Abstract
Background
To explore the association of genetic polymorphisms in pre-miRNA 30c-1 rs928508 and pre-miRNA 27a rs895819 with non-small-cell lung cancer prognosis.
Materials and Methods
480 patients from five hospitals were enrolled in this prospective cohort study. They were followed up for five years. The association between genotypes and overall survival was assessed by Cox proportional hazards regression models. A meta-analysis was conducted to provide evidence for the effect of microRNA 27a rs895819 on cancer survival.
Results
G-allele containing genotypes of microRNA 30c-1 polymorphisms and C-allele containing genotypes of microRNA 27a were significantly associated with poorer overall survival. Multivariate Cox regression models indicated that these genetic polymorhpisms were independently predictive factors of poorer overall survival. In stratified analysis, the effect was observed in many strata. The significant joint effect was also observed in our study. Patients with G allele of microRNA 30c-1 rs928508 and C allele of microRNA 27a rs895819 had the poorer overall survival than patients with C allele of rs928508 and T allele of rs895819. The effect of the microRNA 27a rs895819 on non-small cell lung cancer overall survival was supported by the meta-analysis results.
Conclusions
The two single nucleotide polymorphisms in microRNA 30c-1 and microRNA 27a can predict the outcome of non-small cell lung cancer patients and they may decrease the sensitivity to anti-cancer drugs.
Keywords: non-small cell lung cancer, prognosis, microRNA, single nucleotide polymorphism, meta-analysis
INTRODUCTION
Lung cancer is a malignancy worldwide with complicated, multi-factorial aetiology, involving both environmental and genetic factors. Tobaccon exposure is a widely recognized risk factor for development of lung cancer. Morbidity and mortality of lung cancer increase in a constant rate in both genders [1] in China. And lung cancer is the most common cause of cancer death in China [2, 3]. With an increasing number of inpatients and a much low 5-year-survival, lung cancer imposes a heavy burden on both family and society in China [4–6]. Much effort was taken to identify prognostic biomarkers. A recent focus arised in the field of epigenetics, which also includes silencing of target genes with microRNAs (miRNAs).
MiRNAs are a class of 19–25 nt in length, small non-coding RNAs, which normally disturb the stability or translation of the target gene by pairing with the mRNAs [7]. For one single miRNA, it may target to tens of mRNAs. The biological functions of most miRNAs may be marvelous. Genetic variants presented in miRNA genes and processing mechanisms may alter miRNA expression and maturation [8]. Moreover, single nucleotide polymorphisms (SNPs) in miRNAs and its binding site may influence the affinity between miRNAs and mRNAs [9]. Thus genetic polymorphisms in miRNAs may influence the cancer prognosis either by affecting the maturation or by altering the ability to combine with target mRNAs.
Two miRNAs, microRNA 27a and microRNA 30c-1, were observed in many pivotal cancer progressions, such as invasion [10–12] and response to chemotherapy [13–16]. Accumulating evidence has shown that aberrant expression profiles and genetic polymorphisms of microRNA 27a [17–21] and microRNA 30c-1 [22, 23] are associated with cancer survival. rs928508 in pre-miRNA 30c-1 was the SNP site identified in 2010 in Hu's study [22]. And the effect of this polymorphism on lung cancer survival needs to be explored. The role of rs895819 in pre-miRNA 27a in various cancers had been discussed in some studies. But the role in lung cancer survival received little exploration [18, 21]. In this present study, we investigated the effect of the two genetic polymorphisms in pre-miRNA 30c-1 and pre-miRNA 27a on the prognosis of non-small cell lung cancer (NSCLC).
RESULTS
There are no differences between follow-up group and lost to follow-up group (data were shown in Supplementary Table 1). The baseline characteristics of lung cancer patients are listed in Table 1. All of the patients are females. We set 60 as the age category boundary. Patients less than 60 are set as one category and the others are set as another category. More than half of the patients are non-smokers and most of the subjects are NSCLC. No small cell lung cancer is included in our present study. About 67% of the female patients are diagnosed with cancer in a relatively later clinical stage. The majority of patients received chemotherapy or surgery after they are diagnosed with lung cancer.
Table 1. The baseline characteristics of patients.
| Characteristics | No. of patients (%) | Death | % | P-value | MST* | Log-rank P-value |
|---|---|---|---|---|---|---|
| Age | ||||||
| < 60 | 248 (54.6) | 213 | 85.9 | 0.991 | 25.72 | 0.293 |
| ≥ 60 | 206 (45.4) | 177 | 85.9 | 23.65 | ||
| Tabaccon exposure | ||||||
| No | 252 (55.5) | 212 | 84.1% | 0.224 | 25.31 | 0.064 |
| Yes | 202 (44.5) | 178 | 88.1% | 22.85 | ||
| Histological type | ||||||
| AD | 237 (52.2) | 196 | 82.7 | 0.101 | 22.20 | < 0.001 |
| SQU | 164 (36.1) | 148 | 90.2 | 23.54 | ||
| Others | 53 (11.7) | 46 | 86.8 | 36.59 | ||
| Clinical stage | ||||||
| I | 95 (20.9) | 65 | 68.4 | < 0.001 | 43.82 | < 0.001 |
| II | 56 (12.2) | 48 | 85.7 | 23.65 | ||
| III | 263 (57.9) | 240 | 91.3 | 17.38 | ||
| IV | 40 (8.8) | 37 | 92.5 | 19.96 | ||
| Chemotherapy | ||||||
| No | 22 (4.8) | 20 | 90.9 | 0.754# | 14.82 | 0.006 |
| Yes | 432 (95.2) | 370 | 85.6 | 25.09 | ||
| Surgery | ||||||
| No | 123 (27.1) | 112 | 91.1 | 0.054 | 20.59 | 0.006 |
| Yes | 331 (72.9) | 278 | 84.0 | 25.42 |
*MST:median survival time, AD:adenocarcinoma, SQU: squamous carcinoma.
#Fisher exact P value.
