Abstract
G protein nucleolar 3 (GNL3), a nucleolar GTP-binding protein, is highly expressed in progenitor cells, stem cells, and various types of cancer cells. Therefore, it is considered to have an important role in cancer pathogenesis. GNL3 has been reported to play crucial roles in cell proliferation, cell cycle regulation, inhibition of differentiation, ribosome biogenesis, and the maintenance of stemness, genome stability and telomere integrity. Furthermore, GNL3 has recently been shown to be involved in cancer invasion and metastasis. However, the biological significance of GNL3 in the invasion and metastasis of colon cancer remains unclear. This study was performed to address this gap in knowledge. GNL3 expression was upregulated in colon cancer tissue specimens and correlated with tumor differentiation, invasion and metastasis. GNL3 overexpression promoted cell proliferation, invasion, migration and the epithelial-mesenchymal transition (EMT) in colon cancer cells. Moreover, inhibition of the EMT and the Wnt/β-catenin signaling pathway induced by GNL3 knockdown was partially reversed by lithium chloride (LiCl). Based on these data, GNL3 promotes the EMT in colon cancer by activating the Wnt/β-catenin signaling pathway. In summary, GNL3 is upregulated in colon cancer and plays an important role in tumor growth, invasion and metastasis. Strategies targeting GNL3 are potential treatments for colon cancer.
Keywords: GNL3, colon cancer, Wnt/β-catenin, epithelial-mesenchymal transition, cell proliferation, migration, invasion
Introduction
Colorectal cancer is the third most frequent malignant neoplasm and the fourth leading cause of cancer-related death worldwide (1). Colorectal cancer-related death is mainly caused by metastasis. Based on accumulating data obtained over the past few years, the EMT plays a crucial role in tumor metastasis (2,3). The EMT is the process by which epithelial cells lose their epithelial characteristics and gain mesenchymal characteristics. In this way, the EMT endows cells with invasive and migratory properties, prevents apoptosis and senescence, induces stem cell properties, and contributes to immunosuppression (4–7). Furthermore, several signaling pathways are involved in the EMT, such as Wnt/β-catenin (8), transforming growth factor-β (TGF-β) (9), Notch (10), and nuclear factor-κB (NF-κB) signaling pathway (11). The Wnt/β-catenin signaling pathway has a significant impact on the maintenance of stem cell properties and cancer metastasis (12,13). Additionally, the nuclear accumulation of β-catenin is a crucial step in the activation of the Wnt signaling pathway (14). Lithium chloride (LiCl), an inhibitor of glycogen synthase kinase 3β (GSK3β), promotes the nuclear accumulation of β-catenin and activates the Wnt/β-catenin signaling pathway (15,16).
G protein nucleolar 3 (GNL3) was initially identified in the nucleoli of neural stem cells, embryonic stem cells and several cancer cells, and it is preferentially expressed by other stem cell-enriched populations, but is not expressed in differentiated somatic cells (17,18). GNL3 was initially named nucleostemin (NS) (17). GNL3 was subsequently shown to play significant roles in regulating cell proliferation and the cell cycle in various human cancers, such as gastric cancer, colorectal cancer, liver cancer and other malignancies (19,20). Furthermore, GNL3 also plays a crucial role in many other physiological and pathological processes, such as the resistance to apoptosis and cellular aging, cellular self-renewal, the maintenance of stemness, the inhibition of differentiation, ribosome biogenesis, the maintenance of genome stability and telomere integrity, radioresistance, the maintenance of tumor-initiating cells, and the induction of pluripotent stem cells (21–27). Moreover, GNL3 is involved in cancer invasion and metastasis (22,28,29). However, the role of GNL3 in the invasion and metastasis of colon cancer remains unclear.
This study is the first to report that GNL3 plays a crucial role in the invasion and metastasis of colon cancer. Moreover, GNL3 promotes the proliferation, invasion, migration, and EMT in colon cancer cells by activating the Wnt/β-catenin signaling pathway.
Materials and methods
Ethics statement
No human participant was involved in this study, and the use of human tissue samples and all experimental procedures and protocols were approved by the medical ethics review committee of the First Affiliated Hospital of Chongqing Medical University, Chongqing, China. Written informed consent was obtained from all subjects prior to the publication of this study. All procedures involving animals were performed in accordance with the guidelines of the National Institutes of Health (NIH) regarding animal care (Guide for the Care and Use of Laboratory Animals, Department of Health and Human Services, NIH Publication No. 86-23, revised 1985). We made every effort to minimize the animals' suffering and the number of animals experimented on.
Tissue specimens and immunohistochemistry assays
In total, 130 colon cancer samples and their corresponding normal, tumor-adjacent tissues were randomly collected from the Department of Pathology, Chongqing Medical University. An additional 60 fresh surgical specimens (30 colon cancer tissues and corresponding normal, tumor-adjacent tissues) were collected from the Department of Gastrointestinal Surgery, the First Affiliated Hospital of Chongqing Medical University. The fresh specimens were immediately frozen and stored in liquid nitrogen after resection. None of the patients had been treated before the operation.
Immunohistochemical staining was performed as previously described (30). Briefly, tissue sections were deparaffinized in xylene, and antigens were retrieved by heating the tissue sections (95°C, 15 min). Then, 3% hydrogen peroxide was added to the slides and incubated for 15 min before the slides were permeabilized with 0.5% Triton-X-100 (Beyotime, Jiangshu, China) for 15 min. Subsequently, 5% bovine serum albumin (BSA) was used to block the slides for 30 min. Then, the slides were incubated with mouse primary antibodies against human GNL3 (1:200; Santa Cruz Biotechnology, Dallas, TX, USA) overnight at 4°C. Negative controls were incubated without primary antibodies. The next day, the tissue sections were washed three times with PBS for 5 min each and incubated with appropriate HRP-conjugated secondary antibodies (anti-mouse IgG; Zhongshan Jinqiao, Beijing, China). The slides were developed in diaminobenzidine and counterstained with hematoxylin. The percentage of positive cells was graded as follows: 0, <10%; 1, 10–30%; 2, 30–50%; and 3, >50% (31). The patients with colon cancer were subsequently divided into either a negative expression group (a score of 0) or a positive expression group (scores of 1–3).
