Abstract
Livin, a member of the inhibitor of apoptosis protein (IAP) family, is expressed at a high level in lung adenocarcinoma and influences the progression of cancer, and its response to chemotherapy and radiotherapy. Aberrant microRNA (miRNA) expression has also been associated with cancer initiation and development. However, the clinical significance of Livin and its relationship with miRNAs in lung adenocarcinoma are still unclear. In the present study, the expression level of Livin in 90 pairs of lung adenocarcinoma and their adjacent tissues were detected by immunohistochemistry staining. Spearman correlation and Kaplan-Meier, univariate and multivariate analyses were applied to evaluate the correlation between the expression of Livin and clinical characteristics. With the integration of bioinformatics analysis and dual-luciferase reporter gene assays, we identified the miRNA that can target Livin mRNA. The functional effects of miRNA-mediated Livin knockdown were assessed by Cell Counting Kit-8 (CCK-8) and apoptosis assays, and cell cycle analysis. The present study revealed that Livin was upregulated in lung adenocarcinoma tissues and may be associated with the poor prognosis in lung adenocarcinoma patients. The overexpression of Livin is partly caused by the downregulation of miR-198. Further exploration revealed that miRNA-198-mediated silencing of Livin significantly inhibited cell growth and enhanced apoptosis of A549 cells, accompanied by marked upregulation of caspase-3. Finally, we observed that the miR-198 overexpression and Livin neutralization had similar effects on improving cisplatin chemosensitivity in A549 cells. Overall, these findings suggest that Livin has the potential to become a biomarker for predicting the prognosis of lung adenocarcinoma and may provide a promising strategy for assisting chemotherapy of lung adenocarcinoma through the miR-198/Livin/caspase-3 regulatory network.
Keywords: Livin, lung adenocarcinoma, prognosis, microRNAs, apoptosis, cell cycle, chemoresistance
Introduction
Lung cancer is the most common cancer worldwide, as well as the major cause of cancer-related deaths (1). Among all cases, ~80% are classified as non-small cell lung cancer (NSCLC), among which adenocarcinoma accounts for more than 50%. There have been several therapies that have been developed for lung adenocarcinoma; however, the mortality rate of lung adenocarcinoma has not largely improved (2). Therefore, it is of great importance to search for prognostic biomarkers and novel targets to increase the therapeutic effect.
Livin, an inhibitor of apoptosis protein (IAP), is selectively overexpressed in certain tumors including lung adenocarcinoma (3–5), and is correlated with resistance to chemotherapy (6,7) and radiotherapy (8). Such as other IAP family proteins, Livin is able to suppress apoptosis by binding certain caspases via a baculoviral IAP repeat (BIR) domain and a COOH terminal RING finger domain (9,10). Furthermore, various studies have demonstrated that Livin plays an important role in tumorigenesis, metastasis (11) and proliferation (6) of lung adenocarcinoma cell lines. However, whether Livin expression is associated with clinical characteristics and prognosis of lung adenocarcinoma remains elusive. Moreover, little information has been focused on the regulation of Livin.
MicroRNAs (miRNAs) are small, non-coding RNA molecules that post-transcriptionally regulate protein expression by complementary base pairing (12,13). miRNAs can contribute to tumorigenesis, cell migration, invasiveness and metastasis via various signaling pathways, which suggests their potential as therapeutic targets in cancer (14,15). With the development of genetic sequencing technology, differentially expressed miRNAs (DEMs) in lung cancer tissues vs. non-cancerous lung tissues have been detected (16). Nevertheless, the miRNAs that potentially target Livin RNA in lung adenocarcinoma are still unclear.
In the present study, the expression level of Livin in lung adenocarcinoma tissues vs. adjacent tissues was detected and the correlation between the expression of Livin and the clinical outcome was investigated. Bioinformatics analysis suggested that 2 out of 61 DEMs, namely miR-198 and miR-515-5p, were more likely to interact with Livin. Then, we identified the interaction between miRNA-198 and Livin using human lung adenocarcinoma cell line A549. Furthermore, we found that forced expression of miR-198 suppressed tumor cell proliferation and induced apoptosis of lung adenocarcinoma via the downregulation of Livin and the activation of caspase-3. Finally, the effects of the downregulation of Livin and the overexpression of miR-198 on A549 cell sensitivity to the cisplatin was detected using Cell Counting Kit-8 (CCK-8) assays.
Materials and methods
Tissue samples
The paraffin section of lung adenocarcinoma and their pericarcinous tissues (2 cm away from the tumor edge) were obtained from patients who had undergone surgery at the Department of Thoracic Surgery of The First Hospital of China Medical University from July 2004 to June 2009. The follow-up data of 90 cases were cut-off in August 2014, and the overall survival for the corresponding patients was calculated from the day of surgery to the day of death or to the last follow-up. The experiments were approved by the Ethics Committee of the First Hospital of China Medical University.
Immunohistochemistry staining
To assess the relationship between the expression of Livin and prognosis, formalin-fixed paraffin-embedded sections from specimens of lung adenocarcinoma and the adjacent tissues were evaluated by immunohistochemistry staining. The 4–5 µm paraffin-embedded slides were deparaffinized with xylene and rehydrated in graded alcohols. After high-pressure antigen retrieval, the activity of endogenous peroxidase was blocked with 3% hydrogen peroxide in ethanol for 10 min. Subsequently, the slides were washed with phosphate-buffered saline (PBS) and reacted with a rabbit primary antibody against Livin (dilution 1:500; Abcam, San Francisco, CA, USA) for 1 h. Then, incubation followed with an HRP-labeled anti-rabbit IgG (Nichirei, Tokyo, Japan) for 30 min at room temperature. The sections were stained with 3,3-diaminobenzidine (DAB) used as the chromogen. Finally the sections were counterstained with hematoxylin.