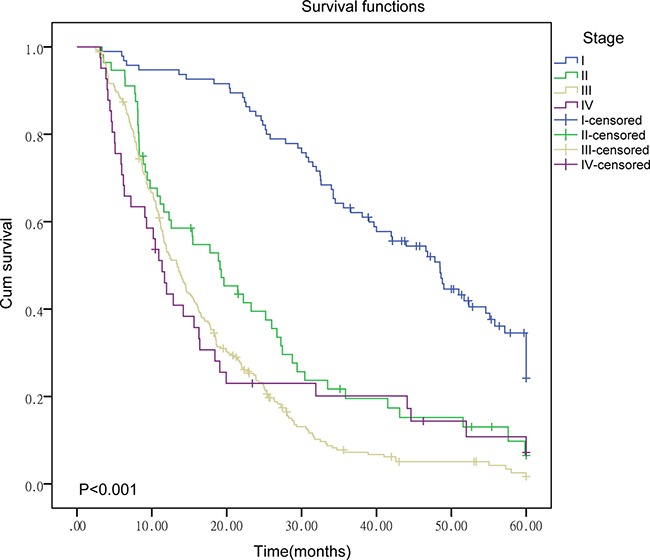
As shown in Table 1, the mortality and median survival time (MST) between patients with different clinical stages are statistically significant (P < 0.001). For late-stage patients, the mortality is higher and the MST is shorter than early-stage patients. Survival curves for patients with different clinical stages are presented in Figure 1. Significant differences in MST between patients with different histological types and therapeutic strategies are also observed (P < 0.006). Patients receiving chemotherapy or surgery can live longer (P = 0.006).
Figure 1. Survival curve of patients in different clinical stages.
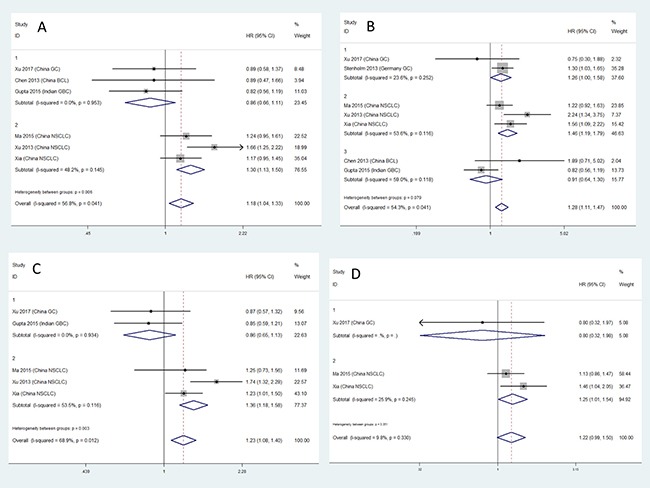
Results for the effect of the two SNPs on cancer overall survival (OS) are listed in Table 2. We observe that G-allele containing genotypes of microRNA 30c-1 rs928508 polymorphisms and C-allele containing genotypes of microRNA 27a rs895819 polymorphisms were significantly associated with poorer survival. Results of multivariate Cox proportional hazards regression models imply that both SNPs were independent predictive factors of poor prognosis. The survival curves are described in Figure 2.
Table 2. The association of polymorphisms in microRNA 30c-1 and microRNA 27a with overall survival.
| SNP | Genotypes | Num | MST | Log-rank P value | HR | 95% CI (P value) | aHR | 95% CI (P value) |
|---|---|---|---|---|---|---|---|---|
| microRNA 30c-1 | AA | 133 | 28.44 | 0.054 | 1.00 | 1.00 | ||
| rs928508 | AG | 229 | 23.20 | 1.28 | 1.01–1.61 (0.041) | 1.30 | 1.03–1.65 (0.027) | |
| GG | 92 | 21.95 | 1.37 | 1.03–1.83 (0.033) | 1.45 | 1.08–1.94 (0.012) | ||
| AG | 229 | 23.20 | 0.592 | 1.00 | 1.00 | |||
| GG | 92 | 21.95 | 1.07 | 0.83–1.39 (0.593) | 1.09 | 0.84–1.43 (0.514) | ||
| AA/AG | 362 | 25.25 | 0.188 | 1.00 | 1.00 | |||
| GG | 92 | 21.95 | 1.18 | 0.92–1.51 (0.190) | 1.24 | 0.96–1.59 (0.097) | ||
| AA | 133 | 28.44 | 0.019 | 1.00 | 1.00 | |||
| AG/GG | 321 | 22.86 | 1.30 | 1.04–1.62 (0.020) | 1.34 | 1.07–1.67 (0.010) | ||
| A allele | 495 | 26.04 | 0.022 | 1.00 | 1.00 | |||
| G allele | 413 | 22.63 | 1.18 | 1.02–1.36 (0.023) | 1.21 | 1.05–1.39 (0.009) | ||
| microRNA 27a | TT | 247 | 26.27 | 0.033 | 1.00 | 1.00 | ||
| rs895819 | CT | 167 | 23.71 | 1.17 | 0.95–1.45 (0.143) | 1.23 | 0.99–1.52 (0.062) | |
| CC | 40 | 18.25 | 1.56 | 1.09–2.22 (0.014) | 1.71 | 1.20–2.45 (0.003) | ||
| CT | 167 | 23.71 | 0.100 | 1.00 | 1.00 | |||
| CC | 40 | 18.25 | 1.35 | 0.94–1.95 (0.102) | 1.44 | 1.00–2.08 (0.050) | ||
| CT/TT | 414 | 25.23 | 0.030 | 1.00 | 1.00 | |||
| CC | 40 | 18.25 | 1.46 | 1.04–2.05 (0.031) | 1.57 | 1.11–2.21 (0.010) | ||
| TT | 247 | 26.27 | 0.041 | 1.00 | 1.00 | |||
| CC/CT | 207 | 22.79 | 1.23 | 1.01–1.50 (0.042) | 1.30 | 1.06–1.50 (0.012) | ||
| T allele | 661 | 25.62 | 0.011 | 1.00 | 1.00 | |||
| C allele | 247 | 22.14 | 1.22 | 1.05–1.43 (0.012) | 1.28 | 1.09–1.49 (0.002) |
*Num: the patients number for each genotype, aHR: adjusted HR for histological type, clinical stage and, receipt of chemotherapy and surgery.
