Abstract
Besides being a key contributor to epithelial-to-mesenchymal transition (EMT) activation and stemness maintenance, zinc finger E-box binding homeobox 1 (ZEB1) is also a crucial inducer of chemoresistance and radioresistance. Unlike the clear mechanism that mediates its effect on EMT and dedifferentiation, the mechanism of how ZEB1 promotes chemo- and radio-resistance remains to be elucidated. It has been previously reported that ZEB1 promotes DNA double-strand break clearance by enhancing the deubiquitylating activity of ubiquitin-specific peptidase (USP)7 on checkpoint kinase 1, which is an important step during DNA repair. It was hypothesized that as a transcriptional suppressor, ZEB1 may be involved in an unbalanced DNA damage response (DDR) by affecting other key components. Therefore, in the present study, the target gene occupancy of ZEB1 was mapped in colorectal cancer cells using the ChIP-on-chip method, revealing positive intervals enriched along the three DDR-associated genes: USP17, chromodomain helicase DNA-binding protein 1-like and double homeobox 4. The E-boxes identified in the binding regions and the enhanced mRNA expression of the three genes following the knockdown of ZEB1 supported the identification of these three genes as downstream target genes of ZEB1. Furthermore, ZEB1 knockdown initiated a chemosensitization effect, induced G1/S arrest and increased apoptosis, which functionally validated the three ZEB1 downstream targets. In summary, the present study identified three DDR-associated genes as ZEB1 downstream targets, and demonstrated that their suppression by ZEB1 contributes to ZEB1-mediated chemoresistance.
Keywords: zinc finger E-box binding homeobox 1, DNA damage response, DNA repair, promoter profiling, chemoresistance, ubiquitin-specific peptidase 17, chromodomain helicase DNA-binding protein 1-like, double homeobox 4
Introduction
Chemo- and radio-resistance are major clinical problems in cancer treatment, and often lead to tumor recurrence and even a fatal outcome. Increasing evidence has demonstrated that the three hallmarks of tumor malignancy, metastasis, neoplastic stem cells and chemo- and radio-resistance are not independent from one another (1,2). Induction of epithelial-to-mesenchymal transition (EMT) concomitantly results in the emergence of cancer stem cells (CSCs) with enhanced tumor-initiating capacity, changes to specific cell-surface markers, increased chemo- and radio-resistance and resistance to apoptosis (3–5).
By suppressing the transcription of E-cadherin and stemness-inhibiting microRNAs, or driving non-CSCs to enter the CSC state in certain types of cancer cells, zinc finger E-box binding homeobox 1 (ZEB1) acts as a key contributor to EMT activation and stemness maintenance (6–8). Additionally, data has indicated that ZEB1 is also crucial for chemoresistance and radioresistance (9). The chemotherapeutic drug gemcitabine has demonstrated a limited success rate in pancreatic cancer; however, a previous study revealed that the stable knockdown of ZEB1 was able to significantly sensitize pancreatic cancer cells to gemcitabine treatment (7). The expression level of ZEB1 in pancreatic cancer cells also correlates with the resistance of these cells to 5-fluorouracil and cisplatin (10). With regard to radiotherapy, comet assays have demonstrated that ZEB1-depleted SUM159 cells experienced more DNA damage following ionizing radiation treatment (11), while overexpression of the major ZEB1 target microRNAs, miR-200c or miR-205, efficiently sensitized various types of cancer cells to irradiation by suppressing DNA repair protein, ubiquitin-conjugating enzyme E2N, and stemness factors (12,13).
Although ZEB1 has a clear mechanism regarding its effect on EMT and dedifferentiation, how it promotes chemo- and radio-resistance remains to be elucidated. Other studies have reported that by directly interacting with ubiquitin-specific peptidase (USP)7 and enhancing its deubiquitylating activity on checkpoint kinase 1 (CHK1) (11), ZEB1 was positively correlated with the protein level of CHK1 and was required for the clearance of DNA double-strand breaks (DSBs). This indicates that ZEB1 plays a role in the DNA damage response (DDR), because CHK1 is an important protein of the DDR that regulates homologous recombination repair and prevents G2 to M transition (14).
Although activation of DDR pathways may decrease tumor development by acting as a barrier to unchecked proliferation in its early stages (15), when aberrantly activated, DDR has been demonstrated to be involved in the development of cancer cell resistance to the lethal effects of genotoxic agents (16). By coordinating cell cycle arrest, apoptosis, senescence and DNA repair pathways, DDR forms a complex network that is responsible for various outcomes following DNA damage (17). However, due to the loss of p53, an apoptosis-inducing DDR regulator, and other unknown mechanisms, tumor cells rely heavily upon the DNA repair pathways and thus, struggle to survive following DNA damage, with increased genome instability as a consequence (18).
We hypothesized that, in addition to stabilizing CHK1, the transcriptional repressor ZEB1 may be involved in an unbalanced DDR by affecting its other key components. To further understand the targets via which ZEB1 fulfills its regulation of DDR, the present study mapped the target gene occupancy of ZEB1 in colorectal cancer cells using a chromatin immunoprecipitation (ChIP)-on-chip method (19), and identified USP17-like family member 2 (DUB3), chromodomain helicase DNA-binding protein 1-like (CHD1L) and double homeobox 4 (DUX4) as three potential downstream genes of ZEB1. Additionally, the results indicate that the suppression of the three genes by ZEB1 mediates a dysregulated DDR, leading to chemoresistance and anti-apoptotic processes.