Cell culture and transfection
The Caco-2, HCT-116, HT-29, LoVo, and SW620 human colon cancer cell lines were obtained from the Type Culture Collection of the Chinese Academy of Sciences, Shanghai, China. HCT-116 and HT-29 cells were both maintained in RPMI-1640 medium (RPMI-1640, Gibco, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin (Beyotime) in a humidified atmosphere containing 95% air and 5% CO2 at 37°C.
GNL3 knockdown plasmids were purchased from Sigma-Aldrich (St. Louis, MO, USA; TRCN0000293740) and GNL3 expression plasmids were purchased from Sino Biological Inc. (Beijing, China; HG12415-UT). The sequences of the GNL3-specific shRNAs used in this study were: GNL3-shRNA, 5′-TTCATCGTATCTCCACTTAAT-3′ and control-shRNA, 5′-AAGCAATACCAGCAATTCATA-3′. Cells were transfected with plasmids using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). HCT-116 cells were transfected with the GNL3 knockdown plasmids and HT-29 cells were transfected with the GNL3 expression plasmids according to the manufacturer's protocols. Briefly, cells (at densities of 3.0×105 and 2.0×105 cells/well, respectively) were seeded in 6-well plates in 2 ml of antibiotic-free growth medium. The cells were transfected on the day after plating using 10 µl of Lipofectamine and 4 µg of plasmid DNA; the cells were incubated with the transfection mixture for 6 h. Then, the cells were transferred to 2 ml of complete growth medium at 37°C in a 5% CO2 incubator. After 48 h of incubation, the cells were selected with the corresponding concentrations of puromycin (Qcbio S&T Co., Shanghai, China) and hygromycin B (Qcbio S&T Co.). The cells were collected for protein analysis and other analyses after a reasonable time.
Cell proliferation and colony formation assays
HCT-116 and HT-29 cells were seeded in triplicate in 96-well plates containing 100 µl of medium at densities of 3×103 cells and collected for the MTT assay at time points ranging from 1–5 days. The cells were treated with 20 µl of MTT (5 mg/ml; Sigma-Aldrich) for 4 h, after which the MTT solution was removed. The crystals were dissolved in 200 µl of DMSO (Sigma-Aldrich). The absorbance of the solution was measured at 570 nm using a spectrophotometer.
HCT-116 and HT-29 cells were treated with 0.25% trypsin and dispersed into a single cell suspension in medium supplemented with 10% FBS (Gibco) prior to use. Cells were seeded into culture dishes at a density of 200 cells/dish and cultured in a 5% CO2 atmosphere at 37°C for 2–3 weeks. When cell clones appeared, they were fixed with 4% paraformaldehyde for 10–30 min. Then, the cells were stained with Giemsa (Beyotime) for 10–30 min and counted under a light microscope.
Cell invasion and migration assays
Three hundred microliters of preheated serum-free medium was added to a cell invasion chamber (Millipore, Billerica, MA, USA) to moisten the extracellular matrix (ECM) membrane. The serum-free medium was removed after 1–2 h. Five hundred microliters of medium supplemented with 10% FBS was added to each well of a 24-well plate that did not contain a chamber. Then, the chamber was placed in the well and 2×105 cells were seeded in the chamber (suspended in serum-free medium). After the cells were incubated in a 5% CO2 atmosphere at 37°C for 24 h, they were stained with 0.1% crystal violet for 10 min. Cells were counted under a light microscope, and images were captured.
HCT-116 and HT-29 cells were separately seeded in a 6-well plate in serum-free medium. Cells grew overnight to confluence. A pipette tip was dragged through the cells to scratch the monolayer, creating a wound with an even width. Then, PBS was used to wash and remove the cellular debris. Cells were allowed to migrate for 24 h. An inverted microscope was used to capture images at 0 and 24 h.
Western blot analysis
Western blot analyses were performed as previously described (32). Briefly, the proteins were extracted and their concentrations were determined using a protein extraction kit (Beyotime) and an enhanced bicinchoninic acid (BCA) protein assay kit (Beyotime), respectively, according to the manufacturer's instructions. Protein samples were separated by SDS-PAGE and transferred to PVDF membranes (Millipore). The transferred PVDF membranes were blocked with 5% non-fat dry milk in PBST for 1.5 h and incubated with primary antibodies against GNL3 (1:200 dilution; Santa Cruz Biotechnology), E-cadherin (1:1,000 dilution; Santa Cruz Biotechnology), N-cadherin (1:1,000 dilution; Epitomics, Burlingame, CA, USA), vimentin (1:1,000 dilution; eBioscience, San Diego, CA, USA), and β-catenin (1:5,000 dilution; Epitomics) overnight at 4°C. Then, the membranes were incubated with the appropriate secondary antibodies for 1 h. Bands were visualized with an enhanced chemiluminescence kit (Beyotime) and quantitated with Quantity One 4.6.2 software. Protein band intensities were normalized to the intensity of GAPDH (1:500 dilution; Beyotime).
Immunofluorescence staining
Cells were seeded in plates and fixed with 4% paraformaldehyde for 15 min. The cells were permeabilized with 0.5% TritonX-100 (Beyotime) for 10 min at room temperature. Then, the cells were blocked with 1% bovine serum albumin (BSA) for 2 h. Afterwards, the cells were incubated with primary antibodies overnight at 4°C. The next day, the appropriate secondary antibodies were incubated with the cells for 2 h in the dark at 4°C, and 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich) was added to stain the nucleus. Finally, the cells were visualized, and images were captured using a fluorescence microscope (Leica, Wetzlar, Germany).
In vivo experiments
Five-week-old male nude BALB/c mice were purchased from the National Biological Industry Base, Laboratory Animal Center of Chongqing Medical University. Approximately 5×106 HCT-116 and HT-29 cells that had been transfected with the GNL3 knockdown plasmids and GNL3 expression plasmids, respectively, were subcutaneously injected into the axillae of nude mice. The sizes of the xenograft tumors were measured weekly. Four weeks later, the nude mice were euthanized and tumor samples were collected.