Figure 2. Survival curve of patients with different genotypes.
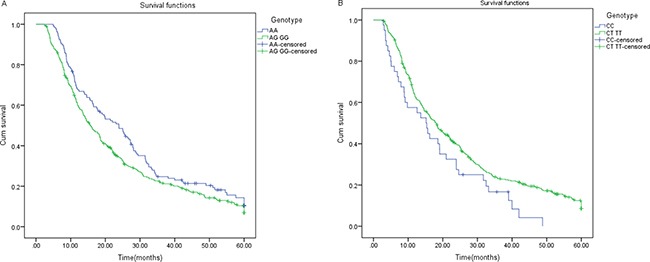
(A) The survival curve of patients with different genotypes of microRNA 30c-1 (P = 0.023). (B) The survival curve of patients with different genotypes of microRNA 27a (P = 0.029).
Results of the stratified analysis are summarized in Supplementary Table 2. The significant effect for microRNA 30c-1 rs928508 and microRNA 27a rs895819 polymorphisms can be observed in many strata. Both the two SNPs can be prognostic in smokers, lung adenocarcinoma patients, squamous cell carcinoma patients, late stage patients and patients receiving chemotherapy.
Results in stratified analysis imply the joint effect of the two SNPs on prognosis. We proceed to analyse the joint effect. Results are shown in Table 3. As shown in Table 3, we observe the joint effect of the two SNPs on cancer prognosis in many strata. The more risk alleles, the poorer OS (larger HRs with smaller P values).
Table 3. The joint effect of the two SNPs.
| Stratum | Model* | Cases | HR | 95% CI | P value |
|---|---|---|---|---|---|
| Smokers | 0 | 39 | 1.00 | ||
| 1 | 71 | 1.29 | 0.83–2.00 | 0.252 | |
| 2 | 67 | 1.66 | 1.07–2.59 | 0.024 | |
| 3 + 4 | 25 | 3.00 | 1.75–5.13 | < 0.001 | |
| AD | 0 | 48 | 1.00 | ||
| 1 | 85 | 1.24 | 0.93–1.86 | 0.289 | |
| 2 | 68 | 1.30 | 0.85–1.98 | 0.226 | |
| 3 + 4 | 36 | 2.20 | 1.35–3.58 | 0.001 | |
| SQU | 0 | 26 | 1.00 | ||
| 1 | 58 | 0.84 | 0.51–1.40 | 0.505 | |
| 2 | 58 | 1.64 | 0.99–2.72 | 0.054 | |
| 3 + 4 | 22 | 1.85 | 1.01–3.39 | 0.047 | |
| III/IV | 0 | 47 | 1.00 | ||
| 1 | 108 | 1.22 | 0.85–1.75 | 0.290 | |
| 2 | 107 | 1.53 | 1.06–2.19 | 0.023 | |
| 3 + 4 | 41 | 2.00 | 1.30–3.09 | 0.002 | |
| Chemotherapy | 0 | 80 | 1.00 | ||
| 1 | 154 | 1.15 | 0.85–1.55 | 0.364 | |
| 2 | 138 | 1.41 | 1.04–1.91 | 0.029 | |
| 3 + 4 | 60 | 1.67 | 1.16–2.41 | 0.006 |
*0:AA of microRNA 30c-1 polymorphisms and TT of microRNA 27a polymorphisms, 1: AG of microRNA 30c-1 polymorphisms and TT of microRNA 27a polymorphisms, or AA of microRNA 30c-1 polymorphisms and CT of microRNA 27a polymorphisms, 2: GG of microRNA 30c-1 polymorphisms and TT of microRNA 27a polymorphisms, or AA of microRNA 30c-1 polymorphisms and CC of microRNA 27a polymorphisms, or AG of microRNA 30c-1 polymorphisms and CT of microRNA 27a polymorphisms, 3: AG of microRNA 30c-1 polymorphisms and CC of microRNA 27a polymorphisms, or GG of microRNA 30c-1 polymorphisms and CT of microRNA 27a polymorphisms, 4:AA of microRNA 30c-1 polymorphisms and CC of microRNA 27a polymorphisms.
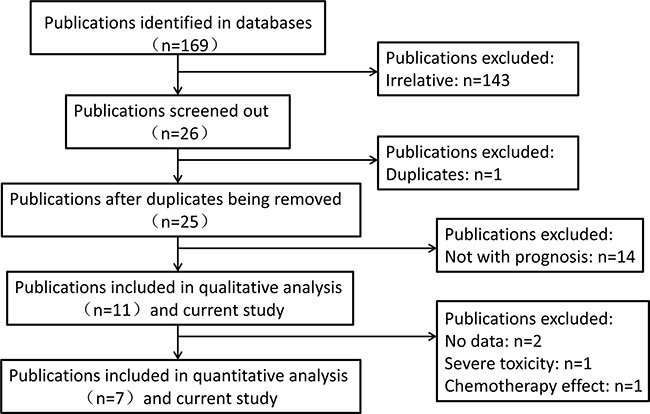
With only two publications, we didn't conduct the meta-analysis for the relationship between cancer prognosis and microRNA 30c-1 rs928508. Publications included in this analysis for microRNA 27a rs895819 are exhibited in Table 4. The workflow of the enrollment is described in Figure 3. Finally, eleven [18, 21, 24–32] eligible publications were screened out. Among them, eight publications focus on the relationship with OS and two focus on recurrence-free survival (RFS). Tumor types of these publications include lung cancer, gastric cancer, breast cancer, B-cell lymphoma, gallbladder cancer and colorectal cancer. Seven [18, 21, 24, 27, 28, 30, 32] publications and our current study were included in quantitative analysis.
