Abstract
Pancreatic cancer stem cells (CSCs) play a crucial role in tumorigenesis and chemoresistance of pancreatic ductal adenocarcinoma. Pancreatic adenocarcinoma up-regulated factor (PAUF), a novel secretory protein, has been shown to contribute to cancer progression and metastasis. Because the clinical relationship between PAUF and pancreatic CSCs is largely unknown, we investigated the associations between the functional role of PAUF and pancreatic CSCs. Pancreatic cancer sphere cultured from the CFPAC-1 cells showed elevated expression of PAUF and pluripotent stemness genes (Oct4, Nanog, Stat3, and Sox2), and the mRNA of PAUF were increased in CD44+CD24+ESA+ pancreatic CSCs. PAUF knockdown (shPAUF) CFPAC-1 diminished the number of spheres and decreased stemness genes and CSC surface markers (CD133, c-MET and ALDH1). In addition, siPAUF CFPAC-1 decreased the mRNA expression of multidrug resistant protein 5 (MRP5) and ribonucleotide reductase M2 (RRM2) and were more vulnerable to gemcitabine and 5-FU than negative control (p<0.05). In conclusion, PAUF was increased in pancreatic CSCs and the suppression of PAUF enhances chemotherapeutic response to gemcitabine and 5FU by decreasing MRP5 and RRM2 in pancreatic cancer cells.
Keywords: cancer stem cells, PAUF, pancreatic cancer, MRP5, RRM2
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is one of the most aggressive malignancies and is the fourth leading cause of cancer-related death in the western world [1, 2]. The difficulty in early detection of PDAC and its resistance to conventional treatment options contribute to its dismal prognosis, as indicated by an overall 5-year survival rate of less than 10% [1, 2]. Many researchers have attempted to elucidate the pathogenesis and carcinogenesis of PDAC. However, the molecular basis for the aggressive nature of PDAC is incompletely understood, and the overall survival of patients has not improved significantly despite advances in treatment modalities and introduction of novel targeted therapies.
The clinical outcomes of PDAC may be poor because conventional therapies are directed at tumor cells that have limited tumorigenic potential instead of targeting the pancreatic cancer stem cells (CSCs) that have been identified in human PDAC [3]. Pancreatic CSCs are identified by a variety of biomarkers, and transplantation assays using immune-deficient mice show that pancreatic CSCs self-renew and propagate the parental tumor. Li et al. has identified populations of CSCs in PDAC that express the cell surface markers CD44+CD24+ESA+or c-Met [3, 4]. Apart from these markers, CD133+ and ALDH1 are regarded as other markers of CSCs in PDAC [5–7]. These CSCs are also characterized by their chemoresistance, as CD133+ pancreatic cancer cells have greater drug resistance to gemcitabine [6], and c-MET inhibitors have been shown to enhance antitumor effects in combination with gemcitabine [3].
Genome-wide analyses have uncovered pancreatic adenocarcinoma up-regulated factor (PAUF), a novel secretory protein associated with pancreatic cancer [8]. Although the mechanism through which PAUF contributes to cancer progression is not clearly established, previous reports suggest it plays critical roles in PDAC progression and metastasis processes, including cell proliferation and modulation of adhesion, migration, and invasion [9–15].
No studies have investigated the association between PAUF and CSCs and how it may contribute to drug sensitivity and resistance. Therefore, the purpose of the present study was to investigate the relationship between PAUF function and pancreatic CSCs. Our experiments provided evidence of PAUF expression in pancreatic CSCs and suggested that PAUF may contributes to multi-drug resistance in the pancreatic CSCs.
RESULTS
Pancreatic cancer spheres cultured from human pancreatic cancer cell lines
We used sphere culture to identify CSCs. This in vitro method involved culturing candidate pancreatic CSCs with serum-free media containing only EGF and bFGF under nonadherent conditions, and the resulting spheres indicated self-renewal consistent with a CSC phenotype. In this study, PDAC spheres cultured from the CFPAC-1 cells showed upregulated expression of the pluripotent stemness genes Oct4, Nanog, Stat3, and Sox2 relative to adherent CFPAC-1 cells (Figure 1A).
Figure 1. PAUF overexpression in pancreatic cancer spheres.
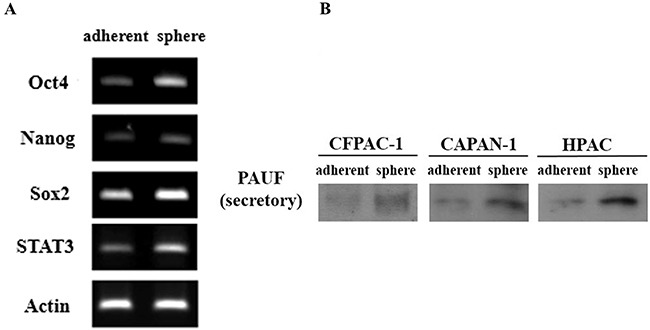
(A) RT-PCR showed that the overexpression of Oct4, Nanog, Stat3, and Sox2 genes in sphere formed from CFPAC-1 cells compare with adherent cells. (B) Up-regulation of secretory PAUF was determined by western blot in CFPAC-1, CAPAN-1 and HPAC spheres (Sp) than adherent (Ad) cells.