Materials and methods
Cell culture
The human colorectal cancer cell line LoVo, which was purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA), was cultured in F12K medium supplemented with 10% heat-inactivated fetal bovine serum (FBS), penicillin (100 U/ml) and streptomycin (100 g/ml), in a humidified 5% CO2 incubator at 37°C.
ChIP
The ChIP was performed following the instructions of an Enzymatic Chromatin IP kit (cat no. 9003; Cell Signaling Technology, Inc., Danvers, MA, USA). Briefly, ~4×107 cells were prepared for each experiment. Formaldehyde (1% concentration) was added to crosslink proteins to DNA for 10 min. Following cell lysis and nuclei collection, the chromatin was fragmented by partial digestion with micrococcal nuclease to obtain chromatin fragments of 1–5 nucleosomes in size (150–900 bp). The crosslinked chromatin preparation was then immunoprecipitated with 5 µg polyclonal ZEB1 antibody (cat no. sc-25388 X; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) or negative control normal rabbit IgG (cat no. 2729; Cell Signaling Technology, Inc.) at 4°C overnight. Elution of chromatin from the crosslinked complex using Protein G magnetic beads was performed with KingFisher Flex Magnetic Particle Processors. After reversing the crosslinks, DNA was purified using the reagents and spin columns provided in the Enzymatic Chromatin IP kit. In addition, a positive control histone H3 rabbit monoclonal antibody (cat no. 4620; Cell Signaling Technology, Inc.) and a primer for its known binding gene, ribosomal protein L30, were also included in the experiment to analyze IP efficiency. Three biological replicates were performed and successful enrichment was validated in each experiment.
Array hybridization, staining and scanning
ChIP DNA was amplified to 7.5 µg, using the high-performance liquid chromatography-purified primers: A, 5′-GTTTCCCAGTCACGG TC(N)9-3′; and B, 5′-GTTTCCCAGTCACGGTC-3′. The dUTP-containing post-amplified samples was then fragmented by uracil-DNA glycosylase and labeled by terminal deoxynucleotidyl transferase (cat no. 72033) with the biotin-like labeling reagent (cat no. 79015) (both from USB Corp.; Affymetrix, Inc., Santa Clara, CA, USA). The labeled DNA was hybridized to six Affymetrix GeneChip Human Promoter 1.0R arrays (three biological replicates using ZEB1 antibody and three using normal rabbit IgG) at 45°C for 16 h. The arrays were washed and stained by R-phycoerythrin streptavidin (cat no. S-866; Molecular Probes; Thermo Fisher Scientific, Inc., Waltham, MA, USA) using Fluidics Station 450 and then scanned using GeneChip Scanner 3000 7G, which was controlled by Affymetrix GeneChip Command Console software (Affymetrix, Inc.).
Array data analysis
Affymetrix Tiling Analysis software (Affymetrix, Inc.) and Integrated Genome Browser were used to select the positive intervals that satisfied the two conditions: that the signal intensity of the ZEB1-binding DNA and the intensity ratio of the ZEB1-binding DNA signal/normal rabbit IgG-binding DNA signal were within the top 1% among all the intervals. The filter criteria ruled out the false-positive regions with a high intensity ratio that resulted from the division of two infinitely small numbers. The positive intervals of each chromosome in bed files were uploaded to the GALAXYP online platform (usegalaxyp.org) to produce a tail-to-head concatenation and then submitted to the Cis-regulatory Element Annotation System (CEAS) to produce nearby gene mapping and motif identification. For each positive ChIP region, CEAS identified the nearest RefSeq gene names and predicted locations within 300 kb upstream or downstream of the gene. Regions within 1 kb upstream from the gene 5′ start site were assumed to be proximal promoters; those within 1 kb downstream from the gene 3′-end were reported to be immediate downstream regulatory elements; while those distributed >1 kb from the gene were assumed to be enhancers (20).
Short hairpin RNA (shRNA) constructs and transfection
The LoVo cell line was selected as a suitable host for transfection. To avoid off-target effects, two different sequences were used to knock down ZEB1. Using Endofectin™ Plus, cells were transfected with plasmids (both from GeneCopoeia, Inc., Rockville, MD, USA) containing short hairpin RNA targeting ZEB1 HSH017963-4-LVRH1GP (OS241004) or HSH017963-5-LVRH1GP (OS397209). Parallel transfection was performed using plasmids with non-targeting shRNA CSHCTR001-1-LVRH1GP (OSNEG20) to generate scrambled control clones. The cells were then selected with puromycin and after 4 weeks, single colonies were analyzed for ZEB1 expression by western blotting assay. The results revealed that the transfections had successfully induced a decrease in the protein level of ZEB1. The shRNA targeted sequences were as follows: ZEB1-knockdown-1, 5′-GGTCAACTATCACTAGTGT-3′; ZEB1-knockdown-2, 5′-TGATCAGCCTCAATCTGCA-3′; and scrambled control, 5′-GCTTCGCGCCGTAGTCTTA-3′. Knockdown is referred to as KD hereinafter.