Table 4. The information of the publications enrolled in this meta-analysis.
| Year | Name | Tumor type | Population | Number | Outcome |
|---|---|---|---|---|---|
| 2017[24] | Xu | GC | China | 939 | OS |
| 2015[27] | Ma | NSCLC | China | 560 | RFS, OS |
| 2013[18] | Xu | NSCLC | China | 576 | OS |
| 2013[28] | Stenholm | GC | Germany | 674 | OS |
| 2012[21] | Yoon | NSCLC | Korea | 388 | RFS |
| 2013[30] | Zhang | BCL | China | 100 | OS |
| 2015[32] | Gupta | GBC | Indian | 606 | OS |
| Current study | Xia | NSCLC | China | 454 | OS |
GC:gastric cancer, NSCLC:non-small cell lung cancer, BCL: B-cell lymphoma, GBC:gallbladder cancer.
Figure 3. The workflow of the enrollment in the meta-analysis.
According to the publications’ results, C-allele containing genotypes of microRNA 27a rs895819 are related to gastrointestinal toxicity in more than 56 years of age, smoking and non-smoking lung cancer patients that received platinum-based chemotherapy [25]. No significant association with disease-free survival was observed in breast cancer [29]. No significant association with 5FU chemotherapy effect was observed in colorectal cancer patients [31]. The results for the association with cancer OS are inconsistent.
Results for meta-analysis are shown in Table 5. CC genotype of microRNA 27a rs895819 is associated with OS. Patients with CC genotype of microRNA 27a rs895819 may have a poorer survival. The forest plots are exhibited in Figure 4. The funnel plots are exhibited in Figure 5. Sensitivity analysis results are exhibited in Figure 6. No significant association is observed in our meta-analysis for the relationship between RFS and rs895819. No publication bias exists in this study.
Table 5. Pooled HR and 95% CIs of microRNA 27a rs895819.
| Outcome | Model | No. of studies | No. of patients | HR (95% CI) | P-value | Heterogeneity (I2, P-value) |
|---|---|---|---|---|---|---|
| OS | CT vs TT | 6 | 3235 | 1.133 (0.923, 1.391) | 0.232 | 56.8%, 0.041 |
| CC vs TT | 7 | 3909 | 1.296 (1.027, 1.636) | 0.029 | 54.3%, 0.041 | |
| CC + CT vs TT | 5 | 3135 | 1.176 (0.916, 1.509) | 0.204 | 68.9%, 0.012 | |
| CC vs CT + TT | 3 | 1953 | 1.220 (0.975, 1.526) | 0.082 | 9.8%, 0.330 | |
| RFS | CT vs TT | 2 | 948 | 1.117 (0.892, 1.400) | 0.335 | 0.0%, 0.891 |
| CC vs TT | 2 | 948 | 0.979 (0.762, 1.256) | 0.866 | 0.0%, 0.558 | |
| CC + CT vs TT | 2 | 948 | 1.091 (0.899, 1.323) | 0.380 | 0.0%, 0.791 |
OS: overall survival, RFS: recurrence-free survival.
Figure 4. The forest plots for the relationship between overall survival and cancer prognosis.
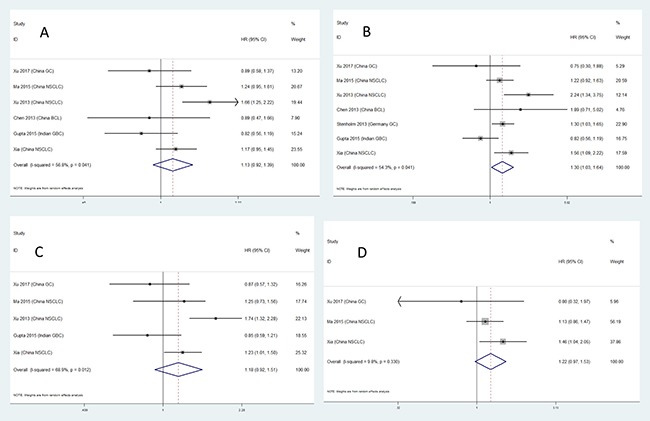
(A) AG vs AA, (B) GG vs AA, (C) AG + GG vs AA, (D) GG vs AA + AG.
Figure 5. The funnel plots for the relationship between overall survival and cancer prognosis.
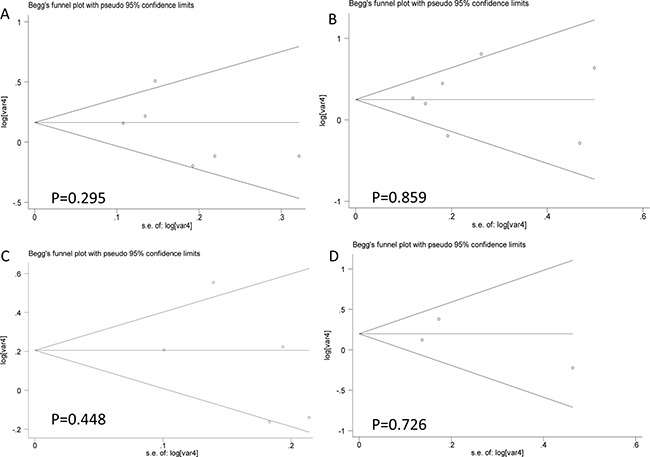
(A) AG vs AA, (B) GG vs AA, (C) AG + GG vs AA, (D) GG vs AA + AG.
Figure 6. Sensitivity analysis results for the relationship between overall survival and cancer prognosis.
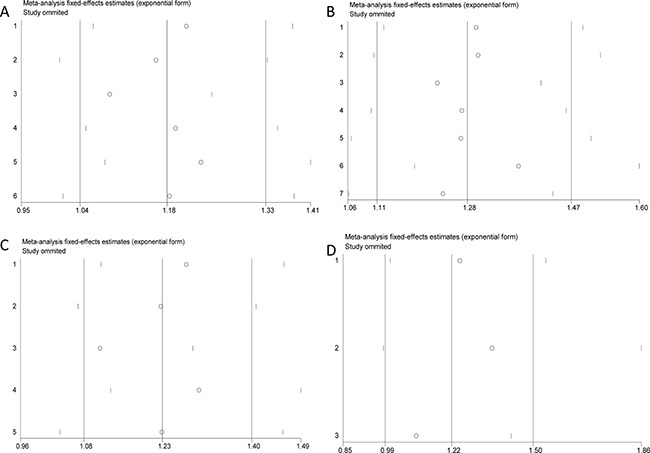
(A) AG vs AA, (B) GG vs AA, (C) AG + GG vs AA, (D) GG vs AA + AG.
