Abstract
Telomeres at the ends of linear chromosomes protect the genome. Telomeres shorten with each round of cell division, placing a finite limit on cell growth. Telomere attrition is associated with cell senescence and apoptosis. Telomerase, a specialized ribonucleoprotein complex, maintains telomeres homeostasis through repeat addition of telomere sequences to the 3′ telomeric overhang. Telomere biology is closely related to cancer and normal aging. Upregulation of telomerase or activation of the alternative pathway of telomere lengthening is a hallmark of cancer cells, making telomerase an attractive target for cancer therapeutics. In this review, we will discuss telomere biology and the prognostic implications of telomere length in acute myeloid leukemia, and review exciting new investigational approaches using telomerase inhibitors in acute myeloid leukemia and other myeloid malignancies.
Keywords: acute myeloid leukemia, myeloid malignancies, telomerase, telomerase inhibitors, telomeres, telomere length
Introduction
Telomeres cap the terminal ends of chromosomes. They are composed of tandem repeats of the noncoding DNA sequence (5′-TTAGGG-3′), ending in a 3′ single-stranded overhang that is associated with bound proteins [O’Sullivan and Karlseder, 2010]. These proteins form a complex called Shelterin that protects 3′ chromosome ends from recognition as double-strand breaks, preventing inadvertent activation of DNA damage response pathways [de Lange, 2009]. Telomeres shorten with each cell division, which eventually triggers senescence, resulting in growth arrest. Cells can occasionally overcome critically short telomere length (TL) (‘telomeric crisis’) through the activation of telomere maintenance mechanisms. Telomere attrition can be viewed as protective. Accumulation of critically short and dysfunctional telomeres, which are recognized by the cell as double-stranded breaks, triggers DNA damage response pathways [particularly the ataxia telangiectasia-mutated (ATM) and ataxia telangiectasia and Rad3 (ATR) kinases pathways] that halt cell proliferation and induce growth arrest or cell death [Maciejowski and de Lange, 2017]. However, critical loss of telomeric DNA in hematopoietic stem cells is also associated with genomic instability and cytogenetic abnormalities, such as the gain or loss of chromosomes and nonreciprocal translocations [Swiggers et al. 2006]. It is not clear why some cells appropriately undergo programmed cell death or senescence, while others become malignant in the face of telomere crisis. Possible explanations include the ability of the cell to maintain some degree of TL, or a dysfunctional or mutant p53 pathway that both contributes to and allows genomic instability to propagate. In patients with myeloid malignancies, those with complex cytogenetic abnormalities/aneuploidy are known to have shorter TL than those with a normal karyotype [Hartmann et al. 2005].
Telomere shortening can be counteracted by activation of telomerase [Collins et al. 1995], a process that can also be exploited by neoplastic cells. The telomerase enzymatic subunit, encoded by telomerase reverse transcriptase (TERT), is transcriptionally silent in most non-neoplastic cells, but reactivation may endow a small population of cells with the ability to survive crisis, at which point they become immortalized [Morales et al. 1999]. It has been proposed that up to 90% of human cancers reactivate telomerase [Kim et al. 1994; Shay and Bacchetti, 1997], while the remainder rely on homologous recombination (HR) to solve the problem of telomere crisis. HR is a much more energy-intensive process for cancer cells, which may slow growth rates (i.e. more indolent cancers); however, the HR pathway may also contribute to treatment resistance (to either chemotherapy or telomerase inhibitors). To add to the complexity, many tumors will have different clones utilizing each pathway (telomerase and HR) to protect telomeres, as intact telomeres are so essential for cell survival. Cancer treatment with chemotherapy (or telomerase-targeted therapies) may influence the clonal predominance of these two pathways.
Perturbations of telomere homeostasis are clearly implicated in the pathogenesis of myeloid disorders, including aplastic anemia, myelodysplastic syndrome (MDS), myeloproliferative neoplasms (MPNs), and acute myeloid leukemia (AML). Herein, we focus on telomere biology in AML. We examine predisposing factors to short TL and associations with leukemogenesis. Current methods for quantifying TL are also discussed. We assess the relationship between TL and molecular genetic abnormalities, prognosis, and treatment in AML patients. Finally, we describe several novel clinical trials examining telomerase inhibitors in various myeloid malignancies (MPNs and MDS) that may ultimately lead to trials in AML as well.
