Abstract
MicroRNAs (miRNAs) are a class of small, non-coding RNAs that regulate gene expression and are involved in cell biological processes. The aberrant expression of miR-195 has been found in various types of human cancer. However, the effect of miR-195 on the initiation and development of laryngeal squamous cell carcinoma (LSCC) remains to be elucidated. Accordingly, in the present study, we detected the expression level of miR-195 in the LSCC and the normal tissues and found that miR-195 were significantly downregulated in the LSCC tissues. Gain-of-function or loss-of-function studies including cell proliferation, wound healing assay, Transwell assay, cell cycle and apoptosis assays were performed to investigate the biological function of miR-195. Luciferase reporter assay and the rescue study confirmed that DCUN1D1 was a target of miR-195. Furthermore, DCUN1D1 expression levels were found to be upregulated in laryngeal tissues and to have a negative correlation with miR-195. We also found that both miR-195 and DCUN1D1 siRNAs can inhibit cell invasion possibly through downregulating Matrix metalloproteinase-2 (MMP-2) and Matrix metalloproteinase-9 (MMP-9) at the post-transcriptional level, which can be attenuated by restoring the expression of DCUN1D1. In summary, these data suggest that low expression of miR-195 contributes to the poor prognosis of LSCC and miR-195 regulates the proliferation and invasion ability of LSCC cells in vitro. miR-195 may suppress growth and invasion of LSCC cells possibly through targeting DCUN1D1, which would provide a candidate target for cancer therapy.
Keywords: miR-195, DCUN1D1, laryngeal squamous cell carcinoma, invasion, MMP-2
Introduction
Laryngeal squamous cell carcinoma (LSCC) is one of the most common types of tumor in the head and neck which accounts for approximately 25% of the total (1,2). During the last decade, great advance has been made in the study on laryngeal carcinoma. The treatment for laryngeal carcinoma is the combination of primary surgery with non-surgical radiotherapy or chemoradiotherapy. However, the 5-year survival rate of laryngeal carcinoma (60%) has not been improved. To identify novel biomarkers that indicate the early stages of laryngeal carcinoma, or specific biomarkers for various individuals, is urgently required for the early detection of laryngeal carcinoma and the development of individualized therapies.
MicroRNAs (miRNAs) are a class of small, non-coding RNAs that regulate gene expression through targeting the 3′-untranslated region of target mRNA (3–5). Increasing evidence indicates that miRNAs have significant roles in various cell biological processes including cell proliferation, cell cycle, cell apoptosis, cell migration and invasion (6,7). In the carcinogenesis and development of laryngeal carcinoma, a number of miRNAs have been reported to be dysregulated such as miR-135, miR-144, miR-205, miR-296-5p, miR-206, miR-375, miR-101 and miR-150 (8–13). Recent studies suggest that miR-195 may act as a suppressor gene in various types of cancers (14–17), indicating that miR-195 may be critical in cancer development. However, the biological roles of miR-195 in laryngeal carcinoma still remain to be established.
In the present study, we predicted DCUN1D1 is the target gene of miR-195 by bioinformatic prediction and investigated the effect of miR-195 on the biological function of LSCC cells and identified the correlation between miR-195 and its target DCUN1D1.
Materials and methods
Patient tissues
The LSCC tissues and adjacent normal tissues were obtained from 122 patients at the Second Hospital of Tianjin Medical University (Tianjin, China) following surgical resection, and stored immediately in liquid nitrogen. The study was approved by the ethics committee of the Second Hospital of Tianjin Medical University and written informed consent was obtained from all patients.
RNA isolation and qRT-PCR
Total RNA was extracted from the collected fresh frozen tissues and cells using the standard TRIzol method. For the detection of miRNA expression, cDNA were prepared by reversely transcription of RNA using one Step PrimeScript miRNA cDNA Synthesis kit (Takara, Dalian, China). A SYBR® Premix Ex Taq™ II kit (Takara) was used for qRT-PCR in an ABI 7500 Real-time PCR system (Applied Biosystems, Foster City, CA, USA). Relative gene expression was calculated using 2−∆∆Ct method. U6 and GAPDH were selected as internal controls for miRNA and mRNA, respectively. miR-195 mimics Mature sequence 5′-UAGCAGCACAGAAAUAUUGGC-3′; DCUN1D1 (forward: 5′-ACTCGATCCAGCCAGCATTA-3′, reverse: 5′-TGTTTGGAGAACTCGCACTG-3′); U6 (forward: 5′-CTCGCTTCGGCAGCACA-3′, reverse: 5′-AACGCTTCACGAATTTGCGT-3′); GAPDH (forward: 5′-CGCTCTCTGCTCCTCCTGTT-3′, reverse: 5′-CCATGGTGTCTGAGCGATGT-3′).
Cell culture and transfections
The human laryngeal cancer cell line (TU-177) and human normal bronchial epithelial cell line (16HBE) were obtained from ATCC (Manassas, VA, USA) and maintained in RPMI supplemented with 10% fetal bovine serum (HyClone, Logan, UT, USA). All the cell lines were cultured in a humidified incubator with 5% CO2 at 37°C. Transfections were done using Lipofectamine 2000/Lipofectamine LTX-Plus reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. miR-195 mimics (forward: 5′-UAGCAGCACAGAAAUAUUGGC-3′, reverse: 5′-CAAUAUUUCUGUGCUGCUAUU-3′); Control (forward: 5′-UUCUCCGAACGUGUCACGU-3′, reverse: 5′-ACGUGACACGUUCGGAGAA-3′). miR-195 inhibitor (5′-GCCAAUAUUUCUGUGCUGCUA-3′), Inhibitor control (5′-CAAUAUUUCUGUGCUGCUAUU-3′).
Oligonucleotide and plasmid transfection
The miRNA mimics and corresponding negative control that were used for transient transfection were designed and synthesized by GenePharma China. The DCUN1D1 cDNA containing the coding sequence was cloned by PCR, and the PCR product was cloned into the pcDNA3.1 vector (Invitrogen). The insert was confirmed by DNA sequencing. The siRNA against DCUN1D1 (DCUN1D1 siRNA) was synthesized by GenePharma China. The sequence of DCUN1D1 siRNA was as follows (forward: 5′-GCAGATGACATGTCTAATT-3′, reverse: 5′-AAUUAGACAUGUCAUCUGC-3′). TU-177 cells were transfected with miRNA mimic, siRNA, and pCDNA3.1-DCUN1D1 using Lipofectamine 2000/Lipofectamine LTX-Plus Reagent (Invitrogen) according to the manufacturer's protocol.
Cell viability assay
Cell viability was determined by thiazolyl blue tetrazolium bromide (MTT) assay. TU-177 cells were seeded into 96-well plates at a density of 4×104 cells/ml and incubated for 24, 48 and 72 h. At each time point, 0.5% MTT solution were respectively added to TU-177 cells, and incubated for 4 h at 37°C. Then the cell supernatants were discarded and 150 µl DMSO were added to dissolve the formazan. The optical density of each group was measured using a microplate reader (Bio-Rad Laboratories, Hercules, CA, USA) at a wavelength of 490 nm.
