Abstract
Germline variations at JAK2, TERT, HBS1L-MYB and MECOM have been found to associate with myeloproliferative neoplasms (MPNs) in European populations. Whether these germline variations are associated with MPNs in Taiwanese population is obscure. Here we aimed to evaluate the association of five germline variations (JAK2 46/1 haplotype tagged by rs12343867, JAK2 intron 8 rs12339666, TERT rs2736100, HBS1L-MYB rs9376092 and MECOM rs2201862) and the risk of MPNs in Taiwanese population. A total of 178 MPN patients (109 essential thrombocythemia, 54 polycythemia vera and 15 primary myelofibrosis) were enrolled into this study. The information of 17033 control subjects was obtained from Taiwan Biobank database. The JAK2 46/1 haplotype, JAK2 rs12339666 and TERT rs2736100 were significantly associated with Taiwanese MPNs (P = 3.6×10-19, 1.9×10-19 and 3.1×10-6, respectively), and JAK2V617F-positive MPNs (n=121) (P = 5.6×10-21, 4.4×10-21 and 8.6×10-7, respectively). In JAK2V617F-negative cases (n=55), only the JAK2 46/1 haplotype and JAK2 rs12339666 remained statistically significant (P= 0.009 and 0.007, respectively). When stratified by disease subtypes, the JAK2 46/1 haplotype and JAK2 rs12339666 were significantly associated with all three MPN subtypes, but TERT rs2736100 was only associated with essential thrombocythemia and polycythemia vera. We did not find any association of these five SNPs with CALR mutations in our cohort. Furthermore, the risk alleles of MECOM rs2201862 and HBS1L-MYB rs9376092 were demonstrated to be negatively associated with the risk of developing polycythemia vera. In conclusion, germline variations at JAK2 (both the 46/1 haplotype and rs12339666) and TERT rs2736100 were associated with MPNs in Taiwanese population.
Keywords: JAK2, TERT, myeloproliferative neoplasms, single nucleotide polymorphism
INTRODUCTION
The classic BCR-ABL1-negative myeloproliferative neoplasms (MPNs) are clonal hematopoietic stem cell diseases, including three major disease entities: polycythemia vera (PV), essential thrombocythemia (ET) and primary myelofibrosis (PMF) [1]. MPNs are characterized by the overproduction of myeloid cells derived from one or more lineages. Among the three subtypes of MPNs, PV and ET are characterized by excessive formation of mature red blood cells and platelets, respectively, while PMF appears as fibrosis of the bone marrow, abnormal megakaryocytic proliferation and clustering, extramedullary hematopoiesis and variable peripheral blood counts. Somatic mutations in JAK2V617F, CALR exon 9 and MPL exon 10 have been demonstrated as three major driver mutations in MPNs, are usually mutually exclusive in the majority of cases, and involve in the activation of JAK-STAT signaling [2, 3]. The frequency of JAK2V617F mutation is over 95% in PV, and about 60% in ET and PMF. MPL exon 10 mutation is found in about 1% of ET and 5% of PMF patients. In ET or PMF patients without gene mutations in either JAK2 or MPL, CALR mutations have been observed in approximately 70% of these patients [4–8].
From both family studies and epidemiological data, inherited factors have been proposed to predispose to MPNs, and it has also been suggested that inherited single-nucleotide polymorphisms (SNPs) within JAK2 are associated with specific MPN subtypes [9–12]. A recent study has indicated that germline variations at JAK2 are linked to the acquisition of JAK2 V617F mutation in MPNs [10]. Several studies have discovered that a specific JAK2 haplotype, called ‘46/1’ or ‘GGCC’ strongly predisposes to JAK2V617F-positive MPNs in Caucasian populations [9–11]. The JAK2 46/1 haplotype is also demonstrated to predispose to MPNs with MPL mutation and PV with JAK2 exon 12 mutations [13, 14]. In addition, two JAK2 SNPs (rs12343867 and rs12340895) are demonstrated to express in complete linkage disequilibrium with JAK2 46/1 haplotype and have been used to tag this haplotype. Moreover, JAK2V617F mutation was also found to arise preferentially on the JAK2 46/1 haplotype [9]. Following these observations, germline variation at TERT rs2736100 has also been found to associate with MPNs in Icelandic population [15], and this finding was then confirmed by others [16–18].
In order to characterize the predisposition of inherited factors towards MPNs, Tapper et al. have performed a genome-wide association study in European populations [17]. After meta-analysis, they found that JAK2 rs12339666 and MECOM rs2201862 are significantly associated with JAK2V617F-negative MPNs. When JAK2V617F-positive cases were included in the analysis, two additional SNPs, TERT rs2736100 and HBS1L-MYB rs9376092, also achieved statistically significance. They also showed that HBS1L-MYB rs9376092 has a stronger effect on JAK2V617F-negative cases with CALR and/or MPL mutations, and has a stronger association with ET rather than with PV in JAK2V617F-positive cases. They concluded that multiple germline variants predispose to MPNs and linked constitutional differences in MYB expression to disease phenotype [17]. Although the association of JAK2 46/1 haplotype with MPNs has been validated across different ethnic groups, whether the other four SNPs (rs12339666, rs2736100, rs9376092 and rs2201862) might also associate with MPNs in Taiwanese population is currently unclear.
High-resolution melting analysis (HRMA) is a closed-tube and polymerase chain reaction (PCR)-based technique for the detection of gene polymorphism and mutations by measuring changes in the melting of a DNA duplex [19]. HRMA has been used for the detection and prescreening of SNPs and/or mutations including JAK2V617F and JAK2 exon 12 mutations in MPNs [20–22]. We have utilized HRMA for rapid and sensitive detection of CALR exon 9 mutations in ET using the CFX Connect real-time system (Bio-Rad Laboratories, Hercules, CA, USA) [23]. Here we aimed to determine the association of the above mentioned five SNPs with MPNs in Taiwanese population using HRMA and/or Sanger sequencing.