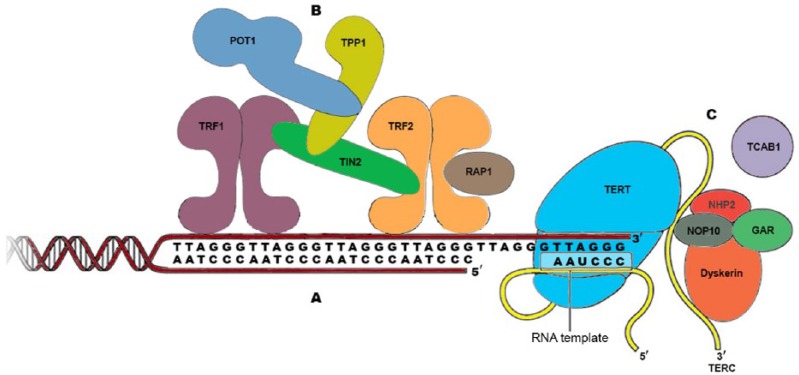
Telomere and telomerase biology
Telomeres are highly conserved in structure and function across species. Telomere function depends on three factors: (a) Telomeric DNA, (b) Shelterin complex, and (c) Telomerase complex (Figure 1). Telomeric DNA is composed of tandem repeats of the sequence 5′-TTAGGG-3′, and includes a double-strand tract of repeats many kilobases long, and a 3′ single-stranded G-rich overhang, measuring a few hundred nucleotides [Maciejowski and de Lange, 2017]. Shelterin is a six-subunit protein complex (comprising TRF1, TRF2, POT1, TPP1, TIN2 and Rap1) that associates specifically with mammalian telomeres and allows cells to distinguish the natural ends of chromosomes from sites of DNA damage (Figure 1). In addition, shelterin protects telomeric DNA from several DNA double-strand break pathways, including the nonhomologous end-joining (NHEJ) pathway that could lead to chromosome end fusions. It protects telomeres by forming the t-loop structure that conceals telomere ends. T-loops are formed through 3′-single-stranded overhang invasions into double-stranded telomeric DNA. TRF1, TRF2, and POT1 of the shelterin complex directly recognize and bind to TTAGGG repeats (POT1 binds to single-stranded repeats), whereas the other proteins, TIN2, TPP1 and Rap1 interconnect the telomere-binding proteins to form the entire complex.
Figure 1.
Structure of human telomere system.
Telomeres comprise three components: (A) Telomeric DNA: long array of double-stranded TTAGGG repeats and a 3′ single-stranded G-rich overhang, which folds back and invades the double-stranded telomeric helix, forming the T loop. (B) Shelterin complex: a six-subunit complex comprising telomeric repeat-binding factor 1 (TRF1), TRF2, repressor/activator protein 1 (RAP1), TRF1-interacting nuclear factor 2 (TIN2), TPP1 (also known as adrenocortical dysplasia protein homolog) and protection of telomeres 1 (POT1). Shelterin has multiple roles in maintaining telomere length homeostasis by forming the T loops, preventing DNA damage response activation, and recruiting the telomerase complex and modulating its activity. (C) Telomerase complex: composed of telomerase reverse transcriptase (TERT), telomerase RNA template component (TERC) and several accessory proteins: dyskerin, nonhistone protein 2 (NHP2), nucleolar protein 10 (NOP10), telomerase Cajal body protein 1 (TCAB1), and GAR1.
Telomeres shorten with each cell division since telomeres cannot be fully duplicated during cell division due to the ‘end replication problem’ [Levy et al. 1992; Olovnikov, 1973; Watson, 1972], whereby lagging strand DNA synthesis cannot be completed all the way to the very end. As subsequent cell divisions lead to critically shortened telomeres (human telomeres shorten by ~50 bps per division), a p53-dependent DNA damage response that triggers cell senescence or apoptosis is elicited [d’Adda di Fagagna et al. 2003]. If cells bypass this pathway and continue to proliferate, extremely short telomeres lose their chromosomal protection, promoting genome instability and leading to either cell death, or potentially, a malignant state.
Telomere attrition can be counteracted by telomerase, which adds TTAGGG repeats to the chromosomal 3′ DNA terminus at the end of chromosomes using an internal RNA template. Telomerase is a ribonucleoprotein enzyme complex consisting of a reverse transcriptase (encoded by TERT), an RNA template (encoded by TERC), and stabilizing accessory proteins including dyskerin (encoded by DKC1), nucleolar protein 10 (NOP10), nonhistone protein 2 (NHP2), GAR1 and telomerase Cajal body protein 1 (TCAB1). These accessory proteins contribute to the biogenesis and nuclear trafficking of telomerase [Maciejowski and de Lange, 2017]. During human development, telomerase activity is downregulated through the silencing of TERT. As a result, most human somatic cells (with the exception of certain stem cells, including hematopoietic stem cells) undergo programmed telomere shortening.
Most human cancers (80–90%) escape telomere crisis by pathologically activating telomerase, one of the hallmarks of cancer allowing replicative immortality [Blackburn et al. 2015; Kim et al. 1994]. The mechanisms underlying reactivation of telomerase in cancer cells are currently being investigated. A small subset of cancer cells extends their telomeres by the alternative lengthening of telomeres (ALT) pathway, which utilizes HR [Bryan et al. 1997].