The degree of immunostaining was evaluated and scored by two independent pathologists blinded to the clinical information. Both cytoplasmic and nuclear immunostaining was considered to be positive staining. The proportion of positive cells was categorized as following: 5%, score 0; >6–25%, score 2; >26–50%, score 3; and >51%, score 4. The other score was set on the basis of the extent of staining: no staining as 1; light yellow staining as 2; medium brown staining as 3; brown staining as 4. The average of the two scores was interpreted as either low expression (average <4) or high expression (average ≥4).
Bioinformatics analysis
The information of miRNA microarray was downloaded from the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under series GSE18692 and GPL4717. The detailed experimental design of the microarray was previously elaborated (16). The known Homo sapien miRNAs registered in miRBase (release 21) were selected to perform analysis of miRNAs with differential expression using the Limma package. Statistically significant differences in DEMs between cancer and control groups were considered to exist at an adjusted p-value <0.05 and |logFC| >1. The statistical tests were carried out with the R program version 3.2.2 (http://www.r-project.org/). Two miRNA-target gene databases, TargetScan (release 6.2) and MicroCosm 5, were used to predict the miRNAs suppressing Livin.
Cell culture
The human lung adenocarcinoma cell line A549 was obtained from the Cell Bank of the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China) and was propagated in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) (both from Invitrogen, Carlsbad, CA, USA), 100 IU/ml penicillin and 100 µg/ml streptomycin, at 37°C in an atmosphere of 5% CO2.
Synthetic RNA oligonucleotides and transient transfection
All the miRNA inhibitors, miRNA mimics, nonsense sequence as miRNA negative control (NC), short hairpin RNA of Livin (sh-Livin) and short hairpin RNA negative control (NC-shRNA) were chemically synthesized by GenePharma (Shanghai, China). All of the transfections in the present study were transient, using JetPRIME reagent (PolyPlus Transfections SA, Illkirch, France) according to the manufacturers protocol. The cells were not harvested for the subsequent assays until 48 h after RNA oligonucleotide transfection.
RNA extraction and quantitative reverse transcription-PCR (qRT-PCR)
Total RNA was extracted from A549 using TRIzol reagent (Takara, Otsu, Japan). Complementary DNA (cDNA) of miR-198 and miR-515-5p were obtained with TransScript® miRNA First-Strand cDNA Synthesis (TransGen Biotech, Beijing, China). The transfection efficiency of miRNAs was assessed by quantitative real-time PCR (qPCR) with SYBR-Green qPCR Master Mix (Takara) on an ABI 7500 Fast System thermocycler (Applied Biosystems, Foster City, CA, USA). All the experiments were conducted in accordance with the manufacturer's instructions. Triplicate reactions were performed and the data were normalized to U6 and calculated with the 2−ΔΔCt method. The involved primers are described as follows: miR-198 forward, 5′-GCCAACTGGTCCAGAGGG-3′; miR-515-5p forward, 5′-TTCTCCAAAAGAAAGCACTTTCTG-3′; U6 forward, 5′-CGCTTCACGAATTTGCGTGTCAT-3′; the universal reverse primers of miRNAs from the kit.
Protein extraction and western blot analysis
Forty-eight hours after transfection, the cells were lysed using cell lysis buffer supplemented with protease inhibitors and phenylmethylsulfonyl fluoride (PMSF) (Beyotime, Zhejiang, China). Total proteins were extracted by centrifugation at 12,000 × g and 4°C for 20 min. The protein concentration was assessed using the BCA protein assay kit (Beyotime) and equal amounts of total proteins were separated in 10% SDS-PAGE and transferred onto nitrocellulose membranes. The membranes were blocked for 1 h with 5% non-fat milk powder in Tris-Buffered saline containing 0.5% Tween-20, and then incubated with a primary antibody overnight at 4°C, followed by washing and incubation with a secondary antibody for 2 h at room temperature. Finally, the membranes were detected by enhanced chemiluminescence (ECL) plus western blot detection reagents (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The antibodies (Thermo Fisher Scientific, Inc.) were used according to the manufacturers instructions, and were as follows: primary antibodies against GAPDH (ab9485), Livin (ab97350), caspase-3 (ab32351) and the HRP-conjugated secondary antibody (ab6721).
Dual-luciferase assays
To confirm whether miR-198 or miR-515-5p can interact with Livin mRNA, pGLO Dual-Luciferase miRNA Target Expression Vector pmirGLO-wt-Livin (wild-type) and pGLO-mut-Livin (mutant type) of miR-198, pGLO-wt-Livin (wild-type) and pGLO-mut-Livin (mutant type) of miR-515-5p were constructed by GeneChem (Shanghai, China). The pGLO Dual-Luciferase miRNA Target Expression Vector plasmids (mutant or wild-type) and the internal control Renilla luciferase plasmid pRL-TK were cotransfected with miRNAs (miR-198 and miR-515-5p mimics, or NC) with a ratio of 20:1, using JetPRIME reagent into A549. The luciferase activity of firefly and Renilla luciferase was assessed 24 h after cotransfection, using the Dual-Luciferase® Reporter Assay System (Promega, Madison, WI, USA), complying with the manufacturer's protocol. To aid comparisons, values for cells with the NC + mut-Livin 3′-UTR group were set equal to 1.
Cell proliferation assays
Cell proliferation was detected using CCK-8 (KeyGen Biotech, Jiangsu, China). The A549 cells in the logarithmic phase with or without transfection were seeded in 96-well plates at a density of 5×103 cells/well. CCK-8 (10 µl/well) was added at various time-points (0, 24, 48 and 72 h) and incubated at 37°C for 2 h. The absorbance of each well at 450 nm was measured using a microplate reader (Bio-Rad, Hercules CA, USA) to assess the number of viable cells. To decrease random errors, all of the procedures were replicated at least three times.
Cell cycle distribution and apoptosis analysis
Flow cytometric analyses were utilized to assess cell apoptosis and cell cycle distribution using a flow cytometer (Bio-Rad). The A549 cells were synchronized before the cell cycle assays. After 48 h of transfection, the cells, which were cultured in 6-well plates and prepared for apoptosis examination and cell cycle distribution, were harvested by trypsin and detected using an Annexin V-FITC/PI apoptosis detection kit and a Cell Cycle Detection kit (both from KeyGen Biotech) respectively, according to the manufacturers recommendations. Each experiment was performed at least in triplicate.
