Abstract
In recent years it was found that the synthesis and biological activity of ribosomes are closely associated with tumor cell growth, tumorigenesis, and malignant transformation. However, the role of regulator of ribosome synthesis 1 (RRS1) in hepatocellular carcinoma (HCC) has not yet been reported. In the present study, we aimed to examine the potential role of RRS1 in tumor cell growth by using a lentivirus-mediated RNA interference (RNAi) system in the HCC cell line SMMC-7721 in vitro. Compared with that of the negative control group (Lv-shCon), the mRNA and protein expression levels of RRS1 in SMMC-7721 cells transfected with Lv-shRRS1 were significantly decreased. Further experiments found that silencing of RRS1 gene expression in SMMC-7721 cells significantly suppressed cell proliferation, inhibited colony formation capacity, increased apoptosis and arrested cells in the G1 phase. These results suggest that the RRS1 gene plays a critical role in cell proliferation, colony formation, cell apoptosis and cell cycle distribution in human HCC cells, and that silencing of RRS1 by RNAi is a promising therapeutic approach for the treatment of HCC, and should be further developed.
Keywords: regulator of ribosome synthesis 1, RNA interference, hepatocellular carcinoma, apoptosis, proliferation
Introduction
Hepatocellular carcinoma (HCC) is the sixth most prevalent cancer globally and the second leading cause of cancer-related death (1). Due to the important metabolic functions of the liver within the body, malignancies in the liver typically lead to life-threatening consequences (2–5). Early diagnosis is essential for the successful treatment of liver cancer; however, there is typically a lack of specific symptoms (2,6,7). Thus, the identification of a novel biomarker of liver cancer may aid in developing new methods for treating HCC (8).
RNAi is a gene-based technology that utilizes shRNA, and is frequently used to suppress the expression of target genes (9–11). RNAi and lentiviral technologies are widely used in genetic research (12–14), and RNAi technology could continue to provide novel methods for the study of human gene function, early cancer diagnostics and targeted therapy (15–17).
Ribosomes, as the sole organelles responsible for protein synthesis, serve essential functions in all cells. The assembly and activity of ribosomes are regulated by complex and precise intracellular pathways. A study indicated that the overexpression of oncogenes and downregulation of tumor suppressor genes may lead to abnormal regulation of ribosome biogenesis and the initiation of protein translation (18). In recent years, abundant research has pointed out that dysfunction of ribosome biogenesis plays a major role in the process of tumorigenesis (18–20). Additionally, it has been shown that patients with quantitative and qualitative changes in their ribosomes were more susceptible to cancer (5). These data indicate that the biogenesis of ribosomes may be involved in the development and progression of cancers.
Regulator of ribosome synthesis 1 (RRS1), which comprises 203 amino acids, was initially discovered in yeast (21), and is a conserved nuclear protein in eukaryotes (22). In a murine model of Huntington's disease, high-level expression of RRS1 was identified in neurons (23). Furthermore, immunohistochemical staining has shown that RRS1 is not solely present in the nucleus, as in yeast, but is also present in the endoplasmic reticulum (22,24), and its expression may be induced by neuronal endoplasmic reticulum stress (25,26). Additionally, secretion-deficient yeast carrying a nonsense mutation in an amino codon in the C-terminal of Rrs1 exhibited attenuated transcriptional suppression of ribosomal (r)RNA and protein (21). Therefore, RRS1 is a key protein connection between ribosome biogenesis and protein secretion (27,28). It has been reported that RRS1 plays an important role in the regulation of ribosome biogenesis, particularly in the maturation of the ribosomal subunits 25S rRNA and 60S rRNA (21,29,30). Based on these critical functions of RRS1, we hypothesized that the expression and activity of RRS1 may be abnormally regulated in cancer cells, in which physiological function changes dramatically. To the best of our knowledge, the expression of RRS1 in most cancer types, including HCC, has not been reported. Therefore, the present study investigated the expression of RRS1 in human HCC, as well as the roles of RRS1 in the HCC cell line SMMC-7721 through the use of lentivirus-mediated RNA interference (RNAi).