Current methods for quantifying telomere length
Studies of telomere biology hinge on the measurement of TL in either single cells or, more commonly, populations of cells. TL is a critical variable in deciding cell fate and biologic function, ranging from aging to carcinogenesis, underscoring the need for detection methods that provide accurate information on the length of telomere repeats in different cell types. Multiple methods have been developed for the study of TL [Aubert et al. 2012]. It is often not clear which method is the most appropriate to address a specific research question. Many studies and diagnostic laboratories apply one of the following methods: (a) Terminal restriction fragment (TRF) length analysis, (b) fluorescence in situ hybridization combined with flow cytometry (flow-FISH), or (c) polymerase chain reaction (PCR) amplification of telomere repeats relative to a single-copy gene by quantitative PCR (qPCR) or monochrome multiplex PCR (MMqPCR). Quantitative FISH (Q-FISH) and single TL analysis (STELA) are also widely employed but are mostly only applicable to small numbers of patient samples.
TRF analysis is considered the ‘gold standard’ for quantifying TL, as it directly estimates the average TL in kilobases. In this method, genomic DNA is digested with restriction enzymes, which specifically excise telomeric and subtelomeric repeats. The DNA digest is resolved according to size by agarose gel electrophoresis, and telomere fragments are visualized either by Southern blotting or in-gel hybridization using a labeled probe specific to telomeric DNA. However, TRF analysis has some limitations: it is time consuming, requires large amounts of DNA (micrograms), may be influenced by ‘gel effects’, and includes subtelomeric DNA length in the measurement, thereby often leading to overestimation of true TL [Aubert et al. 2012]. Alternative methods, such as flow-FISH and PCR-based assays, have become important adjuncts to the more laborious TRF analysis and have the advantage of strictly measuring canonical telomeric sequences (i.e. TTAGGG repeats). Flow-FISH is a method that combines fluorescent in situ hybridization (FISH) with flow cytometry, using labeled peptide nucleic acid (PNA) probes that hybridize to telomere repeats in cells in suspension [Baerlocher et al. 2006]. This method estimates the average TL based on the average telomere content (quantity of telomere repeats) of single cells, expressed as mean fluorescence intensity, and is translated into a kilobase readout based on correlation with TRF analysis. PCR-based procedures including qPCR and MMqPCR have been adapted to quantify TL by determining the average content of telomere sequences in each sample using the ratio of telomere repeat-copy number from a DNA sample to single-copy gene, referred to as the T/S ratio. It calculates the abundance of telomere sequences in comparison with a single genome by PCR amplification using a double standard DNA-binding dye (SYBR Green) [Cawthon, 2009, 2002]. Although a more approximate measure, the benefits of PCR-based assays include the small quantity of DNA (nanograms) required and the high-throughput design, making this a practical technique for larger studies where an absolute measurement is less important. More recently, whole-genome sequencing data along with bioinformatics software applications have also been used to quantitate TL [Parker et al. 2012].
Telomere biology in acute myeloid leukemia
Telomere maintenance is particularly important for pluripotent and multipotent stem cells, including hematopoietic stem cells, and it is critical for tissue homeostasis and regeneration. Telomere shortening is among the hallmarks of aging and malignant cells. Once a telomere is critically short, chromosome ends elicit a double-strand-break-like DNA damage response, resulting in growth arrest and cell death. Perturbations of telomere homeostasis have been implicated in the pathogenesis of aplastic anemia, MDS and AML [Scheinberg et al. 2010; Townsley et al. 2015]. Age-adjusted TL appears to be significantly reduced in patients with myeloid malignancies as compared with matched controls [Brümmendorf and Balabanov, 2006]. Telomere shortening in myeloid neoplasms may result from increased replication required for leukemia development or altered telomere regulatory mechanisms. Telomere attrition in hematopoietic cells can lead to genomic instability and cytogenetic abnormalities, such as gain or loss of chromosomes and nonreciprocal translocations. Such genomic instability plays a major role in the initiation of leukemia as indicated by the development of AML after exposure to cytotoxic chemotherapy or ionizing radiation and the 40–50% of AML patients presenting with abnormal cytogenetics [Mrózek et al. 2004]. Telomeres are significantly shorter in leukemia cells with abnormal cytogenetics compared with those with no cytogenetic abnormality, and patients with complex cytogenetics have the shortest telomeres [Capraro et al. 2011]. Shortened TL and increased telomerase activity also correlate with disease progression and relapse [Wang et al. 2010].