RESULTS
Mutational screening
In this cohort, JAK2V617F mutation was detected in 38 (70.4%) of 54 PV, 76 (69.7%) of 109 ET and 9 (60%) of 15 PMF, respectively. CALR mutations were detected in 17 (15.6%) of 109 ET including four types of CALR mutants: 5 type 1 (p.L367fs*46), 8 type 2 (p.K385fs*47), 3 type 3 (p.L367fs*48) and 1 other type (p.E369fs*50). Two (13.3%) of 15 PMF harbored CALR mutations including two types of CALR mutants: 1 type 34 (p.K385fs*47) and 1 other type (p.L367fs*43). Interestingly, two (1.8%) ET patients with type 1 CALR mutation were also found to harbor JAK2V617F mutation. No CALR mutation was detected in PV. Only one heterozygous MPLW515K mutation was detected in 1 (0.9%) of 109 ET.
Association of the 5 SNPs with MPN patients
By using our HRMA platform, we have successfully genotyped the 5 SNPs including rs12343867 (JAK2 46/1 haplotype), JAK2 rs12339666, TERT rs2736100, MECOM rs2201862 and HBS1L-MYB rs9376092 in more than 95% of samples from MPN patients (Figure 1). Based on the data from Taiwan Biobank, the risk allele frequency (RAF) of MECOM rs2201862 and TERT rs2736100 is significantly lower in the Taiwanese population (0.23 and 0.41, respectively) than that of the Western populations (0.48 and 0.51, respectively) indicating primordially ethnic difference in these 2 SNPs (Table 1). We found that JAK2 rs12343867, JAK2 rs12339666 and TERT rs2736100 were associated with MPNs in Taiwanese population. These 3 SNPs had stronger association with JAK2V617F-positive MPNs when compared to controls. In JAK2V617F-negative MPNs, only JAK2 rs12343867 and JAK2 rs12339666 remained statistically significant. Although JAK2 rs12339666 had a modest population attributable risk (PAR) in Western MPNs (27.5%), it had the highest PAR in Taiwanese MPNs (45.8%). We did not find any association of the 5 SNPs with CALR mutations in our cohort possibly because of small sample sizes (n = 17).
Figure 1. Normalized melt curves and difference curves of the 5 SNPs.
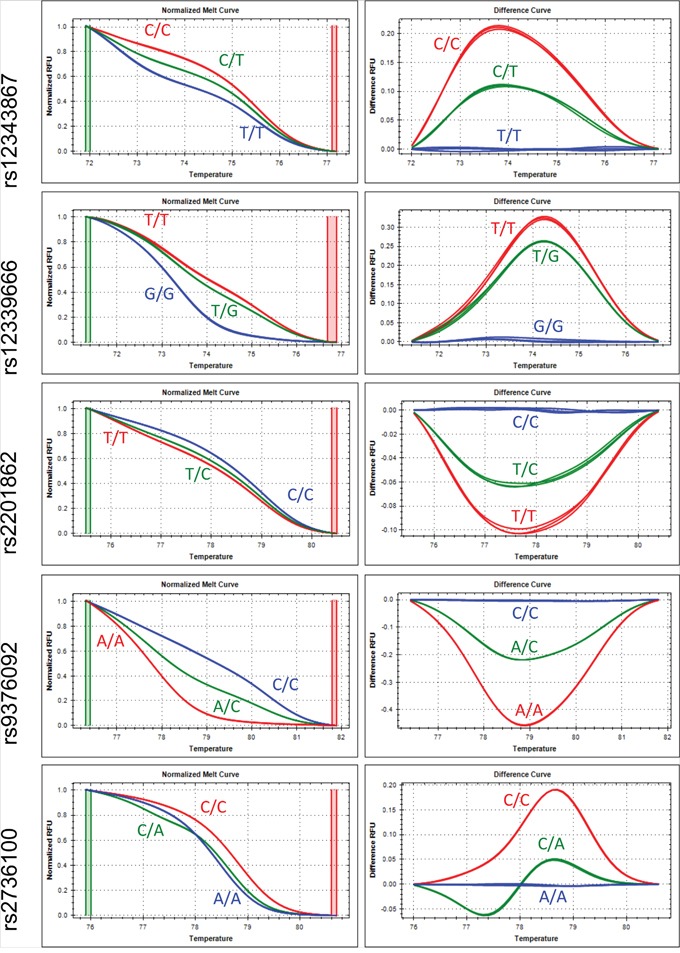
The curves of risk homozygosity, heterozygosity and non-risk homozygosity were shown in red, green and blue colors, respectively.
Table 1. Association of the five SNPs in MPN patients stratified by mutation profiles.
| SNP | Alleles* | Taiwanese RAF# | Gene | All MPN cases (n=178†) | JAK2V617F-positive cases (n=121†) | JAK2V617F-negative cases (n=55) | CALR-mutated cases (n=17) | Western RAF# | Western JAK2V617F-negative and all JAK2V617F positive | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P value | OR (95% CI) | PAR | P value | OR (95% CI) | PAR | P value | OR (95% CI) | PAR | P value | OR (95% CI) | PAR | P value | OR (95% CI) | PAR | |||||
| rs12343867 | C/T | 0.25 | JAK2 46/1 | 3.616 × 10-19 | 2.81 (2.23 - 3.54) | 42.9 | 5.604 × 10-21 | 3.52 (2.67 - 4.63) | 46.6 | 0.009 | 1.70 (1.14 - 2.54) | 36.3 | 0.573 | 1.24 (0.59 - 2.61) | 28.6 | 0.24§ | 6.379 × 10-43 | 3.19 (2.69 - 3.79) | 48.1 |
| rs12339666 | T/G | 0.26 | JAK2 intron 8 | 1.941 × 10-19 | 2.81 (2.23 - 3.53) | 45.8 | 4.365 × 10-21 | 3.50 (2.67 - 4.60) | 49.1 | 0.007 | 1.71 (1.15 - 2.55) | 40.2 | 0.701 | 1.16 (0.55 - 2.44) | 25.1 | 0.26 | 2.287 × 10-62 | 1.76 (1.65–1.88) | 27.5 |
| rs2201862 | T/C | 0.23 | -MECOM | 0.316 | 0.87 (0.66 - 1.15) | -8.3 | 0.345 | 0.85 (0.61 - 1.19) | -10.5 | 0.796 | 0.94 (0.59 - 1.50) | -1.4 | 0.752 | 0.87 (0.38 - 2.02) | 0.3 | 0.48 | 4.075 × 10-10 | 0.84 (0.80–0.89) | 14.3 |
| rs9376092 | A/C | 0.26 | HBS1L-MYB | 0.654 | 1.06 (0.83 - 1.34) | -0.4 | 0.532 | 1.09 (0.82 - 1.45) | 0.6 | 0.912 | 0.98 (0.64 - 1.50) | -2.8 | 0.101 | 1.77 (0.89 - 3.53) | 35.6 | 0.27 | 5.547 × 10-7 | 1.16 (1.09–1.23) | 6.9 |
| rs2736100 | C/A | 0.41 | TERT | 3.115 × 10-6 | 1.64 (1.33 - 2.02) | 28.6 | 8.624 × 10-7 | 1.88 (1.45 - 2.42) | 37.0 | 0.301 | 1.22 (0.84 - 1.78) | 7.6 | 0.444 | 1.30 (0.66 - 2.55) | 17.0 | 0.51 | 3.667 × 10-26 | 1.51 (1.40–1.63) | 42.7 |
Abbreviations: CI, confident interval; MPN, myeloproliferative neoplasm; OR, odds ratio; PAR, population attributable risks; RAF, risk allele frequency; SNP, single nucleotide polymorphism.
