Abstract
The purposes of this study were to associate the genetic polymorphisms in carbonic anhydrase (CA) 9 with uterine cervical cancer and identify the clinical implications. Three single-nucleotide polymorphisms (SNPs), rs2071676 (+201, G/A), rs3829078 (+1081, A/G), and rs1048638 (+1584, C/A), and an 18-base-pair deletion/insertion (376del393) in CA9 were examined. We used the Boyden chamber assay to evaluate the influence of CA9 on the migration of cervical cancers. Tissue microarrays were used to evaluate CAIX immunoreactivity and determine its clinical significance. The results revealed that the CA9 SNP rs1048638 is the only significant polymorphism that increases the risk of cervical cancer in Taiwanese women. We discovered that the CA9 SNP rs1048638 influences the expression of CA9 through the interaction between the 3′-untranslated region (UTR) of exon 11, where the SNP is located, and miR-34a, and influences the migration of cervical cancer cells. Moreover, we demonstrated that CAIX immunoreactivity is related to the occurrence of cervical cancer, and elevated CAIX immunoreactivity is associated with a more advanced stage. In conclusion, the finding that the CA9 SNP rs1048638 exerts its action through duplexes of the miR-34a and CA9 3′-UTRs and plays a vital role in cervical cancer in Taiwanese women may be applicable to translational medicine.
Keywords: carbonic anhydrase, single nucleotide polymorphisms, miR-34a, migration, tissue microarray
INTRODUCTION
Uterine cervical cancer is the fifth most common type of cancer in women in Taiwan. The Health Promotion Administration of the Ministry of Health and Welfare in Taiwan reported its age-standardized incidence rate in 2009 to be 11.86 per 100,000 women. Cervical cancer ranked seventh in cancer deaths in Taiwanese women in 2011. Preinvasive lesions associated with cervical cancer are defined as cervical intraepithelial neoplasia and are currently graded as low or high squamous intraepithelial lesions (LSIL or HSIL). Approximately 10% of LSIL and approximately 20%–30% of HSIL may progress to invasive cancer [1, 2].
The high metabolic rate of tumor cells often results in hypoxia and acidosis in poorly perfused regions; therefore, they develop the ability to function in a more acidic environment than normal cells [3–5]. To date, 13 active and 3 inactive isoforms of carbonic anhydrases (CAs) have been characterized in mammals, and most CAs can efficiently catalyze a reversible hydration reaction: CO2 + H2O D HCO3− + H+ [6, 7]. In contrast to the other isoforms, the membrane-associated CAIX and CAXII isoforms are substantially increased in a hypoxic environment and are considered key pH regulators [6, 7]. CAIX is the most active isoform of CA in the hydration of carbon dioxide [8]. In response to hypoxia in human cancer cells, carbonic anhudrase 9 (CA9) is the most strongly expressed gene [9, 10]. It is overexpressed in many tumors and is associated with cancer progression [11–16].
Single-nucleotide polymorphism is a difference in a single nucleotide in the shared sequence of a gene between the members of a species or paired chromosomes in an individual in at least 1% of a certain population [17]. SNP is probably associated with the development and occurrence of certain diseases such as cancers. When an SNP occurs in a coding sequence, it may change the encoded amino acids in the related protein and is known as nonsynonymous. If an SNP leads to the same amino acid, it is called a synonymous SNP. In addition, an SNP in the 3′-untranslated region (UTR) of a gene may influence biological processes [18]. CA9 is located on chromosome 9p13–p12 and consists of 11 exons. It encodes for the 459-amino-acid protein CAIX [19]. Several studies have suggested that CA9 variations influence the severity and prognosis of several types of cancer, including prostate cancer, urothelial cell carcinoma, and oral cancer [20–22].
According to our review of the relevant literature, few studies have correlated CA9 SNPs with cervical cancer. We hypothesized that CA9 SNPs have an impact on the expression of CAIX. Furthermore, CA9 is overexpressed in many cancers [11–13]. Therefore, we investigated the distribution of the CA9 SNPs rs2071676 (+201, G/A) in exon 1, rs3829078 (+1081, A/G) in exon 7, rs1048638 (+1584, C/A) in the 3′-UTR of exon 11, and an 18-base-pair deletion/insertion (376del393 in exon 1) among patients with cervical cancer and normal controls; moreover, we defined their clinical implications in Taiwanese women. We further delineated the mechanism by which CA9 SNPs may influence the expression of CAIX in cervical cancer and associated the CAIX expression with the clinicopathological variables, cancer recurrence, and survival of patients with cervical cancer.
RESULTS
The age distribution differed significantly between patients with cervical cancer and controls (53.9 ± 11.9 vs. 44.2 ± 10.2, p < 0.001). However, age difference was not correlated with the CA9 SNP distribution (p = 0.181 for rs2071676, p = 0.758 for rs3829078, p = 0.191 for rs1048638, and p = 0.244 for 376del393). The genotype distributions of the SNPs rs2071676, rs3829078, and rs1048638 were in Hardy–Weinberg equilibrium (p = 0.406, χ2 value: 0.689; p = 0.745, χ2 value: 0.106; and p = 0.323, χ2 value: 0.976, respectively).
Association between CA9 gene polymorphism and cervical cancer
A significant difference was observed in the distribution of the CA9 SNP rs1048638 (p = 0.011) between women with cervical cancer and controls (Table 1). However, no such difference was observed in the distribution of rs2071676, rs3829078, and 376del393. The distribution of the CA/AA genotype of the CA9 SNP rs1048638 differed between patients with cervical cancer and controls when the wild genotype CC was used as a reference (p = 0.007). After age was controlled for, women with the CA/AA genotype exhibited a higher risk (adjusted odds ratio [AOR]: 1.92, 95% CI: 1.01–3.65) of cervical cancer compared with those with the wild genotype CC. Although no significant difference was observed among the patients with CA9 376del393, notably, no patients with cervical cancer exhibited 376del393 in both chromosomes (deletion/deletion).
Table 1. Genotype distribution of single nucleotide polymorphisms of carbonic anhydrase 9 gene in patients with uterine cervical cancer or normal women.