Statistical analysis
The data are presented as the means ± standard deviations (SD). The expression levels of GNL3 and clinicopathological features were compared using the Chi-square test. The differences between groups were analyzed using Student's t-test using SPSS 13.0 (SPSS, Inc., Chicago, IL, USA). A Kaplan-Meier analysis was used to obtain the survival curves, and differences were assessed with the log-rank test. P<0.05 was considered to indicate a statistically significant difference.
Results
The correlation between GNL3 expression and clinicopathological characteristics in colon cancer
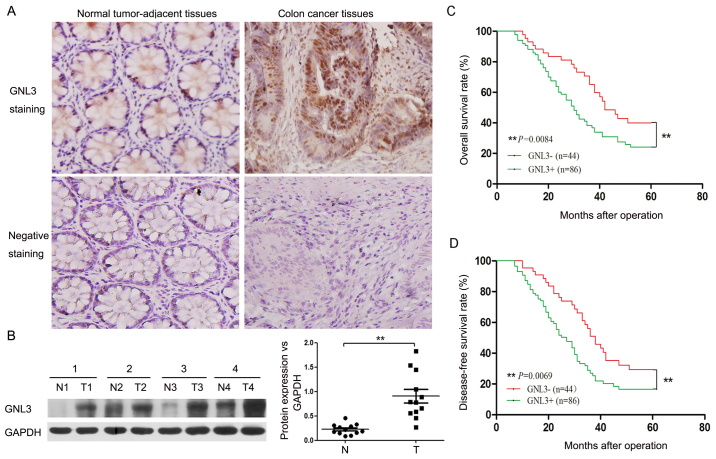
We collected tissue specimens from 130 patients and performed immunohistochemistry assays on these specimens to determine the GNL3 expression level in colon cancer. The rates of GNL3-positive expression in colon cancer tissues and normal, tumor-adjacent tissues were 66.2% (86/130) and 31.5% (41/130), respectively. As expected, GNL3 was expressed at much higher levels in colon cancer tissues than in normal, tumor-adjacent tissues (Fig. 1A). Furthermore, we examined the expression levels of GNL3 in the fresh samples using western blotting. GNL3 was expressed at significantly higher levels in colon cancer tissues than in normal, tumor-adjacent tissues (P<0.01; Fig. 1B). The correlations between the GNL3 expression levels and the clinicopathological features of patients with colon cancer are presented in Table I. The GNL3 expression levels were significantly associated with tumor differentiation (P<0.01), serosal invasion (P<0.05), lymph node metastasis (P<0.05), and vascular invasion (P<0.05) in patients with colon cancer (P<0.05) (Table I).
Figure 1.
GNL3 expression and its clinical significance in colon cancer. (A) Representative images of immunohistochemical staining for GNL3 in normal, tumor-adjacent tissues and colon cancer tissues. Original magnification, ×200. (B) GNL3 levels in normal, tumor-adjacent tissues (N1-4) and colon cancer tissues (T1-4) were detected by western blotting. According to the quantitative analysis, GNL3 was expressed at significantly higher levels in colon cancer tissues than in normal, tumor-adjacent tissues. (C and D) Overall 5-year and disease-free survival curves for GNL3-negative and GNL3-positive patients with colon cancer according to the Kaplan-Meier analysis. The overall and disease-free survival rates of the GNL3-positive groups were markedly reduced compared with the GNL3-negative groups. **P<0.01. The statistical analysis of the data shown in (B) was performed using the paired t-test; the Kaplan-Meier analysis was used to obtain survival curves and differences in the data presented in (C and D) were assessed using the log-rank test. GNL3, G protein nucleolar 3.
Table I.
The correlations between the GNL3 expression levels and the clinicopathological features of patients with colon cancer.
| GNL3 | ||||
|---|---|---|---|---|
| Clinicopathological features | Cases n=130 | Negative n=44 | Positive n=86 | P-value |
| Sex | 0.5169 | |||
| Male | 76 | 24 | 52 | |
| Female | 54 | 20 | 34 | |
| Age (years) | 0.4647 | |||
| <60 | 53 | 16 | 37 | |
| ≥60 | 77 | 28 | 49 | |
| Tumor size (cm) | 0.1059 | |||
| ≤3 cm | 61 | 25 | 36 | |
| >3 cm | 69 | 19 | 50 | |
| Differentiation | 0.0043b | |||
| Poor | 58 | 11 | 47 | |
| Moderate | 36 | 18 | 18 | |
| Well | 36 | 15 | 21 | |
| Serosal invasion | 0.0176a | |||
| Absent | 58 | 26 | 32 | |
| Present | 72 | 18 | 54 | |
| Vascular invasion | 0.0468a | |||
| Absent | 61 | 26 | 35 | |
| Present | 69 | 18 | 51 | |
| Lymph node metastasis | 0.0353a | |||
| Yes | 84 | 23 | 61 | |
| No | 46 | 21 | 25 | |
| Liver metastasis | 0.0830 | |||
| Absent | 94 | 36 | 58 | |
| Present | 36 | 8 | 28 | |
| Stage | 0.1559 | |||
| I + II | 51 | 21 | 30 | |
| III + IV | 79 | 23 | 56 | |
P<0.05
P<0.01; Chi-square test; P<0.05, a significant difference.
A Kaplan-Meier survival analysis was used to determine the correlation between the GNL3 expression levels and the survival of patients with colon cancer as a measure of the prognostic significance of GNL3 expression in these patients. As shown in Fig. 1C and D, the 5-year survival rate of the GNL3-positive groups was markedly reduced compared with that in the GNL3-negative groups (P<0.01; Fig. 1C). The median disease-free survival times were 27.0 and 38.0 months for the GNL3-positive and GNL3-negative groups, respectively (P<0.01; Fig. 1D). Based on these results, the upregulation of GNL3 expression was closely related to a shorter disease-free survival (P<0.01; Fig. 1D).