*Risk-associated/non-risk-associated alleles.
#RAF for healthy controls was used to calculate the PAR.
†Two cases and one case of JAK2V617F-positive essential thrombocythemia were absent in rs12343867 and rs2201862 groups, respectively due to lack of genomic DNA.
§The information was from another JAK2 46/1 tagged SNP rs12340895 instead of rs12343867.
P values were analyzed using Pearson χ2-test (two-tailed) or Fisher's exact test (two-tailed).
To determine whether the 5 SNPs predispose to different subtypes of MPNs, we then analyzed the association of the 5 SNPs with subtypes of MPNs (Table 2). JAK2 rs12343867 and JAK2 rs12339666 were significantly positively associated with all three MPN subtypes; TERT rs2736100 with PV and ET; and MECOM rs2201862 only with MF. Unexpectedly, MECOM rs2201862 and HBS1L-MYB rs9376092 were found to have negative association with Taiwanese PV patients, and both had a significantly negative PAR (-27.8% and -24.9%, respectively). Among these 5 SNPs, JAK2 rs12339666 had the highest PAR especially in PV patients (65.2%). Similar findings were found when we used the genotype distribution to calculate the association of these 5 SNPs with Taiwanese MPN patients (Supplementary Table 1).
Table 2. Association of the five SNPs in MPN patients stratified by disease subtypes.
| SNP | Alleles* | Taiwanese RAF# | Gene | PV (n=54) | ET (n=109†) | MF (n=15) | All ET and MF cases (n=124†) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P value | OR (95% CI) | PAR | P value | OR (95% CI) | PAR | P value | OR (95% CI) | PAR | P value | OR (95% CI) | PAR | ||||
| rs12343867 | C/T | 0.25 | JAK2 46/1 | 7.800 × 10-18 | 5.05 (3.38 - 7.56) | 62.7 | 2.566 × 10-7 | 2.12 (1.58 - 2.83) | 37.0 | 0.007 | 2.60 (1.26 - 5.37) | 14.2 | 1.556 × 10-8 | 2.17 (1.65 - 2.85) | 34.3 |
| rs12339666 | T/G | 0.26 | JAK2 intron 8 | 1.345 × 10-18 | 5.33 (3.54 - 8.02) | 65.2 | 9.319 × 10-7 | 2.03 (1.52 - 2.70) | 37.5 | 0.001 | 3.17 (1.54 - 6.55) | 37.6 | 1.892 × 10-8 | 2.14 (1.63 - 2.81) | 37.5 |
| rs2201862 | T/C | 0.23 | -MECOM | 0.051 | 0.59 (0.34 - 1.01) | -27.8 | 0.539 | 0.90 (0.64 - 1.27) | -4.2 | 0.075 | 1.95 (0.92 - 4.13) | 32.6 | 0.980 | 1.00 (0.73 - 1.38) | 0.3 |
| rs9376092 | A/C | 0.26 | HBS1L-MYB | 0.049 | 0.61 (0.37 - 1.00) | -24.9 | 0.077 | 1.30 (0.97 - 1.73) | 11.3 | 0.610 | 1.22 (0.56 - 2.68) | 2.8 | 0.066 | 1.29 (0.98 - 1.69) | 10.3 |
| rs2736100 | C/A | 0.41 | TERT | 7.957 × 10-4 | 1.90 (1.30 - 2.78) | 42.5 | 7.646 × 10-4 | 1.57 (1.21 - 2.06) | 24.9 | 0.499 | 1.28 (0.62 - 2.62) | 5.8 | 7.039 × 10-4 | 1.54 (1.20 - 1.97) | 22.6 |
CI, confident interval; ET, essential thrombocythemia; MPN, myeloproliferative neoplasm; MF; myelofibrosis; PV, polycythemia vera; OR, odds ratio; PAR, population attributable risks; RAF, risk allele frequency; SNP, single nucleotide polymorphism.
*Risk-associated/non-risk-associated alleles.
#RAF for healthy controls was used to calculate the PAR.
†Two cases and one case of JAK2V617F-positive ET were absent in rs12343867 and rs2201862 groups, respectively due to lack of genomic DNA.
P values were analyzed using Pearson χ2-test (two-tailed) or Fisher's exact test (two-tailed).
Comparison of the minor allele frequency of the 5 SNPs in MPN subtypes
We found that the minor allele frequency (MAF) of JAK2 rs12343867 and JAK2 rs12339666 was significantly higher in PV when all MPNs were included (Table 3) and also in JAK2V617F-positive cases (Table 4). The MAF of MECOM rs2201862 and HBS1L-MYB rs9376092 was significantly higher in MF only when all MPNs were included. The MAF of TERT rs2736100 did not have statistical difference among the three MPN subtypes. In disease subtype comparison, the MAF of JAK2 rs12343867 and JAK2 rs12339666 was significantly higher in PV than ET, and the MAF of MECOM rs2201862 was significantly higher in MF than PV when all MPNs were included (Table 3) and also in JAK2V617F-positive cases (Table 4). The MAF of MECOM rs2201862 and HBS1L-MYB rs9376092 in ET was significantly lower than that of MF and PV, respectively, only when all MPNs were considered.