Colony formation assay
Cells were seeded on 6-well plates at a density of 400 cells per well then cultured at 37°C in a incubator with 5% CO2 for 14 days. The cells were fixed with methanol for 20 min at room temperature and stained with crystal violet solution (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). The colonies of more than 50 cells were counted.
Wound healing assay
Cells were seeded in 6-well plates at a density of 1×105 cells/well and cultured to confluence. Cell monolayer was scraped with a 200 µl pipette tip to generate wounds and then washed with DMEM media twice to remove cell debris. The cells were grown in DMEM supplemented with 3% FBS for additional 48 h. The cell motility was measured by photographing five random fields at the time of 0 and 24 h after wounding.
Transwell invasion assays
TU-177 cells were seeded into the upper chamber of a Transwell insert (Millipore Corp., Billerica, MA, USA) and incubated in an incubator with 5% CO2 at 37°C. Lower chamber contains serum-loaded medium as attractant. The invasive assay was performed using Transwell inserts coated with Matrigel (BD Biosciences, San Diego, CA, USA). After incubation for 24 h, the cells on upper surfaces of the Transwell chambers were removed with cotton swabs, and the invaded cells were fixed with ethanol and stained using 0.05% crystal violet solution. Images were captured under a wide-field microscope (Nikon, Melville, NY, USA).
Flow cytometry assay
For the apoptosis assay, cells were analyzed by flow cytometry using a FITC-Annexin V/PI kit according to the manufacturer's instructions (BD Pharmingen, Franklin Lakes, NJ, USA). In detail, TU-177 cells were seeded into 6-well plates at a density of 4×104 cells/ml and incubated for 24, 48 and 72 h. At each time point, TU-177 cells were collected and resuspended with the PBS buffer. Then cells were incubated with 5 µl FITC-Annexin V and 5 µl PI in the dark at room temperature. The apoptosis rates of cells were analyzed using a FACS Verse flow cytometry (BD Biosciences) according to the fluorescence signals.
Luciferase activity assay
The 3′UTR sequence of DCUN1D1 was amplified and subcloned into the pmirGLO luciferase reporter vector (Promega, Sunnyvale, CA, USA). HEK293T cells (5×104) were seeded in 24-well plates and incubated for 24 h. Thereafter, cells were co-transfected with wild-type (WT) or mutant (Mut) 3′UTR vectors and miR-195 mimics using Lipofectamine 2000. After 48 h, the cells were assayed for luciferase activity using the Dual-Luciferase Reporter Assay System (Promega) following the manufacturer's protocol. The firefly luciferase activities were normalized to Renilla luciferase activity.
Immunohistochemistry assay
Sections (4 µm) were cut from formalin-fixed paraffin-embedded tissue. After dewaxing and rehydration sections were heated in a microwave oven for 2 min in citrate buffer at middle power. Subsequently, sections were subjected to blockade of endogenous peroxidase activity by incubating in 3% H2O2 solution for 25 min. The slides were then incubated with the rabbit polyclonal antibody against DCUN1D1 (Cell Signaling Technology, Inc., Danvers, MA, USA) at 4°C overnight. After sequential incubation with a secondary antibody (Abcam, Cambridge, UK) at room temperature for 1 h, sections were washed and stained with 3,3′-diaminobenzidine, and visualized under a microscope (Nikon, Japan). Brown staining in cytoplasm was considered as positive immunoreactivity, and was evaluated as percentage staining over the whole preparation. DCUN1D1 protein level was classified semiquantitatively combining the proportion and intensity of positively stained immunoreactive cells (18). The percentage of positive-staining cells was scored as follows: 0 (<5% positive cells), 1 (5–50% positive cells), and 2 (>50% positive cells). Staining intensity was scored as follows: 0 (no staining or only weak staining); 1 (moderate staining); and 2 (strong staining). The sum of the staining intensity score and the percentage score was used to define the DCUN1D1 protein expression levels: 0–2, low expression and 3–4, high expression.
Protein analysis
Cells were lysed in RIPA buffer (Beyotime Institute of Biotechnology, Haimen, China) and the protein extracts were quantified using a BCA-protein quantification kit (Beyotime Institute of Biotechnology). Proteins were separated by SDS-PAGE, transferred to PVDF membranes and then blocked with 10% non-fat milk for 1.5 h and incubated overnight at 4°C with primary antibodies against DCUN1D1, caspase-3 (Cell Signaling Technology, Inc.), MMP-9, MMP-2, GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA, USA) at dilutions specified by the manufacturer. After being washed with TBST, the membranes were incubated with the HRP-conjugated secondary antibodies for 120 min at room temperature. Then proteins were detected by chemiluminescence using the ECL kit (Beyotime Institute of Biotechnology) and imaged with a digital chemiscope (Qinxiang, Shanghai, China). Band intensity quantification was calculated by ImageJ software, and GAPDH was used as a loading control.
Statistical analysis
All statistical analyses were carried out using the SPSS 17.0 statistical software package. Experimental data are presented as mean ± SD. An independent Student's two-tailed t-test and one-way ANOVA were performed to compare the difference. The expression of miR-195 and DCUN1D1 related with various clinicopathological characteristics were assessed using the χ2 test or Fisher's exact test. The Spearman's correlation was calculated between the expression levels of miR-195 and DCUN1D1 in LSCC. Survival curves were carried out by the Kaplan-Meier method and Cox regression analysis was used for the univariate and multivariate analysis. P-values <0.05 were considered statistically significant.
Results
miR-195 was downregulated in LSCC and cell lines, and was associated with cancer progression
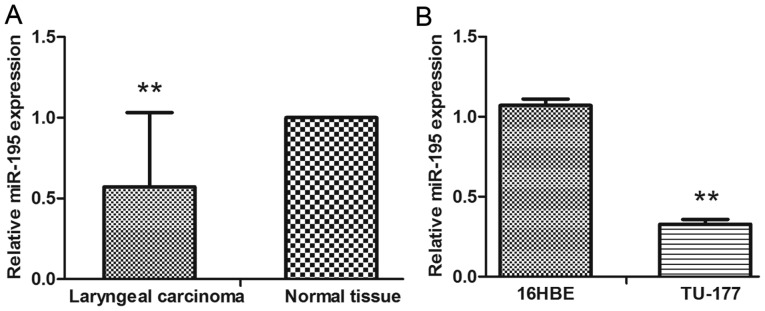
The expression of miR-195 was evaluated using qRT-PCR. We found that the expression levels of miR-195 in LSCC were significantly decreased when compared with the adjacent normal tissue and normal human bronchial epithelial cells (16HBE) (Fig. 1A and B). To determine whether the expression of miR-195 was associated with clinicopathological characteristics of LSCC patients, the data shown in Table I revealed that miR-195 downregulation was frequently found in LSCC with high T stage (P<0.001), N+ (P<0.05), and high clinical stage (P<0.001). Decreased expression of miR-195 predicted poor prognosis of LSCC.