| Variables | Control (n=327) | Cervical cancer(n=123) | p value | OR (95% CI)b | Adjusted OR(95% CI)c |
|---|---|---|---|---|---|
| rs2071676 | |||||
| GGd | 79 (24.2%) | 26 (22.8%) | 0.109 | 1.00 | 1.00 |
| AG | 171 (52.3%) | 56 (43.6%) | 1.00 (0.58-1.70) | 0.72 (0.39-1.34) | |
| AA | 77 (23.5%) | 41 (33.6%) | 1.62 (0.90-2.90) | 1.27 (0.65-2.48) | |
| GGd | 79 (24.2%) | 26 (22.8%) | 0.500 | 1.00 | 1.00 |
| GA/AA | 248 (75.8%) | 97 (77.2%) | 1.19 (0.72-1.96) | 1.13 (0.63-2.00) | |
| GG/GAd | 250 (76.5%) | 82 (66.7%) | 0.035a | 1.00 | 1.00 |
| AA | 77 (23.5%) | 41 (33.3%) | 1.62 (1.03-2.56) | 1.59 (0.93-2.70) | |
| rs3829078 | |||||
| AAd | 297 (90.8%) | 114 (92.7%) | 0.717 | 1.00 | 1.00 |
| AG | 29 (8.9%) | 9 (7.3%) | 0.81 (0.37-1.76) | 0.84 (0.34-2.06) | |
| GG | 1 (0.3%) | 0 (0%) | u.a. | u.a. | |
| AAd | 297 (90.8%) | 114 (92.7%) | 0.533 | 1.00 | 1.00 |
| AG/GG | 30 (9.2%) | 9 (7.3%) | 0.78 (0.36-1.70) | 0.79 (0.32-1.93) | |
| AA/AGd | 326 (99.7%) | 123 (100%) | 0.539 | 1.00 | 1.00 |
| GG | 1 (0.3%) | 0 (0%) | u.a. | u.a. | |
| rs1048638 | |||||
| CCd | 292 (89.3%) | 98 (79.7%) | 0.011a | 1.00 | 1.00 |
| CA | 33 (10.1%) | 25 (20.3%) | 2.26 (1.28-3.98) | 1.98 (1.03-3.79) | |
| AA | 2 (0.6%) | 0 (0%) | u.a. | u.a. | |
| CCd | 292 (89.3%) | 98 (79.7%) | 0.007a | 1.00 | 1.00 |
| CA/AA | 35 (10.7%) | 25 (20.3%) | 2.13 (1.21-3.73) | 1.92 (1.01-3.65) | |
| CC/CAd | 325 (99.4%) | 123 (100%) | 1.000 | 1.00 | 1.00 |
| AA | 2 (0.6%) | 0 (0%) | u.a. | u.a. | |
| 376deletion393 | |||||
| Ins/Insd | 249 (76.1%) | 96 (78.0%) | 0.651 | 1.00 | 1.00 |
| Ins/del | 76 (23.3%) | 27 (22.0%) | 0.92 (0.56-1.52) | 0.86 (0.47-1.57) | |
| Del/del | 2 (0.6%) | 0 (0%) | u.a. | u.a. | |
| Ins/Insd | 249 (76.1%) | 96 (78.0%) | 0.671 | 1.00 | 1.00 |
| Ins/del or del/del | 78 (23.9%) | 27 (22.0%) | 0.90 (0.55-1.48) | 0.84 (0.46-1.52) | |
| Ins/Ins or ins/deld | 325 (99.4%) | 123 (100%) | 1.000 | 1.00 | 1.00 |
| Del/del | 2 (0.6%) | 0 (0%) | u.a. | u.a. |
Statistical analysis: logistic regression model or chi-square or Fisher's exact tests, ap < 0.05.
bComparison between patients with uterine cervical cancer and control women.
cThe adjusted OR with their 95% CI was estimated by logistic regression models after controlling for age between cancer patients and control women.
dUsed as a reference for comparison to evaluate the odds ratio of other genotypes.
OR, odds ratio; 95% CI, 95% confidence interval. Del, deletion; ins, insertion; u.a., unavailable.
Analysis of allele frequencies of CA9 polymorphisms in women with cervical cancer and controls
The minor allele frequencies of CA9 SNPs analyzed in this study were 0.49 for rs2071676, 0.05 for rs3829078, and 0.06 for rs1048638, which are similar to those in HCB recorded in the National Center for Biotechnology Information SNP database. Only the mutant allele A in the CA9 SNP rs1048638 tended to increase the risk of cervical cancer (p = 0.017; AOR: 1.75; Table 2). Other CA9 SNPs and 376del393 did not carry this risk.
Table 2. Allele distribution of single nucleotide polymorphisms (SNPs) of carbonic anhydrase 9 in patients with uterine cervical cancer or normal women.
| Variables | Normal (n=327) | Cervical cancer(n=123) | p value | OR (95% CI)b | Adjusted OR(95% CI)c |
|---|---|---|---|---|---|
| rs2071676 | |||||
| Gd | 329 | 108 | 0.087 | 1.00 | 1.00 |
| A | 325 | 138 | 1.29 (0.96-1.74) | 1.16 (0.82-1.63) | |
| rs3829078 | |||||
| Ad | 623 | 237 | 0.483 | 1.00 | 1.00 |
| G | 31 | 9 | 0.76 (0.36-1.63) | 0.75 (0.32-1.80) | |
| rs1048638 | |||||
| Cd | 617 | 221 | 0.017a | 1.00 | 1.00 |
| A | 37 | 25 | 1.89 (1.11-3.21) | 1.75 (0.95-3.22) | |
| 376deletion393 | |||||
| Insertiond | 574 | 219 | 0.604 | 1.00 | 1.00 |
| Deletion | 80 | 27 | 0.89 (0.56-1.41) | 0.83 (0.47-1.44) |
Statistical analysis: logistic regression model or chi-square test, ap < 0.05.
bComparison between patients with uterine cervical cancer and control women.
cThe adjusted OR with their 95% CI was estimated by logistic regression models after controlling for age between cancer patients and control women.
dused as a reference to evaluate the odds ratio. OR, odds ratio; 95% CI, 95% confidence interval.
Role of microRNA in the CA9 SNP rs1048638
The mutant allele A in the CA9 SNP rs1048638 was a crucial factor for cervical cancer in the analysis of the genotype and allele distribution in Taiwanese women. We hypothesize that microRNAs (miRNAs) interact with the 3′-UTR of exon 11 of CA9, where the CA9 SNP rs1048638 is located. We analyzed the CA9 3′-UTR in miRNA databases by using miRanda and TargetScan for targeting CA9 miRNAs and found that rs1048638 was located in the target sequence of miR-34a. These results were further supported by the results of assessing the optimal minimal free energy duplexes of miR-34a and CA9 3′-UTRs. The A allele of CA9 rs1048638 led to a substantially lower energy (−12.9 kcal/mol) than the C allele (−17.4 kcal/mol) with miR-34a hybridization, as previously described [23]. Collectively, these results suggest that miR-34a inhibits CA9 expression by binding to the predicted binding sites on the 3′-UTR of exon 11 of CA9 mRNA with the wild type C but not the mutant A in the CA9 SNP rs1048638.
Elevated CAIX expression associated with increased cell migration and invasiveness in cervical cancer cells
To confirm the importance of the rs1048638 polymorphism in the regulation of CA9 expression by miR-34a, we examined the rs1048638 genotypes of three cervical cancer cell lines (HeLa, SiHa, and Caski). The mRNA expression of CA9 was the highest in Caski and the lowest in SiHa cervical cancer cells (Figure 1A). Similarly, the protein expression of CAIX was the highest in Caski and the lowest in SiHa cell lines (Figure 1B). Therefore, the overexpression of CA9 in SiHa cervical cancer cells was analyzed. After CA9 transfection, the mRNA and protein expression of CAIX was confirmed using reverse transcription polymerase chain reaction and a Western blot assay (Figure 1C and 1D). The overexpression of CAIX significantly increased the migratory and invasive ability of SiHa cell lines (Figure 1E and 1F). These results indicate a functional role of CAIX in promoting cell motility in cervical cancer.
