phenotypic examination of carriers of the protective variant revealed no association with other disease states or impaired reproductive health. These findings suggest that reducing IGF2 isoform 2 expression in relevant tissues has potential as a new therapeutic strategy for T2D, even beyond the Latin American population, with no major adverse effects on health or reproduction.
Introduction
Type 2 diabetes (T2D) affects 415 million people worldwide and is predicted to be the seventh leading cause of death by 2030 (1). T2D is also the leading cause of preventable blindness (2) and end-stage renal disease (3) and is a major risk factor for heart attack and stroke (4).
An individual’s risk of developing T2D is influenced by a combination of lifestyle, environmental, and genetic factors. Uncovering the genetic contributors to diabetes holds promise for clinical impact by revealing new therapeutic targets aimed at the molecular and cellular mechanisms that lead to disease. Genome-wide association studies performed during the past decade have uncovered more than 100 regions associated with T2D (5–12). Although these studies have provided a better understanding of T2D genetics, the majority of identified variants fall outside protein-coding regions, leaving the molecular mechanism by which these variants confer altered disease risk obscure. Consequently, T2D genome-wide association studies have identified few loci with clear therapeutic potential.
The identification of loss-of-function variants associated with reduced risk of disease is of particular interest, as their protective genetic effect can be potentially recapitulated by pharmacological inhibition. Furthermore, if carriers of protective, loss-of-function variants are otherwise healthy, this suggests that specific pharmacological perturbation of the effector protein could confer benefit without significant adverse health effects (13).
Genetic explorations in traditionally understudied populations have succeeded in identifying novel T2D variants in Mexican populations (6,14), as well as in East Asians (15), Greenlanders (16), and African Americans (8). In Mexico, T2D is one of the leading causes of death and has a prevalence twice that of non-Hispanic whites in the U.S. and is among the highest worldwide (17,18). Although different environmental and lifestyle risk factors in Mexico partially explain the increased prevalence of T2D, unique genetic influences also contribute (6,14). Here, we explored protein-coding variants present at higher frequency in people of Latino descent to shed further light on genetic risk factors for T2D in Mexico. We identified a novel T2D association with a protective, splice-acceptor variant that disrupts expression of IGF2 isoform 2, providing a clear hypothesis for future mechanism of action and therapeutic inquiries.
Research Design and Methods
Study Participants
This study was performed as part of the Slim Initiative in Genomic Medicine for the Americas (SIGMA) Type 2 Diabetes Consortium, whose goal is to improve the understanding of the genetic basis of T2D in Mexican and Latin American populations. The discovery data set consisted of four studies from Mexico or Mexicans living in the U.S. comprising a total of 4,210 case and 4,786 control subjects, which resulted in a final sample size of 4,052 case and 4,606 control subjects after quality control of the genotyping data (Table 1, details of these studies are provided in the Supplementary Data). All participants from the discovery and replication data sets provided informed consent for conducting this study. Their respective local ethics committees approved all contributing studies.
Table 1.
Study cohorts comprising the SIGMA T2D exome chip project data set
| Study | Sample location | Study design | Group | n | Percent male | Age (years) | Age of onset (years) | BMI (kg/m2) | Fasting plasma glucose (mmol/L) |
|---|---|---|---|---|---|---|---|---|---|
| UNAM/INCMNSZ Diabetes Study (UIDS) |
Mexico City, Mexico |
Prospective cohort |
Control |
1,164 |
41.3 |
55.4 (9.4) |
— |
28.2 (3.9) |
4.8 (0.5) |
| T2D |
835 |
40.1 |
56.3 (12.4) |
44.2 (11.4) |
28.6 (4.6) |
9.8 (4.5) |
|||
| Diabetes in Mexico Study (DMS) |
Mexico City, Mexico |
Prospective cohort |
Control |
486 |
25.3 |
52.6 (7.8) |
— |
28.1 (4.5) |
5 (0.4) |
| T2D |
715 |
32.3 |
55.9 (11) |
47.7 (10.4) |
29.0 (5.6) |
8.8 (3.9) |
|||
| Mexico City Diabetes Study (MCDS) |
Mexico City, Mexico |
Prospective cohort |
Control |
671 |
38.3 |
62.2 (7.7) |
— |
29.4 (4.6) |
5 (0.6) |
| T2D |
315 |
40.3 |
63.9 (7.5) |
54.7 (9.7) |
30.0 (5.3) |
8.8 (4) |
|||
| Multiethnic Cohort Study of Diet and Cancer (MEC) | Los Angeles, CA | Case-control | Control |
2,285 |
48.5 |
59.2 (7) |
— |
26.6 (3.9) |
— |
| T2D | 2,187 | 47.6 | 59.1 (6.9) | — | 29.9 (5.3) | — |
Data are mean (SD), unless stated otherwise. UNAM/INCMNSZ, Universidad Nacional Autónoma de México/Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán.
Genotyping and Quality Control
The genotyping of the discovery sample was done using the Exome Illumina array at the Broad Institute Genomics Platform (Cambridge, MA), which received, quality controlled, and tracked DNA samples for Exome array processing. The exome array was designed to cover rare and low-frequency coding variants identified through whole-exome sequencing studies of 12,031 individuals from different populations including 362 individuals of Hispanic ancestry. Details on the genotyping of the different discovery and replication cohorts are provided in the Supplementary Data.
A total of 3,732 samples genotyped by the exome array also underwent whole-exome sequencing (14) and were used to create a population-specific reference panel in order to fine-map the association at the IGF2 locus (Supplementary Figs. 1 and 2 and Supplementary Data).
In Vitro Splicing Assay
IGF2 minigenes, including the first three exons and two introns of the IGF2 gene (chr11:2150342–2156088, Hg19), and containing either the G or A allele of rs149483638, were synthesized by GENEWIZ and subcloned into the mammalian expression vector pcDNA3.1. A stop codon was introduced at the end of exon 3 to stop translation of the expressed protein. Human embryonic kidney 293 cells were transfected with either minigene using TransIT transfection reagent (Mirus Bio). RNA was extracted from the cells 24 h posttransfection using the RNeasy Extraction Kit (Qiagen), and 1 μg of RNA was reverse-transcribed into cDNA using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems).
We used two probes to detect IGF2 expression by droplet digital PCR (ddPCR): one probe that targets exon 3 and recognizes all IGF2 isoforms (custom probe 10031276; Bio-Rad), and one probe that targets the exon 1-2 junction and recognizes only isoforms with exon 1-2 splicing (cat. # Hs04188275; Life Technologies). A probe targeting ACTB (cat. # 10031255; Bio-Rad) was used as an endogenous control for both IGF2 probes. Reaction mixtures consisted of 1 µL of cDNA (diluted 200× from the RT-PCR reaction), 1× of Supermix (Version 1) for Probes (Bio-Rad), 1× of each probe (IGF2 specific and ACTB specific), and water to a final volume of 20 µL. Each reaction was partitioned into droplets using a QX200 automatic droplet generator (Bio-Rad). The droplets then underwent PCR as follows: 95°C for 10 min, 40 cycles of 94°C for 30 s and 60°C for 1 min, followed by 98°C for 10 min. The QX200 droplet reader was then used to measure the fluorescence of each of the two fluorophores corresponding to the ACTB and IGF2 probes. After subtracting the background IGF2 signal detected in untransfected cells (which was minimal), IGF2 was normalized to ACTB within each sample. The level of exon 1-2 splicing is presented relative to the total IGF2 for that sample, as determined by the exon 3 probe.