Figure 1.
The abberent expression of miR-195 in LSCC tissues and cell lines. (A) Quantitative real-time PCR was used to evaluate the miR-195 expression in 122 paired LSCC and the normal tissues. (B) Quantitative real-time PCR was used to evaluate the miR-195 expression in LSCC cell line TU-177, and human normal bronchial epithelial cell line 16HBE. Data are expressed as the mean value ± SD. **P<0.01.
Table I.
Correlation between miR-195, DCUN1D1 expression and clinicopathological parameter in 122 LSCC patients.
| Expression of miR-195 | Expression of DCUN1D1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Parameters | n | Low (n, %) | High (n, %) | χ2 | P-value | Low (n, %) | High (n, %) | χ2 | P-value |
| Age (years) | |||||||||
| ≤60 | 69 | 35 (50.7) | 34 (49.3) | 0.033 | 0.855 | 33 (47.8) | 36 (52.2) | 0.078 | 0.780 |
| >60 | 53 | 26 (49.1) | 27 (50.9) | 24 (45.3) | 29 (54.7) | ||||
| Sex | |||||||||
| Female | 42 | 22 (52.4) | 20 (47.6) | 0.145 | 0.703 | 19 (45.2) | 23 (54.8) | 0.057 | 0.812 |
| Male | 80 | 39 (48.8) | 41 (51.2) | 38 (47.5) | 42 (52.5) | ||||
| Primary site | |||||||||
| Glottis | 61 | 31 (50.8) | 30 (49.2) | 0.871 | 0.647 | 22 (36.1) | 39 (63.9) | 6.917 | 0.031 |
| Supraglottis | 42 | 19 (45.2) | 23 (54.8) | 22 (52.4) | 20 (47.6) | ||||
| Subglottis | 19 | 11 (57.9) | 8 (42.1) | 13 (68.4) | 6 (31.6) | ||||
| Differentiation level | |||||||||
| High | 77 | 38 (49.4) | 39 (50.6) | 0.035 | 0.851 | 33 (42.9) | 44 (57.1) | 1.252 | 0.263 |
| Medium-low | 45 | 23 (51.1) | 22 (48.9) | 24 (53.3) | 21 (46.7) | ||||
| T stage | |||||||||
| T1+T2 | 30 | 1 (3.3) | 29 (96.7) | <0.001a | 20 (66.7) | 10 (33.3) | 6.358 | 0.012 | |
| T3+T4 | 92 | 60 (65.2) | 32 (34.8) | 37 (40.2) | 55 (59.8) | ||||
| Lymph node metastases | |||||||||
| N0 | 81 | 35 (43.2) | 46 (56.8) | 4.445 | 0.035 | 54 (66.7) | 27 (33.3) | <0.001a | |
| N+ | 41 | 26 (63.4) | 15 (36.6) | 3 (7.3) | 38 (92.7) | ||||
| Distant metastasis | |||||||||
| M0 | 118 | 57 (48.3) | 61 (51.7) | 0.059a | 57 (48.3) | 61 (51.7) | 0.077a | ||
| M1 | 4 | 4 (100.0) | 0 (0.0) | 0 (0.0) | 4 (100.0) | ||||
| Clinical stage | |||||||||
| I+II | 23 | 1 (4.3) | 22 (95.7) | <0.001a | 18 (78.3) | 5 (21.7) | 11.326 | 0.001 | |
| III+IV | 99 | 60 (60.6) | 39 (39.4) | 39 (39.4) | 60 (60.6) | ||||
| Preoperative smoking | |||||||||
| No | 23 | 15 (65.2) | 8 (34.8) | 2.625 | 0.105 | 8 (34.8) | 15 (65.2) | 1.623 | 0.203 |
| Yes | 99 | 46 (46.5) | 53 (53.5) | 49 (49.5) | 50 (50.5) | ||||
Fisher's exact test.
Upregulated DCUN1D1 expression was associated with cancer progression and was inversely correlated with miR-195 expression in LSCC
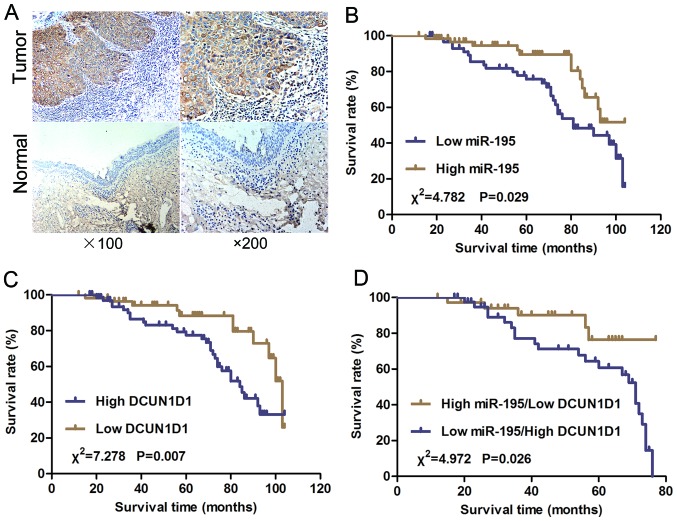
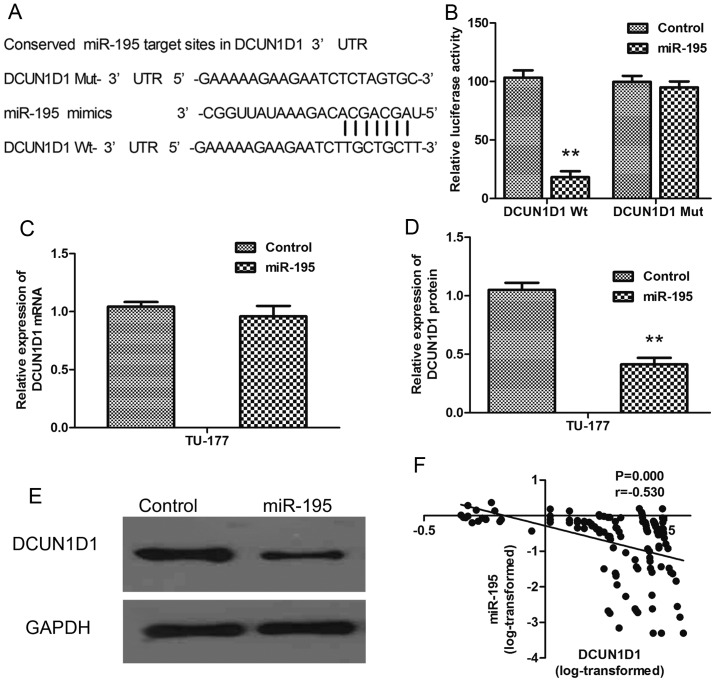
We examined the expression of the DCUN1D1 protein in paraffin sections of LSCC and nonmalignant samples using immunohistochemistry. The DCUN1D1 staining in LSCC adjacent nonmalignant tissues was generally of reduced intensity (Fig. 2A). To understand the roles of DCUN1D1 in LSCC progression, we analyzed the association of the expression of DCUN1D1 with clinicopathological characteristics of LSCC patients. As shown in Table I, DCUN1D1 upregulation was frequently found in LSCC with high T stage (P=0.012), N+ (P<0.001), and high clinical stage (P=0.001). Increased expression of DCUN1D1 predicted poor prognosis of LSCC. Spearman's correlation analysis showed expression levels of DCUN1D1 were inversely correlated with miR-195 expression levels in LSCC tissues (Table II, Fig. 6F; r= −0.530, P<0.001). Therefore, high DCUN1D1 mRNA expression in LSCC tissues was correlated with decreased miR-195 expression.