Figure 1. The mRNA and protein expressions of carbonic anhydrase IX (CAIX) in different cancer cell lines.
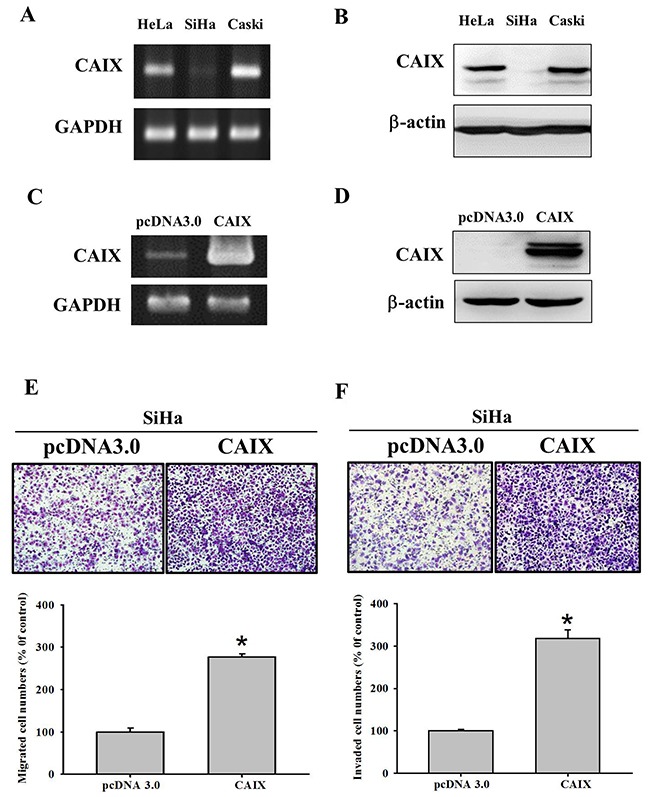
(A) The mRNA and (B) protein expressions of CAIX in SiHa cervical cell lines were lowest among SiHa, HeLa and Caski cancer cell lines of uterine cervix. The (C) mRNA and (D) protein expressions of CAIX were elevated in SiHa cell lines with CAIX gene transfection. The mRNA and protein expressions of CAIX were detected by RT-PCR and Western blot analysis. GAPDH and β-actin were separately used as internal controls. The (E) migratory and (F) invasive abilities of pcDNA3.0 and pcDNA3.0-CAIX SiHa cells were evaluated using Boyden chamber migration and Matrigel invasion assays. Differences are presented as the mean of triplicate experiments compared with control cells. *p < 0.05 compared with control cells.
Influence of CAIX expression on cell migration and invasiveness through miR-34a in cervical cancer cells
After transfection of HeLa and SiHa cervical cancer cells with an miR-34a mimic and miR-34a inhibitor, the expression of miR-34a was determined to be increased and reduced, respectively, in these cancer cells by using real-time polymerase chain reaction (p < 0.001 and p < 0.001 for HeLa cells, Figure 2A and 2B; p < 0.001 and p < 0.001 for SiHa cells, Figure 2C and 2D). Moreover, after transfection of HeLa and SiHa cervical cancer cells with the miR-34a inhibitor, the expression of CAIX was elevated (Figure 2E). By contrast, after transfection with the miR-34a mimic, the expression of CAIX was reduced (Figure 2F). We further evaluated the influence of the miR-34a inhibitor on the migration and invasiveness of HeLa and SiHa cells and found them to be increased (Figure 2G and 2H). This result indicates that the miR-34a inhibitor may inhibit the suppression of CAIX expression by miR-34a and then increase the expression of CAIX. Consequently, the migration and invasiveness ability were elevated in both cancer cell lines.
Figure 2. Effect of miR-34a inhibitor and mimic on the protein expressions of carbonic anhydrase IX (CAIX), migratory and invasive abilities in different cancer cell lines.
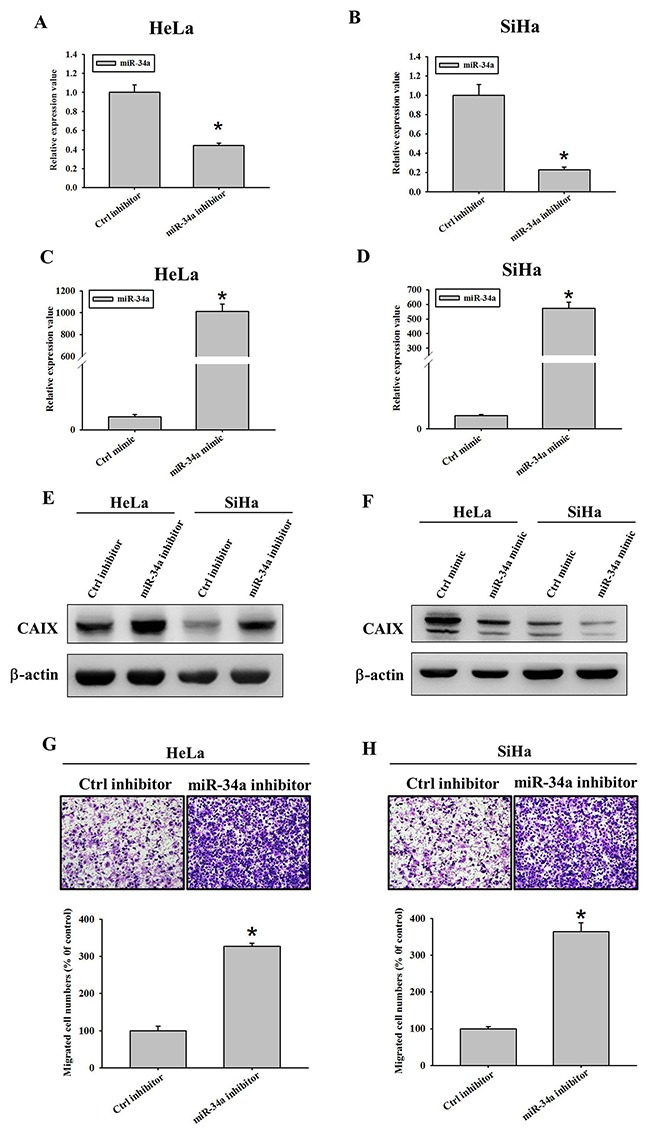
Reduced expressions of miR-34a after miR-34a inhibitor transfection into (A) HeLa and (B) SiHa cancer cell lines of uterine cervix. Elevated expressions of miR-34a after miR-34a mimic transfection into (C) HeLa and (D) SiHa cancer cell lines. The RNA levels were determined by Real time PCR 24 hours after miR-34a mimics and inhibitors transfection into these cancer cells. (E) Elevated expressions of CAIX protein after 200 nM miR-34a inhibitor transfection into HeLa and SiHa cancer cell lines of uterine cervix. (F) Reduced expressions of CAIX protein after 100 nM miR-34a mimic transfection into HeLa and SiHa cancer cell lines. The protein levels were determined by Western blot 24 hours after miR-34a inhibitors and mimics transfection into these cancer cells. (G-H) Increased migratory abilities in (G) HeLa and (H) SiHa cervical cancer cells with miR-34a inhibitor, as compared to their negative control cells without inhibitor by Boyden chamber assay.