Visceral Adipose and Liver Tissue Collection
Visceral adipose and liver tissue samples were collected from subjects undergoing bariatric surgery for severe obesity (BMI >40 kg/m2, or >35 kg/m2 with comorbid entities) or elective surgery in nonobese patients. Patients were selected for bariatric surgery after 6 months of rigorous lifestyle intervention. All individuals were Mexican Mestizos older than 18 years, carefully selected from the Integral Clinic of Surgery for Obesity and Metabolic Diseases or General Surgery Department at the Tláhuac Hospital in Mexico City. Tissue samples were obtained at the beginning of the surgery with harmonic scalpel in all cases as follows: visceral fat was obtained from the greater omentum at the middle of the greater curvature of the stomach, and liver biopsy was obtained at the distal end of the left hepatic lobe, just above the spleen. Visceral adipose and liver tissue samples were frozen immediately after removal. The protocol for collecting visceral adipose and liver tissue samples was approved by the respective local research and ethics committees and all patients signed an informed consent. Genomic DNA was purified from whole-blood samples and the genotyping of the rs149483638 variant was performed as described in the Diabetes in Mexico Study 2 (DMS2) cohort. RNA was isolated at the Broad Institute Genomics Platform (Supplementary Data).
RNA-seq Analysis of Adult and Embryonic Stem Cell–Derived Cell Lines
RNA-seq data sets for embryonic stem cell–derived human pancreatic progenitor cells (19); embryonic stem cell–derived neuronal progenitor, trophoblast, mesendoderm, and mesenchymal cells; and adult liver (20) and adult pancreatic islets (21) were aligned using STAR (22) against the hg19 reference genome, allowing for up to 10 mismatches and disallowing multimapping. Exon expression level was calculated in reads per kilobase million as described in Mortazavi et al. (23).
The expression of IGF2 exon 2 across adult human tissues was queried using RNA sequencing data from the Genotype-Tissue Expression (GTEx) Consortium (24) spanning 54 tissue types and 550 individuals (dbGaP Accession phs000424.v5.p1). The sample collection, sequencing, and data processing have been described previously (24). For these analyses, the exon-level quantifications were generated using RNA-SeQC (25) with GENCODE version 18 reference annotations.
IGF2 Isoform Expression In Vivo by ddPCR
For the tissue samples, we used reverse-transcriptase ddPCR (RT-ddPCR) (Bio-Rad) to measure the expression of IGF2 using probes that targeted all IGF2 isoforms (assay Hs01005963; Life Technologies) and the specific isoform disrupted by the splice-site variant (assay Hs04188276; Life Technologies). Each assay was run separately, with an assay targeting G2E3 used as an endogenous control, which was selected for stability across different samples and for showing levels of expression similar to IGF2 isoform 2 (forward primer: GTCCACACACCCTTTGAAAGTT; reverse primer: CAGGTTTATGACACAGGATGCTA; probe: CACCAAGGGTTTTCAGACCCTGC, HEX-labeled). In adipose tissue, we used 30 ng of RNA to quantify exon 2 of IGF2 and 5 ng to quantify total IGF2 expression. In liver, we used 20 ng of RNA to quantify exon 2 of IGF2 and 15 ng to quantify total IGF2 expression. We used 1× of IGF2 assay, 1× of G2E3 assay primer probe mix (20× mixture containing 18 μmol/L of forward and reverse primers each and 5 μmol/L of fluorescent probe), 1× of 2× One-Step RT-ddPCR Supermix (Bio-Rad), 1 mmol/L manganese acetate (Bio-Rad), and water to a final volume of 20 μL. Each reaction was partitioned into thousands of nanoliter-sized droplets using a QX200 manual or automatic droplet generator (Bio-Rad). The droplets underwent PCR as follows: 60°C for 30 min, 95°C for 5 min, 50 cycles of 94°C for 30 s and 60°C for 1 min, followed by 98°C for 10 min. Following PCR, the fluorescence from each of the two fluorophores corresponding to IGF2 and G2E3 was read by a QX200 droplet reader (Bio-Rad), yielding precise, digital counts of the number of droplets containing the RNA targeted by each assay. Data were processed using QuantaSoft software (Bio-Rad), which estimates the absolute concentration of input RNA templates by Poisson-correcting the fraction of droplets that are positive for each amplicon. We used the ratio of IGF2 concentration to control G2E3 concentration as the normalized IGF2 expression value for downstream analyses.
Plasma IGF2 Measurements
Total, circulating IGF2 levels were measured in plasma from 120 individuals, 40 per genotype at rs149483638, which were matched by ancestry, BMI, age, sex, and T2D status. IGF2 measurements were performed by the Vanderbilt University Medical Center Hormone Assay & Analytical Services Core, using a Millipore Human IGF-I, II Magnetic Bead Panel (cat. # HIGFMAG-52K). The assay was read on a Luminex MAGPIX instrument. The association results were compared using linear regression adjusting for the first two principal components, BMI, age, sex, and T2D status.
Statistics
We used efficient mixed-model association (EMMAX) to test the genetic variants for association with T2D adjusted by age, BMI, and sex, while controlling for sample structure (26). Odds ratios (ORs) were estimated using logistic regression models on T2D status, adjusting for age, BMI, and ancestry as specified in the Supplementary Data. The experiment-wide statistical significance threshold was set to P < 5 × 10−8 to adjust for the number of variants evaluated.
For functional analyses, statistical analyses were performed using linear and logistic regression and nonparametric tests, and P < 0.05 was considered significant for these functional studies.
Integration of Data and Imputation
For the credible set analysis, we first built two data sets. One data set was comprised of 4,478 samples that had been genotyped by exome chip and OMNI2.5. The other data set comprised another subset of 3,732 samples genotyped by exome chip, OMNI2.5, and whole-exome sequencing, which we integrated to build a population-specific reference panel for protein-coding variation. We kept all the variants with a minor allele frequency (MAF) higher than 0.001 for both data sets. We phased both data sets with SHAPEIT2 (27) (version 2.5,). We then imputed the 1000 Genomes (phase 3, released June 2014) into both data sets separately. We also imputed the whole-exome variants with the population-specific reference panel described above into the samples that did not undergo whole-exome sequencing. We used impute 2 information score >0.8 as a postimputation quality control. We then performed the association analysis separately in each cohort using SNPTEST and adjusted for BMI, age, sex, and the first two principal components to adjust for population stratification. We then meta-analyzed both results using METAL (28).
Results
We performed association analysis between T2D and each of the 158,892 nonmonomorphic variants genotyped in the Illumina exome array that passed stringent quality control in 4,210 T2D case and 4,786 control subjects from four different cohorts in Mexico or Mexicans living in the U.S. (Table 1 and Supplementary Data). The top genome-wide significant (P < 5 × 10−8) signals replicated previously reported variants, including those at TCF7L2 and KCNQ1 (29,30), with consistent effect sizes and directions of effect (Fig. 1A and Supplementary Table 1), and confirmed the association of variants in SLC16A11, originally identified in a genome-wide study of the same subjects included in the present analysis (6).