Stratified analysis results are shown in Table 6. In this study, we conducted the stratified analysis according to tumor types. The C-allele containing genotypes of microRNA 27a rs895819 are associated with poorer NSCLC prognosis. CC genotype of microRNA 27a rs895819 is associated with poorer gastric cancer prognosis. The forest plots are exhibited in Figure 7.
Table 6. Stratified analysis of the relationship between microRNA 27a rs895818 and overall survival.
| Model | Subgroup | No. of studies | No. of patients | HR (95% CI) | P- value |
|---|---|---|---|---|---|
| CT vs TT | NSCLC | 3 | 1590 | 1.298 (1.125, 1.498) | < 0.001 |
| CC vs TT | NSCLC | 3 | 1590 | 1.457 (1.187, 1.787) | < 0.001 |
| GC | 2 | 1613 | 1.260 (1.004, 1.583) | 0.046 | |
| CT + CC vs TT | NSCLC | 3 | 1590 | 1.364 (1.177, 1.581) | < 0.001 |
| CC vs CT + TT | NSCLC | 2 | 1014 | 1.247 (1.010, 1.539) | 0.040 |
NSCLC: non-small cell lung cancer, GC: gastric cancer.
Figure 7. Forest plots for stratified analysis.
(A) AG vs AA, (B) GG vs AA, (C) AG + GG vs AA, (D) GG vs AA + AG.
DISCUSSION
Once detected, the two miRNAs were observed to be involved in pivotal processes that may lead to poor cancer prognosis. Downregulation of microRNA 30c can promote the invasion of non-small cell lung cancer [10]. Downregulation of microRNA 30c was observed in drug-resistant cancer cells [13]. The polymorphisms in microRNA 30c-1 can alter the expression of mature microRNA 30c. The expression of pre-miR-30c and mature microRNA -30c is higher for AG/GG than AA [33]. All above indicated that the SNP of microRNA 30c-1 may influnce the cancer prognosis by altering the expression of mature microRNA 30c-1. In our study, we observed the poorer OS of patients with G-allele containing genotypes of microRNA 30c-1.
MicroRNA 27a could function to reverse multiple drug resistance by inhibiting FZD7/beta-catenin pathway in hepatocellular carcinoma and therefore promote the therapeutic effect in patients receiving chemotherapy in Chen's study [15]. The similar observation presented in Noratto's study [16] and the role of microRNA 27a was played through microRNA-27a-ZBTB10-Sp axis in colon cancer. The downregulation of microRNA 27a was essential for angiogenesis. And VE-cadherin was observed to be the dominant target of microRNA 27a in vivo and in vitro [34]. Rs10719 T to G substitution in Drosha 3′UTR was observed to result in the disruption of binding activity with microRNA 27a/b [35]. And A allele of microRNA 27a rs11671784 was related to decreased microRNA 27a expression [36]. All the results above indicated the effect of microRNA 27a SNP on therapeutic effect and survival. In our present study, C-allele containing genotypes of microRNA 27a were associated with poorer OS. Results are supported by the meta-analysis results.
In a previous study containing a relatively larger sample size, the protective effect of AG or GG of microRNA 30c-1 was observed on NSCLC survival, especially in older, early-stage and surgically resected patients [22]. In our study, the risk effect of AG or GG of microRNA 30c-1 was observed on NSCLC survival, especially in late-stage and squamous cell carcinoma patients. The effect of the SNP is not consistent in younger, smokers, adenocarcinoma and patients with chemotherapy. The variant allele-containing genotypes of microRNA 27a may function as risk factor for OS in gastric cancer patients in Germany as described in a previous study [28]. In Yoon's study [21], no significant effect of microRNA 27a polymorphisms on recurrence-free survival for NSCLC was observed. Xu [18] observed a risk effect of G-allele containing genotypes of microRNA 27a on OS in Chinese NSCLC patients, which is consistent with our results. Moreover, we observed the joint effect of the two SNPs on cancer prognosis in many strata.
There are some strengths and limitations in our study. One of the strengths is that this is a prospective cohort study indicating that our results are more reliable. Another one is that all of the patients are not included in a single hospital, which indicates the representativity of our study. The third one is that, to the best of our knowledge, this is the first meta-analysis concerning the association between microRNA 27a rs895819 and cancer prognosis. The forth one is that the meta-analysis is reliable and stable. One limitation of this study is that the sample size in each stratum is small. Another one is that the majority of included patients are late-stage. This may introduce biases in our study. The third one is that the number of publications included in this meta-analysis is small.
MATERIALS AND METHODS
Study population and follow-up
This study was a prospective cohort study and approved by the institutional review board of China Medical University and all subjects signed a written informed consent form. All subjects were females and they were from unrelated ethnic Han Chinese. Estimates of the exposure to environmental factors were reported in previous study [37]. 480 patients were recruited during March 2010 to March 2013 at five Liaoning hospitals. All patients were histologically confirmed when they were enrolled. All subjects were interviewed and venous blood sample was obtained from each subject. Detailed baseline information including age, gender, tobaccon exposure, clinical stage, histological type, receipt of chemotherapy and receipt of surgery has been collected.
Subjects received telephone follow-up every three months after being diagnosed with lung cancer. We adopted at least one of the following methods to confirm the date of death: 1) data from Shenyang Center for Disease Control and Prevention (CDC) registry system for cause of death; 2) inpatient and outpatient medical records; 3) Death Registry System of Shenyang Public Security Bureau. The patients were followed up to death or April 2015. Totally, we collected complete survival data of 454 patients. The others are lost to follow-up. To avoid information bias, we adopted two methods. One is that all interviewers received training before the study was conducted. Another one is that two individuals record the data independently and the third one check up the data for any contradiction. The MST is 24.7 months in the ongoing study.
Genotyping
Genomic DNA was extracted from peripheral blood samples using the phenol-chloroform method. The TaqMan allelic discrimination method was used to genotype the two SNPs. The samples were read and analyzed from the ABI 7500 Fast Sequence Detection System (Applied Biosystems, USA). The average genotype call rates for the two SNPs was 99.5%. About 10% of the samples were randomly selected for confirmation by repeat genotyping, and the results were 100% concordant.