Drug sensitivity assays
Before each experiment, the chemotherapeutic drug cisplatin (F.H. Faulding & Co, Mulgrave Victoria, Australia) was dissolved in 0.9% sodium chloride, and then diluted with fresh medium. The concentration of cisplatin was set as 0.01, 0.1 and 1 µmol/l. The cell proliferation rate 48 h after transfection and drug treatment was detected by CCK-8 assay kits. Furthermore, the cell proliferation rate treated with 1 µmol/l of cisplatin was assessed every 24 h for a period of 72 h.
Statistical analysis
The results of immunohistochemical staining were analyzed using Chi-square test, Kaplan-Meier analysis, Spearman rank correlation and Cox regression analysis. The result of the Kaplan-Meier survival analysis was compared with the log-rank test. Other experiments were analyzed using Student's t-test and expressed as the mean ± standard deviation (SD). Statistical analyses were processed using SPSS 23.0 software (SPSS, Inc., Chicago, IL, USA) and p<0.05 was considered as statistically significant.
Results
Livin is overexpressed in lung adenocarcinoma tissues
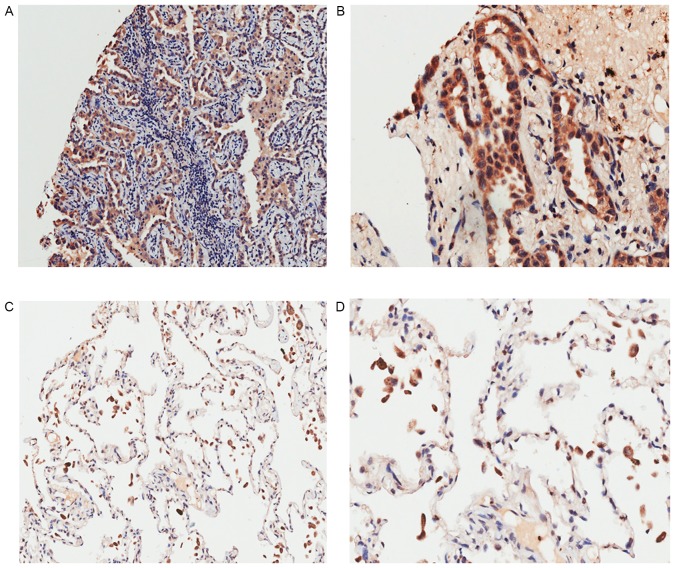
The expression of Livin in cancerous and pericancerous tissues is shown in Fig. 1 and Table I. According to the immunohistochemical results, Livin was mainly expressed in the cytoplasm and notably overexpressed in cancerous tissues (p<0.05). Spearman's rank correlation analysis was performed to display the correlations between the expression of Livin and the clinicopathological characteristics such as sex, age, tumor-node-metastasis (TNM) stage and pathological grade (Table II). Notably, it was determined that the expression of Livin in patients who suffer from lymph node metastasis was elevated (p<0.05). Survival analysis revealed that lung adenocarcinoma patients with lower expression of Livin had a higher survival rate than those with higher Livin expression (Fig. 2; p<0.05). Moreover, univariate analyses indicated that Livin expression, along with T, N and TNM stage and grade, all play important roles in the prognosis of lung adenocarcinoma. Furthermore, these variables with statistical significance in the univariate analyses were included in a multivariate regression analysis, and Livin expression and N stage were identified as significant independent prognostic factors of lung adenocarcinoma prognosis (Table III).
Figure 1.
Immunohistochemistry staining of Livin protein. High expression of Livin in lung adenocarcinoma tissues: (A) magnification, ×100 and (B) magnification, ×200; low expression of Livin in adjacent tissues: (C) magnification, ×100 and (D) magnification, ×200.
Table I.
Differential expression of Livin in cancerous and pericancerous tissues.
| Livin expression | |||||
|---|---|---|---|---|---|
| Tissues | n | High(%) | Low(%) | Chi-square value | P-value |
| Cancerous | 90 | 52 (57.8) | 38 (42.2) | 34.545 | <0.0001a |
| Pericarcinous | 90 | 14 (15.6) | 76 (84.4) | ||
Statistically significant (p<0.05).
Table II.
Correlation between Livin expression and clinicopathological characteristics.
| Livin expression | |||||
|---|---|---|---|---|---|
| Variables | High | Low | Total | rs | P-value |
| Sex | −0.059 | 0.579 | |||
| Female | 25 | 16 | 41 | ||
| Male | 27 | 22 | 49 | ||
| Age (years) | −0.104 | 0.327 | |||
| ≤60 | 26 | 15 | 41 | ||
| >60 | 26 | 23 | 49 | ||
| Grade | 0.161 | 0.129 | |||
| 1 | 4 | 8 | 12 | ||
| 2 | 43 | 27 | 70 | ||
| 3 | 5 | 3 | 8 | ||
| T stage | 0.087 | 0.413 | |||
| T1 | 9 | 8 | 17 | ||
| T2 | 28 | 22 | 50 | ||
| T3 | 11 | 6 | 17 | ||
| T4 | 4 | 2 | 6 | ||
| N stage | 0.239 | 0.023a | |||
| N0 | 19 | 23 | 42 | ||
| N1 | 16 | 8 | 24 | ||
| N2 | 12 | 6 | 18 | ||
| N3 | 5 | 1 | 6 | ||
| M stage | 0.033 | 0.755 | |||
| M0 | 50 | 37 | 87 | ||
| M1 | 2 | 1 | 3 | ||
| TNM stage | 0.175 | 0.099 | |||
| I | 16 | 15 | 31 | ||
| II | 11 | 13 | 24 | ||
| III | 23 | 9 | 32 | ||
| IV | 2 | 1 | 3 | ||
Statistically significant (p<0.05). TNM, tumor-node-metastasis.