Association between CAIX expression and clinicopathological variables, cancer recurrence, and survival of patients with cervical cancer
The H score of the CAIX immunoreactivity of cancer tissues in 149 patients with cervical cancer was significantly higher than that of normal tissues in 29 normal controls (median: 0.5 vs. 0; p = 0.012). Patients whose tissues exhibited higher H scores of CAIX immunoreactivity had a more advanced stage (>stage I) compared with those with lower H scores (p = 0.014; Table 3). Nonsignificant associations between higher H scores of CAIX and higher recurrence probability and between higher H scores and shorter survival (p = 0.630 and p = 0.197; based on Kaplan–Meier curves) were observed.
Table 3. The association of carbonic anhydrase IX (CAIX) immunoreactivity in 149 cancer tissue microarrays with clinicopathological parameters of uterine cervical cancer.
| Clinicopathological variablesb | CAIXc | p values | OR and 95% CI | |
|---|---|---|---|---|
| (+) | (-) | |||
| Stage | 0.014a | |||
| I | 8 | 78 | 1.00 | |
| Others (II + III + IV) | 12 | 36 | 3.25 (1.10-9.94) | |
| Pathologic type | 1.000 | |||
| squamous cell carcinoma | 22 | 99 | 1.00 | |
| adenocarcinoma | 4 | 21 | 0.86 (0.19-2.92) | |
| Depth of stromal invasion | 0.913 | |||
| ≤10 mm | 7 | 46 | 1.00 | |
| >10 mm | 10 | 62 | 1.06 (0.33-3.54) | |
| Tumor grade | 1.000 | |||
| well | 3 | 17 | 1.00 | |
| moderate or poor | 19 | 85 | 1.27 (0.32-7.40) | |
| Parametrial invasion | 0.576 | |||
| no invasion | 19 | 93 | 1.00 | |
| invasion | 7 | 26 | 1.32 (0.42-3.73) | |
| Vaginal invasion | 0.232 | |||
| no invasion | 20 | 103 | 1.00 | |
| invasion | 6 | 16 | 1.93 (0.55-6.01) | |
| Pelvic lymph node metastasis | 0.161 | |||
| negative | 18 | 97 | 1.00 | |
| positive | 8 | 22 | 1.96 (0.65-5.49) | |
Statistical analysis: chi-square or Fisher's exact test,
ap < 0.05.
bSome clinicopathological data could not be collected from the patients with cervical cancer due to incomplete records of medical charts.
c(+): positive immunoreactivity; (-): negative immunoreactivity. The median value of all H scores of CAIX in 149 cervical cancer cores was determined as the cutoff point to separate CAIX positive from CAIX negative tissue cores. Semiquantitative H score of CAIX immunoreactivity was calculated by multiplying the proportion score of stained cells by their immunoreactivity intensity.
OR: odds ratio; CI: confidence interval.
DISCUSSION
In our study, we demonstrated that the genotype CA/AA increases the susceptibility of Taiwanese women to cervical cancer by using the wild homozygous genotype CC as a reference in the CA9 SNP rs1048638. In addition, the mutant homozygous genotype AA was demonstrated to increase the susceptibility of Taiwanese women to invasive cervical cancer by using GG/GA as a reference in rs2071676. Furthermore, we observed that only one mutant allele A is sufficiently strong to increase the risk of cervical cancer in the CA9 SNP rs1048638 based on the analysis of the allele distribution for CA9 SNPs. However, two mutant alleles were necessary to increase the risk of cervical invasive cancer in the CA9 SNP rs2071676. Paired allele mutation (homozygous mutant AA) was required for an increased risk of cervical cancer in comparison with GG/GA in the CA9 SNP rs2071676.
Bioinformatics analysis was performed for the three SNPs, rs2071676, rs3829078, rs1048638, and the deletion mutation 376del393 in CA9 in our study. The CA9 SNP rs2071676 is located on exon 1 in chromosome 9p13.3, and a nucleotide change from G to the mutant A may lead to a nonsynonymous function (amino acid change from valine to methionine) [24]. This polymorphism is correlated with the signal peptide of CAIX and may influence its function [24]. The SNP rs3829078 is a known SNP of A1019G (Gln326Arg) located on exon 7 and also has a nonsynonymous function [24]. It affects the nearby substrate binding site of the CA active domain of CAIX [25, 26]. However, in our study, we did not identify a role of the SNP rs3829078 in the occurrence of cervical cancer. The SNP rs1048683 is located on the 3′-UTR region and near the cis elements of the human poly(A) site of chromosome 11, and an SNP in the 3′-UTR of a gene may influence biological processes [18, 24]. We found that the mutant allele A was involved in the development of cervical cancer. This SNP implication may result from miRNA–mRNA interaction. We found that two miRNAs, miR-34a and miR-449a, were involved in the rs1048638 site by using the TargetScanHuman prediction server. In the prediction of miRNA/target duplex interactions between miR-34a and CA9 among different alleles, we found a 21.2% difference in minimum free energy between the C and A alleles by using the Bielefeld bioinformatics server [27, 28]. This explains the decreased binding of miR-34a with the 3′-UTR of exon 11 of CA9, where the CA9 SNP rs1048638 is located, in women with cervical cancer with the mutant allele A in the CA9 SNP rs1048638 and the elevated CAIX expression in Taiwanese women.
CA9 376del393 is a known deletion mutation of “DEEDLP” (“AspGluGluAspLeuPro”) in the proteoglycan domain and is located on chromosome 1. It may affect the proteoglycan domain of CAIX, which acts as an excellent catalyst for CO2 hydration at acidic pH values [29]. A distinctive feature of CAIX among all known CAs is the presence of a proteoglycan region of 58 amino acids, which is situated extracellularly at the amino-terminal region of the protein, in front of the CA catalytic domain [29]. This domain is rich in acidic amino acid residues with 8 aspartic acid and 18 glutamic acid residues constituting a total of 58 residues [30]. The presence of 26 COOH side chains from these acidic amino acid residues within the proteoglycan domain enables this domain to function in a buffer-like manner because the carboxylate/carboxylic acid conjugate bases are excellent biological buffers. The domain provides an intrinsic buffer promoting efficient CO2 hydration at acidic pH values, which is observed in hypoxic tumors. However, in our study, 376del393 did not exhibit a significant role in the susceptibility of Taiwanese women to cervical cancer. Our finding is in agreement with that of Chien et al. [20]. They reported that 376del393 is not associated with the susceptibility to oral cancer. Based on our findings that no patients with cervical cancer had 376del393 in both homologous chromosomes and those of de Martino et al. that the novel deletion 376del393 was present only in two patients with renal cell carcinoma [24], 376del393 appears to protect individuals from developing cancer.