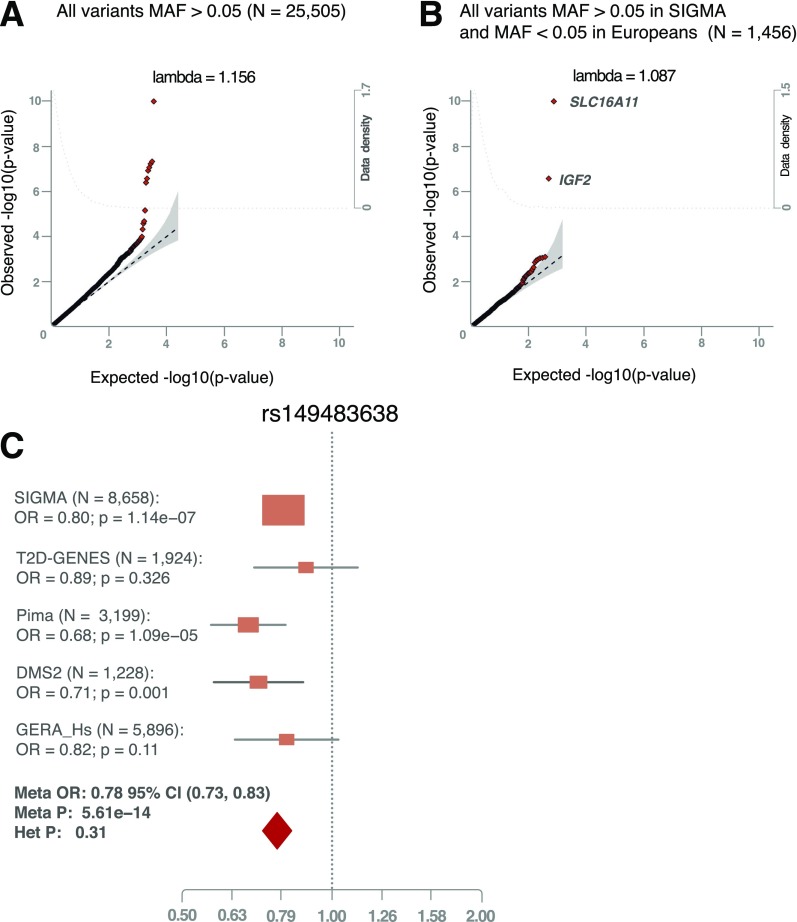
Figure 1.
Discovery and replication of the rs149483638 T2D protective variant. Quantile-quantile plot for all common variants (A) and for Mexican population–specific variants (B). The plot shows the two most significant variants that have low frequency in Europeans but higher frequency in the Mexican population. C: Forest plot for the meta-analysis of rs149483638 variant in IGF2. We replicated the rs149483638 association in four independent data sets: 1,007 T2D case and 917 control subjects of Hispanic origin from T2D-GENES (MAF = 0.12, OR = 0.89, P = 0.3), 1,519 T2D case and 1,680 control subjects of full-heritage American Indian ancestry from the Pima cohort (MAF = 0.14, OR = 0.68, P = 1.09 × 10−5), 427 case and 751 control subjects of self-identified indigenous individuals from different ethnic groups in Mexico (DMS2 cohort) (MAF = 0.36, OR = 0.71, P = 0.001), and 1,064 case and 4,832 control subjects from the subset of subjects with Latino ancestry from the GERA cohort (MAF = 0.06, OR = 0.82, P = 0.11).
To identify variants enriched in the Mexican population, we next focused our analysis on variants of low or rare frequency in Europeans (MAF <0.05), but common (MAF >0.05) in Mexicans (Fig. 1B). Of novel findings in this analysis, a single nucleotide polymorphism predicted to disrupt a canonical splice acceptor site in IGF2 achieved the highest statistical significance (rs149483638, MAF = 0.17, OR = 0.80, P = 1.6 × 10−7). Heterozygous carriers of this variant have a 22% decreased risk of T2D, and risk in homozygous carriers is reduced by 40%. We did not find associations between rs149483638 and other glycemic or metabolic traits (Supplementary Table 2). This variant is rare in individuals of European ancestry (MAF = 0.0002) and at low frequency in individuals of East Asian (MAF = 0.01) or African ancestry (MAF = 0.001) (31) (http://exac.broadinstitute.org). This variant showed a stronger association with T2D when adjusting for population stratification using principal components, as the protective T allele was more frequent in individuals with higher Native American ancestry, which is also a risk factor for T2D. Thus, we identified a protective genetic factor for T2D, present in 17% of a Latino population.
We performed several analyses that suggest rs149483638 is the most likely causal variant for the protective signal. First, we confirmed that other rare variants do not explain the association through a phenomenon called “synthetic association” (32) (Supplementary Figs. 1–3 and Supplementary Data). Second, we established that known T2D variants at the nearby KCNQ1 locus (6,30,33) do not explain the association signal, as the two independently associated variants at the KCNQ1 locus are in weak linkage disequilibrium with rs149483638 in our data set, (r2 with rs139647931 = 0.026, r2 with rs2237897 = 0.028), and the T2D association with rs149483638 remains significant after conditioning for these two variants (OR = 0.81, P = 6.9 × 10−6). Last, we carried out an analysis to identify the most likely causal variant(s). To do so, we first integrated whole-exome sequencing data, available for a subset of 3,732 samples, with exome chip and genotyping array data from OMNI2.5 and performed imputation with 1000 Genomes phase 3 reference panel in all the samples (Supplementary Figs. 1 and 2 and Supplementary Data). We then used a Bayesian approach to prioritize and rank variants according to likelihood of being causal (Supplementary Data). This analysis identified the splice acceptor variant (rs149483638) as having the highest probability of being causal for the T2D-protective association (Supplementary Fig. 2 and Supplementary Table 3).
We then sought to replicate the rs149483638 association in four independent data sets: T2D case and control subjects of Hispanic origin from the Type 2 Diabetes Genetic Exploration by Next-generation sequencing in multi-Ethnic Samples (T2D-GENES) Consortium (14) (MAF = 0.12, OR = 0.89, P = 0.3), individuals of full-heritage American Indian ancestry from the Pima cohort (34) (MAF = 0.14, OR = 0.68, P = 0.1 × 10−5), self-identified indigenous individuals from different ethnic groups in Mexico (DMS2 cohort) (14) (MAF = 0.36, OR = 0.71, P = 0.001) (Supplementary Data), and a subsample of Hispanic individuals from the Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort (35) (MAF = 0.06, OR = 0.82, P = 0.11). A meta-analysis of the discovery and these replication studies produced a genome-wide significant association (OR = 0.78, P = 5.6 × 10−14) (Fig. 1C). We also tested the association of rs149483638 with diabetes incidence in the subset of 616 Hispanic or American Indian individuals with prediabetes that were followed for an average of 3 years in the Diabetes Prevention Program (DPP) (36). The direction of effect was consistent with the results in other data sets, but was not statistically significant (hazard ratio = 0.76, P = 0.24) (Supplementary Table 4), possibly because of lower power in this data set due to its smaller sample size and/or the inclusion of individuals with prediabetes who are at high risk for T2D at baseline. As an additional replication, and to further confirm that the findings are not due to potential population stratification, we analyzed this variant in the San Antonio Families studies, using a family-based association approach (37–39) (N = 2,980); results are consistent with those obtained through the population-based analyses (z = −2.3, P = 0.02) (Supplementary Table 5). The overall meta-analyses including these two last data sets further strengthened the observed association between rs149483638 and T2D (overall P = 4.8 × 10−14) (Supplementary Table 4).