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from the cells was extracted and reverse transcribed into cDNA according to the protocols of the Total RNA kit I (Omega Bio-Tek, Inc., Norcross, GA, USA) and First Strand cDNA Synthesis kit (GeneCopoeia, Inc.). The cDNA template (100 ng) was amplified using 0.2 µM primers and the 20 µl SYBR-Green-based qPCR reaction system (GeneCopoeia, Inc.). A Bio-Rad IQ 5 instrument (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used to perform the reaction and detect the fluorescent signals. Standard curves were created to confirm the amplification efficiency of each gene, while melting curves ensured the specificity of the amplification. Normalization to the reference gene (β-actin) was performed for each sample. Fold changes between the mRNA level of the ZEB1-KD groups and the scrambled control group were calculated according to the 2−ΔΔCq method (21). The sequences of primers were as follows (5′-3′): USP17 forward, AGGTGAGTGGCAGTTCAACC and reverse, GGAAGCTTCTTCCTGGGAGC; CHD1L forward, ACTAGCATTCCTGTATTCTGGGG and reverse, CACGCTCATAGCTGTAGCCTC; DUX4 forward, CGATGGCCCTCCCGACA and reverse, GGCGTGACCTCTCATTCTGA; and β-actin forward, CCTAGAAGCATTTGCGGTGG and reverse, GAGCTACGAGCTGCCTGACG.
Cell proliferation assay
Cells were seeded into 96-well plates at 5,000 cells/200 µl/well, treated with increasing concentrations of the DNA-damaging agents cisplatin (cat no. P4394) and etoposide (cat no. E1383) (both from Sigma-Aldrich; Merck Millipore, Darmstadt, Germany), and then cultured for 24 h. Cell proliferation was assessed using Cell Counting Kit-8 (Boster Biological Technology, Ltd., Wuhan, China), and IC50 values were calculated.
Flow cytometric analysis
Cells were seeded in 6-well plates at 5×105 cells/2.5 ml/well and treated with DNA-damaging agents at the indicated concentrations for 24 h. For cell cycle analysis, cell suspensions were harvested, washed with PBS, and resuspended in propidium iodide (PI) stain buffer [0.1% (v/v) Triton X-100, 10 µg/ml PI and 100 µg/ml DNase-free RNaseA; Sigma-Aldrich; Merck Millipore] for 30 min in the dark. The DNA contents of the cells were analyzed using a BD FACSCalibur (BD Biosciences, Franklin Lakes, NJ, USA). For apoptosis analysis, the cells were stained with PI and allophycocyanin-conjugated Annexin V and the FACSCalibur was used to detect early-stage apoptotic cells.
Statistical analysis
Data are presented as the mean ± standard deviation from at least three independent experiments. When data followed a normal distribution, the statistical significance between experimental values was assessed by unpaired Student's t-test using SPSS software, and P<0.05 was considered to indicate a statistically significant difference.
Results
USP17, CHD1L and DUX4 as putative downstream targets of ZEB1
The Human promoter 1.0R array is tiled with >25,500 human promoter regions. Each region covers ~7.5 kb upstream to 2.45 kb downstream of the 5′ transcription start sites of >1,300 cancer-associated genes. The DNA fractions that co-immunoprecipitated with ZEB1 were hybridized to the array, in order to identify putative downstream targets of ZEB1 (normal rabbit IgG-binding DNA was used as the background).
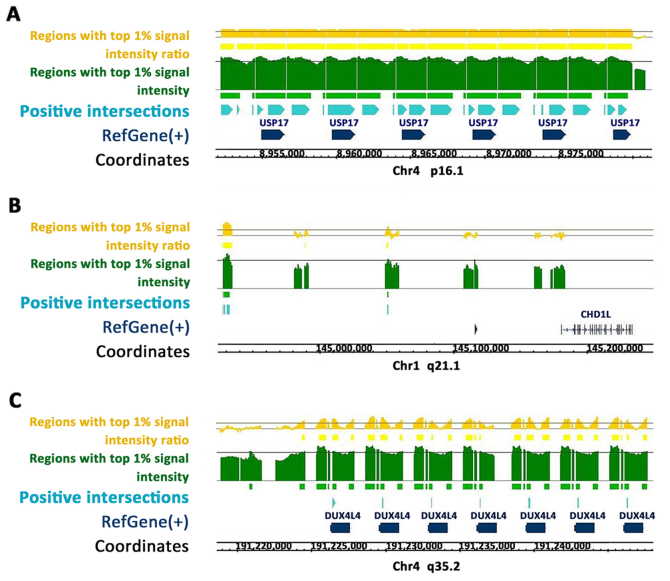
Using the Homo sapiens hg18 genome (Refgene; NCBI Build 36.1; March 2006), continuous positive regions enriched along the USP17 (Fig. 1A), CHD1L (Fig. 1B) and DUX4 (Fig. 1C) gene sequences were identified. CEAS predicted ZEB1 binding sites located at the enhancer, promoter and immediate downstream regions of the USP17 gene; at the enhancer, intron and exon regions of the DUX4 gene; and at the enhancer regions of the CHD1L gene.