Searching strategy
This meta-analysis was carried out in accordance with the guidelines of the meta-analysis of the Observational Studies in Epidemiology group (MOOSE) [38]. Databases including PubMed, SCIE, WanFang and CNKI were searched to identify the publications concerning the association with microRNA 30c-1 rs928508 and microRNA 27a rs895819. The strategies are as follows: all fields or subjects or full-text or MeSH or keywords contain “microRNA 30c-1 OR miRNA 30c-1 OR miR 30c-1 OR rs928508 OR microRNA 27a OR miRNA 27a OR miR 27a OR rs895819” OR “cancer OR carcinoma OR tumor OR neoplasm” OR “prognosis OR survival”. The reference lists were searched, as well. The last time for search is June, 2017.
Including criteria: 1) focus on the association with either rs928508 or rs895819 and cancer prognosis; 2) HR and 95%CI are available; 3) publications written in English or Chinese.
Excluding criteria: 1) duplicated publications or data; 2) meeting abstract with vague information.
Data extraction
Data extraction and logging were conducted by two authors individually. The third author will take part in if the data are inconsistent. Information including publication year, first author name, tumor type, study population, sample size, outcome, HRs and 95% CIs was extracted.
Statistical analysis
Goodness-of-fit chi-square test is used to compare the differences in the distribution of deaths between groups with different baseline characteristics. The OS is calculated from the date at diagnosis to the date of last follow-up or death. Median survival time and survival curves were estimated by Kaplan-Meier method and analyzed by the means of log-rank test. Univariate and multivariate Cox proportional hazards regression models were used to estimate the crude and adjusted HRs and 95% CIs. Stratified analysis was adopted to control the confounding bias. Q statistics were used to assess heterogeneity. P value less than 0.05 is considered heterogeneous and the random-effects model will be used to calculate the pooled HRs and 95% CIs. Otherwise, the fixed-effects model will be used. I2 statistic was used to measure the percentage of the variation that is due to heterogeneity rather than to chance. Stratified analysis was conducted according to the tumor types. Funnel plots and Begg's test were used to evaluate the publication bias. Sensitivity analysis was conducted to evaluate the stability of the study. Statistically significant P-value is less than 0.05. All of the statistical analyses were performed in SPSS 17.0 and all P-values are two-sided. In our study, we selected six genetic models for cohort study and four genetic models for meta-analysis. The Bonferroni P value for multiple-comparison is 0.008 for the estimation of the association between SNPs and cancer prognosis. Bonferroni P value for joint effect estimation is 0.01. Bonferroni P value for meta-analysis is 0.006.
CONCLUSIONS
The two SNPs of microRNA 30c-1 and microRNA 27a may decrease the sensitivity to anti-cancer drugs and are predictive of non-small cell lung cancer patients survival. The observations in our study imply the role of microRNA 30c-1 and microRNA 27a polymorphisms in NSCLC patients prognosis.
SUPPLEMENTARY MATERIALS FIGURES AND TABLES
Acknowledgments
We really thank for the help from Quan Xiaowei, Li Hang and Qu Ruoyi, Department of Epidemiology, China Medical University.
Abbreviations
- SNP
single nucleotide polymorphism
- NSCLC
non-small cell lung cancer
- HR
hazard ratio
- CI
confidential interval
- MST
median survival time
- OS
overall survival
- RFS
recurrence-free survival
- FZD7
frizzled class receptor 7
- ZBTB10
zinc finger and BTB domain containing 10
- CDC
center for disease control and prevention
- AD
adenocarcinoma
- SQU
squamous carcinoma
- GC
gastric cancer
- BCL
B-cell lymphoma
- GBC
gallbladder cancer
Author contributions
XLZ designed the experiment and drafted the article, LXL, RYW and ZHB included and excluded the participants, LXL, RYW and ZHB conducted the epidemiological investigation and collected the blood samples, LXL, ZHB and ZYX conducted the follow-up, XLZ and RYW conducted the DNA extraction and genotyping, XLZ, YZH and RYW conducted the data extraction and logging, XLZ and YZH conducted the statistical analyses, YZH and LXL revised the article, ZBS provided the fund and gave final approval of the version to be published.
CONFLICTS OF INTERESTS
The authors declare no conflicts of interest.
FUNDING
This study is supported by grant no.81272293 and no.81102194 from National Natural Science Foundation of China.
REFERENCES
- 1.Bray F, Jemal A, Grey N, Ferlay J, Forman D. Global cancer transitions according to the Human Development Index (2008–2030): a population-based study. Lancet Oncol. 2012;13:790–801. doi: 10.1016/S1470-2045(12)70211-5. http://doi.org/10.1016/s1470-2045(12)70211-5. [DOI] [PubMed] [Google Scholar]
- 2.Lyon F. 2013. International Agency for Research on Cancer.