Figure 2.
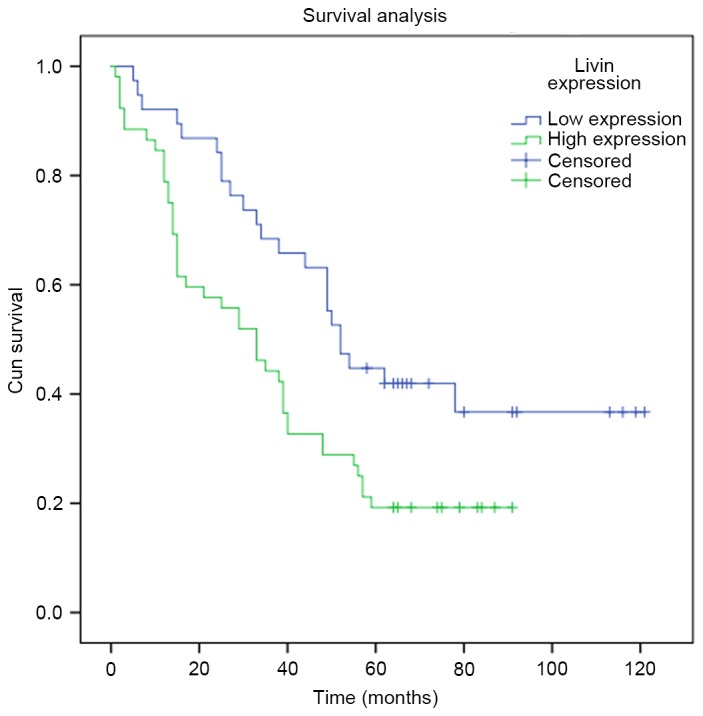
Kaplan-Meier analysis of the overall survival (months) in 90 lung adenocarcinoma patients with different Livin expression (log-rank test; p<0.05).
Table III.
Univariate and multivariate analyses of the factors correlated with overall survival of lung adenocarcinoma patients.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Livin expression | 0.466 | 0.281–0.773 | 0.003a | 0.568 | 0.339–0.950 | 0.031a |
| Sex | 1.393 | 0.866–2.243 | 0.172 | |||
| Age | 1.107 | 0.690–1.776 | 0.674 | |||
| T stage | 1.541 | 1.135–2.092 | 0.006a | 1.170 | 0.799–1.715 | 0.419 |
| N stage | 1.788 | 1.404–2.276 | <0.001a | 1.565 | 1.107–2.211 | 0.011a |
| M stage | 0.700 | 0.171–2.863 | 0.620 | |||
| TNM stage | 1.562 | 1.223–1.995 | <0.001a | 1.031 | 0.679–1.564 | 0.886 |
| Grade | 1.969 | 1.218–3.182 | 0.006a | 1.697 | 0.953–3.022 | 0.072 |
Statistically significant (p<0.05). HR, hazard ratio; CI, confidence interval; TNM, tumor-node-metastasis.
Prediction of putative miRNAs targeting Livin
Forty-five putative miRNAs generated by two databases, were identified as the possible miRNAs targeting Livin (Table IV). Among the 45 miRNAs, only two of them, miR-198 and miR-515-5p were significantly differentially expressed in lung adenocarcinoma tissues vs. non-cancerous lung tissues according to the miRNA microarray analysis (Fig. 3).
Table IV.
The possible miRNAs targeting Livin.
| hsa-miR-940 | hsa-miR-323-5p |
| hsa-miR-548b-3p | hsa-miR-24 |
| hsa-miR-198 | hsa-miR-324-5p |
| hsa-miR-25* | hsa-miR-148b* |
| hsa-miR-214 | hsa-miR-652 |
| hsa-miR-455-3p | hsa-miR-525-5p |
| hsa-miR-516a-5p | hsa-miR-582-5p |
| hsa-miR-296-3p | hsa-miR-183 |
| hsa-miR-141 | hsa-miR-139-5p |
| hsa-miR-188-5p | hsa-miR-486-3p |
| hsa-miR-515-3p | hsa-miR-551a |
| hsa-miR-552 | hsa-miR-598 |
| hsa-miR-572 | hsa-miR-601 |
| hsa-miR-593* | hsa-miR-541 |
| hsa-miR-596 | hsa-miR-296-5p |
| hsa-miR-149* | hsa-miR-432* |
| hsa-miR-548d-3p | hsa-miR-615-3p |
| hsa-miR-942 | hsa-miR-654-5p |
| hsa-miR-874 | hsa-miR-660 |
| hsa-miR-222* | hsa-miR-7 |
| hsa-miR-519e | hsa-miR-512-3p |
| hsa-miR-423-5p | hsa-miR-589* |
| hsa-miR-662 |
Figure 3.
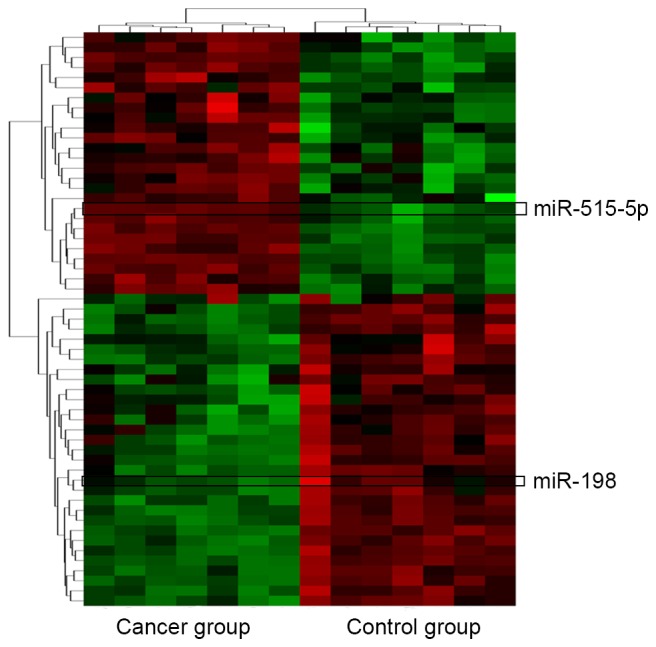
Bioinformatics analysis. Cluster analysis for the differentially expressed miRNAs between lung adenocarcinoma tissues (cancer group) and adjacent tissues (control group). Sixty-one significant differentially expressed miRNAs (|logFC| >1, adjusted p<0.05) are highlighted in green (downregulation) and red (upregulation).