PAUF overexpression in pancreatic CSCs
We previously reported PAUF as a novel secretory protein associated with pancreatic cancer [8]. To show the relationship between the PAUF and pancreatic CSCs, secretory PAUF expression were detected in spheres and adherent CFPAC-1 cells by western blot analysis. As Figure 1B, PAUF expressions were upregulated in spheres than adherent CFPAC-1 cells. Furthermore, in other pancreatic cancer cells such as CAPAN-1 and HPAC cells, PAUF expression were upregulated in spheres than adherent cells (Figure 1B).
One of the other enrichment methods of CSCs is fluorescence-activated cell sorting (FACS) through cell surface marker. Based on previous reports that identified the cell surface markers CD44+CD24+ESA+ in a CSC population of human PDAC, we sorted the CFPAC-1 cell line into CD44+CD24+ESA+ and CD44-CD24-ESA- cells using FACS (Figure 2A), and validated that the previous reported stemness gene expression level using RT-PCR (Figure 2B). Oct4, Nanog, Stat3, Sox2, CD133, c-Met, beta-catenin et al. genes expression levels were upregulated in CD44+CD24+ESA+ than in CD44-CD24-ESA- CFPAC-1 cells. As well as, PAUF mRNA content was higher in CD44+CD24+ESA+ than in CD44-CD24-ESA- CFPAC-1 cells (Figure 2B, 2C). Furthermore, in the other pancreatic cancer cell line of HPAC, the expressions of stemness related genes and PAUF were upregulated in CD44+CD24+ESA+ than in CD44-CD24-ESA- HPAC cells (Figure 2D, 2E).
Figure 2. PAUF overexpression in CD24+/CD44+/ESA+ pancreatic CSCs.
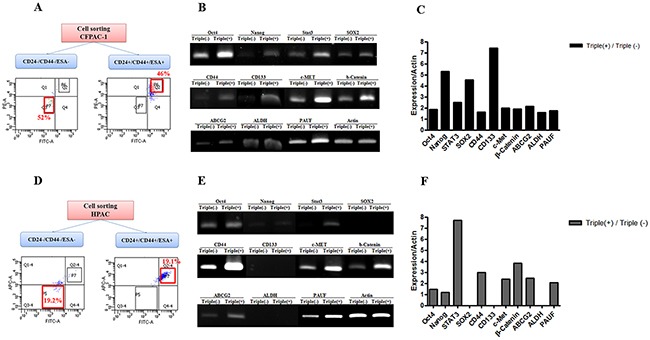
(A) CFPAC-1 cells were sorted using surface marker antibodies. CD24+/CD44+/ESA+ cells were 46% and CD24-/CD44-/ESA- cells were 52%. (B) RT-PCR demonstrated that the mRNA expression of stemness-related genes and PAUF was higher in CD24+/CD44+/ESA+ pancreatic CSCs sorted from CFPAC-1 cells than in CD24-/CD44-/ESA- CFPAC-1 cells. (C) Bar graph demonstrated that quantitative analysis of RT-PCR data. (D) HPAC-1 cells were sorted using surface marker antibodies. CD24+/CD44+/ESA+ cells were 19.1% and CD24-/CD44-/ESA- cells were 19.2%. (E) RT-PCR demonstrated that the mRNA expression of stemness-related genes and PAUF was higher in CD24+/CD44+/ESA+ pancreatic CSCs sorted from HPAC-1 cells than in CD24-/CD44-/ESA- HPAC-1 cells. (F) Bar graph demonstrated that quantitative analysis of RT-PCR data.
PAUF knockdown effects on pancreatic cancer stem cells in pancreatic cancer cell lines
To further access the specific role of PAUF in pancreatic CSCs function, we stably knocked down PAUF expression using shRNA targeting regions of PAUF.(Figure 3A) The shPAUF CFPAC-1 cells showed reduced cell proliferation, migration ability and in vivo xenograft tumor formation. Migration ability also decreased 0.67-fold in shPAUF CFPAC-1 cells compared with NC CFPAC-1 cells (mean ± SD); 187.71 ± 41.33 cells/microscopic field in shPAUF vs. 278.67 ± 28.66 cells/microscopic field in NC (Figure 3B). To investigate the effect of PAUF knockdown on the xenograft tumor formation in vivo, both shPAUF and NC CFPAC-1 cells were injected subcutaneously into nude mice. After 48 days, shPAUF CFPAC-1 cells formed 0.44-fold smaller tumor compared with NC CFPAC-1 cells (mean ± SD); the average tumor size was 114.5 ± 30.12 mm3 in shPAUF-xenografts vs. 258.0 ± 103.0 mm3 in NC-xenograft mice (Figure 3C). The effect of PAUF knockdown on CSCs-related characteristics was determined using sphere formation and soft agar assay. The number of sphere formation was markedly diminished in shPAUF CFPAC-1 cells compared with NC CFPAC-1 cells. (Figure 3D). Furthermore, the soft agar assay showed that the number of colonies was also 0.54-fold lower in the shPAUF CFPAC-1 cells (mean ± SD); 106.25 ± 0.24 colonies in shPAUF vs. 195.25 ± 0.10 colonies in NC (Figure 3E).
Figure 3. PAUF knockdown effects on pancreatic cancer stem cells in pancreatic cancer cell lines.