Materials and methods
Human tissue samples, cell lines, reagents and antibodies
Human HCC tissues and corresponding paracancerous tissues were obtained from 18 patients with HCC who first underwent surgery at the Beijing 302 Hospital (Beijing, China) between January 2013 and June 2013. All of the human tissue samples were collected with the consent of patients or their legal representatives and the approval of the local medical ethics committee. All samples were immediately frozen in liquid nitrogen and stored at −80°C to reduce RNA degradation. The human liver cancer cell lines SMMC-7721, HepG2, Huh-7 and Hep3B, and the human embryonic kidney cell line 293T, were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and cultured in RPMI-1640 medium containing 10% fetal bovine serum, 100 µg/ml streptomycin and 100 IU/ml penicillin at 37°C in 95% humidity and 5% CO2. Rabbit anti-RRS1 polyclonal antibodies were purchased from Abcam (cat. no. AB188161, Shanghai, China). Mouse anti-GAPDH monoclonal antibodies were obtained from Santa Cruz Biotechnology, Inc. (cat. no. sc-32233, Dallas, TX, USA). Anti-rabbit and anti-mouse IgG secondary antibodies were from Santa Cruz Biotechnology, Inc. (cat. no. sc-2004). An APC Annexin V apoptosis detection kit was from eBioscience (San Diego, CA, USA). Propidium iodide (PI) was from Sigma-Aldrich (St. Louis, MO, USA).
Lentivirus-mediated short hairpin (sh)RNA delivery
An RRS1 shRNA sequence was inserted into a pGCSIL-GFP lentivirus RNAi expression system (GeneChem Co., Ltd., Shanghai, China). The effective targeting sequence of RRS1 (GCT GCC TTC ATT GAG TTT A) was selected by western blot analysis. A non-silencing shRNA sequence (TTC TCC GAA CGT GTT CAC GT) was used as a negative control, which does not target any gens in humans, mice or rats as determined by screening with NCBI RefSeq. The shRNA vectors were co-transfected with the lentiviral packaging plasmids pHelper1.0 and pHelper2.0 into 293T cells using Lipofectamine 2000 (Invitrogen, Shanghai, China) to generate the respective lentiviruses. Viral stocks were collected from the 293T cells 3 days after infection and were used to infect SMMC-7721 cells. The SMMC-7721 cells were infected with the RRS1-siRNA-lentivirus (Lv-shRRS1 group) or negative control lentivirus (Lv-shCon group) according to the recommended multiplicity of infection (MOI). The stem-loop-stem oligos (shRNAs) were synthesized, annealed, and ligated into the pGCSIL-GFP lentivirus RNAi expression system. The lentiviral-based shRNA-expressing vectors were confirmed by DNA sequencing.
After lentiviral infection, the SMMC-7721 cells were observed under a fluorescence microscope (MicroPublisher 3.3RTV; Olympus Corp., Tokyo, Japan) at 3 days post-infection. The lentiviral transfection experiments were performed as in a previous study (31).
Reverse transcription-quantitative PCR (RT-qPCR) analysis
Total RNA was isolated from cell lines or frozen tissues using TRIzol reagent (Invitrogen) and reverse transcribed with a PrimeScript® RT reagent kit (Takara, Dalian, China). cDNA was normalized with GAPDH. PCR was performed via a two-step method using a SYBR® premix Ex Taq™ II kit (Takara). The RT-qPCR comprised an initial denaturation at 95°C for 15 sec, then 45 cycles at 95°C for 5 sec and 60°C for 30 sec. The melting curve was monitored with the following conditions: 60 sec at 95°C, 60 sec at 55°C, followed by a temperature range from 55°C to 95°C increased by 0.5°C every 4 sec. PCR reactions were carried out in triplicate. GAPDH was used as internal control. The relative amount of each cDNA was determined using the 2−∆∆Cq method (32). The primer sequences used for RRS1 and GAPDH were as follows: RRS1 forward, CCGAAAAGGGGTTGAAACTTCC, and reverse, CCCTACCGGACACCAGAGTAA; and GAPDH forward, TGACTTCAACAGCGACACCCA, and reverse, CACCCTGTTGCTGTAGCCAAA. The PCR products of the RRS1 and GAPDH primers were 153 and 121 bp in length, respectively.