Livin is a direct target of miR-198
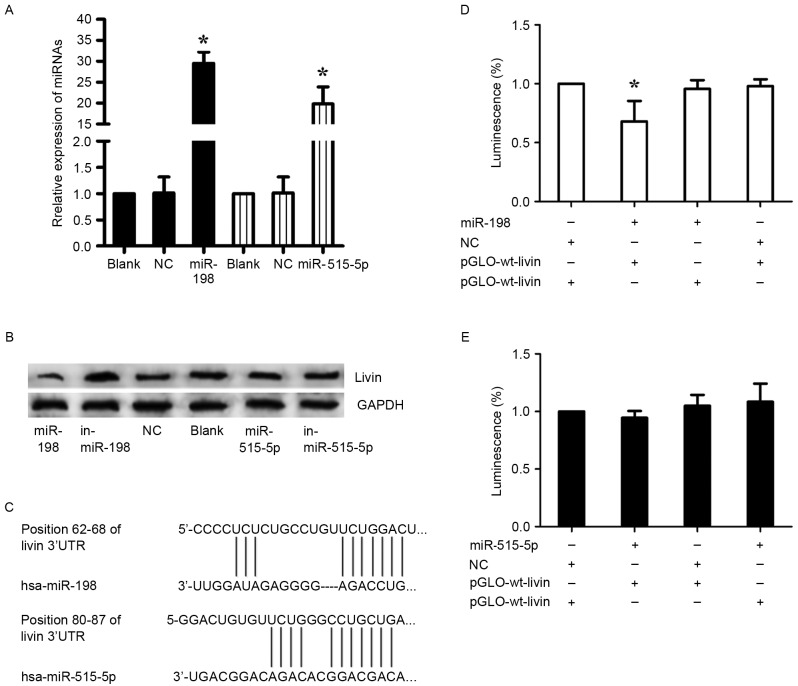
The aberrant expression of miR-198 and miR-515-5p in lung adenocarcinoma was reported as previously described (17,18). To confirm the prediction, miR-198 mimics and inhibitor (in-miR-198), miR-515-5p mimics and inhibitor (in-miR-515-5p), nonsense sequence as NC were transfected into A549 cells. Before the transfection, the transfection efficiency was detected by RT-qPCR (Fig. 4A). In consequence, we found that miR-198 expression revealed a completely contrary trend to Livin expression, whereas this phenomenon was not observed in the regulation of miR-515-5p, inferring that miR-198 may be a specific miRNA targeting Livin in A549 cells (Fig. 4B). Furthermore, luciferase reporter assays were employed to illuminate the interaction of miR-198 or miR-515-5p with Livin mRNA, and the predicted binding sites are shown in Fig. 4C. Either pGLO-wt-Livin or pGLO-mut-Livin of miR-198 and pRL-TK were cotransfected into A549 cells with miR-198 or NC. As with miR-198, the luciferase reporter assays of miR-515-5p were performed. A significant decrease of the relative luciferase activity was observed between the miR-198 + pGLO-wt-Livin compared to the other groups (p<0.05) (Fig. 4D). In stark contrast, there was no significant change in the relative luminescence intensities in the miR-515-5p + luciferase reporter gene groups (Fig. 4E). Therefore, it is miR-198, but not miR-515-5p that bound directly to the 3′-UTR of Livin mRNA and negatively regulated the expression of Livin in A549 cells.
Figure 4.
Direct regulation of Livin by miR-198 in A549 cells. (A) The expression of miR-198 and miR-515-5p was evaluated by qRT-PCR in A549 cells 48 h after transfection to confirm the transfection efficiency. U6 was used as an internal control; *p<0.05. (B) The effects of miRNA mimics and inhibitors on Livin expression in A549 cells was detected using western blotting 48 h after transfection. GAPDH was used as a loading control. (C) The putative miR-198 and miR-515-5p binding sites in the 3′-UTR of Livin mRNA. (D and E) Dual-luciferase reporter assays using vectors encoding the putative miR-198 and miR-515-5p target sites in the Livin 3′-UTR for both the wild-type and mutant type. Normalized data were calculated as Renilla/firefly luciferase activity. In the miR-515-5p group, no significant change was observed. In the miR-198 group, the relative luciferase activity was clearly lower in cells cotransfected with the miR-198 mimics and pGLO-wt-Livin; *p<0.05 (the values for the pGLO-mut-Livin + NC group compared with pRL-TK vector were set equal to 1).