Figure 2.
Expression of DCUN1D1 protein and Kaplan-Meier survival curve. (A) Immunohistochemistry staining of DCUN1D1 in LSCC and the normal tissues. (B) Kaplan-Meier curves with log-rank tests show that the overall survival of patients with high miR-195 expression (T/N fold change >0.436) was longer than those with low miR-195 expression (T/N fold change <0.436). The median miR-195 expression level (T/N=0.436) in the LSCC samples was chosen as the cut-off point. (C) Kaplan-Meier curves with log-rank tests show that the overall survival of patients with low DCUN1D1 expression was statistically significantly longer than those with high DCUN1D1 expression. (D) The overall survival of patients with high miR-195 expression accompanied by low expression of DCUN1D1 was longer than those with low miR-195 expression accompanied by high DCUN1D1 expression.
Table II.
Correlation between miR-195 and DCUN1D1.
| Expression of DCUN1D1 | ||||||
|---|---|---|---|---|---|---|
| Expression of miR-195 | n | Low (n, %) | High (n, %) | rs | χ2 | P-value |
| Low (n, %) | 61 | 20 (32.8) | 41 (67.2) | −0.279 | 9.516 | 0.002 |
| High (n, %) | 61 | 37 (60.7) | 24 (39.3) | |||
Figure 6.
miR-195 directly targets DCUN1D1. (A) Sequence alignment of miR-195 and seed match region in the 3′-UTR of DCUN1D1 as well as the mutated 3′-UTR of DCUN1D1 are shown. (B) The 3′-UTR sequence of DCUN1D1 was amplified and subcloned into the pmirGLO luciferase reporter vector. HEK293T cells were co-transfected with wild-type (WT) or mutant (Mut) 3′UTR vectors and miR-195 mimics using Lipofectamine 2000. After 48 h, the cells were assayed for luciferase activity using the Dual-Luciferase Reporter Assay System. The firefly luciferase activities were normalized to Renilla luciferase activity. (C) Quantitative real-time PCR was used to evaluate the DCUN1D1 mRNA expression in TU-177 cells transfected with miR-195 or control. (D and E) Western blot was used to evaluate the protein expression of DCUN1D in TU-177 cells transfected with miR-195 or control. Data are expressed as the mean value ± SD. **P<0.01. (F) DCUN1D1 mRNA was inversely associated with miR-195 in 122 pairs of LSCC tissues using linear regression models.
Decreased miR-195 expression and increased DCUN1D1 expression were associated with poor overall survival in LSCC patients
We further analyzed the correlation of miR-195 expression and postoperative overall survival of LSCC patients. We used the mean fold change (T/N=0.436) in miR-195 expression chosen as the cut-off point and the patients were divided into a high miR-195 expression group (T/N fold change >0.436) and a low miR-195 expression group (T/N fold change <0.436). The overall survival of patients with high miR-195 expression was longer than those with low miR-195 expression (Fig. 2B; P=0.029). The overall survival of patients with low DCUN1D1 expression was statistically significantly longer than those with high DCUN1D1 expression (Fig. 2C; P=0.007). Furthermore, the overall survival of patients with high miR-195 expression accompanied by low expression of DCUN1D1 was longer than those with low miR-195 expression accompanied by high DCUN1D1 expression (Fig. 2D; P=0.026). A univariate and multivariable Cox proportional hazards analysis were performed. The final multivariable Cox regression model showed that the expression of miR-195 (P=0.041, relative risk=0.358) and the expression of DCUN1D1 (P=0.021, relative risk=4.253) were associated with poor prognosis according to overall survival in LSCC (Table III), suggesting that miR-195 and DCUN1D1 might be used as independent prognostic factors for LSCC.
Table III.
Prognostic value of miR-195 expression and DCUN1D1 expression for the overall survival in univariate and multivariate analyses by Cox regression.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Covariant | Exp (B) | 95% CI | P-value | Exp (B) | 95% CI | P-value |
| miR-195 | 0.476 | 0.241–0.942 | 0.033 | 0.358 | 0.134–0.959 | 0.041 |
| DCUN1D1 | 2.588 | 1.264–5.297 | 0.009 | 4.253 | 1.243–14.557 | 0.021 |
| Differentiation level | 2.014 | 1.065–3.810 | 0.031 | |||
| T stage | 3.344 | 1.184–9.449 | 0.023 | |||
| Lymph node metastasis | 2.809 | 1.474–5.350 | 0.002 | |||
| Distant metastasis | 3.071 | 0.920–10.253 | 0.068 | |||
| Clinical stage | 3.586 | 0.859–14.976 | 0.080 | |||
miR-195 inhibits the proliferation and colony formation ability of LSCC cells
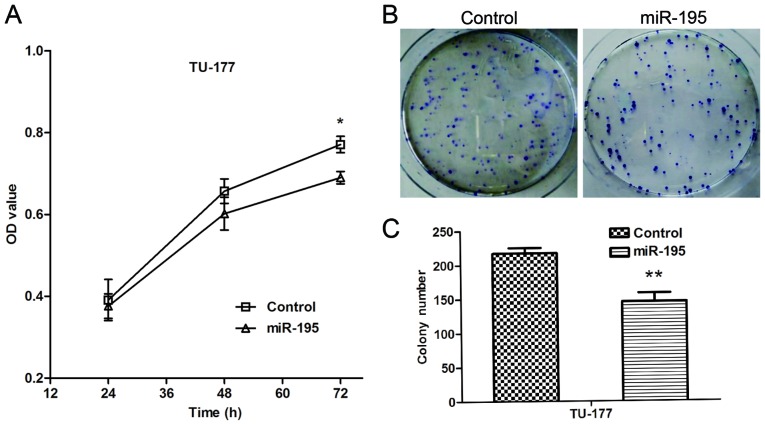
To investigate the biological functions of miR-195 in LSCC cells, we elevated the expression of miR-195 in the TU-177 cells. The MTT assay was performed to evaluate the effect of miR-195 on the proliferation of TU-177 cells. The results indicated that ectopic expression of miR-195 significantly reduced proliferation of TU-177 cells (Fig. 3A). The colony formation assay was performed to determine forced expression of miR-195 on the proliferation of TU-177 cells. The results indicated miR-195 significantly inhibited colony formation ability of TU-177 cells (Fig. 3B and C). Taken together, these data show that miR-195 may function as a negative regulator on the proliferation of LSCC cells.