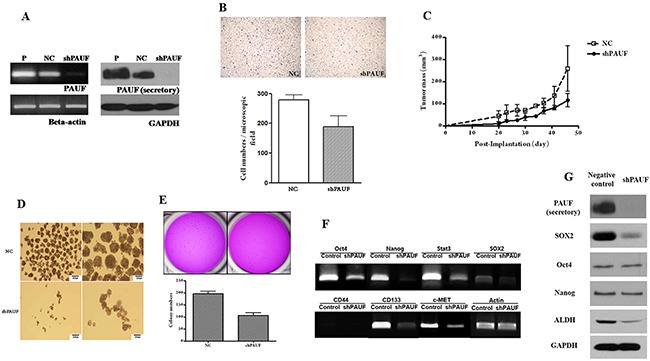
(A) PAUF knockdown CFPAC-1 cells were established by shRNA transfection, and shPAUF CFPAC-1 cells were confirmed using RT-PCR and western blot. B-actin and GAPDH served as a loading control. (B) Migration assays showed that shPAUF CFPAC-1 cells had decreased migration ability compared with NC CFPAC-1 cells; 187.71 ± 41.33 cells/microscopic field in shPAUF vs. 278.67 ± 28.66 cells/microscopic field in NC (C) in vivo tumor cell xenograft assay showed shPAUF CFPAC-1 cells formed smaller size of tumor compared with NC CFPAC-1 cells; the average tumor size 114.5 ± 30.12 mm3 in shPAUF vs. 258.0 ± 103.0 mm3 in NC. (D) Sphere formation assays showed that shPAUF CFPAC-1 cells had a lower sphere-forming ability compared with NC CFPAC-1 cells. (E) Soft agar assays showed that shPAUF CFPAC-1 cells had lower number of colonies compared with NC CFPAC-1 cells; 106.25 ±0.24 colonies in shPAUF vs. 195.25 ± 0.10 colonies in NC (F) RT-PCR demonstrated that the expression of pluripotent stemness genes and cancer stem cell-related genes were reduced in shPAUF CFPAC-1 cells compared with NC CFPAC-1 cells. (G) Western blot showed a lower protein expression of PAUF and cancer stem cell-related proteins in shPAUF CFPAC-1 cells compared with NC CFPAC-1 cells.
The mRNA and protein expression of pluripotent stemness genes in shPAUF CFPAC-1 was measured using RT-PCR and western blot, respectively. The expression of stemness-related genes (including Oct4, Nanog, and Sox2), and CSC surface markers (such as CD44, CD133, and c-Met) were lower in the shPAUF CFPAC-1 than in the control CFPAC-1 cells (Figure 3F). Protein expression of PAUF, Sox2, and ALDH were lower in shPAUF CFPAC-1 than in control cells (Figure 3G).
Effects of PAUF on chemoresponse of pancreatic cancer cell lines
Past studies have suggested that pancreatic CSCs are more resistant to anti-cancer chemotherapy agents such as gemcitabine and 5-FU. To determine the role of PAUF in chemoresponse, we performed cell survival assays to validate cytotoxic effects of 5-FU or gemcitabine against PAUF-associated pancreatic cancer. Two transient siPAUF RNAs were used for this experiment.
Both siPAUF CFPAC-1 and siPAUF cells were more vulnerable to the two cytotoxic drugs than the control CFPAC-1 cells (p<0.05, Figure 4A–4B). Furthermore, mRNA expression of PAUF, multidrug resistant protein 5 (MRP5) and ribonucleotide reductase M2 (RRM2) was lower in siPAUF CFPAC-1 cells than in control cells (Figure 4C). These responses for knockdown effect of PAUF were in same manner in AsPC-1 cells (Figure 4D–4F).
Figure 4. Effects of PAUF on chemoresponse of pancreatic cancer cell lines.
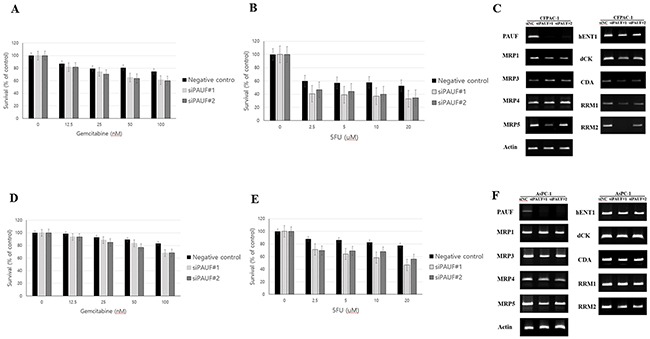
(A-B) PAUF knockdown effects on the cytotoxicity of 72 hours inoculation with gemcitabine and 5FU in CFPAC-1 cells. MTT assay showed significantly increased antitumor effect of gemcitabine (A) and 5FU (B) in PAUF-silenced CFPAC-1 cells compared with negative control CFPAC-1. Data are shown as the mean ± SD. (C) RT-PCR demonstrated mRNA expression of MRP5 and RRM2 was lower in the siPAUF CFPAC-1 cells than in the negative control CFPAC-1 cells. (D-E) PAUF knockdown effects on the cytotoxicity of 72 hours inoculation with gemcitabine and 5FU in AsPC-1 cells. MTT assay showed significantly increased antitumor effect of gemcitabine (D) and 5FU (E) in PAUF-silenced AsPC-1 cells compared with negative control CFPAC-1. (F) RT-PCR demonstrated mRNA expression of MRP5 and RRM2 was lower in the siPAUF AsPC-1 cells than in the negative control AsPC-1 cells
DISCUSSION
Recent studies have demonstrated the presence of pancreatic CSCs, which are capable of self-renewal and production of differentiated progeny, in PDAC. Although the isolation and culturing of pancreatic CSCs remains difficult, the role of CSCs in development, maintenance, and metastasis of PDAC is increasingly recognized. In this study, we cultured the pancreatic cancer cells in serum-free media and obtained pancreatic cancer sphere cells, which had self-renewal capabilities. Relative to the adherent cells, PDAC spheres showed increased mRNA and protein expression of PAUF and the pluripotent stemness genes Oct4, Nanog, Sox2 and STAT3. Additionally, CD44+CD24+ESA+ pancreatic CSCs had increased mRNA expression of PAUF.