GNL3 expression levels in colon cancer cell lines
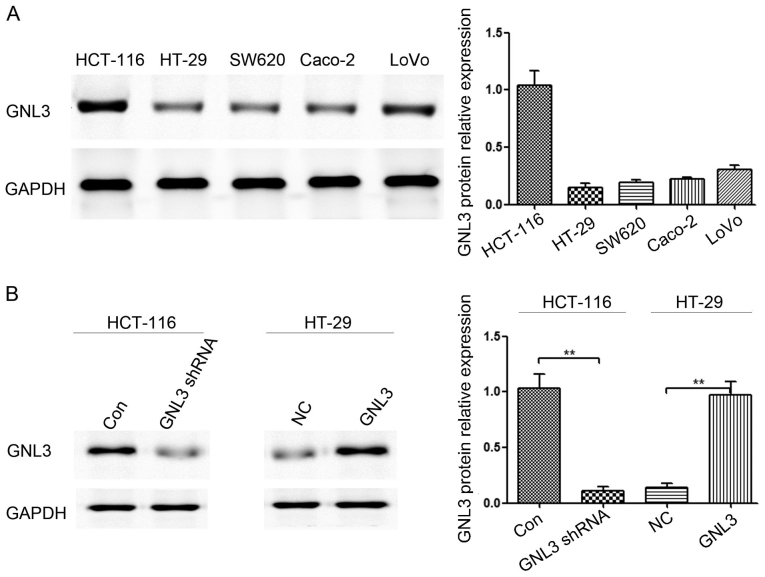
Western blot assays were used to detect the GNL3 levels in five colon cancer cell lines, and the results showed different levels of GNL3 expression in Caco-2 (0.22±0.02), HCT-116 (1.04±0.13), LoVo (0.31±0.05), HT-29 (0.15±0.04), and SW620 (0.19±0.02) cells (Fig. 2A). Therefore, we selected HCT-116 and HT-29 cells for transfection. We used western blot assays to test the GNL3 expression levels in HCT-116 and HT-29 cells that had been transfected with GNL3 knockdown and expression plasmids, respectively, as well as cells transfected with the corresponding control plasmids. GNL3 expression was decreased in HCT-116 cells transfected with GNL3 knockdown plasmids (P<0.01; Fig. 2B) and increased in HT-29 cells transfected with GNL3 expression plasmids (P<0.01; Fig. 2B).
Figure 2.
GNL3 expression was assessed in different colon cancer cell lines using western blotting. (A) GNL3 levels in five colon cancer cell lines: HCT-116, HT-29, SW620, Caco-2 and LoVo cells. (B) GNL3 expression was significantly reduced in GNL3 knockdown cells (GNL3 shRNA) compared with that in the control cells (Con) and was substantially increased in GNL3-overexpressing cells (GNL3) compared with that in the negative control cells (NC). **P<0.01; Student's t-test. GNL3, G protein nucleolar 3.
GNL3 promotes colon cancer cell proliferation and colony formation
As previously reported, GNL3 plays an important role in regulating the proliferation of various stem cells and cancer cells. Therefore, we postulated that GNL3 plays a similar role in regulating the proliferation of colon cancer cells. We performed MTT cell proliferation assays to assess their proliferative capacity. GNL3 knockdown inhibited the proliferation of HCT-116 cells (P<0.05; Fig. 3A) and GNL3 overexpression promoted the proliferation of HT-29 cells (P<0.05 or P<0.01; Fig. 3B). Colony formation assays were performed to further examine the effects of GNL3 on the formation of colon cancer cell colonies. Cells were transfected with the vectors described above. As shown in Fig. 3C, GNL3 knockdown significantly inhibited the ability of HCT-116 cells to form colonies (P<0.05; Fig. 3C), and GNL3 overexpression remarkably enhanced the capacity of HT-29 cells to form colonies (P<0.05; Fig. 3C). In conclusion, GNL3 overexpression promoted colon cancer cell proliferation and colony formation in vitro.
Figure 3.
Effects of GNL3 knockdown and overexpression on cell proliferation and colony formation in vitro and tumor growth in nude mice in vivo. (A) In HCT-116 cells, the proliferation rates of the GNL3 knockdown group (GNL3 shRNA) were lower than the control group (Con). (B) In HT-29 cells, the proliferation rates of the GNL3-overexpressing group (GNL3) were much higher than in the negative control group (NC). (C) GNL3 knockdown inhibited the formation of HCT-116 cell colonies, and GNL3 overexpression promoted the formation of HT-29 cell colonies. (D and E) Representative xenograft tumors are shown and the tumor volume was measured weekly. *P<0.05 and **P<0.01; Student's t-test. GNL3, G protein nucleolar 3.
GNL3 promotes tumor growth in vivo
We further tested whether GNL3 affected tumorigenesis in vivo using a xenograft model in nude mice. Nude mice were transplanted with HCT-116 and HT-29 cells that had been transfected with the vectors described above. Four weeks later, the xenograft tumors in the HCT-116 knockdown groups were smaller than the tumors in the control groups (Fig. 3D), and the tumors in the HT-29 overexpression groups were larger than the tumors in the corresponding controls (Fig. 3E). Moreover, the average tumor volume exhibited similar trends in the GNL3 knockdown groups (P<0.01; Fig. 3D) and GNL3 overexpression groups (P<0.05; Fig. 3E) as those in the corresponding control groups. Thus, GNL3 promoted tumor growth in vivo.