Figure 1.
Positive ZEB1-bound regions enriched along USP17, CHD1L and DUX4. Bright blue regions represent positive ZEB1-bound regions with signal intensity and intensity ratio in the top 1% among all the intervals. (A) Positive regions along the USP17 RefSeq. (B) Positive regions along the CHD1L RefSeq. (C) Positive regions along the DUX4 RefSeq. RefSeq, reference sequences; ZEB1, zinc finger E-box-binding homeobox 1; USP17, ubiquitin-specific peptidase 17; CHD1L, chromodomain helicase DNA-binding protein 1-like; DUX4, double homeobox 4.
E-box elements and predicted ZEB1-binding motifs are present within the positive regions
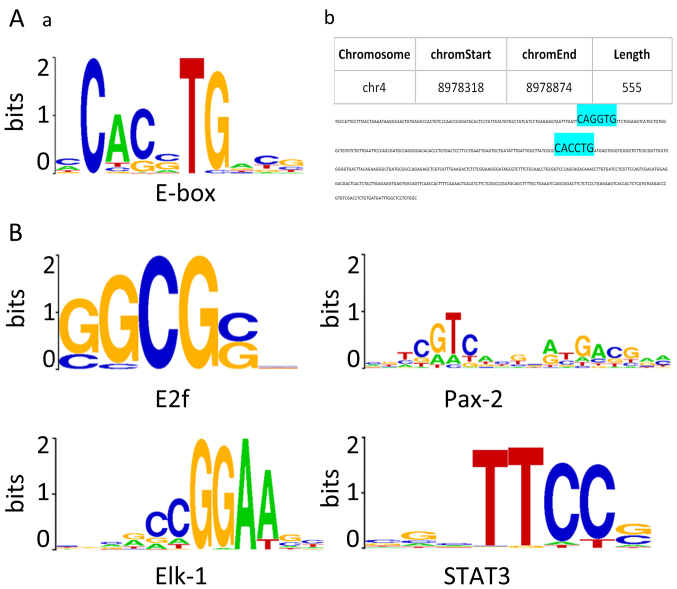
As ZEB1 is a well-known E-box binding transcriptional repressor, the sequences of each positive region from the three putative target genes (in FASTA format) were screened for the presence of E-box elements (5′-CANNTG-3′). As shown in Fig. 2A-a, CACCTG, CACGTG, CAGCTG, CAGGTG were the four most canonical E-box sequences. Duplication of the sequences, particularly in an AGGTG-Nn-CACCT structure, have been reported to possess strong ZEB1-binding affinity (22,23). In 24 positive intervals along the USP17 gene, 10 CACCTG sequences, 1 CAGGTG sequence and 12 CAGCTG sequences, and, notably, an AGGTG-Nn-CACCT duplication were identified (Fig. 2A). In 12 upstream positive regions of the CHD1L gene, two CACCTG sequences were detected. The 7 positive regions along the DUX4 gene were identified to contain CAGCTG sequences.
Figure 2.
ZEB1-binding motifs. (A) a, E-box motif; b, AGGTG-Nn-CACCT structure (text in blue) identified in one of the positive regions along the USP17 gene. (B) Four predicted ZEB1-binding motifs with the highest significance. ZEB1, zinc finger E-box-binding homeobox 1; USP17, ubiquitin-specific peptidase 17.
In addition to E-boxes, CEAS identified ~40 predicted ZEB1-binding motifs using the default algorithm, arranged in ascending order of p-value. The top four ranked sequences predicted to be ZEB1-interacting DNA motifs following CEAS analysis are presented in Fig. 2B, namely E2f, Pax-2, Elk-1 and STAT3 motifs. The FASTA files were also searched for the de novo predicted ZEB1-binding elements, and identified those consensus binding sites within the positive regions along the three genes.
Stable depletion of ZEB1 enhances the expression of the putative target genes
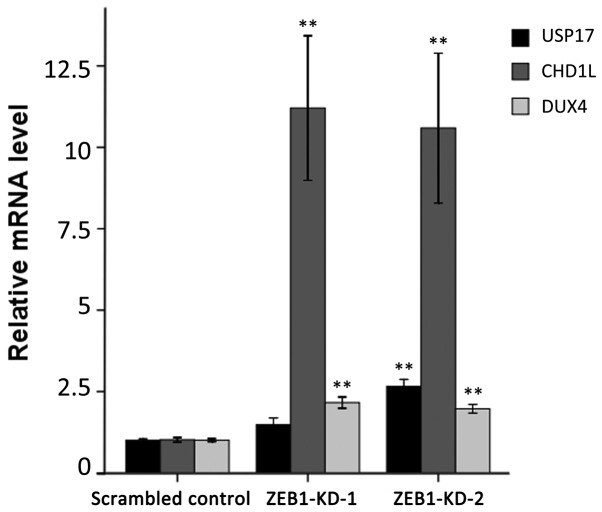
To confirm the ChIP-on-chip data, the mRNA level of the three genes was examined for their responses to ZEB1 knockdown by RT-qPCR. As demonstrated in Fig. 3, the three genes exhibited significantly enhanced expression following ZEB1 depletion compared with the scrambled control group. The relative expression levels of USP17, CHD1L and DUX4 reached 1.48-, 11.21- and 2.16-fold in ZEB1-KD-1 cells, and 2.66-, 10.59- and 1.97-fold in ZEB1-KD-2 cells, respectively.