There may also be an association between TL and specific mutations and mutation classes in AML patients. In a single institution study, there was a suggestion of increased survival at 6 months in AML patients with longer TL, a difference that diminished over time [Watts et al. 2016]. This trend did not achieve statistical significance, possibly due to limited sample size (n = 67) and short overall survival of this poor-risk/refractory AML cohort. In the same study, a group of commonly mutated DNA modifying enzymes (IDH1/2, DNMT3A, TET2) showed a trend towards longer TL, while mutations in FLT3 and other signaling mutations were associated with shorter TL [Watts et al. 2016]. Further studies of TL and associations with somatic mutations and outcome are indicated. Another study in acute promyelocytic leukemia (APL) measured TL in 187 patients enrolled across four intergroup trials [Baljevic et al. 2016]. At diagnosis of APL, TL was statistically significant predictor of both disease-free survival and overall survival.
A recent study by Gerbing and colleagues evaluated whether TL can be used as a marker of blood count recovery, given that defects in telomere maintenance are known risk factors for bone marrow failure and aplastic anemia. They hypothesized that short TL could be associated with delayed neutrophil recovery. In this study, bone marrow samples were obtained from pediatric patients with de novo AML from a Children’s Oncology Group study (AAML0531), who recovered as expected (within 30 days) after each chemotherapy course (n = 62), and from those who experienced significant delays in recovery after chemotherapy (n = 53). qPCR was used to measure telomere content (TC), a proxy for TL. TC was measured on each subject and the groups were categorized by quartile, from shortest to longest. Patients in the shortest telomere content quartile had statistically significant prolonged neutropenia, especially during the last two courses of chemotherapy. In an adjusted analysis including age at diagnosis, telomere content remained independently predictive of prolonged time to neutrophil recovery after the fourth (p = 0.002) and fifth courses of chemotherapy (p = 0.009). Patients were screened for acquired and germline mutations in four telomere maintenance genes (TERT, TERC, DKC1, and TINF2) commonly associated with telomere-associated disorders, and there was no enrichment for rare or novel variants in the patients with delayed neutrophil recovery. Overall, this study suggests that less telomere content after AML induction and subsequent chemotherapy cycles may predict for delayed count recovery following further chemotherapy courses, and potentially identify patients at high risk for severe myelosuppression and related toxicities such as sepsis [Gerbing et al. 2016].
Mutations in genes encoding components of the telomerase complex result in deficient telomerase function and telomere attrition, predisposing to cancer [Townsley et al. 2015]. The association between constitutional telomerase gene hypomorphic variants and risk of adult AML has been well described. In one study, bone marrow samples from 594 patients with AML were examined for variations in TERT and TERC genes. Analysis of patient samples for the most common gene variant (A1062T) in TERT revealed a prevalence three times higher than controls. The introduction of TERT mutants into telomerase-deficient cells resulted in loss of enzymatic activity by haploinsufficiency. This study demonstrated that inherited mutations in TERT that reduce telomerase activity are risk factors for AML [Calado et al. 2009]. The authors suggested that short and dysfunctional telomeres in the hematopoietic compartment may contribute to genomic instability and predispose to leukemia. More recently, two TERT promoter polymorphisms have been identified that are associated with increased AML risk and reduced survival [Mosrati et al. 2015]. Interestingly, in a 168-patient pediatric study, mutations in TERT and TERC were infrequent and not associated with AML, suggesting a significant latency period before the development of AML that is associated with constitutional telomerase mutations [Aalbers et al. 2013].
Most, but not all cancers have relatively short telomeres but high levels of telomerase activity compared with normal cells. In experimental models of AML, telomerase activity is required for leukemia maintenance. Enhanced telomerase activity likely represents an important adaptive mechanism that allows leukemia cells to continue to replicate despite accelerated telomere shortening. Acute leukemia-causing fusion genes MLL-AF4 and AML1-ETO have been reported to upregulate TERT expression [Gessner et al. 2010]. AML is known to originate from small populations of leukemic stem cells (LSCs) that have extensive self-renewing capacity but tend to be resistant to chemotherapy, causing relapse [Bonnet and Dick, 1997; Lapidot et al. 1994]. In a landmark study, the role of telomerase in AML LSC function and maintenance was demonstrated using mice depleted of the RNA component of telomerase (Terc-/-) in a well-defined transgenic MLL-AF9 AML model. In this study, telomerase deficiency significantly compromised the self-renewal capacity of LSCs [Bruedigam et al. 2014]. Although AML developed with full penetrance, suggesting that telomerase is not essential for leukemia initiation, telomerase deficiency significantly reduced leukemia burden and the frequency of functional LSCs as measured by limiting dilution analysis and secondary transplantation assays. Terc-/- LSCs demonstrated chromosomal instability, marked by increased end-to-end chromosome fusions, which resulted in increased apoptosis and induction of the p53 regulatory network. These results demonstrated that in already established leukemias, telomerase loss can induce chromosomal instability and p53 activation, resulting in loss of LSC function and leukemia regression. Functional studies using a TERT-shRNA (small hairpin RNA) (knockdown) in the MM6 human AML cell line also inhibited LSC function in vitro and in xenograft models. To investigate pharmacological inhibition of telomerase in AML, the investigators used imetelstat, a lipid-conjugated 13-mer oligonucleotide sequence that is complementary to and binds the RNA template of telomerase. In a patient-derived AML xenograft experiment, imetelstat prevented AML development, and leukemia progression was significantly delayed for weeks after suspension of treatment [Bruedigam et al. 2014]. This important study provided proof-of-concept evidence that telomerase inhibition may be a stem-cell targeted novel AML therapy. Further preclinical studies looking at combinations of imetelstat and standard AML chemotherapy and hypomethylating agents are underway.