In this study, cervical cancer tissues exhibited higher immunohistochemical expression of CAIX compared with normal tissues. Moreover, patients whose tissues exhibited higher H scores of CAIX immunoreactivity had more advanced stages (>stage I, beyond uterus) compared with those with lower H scores. Transfection with themiR-34a mimic led to changes in the miRNA–mRNA duplex interaction in the CA9 SNP rs1048638 and reduced the CAIX expression, which reduced the migratory abilities of HeLa and SiHa cervical cancer cells. By contrast, the miR-34a inhibitor increased CAIX expression and therefore elevated its migration. Previous studies have demonstrated that miR-34a contributes to the development and progression of cervical cancer [31–33]. Moreover, Imani et al. demonstrated that miR-34a inhibits breast cancer cell migration by targeting epithelial-to-mesenchymal transition–inducing transcription factors [34]. In addition, the overexpression of CAIX in parental SiHa cells resulted in a significant elevation of the migratory and invasive abilities of SiHa cervical cancer cells. However, no patient with higher CAIX immunoreactivity exhibited deeper stromal invasion or more lymph node metastasis. Furthermore, a nonsignificant association was observed between a higher H score and shorter survival. Woelber et al. demonstrated that enhanced CAIX expression is an important feature of carcinogenesis in cervical cancer [35]. In their analysis, moderate or strong intratumoral CAIX expression was significantly associated with an advanced tumor stage, a higher depth of invasion, an undifferentiated tumor grade, and a higher preoperative serum squamous cell carcinoma level. Furthermore, the patients with cervical cancer with advanced stages (>stage I; CAIX expression: moderate or strong: 63 patients, none or weak: 22) exhibited stronger CAIX expression (p = 0.027; we calculated the p value based on their data) than those with stage I cervical cancer (moderate or strong: 74 patients, none or weak: 62). In agreement with our findings, Woelber et al. could not find a significant association between CAIX expression and patient survival. High CAIX expression has been reported to be more frequent in patients with cervical cancer with lymph node metastasis [36, 37]. However, Liao et al. demonstrated that immunohistochemical CAIX expression is an independent prognostic factor for both progression-free survival and overall survival in patients with high-risk, early-stage cervical cancer who received adjuvant pelvic radiotherapy with or without radiosensitizing chemotherapy [38]. Our study has some limitations. First, it lacked animal investigation, which could provide additional support to our findings. Furthermore, the molecular functional role of the binding between CA9 3′-UTR mRNA and miR-34a binding in cervical cancer requires further investigation.
In conclusion, our study reveals that the CA9 SNP rs1048638 is the only significant polymorphism that increases the risk of cervical cancer among four CA9 gene polymorphisms, rs2071676 (+201, G/A), rs3829078 (+1081, A/G), rs1048638 (+1584), and an 18-base pair deletion/insertion, in Taiwanese women. We discovered that the CA9 SNP rs1048638 influences the expression of CA9 through the interaction between the 3′-UTR of exon 11, where the SNP is located, and miR-34a. Elevated expression of CAIX was observed after transfection of HeLa and SiHA cervical cancer cells with the miR-34a inhibitor, which increased the migratory abilities of these cells through the inhibition of the microRNA–mRNA (miR-34a with 3′-UTR containing allele C of CA9 SNP rs1048638) duplex interaction. We demonstrated that CAIX immunoreactivity is related to the occurrence of uterine cervical cancer, and elevated CAIX immunoreactivity is associated with a more advanced stage of cervical cancer. These findings may be applicable to translational medicine.
MATERIALS AND METHODS
Population
Four hundred fifty Taiwanese women, including 123 patients with uterine cervical cancer and 327 control Taiwanese women, were consecutively recruited into this study. The studied subjects all live in Mid-Taiwan where our hospital is located. Patients with cervical cancer were clinically staged I (n=79) or others (stage II, III and IV; n=44) based on the International Federation of Gynecology and Obstetrics Classification and received routine protocols at the Department of Obstetrics and Gynecology in Chung Shan Medical University Hospital, Taiwan, between January 1, 1999 and August 31, 2010. The histological types of the 123 cervical cancer tissues included 93 squamous cell carcinoma and 30 adenocarcinoma. Their grading were 31 grade 1, 73 grade 2 and 19 grade 3. Meanwhile, 327 control women without previous cancer history, who had normal Papanicolaou smear and were further verified using colposcopy in general examination at outpatient department in Chung Shan Medical University Hospital, were enrolled. The ages of women with cervical cancer and normal women were 53.9 ± 11.9, and 44.2 ± 10.2 (mean ± SD) years old, respectively. The study was approved by Institutional Review Board, Chung Shan Medical University Hospital (CSMUH No: CS11152). Each participant completed the informed consent.
Selection of CA9 gene polymorphisms
Over 30 SNPs have been documented in the 11 exons region of the CA9 gene in NCBI (National Center for Biotechnology Information), database SNP (dbSNP). We selected one SNP rs2071676 (+201, G/A) in exon 1, one SNP rs3829078 (+1081, A/G) in exon 7, one SNP rs1048638 (+1584, C/A) in the 3’-UTR of exon 11 as well as one polymorphism of an 18-base pair deletion/insertion (376deltion393 in exon 1) of CA9 gene based on Chinese HapMap (Han Chinese in Beijing, China) data and the studies of Chien et al. [20]. The minor allele frequencies of these polymorphisms are ≥ 5%.
Blood samples collection and genomic DNA extraction
One hundred and twenty-three blood specimens were collected from patients with cervical cancer. Overall, 327 blood specimens were collected from control women. Genomic DNA was extracted from EDTA anticoagulated venous blood using a QIAamp DNA blood mini kits (Qiagen, Valencia, Valencia, CA, USA) as described in detail previously [39, 40]. DNA was dissolved in Tris ethylene buffer (10 mmol/L Tris and 1 mmol/L EDTA; pH 7.8), and thereafter quantified by a measurement of OD260. Final preparation was stored at -20°C and applied as a template in polymerase chain reaction (PCR).
Determination of single nucleotide polymorphisms of CA9 using real time polymerase chain reaction and genotyping
The allelic discrimination of the CA9 SNPs rs2071676 (Assay ID: C_25472146_10), rs3829078 (Assay ID: C_27507259_10) and rs1048638 (Assay ID: C_1294917_10) was assessed with the ABI StepOne Real-Time PCR System (Applied Biosystems, Foster City, CA, USA), and analyzed by the TaqMan assay using SDS vers. 3.0 software as previously described [20]. The 376del393 SNP was detected with PCR. The PCR products were then electrophoresed through 3% agarose gels and stained with ethidium bromide. The primer sequences and probes for analysis of the CA9 gene polymorphisms were described in our previous study [20].