Table 3. Comparison of the minor allele frequency of the 5 SNPs in MPN patients.
| SNP | Gene | Alleles* | PV MAF (n=54) | ET MAF (n=109†) | MF MAF (n=15) | PV vs. ET | PV vs. MF | ET vs. MF | PV vs. ET vs. MF | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P value | OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | ||||||
| rs12343867 | JAK2 46/1 | C/T | 0.63 | 0.42 | 0.47 | <0.001 | 2.39 (1.48-3.84) | 0.108 | 1.94 (0.86-4.40) | 0.598 | 0.81 (0.38-1.75) | 0.001 |
| rs12339666 | JAK2 intron 8 | T/G | 0.66 | 0.42 | 0.53 | <0.001 | 2.63 (1.63-4.25) | 0.213 | 1.68 (0.74-3.81) | 0.249 | 0.64 (0.30-1.37) | <0.001 |
| rs2201862 | -MECOM | T/C | 0.15 | 0.21 | 0.37 | 0.179 | 0.65 (0.35-1.22) | 0.008 | 0.30 (0.12-0.75) | 0.056 | 0.46 (0.20-1.04) | 0.030 |
| rs9376092 | HBS1L-MYB | A/C | 0.18 | 0.31 | 0.30 | 0.009 | 0.47 (0.27-0.83) | 0.135 | 0.50 (0.20-1.26) | 0.895 | 1.06 (0.46-2.43) | 0.031 |
| rs2736100 | TERT | C/A | 0.56 | 0.52 | 0.47 | 0.429 | 1.21 (0.76-1.92) | 0.340 | 1.48 (0.66-3.34) | 0.595 | 1.23 (0.57-2.64) | 0.571 |
CI, confident interval; ET, essential thrombocythemia; MAF, minor allele frequency; MPN, myeloproliferative neoplasm; MF; myelofibrosis; PV, polycythemia vera; OR, odds ratio; SNP, single nucleotide polymorphism.
*Risk-associated/non-risk-associated alleles.
†Two cases and one case with ET were absent in rs12343867 group and rs2201862 group, respectively due to lack of sample.
P values were analyzed using Pearson χ2-test (two-tailed).
Table 4. Comparison of the minor allele frequency of the 5 SNPs in JAK2V617F-positive MPN patients.
| SNP | Gene | Alleles* | PV MAF (n=38) | ET MAF (n=74†) | MF MAF (n=9) | PV vs. ET | PV vs. MF | ET vs. MF | PV vs. ET vs. MF | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P value | OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | ||||||
| rs12343867 | JAK2 46/1 | C/T | 0.72 | 0.45 | 0.50 | <0.001 | 3.18 (1.75-5.80) | 0.067 | 2.62 (0.91-7.50) | 0.696 | 0.82 (0.31-2.19) | <0.001 |
| rs12339666 | JAK2 intron 8 | T/G | 0.76 | 0.46 | 0.50 | 1.437 × 10-5 | 3.79 (2.04-7.05) | 0.027 | 3.22 (1.11-9.34) | 0.745 | 0.85 (0.32-2.26) | 7.334 × 10-5 |
| rs2201862 | -MECOM | T/C | 0.14 | 0.22 | 0.33 | 0.206 | 0.62 (0.29-1.31) | 0.087 | 0.34 (0.11-1.09) | 0.250 | 0.55 (0.19-1.58) | 0.161 |
| rs9376092 | HBS1L-MYB | A/C | 0.22 | 0.31 | 0.22 | 0.170 | 0.64 (0.34-1.21) | 1.000 | 1.01 (0.29-3.47) | 0.439 | 1.58 (0.49-5.06) | 0.363 |
| rs2736100 | TERT | C/A | 0.59 | 0.56 | 0.44 | 0.654 | 1.14 (0.65-1.99) | 0.256 | 1.81 (0.64-5.11) | 0.349 | 1.60 (0.60-4.27) | 0.524 |
CI, confident interval; ET, essential thrombocythemia; MAF, minor allele frequency; MPN, myeloproliferative neoplasm; MF; myelofibrosis; PV, polycythemia vera; OR, odds ratio; SNP, single nucleotide polymorphism.
*Risk-associated/non-risk-associated alleles.
†Two cases and one case with ET were absent in rs12343867 group and rs2201862 group, respectively due to lack of sample.
P values were analyzed using Pearson χ2-test (two-tailed) or Fisher's exact test (two-tailed).
Linkage of JAK2V617F mutation and JAK2 46/1 haplotype
We also evaluated the location of JAK2V617F mutation in heterozygous JAK2 46/1 haplotype cases (Table 5). 38 patients (9 PV and 27 ET) were found to have heterozygous JAK2V617F mutation in our cohort. Consistent with previous studies [9–11, 24], the frequency of JAK2V617F mutation was significantly higher on the 46/1 allele (86%, P = 7.428 × 10-6) in this cohort. Similar findings were also found when PV and ET patients were evaluated separately.
Table 5. Location of JAK2V617F mutation in heterozygous JAK2 46/1 haplotype MPN patients.
| Category | Case no. | On 46/1 allele | P value |
|---|---|---|---|
| All MPNs | 37 | 32 (86%) | 7.428 × 10-6 |
| PV | 9 | 8 (89%) | 0.039 |
| ET | 27 | 23 (85%) | 3.107 × 10-4 |
ET, essential thrombocythemia; MPN, myeloproliferative neoplasm; no., number; PV, polycythemia vera.