GNL3 promotes the invasion and migration of colon cancer cells
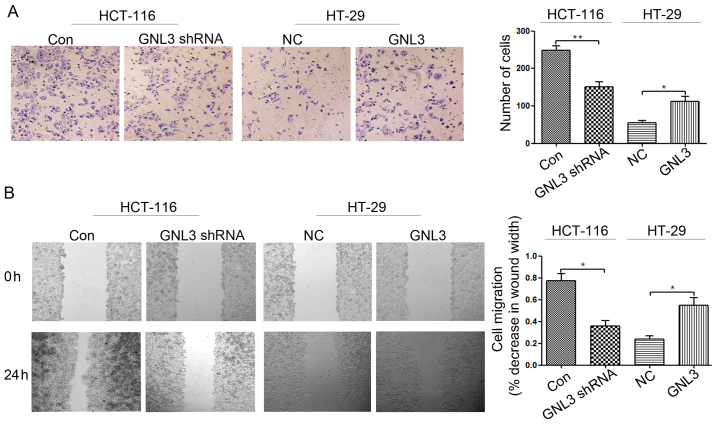
Cell invasion assays were performed using cell invasion chambers to assess the effects of GNL3 on colon cancer cell invasion. Cells that migrated through the ECM membrane are shown in Fig. 4A. Following GNL3 knockdown, the invasion of HCT-116 cells was significantly inhibited compared with the paired control groups (P<0.01; Fig. 4A). However, GNL3 overexpression significantly increased the invasion of HT-29 cells compared with the corresponding control groups (P<0.05; Fig. 4A). In vitro scratch wound-healing assays were performed to assess the effects of GNL3 on colon cancer cell migration. As shown in Fig. 4B, cells in which GNL3 expression was knocked down migrated significantly slower than the paired control groups (P<0.05; Fig. 4B). However, GNL3 overexpression induced significantly faster migration in the transfected cells than in the corresponding control groups (P<0.05; Fig. 4B). Based on these data, GNL3 promotes the invasion and migration of colon cancer cells.
Figure 4.
Effects of GNL3 knockdown and overexpression on cell invasion and migration in vitro. (A) Following GNL3 knockdown (GNL3 shRNA), the invasive capacity of HCT-116 cells was decreased compared with those of control cells (Con). Following GNL3 overexpression (GNL3), the invasive capacity of HT-29 cells was increased compared with negative control cells (NC). (B) GNL3 knockdown inhibited the migration of HCT-116 cells, and GNL3 overexpression promoted the migration of HT-29 cells. *P<0.05 and **P<0.01; Student's t-test. GNL3, G protein nucleolar 3.
GNL3 promotes EMT in colon cancer cells by activating the Wnt/β-catenin signaling pathway
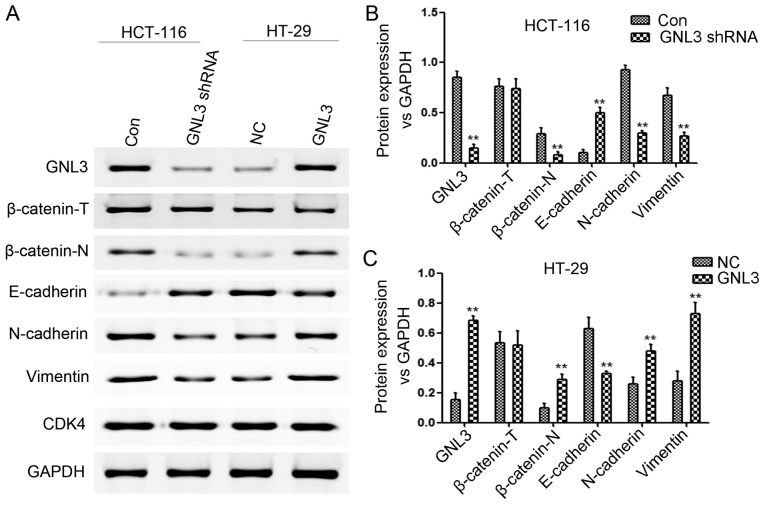
We performed western blot assays to quantitate the expression levels of total β-catenin (β-catenin-T), nuclear β-catenin (β-catenin-N), GNL3, E-cadherin, N-cadherin, and vimentin in HCT-116 and HT-29 cells transfected with GNL3 knockdown vectors or GNL3 expression vectors, respectively, or paired control vectors to confirm that GNL3 promoted EMT in colon cancer cells by activating the Wnt/β-catenin signaling pathway. GNL3 knockdown in HCT-116 cells increased E-cadherin expression (P<0.01) and decreased β-catenin-N (P<0.01), GNL3 (P<0.01), N-cadherin (P<0.01) and vimentin expression (P<0.01) (Fig. 5A and B). However, GNL3 overexpression in HT-29 cells decreased E-cadherin expression (P<0.01) and increased β-catenin-N (P<0.01), GNL3 (P<0.01), N-cadherin (P<0.01) and vimentin expression (P<0.01) (Fig. 5A and C). No significant differences in the β-catenin-T levels were observed in each group (P>0.05; Fig. 5A-C).
Figure 5.
Effects of GNL3 knockdown and overexpression on the levels of the EMT-related markers, β-catenin and GNL3 in vitro. (A and B) In HCT-116 cells, GNL3 knockdown (GNL3 shRNA) increased E-cadherin levels and reduced N-cadherin, vimentin, nuclear β-catenin (β-catenin-N) and GNL3 levels compared with controls (Con). (A and C) In HT-29 cells, GNL3 overexpression (GNL3) increased the N-cadherin, vimentin, β-catenin-N and GNL3 levels and reduced the E-cadherin levels compared with the negative controls (NC). **P<0.01; Student's t-test. GNL3, G protein nucleolar 3.
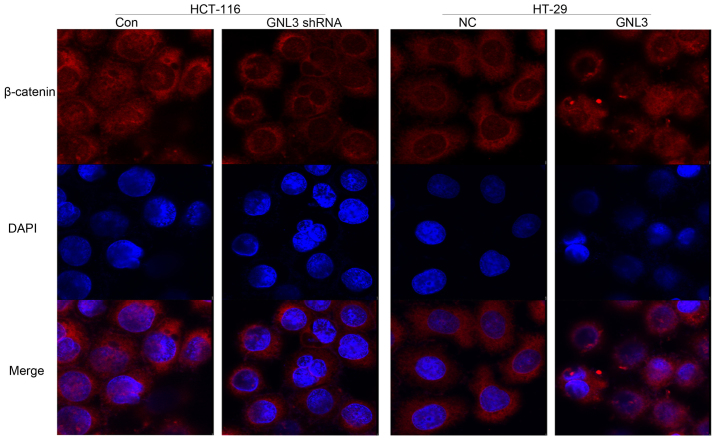
Moreover, nuclear β-catenin immunofluorescence staining was reduced upon GNL3 knockdown in HCT-116 cells and increased upon GNL3 overexpression in HT-29 cells (Fig. 6).