Figure 3.
miR-195 inhibits the proliferation and colony formation of TU-177 cells in vitro. (A) The growth curves of cells were examined by MTT assays. (B and C) Effects of miR-195 overexpression on the colony formation of TU-177 cells were determined by colony formation assay. Data are expressed as the mean value ± SD. *P<0.05, **P<0.01.
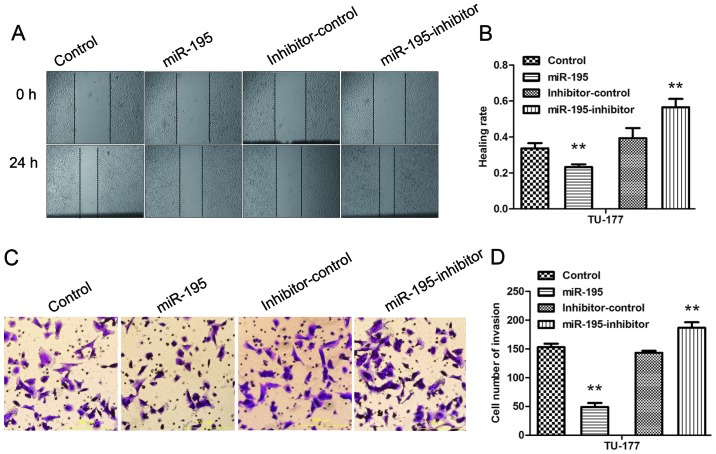
miR-195 inhibits the migration and invasion ability of LSCC cells, and inhibition of miR-195 partially reverses the effects
The wound healing assay was performed to evaluate the effect of miR-195 on the migration ability of TU-177 cells. The results indicated that transfection of mimics or inhibitors for miR-195 could significantly inhibit or promote migration ability of TU-177 cells (Fig. 4A and B). The Transwell assay was conducted to investigate effect of miR-195 on the invasion ability of TU-177 cells. The results showed that transfection of mimics or inhibitors for miR-195 could significantly reduce or increase the invasion ability of TU-177 cells (Fig. 4C and D). Thus, we confirmed that miR-195 inhibitor could reverse the effects of over-expression of miR-195 on the migration and invasion abilities of LSCC cells.
Figure 4.
miR-195 inhibits the migration and invasion of TU-177 cells in vitro. (A and B) Effects of miR-195 overexpression on the migration of TU-177 cells were determined by wound healing assay. (C and D) Transwell assay was performed to evaluate the invasion of TU-177 cells. Data are expressed as the mean value ± SD. **P<0.01.
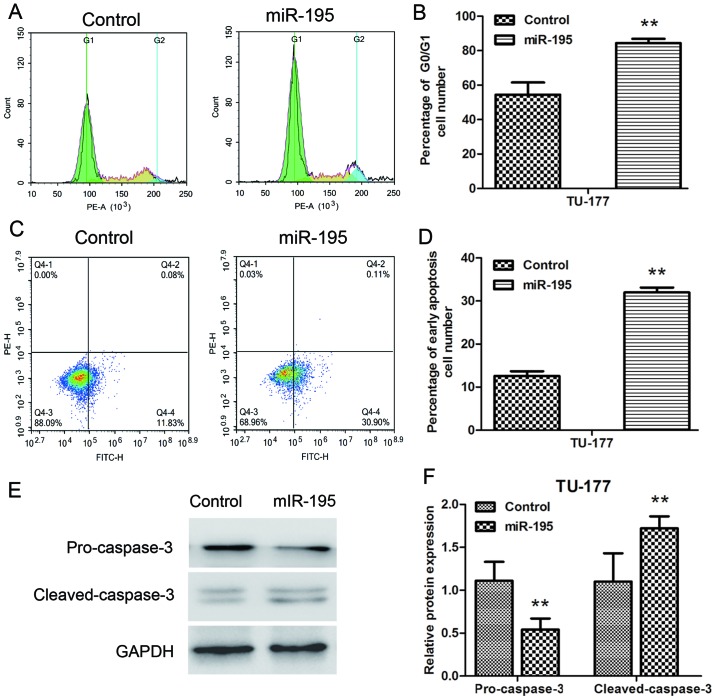
miR-195 induces G1 cell cycle arrest and promoted apoptosis in LSCC cells
We further analyzed the effect of miR-195 on cell cycle and apoptosis of TU-177 cells by flow cytometry. The cell cycle distribution results indicated that miR-195 overexpression significantly blocked the G1 phase of TU-177 cells (Fig. 5A and B). Moreover, miR-195 overexpression significantly increased early apoptotic cell number of TU-177 cells (Fig. 5C and D). In addition, the effect of miR-195 on endogenous expression levels of pro-caspase-3 and cleaved-caspase-3 were assessed by western blotting. Transfection of miR-195 significantly reduced the expression of pro-caspase-3 and upregulated the expression of cleaved-caspase-3 at the protein level in TU-177 cells (Fig. 5E and F). These results indicated that overexpression of miR-195 mainly inhibits the growth of TU-177 cells by inducing G1 phase cell cycle arrest and apoptosis.
Figure 5.
miR-195 promotes apoptosis and induces cell cycle arrest in TU-177 cells in vitro. (A and B) Flow cytometry and PI staining were used to detect the cell cycle distribution of TU-177 cells. (C and D) Flow cytometry and Annexin V/PI staining were used to detect apoptosis of TU-177 cells. (E and F) Endogenous pro-caspase-3 protein levels are reduced and cleaved caspase-3 protein levels are upregulated in TU-177 cells transfected with miR-195 mimics. Data are expressed as the mean value ± SD. **P<0.01.
DCUN1D1 is directly targeted by miR-195
In this study, DCUN1D1 was predicted to be a potential target of miR-195 after computational analysis using three microRNA target gene prediction software (TargetScan, miRDB, DIANAmT) (Fig. 6A). To confirm whether the 3′UTR of DCUN1D1 was a functional target of miR-195, we constructed luciferase reporter plasmids containing pGL3-DCUN1D1-3′UTR Wt and pGL3-DCUN1D1-3′UTR Mut and carried out luciferase reporter assay. The results indicated that significant decreases in luciferase activities were observed in the presence of miR-195 in the HEK293T cells cotranfected with pGL3-DCUN1D1-3′UTR Wt (P<0.01), but not with pGL3-DCUN1D1-3′UTR Mut (Fig. 6B). Furthermore, qRT-PCR and western blot analysis showed a notable downregulation of DCUN1D1 expression treated with overexpression of miR-195 in TU-177 cells at the post-transcriptional level. (Fig. 6C-E). Above all, DCUN1D1 was the direct target of miR-195.