Telomerase inhibitors in the myeloid malignancies
Clinical efficacy of telomerase inhibition with imetelstat (given as an intravenous infusion) has already been demonstrated in myeloid malignancies such as myelofibrosis (MF) and essential thrombocythemia (ET) (Table 1) [Baerlocher et al. 2015; Tefferi et al. 2015]. A recent multi-institutional phase II trial of imetelstat (7.5 mg/kg or 9.4 mg/kg weekly until platelet count control followed by less frequent maintenance dosing) as second-line therapy in patients with ET showed hematologic responses with platelet-lowering activity in all 18 patients enrolled with a median follow up of 17 months. A total of 89% (16) of those patients had a complete hematologic response [Baerlocher et al. 2015]. Additionally, 88% of patients with a JAK2 V617F mutation demonstrated a partial molecular response (mutant allele burden was reduced by median 71% at 3 months of treatment). CALR and MPL mutant allele burdens were also reduced by 15–66%. Patients reported mild-to-moderate adverse events during treatment, including grade 3 or higher neutropenia in 22% of patients and grade 3 or higher anemia, headache, abnormal liver function tests, and syncope in 11% of patients, respectively.
Table 1.
Completed and ongoing clinical trials of imetelstat in myeloid malignancies.
| ClinicalTrials.gov identifier/phase | Indication | Objective | Start/completion date | Design | Results | Sponsor/ reference |
|---|---|---|---|---|---|---|
|
NCT01243073/ phase II |
Essential thrombocythemia | Safety and efficacy | December 2010/April 2015 | Open label, single group | 18 patients, all with positive hematologic response Positive molecular response in most patients with JAK2 V617 mutation | Geron Baerlocher et al. [2015] |
|
NCT01731951/ phase II |
Primary/secondary myelofibrosis | Efficacy | October 2012/January 2019 | Open label, parallel, active, not recruiting | Complete or partial remission in 21% of patients Bone marrow fibrosis is reversed in few patients | Geron/Janssen Tefferi et al. [2015] |
| Mayo Clinic pilot study | Refractory anemia with ring sideroblasts with (RARS-T) or without (RARS) thrombocytosis | Safety and efficacy | N/A | Open label, single group | 3/8 transfusion-dependent patients achieved transfusion independence that lasted for a median of 28 weeks | Geron/Janssen Tefferi et al. [2016] |
|
NCT02598661/ phase III IMerge study |
Myelodysplastic syndromes | Safety and efficacy | November 2015/May 2019 | Randomized, double-blind | Recruiting participants | Geron/Janssen |
|
NCT02426086/ phase II IMbark study |
Myelofibrosis patients previously treated with JAK inhibitor | Safety and efficacy | June 2015/March 2018 | Randomized, single-blind, multicenter | Recruiting participants | Geron/Janssen |
RARS, refractory anemia with ring sideroblasts; JAK, Janus kinase.
A single-institution pilot phase II study of imetelstat in patients with primary or secondary MF demonstrated complete or partial responses in 7 out of 33 patients enrolled (21%) with a median duration of complete response of 18 months (13 to at least 20) and partial response of 10 months (7 to at least 10) [Tefferi et al. 2015]. For this study, two different dosing schedules were assessed at the 9.4 mg/kg dose level (once every 3 weeks and weekly for four doses followed by once every 3 weeks). Bone marrow fibrosis was reversed in all four patients with complete response. Mutations in genes encoding RNA splicing machinery (i.e. SF3B1 or U2AF1) were significantly correlated with improved responses to imetelstat (38% versus 4%). Interestingly, no changes in TL were observed in patients receiving imetelstat, and initial length of telomeres did not predict a clinical response, raising intriguing questions about the mechanism of action of imetelstat and if telomerase inhibition is merely an off-target effect [Armanios and Greider, 2015]. However, it is also possible that imetelstat’s anti-MF activity is due to on-target activity, but that effects on downstream pathways regulated by telomerase, such as DNA repair enzymes, are actually driving the response. Interestingly, in a mouse model of telomere dysfunction, mice had reduced expression of genes required for 3′ mRNA splice site recognition [Colla et al. 2015]. These mice had aberrant splicing and developed leukopenia, dysplasia and 5% progressed to AML. Mechanistically, telomerase deficiency was shown to activate ATR-mediated DNA damage responses, resulting in reduced expression of SRSF2 and other splicing factors, which was reversible by telomerase induction and ATR inhibition. These findings suggest a complex interplay between the telomerase complex, RNA splicing, DNA damage and myeloid differentiation/neoplasia that is not yet fully understood.