Impact of miR-198 restoration on the proliferation, apoptosis and cell cycle of A549 cells
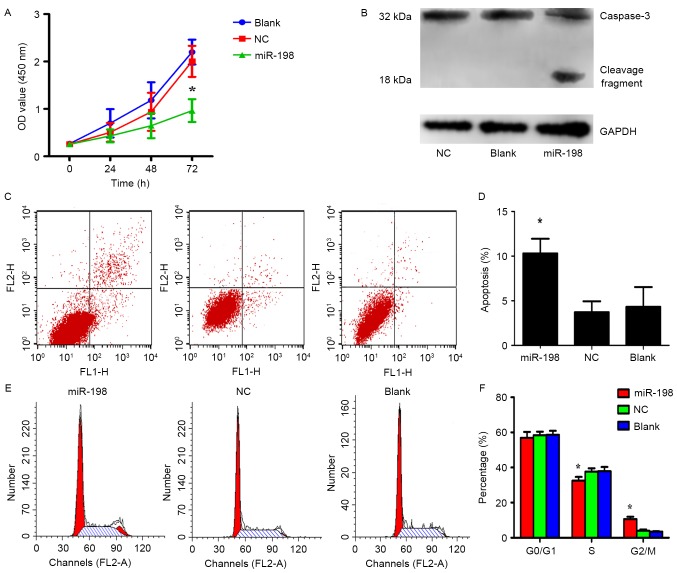
The effects of overexpressing or silencing Livin in A549 cells have been previously demonstrated in detail (10,11). To investigate the role of miR-198 in A549 cells, miR-198 and NC were separately transfected into 6-well plates and the CCK-8 assays were performed to detect cell proliferation. The results revealed that the overexpression of miR-198 greatly inhibited cell proliferation in comparison to the control group, respectively (Fig. 5A; p<0.05). In order to elucidate the mechanism of miR-198 in the inhibition of cell proliferation, apoptosis and cell cycle assays were performed with miR-198 or NC transfected A549 cells. Flow cytometric analysis indicated that the percentage of apoptotic cells in the miRNA-198 group was significantly higher than that in both control groups (Fig. 5C and D; p<0.05). Furthermore, we found that the miR-198 group had a lower percentage of S phase cells and a higher ratio of G2/M phase cells than the NC or blank groups (Fig. 5E and F; p<0.05). This result indicated that miR-198 induced cell cycle arrest in the G2/M phase. Nevertheless, proliferation which is determined by the S + G2/M phases appeared to not be affected, possibly due to the synchronization of the cells before the experiments. From this point of view, the diminished cell growth was attributed to apoptosis due to synchronization of the cells prior to the experiment. These data demonstrated that low expression of miR-198 exerts tumor-promoting effects in A549 cells, therefore their re-expression may benefit epigenetic cancer therapy, consistent with the outcomes of silencing Livin.
Figure 5.
Effect of miR-198 on the A549 cell line. (A) Cell proliferation was determined using CCK-8 assays after transfection with miR-198. The results revealed that miR-198 inhibited cell growth compared with the control groups; *p<0.05. (B) Caspase activation was revealed after miR-198 transfection for 48 h. (C and D) Double staining with Annexin V-FITC and PI was used to assess apoptosis after miR-198 transfection for 48 h. The results revealed that the apoptotic rate in the miR-198 group was higher than that of the control groups; *p<0.05. (E and F) Flow cytometric analysis of the cell cycle revealed that the miR-198 group had a lower proportion of S phase cells and a higher proportion of G2/M phase cells than in the control groups, which means that upregulation of miR-198 may cause cell cycle arrest, *p<0.05.
Livin has been demonstrated to be capable of inhibiting apoptosis by binding to caspase family members directly with its BIR domain, and then suppressing caspase activities (9). In order to further ascertain whether miR-198 induces apoptosis through downregulation of Livin, miR-198 was overexpressed in A549 cells and the expression of caspase-3, a member of the caspase family, was observed. Western blot analysis revealed that the activation of caspase-3 significantly increased in the miR-198 groups, implying that miR-198 may inhibit cell proliferation by decreasing the expression of Livin which activates caspase-3 directly (Fig. 5B).
In addition, we determined the correlation between Livin and lymph node status, which had been reported by Lin et al. Thus, we attempted to explore whether miR-198 was related to cell migration and invasion. Preliminary experiments revealed that miR-198 did not markedly impact cell migration and invasion, therefore, we finally chose to investigate proliferation and apoptosis, which may be primarily associated with miR-198.
The effect of miR-198 overexpression and silencing of Livin on cisplatin chemosensitivity in A549 cells
Livin RNA interference has been identified to enhance the chemosensitivity to chemotherapeutic drugs in NSCLC cells (6,7). To investigate whether miR-198 ectopic expression could increase the sensitivity of A549 cells to cisplatin, sh-Livin or NC-shRNA were cotransfected with miR-198 or NC into A549 cells. The proliferation rate of these cells was assessed by CCK-8 assays. As shown in Fig. 6A and B, the proliferation rate in dose- and time-dependent manners over a 72-h treatment period were observed. On the one hand, from the dose-survival bar chart, it was evident that cisplatin had a dose-dependent suppressive effect on cell growth. Both miR-198 and sh-Livin may enhance the chemosensitivity of A549 cells to cisplatin significantly, and have a similar efficiency (Fig. 6A). On the other hand, we found that the proliferation rate was significantly slower in sh-Livin + NC and NC-shRNA + miR-198 group by the end of the 72-h treatment time (Fig. 6B). Therefore, the combination of chemotherapy and silencing of Livin or miR-198 ectopic expression may enhance the treatment of cisplatin-resistant lung adenocarcinoma patients.
Figure 6.
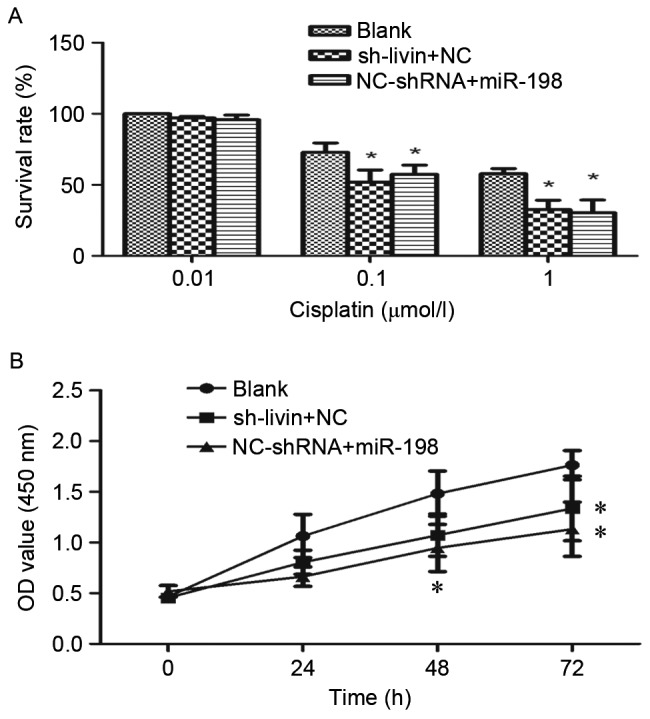
Influence of miR-198 overexpression and silencing of Livin on A549 cell sensitivity to cisplatin. (A) Evidently the survival rate was dose-dependent, and both miRNA-198 and Livin-shRNA enhanced the sensitivity to cisplatin. (B) The proliferation curve of NC-shRNA + miRNA-198 and Livin shRNA + NC groups with the treatment of 1 µmol/l of cisplatin. The proliferation rate of the blank group was significantly higher than that of the NC-shRNA + miRNA-198 and Livin-shRNA + NC groups. However, A549 cells cotransfected with Livin-shRNA and NC had a similar effect on the sensitivity to cisplatin as miR-198 with NC-shRNA (*p<0.05).