Figure 6.
Immunofluorescence staining for β-catenin. The β-catenin proteins labeled in red and the nuclei were stained with DAPI and are labeled in blue. GNL3 knockdown (GNL3 shRNA) in HCT-116 cells reduced the nuclear β-catenin levels compared with the controls (Con). However, GNL3 overexpression (GNL3) in HT-29 cells increased the nuclear β-catenin levels compared with the negative controls (NC). Original magnification, ×400.
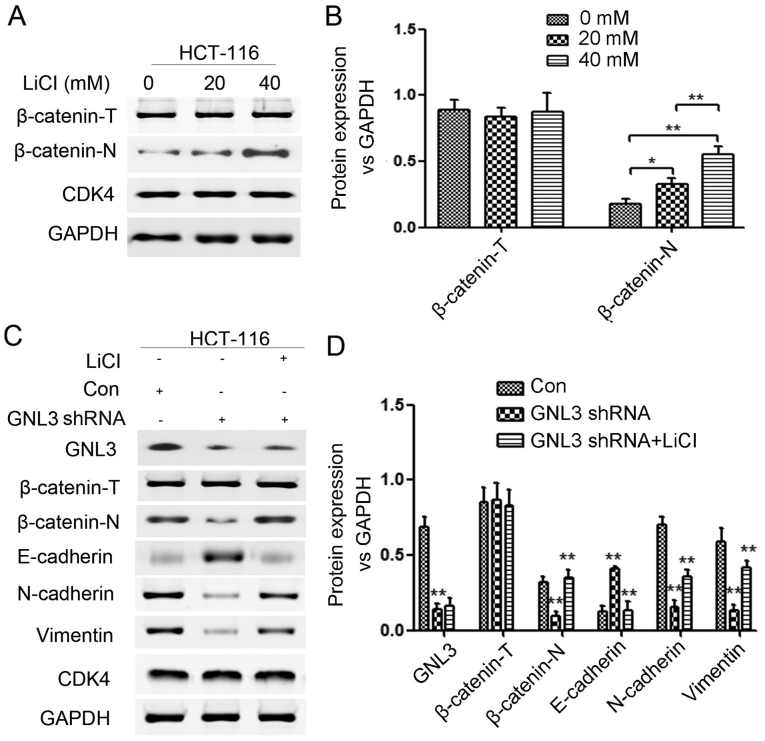
We further evaluated whether the modulatory effects of GNL3 on EMT in colon cancer cells were attributable to the Wnt/β-catenin signaling pathway. LiCl was used to promote the nuclear accumulation of β-catenin (β-catenin-N) and activate the Wnt/β-catenin signaling pathway in HCT-116 cells transfected with GNL3 knockdown plasmids. The control group (Con), GNL3 knockdown group (GNL3 shRNA), and LiCl-treated GNL3 knockdown group (GNL3 shRNA+LiCl) were evaluated. Western blot assays were performed to examine the expression levels of proteins related to the EMT and Wnt/β-catenin signaling pathway. First, we determined the appropriate concentration of LiCl needed to activate the Wnt/β-catenin signaling pathway and found that 20 mM (P<0.05) and 40 mM (P<0.01) were suitable concentrations (Fig. 7A and B). We found that β-catenin-N, GNL3, N-cadherin and vimentin were all remarkably downregulated and E-cadherin was upregulated in GNL3 knockdown groups compared with the levels in the control groups (P<0.01; Fig. 7C and D). We also found that β-catenin-N, N-cadherin and vimentin expression were all upregulated and E-cadherin expression was downregulated in the GNL3 shRNA+LiCl groups compared with the levels in the GNL3 shRNA groups (P<0.01; Fig. 7C and D). The inhibition of the EMT and Wnt/β-catenin signaling pathway caused by GNL3 knockdown was partially reversed by LiCl. In other words, GNL3 actually regulated EMT in colon cancer cells by activating the Wnt/β-catenin signaling pathway; otherwise, LiCl would not have partially restored the activity of the suppressed Wnt/β-catenin signaling pathway and EMT. In summary, GNL3 promotes the EMT in colon cancer cells by activating Wnt/β-catenin signaling pathway.
Figure 7.
Treatment with suitable concentrations of LiCl activates the Wnt/β-catenin signaling pathway, and LiCl partially reverses the GNL3 knockdown-induced inhibition of the EMT and Wnt/β-catenin signaling pathway in HCT-116 cells. (A and B) Both 20 and 40 mM LiCl were appropriate concentrations to activate the Wnt/β-catenin signaling pathway. (C and D) GNL3 knockdown (GNL3 shRNA) increased E-cadherin expression and decreased N-cadherin, vimentin, nuclear β-catenin (β-catenin-N) and GNL3 expression compared with the controls (Con). LiCl increased N-cadherin, vimentin and β-catenin-N expression and reduced E-cadherin expression compared with the GNL3 shRNA group. *P<0.05 and **P<0.01; Student's t-test. GNL3, G protein nucleolar 3.
Discussion
Metastasis is the main cause of death in patients with colon cancer. The EMT plays a crucial role in tumor metastasis (33). Although GNL3 has been shown to be involved in cancer metastasis, the role of GNL3 in colon cancer metastasis remains unclear (22,29,34). Herein, GNL3 was expressed at much higher levels in colon cancer tissues than in normal, tumor-adjacent tissues, and GNL3 levels were related to the prognosis of patients with colon cancer. GNL3 promotes the proliferation, invasion, migration and EMT in colon cancer cells. Furthermore, GNL3 promotes EMT in colon cancer by activating the Wnt/β-catenin signaling pathway. These results are consistent with our previous hypotheses.