Clinical and prognostic implications of the five SNPs and the use of cytoreductive therapy
In MPN patients harbored the risk alleles of rs12343867C (JAK2 46/1) and rs12339666T (JAK2 intron 8), more patients were found to have splenomegaly at diagnosis (C vs T: 60% vs 40%, P = 0.007; T vs G: 61% vs 39%, P = 0.011, respectively), and fewer patients had overall hemorrhagic complications (C vs T: 36% vs 64%, P = 0.007; T vs G: 38.6% vs 61.4%, P = 0.012, respectively) (Supplementary Table 2). MPN patients harbored the risk allele of MECOM rs2201862T were found to have significantly less myelofibrosis transformation (T vs C: 42.9% vs 57.1%, P = 0.032). In MPN patients carried the risk allele of HBS1L-MYB rs9376092A, fewer patients had overall thrombotic complications (A vs C: 37% vs 63%, P = 0.012). Besides, those MPN patients with homozygous rs9376092A risk allele had encountered significantly fewer overall hemorrhagic complications (AA vs AC vs CC: 6.8% vs 52.3% vs 40.9%, P = 0.043). In patients harbored the TERT rs2736100C risk allele, significantly more patients were diagnosed at age less than 60-year-old (C vs A: 59.2% vs 40.8%, P = 0.008). Regarding to the use of cytoreductive therapy in MPN patients, significantly more patients with age equal to or higher than 60-year-old were treated with cytoreductive therapy (yes vs no: 87.5% vs 12.5%, P = 0.043).
DISCUSSION
In our study, we evaluated five MPN-associated SNPs in Taiwanese MPN patients, and identified significant associations of 3 of the 5 SNPs, rs12343867 (JAK2 46/1), rs12339666 (JAK2 intron 8) and rs2736100 (TERT) in our cohorts. Our results showed that the associations of these 3 germline variations with MPNs in our cohort are similar to those observed in Western populations including stronger associations for rs12343867 and rs12339666 in PV patients. Consistent with the Western populations, the JAK2 46/1 haplotype rs12343867 and JAK2 intron 8 rs12339666 had the greatest effect on JAK2V617F-positive and JAK2V617F-negative MPN in Taiwanese population. Our data also confirmed the associations of germline variations at JAK2 and TERT with MPNs in Taiwanese population.
Furthermore, both MECOM rs2201862 and HBS1L-MYB rs9376092 were not found to be a risk factor for Taiwanese MPNs in our study, although they have been demonstrated to have moderate association with Western MPNs. This discrepancy in MECOM rs2201862 might be explained by the difference in the RAF for healthy controls between the Western (0.48) and Taiwanese populations (0.23) (Table 1). For HBS1L-MYB rs9376092, we might not be able to detect the modest association of this SNP with Taiwanese MPNs due to a relatively small patient size in our study. However, both MECOM rs2201862 and HBS1L-MYB rs9376092 were found to have negative association with PV in our cohort. These findings argue that different genetic background might account for the discrepancy seen between Western and Taiwanese populations in these 2 SNPs rather than sample size. Our observations suggested that MECOM rs2201862 and HBS1L-MYB rs9376092 might have different contributions to the development of MPNs in different ethnic groups.
Regarding to the clinical and prognostic implications of the 5 SNPs in MPN patients, our results showed that MPN patients harbored the risk alleles of rs12343867_C (JAK2 46/1) and rs12339666_T (JAK2 intron 8) were more likely to have splenomegaly at diagnosis and were less likely to encounter hemorrhagic complications. Besides, those MPN patients with homozygous rs9376092_A (HBS1L-MYB) risk allele were also found to have fewer hemorrhagic complications. Furthermore, MPN patients carried the risk allele of rs9376092_A were also found to have fewer thrombotic complications. Only the risk allele of MECOM rs2201862_T was found to have protective effect on myelofibrosis transformation. To the best of our knowledge, these clinical and prognostic implications of the above-mentioned 4 SNPs have never been reported in MPNs. Whether the harboring of risk allele(s) of these 4 SNPs might have any therapeutic implications requires further study. Moreover, the cause(s) of the protective effects of these 4 risk alleles against hemorrhagic and/or thrombotic complications, or myelofibrosis transformation are not yet clear, and further study is also warranted to confirm our observation. In contrast to the findings of Krahling et al. from Hungarian population that TERT rs2736100_C risk allele predisposes to the development of MPNs with the co-occurrence of solid tumors, especially with the usage of cytoreductive treatment [25], we found that MPN patients harboring the TERT rs2736100_C risk allele were only associated with younger age at diagnosis. In addition, the use of cytoreductive therapy was not associated with the risk of secondary solid cancer in our cohort, but was more frequently seen in older patients indicating the compliance with the current treatment recommendation. More studies from different populations may be needed to clarify the discordance between our results from that of Krahling et al.
In addition to the 5 SNPs screened in our study, Hinds et al. has recently identified additional germline variants at TERT, SH2B3, TET2, ATM, CHEK2, PINT and GFI1B that predispose to JAK2V617F clonal hematopoiesis and MPNs in another Western population [26]. In their study, two TERT loci, rs7705526 and rs2853677, were found to be the second most statistically significant SNPs following the JAK2 46/1 haplotype. They also indicated that TERT rs2736100 is associated with MPNs, which is consistent with our findings in this study. The biological function of TERT rs2736100 has been reported to relate to myeloproliferative phenotypes, including increased red blood cell, platelet counts and white blood cells of the myeloid lineage [15]. Moreover, another TET2 rs3733609 C/T genotype was recently found to associate with JAK2V617F-positive MPNs in a Chinese cohort and related to a proliferative potential on erythroid lineage [27]. The RAF of TET2 rs3733609 in healthy subjects in the Taiwanese population (6.68%, data from the Taiwan Biobank) is similar to that in the Chinese population (6.35%, data from [27]), but different from that in the European populations (1%, data from the 1000 Genomes Project). Furthermore, it is also noteworthy that other SNPs at JAK2 such as rs4495487 in the Japanese population and rs12342421 in the Hong Kong Chinese population have also been found to associate with MPNs [28, 29], indicating that SNPs at JAK2 are important in MPNs as high risk factors. Because we did not evaluate JAK2 rs4495487, JAK2 rs12342421 and TET2 rs3733609 in this study, whether these germline variants may also predispose to MPNs in Taiwanese population require further study.
In conclusion, we identified genetic predisposition of germline variations at JAK2 46/1, JAK2 intron 8 and TERT to MPNs in the Taiwanese population. We also revealed negative association of germline variants at MECOM and HBS1L-MYB with PV in the Taiwanese population. Our study is limited by the evaluation of only 5 SNPs that have been known to associate with MPNs in Western populations. Additional germline variations that predispose to MPNs may be further discovered by genome-wide association study in Taiwanese population. Finally, the contribution of these germline variations to clonal hematopoiesis in the patients with myeloproliferative phenotypes will be worthwhile to investigate in the future.