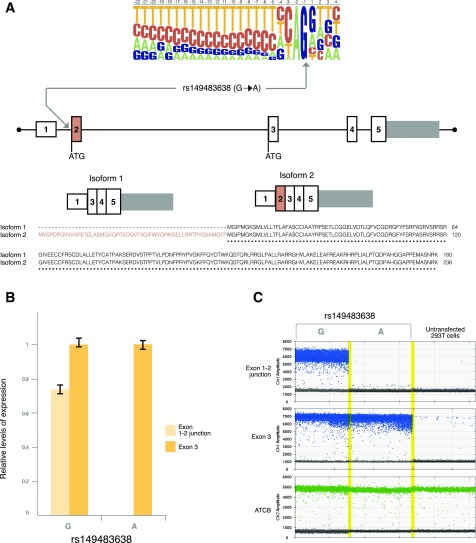
Having confirmed that the rs149483638 is driving the association for T2D protection, we performed experiments to understand the mechanism through which this beneficial metabolic action occurs. Using in silico analyses, we found that the protective A allele of rs149483638 variant (allele defined in the reverse strand, in which IGF2 is expressed) is predicted to disrupt a canonical splice-site acceptor controlling inclusion of exon 2 in IGF2 isoform 2 (P01344–3, Uniprot). Compared with isoform 1, IGF2 isoform 2 has 56 additional N-terminal amino acids, encoded by exon 2. Therefore, the A allele is predicted to specifically disrupt expression of isoform 2 (Fig. 2A). IGF2 isoform 2 is lowly expressed in most adult tissues (24), showing the highest expression in pancreatic islets and liver and adipose tissue, where it represents approximately 1–2% of total IGF2 transcripts (Supplementary Figs. 5 and 6).
Figure 2.
rs149483638 prevents splicing in vitro. A: This variant is located at a canonical splice acceptor site and is predicted to cause skipping of exon 2 of IGF2 isoform 2. B: 293T cells were transfected with IGF2 minigenes containing the first three exons and two introns of the IGF2 gene, and either allele of the rs149483638 C>T variant (G>A in the reverse strand) and cDNA were analyzed by ddPCR. This analysis revealed no expression of the IGF2 exon 1-2 junction in cells transfected with the minigene containing the T2D-protective rs149483638 A allele. This was in contrast to the high levels of exon 1-2 splicing detected in cells transfected with the G allele. C: One-dimensional plots of the ddPCR droplets plotted in B. No IGF2 transcript was detected in untransfected samples. ACTB was used as an internal control.
To determine if rs149483638 affects splicing as predicted, we measured exon 1-2 splicing in human cells transfected with IGF2 minigenes consisting of the first three exons and two introns of IGF2 (chr11:2150342–2156088) and containing either the G or A allele of rs149483638. In contrast to the high level of exon 1-2 splicing detected from the G allele, no exon 1-2 splicing was detected in samples expressing the IGF2 minigene containing the A allele (Fig. 2B and C), indicating a specific effect of the rs14983836 variant on IGF2 isoform 2 splicing.
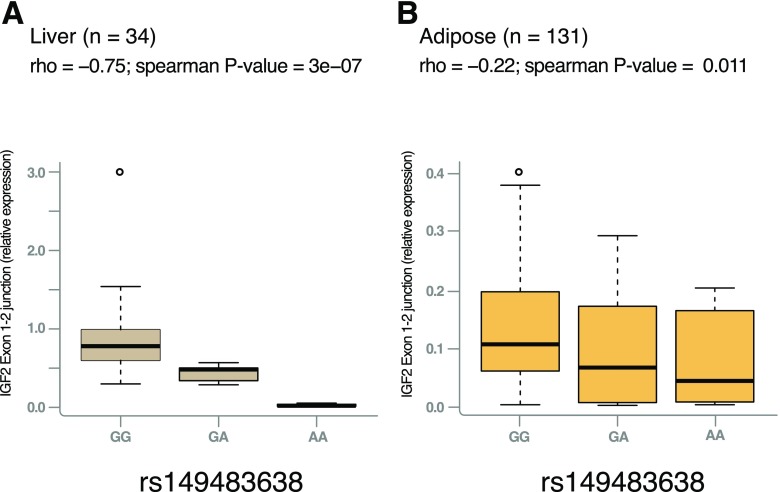
To assess whether the alternative allele at rs149483638 alters transcript expression in vivo, we collected 34 liver and 133 adipose tissue samples from Mexican rs149483638 variant carriers and noncarriers and analyzed expression of IGF2 isoform 2 by measuring levels of the exon 1-2 splice junction using ddPCR. The dosage of the A allele was negatively correlated with expression of IGF2 isoform 2 in both liver (ρ = −0.75, Spearman P = 3.2 × 10−7) and adipose tissue (ρ = −0.22, Spearman P = 0.01) (Fig. 3). In contrast, no significant correlation was detected for IGF2 isoform 1 expression, as measured by exon 3 (common to both isoforms but representative of isoform 1, which constitutes ∼98% of IGF2 in these tissues (Supplementary Fig. 7A and B). Similarly, we observed no association between rs149483638 genotype and circulating levels of total IGF2, which is expected to reflect the majority isoform, isoform 1 (Supplementary Fig. 7C). Together, in vitro and in vivo studies indicate that the T2D-protective A allele cause a reduction of the expression of IGF2 isoform 2 via disruption of exon 1-2 splicing.
Figure 3.
rs149483638 prevents splicing between exon 2 in liver and in adipose tissue. The dosage of the T2D-protective A allele is correlated with lower expression of IGF2 isoform 2 (as measured by expression levels of the exon 1-2 junction) in liver (GG, n = 21; GA, n = 9; AA, n = 4) (A) and in adipose tissue (GG, n = 83; GA, n = 43; AA, n = 5) (B).
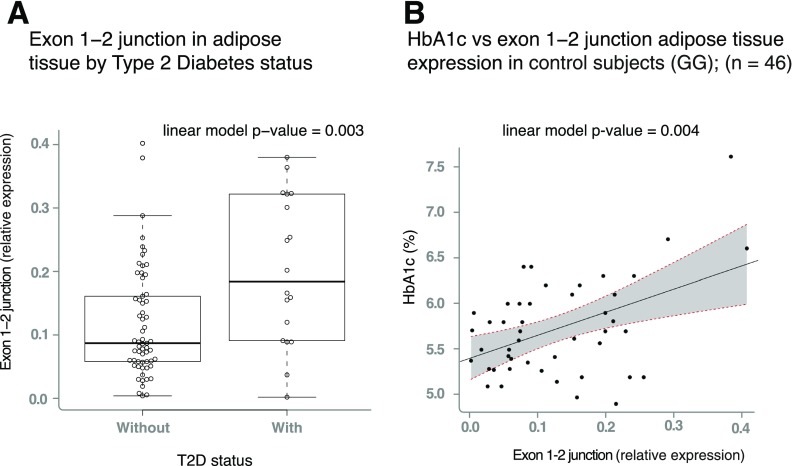
Collectively, our results suggested that decreased expression of IGF2 isoform 2 is associated with decreased risk of T2D. We formally tested the association between expression of IGF2 isoform 2 and T2D status and glycemic traits relevant to T2D in homozygous noncarriers (GG) and observed reduced expression of the isoform 2 in visceral adipose tissue in individuals without diabetes compared with those with T2D (P = 0.003) (Fig. 4A). This finding provides a link between the genetic association, gene expression, and T2D risk, suggesting that a “dose-response” curve may exist between IGF2 isoform 2 expression and T2D risk.