Figure 3.
Effect of ZEB1 silencing on the mRNA expression of USP17, CHD1L and DUX4. Each bar represents the mean ± standard deviation of three independent experiments, each performed in triplicate. **P<0.01 vs. scrambled control group. ZEB1, zinc finger E-box-binding homeobox 1; USP17, ubiquitin-specific peptidase 17; CHD1L, chromodomain helicase DNA-binding protein 1-like; DUX4, double homeobox 4.
ZEB1 knockdown induces sensitivity to genotoxic agents
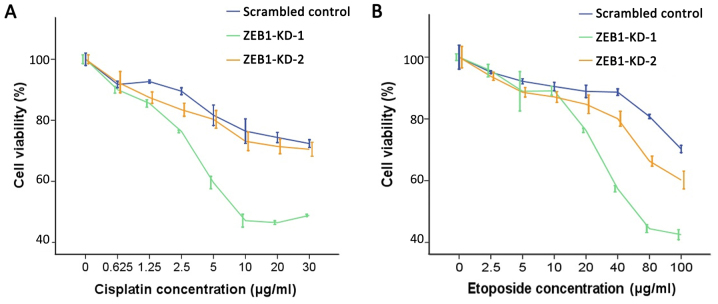
Overexpression of USP17 and CHD1L was previously reported to cause alterations to the DDR, resulting in increased sensitivity to DNA-damaging agents (24,25). Therefore, it was investigated whether silencing of ZEB1 may induce similar effects. Data demonstrated that when ZEB1 was knocked down by two specific shRNA constructs, cell proliferation was significantly impaired and cells were more susceptible to cisplatin and etoposide-induced DNA damage. As demonstrated in Fig. 4, ZEB1 knockdown enhanced the inhibitory effect of cisplatin and etoposide on cell proliferation. Compared with the scrambled control group, the two ZEB1-knockdown LoVo cell lines exhibited decreased IC50 values for cisplatin and etoposide as shown in Table I.
Figure 4.
Cytotoxic effect of cisplatin and etoposide on LoVo cells. Cell viability of scrambled control cells and ZEB1-KD cells was assessed 24 h after the addition of various doses of (A) cisplatin or (B) etoposide. Data represent the values ± standard deviation of four independent experiments. ZEB1, zinc finger E-box-binding homeobox 1.
Table I.
IC50 values for cisplatin and etoposide in scrambled control and ZEB1-knockdown cells (µg/ml).
| Genotoxic agents | ||
|---|---|---|
| Group | Cisplatin | Etoposide |
| Scrambled control | 52.26 | 206.64 |
| ZEB1-KD-1 | 21.03 | 73.90 |
| ZEB1-KD-2 | 48.62 | 128.38 |
ZEB1, zinc finger E-box binding homeobox 1.
Silencing of ZEB1 blocks G1/S transition and increases apoptosis
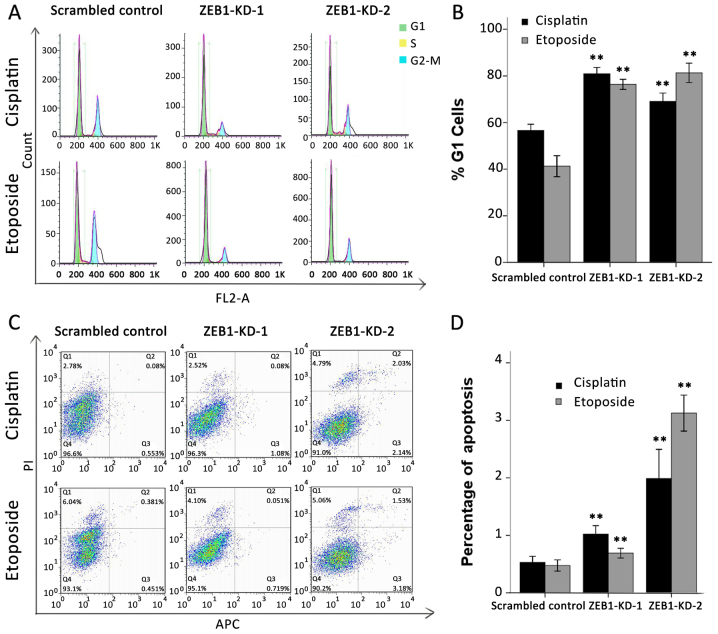
A disabled G1/S checkpoint, together with its mediated anti-apoptotic effect, is another characteristic of an unbalanced DDR network (26). It has been previously reported that overexpression of USP17 or DUX4 may lead to G1/S arrest and a higher apoptotic rate (27,28). Therefore, the present study evaluated whether ZEB1 silencing impacted the cell cycle and apoptosis in a similar way. Cells were synchronized in G1 by serum starvation, then treated with cisplatin at 0.5 µg/ml or etoposide at 3 µg/ml for 24 h and analyzed using flow cytometry. As expected, >70% of ZEB1-depleted cells failed to exit G1 and progress into the S phase following drug treatment. By contrast, only 57 and 42% of scrambled shRNA-transfected cells accumulated in the G1 phase following cisplatin or etoposide treatment, respectively (Fig. 5A). The results of three independent experiments are summarized in Fig. 5B. With regard to the apoptosis assay, the proportion of cells undergoing early apoptosis was also markedly upregulated in response to ZEB1 depletion. Cisplatin and etoposide treatment induced early apoptosis in only 0.53 and 0.48% of scrambled control cells, respectively; however, the same treatment induced early apoptosis in 1.02 and 0.69% of ZEB1-KD-1 cells and 1.99 and 3.13% ZEB1-KD-2 cells (Fig. 5C). The results of three independent experiments are summarized in Fig. 5D.