We recently identified PAUF, (also known as a paralog of ZG16B), a novel 27-kDa secretory protein highly expressed in human PDAC [8]. As an autocrine factor, PAUF is involved in altered migration, invasion, proliferation, and metastasis [9, 11, 16]. PAUF also acts as a paracrine factor by causing stromal changes in the tumor microenvironment that promotes tumor evasion of the immune surveillance system [12]. PAUF also modulates permeability of the endothelial cells and vasculature to promote tumor angiogenesis [17]. Although only a few preclinical studies have investigated anti-PAUF monoclonal antibodies, PAUF-specific RNA aptamers (P12FR2), and adenoviral PAUF-targeting trans-splicing ribozymes (TSR) for PDAC, PAUF-targeted therapies may be an alternative approach for cancer treatment [10, 14, 15]. The above-mentioned studies suggest functional roles of PAUF in pancreatic cancer; however, little is known about the clinical relationship between PAUF and pancreatic CSCs. Therefore, we show the association between PAUF expression and characteristics of CSCs to determine the contribution of PAUF activity to chemoresponse in pancreatic CSCs,
Of clinical importance, recent research suggests CSCs are resistant to conventional chemotherapy and radiation [4]. Enhanced expression of ATP-binding cassette (ABC) family of transporters has been shown to be characteristic of side population cells with CSC features. These side population cells are more resistant to chemotherapeutic agents and may play a key role in tumor progression, recurrence, and metastasis [18, 19]. To identify the relationship between chemoresponse and the functional role of PAUF, we conducted cytotoxic studies with gemcitabine and 5-FU in siPAUF CFPAC-1/siPAUF AsPC-1 cells and negative controls. Our study showed that siPAUF CFPAC-1/siPAUF AsPC-1 cells were significantly more vulnerable to both cytotoxic drugs. Similar to our results, recent study reported that silencing of PAUF leads to increase the sensitivity to gemcitabine in BxPC-3 cells and this mechanism may involve drug transporter such as MRP2, MRP3, and MDR1 genes [20]. However, because both chemosensitivity and chemoresistance can occur through intracellular enzymes regardless of drug transporters, we measured the mRNA expression of genes involved in drug metabolism (such as dCK, CDA, RRM1, and RRM2) in addition to transporters (including MRP3, MRP4, MRP5, and hENT1). In the present study, the both siPAUF CFPAC-1 and siPAUF AsPC-1 cells decreased mRNA expression of MRP5 and RRM2. MRP5 (also known as ABCC5) is a member of the multidrug resistance-associated protein subfamily of ABC transporters and confers resistance against chemotherapeutic drugs such as etoposide, 5-FU, and gemcitabine [21–23]. RRM2 is the catalytic subunit of ribonucleotide reductase, a dimeric enzyme that supplies deoxynucleotides essential for DNA replication and cell growth. The overexpression of RRM2 has been associated with gemcitabine resistance and increased cellular invasiveness in vitro [24], [25] and high expression of RRM2 in pancreatic tumors is associated with reduced overall survival after resection [26].
Taken together, our study indicated that PAUF signaling was required for the formation of pancreatic CSCs. In addition, the suppression of PAUF may contribute to increase drug sensitivity through mechanisms involving MRP5 and RRM2, thus making it a novel target for treatment of PDAC. However, our study had some limitations. First, we could not demonstrate a PAUF mediated signal transduction mechanism by which PAUF induces CSCs phenotypes. Second, we used pancreatic cancer cell lines rather than patient-derived pancreatic cancer cells and only investigated the effect of PAUF knockdown in PDAC cancer cell lines rather than in pancreatic CSCs derived from human tissues. Third, we did not perform the experimental study using in vivo models of human PDAC, which are required to prove a causal relationship between PAUF and target proteins such as MRP5 and RRM2. Thus, the possible connection between PAUF, drug transporters, and cell signaling pathways should be investigated in future studies.
In conclusion, we showed that PAUF is a novel protein overexpressed in human pancreatic CSCs that may contribute to chemosensitivity by decreasing MRP5 and RRM2. Thus, PAUF may constitute a therapeutic target to enhance the drug sensitivity of pancreatic CSCs.