- 3.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. 2012. GLOBOCAN 2012 V1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. [DOI] [PubMed]
- 4.She J, Yang P, Hong Q, Bai C. Lung cancer in China: challenges and interventions. Chest. 2013;143:1117–26. doi: 10.1378/chest.11-2948. http://doi.org/10.1378/chest.11-2948. [DOI] [PubMed] [Google Scholar]
- 5.Cai Y, Xue M, Chen W, Hu M, Miao Z, Lan L, Zheng R, Meng Q. Expenditure of hospital care on cancer in China, from 2011 to 2015. Chin J Cancer Res. 2017;29:253–62. doi: 10.21147/j.issn.1000-9604.2017.03.11. http://doi.org/10.21147/j.issn.1000-9604.2017.03.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X, Liu S, Liu Y, Du J, Fu W, Zhao X, Huang W, Zhao X, Liu G, Mao Z, Hu TW. Economic Burden for Lung Cancer Survivors in Urban China. Int J Environ Res Public Health. 2017:14. doi: 10.3390/ijerph14030308. http://doi.org/10.3390/ijerph14030308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. http://doi.org/10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 8.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. http://doi.org/10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 9.Jia-Ping C, Zhi-Bin HU, Tian T, Xiao-Yi Z, Rui-Fen M, Guang-Fu JIN, Hong-Xia MA, Hong-Bing S. Single nucleotide polymorphism associated with mature miR-196a2 influences the expression of Lymphocyte-specific protein 1 (LSP1) gene. Acta Universitatis Medicinalis Nanjing. 2009;29:762–6. [Google Scholar]
- 10.Chen Y. Down-regulation of MiR-30c promotes the Invasion of Non-Small Cell Lung Cancer by Targeting MTA1 (vol 32, pg 476, 2013) Cellular Physiology and Biochemistry. 2013;32:1124. doi: 10.1159/000354452. http://doi.org/10.1159/000354512. [DOI] [PubMed] [Google Scholar]
- 11.Jiang X, Du L, Duan W, Wang R, Yan K, Wang L, Li J, Zheng G, Zhang X, Yang Y, Wang C. Serum microRNA expression signatures as novel noninvasive biomarkers for prediction and prognosis of muscle-invasive bladder cancer. Oncotarget. 2016;7:36733–42. doi: 10.18632/oncotarget.9166. http://doi.org/10.18632/oncotarget.9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong P, Ihira K, Xiong Y, Watari H, Hanley SJ, Yamada T, Hosaka M, Kudo M, Yue J, Sakuragi N. Reactivation of epigenetically silenced miR-124 reverses the epithelial-to-mesenchymal transition and inhibits invasion in endometrial cancer cells via the direct repression of IQGAP1 expression. Oncotarget. 2016;7:20260–70. doi: 10.18632/oncotarget.7754. http://doi.org/10.18632/oncotarget.7754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang Y, Shen H, Cao Y, Li H, Qin R, Chen Q, Long L, Zhu XL, Xie CJ, Xu WL. Involvement of miR-30c in resistance to doxorubicin by regulating YWHAZ in breast cancer cells. Braz J Med Biol Res. 2014;47:60–9. doi: 10.1590/1414-431X20133324. http://doi.org/10.1590/1414-431×20133324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu YF, Zhang H, Su D, Mo ML, Song P, Zhang F, Zhang SC. miR-30b and miR-30c expression predicted response to tyrosine kinase inhibitors as first line treatment in non-small cell lung cancer. Chin Med J (Engl) 2013;126:4435–9. http://doi.org/10.3760/cma.j.issn.0366-6999.20131112. [PubMed] [Google Scholar]
- 15.Chen Z, Ma T, Huang C, Zhang L, Lv X, Xu T, Hu T, Li J. MiR-27a modulates the MDR1/P-glycoprotein expression by inhibiting FZD7/beta-catenin pathway in hepatocellular carcinoma cells. Cell Signal. 2013;25:2693–701. doi: 10.1016/j.cellsig.2013.08.032. http://doi.org/10.1016/j.cellsig.2013.08.032. [DOI] [PubMed] [Google Scholar]
- 16.Noratto GD, Jutooru I, Safe S, Angel-Morales G, Mertens-Talcott SU. The drug resistance suppression induced by curcuminoids in colon cancer SW-480 cells is mediated by reactive oxygen species-induced disruption of the microRNA-27a-ZBTB10-Sp axis. Molec Nutr Food Res. 2013;57:1638–48. doi: 10.1002/mnfr.201200609. http://doi.org/10.1002/mnfr.201200609. [DOI] [PubMed] [Google Scholar]
- 17.Huang D, Wang H, Liu R, Li H, Ge S, Bai M, Deng T, Yao G, Ba Y. miRNA27a Is a Biomarker for Predicting Chemosensitivity and Prognosis in Metastatic or Recurrent Gastric Cancer. J Cell Biochem. 2014;115:549–56. doi: 10.1002/jcb.24689. http://doi.org/10.1002/jcb.24689. [DOI] [PubMed] [Google Scholar]
- 18.Xu J, Yin Z, Shen H, Gao W, Qian Y, Pei D, Liu L, Shu Y. A Genetic Polymorphism in pre-miR-27a Confers Clinical Outcome of Non-Small Cell Lung Cancer in a Chinese Population. Plos One. 2013;8 doi: 10.1371/journal.pone.0079135. http://doi.org/10.1371/journal.pone.0079135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taheriazam A, Bahador R, Karbasy SH, Jamshidi SM, Torkaman A, Yahaghi E, Shakeri M. Down-regulation of microRNA-26a and up-regulation of microRNA-27a contributes to aggressive progression of human osteosarcoma. Diagn Pathol. 2015;10:166. doi: 10.1186/s13000-015-0400-3. http://doi.org/10.1186/s13000-015-0400-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Nakata W, Uemura M, Sato M, Fujita K, Jingushi K, Ueda Y, Kitae K, Tsujikawa K, Nonomura N. Expression of miR-27a-3p is an independent predictive factor for recurrence in clear cell renal cell carcinoma. Oncotarget. 2015;6:21645–54. doi: 10.18632/oncotarget.4064. http://doi.org/10.18632/oncotarget.4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoon KA, Yoon H, Park S, Jang HJ, Zo JI, Lee HS, Lee JS. The prognostic impact of microRNA sequence polymorphisms on the recurrence of patients with completely resected non-small cell lung cancer. J Thorac Cardiovasc Surg. 2012;144:794–807. doi: 10.1016/j.jtcvs.2012.06.030. http://doi.org/10.1016/j.jtcvs.2012.06.030. [DOI] [PubMed] [Google Scholar]