Knockdown of DCUN1D1 inhibits the proliferation of LSCC cells
The MTT assay was performed to evaluate the effect of DCUN1D1 siRNA on the proliferation of TU-177 cells. The results indicated that ectopic expression of DCUN1D1 siRNA significantly reduced proliferation of LSCC cells (Fig. 7).
Figure 7.
Knockdown of DCUN1D1 inhibits the proliferation of TU-177 cells in vitro. The growth curves of cells were examined by MTT assays. Data are expressed as the mean value ± SD. *P<0.05.
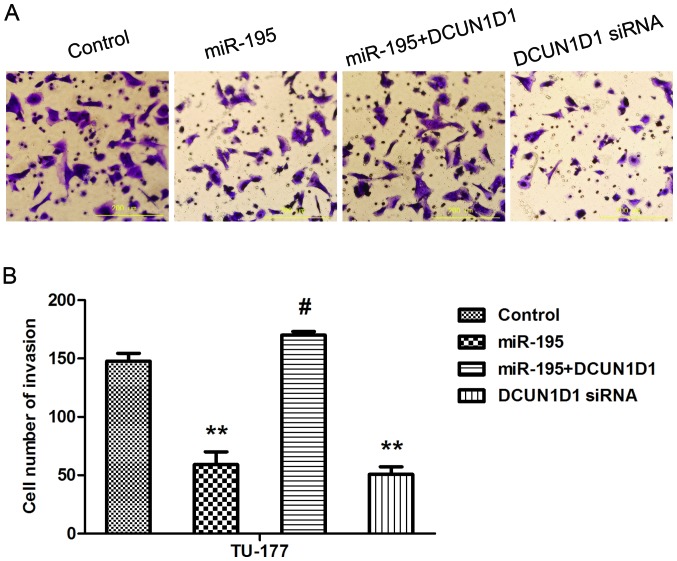
Overexpression of DCUN1D1 restores invasive capacity of LSCC cells and knockdown of DCUN1D1 simulates the effect of miR-195
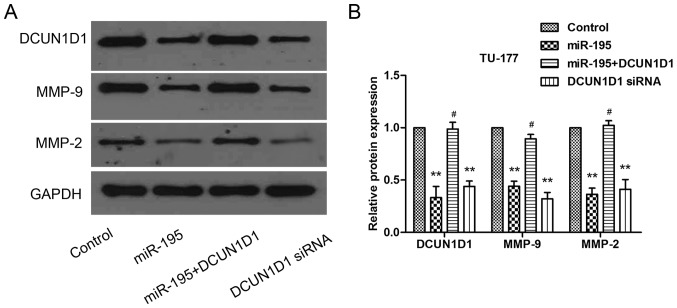
To confirm that miR-195 inhibited the metastasis of LSCC cells by targeting DCUN1D1, we next performed cell invasion assays in TU-177 cells that were co-transfected with miR-195 and pcDNA3.1-DCUN1D1, which lacked the 3′UTR of DCUN1D1 and therefore cannot be restrained by miR-195. The results showed that forced DCUN1D1 expression promoted the invasive capability of TU-177 cells. Importantly, this restoration of DCUN1D1, partially, but significantly, rescued the invasion capability of miR-195-transfected cells. Knockdown of DCUN1D1 exerted similar effect as miR-195 (Fig. 8A and B). Besides, the DCUN1D1, MMP-9 and MMP-2 protein expression levels confirmed by western blot analysis were consistent with Transwell assay (Fig. 9A and B). Taken together, these data suggested that the effects of miR-195 on cell invasion were partly regulated by DCUN1D1.
Figure 8.
Overexpression of DCUN1D1 restores TU-177 cell invasive capacity and knockdown of DCUN1D1 simulates the effect of miR-195. (A and B) Transwell assay was performed to evaluate the invasion of TU-177 cells transfected with miR-195, DCUN1D1 siRNA or co-transfected with miR-195 and pcDNA3.1/DCUN1D1 in TU-177 cells. Invasion of TU-177 cells into the scratched area was monitored at the indicated time. Overexpression of DCUN1D1 can restore invasion capacity and DCUN1D1 knockdown can mimic the suppression of invasion activity induced by miR-195 in TU-177 cells. Data are expressed as the mean value ± SD. **P<0.01 (control vs. miR-195; control vs. DCUN1D1 siRNA), #P<0.01 (miR-195 vs. miR-195+DCUN1D1).
Figure 9.
Western blot analysis was performed to confirm the re-expression of DCUN1D1 and other related proteins. (A and B) Western blot was used to evaluate the expression levels of DCUN1D1, MMP-9 and MMP-2 proteins transfected with miR-195, DCUN1D1 siRNA or co-transfected with miR-195 and pcDNA3.1-DCUN1D1 in TU-177 cells. Data are expressed as the mean value ± SD. *P<0.01 (control vs. miR-195; control vs. DCUN1D1 siRNA), #P<0.01 (miR-195 vs. miR-195+DCUN1D1). #P<0.01; **P<0.05.
Discussion
Extensive studies have demonstrated that miRNAs play important roles in tumor initiation, progression, and metastasis particularly through regulating their corresponding target genes (19,20). One of them, miR-195 has been regarded as a tumor suppressor in various cancers including non-small cell lung cancer (NSCLC) (21), colorectal cancer (22), gastric cancer (23), hepatocellular carcinoma (24), and prostate cancer (25).