MATERIALS AND METHODS
Cell culture
Four PDAC cell lines (AsPC-1, Capan-1, CFPAC-1, and HPAC) were obtained from ATCC(American Type Culture Collection). All cells were grown in the appropriate conditioned medium and maintained in an atmosphere of 5% CO2/95% air at 37°C. We primarily used CFPAC-1, which expresses high endogenous levels of PAUF, for our experiments. In order to form cancer spheres, single cells were cultured for 7 days in Dulbecco's Modified Eagle Medium: Nutrient Mixture 12 (DMEM/F12,1:1; Gibco) containing 0.5% fetal bovine serum (FBS) (Hyclone), 0.5% bovine serum albumin fraction V (Gibco), insulin-transferrin-selenium A (Gibco), 10 ng/ml human epidermal growth factor (hEGF; R&D systems, Wiesbaden-Nordenstadt, Germany), 10 ng/ml human fibroblast growth factor (hFGF; R&D), and 10 ng/ml human leukemia inhibitory factor (hLIF; R&D) at a density of 1×103 cells/ml in an ultralow attachment plate (Corning, Corning, NY, USA). Growth factors were added every 3 days. For preparation of secretory proteins, culture medium was changed to serum-free medium in the post sphere culture after 5 days and cultured for an additional 2 days.
Semi-quantitative PCR (reverse transcription PCR)
The total RNA from cancer cells was extracted using a RNA easy extraction kit (Qiagen) according to the manufacturer's instructions. To quantify the relative expression of the genes, we used β-actin primer as a control. The PCR primers used were PAUF sense, 5’-cacctgggcagggaagatgta-3’; PAUF antisense, 5’-gctcagtggtcggctcctct-3’; β-actin sense, 5’-ggcatc ctcaccctgaagta –3’; and β-actin antisense, 5’-ggggtgttgaa ggtctcaaa-3’. The PCR primer sequences for the expression of genes related to pancreatic CSCs are described in Table 1.
Table 1. The sequences of PCR primer for the expression of known pancreatic CSCs related molecules.
| Name | Sequence |
|---|---|
| ß-catenin (sense) | GTATGAGTGGGAACAGGGATTT |
| ß-catenin (antisense) | CCTGGTCCTCGTCATTTAGC |
| SOX2 (sense) | CACAACTCGGAGATCAGCAA |
| SOX2 (antisense) | GTTCATGTGCGCGTAACTGT |
| NANOG (sense) | ACTGTCTCTCCTCTTCCTTCCT |
| NANOG (antisense) | AGAGTAAAGGCTGGGGTAGGTA |
| OCT4 (sense) | AAG TGG GTG GAG GAA GCT |
| OCT4 (antisense) | CGA GGA GTA CAG TGC AGT |
| STAT3 (sense) | GTCTGGCTGGACAATATCAT |
| STAT3 (antisense) | TTGGGAATGTCAGGATAGAG |
| CD44 (sense) | CACCATTTCAACCACACCAC |
| CD44 (antisense) | GGTGTTGTCCTTCCTTGCAT |
| CD133 (sense) | GCTCAGACTGGTAAATCCCC |
| CD133 (antisense) | GACTCGTTGCTGGTGAATTG |
| c-MET (sense) | CAA TGT GAG ATG TCT CCA GC |
| c-MET (antisense) | CCT TGT AGA TTG CAG GCA GA |
| ALDH1A1 (sense) | AGCAGGAGTGTTTACCAAAGA |
| ALDH1A1 (antisense) | CCCAGTTCTCTTCCATTTCCAG |
| ABCG2 (sense) | CAC TGA TCC TTC CAT CTT GT |
| ABCG2 (antisense) | TAT GAG TGG CTT ATC CTG CT |
Establishment of stable PAUF knockdown cell line using shRNA
The shRNA-expressing plasmid targeting human PAUF and negative control plasmid were purchased from SABiosciences. The human PAUF shRNA sequence was 5’-ACACCAGCAAGGACCGCTATT -3’ and the control shRNA sequence 5’-GGAATCTCATTCGATGCATAC -3’. For shRNA transfection, CFPAC-1 cells were plated into 6-well plates at a density of 5×104 cells/well the day before transfection. Transfection was performed using Lipofectamine2000 reagent according to manufacturer's instructions, and stable knockdown clones were selected using neomycin.
Transient transfection of small interfering RNA (siRNA) targeted for PAUF
The two sets of 25-nucleotide stealth RNAi-targeting PAUF were synthesized customarily by Invitrogen. Stealth RNAi duplexes, which have a similar GC content to that of the duplex siRNA obtained from Invitrogen, were used as negative controls. Stealth RNAi-targeting PAUF were transfected into CFPAC-1 cells using Lipectamine™ RNAi Max transfection agent (Invitrogen) according to the manufacturer's protocol. Cells were harvested 72 h post-transfection and subjected to total RNA extraction and a migration and invasion assay. Secreted protein was prepared from culture medium 48 h post-transfection.
MTT assay
After incubation at 37°C overnight, cells were treated with various concentrations of gemcitabine or fluorouracil (5-FU) in complete growth media and incubated for 72 h at 37°C. A 3-(4,5-dimenthelthiazol-2-ly)-2,5-diphenyltetrasolium bromide-based assay (absorbance 570 nm) was used to measure the number of metabolically active cells.