We used immunohistochemistry and western blot assays to determine the GNL3 expression levels in 130 colon cancer tissue specimens as a first step in elucidating the function and mechanism by which GNL3 regulates colon cancer metastasis. GNL3 was expressed at significantly higher levels in tumor tissues than in normal, tumor-adjacent tissues. The GNL3 expression levels were closely related to tumor differentiation, serosal invasion, lymph node metastasis and vascular invasion. Moreover, high GNL3 expression levels resulted in lower 5-year and disease-free survival rates for patients with colon cancer. These results are consistent with the findings of a previous study (29). In hepatocellular carcinoma (HCC), GNL3 expression levels are also closely related to tumor differentiation and metastasis, and high GNL3 expression levels predict a worse prognosis for patients with HCC. Based on these results, high GNL3 expression levels show significant associations with the clinicopathological characteristics and predict a poorer prognosis for patients with colon cancer.
Next, we tested the effects of GNL3 on colon cancer cell proliferation and colony formation. GNL3 overexpression increased cell proliferation and colony formation; GNL3 knockdown exerted the opposite effect. Furthermore, we used a xenograft model in nude mice and found that the xenograft tumors in the GNL3-overexpressing groups were larger than the tumors in the control groups. In contrast, GNL3 knockdown groups exhibited the opposite trend. These data were similar to those in previously published studies. As shown in previous studies (17,29), GNL3 regulates cell proliferation in p53-dependent and p53-independent manner. The Wnt/β-catenin signaling pathway is involved in regulating cell proliferation (35). Moreover, GNL3 was recently shown to promote the proliferation of human glioma through the Wnt/β-catenin pathway (36).
Then, we examined the effects of GNL3 on the invasion and migration of colon cancer cells using cell invasion assays and scratch wound-healing assays. GNL3 overexpression promoted the invasion and migration of colon cancer cells, whereas GNL3 knockdown inhibited invasion and migration. Then, we tested the expression levels of EMT-related markers using western blot assays and showed that GNL3 overexpression reduced E-cadherin expression and increased N-cadherin and vimentin expression. In contrast, GNL3 knockdown exerted the opposite effects. These data are similar to the results from a previous study (22). GNL3 overexpression activates EMT via the TGF-β signaling pathway to induce HeLa cell metastasis (22). However, the TGF-β signaling pathway is different from the pathway examined in our study. Because of the complexity of the EMT, the EMT utilizes several signaling pathways, as we previously described. Different tumor cells may utilize different signaling pathways to induce EMT.
Finally, we performed western blot analyses and immunofluorescence staining to evaluate the levels and localization of β-catenin in colon cancer cells and to clarify the mechanism by which GNL3 regulates EMT in colon cancer cells. Furthermore, we used LiCl to reverse the GNL3 knockdown-induced suppression of the EMT and Wnt/β-catenin signaling pathway. Then, GNL3 over-expression increased the β-catenin-N levels, and GNL3 knockdown reduced the β-catenin-N levels. Moreover, the suppression of EMT and Wnt/β-catenin signaling pathway in the GNL3 shRNA groups was partially restored by the LiCl treatment. Based on these results, GNL3 induces the nuclear translocation of β-catenin, thereby activating the Wnt/β-catenin signaling pathway and inducing EMT in colon cancer cells. β-catenin may be located downstream of GNL3, and we conclude that GNL3 promotes the EMT in colon cancer by activating the Wnt/β-catenin signaling pathway.
In conclusion, GNL3 promotes EMT by activating the Wnt/β-catenin signaling pathway and thus plays an important role in the invasion and metastasis of colon cancer. Our findings not only expand our understanding of the molecular mechanisms underlying GNL3 function and the invasion and metastasis of colon cancer but also suggest a potential target or strategy for treating colon cancer.
Acknowledgements
This study was supported by the National Key Clinical Specialties Construction Program of China ([No. 2012]649) and the National Natural Science Foundation of China (No. 81272753).
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Nieto MA. Epithelial-mesenchymal transitions in development and disease: Old views and new perspectives. Int J Dev Biol. 2009;53:1541–1547. doi: 10.1387/ijdb.072410mn. [DOI] [PubMed] [Google Scholar]
- 3.Yang Y, Liu Q, Zhang H, Zhao H, Mao R, Li Z, Ya S, Jia C, Bao Y. Silencing of GP73 inhibits invasion and metastasis via suppression of epithelial-mesenchymal transition in hepatocellular carcinoma. Oncol Rep. 2017;37:1182–1188. doi: 10.3892/or.2017.5351. [DOI] [PubMed] [Google Scholar]
- 4.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 5.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 6.Yang J, Weinberg RA. Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 7.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaudhury A, Hussey GS, Ray PS, Jin G, Fox PL, Howe PH. TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat Cell Biol. 2010;12:286–293. doi: 10.1038/ncb2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Timmerman LA, Grego-Bessa J, Raya A, Bertrán E, Pérez-Pomares JM, Díez J, Aranda S, Palomo S, McCormick F, Izpisúa-Belmonte JC, et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004;18:99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huber MA, Azoitei N, Baumann B, Grünert S, Sommer A, Pehamberger H, Kraut N, Beug H, Wirth T. NF-kappaB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J Clin Invest. 2004;114:569–581. doi: 10.1172/JCI200421358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leung CO, Mak WN, Kai AK, Chan KS, Lee TK, Ng IO, Lo RC. Sox9 confers stemness properties in hepatocellular carcinoma through Frizzled-7 mediated Wnt/β-catenin signaling. Oncotarget. 2016;7:29371–29386. doi: 10.18632/oncotarget.8835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan X, Sun X, Shi X, Wang H, Wu G, Jiang C, Yu D, Zhang W, Xue B, Ding Y. USP39 promotes colorectal cancer growth and metastasis through the Wnt/β-catenin pathway. Oncol Rep. 2017;37:2398–2404. doi: 10.3892/or.2017.5454. [DOI] [PubMed] [Google Scholar]