In the present study, we found that miR-195 was downregulated significantly in LSCC tissue and cell lines compared to the normal tissue and human bronchial epithelial cells which is consistent with a previous study (26). We also found that miR-195 downregulation was frequently found in LSCC with high T stage, N+, and high clinical stage. However, the role of miR-195 in LSCC remains unknown. For the first time, overexpression of miR-195 was used to observe its effect on LSCC cells by transfection technique. As expected, miR-195 inhibited the proliferation, colony formation, migration and invasion ability and induced G1 cell cycle arrest and promoted apoptosis in LSCC cell lines.
miRNAs perform biological functions by directly binding to the 3′UTR of mRNAs and inhibits protein translation. For instance, CARMA3 and Bcl-2 are direct targets of miR-195 in colorectal cancer (27,28). IGF1R and HDGF are direct targets of miR-195 in NSCLC (29). We predicted the target gene of miR-195 using bioinformatics analysis and selected DCUN1D1 as the potential target gene. DCUN1D1 is also known as squamous cell carcinoma-related oncogene (SCCRO). It is a novel gene initially identified as a result of 3q26-27 locus amplification of in head and neck squamous cell carcinoma (30). DCUN1D1 can enhance recruitment of Ubc12 to Cul1 to promote neddylation in the nucleus. DCUN1D1 has an essential role in neddylation in vivo involving nuclear localization of neddylation components and recruitment and proper positioning of Ubc12 (31). DCUN1D1 was previously reported to be associated with tumor progression and development of brain metastasis in patients with NSCLC (32). DCUN1D1 overexpression was associated with an aggressive clinical course in primary SCC of the lung. It functions as an oncogene which can transform cells of both fibroblastic and keratinocytic lineage (33). Luciferase reporter assays suggested that DCUN1D1 was one of the downstream targets of miR-195. We found that miR-195 significantly downregulated DCUN1D1 expression at the post-transcriptional level in LSCC cells. We also found that DCUN1D1 was overexpressed in human laryngeal carcinoma tissues when compared with its adjacent normal tissues and negatively correlated with the levels of miR-195. In addition, DCUN1D1 upregulation was also frequently found in LSCC with high T stage, N+, and high clinical stage. Our results indicate that miR-195, as a tumor suppressor miRNA, is also a negative regulator of the oncogene DCUN1D1.
More than 90% of carcinoma-associated mortality is caused by distant metastases (34). Cancer cell invasion and metastasis is a complicated, multi-step procedure involving interactions between invading cells, the extracellular matrix, and other stromal elements (35). Proteolytic enzymes produced by tumor cells are essential for cancer cells to degrade the extracellular matrix and penetrate lymphatic or blood vessel walls to extend to regional or distant sites. Matrix metalloproteinases (MMP), a family of zinc-dependent endopeptidases, play a distinguished role in extracellular matrix invasion and angiogenesis (36,37). The aberrant expression or activation of MMP-2 and MMP-9 has been linked to promotion of tumor invasion or metastasis in many different human tumors (38). DCUN1D1 can induce MMP-2 transcription indirectly through the upregulation of activator protein 2 (AP2) in a process requiring p53. It may be a marker for invasion and metastasis in squamous cell carcinoma (39). Recent study revealed that miR-218 inhibited invasion in cervical cancer by targeting DCUN1D1. DCUN1D1 knockdown was confirmed to significantly decrease the invasiveness of cervical cancer cells (40). However, this effect of DCUN1D1 has not been studied in LSCC. Our study indicates that DCUN1D1 knockdown can successfully simulate the anti-proliferative, anti-invasion effects of miR-195 on LSCC cells. Moreover, introduction of DCUN1D1 can reduce invasion inhibition induced by miR-195. We also found that both miR-195 and DCUN1D1 siRNA can reduce invasion involving the post-transcriptional downregulation of MMP-2 and MMP-9. These effects can be attenuated by introduction of DCUN1D1.
In conclusion, DCUN1D1 is a potential target of miR-195 and miR-195 may suppress growth and invasion of LSCC cells possibly through targeting DCUN1D1, which would provide a candidate target for cancer therapy.
Acknowledgements
The study was supported by the science and technology fund of Tianjin Health Bureau (2015KZ106).
References
- 1.Chen Z, Jin Y, Yu D, Wang A, Mahjabeen I, Wang C, Liu X, Zhou X. Down-regulation of the microRNA-99 family members in head and neck squamous cell carcinoma. Oral Oncol. 2012;48:686–691. doi: 10.1016/j.oraloncology.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Borel C, Deutsch S, Letourneau A, Migliavacca E, Montgomery SB, Dimas AS, Vejnar CE, Attar H, Gagnebin M, Gehrig C, et al. Identification of cis- and trans-regulatory variation modulating microRNA expression levels in human fibroblasts. Genome Res. 2011;21:68–73. doi: 10.1101/gr.109371.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, function and role in cancer. Curr Genomics. 2010;11:537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davidson B, Tropé CG, Reich R. The clinical and diagnostic role of microRNAs in ovarian carcinoma. Gynecol Oncol. 2014;133:640–646. doi: 10.1016/j.ygyno.2014.03.575. [DOI] [PubMed] [Google Scholar]
- 7.Nugent M. MicroRNA function and dysregulation in bone tumors: The evidence to date. Cancer Manag Res. 2014;6:15–25. doi: 10.2147/CMAR.S53928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li M, Tian L, Ren H, Chen X, Wang Y, Ge J, Wu S, Sun Y, Liu M, Xiao H. MicroRNA-101 is a potential prognostic indicator of laryngeal squamous cell carcinoma and modulates CDK8. J Transl Med. 2015;13:271. doi: 10.1186/s12967-015-0626-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu JY, Lu JB, Xu Y. MicroRNA-153 inhibits the proliferation and invasion of human laryngeal squamous cell carcinoma by targeting KLF5. Exp Ther Med. 2016;11:2503–2508. doi: 10.3892/etm.2016.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu S, Jia S, Xu P. MicroRNA-9 as a novel prognostic biomarker in human laryngeal squamous cell carcinoma. Int J Clin Exp Med. 2014;7:5523–5528. [PMC free article] [PubMed] [Google Scholar]
- 11.Wu X, Cui CL, Chen WL, Fu ZY, Cui XY, Gong X. miR-144 suppresses the growth and metastasis of laryngeal squamous cell carcinoma by targeting IRS1. Am J Transl Res. 2016;8:1–11. [PMC free article] [PubMed] [Google Scholar]
- 12.Yu WF, Wang HM, Lu BC, Zhang GZ, Ma HM, Wu ZY. miR-206 inhibits human laryngeal squamous cell carcinoma cell growth by regulation of cyclinD2. Eur Rev Med Pharmacol Sci. 2015;19:2697–2702. [PubMed] [Google Scholar]