Flow cytometry and cell sorting
Cultured cells were detached with accutase solution (Sigma-Aldrich, Inc., St. Louis, MO, USA) and washed in phosphate-buffered saline (PBS) containing 0.5% FBS. Single cells were stained for 20 min on ice in the dark, washed twice in PBS containing 0.5% FBS, and fixed in 2% paraformaldehyde. Flow cytometry analysis was performed using a FACSCalibur system (BS Biosciences, San Jose, CA), and cell sorting was performed using FACSAria II (BD Immunocytochemistry System, Franklin Lakes, NJ). Antibodies against CD24 (anti-CD24-PE, BD), CD44 (anti-CD44-APC, BD), and ESA (anti-ESA-FITC, BD) were used. FITC-mouse IgG2b, κ isotype control (BD), rat IgG1 κ isotype control FITC (eBioscience), PE-mouse IgG2a, κ isotype control (BD), and APC-mouse IgG2b, κ isotype control (BD) were used as controls.
Protein extraction and western blot
Cells were prepared in lysis buffer containing 50 mM HEPES (pH 7.2), 150 mM NaCl, 25 mM beta-glycerophosphate, 25 mM NaF, 5 mM EGTA, 1 mM EDTA, 1% NP-40, 1 mM sodium orthovanadate, 0.1 mM phenylmethanesulfonylfluoride (PMSF), and a protease inhibitor cocktail (Roche Diagnostics). For secretory protein preparation, culture medium was centrifuged and the cellular components and debris were discarded. Culture medium was concentrated by the addition of ice-cold acetone, and the precipitated protein was resuspended with lysis buffer. Proteins were separated on SDS-PAGE and transferred to 0.45-μm Immobilon P-transfer membrane (Millipore). Membrane was blocked in 5% (w/v) non-fat milk and probed with the following primary antibodies: anti-human PAUF antibody (2009 Cancer Science), Sox2 (Cell Signaling Technology), Oct4 (Cell Signaling Technology), Nanog (Cell Signaling Technology), aldehyde dehydrogenase (ALDH) (BD), Cdk6 (Santa Cruz), cyclin D3 (Santa Cruz), Snail (Cell Signaling Technology), slug (Cell Signaling Technology), twist (Cell Signaling Technology) and glyceraldehyde phosphate dehydrogenase (GAPDH) (Santa Cruz). Immunoreactive material was then visualized using SuperSignal West Pico Chemiluminescence Substrate [27] according to the manufacturer's instructions.
Soft agar assay
A suspension of 500 single cells containing 0.3% agar medium was overlaid on 0.6% agar medium in 24-well plates. Each well was covered with complete medium, and the plates were incubated for 4 weeks. Colonies were stained with crystal violet and counted. Experiments were done in triplicate.
Cell proliferation, migration and invasion assay
For cell proliferation, 2 × 103 cells were seeded per sell into 24 well plates. Every 24 h, the number of cells was counted. The experimental was done in triplicate to determine the number of cells at each time point. The migration assays were performed in 6-well Transwell plates (Costar, Cambridge, MA). Cells (1 × 105) were seeded in triplicate in the upper compartment. The media incubated the NIH/3T3 with DMEM for 24 hours was added to the lower wells. Following 24-hour incubation at 37C in a 5% CO2 humidified incubator the cells in the upper chamber and on the surface of the filter were completely removed by wiping with a moist cotton swab. Cells that had migrated through the filter and adhered to the outer surface were fixed with methanol and stained with Toluidine Blue. Invading cells were examined, counted and photographed by microscopy at 100× magnification. The invasive abilities were tested by Matrigel invasion assay. Assays were performed in 6-well Transwell plates (Costar, Cambridge, MA) with an 8 μm pore size were coated with Matrigel (Becton Dickinson). Cells (1 × 105) were then seeded in triplicate in the upper compartment chamber coated with Matrigel. The media incubated the NIH/3T3 with DMEM for 24 hours was added to the lower wells. Following 48-hour incubation at 37C in a 5% CO2 humidified incubator the cells in the upper chamber and on the surface of the filter were completely removed by wiping with a moist cotton swab. Cells that had invaded the Matrigel, migrated through the filter and adhered to the outer surface were fixed with methanol and stained with Toluidine Blue. Invading cells were examined, counted and photographed by microscopy at 100× magnification.
In vivo tumorigenic assay
Cells were resuspended at a cell density of 3x 106 cells in 150 μl of serum-free culture medium and injected subcutaneously into the BALB/c nude female mice (6 weeks of age). Tumor formation was monitored twice a week measuring the width and length. Tumor volumes were calculated by the formula V (mm3) = A x B2, where A is the largest dimension and B is the perpendicular diameter. Tumor xenografts were recovered from mice, fixed in 4% paraformaldehyde, and embedded in paraffin.
Statistical analysis
Mann-Whitney U test and independent t-test were used to compare cell survival between siPAUF and control groups. Statistical calculations were performed using SPSS (version 12.0 for Windows; SPSS, Inc., Chicago, IL, USA). Values of p < 0.05 were considered statistically significant. For quantitative analysis western blot and RT-PCR data, we used ImageJ softwere (U.S. National Institutes of Health, Bethesda, MD, https://imagej.nih.gov/ij).
Acknowledgments
This research was supported by Basic Science Research Program through the National Research Foundation of Korea(NRF) funded by the Ministry of Education (Jae Hee Cho, No. NRF-2017R1D1A1B04034547). In addition, this research was supported by a grant (Si Young Song) of the Korean Heath Technology R&D Project (A111075), Ministry of Health & Welfare and by a grant of Korean Heath Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI14C1324).