Treatment-related adverse events on the MF study included grade 4 thrombocytopenia (in 18% of patients), grade 4 neutropenia (in 12%), grade 3 anemia (in 30%), grade 1–2 hyperbilirubinemia (in 12%), grade 1–2 elevated alkaline phosphatase (in 21%), and grade 1–2 elevated aspartate aminotransferase (in 27%). Due to concerns about hepatotoxicity and potential chronic liver injury, the FDA temporarily placed imetelstat clinical trials on a complete hold. However, the FDA has since lifted the clinical hold after the makers of imetelstat showed resolution of liver function abnormalities in 14 of 18 follow-up patients (and improvement in 3 of the other 4) treated on the ET trial. Of note, the ET patients had been treated with higher cumulative doses of imetelstat than those on the MF study.
Imetelstat has also been examined in patients with refractory anemia with ring sideroblasts, with (RARS-T) or without (RARS) thrombocytosis. RARS-T is now termed MDS/MPN with ring sideroblasts and thrombocytosis per the 2016 World Health Organization classification of myeloid malignancies. In MF [Tefferi et al. 2015], clinical and molecular remissions were significantly higher in patients with SF3B1 or U2AF1 spliceosome mutations, providing a rationale for the RARS(-T) study as spliceosome mutations are essentially always associated with these MDS/MPN subtypes. Patients with RARS(-T) were treated with imetelstat (dose-reduced to 7.5 mg/kg every 4 weeks) for a median duration of 13.7 months. Five out of the nine patients had RARS-T, three had RARS, and one had MDS/MPN overlap syndrome. Three (38%; two RARS, one RARS-T) of the eight patients who had been transfusion dependent before treatment, achieved transfusion independence for a median of 28 weeks. In one of these patients, leukocytosis and thrombocytosis also resolved in this time period. Another patient had decrease in spleen size and decrease in transfusion requirements. Two additional patients with thrombocytosis and leukocytosis had normalization of blood counts. Treatment-emergent grade 4 neutropenia and thrombocytopenia were observed in 22% and 11% of patients, respectively. Grade 3 anemia was noted in 67% of patients; however, 30% of patients had baseline grade 3 anemia. None of the grade 3/4 hematologic toxicities lasted more than 4 weeks in duration. A total of 67% of patients experienced liver function test abnormalities, but none experienced grade 3 or higher hepatotoxicity and these events were reversible upon study drug discontinuation. All three patients who achieved transfusion independence and a fourth patient with decrease in spleen size were SF3B1 mutated. Interestingly, post-treatment analysis showed no effect on mutational burden in these patients [Tefferi et al. 2016].
In aggregate, these studies have shown encouraging clinical responses. A large international randomized clinical trial (IMbark) including more than 200 patients with MF is currently underway to further study the efficacy and safety of imetelstat in a larger population (Table 1). Correlative studies on this trial will also be essential to better understand the mechanism of action of imetelstat in myeloid malignancies and offer a rationale to expand this therapy into AML. Telomerase-targeted dendritic cell-based vaccine strategies have also been assessed in AML, as a maintenance therapy for patients in remission. This immunotherapy approach was demonstrated to be safe and feasible and to potentially improve recurrence-free survival [Khoury et al. 2017].
Conclusion
Altered telomere homeostasis is clearly relevant to AML pathogenesis and a potential drug target in myeloid malignances. As a biomarker, TL may be associated with survival and specific somatic mutations in AML, such as IDH1/2 and FLT3-ITD, but these findings need to be further explored in larger cohorts. Short TL at AML diagnosis can predict prolonged myelosuppression after chemotherapy, potentially allowing for dose modifications and tailored supportive care as TL becomes easier to measure on standardized assays. Therapeutically, telomerase inhibitors have shown preliminary signs of activity in early phase clinical trials. Hepatic toxicity may be dose-limiting. It is not yet clear if mutational status will predict response to telomerase inhibitors. There is evidence that spliceosome mutations may sensitize cells to telomerase inhibition in MF, which raises the intriguing possibility of combining telomere therapies with spliceosome inhibitors. Murine leukemia models have demonstrated that genetic silencing of p53 can rescue a TERC knockout phenotype [Bruedigam et al. 2014], suggesting that p53 mutations could lead to telomerase inhibitor resistance, but this has not been evaluated in a clinical population. In summary, given the preliminary activity and relative tolerance of imetelstat in MDS and MPNs, and telomerase’s critical role in maintaining self-renewal of LSCs and proliferation capacity, telomerase inhibitors merit further study in myeloid malignancies including AML, with trial design focused on both safety and responsiveness across molecular subtypes.