Figure 5.
Effect of ZEB1 silencing on the cell cycle and apoptosis. Cells were synchronized in G1 by serum starvation, then treated with cisplatin at 0.5 µg/ml or etoposide at 3 µg/ml for 24 h prior to flow cytometric analysis. (A) Silencing of ZEB1 caused G1/S block in LoVo cells. (B) Data are summarized and each bar represents the mean ± standard deviation of three independent experiments. **P<0.01 vs. scrambled control group. (C) Representative images demonstrate that ZEB1-silencing increased the number of early apoptotic LoVo cells. (D) Quantification of the apoptotic rate in scrambled control cells and ZEB1-KD cells. Data are presented as the mean ± standard deviation of three independent experiments. **P<0.01 vs. scrambled control group. ZEB1, zinc finger E-box-binding homeobox 1.
Discussion
To investigate the repertoire of ZEB1 in therapy resistance, and particularly in the DDR process, the current study mapped the DNA binding sites of ZEB1 in >1,300 cancer-associated genes using the ChIP-on-chip method, and identified positive intervals enriched along three DDR genes: USP17, CHD1L and DUX4. To confirm that the potential targets were not detected as a result of nonspecific binding of the ZEB1 antibody and cross-linked protein-DNA complexes, each positive region was searched for E-box elements, and changes in the mRNA expression of the three newly identified ZEB1 downstream genes were analyzed following ZEB1 knockdown. Collectively, the E-boxes (particularly the AGGTG-Nn-CACCT duplication) identified in the binding regions, and the enhanced mRNA expression of these genes, supported the prediction that USP17, CHD1L and DUX4 are ZEB1 target genes. Besides, the top four ranked predicted ZEB1-interacting motifs detected in the positive intervals along the three genes also slightly supported them as ZEB1 downstream targets. The ZEB1-binding activity of the predicted motifs will be confirmed later by pull-down assay using recombinant ZEB1 protein and oligonucleotides of the motifs, which will provide new insight into transcriptional regulation by ZEB1.
In order to functionally validate the three ZEB1 downstream target genes, their detailed functions in the DDR were reviewed, and whether the recovery of their expression by ZEB1 silencing may initiate similar biological effects was examined. Generally, previous studies have reported that the three potential targets of ZEB1 inhibit DNA repair by multiple mechanisms, and induce apoptosis and cell cycle arrest. Considering that USP17 suppresses the repair of DSBs (24) and DUX4 may inhibit interstrand crosslink (ICL) repair by indirectly suppressing proliferating cell nuclear antigen (PCNA) (28,29), the current study investigated the results of treating ZEB1-silenced cells with two DSB-causing agents, cisplatin (which creates DSBs as a result of inhibition of the ICL repair process) (30) and etoposide (31), and confirmed the chemosensitization effect of ZEB1 knockdown. We speculate that this sensitization effect was partially dependent on the subsequent upregulation of the potential ZEB1 downstream targets, particularly USP17. As a deubiquitylating enzyme, USP17 decreases the DNA damage-induced monoubiquitination of H2A histone family member X (H2AX), thus hindering the recruitment of the critical DNA repair effector proteins (tumor protein p53-binding protein 1 and ubiquitin interaction motif-containing 1/BRCA1) that promote the two major repair ways (non-homologous end joining and homologous recombination) to help cells overcome DSBs (24,32). In the present study, it is possible that the upregulation of USP17 resulting from ZEB1 silencing increased the deubiquitylation of H2AX, leading to impaired DSB repair.
In addition to USP17, the upregulation of CHD1L and DUX4 were also likely to be involved in chemosensitization. CHD1L (also termed ALC1) has long been defined as an oncogene as it disrupts the cell death program and activates the AKT serine/threonine kinase 1 pathway (33). In the DDR, CHD1L has a dual role. A previous study demonstrated that CHD1L has PAR-dependent chromatin remodeling activity that confers a more relaxed chromatin structure which, on one hand, facilitates DNA repair by enhancing nucleosome displacement and recruitment of DNA repair factors, while, on the other hand, it also increases the interaction of genotoxic agents with DNA, causing further damage. Thus, for chemotherapeutic drugs that preferentially act on relaxed chromatin, CHD1L may intensify their cytotoxic effect. In a previous study using cells with CHD1L overexpression, phleomycin exposure increased H2AX phosphorylation and increased comet tail lengths in a comet assay, implying an increase in DNA damage (25). It is not clear whether the increase in CHD1L expression in the current study affected drug-DNA contact or DNA repair factor recruitment. Further efforts will investigate how transient transfection of CHD1L siRNA in ZEB1-knockdown cells affects their response to cisplatin and etoposide.