Acronyms list
- PDAC
Pancreatic ductal adenocarcinoma
- CSCs
Cancer stem cells
- PAUF
Pancreatic adenocarcinoma up-regulated factor
- Oct4
Octamer-binding transcription factor 4
- Stat3
Signal transducer and activator of transcription 3
- Sox2
Sex determining region Y-box 2
- ESA
Epithelial-specific antigen
- ALDH
Aldehyde dehydrogenase
- PTEN
Phosphatase and tensin homolog
- Lef1
Lymphoid enhancer-binding factor 1
- Dvl2
Dishevelled segment polarity protein 2
- MRP
Multidrug resistant protein
- RRM
Ribonucleotide reductase M
- ATCC
American Type Culture Collection
- hEGF
Human epidermal growth factor
- hFGF
Human fibroblast growth factor
- hLIF
Human leukemia inhibitory factor
- FACS
Fluorescence-activated cell sorting
- CDK6
Cyclin-dependent kinase 6
- dCK
Deoxycytidine kinase
- ENT1
Equilibrative nucleoside transporter 1
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Cho JH, Bang S, Park SW, Chung JB, Song SY. BRCA2 mutations as a universal risk factor for pancreatic cancer has a limited role in Korean ethnic group. Pancreas. 2008;36:337–340. doi: 10.1097/MPA.0b013e31815c75ea. [DOI] [PubMed] [Google Scholar]
- 2.Zhan H, Xu J, Wu D, Zhang T, Hu S. Pancreatic cancer stem cells: new insight into a stubborn disease. Cancer Lett. 2014;357:429–437. doi: 10.1016/j.canlet.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Li C, Wu JJ, Hynes M, Dosch J, Sarkar B, Welling TH, Pasca di Magliano M, Simeone DM. c-Met is a marker of pancreatic cancer stem cells and therapeutic target. Gastroenterology. 2011;141:2218–2227. doi: 10.1053/j.gastro.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 5.Kim MP, Fleming JB, Wang H, Abbruzzese JL, Choi W, Kopetz S, McConkey DJ, Evans DB, Gallick GE. ALDH activity selectively defines an enhanced tumor-initiating cell population relative to CD133 expression in human pancreatic adenocarcinoma. PLoS One. 2011;6:e20636. doi: 10.1371/journal.pone.0020636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Hong SP, Wen J, Bang S, Park S, Song SY. CD44-positive cells are responsible for gemcitabine resistance in pancreatic cancer cells. Int J Cancer. 2009;125:2323–2331. doi: 10.1002/ijc.24573. [DOI] [PubMed] [Google Scholar]
- 8.Kim SA, Lee Y, Jung DE, Park KH, Park JY, Gang J, Jeon SB, Park EC, Kim YG, Lee B, Liu Q, Zeng W, Yeramilli S, et al. Pancreatic adenocarcinoma up-regulated factor (PAUF), a novel up-regulated secretory protein in pancreatic ductal adenocarcinoma. Cancer Sci. 2009;100:828–836. doi: 10.1111/j.1349-7006.2009.01106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee Y, Kim SJ, Park HD, Park EH, Huang SM, Jeon SB, Kim JM, Lim DS, Koh SS. PAUF functions in the metastasis of human pancreatic cancer cells and upregulates CXCR4 expression. Oncogene. 2010;29:56–67. doi: 10.1038/onc.2009.298. [DOI] [PubMed] [Google Scholar]
- 10.Kim YH, Sung HJ, Kim S, Kim EO, Lee JW, Moon JY, Choi K, Jung JE, Lee Y, Koh SS, Rhee SG, Heo K, Kim IH. An RNA aptamer that specifically binds pancreatic adenocarcinoma up-regulated factor inhibits migration and growth of pancreatic cancer cells. Cancer Lett. 2011;313:76–83. doi: 10.1016/j.canlet.2011.08.027. [DOI] [PubMed] [Google Scholar]
- 11.Lee YS, Kim SJ, Min HJ, Jo JY, Park EH, Koh SS. PAUF promotes adhesiveness of pancreatic cancer cells by modulating focal adhesion kinase. Exp Mol Med. 2011;43:291–297. doi: 10.3858/emm.2011.43.5.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park HD, Lee Y, Oh YK, Jung JG, Park YW, Myung K, Kim KH, Koh SS, Lim DS. Pancreatic adenocarcinoma upregulated factor promotes metastasis by regulating TLR/CXCR4 activation. Oncogene. 2011;30:201–211. doi: 10.1038/onc.2010.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho IR, Koh SS, Malilas W, Srisuttee R, Moon J, Choi YW, Horio Y, Oh S, Chung YH. SIRT1 inhibits proliferation of pancreatic cancer cells expressing pancreatic adenocarcinoma up-regulated factor (PAUF), a novel oncogene, by suppression of beta-catenin. Biochem Biophys Res Commun. 2012;423:270–275. doi: 10.1016/j.bbrc.2012.05.107. [DOI] [PubMed] [Google Scholar]