- 22.Hu Z, Shu Y, Chen Y, Chen J, Dong J, Liu Y, Pan S, Xu L, Xu J, Wang Y, Dai J, Ma H, Jin G, et al. Genetic polymorphisms in the precursor MicroRNA flanking region and non-small cell lung cancer survival. Am J Respir Crit Care Med. 2011;183:641–8. doi: 10.1164/rccm.201005-0717OC. http://doi.org/10.1164/rccm.201005-0717OC. [DOI] [PubMed] [Google Scholar]
- 23.Mu YP, Su XL. Polymorphism in pre-miR-30c contributes to gastric cancer risk in a Chinese population. Med Oncol. 2012;29:1723–32. doi: 10.1007/s12032-011-0115-6. http://doi.org/10.1007/s12032-011-0115-6. [DOI] [PubMed] [Google Scholar]
- 24.Xu Q, Chen TJ, He CY, Sun LP, Liu JW, Yuan Y. MiR-27a rs895819 is involved in increased atrophic gastritis risk, improved gastric cancer prognosis and negative interaction with Helicobacter pylori. Sci Rep. 2017;7:41307. doi: 10.1038/srep41307. http://doi.org/10.1038/srep41307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang C, Li XP, Gong WJ, Wu NY, Tang J, Yin JY, Li X, Zhang W, Zhou HH, Liu ZQ. Age-related common miRNA polymorphism associated with severe toxicity in lung cancer patients treated with platinum-based chemotherapy. Clin Exp Pharmacol Physiol. 2016 doi: 10.1111/1440-1681.12704. http://doi.org/10.1111/1440-1681.12704 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 26.Jiang J, Jia ZF, Cao DH, Wu YH, Sun ZW, Cao XY. Association of the miR-146a rs2910164 polymorphism with gastric cancer susceptibility and prognosis. Future Oncol. 2016;12:2215–26. doi: 10.2217/fon-2016-0224. http://doi.org/10.2217/fon-2016-0224. [DOI] [PubMed] [Google Scholar]
- 27.Ma JY, Yan HJ, Yang ZH, Gu W. Rs895819 within miR-27a might be involved in development of non small cell lung cancer in the Chinese Han population. Asian Pac J Cancer Prev. 2015;16:1939–44. doi: 10.7314/apjcp.2015.16.5.1939. [DOI] [PubMed] [Google Scholar]
- 28.Stenholm L, Stoehlmacher-Williams J, Al-Batran SE, Heussen N, Akin S, Pauligk C, Lehmann S, Senff T, Hofheinz RD, Ehninger G, Kramer M, Goekkurt E. Prognostic role of microRNA polymorphisms in advanced gastric cancer: a translational study of the Arbeitsgemeinschaft Internistische Onkologie (AIO) Ann Oncol. 2013;24:2581–8. doi: 10.1093/annonc/mdt330. http://doi.org/10.1093/annonc/mdt330. [DOI] [PubMed] [Google Scholar]
- 29.Zhang N, Huo Q, Wang X, Chen X, Long L, Jiang L, Ma T, Yang Q. A genetic variant in pre-miR-27a is associated with a reduced breast cancer risk in younger Chinese population. Gene. 2013;529:125–30. doi: 10.1016/j.gene.2013.07.041. http://doi.org/10.1016/j.gene.2013.07.041. [DOI] [PubMed] [Google Scholar]
- 30.Zhang B, Chen J. microRNA genes rs895819, rs6505162 and rs2292832 SNP variation with B cell lymphoma patients survival relationship research. Chongqing Med. 2013;42:1801–3. [Google Scholar]
- 31.Wu R, Hua D, Wang W. Tumor: Soochow University; 2014. The association of miR-SNP with the susceptibility of colorectal cancer and response to chemotherapy. [Google Scholar]
- 32.Gupta A, Sharma A, Yadav A, Rastogi N, Agrawal S, Kumar A, Kumar V, Misra S, Mittal B. Evaluation of miR-27a, miR-181a, and miR-570 Genetic Variants with Gallbladder Cancer Susceptibility and Treatment Outcome in a North Indian Population. Molec Diagn Ther. 2015;19:317–27. doi: 10.1007/s40291-015-0159-y. http://doi.org/10.1007/s40291-015-0159-y. [DOI] [PubMed] [Google Scholar]
- 33.Chen JP, Liu Y, Hu ZB, Shen HB. Single nucleotide polymorphism in flanking region of miR-30c influences the maturing process of miR-30c in lung carcinoma. [Article in Chinese]. Zhonghua Zhong Liu Za Zhi. 2012;34:664–8. doi: 10.3760/cma.j.issn.0253-3766.2012.09.005. http://doi.org/10.3760/cma.j.issn.0253-3766.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 34.Young JA, Ting KK, Li J, Moller T, Dunn L, Lu Y, Moses J, Prado-Lourenco L, Khachigian LM, Ng M, Gregory PA, Goodall GJ, Tsykin A, et al. Regulation of vascular leak and recovery from ischemic injury by general and VE-cadherin-restricted miRNA antagonists of miR-27. Blood. 2013;122:2911–9. doi: 10.1182/blood-2012-12-473017. http://doi.org/10.1182/blood-2012-12-473017. [DOI] [PubMed] [Google Scholar]
- 35.Yuan L, Chu H, Wang M, Gu X, Shi D, Ma L, Zhong D, Du M, Li P, Tong N, Fu G, Qin C, Yin C, et al. Genetic Variation in DROSHA 3'UTR Regulated by hsa-miR-27b Is Associated with Bladder Cancer Risk. Plos One. 2013;8 doi: 10.1371/journal.pone.0081524. http://doi.org/10.1371/journal.pone.0081524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Q, Jie Z, Ye S, Li Z, Han Z, Wu J, Yang C, Jiang Y. Genetic variations in miR-27a gene decrease mature miR-27a level and reduce gastric cancer susceptibility. Oncogene. 2014;33:193–202. doi: 10.1038/onc.2012.569. http://doi.org/10.1038/onc.2012.569. [DOI] [PubMed] [Google Scholar]
- 37.Yin Z, Zhou B, He Q, Li M, Guan P, Li X, Cui Z, Xue X, Su M, Ma R, Bai W, Xia S, Jiang Y, et al. Association between polymorphisms in DNA repair genes and survival of non-smoking female patients with lung adenocarcinoma. BMC Cancer. 2009;9 doi: 10.1186/1471-2407-9-439. http://doi.org/10.1186/1471-2407-9-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, Moher D, Becker BJ, Sipe TA, Thacker SB, Grp M. Meta-analysis of observational studies in epidemiology - A proposal for reporting. JAMA. 2000;283:2008–12. doi: 10.1001/jama.283.15.2008. http://doi.org/10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