Previous research regarding DUX4 has predominantly focused on facioscapulohumeral muscular dystrophy, a progressive disease involving the continuous degeneration of muscle cells and tissue. DUX4, which is normally transcriptionally silenced, was identified to express and encode a transcriptional activator of several genes in the p53 pathway, including p21 and caspase-3, inducing G1/S arrest and apoptosis in myoblasts (28). It is unclear whether DUX4 activates these molecules in cancer cells in the same manner as it does in myoblasts. If so, DUX4 may be a strong inhibitor of DNA repair, as the downstream target p21 suppresses various DNA repair pathways, predominantly by promoting PCNA degradation or by disrupting its recruitment of DNA repair factors (34). Cisplatin is a well-established ICL inducer and PCNA is an activator of the ICL repair pathway (35); thus, it is reasonable to speculate that the DUX4-p21-PCNA chain is involved in increasing the susceptibility of ZEB1-knockdown cells to cisplatin. Future efforts will be made to verify the DUX4-p21 interaction in cancer cells.
Furthermore, the present study demonstrated an increased apoptosis rate and G1/S arrest following cisplatin and etoposide treatment in ZEB1-knockdown cells. This suggests that USP17 and DUX4 are downstream targets of ZEB1, as both USP17 and DUX4 have been previously reported to facilitate apoptosis and induce G1 arrest (27,28). CHD1L was demonstrated to promote cell proliferation, inhibit programmed cell death and accelerate G1/S transition, and we hypothesize that the pro-apoptotic and cell cycle-blocking effects of USP17 and DUX4 counteracted the influence of CHD1L on the two processes, resulting in the final outcome observed in the study. Intriguingly, in the DDR, cell cycle arrest is often considered as a means of self-preservation that allows time for DNA repair and cell survival (31). Indeed, either more active repair or increased DNA damage that requires more time to be dealt with may lead to cell cycle arrest. However, as apoptosis was increased and the enhanced cytotoxic effect of genotoxic agents on ZEB1-knockdown cells strongly indicated an increase in unrepaired DNA lesions, we hypothesize that the G1 arrest indicated further DNA impairment.
In conclusion, three DDR-associated genes were identified as ZEB1 downstream targets and the present study demonstrated that their suppression by ZEB1 may contribute to ZEB1-mediated chemoresistance. This study provides a foundation for a more detailed understanding of the regulation of DDR by ZEB1, and suggests that inhibiting ZEB1 may be a promising treatment for cancer, as it targets three vital malignancy-associated events simultaneously: EMT, stemness and drug resistance.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (grant no. 81273636).
References
- 1.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: An emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitra A, Mishra L, Li S. EMT, CTCs and CSCs in tumor relapse and drug-resistance. Oncotarget. 2015;6:10697–10711. doi: 10.18632/oncotarget.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pattabiraman DR, Weinberg RA. Tackling the cancer stem cells - what challenges do they pose? Nat Rev Drug Discov. 2014;13:497–512. doi: 10.1038/nrd4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koren E, Fuchs Y. The bad seed: Cancer stem cells in tumor development and resistance. Drug Resist Updat. 2016;28:1–12. doi: 10.1016/j.drup.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, Choi H, El Rayes T, Ryu S, Troeger J, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sánchez-Tilló E, Lázaro A, Torrent R, Cuatrecasas M, Vaquero EC, Castells A, Engel P, Postigo A. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene. 2010;29:3490–3500. doi: 10.1038/onc.2010.102. [DOI] [PubMed] [Google Scholar]
- 7.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 8.Chaffer CL, Marjanovic ND, Lee T, Bell G, Kleer CG, Reinhardt F, D'Alessio AC, Young RA, Weinberg RA. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell. 2013;154:61–74. doi: 10.1016/j.cell.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang P, Sun Y, Ma L. ZEB1: At the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle. 2015;14:481–487. doi: 10.1080/15384101.2015.1006048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arumugam T, Ramachandran V, Fournier KF, Wang H, Marquis L, Abbruzzese JL, Gallick GE, Logsdon CD, McConkey DJ, Choi W. Epithelial to mesenchymal transition contributes to drug resistance in pancreatic cancer. Cancer Res. 2009;69:5820–5828. doi: 10.1158/0008-5472.CAN-08-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang P, Wei Y, Wang L, Debeb BG, Yuan Y, Zhang J, Yuan J, Wang M, Chen D, Sun Y, et al. ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nat Cell Biol. 2014;16:864–875. doi: 10.1038/ncb3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cortez MA, Valdecanas D, Zhang X, Zhan Y, Bhardwaj V, Calin GA, Komaki R, Giri DK, Quini CC, Wolfe T, et al. Therapeutic delivery of miR-200c enhances radiosensitivity in lung cancer. Mol Ther. 2014;22:1494–1503. doi: 10.1038/mt.2014.