- 14.Kim SJ, Chang S, Lee Y, Kim NY, Hwang Y, Min HJ, Yoo KS, Park EH, Kim S, Chung YH, Park YW, Koh SS. A PAUF-neutralizing antibody targets both carcinoma and endothelial cells to impede pancreatic tumor progression and metastasis. Biochem Biophys Res Commun. 2014;454:144–150. doi: 10.1016/j.bbrc.2014.10.056. [DOI] [PubMed] [Google Scholar]
- 15.Kim YH, Moon JY, Kim EO, Lee SJ, Kang SH, Kim SK, Heo K, Lee Y, Kim H, Kim KT, Kim D, Song MS, Lee SW, et al. Efficient targeting and tumor retardation effect of pancreatic adenocarcinoma up-regulated factor (PAUF)-specific RNA replacement in pancreatic cancer mouse model. Cancer Lett. 2014;344:223–231. doi: 10.1016/j.canlet.2013.10.028. [DOI] [PubMed] [Google Scholar]
- 16.Cho IR, Koh SS, Min HJ, Kim SJ, Lee Y, Park EH, Ratakorn S, Jhun BH, Oh S, Johnston RN, Chung YH. Pancreatic adenocarcinoma up-regulated factor (PAUF) enhances the expression of beta-catenin, leading to a rapid proliferation of pancreatic cells. Exp Mol Med. 2011;43:82–90. doi: 10.3858/emm.2011.43.2.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim SJ, Lee Y, Kim NY, Hwang Y, Hwang B, Min JK, Koh SS. Pancreatic adenocarcinoma upregulated factor, a novel endothelial activator, promotes angiogenesis and vascular permeability. Oncogene. 2013;32:3638–3647. doi: 10.1038/onc.2012.366. [DOI] [PubMed] [Google Scholar]
- 18.Haraguchi N, Utsunomiya T, Inoue H, Tanaka F, Mimori K, Barnard GF, Mori M. Characterization of a side population of cancer cells from human gastrointestinal system. Stem Cells. 2006;24:506–513. doi: 10.1634/stemcells.2005-0282. [DOI] [PubMed] [Google Scholar]
- 19.Yao J, Cai HH, Wei JS, An Y, Ji ZL, Lu ZP, Wu JL, Chen P, Jiang KR, Dai CC, Qian ZY, Xu ZK, Miao Y. Side population in the pancreatic cancer cell lines SW1990 and CFPAC-1 is enriched with cancer stem-like cells. Oncol Rep. 2010;23:1375–1382. doi: 10.3892/or_00000774. [DOI] [PubMed] [Google Scholar]
- 20.Gao CC, Xu XL, Li F, Gong BG, Liu S, Cui YQ, Sun HC, Xu PY, Zheng YM, Jiang H. Silencing pancreatic adenocarcinoma upregulated factor (PAUF) increases the sensitivity of pancreatic cancer cells to gemcitabine. Tumour Biol. 2015;37:7555–7564. doi: 10.1007/s13277-015-4641-2. [DOI] [PubMed] [Google Scholar]
- 21.Hagmann W, Jesnowski R, Faissner R, Guo C, Lohr JM. ATP-binding cassette C transporters in human pancreatic carcinoma cell lines. Upregulation in 5-fluorouracil-resistant cells. Pancreatology. 2009;9:136–144. doi: 10.1159/000178884. [DOI] [PubMed] [Google Scholar]
- 22.Pratt S, Shepard RL, Kandasamy RA, Johnston PA, Perry W, 3rd, Dantzig AH. The multidrug resistance protein 5 (ABCC5) confers resistance to 5-fluorouracil and transports its monophosphorylated metabolites. Mol Cancer Therap. 2005;4:855–863. doi: 10.1158/1535-7163.MCT-04-0291. [DOI] [PubMed] [Google Scholar]
- 23.Oguri T, Achiwa H, Sato S, Bessho Y, Takano Y, Miyazaki M, Muramatsu H, Maeda H, Niimi T, Ueda R. The determinants of sensitivity and acquired resistance to gemcitabine differ in non-small cell lung cancer: a role of ABCC5 in gemcitabine sensitivity. Mol Cancer Therap. 2006;5:1800–1806. doi: 10.1158/1535-7163.MCT-06-0025. [DOI] [PubMed] [Google Scholar]
- 24.Duxbury MS, Ito H, Zinner MJ, Ashley SW, Whang EE. RNA interference targeting the M2 subunit of ribonucleotide reductase enhances pancreatic adenocarcinoma chemosensitivity to gemcitabine. Oncogene. 2004;23:1539–1548. doi: 10.1038/sj.onc.1207272. [DOI] [PubMed] [Google Scholar]
- 25.Liu X, Lai L, Wang X, Xue L, Leora S, Wu J, Hu S, Zhang K, Kuo ML, Zhou L, Zhang H, Wang Y, et al. Ribonucleotide reductase small subunit M2B prognoses better survival in colorectal cancer. Cancer Res. 2011;71:3202–3213. doi: 10.1158/0008-5472.CAN-11-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisher SB, Patel SH, Bagci P, Kooby DA, El-Rayes BF, Staley CA, 3rd, Adsay NV, Maithel SK. An analysis of human equilibrative nucleoside transporter-1, ribonucleoside reductase subunit M1, ribonucleoside reductase subunit M2, and excision repair cross-complementing gene-1 expression in patients with resected pancreas adenocarcinoma: implications for adjuvant treatment. Cancer. 2013;119:445–453. doi: 10.1002/cncr.27619. [DOI] [PubMed] [Google Scholar]
- 27.Dornhorst AC, Harrison K, Pierce JW. Observations on the normal oesophagus and cardia. Lancet. 1954;266:695–698. doi: 10.1016/s0140-6736(54)92106-6. [DOI] [PubMed] [Google Scholar]