MATERIALS AND METHODS
Patients
A total of 178 Taiwanese MPN patients (109 ET, 54 PV and 15 PMF) seen at MacKay Memorial Hospital from Oct 2009 to Oct 2015 were enrolled into this study. The diagnosis of MPNs was based on the 2008 World Health Organization classification. Genetic testing in MPN patients has been approved by the Institutional Review Board of MacKay Memorial Hospital. Written informed consent was obtained from patients or was waived for deceased patients when deidentified leftover samples were used as approved by the Institutional Review Board.
SNP information in Taiwanese and Western populations
The information of 5 SNPs (rs12343867, rs12339666, rs2736100, rs9376092 and rs2201862) in Han Chinese population in Taiwan were obtained from publicly available Taiwan Biobank database (https://taiwanview.twbiobank.org.tw, last accessed on 23th August 2016). The SNPs in Taiwan Biobank database were screened by whole genome sequencing using Ion Proton (497 healthy subjects) and Illumina Hiseq (500 healthy subjects) next-generation sequencing platforms, and genome-wide genotyping using the Affymetrix Axiom-Taiwan Biobank Array Plate (16036 healthy subjects), which is a customized hybridization-based oligonucleotide array with approximately 653,291 SNPs. The number of 46/1 alleles (349 in cases and 720 in the Wellcome Trust Case Control Consortium controls) and the number of non-46/1 alleles (346 in cases and 2280 in the Wellcome Trust Case Control Consortium controls) for another JAK2 46/1 tagged SNP rs12340895 instead of rs12343867 in Western population were adopted from Jones et al. [9], and were used to calculate and compared to rs12343867 since the implication of 46/1 haplotype is identical in all JAK2 46/1 tagged SNPs. The information of the other SNPs in Western population was adopted from Tapper et al. [17] (Table 1).
Mutational screening and SNP genotyping
All patients were screened for JAK2V617F, CALR and MPL mutations by Sanger sequencing and/or allele-specific PCR as previously described [23, 30]. Genomic DNA derived from peripheral blood, bone marrow, and/or buccal swab were extracted using the high pure PCR template preparation kit (Roche diagnostic GmbH, Mannheim, Germany) according to the manufacturer's instructions. The concentration of gDNA ranged between 16 and 20 ng/μL (in ddH2O) for peripheral blood and bone marrow, and between 6 and 20 ng/μL (in ddH2O) for buccal swab. The purity of gDNA was an A260/A280 ratio ranging between 1.7 and 2.2, and an A260/A230 ratio larger than 1.5 when measured using NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). PCR amplicon of positive controls was run in a 2% agarose gel to check for the quality. The appropriated annealing temperatures for HRMA were optimized using a temperature gradient. Any aberrant amplification plots of gDNA samples detected by HRMA were re-analyzed using Sanger sequencing to determine the genotypes of the samples.
HRMA and/or Sanger sequencing were used for genotyping the 5 SNPs: rs12343867, rs12339666, rs2201862, rs9376092, and rs2736100. For HRMA, genomic DNA was amplified using the Precision Melt Supermix (Bio-Rad, Hercules, CA, USA) in the CFX ConnectTM Real-Time System (Bio-Rad) according to the manufacturer's instructions [13]. General primers were designed using Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/), while melting temperature-shift primers were designed as previously described by other [31]. Primer sequences and annealing temperature for each primer are shown in Supplementary Table 3. For each HRMA experiment, positive genotypes that have been confirmed by Sanger sequencing were also included that the genotype of DNA samples found to have distinct melting curves from wild type could be compared and annotated.
For Sanger sequencing, a target SNP of individual genomic DNA was amplified using the GoTaq® Green Master Mix (Promega, Madison, WI, USA) followed by a pre-sequencing step using the USB ExoSAP-IT and a sequencing step using the BigDye® v3.1 (Applied Biosystems™, Carlsbad, CA, USA) in the ABI Veriti 96 Well Thermal Cycler (Applied Biosystems™) according to the manufacturer's instructions. Sequences were aligned using the CodonCode Aligner (Centerville, MA, USA).
Linkage of JAK2V617F and JAK2 46/1 haplotyope
Linkage of JAK2V617F and JAK2 46/1 tagged SNP rs12343867 was detected using allele-specific PCR as previously described [9]. All novel single-nucleotide variant that was only detected once was treated as artifact and was excluded.
Clinical and prognostic implications of the five SNPs and the use of cytoreductive therapy
The association of the 5 SNPs and the use of cytoreductive therapy with several clinical and prognostic parameters including age at diagnosis, sex, presence of splenomegaly at diagnosis, survival status, presence of secondary solid cancer before or after MPN diagnosis, presence of myelofibrosis or acute leukemia transformation, and overall hemorrhagic or thrombotic complication were evaluated. Clinical information was captured by retrospective chart review.
Statistical analysis
We used Pearson Chi-square (two-tailed) or Fisher's exact test (two-tailed) to analyze difference between patients and healthy controls. Chi-square was used to calculate odds ratios (ORs) and 95% confidence intervals. Exact binomial distribution (two-tailed) was used to calculate deviations from expected allelic ratios of 50%. PAR was calculated as 1 − (1/[p2 ORhomo + 2p {1 − p} ORhetero+ {1 − p}2]) for all SNPs in all populations, and 1 − (1/[p2 ORallele + 2p {1 − p} ORallele+ {1 − p}2]) was used for JAK2 46/1 tagged SNP in Western population. “p” is the RAF in the controls. ORhomo and ORhetero are OR associated with risk homozygotes and heterozygotes, respectively, relative to non-risk homozygotes, and ORallele is OR associated with risk allele relative to non-risk allele for JAK2 46/1 tagged SNP in Western population [17, 32]. Statistical analyses were performed using the Statistical Package of Social Sciences software (version 22.0; IBM, New York, NY, USA). A two-side P value of less than 0.05 was defined as significant difference.