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang P, Wang L, Rodriguez-Aguayo C, Yuan Y, Debeb BG, Chen D, Sun Y, You MJ, Liu Y, Dean DC, et al. miR-205 acts as a tumour radiosensitizer by targeting ZEB1 and Ubc13. Nat Commun. 2014;5:5671. doi: 10.1038/ncomms6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Srivastava M, Raghavan SC. DNA double-strand break repair inhibitors as cancer therapeutics. Chem Biol. 2015;22:17–29. doi: 10.1016/j.chembiol.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 15.Tian H, Gao Z, Li H, Zhang B, Wang G, Zhang Q, Pei D, Zheng J. DNA damage response - a double-edged sword in cancer prevention and cancer therapy. Cancer Lett. 2015;358:8–16. doi: 10.1016/j.canlet.2014.12.038. [DOI] [PubMed] [Google Scholar]
- 16.Skvortsov S, Debbage P, Lukas P, Skvortsova I. Crosstalk between DNA repair and cancer stem cell (CSC) associated intracellular pathways. Semin Cancer Biol. 2015;31:36–42. doi: 10.1016/j.semcancer.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 17.Fokas E, Prevo R, Hammond EM, Brunner TB, McKenna WG, Muschel RJ. Targeting ATR in DNA damage response and cancer therapeutics. Cancer Treat Rev. 2014;40:109–117. doi: 10.1016/j.ctrv.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Benada J, Macurek L. Targeting the checkpoint to kill cancer cells. Biomolecules. 2015;5:1912–1937. doi: 10.3390/biom5031912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weigelt K, Moehle C, Stempfl T, Weber B, Langmann T. An integrated workflow for analysis of ChIP-chip data. Biotechniques. 2008;45:131–140. doi: 10.2144/000112819. [DOI] [PubMed] [Google Scholar]
- 20.Ji X, Li W, Song J, Wei L, Liu XS. CEAS: Cis-regulatory element annotation system. Nucleic Acids Res. 2006;34:W551–W554. doi: 10.1093/nar/gkl322. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Sekido R, Murai K, Funahashi J, Kamachi Y, Fujisawa-Sehara A, Nabeshima Y, Kondoh H. The delta-crystallin enhancer-binding protein delta EF1 is a repressor of E2-box-mediated gene activation. Mol Cell Biol. 1994;14:5692–5700. doi: 10.1128/MCB.14.9.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Remacle JE, Kraft H, Lerchner W, Wuytens G, Collart C, Verschueren K, Smith JC, Huylebroeck D. New mode of DNA binding of multi-zinc finger transcription factors: deltaEF1 family members bind with two hands to two target sites. EMBO J. 1999;18:5073–5084. doi: 10.1093/emboj/18.18.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delgado-Díaz MR, Martín Y, Berg A, Freire R, Smits VA. Dub3 controls DNA damage signalling by direct deubiquitination of H2AX. Mol Oncol. 2014;8:884–893. doi: 10.1016/j.molonc.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahel D, Horejsí Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furgason JM, Bahassi M. Targeting DNA repair mechanisms in cancer. Pharmacol Ther. 2013;137:298–308. doi: 10.1016/j.pharmthera.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 27.Burrows JF, McGrattan MJ, Rascle A, Humbert M, Baek KH, Johnston JA. DUB-3, a cytokine-inducible deubiquitinating enzyme that blocks proliferation. J Biol Chem. 2004;279:13993–14000. doi: 10.1074/jbc.M311291200. [DOI] [PubMed] [Google Scholar]
- 28.Xu H, Wang Z, Jin S, Hao H, Zheng L, Zhou B, Zhang W, Lv H, Yuan Y. Dux4 induces cell cycle arrest at G1 phase through upregulation of p21 expression. Biochem Biophys Res Commun. 2014;446:235–40. doi: 10.1016/j.bbrc.2014.02.105. [DOI] [PubMed] [Google Scholar]
- 29.Karimian A, Ahmadi Y, Yousefi B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair (Amst) 2016;42:63–71. doi: 10.1016/j.dnarep.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 30.Ciccia A, Elledge SJ. The DNA damage response: Making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber AM, Ryan AJ. ATM and ATR as therapeutic targets in cancer. Pharmacol Ther. 2015;149:124–138. doi: 10.1016/j.pharmthera.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 32.Aparicio T, Baer R, Gautier J. DNA double-strand break repair pathway choice and cancer. DNA Repair (Amst) 2014;19:169–175. doi: 10.1016/j.dnarep.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng W, Su Y, Xu F. CHD1L: A novel oncogene. Mol Cancer. 2013;12:170. doi: 10.1186/1476-4598-12-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung YS, Qian Y, Chen X. Examination of the expanding pathways for the regulation of p21 expression and activity. Cell Signal. 2010;22:1003–1012. doi: 10.1016/j.cellsig.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohleder F, Huang J, Xue Y, Kuper J, Round A, Seidman M, Wang W, Kisker C. FANCM interacts with PCNA to promote replication traverse of DNA interstrand crosslinks. Nucleic Acids Res. 2016;44:3219–3232. doi: 10.1093/nar/gkw037. [DOI] [PMC free article] [PubMed] [Google Scholar]