Footnotes
Funding: This work was supported by American Society of Hematology HONORS award (AK) and University of Miami Clinical and Translational Science Institute (CTSI) KL2 award (KL2TR000461) (JW).
Conflict of interest statement: The authors declare that there is no conflict of interest.
Contributor Information
Ashwin Kishtagari, Department of Translational Hematology and Oncology Research, Taussig Cancer Institute, Cleveland, OH, USA.
Justin Watts, Division of Hematology, Department of Medicine, University of Miami, Miller School of Medicine, Sylvester Comprehensive Cancer Center, 1475 NW 12th Avenue, Miami, FL 33136-1002, USA.
References
- Aalbers A., Calado R., Young N., Zwaan C., Wu C., Kajigaya S., et al. (2013) Telomere length and telomerase complex mutations in pediatric acute myeloid leukemia. Leukemia 27: 1786–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios M., Greider C. (2015) Treating myeloproliferation–on target or off? N Engl J Med 373: 965–966. [DOI] [PubMed] [Google Scholar]
- Aubert G., Hills M., Lansdorp P. (2012) Telomere length measurement—caveats and a critical assessment of the available technologies and tools. Mutat Res Mol Mech Mutagen 730: 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerlocher G., Oppliger Leibundgut E., et al. (2015) Telomerase inhibitor imetelstat in patients with essential thrombocythemia. N Engl J Med 373: 920–928. [DOI] [PubMed] [Google Scholar]
- Baerlocher G., Vulto I., de Jong G., et al. (2006) Flow cytometry and FISH to measure the average length of telomeres (flow FISH). Nat Protoc 1: 2365–2376. [DOI] [PubMed] [Google Scholar]
- Baljevic M., Dumitriu B., Lee J., et al. (2016) Telomere length recovery: a strong predictor of overall survival in acute promyelocytic leukemia. Acta Haematol 136: 210–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn E., Epel E., Lin J. (2015) Human telomere biology: a contributory and interactive factor in aging, disease risks, and protection. Science 350: 1193–1198. [DOI] [PubMed] [Google Scholar]
- Bonnet D., Dick J. (1997) Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 3: 730–737. [DOI] [PubMed] [Google Scholar]
- Bruedigam C., Bagger F., Heidel F., et al. (2014) Telomerase inhibition effectively targets mouse and human AML stem cells and delays relapse following chemotherapy. Cell Stem Cell 15: 775–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brümmendorf T., Balabanov S. (2006) Telomere length dynamics in normal hematopoiesis and in disease states characterized by increased stem cell turnover. Leukemia 20: 1706–1716. [DOI] [PubMed] [Google Scholar]
- Bryan T., Englezou A., Dalla-Pozza L., Dunham M., Reddel R. (1997) Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med 3: 1271–1274. [DOI] [PubMed] [Google Scholar]
- Calado R., Regal J., Hills M., Yewdell W., Dalmazzo L., Zago M., et al. (2009) Constitutional hypomorphic telomerase mutations in patients with acute myeloid leukemia. Proc Natl Acad Sci U S A 106: 1187–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capraro V., Zane L., Poncet D., Perol D., Galia P., Preudhomme C., et al. (2011) Telomere deregulations possess cytogenetic, phenotype, and prognostic specificities in acute leukemias. Exp Hematol 39: 195–202. [DOI] [PubMed] [Google Scholar]
- Cawthon R. (2002) Telomere measurement by quantitative PCR. Nucleic Acids Res 30: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthon R. (2009) Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res 37: e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colla S., Ong D., Ogoti Y., Marchesini M., Mistry N., Clise-Dwyer K., et al. (2015) Telomere dysfunction drives aberrant hematopoietic differentiation and myelodysplastic syndrome. Cancer Cell 27: 644–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K., Kobayashi R., Greider C. (1995) Purification of tetrahymena telomerase and cloning of genes encoding the two protein components of the enzyme. Cell 81: 677–686. [DOI] [PubMed] [Google Scholar]
- D’Adda di, Fagagna F., Reaper P., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., et al. (2003) A DNA damage checkpoint response in telomere-initiated senescence. Nature 426: 194–198. [DOI] [PubMed] [Google Scholar]
- De Lange T. (2009) How telomeres solve the end-protection problem. Science 326: 948–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbing R., Alonzo T., Sung L., Gamis A., Meshinchi S., Plon S., et al. (2016) Shorter remission telomere length predicts delayed neutrophil recovery after acute myeloid leukemia therapy: a report from the Children’s Oncology Group. J Clin Oncol 34: 3766–3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessner A., Thomas M., Castro P., Büchler L., Scholz A., Brümmendorf T., et al. (2010) Leukemic fusion genes MLL/AF4 and AML1/MTG8 support leukemic self-renewal by controlling expression of the telomerase subunit TERT. Leukemia 24: 1751–1759. [DOI] [PubMed] [Google Scholar]