The influence of CA9 SNP rs1048638 on the immunohistochemical expression of CAIX protein and the clinical implication of CAIX protein in cervical cancer based on the tissue microarrays
In order to detect the influence of CA9 SNP rs1048638 on the expression of CAIX protein, we simultaneously collected the tissue and blood samples to relate the allele distribution of CA9 SNP (+1584, C/A) with the immunohistchemical expression of CAIX from 25 patients among 123 patients with cervical cancer, whom we recruited previously. Moreover, we established cervical tissue microarrays, consisting of normal and cancer tissues, to evaluate the clinical implication of CAIX in cervical cancer. Tissue microarray sections (MaxArray tissue cores, Zymed Laboratories Inc.) were incubated with anti-CAIX antibody (1:300 dilution; NovocastraTM Liquid Mouse Monoclonal Antibody Carbonic Anhydrase IX, Product Code: NCL-L-CAIX; Newcastle Upon Tyne NE12 8EW United Kingdom). A semi-quantitative H score of CAIX immunoreactivity was determined by multiplying the proportional score of stained cells by their immunoreactivity [41–44].
Isolation of RNA, reverse transcription-polymerase chain reaction (RT-PCR), quantitative real time PCR and western blotting
Total cellular RNA was extracted from SiHa, HeLa and Caski cervical cancer cells using RareRNA (Genepure Technology, Taiwan) according to the manufacturer's protocol. For PCR amplification, the primer sets were described in our previous study [14]. For western blotting, an equivalent amount of total protein was processed by 12% SDS-PAGE and electroblotted onto Hybond ECL PVDF membranes, and incubated with primary antibodies against human CAIX (1:1000 dilution; NovocastraTM Liquid Mouse Monoclonal Antibody Carbonic Anhydrase IX) and β-actin (Sigma, St. Louis, MO, A5441).
The influence of CAIX expression on the migration and invasiveness of cervical cancer cells
Cell migration and invasiveness assays were performed using modified Boyden chambers with 25 × 80 mm porous (12 μm size) polycarbonate membrane or 10× Matrigel precoated polycarbonate membrane (for cell invasiveness) using a 48 well chamber from Neuro Probe, Inc. The cells, which passed through the membrane, were stained with 20% Giemsa stain (Merck, Darmstadt, Germany) as previously described [14]. The cells on per well were in full photograph in microscopic fields at a magnification of ×40 and then were counted for the total migrated or invasive cells in triplicate.
Statistical analysis
The independent Student's t test was used to analyze the age distribution of studied population, including patients with cervical cancer and control women. Chi-square or Fisher's exact tests were used to examine the relationships among frequencies of CA9 gene SNPs or allele and incidence of cervical cancer between patients with cervical cancer and control women. Logistic regression model was used to analyze the comparisons of SNPs genotypes or alleles of CA9 gene after controlling the age variable. The Mann-Whitney test was used to compare the CAIX expression between cervical cancer tissues and normal tissues as well as between cancer tissues of cervical cancer patients with wild homozygous CA9 SNP rs1048638 (CC) and those with heterozygous SNP (CA). Chi-square or Fisher's exact tests were also used to associate the expression of CAIX protein (higher or lower H score) with various clinicopathological parameters, such as clinical staged I or others (stage II, III and IV), histopathologic types including squamous cell carcinoma (SCC) or adenocarcinoma, cell grading (well and moderate or poor differentiation), invasion depth of cervical stroma (≤ 10 mm or>10 mm stromal invasion depth), parametrium and vagina invasion and lymph node metastasis found during radical abdominal hysterectomy in cervical cancer patients. The median H score of CAIX immunoreactivity of cervical cancer tissues was selected as a cutoff level to separating the cancer tissues with higher expression (strong staining) from those with lower expression (weak staining). Kaplan-Meier curves were plotted for the cervical cancer patients based on the CAIX expression for the probability of recurrence or overall survival between primary surgery and death or recurrence or the end of the study (May 31, 2012). Odds ratio (OR) and adjusted odds ratio (AOR; controlling for age) and their 95% confidence interval (CI) were calculated by WinPepi software, version 10.0. or SPSS, version 12.0. A significant difference was defined by p < 0.05.
Acknowledgments
This study was supported by research grants from Ministry of Science and Technology (MOST 105-2314-B-040-016-MY2) and Chung Shan Medical University Hospital (CSH-2016-D-002).
Abbreviations
- CAIX
Carbonic anhydrase IX
- HSIL
High squamous intraepithelial lesions
- LSIL
Low squamous intraepithelial lesions
- SNP
Single nucleotide polymorphism
- UTR
Untranslated region
Author contributions
SFY and PHW conceived and designed the experiments; YFL, CWC, WEY and WLL performed the experiments; YFL and JLK analyzed the data; SFY, YFL and PHW wrote the paper.
CONFLICTS OF INTEREST
The authors declared no conflicts of interest.
REFERENCES
- 1.Bharti AC, Shukla S, Mahata S, Hedau S, Das BC. Anti-human papillomavirus therapeutics: facts & future. Indian J Med Res. 2009;130:296–310. [PubMed] [Google Scholar]
- 2.Baak JP, Kruse AJ, Robboy SJ, Janssen EA, van Diermen B, Skaland I. Dynamic behavioural interpretation of cervical intraepithelial neoplasia with molecular biomarkers. J Clin Pathol. 2006;59:1017–28. doi: 10.1136/jcp.2005.027839. https://doi.org/10.1136/jcp.2005.027839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neri D, Supuran CT. Interfering with pH regulation in tumours as a therapeutic strategy. Nat Rev Drug Discov. 2011;10:767–77. doi: 10.1038/nrd3554. https://doi.org/10.1038/nrd3554. [DOI] [PubMed] [Google Scholar]
- 4.Ho HY, Lin CW, Chien MH, Reiter RJ, Su SC, Hsieh YH, Yang SF. Melatonin suppresses TPA-induced metastasis by downregulating matrix metalloproteinase-9 expression through JNK/SP-1 signaling in nasopharyngeal carcinoma. J Pineal Res. 2016;61:479–92. doi: 10.1111/jpi.12365. https://doi.org/10.1111/jpi.12365. [DOI] [PubMed] [Google Scholar]
- 5.Su SC, Hsieh MJ, Yang WE, Chung WH, Reiter RJ, Yang SF. Cancer metastasis: Mechanisms of inhibition by melatonin. J Pineal Res. 2017;62:e12370. doi: 10.1111/jpi.12370. https://doi.org/10.1111/jpi.12370. [DOI] [PubMed] [Google Scholar]