Figure 4.
Expression of IGF2 isoform 2 is associated with T2D and HbA1c. A: Boxplots representing the expression of IGF2 isoform 2 across T2D case and control subjects in individuals homozygous for the G common allele. The linear model P value represents the association between IGF2 isoform 2 expression, adjusted by age, BMI, and sex. B: IGF2 isoform 2 positively correlates with higher HbA1c in participants without diabetes. The gray area limited by the dashed red lines represents the 95% CI of the slope of the linear regression. Patients with HbA1c above 6.5% did not have T2D according to the diagnostic criteria of Mexico at the time of extraction, as HbA1c was not considered a criterion in Mexico at the time of extraction. Therefore, none of the subjects were receiving any glucose-lowering treatment. For clarity, as the genotype is strongly associated with IGF2 isoform 2 expression, only individuals carrying the GG genotype are plotted in A and B.
Furthermore, expression of IGF2 isoform 2 in visceral adipose tissue is positively correlated with plasma glycated hemoglobin (HbA1c) in individuals without T2D, or untreated subjects with T2D, in homozygous noncarriers (GG) (P = 0.004) (Fig. 4B). We did not detect significant associations between IGF2 isoform 2 expression and glycemic traits or T2D status in the liver, possibly due to smaller sample size and, therefore, reduced statistical power for this tissue. We also did not find associations between the expression of isoform 1 and HbA1c or T2D in either adipose tissue or liver, suggesting the protective effect is specific to IGF2 isoform 2 (Supplementary Fig. 7D and E). Overall, these results suggest that pharmacological inhibition of IGF2 isoform 2 levels or activity could recapitulate the protective effect of the rs149483638 variant.
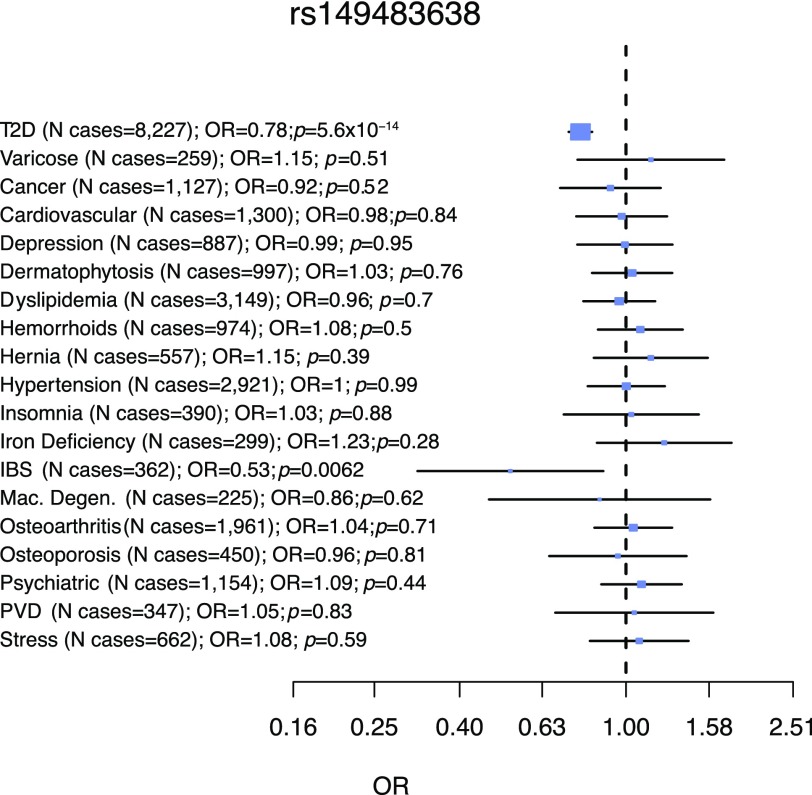
To assess potential negative effects of isoform 2 perturbation, we screened available data sets containing information on humans homozygous for the A allele of rs149483638. In the Exome Aggregation Consortium database (31) (ExAC) (http://exac.broadinstitute.org), we observed that there were 240 AA homozygotes (isoform 2 knockouts) within the Latin American population, all of whom were free of severe clinically recognized pediatric diseases. Furthermore, within the discovery and replication cohorts, we identified 293 AA homozygous individuals for whom clinical history of other diseases and fertility records were available and compared them to up to 6,407 GG homozygous individuals. We found that A allele homozygotes show reduced risk for T2D (OR = 0.63, P = 0.004) but do not exhibit increased prevalence of other diseases and have indistinguishable reproductive performance based on number of children and percentage of individuals with children (Supplementary Table 6 and Supplementary Fig. 9). We also performed a phenome-wide association analysis in the GERA cohort, which revealed that rs149483638 is not associated with increased risk for any of the 18 available relevant medical conditions (Supplementary Table 7 and Fig. 5). Together, these data suggest that reduced activity or levels of IGF2 isoform 2 do not a have a negative impact on general health or fertility.
Figure 5.
Phenome-wide analysis of rs149483638 variant. The protective variant was tested for association across 18 different disease traits previously categorized in the subsample of GERA cohort of Latino ancestry (5,896 individuals). Although the rs149483638 variant was associated with reduced risk of T2D, there was no significant association seen for other 18 conditions. Association analyses were done by logistic regression analyses, considering additive model, and correcting for age, BMI, sex, and the first two principal components to correct for population stratification. IBS, irritable bowel syndrome; Mac. Degen., macular degeneration; Psychiatric, any psychiatric condition; PVD, peripheral vascular disease; Stress, acute reaction to stress.
Discussion
Human genetics has proved successful in uncovering genetic risk factors for both Mendelian and complex diseases. In addition to identifying individuals at increased risk for disease, knowledge of genetic variants that influence disease risk can be translated for clinical impact by identifying new potential therapeutic targets and improving the success rate of drug development. In particular, loss-of-function variants that are protective for disease are attractive drug development targets, as their reduced function can be mimicked by pharmacological inhibition (40). Through whole-genome screening of protein-coding variants in Latin Americans, we have identified a protective T2D variant that disrupts a protein-coding exon of IGF2, leading to lower IGF2 isoform 2 expression. Even in the absence of the protective variant, lower expression of IGF2 isoform 2 is observed in subjects without T2D compared with those with T2D and correlates with lower HbA1c levels in individuals with diabetes. Importantly, we found no genetic evidence that loss of IGF2 isoform 2 has a major negative impact on human health or reproduction. These findings suggest that reducing IGF2 isoform 2 levels could provide therapeutic benefit for patients with T2D without adverse side effects.