SUPPLEMENTARY MATERIALS TABLES
Acknowledgments
We are grateful to Drs. Kuei-Fang Chou, Po-Nien Liao and Guan-Jhe Cai for their help in patient enrollment and collecting clinical specimens. This study was supported by grants from the Ministry of Science and Technology of Taiwan to KHL (grant numbers: MOST 102-2314-B-195-015-MY2 and MOST 104-2314-B-195-017-MY3) and YYK (grant number: MOST 103-2314-B-002-168 and MOST 105-2314-B-002-185-MY2), and the intramural grants from the Department of Medical Research of MacKay Memorial Hospital to KHL and YHC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Barbui T, Thiele J, Vannucchi AM, Tefferi A. Rationale for revision and proposed changes of the WHO diagnostic criteria for polycythemia vera, essential thrombocythemia and primary myelofibrosis. Blood Cancer J. 2015;5:e337. doi: 10.1038/bcj.2015.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tefferi A, Vainchenker W. Myeloproliferative neoplasms: molecular pathophysiology, essential clinical understanding, and treatment strategies. J Clin Oncol. 2011;29:573–582. doi: 10.1200/JCO.2010.29.8711. [DOI] [PubMed] [Google Scholar]
- 3.Tefferi A. Myeloproliferative neoplasms: a decade of discoveries and treatment advances. Am J Hematol. 2016;91:50–58. doi: 10.1002/ajh.24221. [DOI] [PubMed] [Google Scholar]
- 4.Lundberg P, Karow A, Nienhold R, Looser R, Hao-Shen H, Nissen I, Girsberger S, Lehmann T, Passweg J, Stern M, Beisel C, Kralovics R, Skoda RC. Clonal evolution and clinical correlates of somatic mutations in myeloproliferative neoplasms. Blood. 2014;123:2220–2228. doi: 10.1182/blood-2013-11-537167. [DOI] [PubMed] [Google Scholar]
- 5.Klampfl T, Gisslinger H, Harutyunyan AS, Nivarthi H, Rumi E, Milosevic JD, Them NC, Berg T, Gisslinger B, Pietra D, Chen D, Vladimer GI, Bagienski K, et al. Somatic mutations of calreticulin in myeloproliferative neoplasms. N Engl J Med. 2013;369:2379–2390. doi: 10.1056/NEJMoa1311347. [DOI] [PubMed] [Google Scholar]
- 6.Nangalia J, Massie CE, Baxter EJ, Nice FL, Gundem G, Wedge DC, Avezov E, Li J, Kollmann K, Kent DG, Aziz A, Godfrey AL, Hinton J, et al. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N Engl J Med. 2013;369:2391–2405. doi: 10.1056/NEJMoa1312542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim KH, Chang YC, Chen CG, Lin HC, Wang WT, Chiang YH, Cheng HI, Su NW, Lin J, Chang YF, Chang MC, Hsieh RK, Kuo YY, et al. Frequent CALR exon 9 alterations in JAK2 V617F-mutated essential thrombocythemia detected by high-resolution melting analysis. Blood Cancer J. 2015;5:e295. doi: 10.1038/bcj.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bilbao-Sieyro C, Florido Y, Gomez-Casares MT. CALR mutation characterization in myeloproliferative neoplasms. Oncotarget. 2016;7:52614–52617. doi: 10.18632/oncotarget.10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones AV, Chase A, Silver RT, Oscier D, Zoi K, Wang YL, Cario H, Pahl HL, Collins A, Reiter A, Grand F, Cross NC. JAK2 haplotype is a major risk factor for the development of myeloproliferative neoplasms. Nat Genet. 2009;41:446–449. doi: 10.1038/ng.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kilpivaara O, Mukherjee S, Schram AM, Wadleigh M, Mullally A, Ebert BL, Bass A, Marubayashi S, Heguy A, Garcia-Manero G, Kantarjian H, Offit K, Stone RM, et al. A germline JAK2 SNP is associated with predisposition to the development of JAK2(V617F)-positive myeloproliferative neoplasms. Nat Genet. 2009;41:455–459. doi: 10.1038/ng.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olcaydu D, Harutyunyan A, Jager R, Berg T, Gisslinger B, Pabinger I, Gisslinger H, Kralovics R. A common JAK2 haplotype confers susceptibility to myeloproliferative neoplasms. Nat Genet. 2009;41:450–454. doi: 10.1038/ng.341. [DOI] [PubMed] [Google Scholar]
- 12.Jones AV, Cross NC. Inherited predisposition to myeloproliferative neoplasms. Ther Adv Hematol. 2013;4:237–253. doi: 10.1177/2040620713489144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones AV, Campbell PJ, Beer PA, Schnittger S, Vannucchi AM, Zoi K, Percy MJ, McMullin MF, Scott LM, Tapper W, Silver RT, Oscier D, Harrison CN, et al. The JAK2 46/1 haplotype predisposes to MPL-mutated myeloproliferative neoplasms. Blood. 2010;115:4517–4523. doi: 10.1182/blood-2009-08-236448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olcaydu D, Skoda RC, Looser R, Li S, Cazzola M, Pietra D, Passamonti F, Lippert E, Carillo S, Girodon F, Vannucchi A, Reading NS, Prchal JT, et al. The 'GGCC' haplotype of JAK2 confers susceptibility to JAK2 exon 12 mutation-positive polycythemia vera. Leukemia. 2009;23:1924–1926. doi: 10.1038/leu.2009.110. [DOI] [PubMed] [Google Scholar]