- Hartmann U., Brümmendorf T., Balabanov S., Christian T., Illme T., Schnaich M. (2005) Telomere length and hTERT expression in patients with acute myeloid leukemia correlates with chromosomal abnormalities. Haematologica 90: 307–316. [PubMed] [Google Scholar]
- Khoury H., Collins R., Blum W., Stiff P., Elias L., Lebkowski J., et al. (2017) Immune responses and long-term disease recurrence status after telomerase-based dendritic cell immunotherapy in patients with acute myeloid leukemia. Cancer 123: 3061–3072. [DOI] [PubMed] [Google Scholar]
- Kim N., Piatyszek M., Prowse K., Harley C., West M., Ho P., et al. (1994) Specific association of human telomerase activity with immortal cells and cancer. Science 266: 2011–2015. [DOI] [PubMed] [Google Scholar]
- Lapidot T., Sirard C., Vormoor J., Murdoch B., Hoang T., Caceres-Cortes J., et al. (1994) A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 367: 645–648. [DOI] [PubMed] [Google Scholar]
- Levy M., Allsopp R., Futcher A., Greider C., Harley C. (1992) Telomere end-replication problem and cell aging. J Mol Biol 225: 951–960. [DOI] [PubMed] [Google Scholar]
- Maciejowski J., de Lange T. (2017) Telomeres in cancer: tumour suppression and genome instability. Nat Rev Mol Cell Biol 18: 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales C., Holt S., Ouellette M., Kaur K., Yan Y., Wilson K., et al. (1999) Absence of cancer-associated changes in human fibroblasts immortalized with telomerase. Nat Genet 21: 115–118. [DOI] [PubMed] [Google Scholar]
- Mosrati M., Willander K., Falk I., Hermanson M., Höglund M., Stockelberg D., et al. (2015) Association between TERT promoter polymorphisms and acute myeloid leukemia risk and prognosis. Oncotarget 6: 25109–25120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrózek K., Heerema N., Bloomfield C. (2004) Cytogenetics in acute leukemia. Blood Rev 18: 115–136. [DOI] [PubMed] [Google Scholar]
- Olovnikov A. (1973) A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol 41: 181–190. [DOI] [PubMed] [Google Scholar]
- O’Sullivan R., Karlseder J. (2010) Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol 11: 171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M., Chen X., Bahrami A., Dalton J., Rusch M., Wu G., et al. (2012) Assessing telomeric DNA content in pediatric cancers using whole-genome sequencing data. Genome Biol 13: R113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinberg P., Cooper J., Sloand E., Wu C., Calado R., Young N. (2010) Association of telomere length of peripheral blood leukocytes with hematopoietic relapse, malignant transformation, and survival in severe aplastic anemia. JAMA 304: 1358–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay J., Bacchetti S. (1997) A survey of telomerase activity in human cancer. Eur J Cancer 33: 787–791. [DOI] [PubMed] [Google Scholar]
- Swiggers S., Kuijpers M., De Cort M., Beverloo M., Zijlmans J. (2006) Critically short telomeres in acute myeloid leukemia with loss or gain of parts of chromosomes. Genes Chromosom Cancer 45: 247–256. [DOI] [PubMed] [Google Scholar]
- Tefferi A., Al-Kali A., Begna K., Patnaik M., Lasho T, Wang X., et al. (2016) Telomerase inhibitor imetelstat therapy in refractory anemia with ring sideroblasts with or without thrombocytosis. Blood Cancer J 126: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A., Lasho T., Begna K., Patnaik M., Zblewski D., Finke C., et al. (2015) A pilot study of the telomerase inhibitor imetelstat for myelofibrosis. N Engl J Med 373: 908–919. [DOI] [PubMed] [Google Scholar]
- Townsley D., Dumitriu B., Young N. (2015) Bone marrow failure and the telomeropathies. Blood 124: 2775–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Fang M., Sun X., Sun J. (2010) Telomerase activity and telomere length in acute leukemia: correlations with disease progression, subtypes and overall survival. Int J Lab Hematol 32: 230–238. [DOI] [PubMed] [Google Scholar]
- Watson J. (1972) Origin of concatemeric T7 DNA. Nat New Biol 239: 197–201. [DOI] [PubMed] [Google Scholar]
- Watts J., Dumitriu B., Hilden P., Kishtagari A., Rapaport F., Chen C., et al. (2016) Telomere length and associations with somatic mutations and clinical outcomes in acute myeloid leukemia. Leuk Res 49: 62–65. [DOI] [PMC free article] [PubMed] [Google Scholar]