Although a role for IGF2 in T2D and related glycemic traits has been previously suggested, our findings validate IGF2 as a gene relevant to T2D pathophysiology in human populations. Recent human genetic studies in Latino and African American populations identified T2D risk associations at the INS-IGF2 locus (6,8); however, the effector gene responsible for the modified T2D risk was not identified. Here, we identified a protective signal for which the most probable causal variant is functionally validated as having an impact on IGF2 isoform 2 expression.
It is unclear why the allele frequency of rs149483638 shows so much variability across populations. Even within different Latin American populations, the MAF ranges from ∼5% in Puerto Ricans to ∼23% in Peruvians. In Mexicans, the frequency increases with the percentage of Native American ancestry. Future analyses should clarify whether this protective variant underwent positive selection or its variable frequency is a result of genetic drift (41).
IGF2 has previously been implicated in T2D due to well-established metabolic functions of isoform 1. IGF2 is a peptide hormone with 47% amino acid sequence identity to insulin that regulates growth and metabolism through binding with insulin receptor, insulin-like growth factor 1 receptor, and insulin-like growth factor 2 receptor (42). IGF2 regulates fetal development and differentiation and has an important role in embryonic growth (43). In the adult, IGF2 is expressed in several tissues, with highest levels in liver, where it is synthesized and released into the periphery. In the pancreas, IGF2 promotes β-cell proliferation and survival (44). Dysregulation of IGF2 expression has been reported in several metabolic diseases, including growth disorders (45), obesity (46), and diabetes (47).
In mice, which only express isoform 1, Igf2 inactivation promotes brown preadipocyte differentiation, protecting from insulin resistance (48). On the other hand, Igf2 isoform 1 overexpression in murine pancreatic β-cells causes β-cell dysfunction that ultimately leads to hyperglycemia (49,50). In humans, IGF2 has also been indirectly implicated in T2D due to the association of variants in IGF2 mRNA binding protein 2 (encoded by IGF2BP2) with T2D (51,52). The protein encoded by IGF2BP2, IMP2, regulates IGF2 mRNA levels, and mice deficient for Igf2bp2 are resistant to diet-induced obesity and show higher glucose tolerance and insulin sensitivity (52,53). Although these studies support a potential role for IGF2 in T2D pathogenesis, they are based on the function of IGF2 isoform 1, while the function(s) of IGF2 isoform 2—the isoform implicated in our study—remains unknown. Future studies are needed to elucidate how IGF2 isoform 2 differs from isoform 1 in its regulation and its effects on human cellular metabolism and physiology.
Though the molecular and cellular links between reduced expression of IGF2 isoform 2, glucose regulation, and T2D have not yet been established, the observations reported here suggest inhibition of IGF2 isoform 2 might be a potential strategy for prevention or treatment of T2D. Previously identified loss-of-function mutations that cause beneficial metabolic phenotypes have spurred the development of recently approved drugs. One such example is that of sodium–glucose cotransporter 2 inhibitors, a new class of oral antidiabetes agents that lower blood glucose and, for at least one agent in the class, also reduce cardiovascular events among individuals at high risk for such events (54). These agents mimic physiology observed in familial renal glucosuria, a condition caused by loss-of-function mutations in SLC5A2, which encodes sodium–glucose cotransporter 2 (55). Therefore, perturbation of IGF2 isoform 2, which protects against T2D without observable adverse phenotypes in humans, has potential for development as a novel metabolic therapy. Our findings that reduced expression of IGF2 isoform 2 is correlated with lower prevalence of T2D in individuals who do not carry the protective allele further suggests that such a treatment could provide therapeutic benefit beyond Latin American populations.
Alternative splicing is a major source of proteome diversity, and missplicing of genes is a cause of several Mendelian diseases (56). Here, we demonstrate that disruption of a canonical splice-acceptor site is also associated with altered risk of a complex disease. This mechanism of variant action suggests a specific pharmacological strategy for T2D, namely, inducing exon skipping of IGF2 exon 2 to prevent IGF2 isoform 2 expression. Indeed, induction of exon-skipping through use of modified antisense oligonucleotides has been successfully applied as a molecular therapy for a form of Duchenne muscular dystrophy caused by a stop-coding mutation in exon 51 of DMD gene, and two drugs based on this idea are currently in advanced clinical trials (57). Overall, our identification of a T2D-protective splice variant in IGF2 suggests that modulating IGF2 isoform splicing, possibly in accessible hepatic tissue, could be a possible strategy for preventing or treating T2D and opens a new line of investigation to characterize the mechanism through which disruption of IGF2 isoform 2 protects against T2D.
In conclusion, genetic and functional evidence suggest a specific IGF2 isoform as functionally relevant for the T2D physiology. Loss of function of this isoform is associated with reduced risk of T2D and shows no evidence of increased risk for other diseases, highlighting this isoform as a potential therapeutic target for T2D. Our results open a new line of investigation to characterize the mechanism through which disruption of IGF2 isoform 2 protects against T2D.
Supplementary Material
Article Information
Acknowledgments. Researchers of the DMS2 study thank Olaf Iván Corro Labra and José Luis de Jesus García Ruíz from the Comisión Nacional para el Desarrollo de los Pueblos Indígenas for their support in sample collection, for which they were not compensated. The authors also thank Saúl Cano-Colín (Universidad Nacional Autónoma de Mexico) for his technical assistance in the genotyping of rs149483638 variant, Vicky Kaur (Massachusetts General Hospital) for her technical assistance in collecting the plasma samples for measuring IGF2 circulating levels, and Joan Bacardí (freelance designer at Pollomeanschicken) for his assistance in the preparation of figures. This article is dedicated to the memories of our colleagues Laura Riba, Hanna E. Abboud, and Brian E. Henderson.