- 14.Mao Y, Xu J, Li Z, Zhang N, Yin H, Liu Z. The role of nuclear β-catenin accumulation in the Twist2-induced ovarian cancer EMT. PLoS One. 2013;8:e78200. doi: 10.1371/journal.pone.0078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Azzolin L, Panciera T, Soligo S, Enzo E, Bicciato S, Dupont S, Bresolin S, Frasson C, Basso G, Guzzardo V, et al. YAP/TAZ incorporation in the β-catenin destruction complex orchestrates the Wnt response. Cell. 2014;158:157–170. doi: 10.1016/j.cell.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 16.Stambolic V, Ruel L, Woodgett JR. Lithium inhibits glycogen synthase kinase-3 activity and mimics wingless signalling in intact cells. Curr Biol. 1996;6:1664–1668. doi: 10.1016/S0960-9822(02)70790-2. [DOI] [PubMed] [Google Scholar]
- 17.Tsai RY, McKay RD. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 2002;16:2991–3003. doi: 10.1101/gad.55671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai RY. Turning a new page on nucleostemin and self-renewal. J Cell Sci. 2014;127:3885–3891. doi: 10.1242/jcs.154054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu SJ, Cai ZW, Liu YJ, Dong MY, Sun LQ, Hu GF, Wei YY, Lao WD. Role of nucleostemin in growth regulation of gastric cancer, liver cancer and other malignancies. World J Gastroenterol. 2004;10:1246–1249. doi: 10.3748/wjg.v10.i9.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zia-Jahromi N, Hejazi SH, Panjepour M, Parivar K, Gharagozloo M. Comparison of nucleostemin gene expression in CD133+ and CD133− cell population in colon cancer cell line HT29. J Cancer Res Ther. 2014;10:68–72. doi: 10.4103/0973-1482.131375. [DOI] [PubMed] [Google Scholar]
- 21.Meng L, Lin T, Peng G, Hsu JK, Lee S, Lin SY, Tsai RY. Nucleostemin deletion reveals an essential mechanism that maintains the genomic stability of stem and progenitor cells. Proc Natl Acad Sci USA. 2013;110:11415–11420. doi: 10.1073/pnas.1301672110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okamoto N, Yasukawa M, Nguyen C, Kasim V, Maida Y, Possemato R, Shibata T, Ligon KL, Fukami K, Hahn WC, et al. Maintenance of tumor initiating cells of defined genetic composition by nucleostemin. Proc Natl Acad Sci USA. 2011;108:20388–20393. doi: 10.1073/pnas.1015171108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin T, Ibrahim W, Peng CY, Finegold MJ, Tsai RY. A novel role of nucleostemin in maintaining the genome integrity of dividing hepatocytes during mouse liver development and regeneration. Hepatology. 2013;58:2176–2187. doi: 10.1002/hep.26600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng L, Lin T, Tsai RY. Nucleoplasmic mobilization of nucleostemin stabilizes MDM2 and promotes G2-M progression and cell survival. J Cell Sci. 2008;121:4037–4046. doi: 10.1242/jcs.037952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai RY, McKay RD. A multistep, GTP-driven mechanism controlling the dynamic cycling of nucleostemin. J Cell Biol. 2005;168:179–184. doi: 10.1083/jcb.200409053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamashita M, Nitta E, Nagamatsu G, Ikushima YM, Hosokawa K, Arai F, Suda T. Nucleostemin is indispensable for the maintenance and genetic stability of hematopoietic stem cells. Biochem Biophys Res Commun. 2013;441:196–201. doi: 10.1016/j.bbrc.2013.10.032. [DOI] [PubMed] [Google Scholar]
- 27.Zhu Q, Yasumoto H, Tsai RY. Nucleostemin delays cellular senescence and negatively regulates TRF1 protein stability. Mol Cell Biol. 2006;26:9279–9290. doi: 10.1128/MCB.00724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang G, Zhang Q, Zhang Q, Yin L, Li S, Cheng K, Zhang Y, Xu H, Wu W. Expression of nucleostemin, epidermal growth factor and epidermal growth factor receptor in human esophageal squamous cell carcinoma tissues. J Cancer Res Clin Oncol. 2010;136:587–594. doi: 10.1007/s00432-009-0693-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu B, Hua L, Ni W, Wu M, Yan D, Chen Y, Lu C, Chen B, Wan C. Nucleostemin/GNL3 promotes nucleolar polyubiquitylation of p27(kip1) to drive hepatocellular carcinoma progression. Cancer Lett. 2017;388:220–229. doi: 10.1016/j.canlet.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 30.Li W, Wang Z, Zha L, Kong D, Liao G, Li H. HMGA2 regulates epithelial-mesenchymal transition and the acquisition of tumor stem cell properties through TWIST1 in gastric cancer. Oncol Rep. 2017;37:185–192. doi: 10.3892/or.2016.5255. [DOI] [PubMed] [Google Scholar]
- 31.Xu MZ, Yao TJ, Lee NPY, Ng IO, Chan YT, Zender L, Lowe SW, Poon RT, Luk JM. Yes-associated protein is an independent prognostic marker in hepatocellular carcinoma. Cancer. 2009;115:4576–4585. doi: 10.1002/cncr.24495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zha L, Zhang J, Tang W, Zhang N, He M, Guo Y, Wang Z. HMGA2 elicits EMT by activating the Wnt/β-catenin pathway in gastric cancer. Dig Dis Sci. 2013;58:724–733. doi: 10.1007/s10620-012-2399-6. [DOI] [PubMed] [Google Scholar]
- 33.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 34.Lee M, Williams KA, Hu Y, Andreas J, Patel SJ, Zhang S, Crawford NP. GNL3 and SKA3 are novel prostate cancer metastasis susceptibility genes. Clin Exp Metastasis. 2015;32:769–782. doi: 10.1007/s10585-015-9745-y. [DOI] [PubMed] [Google Scholar]
- 35.Xie Y, Wang B. Downregulation of TNFAIP2 suppresses proliferation and metastasis in esophageal squamous cell carcinoma through activation of the Wnt/β-catenin signaling pathway. Oncol Rep. 2017;37:2920–2928. doi: 10.3892/or.2017.5557. [DOI] [PubMed] [Google Scholar]
- 36.Bao Z, Wang Y, Yang L, Wang L, Zhu L, Ban N, Fan S, Chen W, Sun J, Shen C, et al. Nucleostemin promotes the proliferation of human glioma via Wnt/beta-Catenin pathway. Neuropathology. 2016;36:237–249. doi: 10.1111/neup.12265. [DOI] [PubMed] [Google Scholar]