- 6.Supuran CT. Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nat Rev Drug Discov. 2008;7:168–81. doi: 10.1038/nrd2467. https://doi.org/10.1038/nrd2467. [DOI] [PubMed] [Google Scholar]
- 7.Supuran CT. Carbonic anhydrase inhibitors. Bioorg Med Chem Lett. 2010;20:3467–74. doi: 10.1016/j.bmcl.2010.05.009. https://doi.org/10.1016/j.bmcl.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Hilvo M, Baranauskiene L, Salzano AM, Scaloni A, Matulis D, Innocenti A, Scozzafava A, Monti SM, Di Fiore A, De Simone G, Lindfors M, Janis J, Valjakka J, et al. Biochemical characterization of CA IX, one of the most active carbonic anhydrase isozymes. J Biol Chem. 2008;283:27799–809. doi: 10.1074/jbc.M800938200. https://doi.org/10.1074/jbc.M800938200. [DOI] [PubMed] [Google Scholar]
- 9.de Zubicaray GI, McMahon KL, Eastburn MM, Finnigan S, Humphreys MS. fMRI evidence of word frequency and strength effects during episodic memory encoding. Brain Res Cogn Brain Res. 2005;22:439–50. doi: 10.1016/j.cogbrainres.2004.10.002. https://doi.org/10.1016/j.cogbrainres.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Scheurer SB, Rybak JN, Rosli C, Neri D, Elia G. Modulation of gene expression by hypoxia in human umbilical cord vein endothelial cells: A transcriptomic and proteomic study. Proteomics. 2004;4:1737–60. doi: 10.1002/pmic.200300689. https://doi.org/10.1002/pmic.200300689. [DOI] [PubMed] [Google Scholar]
- 11.Tureci O, Sahin U, Vollmar E, Siemer S, Gottert E, Seitz G, Parkkila AK, Shah GN, Grubb JH, Pfreundschuh M, Sly WS. Human carbonic anhydrase XII: cDNA cloning, expression, and chromosomal localization of a carbonic anhydrase gene that is overexpressed in some renal cell cancers. Proc Natl Acad Sci U S A. 1998;95:7608–13. doi: 10.1073/pnas.95.13.7608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Svastova E, Hulikova A, Rafajova M, Zat’ovicova M, Gibadulinova A, Casini A, Cecchi A, Scozzafava A, Supuran CT, Pastorek J, Pastorekova S. Hypoxia activates the capacity of tumor-associated carbonic anhydrase IX to acidify extracellular pH. FEBS Lett. 2004;577:439–45. doi: 10.1016/j.febslet.2004.10.043. https://doi.org/10.1016/j.febslet.2004.10.043. [DOI] [PubMed] [Google Scholar]
- 13.Swietach P, Wigfield S, Cobden P, Supuran CT, Harris AL, Vaughan-Jones RD. Tumor-associated carbonic anhydrase 9 spatially coordinates intracellular pH in three-dimensional multicellular growths. J Biol Chem. 2008;283:20473–83. doi: 10.1074/jbc.M801330200. https://doi.org/10.1074/jbc.M801330200. [DOI] [PubMed] [Google Scholar]
- 14.Yang JS, Lin CW, Chuang CY, Su SC, Lin SH, Yang SF. Carbonic anhydrase IX overexpression regulates the migration and progression in oral squamous cell carcinoma. Tumour Biol. 2015;36:9517–24. doi: 10.1007/s13277-015-3692-8. https://doi.org/10.1007/s13277-015-3692-8. [DOI] [PubMed] [Google Scholar]
- 15.Yang JS, Chen MK, Yang SF, Chang YC, Su SC, Chiou HL, Chien MH, Lin CW. Increased expression of carbonic anhydrase IX in oral submucous fibrosis and oral squamous cell carcinoma. Clin Chem Lab Med. 2014;52:1367–77. doi: 10.1515/cclm-2014-0129. https://doi.org/10.1515/cclm-2014-0129. [DOI] [PubMed] [Google Scholar]
- 16.Lin CW, Yang WE, Lee WJ, Hua KT, Hsieh FK, Hsiao M, Chen CC, Chow JM, Chen MK, Yang SF, Chien MH. Lipocalin 2 prevents oral cancer metastasis through carbonic anhydrase IX inhibition and is associated with favourable prognosis. Carcinogenesis. 2016;37:712–22. doi: 10.1093/carcin/bgw050. https://doi.org/10.1093/carcin/bgw050. [DOI] [PubMed] [Google Scholar]
- 17.Shastry BS. SNP alleles in human disease and evolution. J Hum Genet. 2002;47:561–6. doi: 10.1007/s100380200086. https://doi.org/10.1007/s100380200086. [DOI] [PubMed] [Google Scholar]
- 18.Shastry BS. SNPs: impact on gene function and phenotype. Methods Mol Biol. 2009;578:3–22. doi: 10.1007/978-1-60327-411-1_1. https://doi.org/10.1007/978-1-60327-411-1_1. [DOI] [PubMed] [Google Scholar]
- 19.Klatte T, Rao PN, de Martino M, LaRochelle J, Shuch B, Zomorodian N, Said J, Kabbinavar FF, Belldegrun AS, Pantuck AJ. Cytogenetic profile predicts prognosis of patients with clear cell renal cell carcinoma. J Clin Oncol. 2009;27:746–53. doi: 10.1200/JCO.2007.15.8345. https://doi.org/10.1200/JCO.2007.15.8345. [DOI] [PubMed] [Google Scholar]
- 20.Chien MH, Yang JS, Chu YH, Lin CH, Wei LH, Yang SF, Lin CW. Impacts of CA9 gene polymorphisms and environmental factors on oral-cancer susceptibility and clinicopathologic characteristics in Taiwan. PLoS One. 2012;7:e51051. doi: 10.1371/journal.pone.0051051. https://doi.org/10.1371/journal.pone.0051051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fraga A, Ribeiro R, Coelho A, Vizcaino JR, Coutinho H, Lopes JM, Principe P, Lobato C, Lopes C, Medeiros R. Genetic polymorphisms in key hypoxia-regulated downstream molecules and phenotypic correlation in prostate cancer. BMC Urol. 2017;17:12. doi: 10.1186/s12894-017-0201-y. https://doi.org/10.1186/s12894-017-0201-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang SS, Liu YF, Ou YC, Chen CS, Li JR, Yang SF. Impacts of CA9 gene polymorphisms on urothelial cell carcinoma susceptibility and clinicopathologic characteristics in Taiwan. PLoS One. 2013;8:e82804. doi: 10.1371/journal.pone.0082804. https://doi.org/10.1371/journal.pone.0082804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hua KT, Liu YF, Hsu CL, Cheng TY, Yang CY, Chang JS, Lee WJ, Hsiao M, Juan HF, Chien MH, Yang SF. 3’UTR polymorphisms of carbonic anhydrase IX determine the miR-34a targeting efficiency and prognosis of hepatocellular carcinoma. Sci Rep. 2017;7:4466. doi: 10.1038/s41598-017-04732-3. https://doi.org/10.1038/s41598-017-04732-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Martino M, Klatte T, Seligson DB, LaRochelle J, Shuch B, Caliliw R, Li Z, Kabbinavar FF, Pantuck AJ, Belldegrun AS. CA9 gene: single nucleotide polymorphism predicts metastatic renal cell carcinoma prognosis. J Urol. 2009;182:728–34. doi: 10.1016/j.juro.2009.03.077. https://doi.org/10.1016/j.juro.2009.03.077. [DOI] [PubMed] [Google Scholar]