- 15.Oddsson A, Kristinsson SY, Helgason H, Gudbjartsson DF, Masson G, Sigurdsson A, Jonasdottir A, Jonasdottir A, Steingrimsdottir H, Vidarsson B, Reykdal S, Eyjolfsson GI, Olafsson I, et al. The germline sequence variant rs2736100_C in TERT associates with myeloproliferative neoplasms. Leukemia. 2014;28:1371–1374. doi: 10.1038/leu.2014.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jager R, Harutyunyan AS, Rumi E, Pietra D, Berg T, Olcaydu D, Houlston RS, Cazzola M, Kralovics R. Common germline variation at the TERT locus contributes to familial clustering of myeloproliferative neoplasms. Am J Hematol. 2014;89:1107–1110. doi: 10.1002/ajh.23842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tapper W, Jones AV, Kralovics R, Harutyunyan AS, Zoi K, Leung W, Godfrey AL, Guglielmelli P, Callaway A, Ward D, Aranaz P, White HE, Waghorn K, et al. Genetic variation at MECOM, TERT, JAK2 and HBS1L-MYB predisposes to myeloproliferative neoplasms. Nat Commun. 2015;6:6691. doi: 10.1038/ncomms7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trifa AP, Banescu C, Tevet M, Bojan A, Dima D, Urian L, Torok-Vistai T, Popov VM, Zdrenghea M, Petrov L, Vasilache A, Murat M, Georgescu D, et al. TERT rs2736100 A>C SNP and JAK2 46/1 haplotype significantly contribute to the occurrence of JAK2 V617F and CALR mutated myeloproliferative neoplasms - a multicentric study on 529 patients. Br J Haematol. 2016;174:218–226. doi: 10.1111/bjh.14041. [DOI] [PubMed] [Google Scholar]
- 19.Vossen RH, Aten E, Roos A, den Dunnen JT. High-resolution melting analysis (HRMA): more than just sequence variant screening. Hum Mutat. 2009;30:860–866. doi: 10.1002/humu.21019. [DOI] [PubMed] [Google Scholar]
- 20.Er TK, Lin SF, Chang JG, Hsieh LL, Lin SK, Wang LH, Lin CW, Chang CS, Liu TC. Detection of the JAK2 V617F missense mutation by high resolution melting analysis and its validation. Clin Chim Acta. 2009;408:39–44. doi: 10.1016/j.cca.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 21.Qian J, Lin J, Yao DM, Chen Q, Xiao GF, Ji RB, Li Y, Yang J, Qian Z. Rapid detection of JAK2 V617F mutation using high-resolution melting analysis with LightScanner platform. Clin Chim Acta. 2010;411:2097–2100. doi: 10.1016/j.cca.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 22.Rapado I, Grande S, Albizua E, Ayala R, Hernandez JA, Gallardo M, Gilsanz F, Martinez-Lopez J. High resolution melting analysis for JAK2 Exon 14 and Exon 12 mutations: a diagnostic tool for myeloproliferative neoplasms. J Mol Diagn. 2009;11:155–161. doi: 10.2353/jmoldx.2009.080110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim KH, Lin HC, Chen CG, Wang WT, Chang YC, Chiang YH, Lin CS, Su NW, Su YW, Lin J, Chang YF, Chang MC, Hsieh RK, et al. Rapid and sensitive detection of CALR exon 9 mutations using high-resolution melting analysis. Clin Chim Acta. 2015;440:133–139. doi: 10.1016/j.cca.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Hu T, Wu Z, Kang Z, Liu W, Guan M. The JAK2 46/1 haplotype is a risk factor for myeloproliferative neoplasms in Chinese patients. Int J Hematol. 2012;96:611–616. doi: 10.1007/s12185-012-1169-8. [DOI] [PubMed] [Google Scholar]
- 25.Krahling T, Balassa K, Kiss KP, Bors A, Batai A, Halm G, Egyed M, Fekete S, Remenyi P, Masszi T, Tordai A, Andrikovics H. Co-occurrence of myeloproliferative neoplasms and solid tumors is attributed to a synergism between cytoreductive therapy and the common TERT polymorphism rs2736100. Cancer Epidemiol Biomarkers Prev. 2016;25:98–104. doi: 10.1158/1055-9965.EPI-15-0805. [DOI] [PubMed] [Google Scholar]
- 26.Hinds DA, Barnholt KE, Mesa RA, Kiefer AK, Do CB, Eriksson N, Mountain JL, Francke U, Tung JY, Nguyen HM, Zhang H, Gojenola L, Zehnder JL, et al. Germ line variants predispose to both JAK2 V617F clonal hematopoiesis and myeloproliferative neoplasms. Blood. 2016;128:1121–1128. doi: 10.1182/blood-2015-06-652941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen XH, Sun NN, Yin YF, Liu SF, Liu XL, Peng HL, Dai CW, Xu YX, Deng MY, Luo YY, Zheng WL, Zhang GS. A TET2 rs3733609 C/T genotype is associated with predisposition to the myeloproliferative neoplasms harboring JAK2(V617F) and confers a proliferative potential on erythroid lineages. Oncotarget. 2016;7:9550–9560. doi: 10.18632/oncotarget.7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohyashiki JH, Yoneta M, Hisatomi H, Iwabuchi T, Umezu T, Ohyashiki K. The C allele of JAK2 rs4495487 is an additional candidate locus that contributes to myeloproliferative neoplasm predisposition in the Japanese population. BMC Med Genet. 2012;13:6. doi: 10.1186/1471-2350-13-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koh SP, Yip SP, Lee KK, Chan CC, Lau SM, Kho CS, Lau CK, Lin SY, Lau YM, Wong LG, Au KL, Wong KF, Chu RW, et al. Genetic association between germline JAK2 polymorphisms and myeloproliferative neoplasms in Hong Kong Chinese population: a case-control study. BMC Genet. 2014;15:147. doi: 10.1186/s12863-014-0147-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin HC, Chen CG, Chang MC, Wang WT, Kao CW, Lo AC, Su NW, Chang YC, Chiang YH, Chou KF, Liao PN, Cai GJ, Cheng HI, et al. JAK2 V617F mutation in adult Taiwanese patients with essential thrombocythemia: more prevalent in old patients and correlated with higher hemoglobin level and higher leukocyte count. Int J Gerontol. 2013;7:40–44. [Google Scholar]
- 31.Wang J, Chuang K, Ahluwalia M, Patel S, Umblas N, Mirel D, Higuchi R, Germer S. High-throughput SNP genotyping by single-tube PCR with Tm-shift primers. BioTechniques. 2005;39:885–893. doi: 10.2144/000112028. [DOI] [PubMed] [Google Scholar]
- 32.Chang YC, Liu PH, Lee WJ, Chang TJ, Jiang YD, Li HY, Kuo SS, Lee KC, Chuang LM. Common variation in the fat mass and obesity-associated (FTO) gene confers risk of obesity and modulates BMI in the Chinese population. Diabetes. 2008;57:2245–2252. doi: 10.2337/db08-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