- 13.Zhong G, Xiong X. miR-205 promotes proliferation and invasion of laryngeal squamous cell carcinoma by suppressing CDK2AP1 expression. Biol Res. 2015;48:60. doi: 10.1186/s40659-015-0052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C, Guan H, Wang Y, Chen M, Xu B, Zhang L, Lu K, Tao T, Zhang X, Huang Y. miR-195 inhibits EMT by targeting FGF2 in prostate cancer cells. PLoS One. 2015;10:e0144073. doi: 10.1371/journal.pone.0144073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo Q, Zhang Z, Dai Z, Basnet S, Li S, Xu B, Ge H. Tumor-suppressive microRNA-195-5p regulates cell growth and inhibits cell cycle by targeting cyclin dependent kinase 8 in colon cancer. Am J Transl Res. 2016;8:2088–2096. [PMC free article] [PubMed] [Google Scholar]
- 16.Singh R, Yadav V, Kumar S, Saini N. MicroRNA-195 inhibits proliferation, invasion and metastasis in breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1. Sci Rep. 2015;5:17454. doi: 10.1038/srep17454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao C, Qi L, Chen M, Liu L, Yan W, Tong S, Zu X. microRNA-195 inhibits cell proliferation in bladder cancer via inhibition of cell division control protein 42 homolog/signal transducer and activator of transcription-3 signaling. Exp Ther Med. 2015;10:1103–1108. doi: 10.3892/etm.2015.2633. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Guo J, Wang M, Liu X. MicroRNA-195 suppresses tumor cell proliferation and metastasis by directly targeting BCOX1 in prostate carcinoma. J Exp Clin Cancer Res. 2015;34:91. doi: 10.1186/s13046-015-0209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 20.Baer C, Claus R, Plass C. Genome-wide epigenetic regulation of miRNAs in cancer. Cancer Res. 2013;73:473–477. doi: 10.1158/0008-5472.CAN-12-3731. [DOI] [PubMed] [Google Scholar]
- 21.Liu B, Qu J, Xu F, Guo Y, Wang Y, Yu H, Qian B. MiR-195 suppresses non-small cell lung cancer by targeting CHEK1. Oncotarget. 2015;6:9445–9456. doi: 10.18632/oncotarget.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan YG, Zhang YF, Guo CJ, Yang M, Chen MY. Screening of differentially expressed microRNA in ulcerative colitis related colorectal cancer. Asian Pac J Trop Med. 2013;6:972–976. doi: 10.1016/S1995-7645(13)60174-1. [DOI] [PubMed] [Google Scholar]
- 23.Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D, Wang Y. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol. 2009;24:652–657. doi: 10.1111/j.1440-1746.2008.05666.x. [DOI] [PubMed] [Google Scholar]
- 24.Sohn W, Kim J, Kang SH, Yang SR, Cho JY, Cho HC, Shim SG, Paik YH. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp Mol Med. 2015;47:e184. doi: 10.1038/emm.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J, Ji A, Wang X, Zhu Y, Yu Y, Lin Y, Liu Y, Li S, Liang Z, Xu X, et al. MicroRNA-195-5p, a new regulator of Fra-1, suppresses the migration and invasion of prostate cancer cells. J Transl Med. 2015;13:289. doi: 10.1186/s12967-015-0650-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu ZM, Lin YF, Jiang L, Chen LS, Luo XN, Song XH, Chen SH, Zhang SY. Micro-ribonucleic acid expression profiling and bioinformatic target gene analyses in laryngeal carcinoma. Onco Targets Ther. 2014;7:525–533. doi: 10.2147/OTT.S59871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu L, Chen L, Xu Y, Li R, Du X. microRNA-195 promotes apoptosis and suppresses tumorigenicity of human colorectal cancer cells. Biochem Biophys Res Commun. 2010;400:236–240. doi: 10.1016/j.bbrc.2010.08.046. [DOI] [PubMed] [Google Scholar]
- 28.Wang L, Qian L, Li X, Yan J. MicroRNA-195 inhibits colorectal cancer cell proliferation, colony-formation and invasion through targeting CARMA3. Mol Med Rep. 2014;10:473–478. doi: 10.3892/mmr.2014.2178. [DOI] [PubMed] [Google Scholar]
- 29.Guo H, Li W, Zheng T, Liu Z. MiR-195 targets HDGF to inhibit proliferation and invasion of NSCLC cells. Tumour Biol. 2014;35:8861–8866. doi: 10.1007/s13277-014-2153-0. [DOI] [PubMed] [Google Scholar]
- 30.Singh B, Gogineni SK, Sacks PG, Shaha AR, Shah JP, Stoffel A, Rao PH. Molecular cytogenetic characterization of head and neck squamous cell carcinoma and refinement of 3q amplification. Cancer Res. 2001;61:4506–4513. [PubMed] [Google Scholar]
- 31.Huang G, Kaufman AJ, Ramanathan Y, Singh B. SCCRO (DCUN1D1) promotes nuclear translocation and assembly of the neddylation E3 complex. J Biol Chem. 2011;286:10297–10304. doi: 10.1074/jbc.M110.203729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoo J, Lee SH, Lym KI, Park SY, Yang SH, Yoo CY, Jung JH, Kang SJ, Kang CS. Immunohistochemical expression of DCUN1D1 in non-small cell lung carcinoma: Its relation to brain metastasis. Cancer Res Treat. 2012;44:57–62. doi: 10.4143/crt.2012.44.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarkaria I, O-charoenrat P, Talbot SG, Reddy PG, Ngai I, Maghami E, Patel KN, Lee B, Yonekawa Y, Dudas M, et al. Squamous cell carcinoma related oncogene/DCUN1D1 is highly conserved and activated by amplification in squamous cell carcinomas. Cancer Res. 2006;66:9437–9444. doi: 10.1158/0008-5472.CAN-06-2074. [DOI] [PubMed] [Google Scholar]
- 34.Gupta GP, Massagué J. Cancer metastasis: Building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Liotta LA, Steeg PS, Stetler-Stevenson WG. Cancer metastasis and angiogenesis: An imbalance of positive and negative regulation. Cell. 1991;64:327–336. doi: 10.1016/0092-8674(91)90642-C. [DOI] [PubMed] [Google Scholar]
- 36.Folkman J. What is the evidence that tumors are angiogenesis dependent? J Natl Cancer Inst. 1990;82:4–6. doi: 10.1093/jnci/82.1.4. [DOI] [PubMed] [Google Scholar]
- 37.MacDougall JR, Matrisian LM. Contributions of tumor and stromal matrix metalloproteinases to tumor progression, invasion and metastasis. Cancer Metastasis Rev. 1995;14:351–362. doi: 10.1007/BF00690603. [DOI] [PubMed] [Google Scholar]
- 38.O-Charoenrat P, Rhys-Evans PH, Eccles SA. Expression of matrix metalloproteinases and their inhibitors correlates with invasion and metastasis in squamous cell carcinoma of the head and neck. Arch Otolaryngol Head Neck Surg. 2001;127:813–820. [PubMed] [Google Scholar]
- 39.O-Charoenrat P, Sarkaria I, Talbot SG, Reddy P, Dao S, Ngai I, Shaha A, Kraus D, Shah J, Rusch V, et al. SCCRO (DCUN1D1) induces extracellular matrix invasion by activating matrix metalloproteinase 2. Clin Cancer Res. 2008;14:6780–6789. doi: 10.1158/1078-0432.CCR-08-0719. [DOI] [PubMed] [Google Scholar]
- 40.Jiang Z, Song Q, Zeng R, Li J, Li J, Lin X, Chen X, Zhang J, Zheng Y. MicroRNA-218 inhibits EMT, migration and invasion by targeting SFMBT1 and DCUN1D1 in cervical cancer. Oncotarget. 2016;7:45622–45636. doi: 10.18632/oncotarget.9850. [DOI] [PMC free article] [PubMed] [Google Scholar]