Funding. This work was conducted as part of the Slim Initiative in Genomic Medicine for the Americas (SIGMA), a joint U.S.-Mexico project funded by the Carlos Slim Foundation. The Universidad Nacional Autónoma de México/Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán (UNAM/INCMNSZ) diabetes study was supported by the Consejo Nacional de Ciencia y Tecnología (grants 138826, 128877, and CONACyT-SALUD 2009-01-115250) and a grant from Dirección General de Asuntos del Personal Académico (UNAM, IT 214711). The Diabetes in Mexico Study (DMS) was supported by the Consejo Nacional de Ciencia y Tecnología (grant 86867) and by the Carlos Slim Foundation. The Mexico City Diabetes Study (MCDS) was supported by National Institutes of Health (grant R01HL24799 from the National Heart, Lung, and Blood Institute) and by the Consejo Nacional de Ciencia y Tecnología (grants 2092, M9303, F677-M9407, 251M, 2005-C01-14502, and SALUD 2010-2-151165). The Multiethnic Cohort Study of Diet and Cancer (MEC) was supported by National Institutes of Health (grants CA54281 and CA063464). A.L.W. is supported by National Institutes of Health Ruth L. Kirschstein Institutional National Research Service Award (number F32HG005944). The Diabetes in Mexico Study 2 (DMS2) cohort and the visceral adipose tissue and liver samples collection were supported by the Consejo Nacional de Ciencia y Tecnología (grants SALUD-233970 and 223019, respectively). The Pima longitudinal study is supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases. The DPP Research Group is supported by grant R01DK072041 and by the Intramural Research Program of National Institute of Diabetes and Digestive and Kidney Diseases and the Indian Health Service. The Vanderbilt University Medical Center Hormone Assay & Analytical Services Core is supported by the National Institutes of Health (grants DK059637 and DK020593). J.M.M. was supported by a Sara Borrell Fellowship from the Instituto de Salud Carlos III, grant SEV-2011-00067 of Severo Ochoa Program, EMBO Short-Term Fellowship, European Foundation for the Study of Diabetes/Lilly research fellowship, and the Beatriu de Pinós fellowship from the Agency for Management of University and Research Grants. The Genotype-Tissue Expression (GTEx) project was supported by the Common Fund of the Office of the Director of the National Institutes of Health. Additional funds were provided by the National Cancer Institute, National Human Genome Research Institute, National Heart, Lung, and Blood Institute, National Institute on Drug Abuse, National Institute of Mental Health, and National Institute of Neurological Disorders and Stroke. Donors were enrolled at Biospecimen Source Sites funded by National Cancer Institute\SAIC-Frederick, Inc. (SAIC-F), subcontracts to the National Disease Research Interchange (10XS170), Roswell Park Cancer Institute (10XS171), and Science Care, Inc. (X10S172). The Laboratory, Data Analysis, and Coordinating Center was funded through a contract (HHSN268201000029C) to Broad Institute, Inc. Biorepository operations were funded through an SAIC-F subcontract to the Van Andel Institute (10ST1035). Additional data repository and project management were provided by SAIC-F (HHSN261200800001E). The Brain Bank was supported by supplements to University of Miami grants DA006227 and DA033684 and to contract N01MH000028. Statistical methods development grants were made to the University of Geneva (MH090941 and MH101814), The University of Chicago (MH090951, MH090937, MH101820, and MH101825), the University of North Carolina at Chapel Hill (MH090936 and MH101819), Harvard University (MH090948), Stanford University (MH101782), Washington University in St. Louis (MH101810), and the University of Pennsylvania (MH101822).
Duality of Interest. J.C.F. received consulting honoraria from Merck and Boehringer Ingelheim. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. J.M.M., K.E., A.H.-C., H.M.-M., L.R., H.E.A., C.P.J., R.A.D., D.M.L., C.L.H., R.L.H., G.I.B., M.B., J.B., R.D., D.M., C.G.-V., C.A.H., C.A.A.-S., T.T.-L., J.Fl., S.B.R.J., L.O., D.A., and J.C.F. conceived, planned, and oversaw the study. J.M.M., K.E., J.Fl., A.H.-C., and H.M.-M. designed, performed, and analyzed most experiments. L.Ch., S.R., V.A., P.F., A.L.W., and C.H. performed additional statistical analyses. Broad Genomics Platform, M.H., S.G., L.Ch., C.H., J.Fl., P.F., and J.M.M. performed the genotyping and quality control of the data. J.M.M., S.B.-G., D.T., G.A.W., and S.B.R.J. designed and performed the phenome-wide analysis. R.G.L., Z.D., J.M.M., and S.B.R.J. performed, analyzed, and oversaw the in vitro splicing experiments. A.D.B., K.T., J.M.M., and S.A.M. performed and oversaw the expression assays in human samples. T.T., I.M., and J.Fe. analyzed the RNA-seq data. R.L.H., M.L.O.-S., R.R.-G., M.R.-T., Y.S.-K., H.G.-O., F.C.-C., F.B.-O., S.P., C.G.-V., C.A.H., C.A.A.-S., T.T.-L., L.O., K.A.J., R.S., J.L., C.Z., A.M.-H., E.J.C., E.M.-C., C.C.-C., M.E.G.-V., I.C.-B., L.M.-H., D.G.-V., U.A., B.E.H., L.R.W., L.L.M., O.A.-C., L.R., C.R.-M., S.I.-A., X.S., J.E.C., W.C.K., and L.J.B. provided patient samples and genetic data. L.Ca., N.B., and M.L.C. provided administrative, technical, or material support. The T2D-GENES Consortium and DPP Research Group provided genetic data. J.M.M., K.E., H.M.-M., G.A.W., R.L.H., J.Fl., S.B.R.J., D.A., and J.C.F. wrote and edited the manuscript. J.C.F. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Data Availability. The genotype and phenotype data are available in the Database of Genotypes and Phenotypes (dbGaP) under accession numbers phs001375, phs001388, phs001393, and phs001429.
Prior Presentation. Preliminary results from this study were presented the 75th Scientific Sessions of the American Diabetes Association, Boston, MA, 5–9 June 2015.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db17-0187/-/DC1.
Deceased.
Members of the Diabetes Prevention Program Research Group and T2D-GENES Consortium are provided in Supplementary Data online.
References
- 1.World Health Organization. Global Status Report on Noncommunicable Diseases 2010. Geneva, Switzerland, World Health Organization, 2011 [Google Scholar]
- 2.Kohner EM, Barry PJ. Prevention of blindness in diabetic retinopathy. Diabetologia 1984;26:173–179 [DOI] [PubMed] [Google Scholar]
- 3.Mogensen CE. Preventing end-stage renal disease. Diabet Med 1998;15(Suppl. 4):S51–S56 [DOI] [PubMed] [Google Scholar]
- 4.Laakso M. Hyperglycemia and cardiovascular disease in type 2 diabetes. Diabetes 1999;48:937–942 [DOI] [PubMed] [Google Scholar]
- 5.Mahajan A, Go MJ, Zhang W, et al.; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium; Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium; Mexican American Type 2 Diabetes (MAT2D) Consortium; Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples (T2D-GENES) Consortium . Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet 2014;46:234–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams AL, Jacobs SB, Moreno-Macías H, et al.; SIGMA Type 2 Diabetes Consortium . Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature 2014;506:97–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris AP, Voight BF, Teslovich TM, et al.; Wellcome Trust Case Control Consortium; Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators; Genetic Investigation of ANthropometric Traits (GIANT) Consortium; Asian Genetic Epidemiology Network–Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium . Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 2012;44:981–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng MC, Shriner D, Chen BH, et al.; FIND Consortium; eMERGE Consortium; DIAGRAM Consortium; MuTHER Consortium; MEta-analysis of type 2 DIabetes in African Americans Consortium . Meta-analysis of genome-wide association studies in African Americans provides insights into the genetic architecture of type 2 diabetes. PLoS Genet 2014;10:e1004517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim YJ, Go MJ, Hu C, et al.; MAGIC Consortium . Large-scale genome-wide association studies in East Asians identify new genetic loci influencing metabolic traits. Nat Genet 2011;43:990–995 [DOI] [PubMed] [Google Scholar]