- 25.Whittington DA, Waheed A, Ulmasov B, Shah GN, Grubb JH, Sly WS, Christianson DW. Crystal structure of the dimeric extracellular domain of human carbonic anhydrase XII, a bitopic membrane protein overexpressed in certain cancer tumor cells. Proc Natl Acad Sci U S A. 2001;98:9545–50. doi: 10.1073/pnas.161301298. https://doi.org/10.1073/pnas.161301298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alterio V, Hilvo M, Di Fiore A, Supuran CT, Pan P, Parkkila S, Scaloni A, Pastorek J, Pastorekova S, Pedone C, Scozzafava A, Monti SM, De Simone G. Crystal structure of the catalytic domain of the tumor-associated human carbonic anhydrase IX. Proc Natl Acad Sci U S A. 2009;106:16233–8. doi: 10.1073/pnas.0908301106. https://doi.org/10.1073/pnas.0908301106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006;342:129–38. doi: 10.1385/1-59745-123-1:129. https://doi.org/10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- 28.Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–4. doi: 10.1093/nar/gkl243. https://doi.org/10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Innocenti A, Pastorekova S, Pastorek J, Scozzafava A, De Simone G, Supuran CT. The proteoglycan region of the tumor-associated carbonic anhydrase isoform IX acts as anintrinsic buffer optimizing CO2 hydration at acidic pH values characteristic of solid tumors. Bioorg Med Chem Lett. 2009;19:5825–8. doi: 10.1016/j.bmcl.2009.08.088. https://doi.org/10.1016/j.bmcl.2009.08.088. [DOI] [PubMed] [Google Scholar]
- 30.Pastorek J, Pastorekova S, Callebaut I, Mornon JP, Zelnik V, Opavsky R, Zat’ovicova M, Liao S, Portetelle D, Stanbridge EJ, Závada J, Burny A, Kettmann R. Cloning and characterization of MN, a human tumor-associated protein with a domain homologous to carbonic anhydrase and a putative helix-loop-helix DNA binding segment. Oncogene. 1994;9:2877–88. [PubMed] [Google Scholar]
- 31.Geng D, Song X, Ning F, Song Q, Yin H. MiR-34a Inhibits Viability and Invasion of Human Papillomavirus-Positive Cervical Cancer Cells by Targeting E2F3 and Regulating Survivin. Int J Gynecol Cancer. 2015;25:707–13. doi: 10.1097/IGC.0000000000000399. https://doi.org/10.1097/IGC.0000000000000399. [DOI] [PubMed] [Google Scholar]
- 32.Chandrasekaran KS, Sathyanarayanan A, Karunagaran D. Downregulation of HMGB1 by miR-34a is sufficient to suppress proliferation, migration and invasion of human cervical and colorectal cancer cells. Tumour Biol. 2016;37:13155–66. doi: 10.1007/s13277-016-5261-1. https://doi.org/10.1007/s13277-016-5261-1. [DOI] [PubMed] [Google Scholar]
- 33.Zhang R, Su J, Xue SL, Yang H, Ju LL, Ji Y, Wu KH, Zhang YW, Zhang YX, Hu JF, Yu MM. HPV E6/p53 mediated down-regulation of miR-34a inhibits Warburg effect through targeting LDHA in cervical cancer. Am J Cancer Res. 2016;6:312–20. [PMC free article] [PubMed] [Google Scholar]
- 34.Imani S, Wei C, Cheng J, Khan MA, Fu S, Yang L, Tania M, Zhang X, Xiao X, Zhang X, Fu J. MicroRNA-34a targets epithelial to mesenchymal transition-inducing transcription factors (EMT-TFs) and inhibits breast cancer cell migration and invasion. Oncotarget. 2017;8:21362–79. doi: 10.18632/oncotarget.15214. https://doi.org/10.18632/oncotarget.15214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woelber L, Kress K, Kersten JF, Choschzick M, Kilic E, Herwig U, Lindner C, Schwarz J, Jaenicke F, Mahner S, Milde-Langosch K, Mueller V, Ihnen M. Carbonic anhydrase IX in tumor tissue and sera of patients with primary cervical cancer. BMC Cancer. 2011;11:12. doi: 10.1186/1471-2407-11-12. https://doi.org/10.1186/1471-2407-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim JY, Shin HJ, Kim TH, Cho KH, Shin KH, Kim BK, Roh JW, Lee S, Park SY, Hwang YJ, Han IO. Tumor-associated carbonic anhydrases are linked to metastases in primary cervical cancer. J Cancer Res Clin Oncol. 2006;132:302–8. doi: 10.1007/s00432-005-0068-2. https://doi.org/10.1007/s00432-005-0068-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee S, Shin HJ, Han IO, Hong EK, Park SY, Roh JW, Shin KH, Kim TH, Kim JY. Tumor carbonic anhydrase 9 expression is associated with the presence of lymph node metastases in uterine cervical cancer. Cancer Sci. 2007;98:329–33. doi: 10.1111/j.1349-7006.2007.00396.x. https://doi.org/10.1111/j.1349-7006.2007.00396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao SY, Darcy KM, Randall LM, Tian C, Monk BJ, Burger RA, Fruehauf JP, Peters WA, Stock RJ, Stanbridge EJ. Prognostic relevance of carbonic anhydrase-IX in high-risk, early-stage cervical cancer: a Gynecologic Oncology Group study. Gynecol Oncol. 2010;116:452–8. doi: 10.1016/j.ygyno.2009.10.062. https://doi.org/10.1016/j.ygyno.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su SC, Hsieh MJ, Liu YF, Chou YE, Lin CW, Yang SF. ADAMTS14 Gene Polymorphism and Environmental Risk in the Development of Oral Cancer. PLoS One. 2016;11:e0159585. doi: 10.1371/journal.pone.0159585. https://doi.org/10.1371/journal.pone.0159585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng HL, Liu YF, Su CW, Su SC, Chen MK, Yang SF, Lin CW. Functional genetic variant in the Kozak sequence of WW domain-containing oxidoreductase (WWOX) gene is associated with oral cancer risk. Oncotarget. 2016;7:69384–96. doi: 10.18632/oncotarget.12082. https://doi.org/10.18632/oncotarget.12082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olaussen KA, Dunant A, Fouret P, Brambilla E, Andre F, Haddad V, Taranchon E, Filipits M, Pirker R, Popper HH, Stahel R, Sabatier L, Pignon JP, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006;355:983–91. doi: 10.1056/NEJMoa060570. https://doi.org/10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 42.Al-Haddad S, Zhang Z, Leygue E, Snell L, Huang A, Niu Y, Hiller-Hitchcock T, Hole K, Murphy LC, Watson PH. Psoriasin (S100A7) expression and invasive breast cancer. Am J Pathol. 1999;155:2057–66. doi: 10.1016/S0002-9440(10)65524-1. https://doi.org/10.1016/S0002-9440(10)65524-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Handra-Luca A, Bilal H, Bertrand JC, Fouret P. Extra-cellular signal-regulated ERK-1/ERK-2 pathway activation in human salivary gland mucoepidermoid carcinoma: association to aggressive tumor behavior and tumor cell proliferation. Am J Pathol. 2003;163:957–67. doi: 10.1016/S0002-9440(10)63455-4. https://doi.org/10.1016/S0002-9440(10)63455-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang PH, Ko JL, Yang SF, Lin LY. Implication of human nonmetastatic clone 23 Type 1 and its downstream gene lipocalin 2 in metastasis and patient's survival of cancer of uterine cervix. Int J Cancer. 2011;129:2380–9. doi: 10.1002/ijc.25936. https://doi.org/10.1002/ijc.25936. [DOI] [PubMed] [Google Scholar]