Discussion
To the best of our knowledge, this is the first study elucidating the relationship between Livin and the clinicopathological features in lung adenocarcinoma patients. Livin is a member of the IAPs family, and is also known as baculoviral IAP repeat-containing protein 7 (BIRC7). A high level of Livin has been found in various tumors, including NSCLC (3,5,9) and is considered of great importance in the regulation of cell proliferation, metastasis, the cell cycle and apoptosis in lung adenocarcinoma via the JNKI signaling pathway, the VEGF and MMP pathways or the activity of the caspase family (11,19,20). Notably, our results revealed that a high expression of Livin in cancer tissues was related with the lung adenocarcinoma patient metastasis. Moreover, it has been demonstrated that silencing of Livin in lung adenocarcinoma cell lines enhances the sensitivity of chemotherapy or radiotherapy (6–8,21). Livin takes part in the networks involved in the regulation of cell functions and influences the curative effect of lung adenocarcinoma through cross-communication, making it promising for targeted therapy. In the present study, immunohistochemical staining analysis revealed that the expression of Livin may be associated with the prognosis and lymph node metastasis of lung adenocarcinoma. From this perspective, Livin may function as a therapeutic agent as well as a prognostic indicator for lung adenocarcinoma. However, the number of patients in the present study may be insufficient to draw an definite conclusion. In consideration of this limitation, more samples from lung adenocarcinoma patients at various stages and their follow-up data should be collected. However, it may be necessary to perform in vivo experiments and further investigate the underlying mechanisms of Livin.
Nonetheless, to date, only Yuan et al has reported that the transcriptional signaling pathway of Livin is regulated by β-catenin/TCF signaling, and there is no more research concerning the upstream regulation of Livin in lung adenocarcinoma (22). To explore the molecular mechanism of the aberrant expression of Livin, we employed databases and the results of microRNA (miRNA) microarray to predict the miRNAs involved with Livin regulation. The data indicated that miR-198 and miR-515-5p may be miRNAs that can influence the expression of Livin. The lung adenocarcinoma cell line A549 was selected for the experiments due to the fact that it has the highest expression of Livin compared with other lung adenocarcinoma cell lines (3). The present study, demonstrated that Livin is a target of miR-198 and low expression of miR-198 may be responsible for the upregulation of Livin in lung adenocarcinoma. Furthermore, it is worth noting that the effect of miR-198 upregulation is not only consistent with that of Livin downregulation, but can also activate caspase-3, which is one of the key factors in apoptotic pathways and can be inhibited by Livin. Moreover, we found a new phenomenon where miR-198 can contribute to chemosensitivity by neutralizing Livin. In fact, miR-198 has been found to be closely related with lung carcinoma (17,23), and many oncogenes have been identified as targets of miR-198, such as MCL1 (24), ROCK1 (25), FUT8 (26) and FGFR1 (23).
In general, we demonstrated that Livin, an apoptotic pathway suppressor, can enhance the growth of lung adenocarcinoma, which may be partly due to the downregulation of miR-198. Livin has the potential to be a biomarker for predicting the prognosis of lung adenocarcinoma and focusing on the miR-198/Livin/caspase-3 regulatory network may become a promising strategy for drug resistant lung adenocarcinoma therapy.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant no. 81372876), and the Liaoning Province Natural Science Foundation (grant no. 2013021041).