- 10.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flannick J, Florez JC. Type 2 diabetes: genetic data sharing to advance complex disease research. Nat Rev Genet 2016;17:535–549 [DOI] [PubMed] [Google Scholar]
- 12.Fuchsberger C, Flannick J, Teslovich TM, et al. The genetic architecture of type 2 diabetes. Nature 2016;536:41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plenge RM, Scolnick EM, Altshuler D. Validating therapeutic targets through human genetics. Nat Rev Drug Discov 2013;12:581–594 [DOI] [PubMed] [Google Scholar]
- 14.Estrada K, Aukrust I, Bjørkhaug L, et al.; Sigma Type 2 Diabetes Consortium . Association of a low-frequency variant in HNF1A with type 2 diabetes in a Latino population. JAMA 2014;311:2305–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hara K, Fujita H, Johnson TA, et al.; DIAGRAM Consortium . Genome-wide association study identifies three novel loci for type 2 diabetes. Hum Mol Genet 2014;23:239–246 [DOI] [PubMed] [Google Scholar]
- 16.Moltke I, Grarup N, Jørgensen ME, et al. A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature 2014;512:190–193 [DOI] [PubMed] [Google Scholar]
- 17.Centers for Disease Control and Prevention National Diabetes Fact Sheet: National Estimates and General Information on Diabetes and Prediabetes in the United States. Atlanta, GA, U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, 2011 [Google Scholar]
- 18.Villalpando S, de la Cruz V, Rojas R, et al. Prevalence and distribution of type 2 diabetes mellitus in Mexican adult population: a probabilistic survey. Salud Publica Mex 2010;52(Suppl. 1):S19–S26 [DOI] [PubMed] [Google Scholar]
- 19.Cebola I, Rodríguez-Seguí SA, Cho CH, et al. TEAD and YAP regulate the enhancer network of human embryonic pancreatic progenitors. Nat Cell Biol 2015;17:615–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernstein BE, Stamatoyannopoulos JA, Costello JF, et al.; The NIH Roadmap Epigenomics Mapping Consortium . The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol 2010;28:1045–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morán I, Akerman I, van de Bunt M, et al. Human β cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab 2012;16:435–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 2008;5:621–628 [DOI] [PubMed] [Google Scholar]
- 24.GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015;348:648–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeLuca DS, Levin JZ, Sivachenko A, et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 2012;28:1530–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang HM, Sul JH, Service SK, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet 2010;42:348–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods 2011;9:179–181 [DOI] [PubMed] [Google Scholar]
- 28.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 2007;316:1341–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voight BF, Scott LJ, Steinthorsdottir V, et al.; GIANT Consortium. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 2010;42:579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lek M, Karczewski KJ, Minikel EV, et al.; Exome Aggregation Consortium. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol 2010;8:e1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yasuda K, Miyake K, Horikawa Y, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet 2008;40:1092–1097 [DOI] [PubMed] [Google Scholar]
- 34.Hanson RL, Rong R, Kobes S, et al. Role of established type 2 diabetes-susceptibility genetic variants in a high prevalence American Indian population. Diabetes 2015;64:2646–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kvale MN, Hesselson S, Hoffmann TJ, et al. Genotyping informatics and quality control for 100,000 subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics 2015;200:1051–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knowler WC, Barrett-Connor E, Fowler SE, et al.; Diabetes Prevention Program Research Group . Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med 2002;346:393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitchell BD, Kammerer CM, Blangero J, et al. Genetic and environmental contributions to cardiovascular risk factors in Mexican Americans. The San Antonio Family Heart Study. Circulation 1996;94:2159–2170 [DOI] [PubMed] [Google Scholar]
- 38.Hunt KJ, Lehman DM, Arya R, et al. Genome-wide linkage analyses of type 2 diabetes in Mexican Americans: the San Antonio Family Diabetes/Gallbladder Study. Diabetes 2005;54:2655–2662 [DOI] [PubMed] [Google Scholar]
- 39.Coletta DK, Schneider J, Hu SL, et al. Genome-wide linkage scan for genes influencing plasma triglyceride levels in the Veterans Administration Genetic Epidemiology Study. Diabetes 2009;58:279–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harper AR, Nayee S, Topol EJ. Protective alleles and modifier variants in human health and disease. Nat Rev Genet 2015;16:689–701 [DOI] [PubMed] [Google Scholar]
- 41.Sabeti PC, Reich DE, Higgins JM, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature 2002;419:832–837 [DOI] [PubMed] [Google Scholar]
- 42.Livingstone C, Borai A. Insulin-like growth factor-II: its role in metabolic and endocrine disease. Clin Endocrinol (Oxf) 2014;80:773–781 [DOI] [PubMed] [Google Scholar]
- 43.Morali OG, Jouneau A, McLaughlin KJ, Thiery JP, Larue L. IGF-II promotes mesoderm formation. Dev Biol 2000;227:133–145 [DOI] [PubMed] [Google Scholar]
- 44.Hill DJ, Strutt B, Arany E, Zaina S, Coukell S, Graham CF. Increased and persistent circulating insulin-like growth factor II in neonatal transgenic mice suppresses developmental apoptosis in the pancreatic islets. Endocrinology 2000;141:1151–1157 [DOI] [PubMed] [Google Scholar]
- 45.Sparago A, Cerrato F, Vernucci M, Ferrero GB, Silengo MC, Riccio A. Microdeletions in the human H19 DMR result in loss of IGF2 imprinting and Beckwith-Wiedemann syndrome. Nat Genet 2004;36:958–960 [DOI] [PubMed] [Google Scholar]
- 46.Frystyk J, Skjaerbaek C, Vestbo E, Fisker S, Orskov H. Circulating levels of free insulin-like growth factors in obese subjects: the impact of type 2 diabetes. Diabetes Metab Res Rev 1999;15:314–322 [DOI] [PubMed] [Google Scholar]
- 47.Estil les E, Téllez N, Soler J, Montanya E. High sensitivity of beta-cell replication to the inhibitory effects of interleukin-1beta: modulation by adenoviral overexpression of IGF2 in rat islets. J Endocrinol 2009;203:55–63 [DOI] [PubMed] [Google Scholar]
- 48.Poher AL, Altirriba J, Veyrat-Durebex C, Rohner-Jeanrenaud F. Brown adipose tissue activity as a target for the treatment of obesity/insulin resistance. Front Physiol 2015;6:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Devedjian JC, George M, Casellas A, et al. Transgenic mice overexpressing insulin-like growth factor-II in beta cells develop type 2 diabetes. J Clin Invest 2000;105:731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casellas A, Mallol C, Salavert A, et al. Insulin-like growth factor 2 overexpression induces β-cell dysfunction and increases beta-cell susceptibility to damage. J Biol Chem 2015;290:16772–16785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 2007;316:1336–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saxena R, Voight BF, Lyssenko V, et al.; Diabetes Genetics Initiative of Broad Institute of Harvard and MIT; Lund University; Novartis Institutes of BioMedical Research. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 2007;316:1331–1336 [DOI] [PubMed] [Google Scholar]
- 53.Dai N, Zhao L, Wrighting D, et al. IGF2BP2/IMP2-deficient mice resist obesity through enhanced translation of Ucp1 mRNA and other mRNAs encoding mitochondrial proteins. Cell Metab 2015;21:609–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zinman B, Wanner C, Lachin JM, et al.; EMPA-REG OUTCOME Investigators . Empagliflozin, cardiovascular outcomes, and mortality in type 2 diabetes. N Engl J Med 2015;373:2117–2128 [DOI] [PubMed] [Google Scholar]
- 55.Santer R, Kinner M, Lassen CL, et al. Molecular analysis of the SGLT2 gene in patients with renal glucosuria. J Am Soc Nephrol 2003;14:2873–2882 [DOI] [PubMed] [Google Scholar]
- 56.Scotti MM, Swanson MS. RNA mis-splicing in disease. Nat Rev Genet 2016;17:19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kole R, Krieg AM. Exon skipping therapy for Duchenne muscular dystrophy. Adv Drug Deliv Rev 2015;87:104–107 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.