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Diaz-Garcia CV, Agudo-Lopez A, Perez C, López-Martín JA, Rodríguez-Peralto JL, de Castro J, Cortijo A, Martínez-Villanueva M, Iglesias L, García-Carbonero R, et al. DICER1, DROSHA and miRNAs in patients with non-small cell lung cancer: Implications for outcomes and histologic classification. Carcinogenesis. 2013;34:1031–1038. doi: 10.1093/carcin/bgt022. [DOI] [PubMed] [Google Scholar]
- 3.Hariu H, Hirohashi Y, Torigoe T, Asanuma H, Hariu M, Tamura Y, Aketa K, Nabeta C, Nakanishi K, Kamiguchi K, et al. Aberrant expression and potency as a cancer immunotherapy target of inhibitor of apoptosis protein family, Livin/ML-IAP in lung cancer. Clin Cancer Res. 2005;11:1000–1009. [PubMed] [Google Scholar]
- 4.Li J, Chen P, Li XQ, Bao QL, Dai CH, Ge LP. Elevated levels of survivin and livin mRNA in bronchial aspirates as markers to support the diagnosis of lung cancer. Int J Cancer. 2013;132:1098–1104. doi: 10.1002/ijc.27757. [DOI] [PubMed] [Google Scholar]
- 5.Tanabe H, Yagihashi A, Tsuji N, Shijubo Y, Abe S, Watanabe N. Expression of survivin mRNA and livin mRNA in non-small-cell lung cancer. Lung Cancer. 2004;46:299–304. doi: 10.1016/j.lungcan.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Zhuang L, Shen LD, Li K, Yang RX, Zhang QY, Chen Y, Gao CL, Dong C, Bi Q, Tao JN, et al. Inhibition of livin expression suppresses cell proliferation and enhances chemosensitivity to cisplatin in human lung adenocarcinoma cells. Mol Med Rep. 2015;12:547–552. doi: 10.3892/mmr.2015.3372. [DOI] [PubMed] [Google Scholar]
- 7.Yuan D, Liu L, Xu H, Gu D. The effects on cell growth and chemosensitivity by livin RNAi in non-small cell lung cancer. Mol Cell Biochem. 2009;320:133–140. doi: 10.1007/s11010-008-9915-1. [DOI] [PubMed] [Google Scholar]
- 8.Sun JG, Liao RX, Zhang SX, Duan YZ, Zhuo WL, Wang XX, Wang ZX, Li DZ, Chen ZT. Role of inhibitor of apoptosis protein Livin in radiation resistance in nonsmall cell lung cancer. Cancer Biother Radiopharm. 2011;26:585–592. doi: 10.1089/cbr.2011.0962. [DOI] [PubMed] [Google Scholar]
- 9.Kasof GM, Gomes BC. Livin, a novel inhibitor of apoptosis protein family member. J Biol Chem. 2001;276:3238–3246. doi: 10.1074/jbc.M003670200. [DOI] [PubMed] [Google Scholar]
- 10.Crnkovic-Mertens I, Muley T, Meister M, Hartenstein B, Semzow J, Butz K, Hoppe-Seyler F. The anti-apoptotic livin gene is an important determinant for the apoptotic resistance of non-small cell lung cancer cells. Lung Cancer. 2006;54:135–142. doi: 10.1016/j.lungcan.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 11.Lin X, Li HR, Lin XF, Yu ME, Tu XW, Hua ZD, Lin M, Xu NL, Han LL, Chen YS. Silencing of Livin inhibits tumorigenesis and metastasis via VEGF and MMPs pathway in lung cancer. Int J Oncol. 2015;47:657–667. doi: 10.3892/ijo.2015.3058. [DOI] [PubMed] [Google Scholar]
- 12.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 13.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 14.Kim VN, Nam JW. Genomics of microRNA. Trends in genetics. TIG. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puisségur MP, Mazure NM, Bertero T, Pradelli L, Grosso S, Robbe-Sermesant K, Maurin T, Lebrigand K, Cardinaud B, Hofman V, et al. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 2011;18:465–478. doi: 10.1038/cdd.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu S, Zhang G, Li P, Chen S, Zhang F, Li J, Jiang C, Chen X, Wang Y, Du Y, et al. miR-198 targets SHMT1 to inhibit cell proliferation and enhance cell apoptosis in lung adenocarcinoma. Tumour Biol. 2016;37:5193–5202. doi: 10.1007/s13277-015-4369-z. [DOI] [PubMed] [Google Scholar]
- 18.Pardo OE, Castellano L, Munro CE, Hu Y, Mauri F, Krell J, Lara R, Pinho FG, Choudhury T, Frampton AE, et al. miR-515-5p controls cancer cell migration through MARK4 regulation. EMBO Rep. 2016;17:570–584. doi: 10.15252/embr.201540970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen YS, Li HR, Lin M, Chen G, Xie BS, Xu NL, Lin LF. Livin abrogates apoptosis of SPC-A1 cell by regulating JNKI signaling pathway. Mol Biol Rep. 2010;37:2241–2247. doi: 10.1007/s11033-009-9711-3. [DOI] [PubMed] [Google Scholar]
- 20.Dubrez-Daloz L, Dupoux A, Cartier J. IAPs: More than just inhibitors of apoptosis proteins. Cell Cycle. 2008;7:1036–1046. doi: 10.4161/cc.7.8.5783. [DOI] [PubMed] [Google Scholar]
- 21.Sun J, Liao R, Chen Z, Wang Z, Zhang Q, Hu Y. Study on enhancing sensitivity of SPC-A1 cells to chemotherapy by Livin isoform-specific gene silencing. Zhongguo Fei Ai Za Zhi. 2007;10:461–465. doi: 10.3779/j.issn.1009-3419.2007.06.03. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 22.Yuan D, Liu L, Gu D. Transcriptional regulation of livin by beta-catenin/TCF signaling in human lung cancer cell lines. Mol Cell Biochem. 2007;306:171–178. doi: 10.1007/s11010-007-9567-6. [DOI] [PubMed] [Google Scholar]
- 23.Yang J, Zhao H, Xin Y, Fan L. MicroRNA-198 inhibits proliferation and induces apoptosis of lung cancer cells via targeting FGFR1. J Cell Biochem. 2014;115:987–995. doi: 10.1002/jcb.24742. [DOI] [PubMed] [Google Scholar]
- 24.Gigante M, Pontrelli P, Herr W, Gigante M, D'Avenia M, Zaza G, Cavalcanti E, Accetturo M, Lucarelli G, Carrieri G, et al. miR-29b and miR-198 overexpression in CD8+ T cells of renal cell carcinoma patients down-modulates JAK3 and MCL-1 leading to immune dysfunction. J Transl Med. 2016;14:84. doi: 10.1186/s12967-016-0841-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S, Zhao Y, Wang L. MicroRNA-198 inhibited tumorous behaviors of human osteosarcoma through directly targeting ROCK1. Biochem Biophys Res Commun. 2016;472:557–565. doi: 10.1016/j.bbrc.2016.03.040. [DOI] [PubMed] [Google Scholar]
- 26.Wang M, Wang J, Kong X, Chen H, Wang Y, Qin M, Lin Y, Chen H, Xu J, Hong J, et al. MiR-198 represses tumor growth and metastasis in colorectal cancer by targeting fucosyl transferase 8. Sci Rep. 2014;4:6145. doi: 10.1038/srep06145. [DOI] [PMC free article] [PubMed] [Google Scholar]