Western blot analysis
Protein samples were prepared from SMMC-7721 cells 5 days after infection with RRS1 or control shRNA lentiviruses, and were subjected to 10% SDS-PAGE (20 µg protein per lane), as described by Laemmli (33). The separated proteins were transferred onto polyvinylidene fluoride (PVDF) membranes (EMD Millipore, Billerica, MA, USA) and probed with rabbit anti-RRS1 (1:1,000) or mouse anti-GAPDH (1:5,000) antibodies at room temperature for 2 h, followed by incubation with the anti-rabbit and anti-mouse IgG secondary antibodies (1:5,000) at room temperature for 2 h. GAPDH was used as an internal control. After three washes with 10% bovine serum albumin in phosphate-buffered saline (PBS), immunolabeled proteins on the PVDF membranes were detected with an Enhanced Chemiluminescence kit (Amersham, Uppsala, Sweden) and exposed to X-ray film. Bands on the X-ray films were quantified with an ImageQuant densitometric scanner (Molecular Devices LLC, Sunnyvale, CA, USA).
Cell proliferation assay
Five days after lentiviral infection, SMMC-7721 cells were trypsinized, resuspended, seeded into 96-well plates at a concentration of 2,000 cells per well, and incubated at 37°C in 5% CO2. The number of viable cells was measured at daily intervals (day 1, 2, 3, 4 and 5) using a Cellomics ArrayScan™ VT1 HCS automated reader (Cellomics, Inc., Pittsburgh, PA, USA).
Colony formation assay
SMMC-7721 cells were seeded into 6-well plates (1,000 cell/well; three duplicate wells and cultured at 37°C in 5% CO2. At 72 h after lentiviral infection, cells were grown for 7 days with media replacement every 2 days to enable colony formation. After 7 days, the cells were washed with PBS and fixed with paraformaldehyde for 30 min, then washed with PBS and stained with Giemsa for 20 min. The cells were subsequently washed three times with ddH2O to obtain a clean background. The number of colonies and the cell number in each colony were counted and statistically analyzed according to the method by Zhang et al (34).
Flow cytometric analysis
An apoptosis assay was performed with an APC Annexin V staining kit. When cell confluence reached 85% (5–6 days after infection), the cells were harvested by centrifugation at 10,000 × g for 5 min. The pellets were washed twice with cold PBS, fixed with 70% chilled ethanol, centrifuged at 10,000 × g for 5 min to remove the ethanol, and resuspended in PBS. The suspensions were filtered through 400-mesh membranes and centrifuged at 10,000 × g for 5 min. The cells were resuspended with 1X Annexin V staining buffer and stained with Annexin V-APC at room temperature for 15 min in the dark. Approximately 1.0×105 fixed cells were analyzed with a FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). The assay was carried out in triplicate.
Cell cycle distribution was analyzed with PI staining
In brief, 1.5×105 cells at 4 days post-infection were seeded into 6-cm dishes and cultured for 40 h at 37°C. Cells were harvested by centrifugation at 10,000 × g for 5 min, washed with PBS, and fixed with 70% cold ethanol for 1 h. Cells were then collected by centrifugation at 10,000 × g for 5 min, resuspended in PBS containing 100 µg/ml of DNase-free RNase and 40 µg/ml PI, and incubated for 1 h at 37°C. A total of 1.0×104 fixed cells were analyzed by flow cytometry (FACSCalibur; BD Biosciences).
Statistical analysis
One-way ANOVA and a Student's t-test were used for data analysis. Statistical analysis was performed using SPSS 22.0 software (IBM Corp., Armonk, NY, USA). All values were expressed as the mean ± standard deviation (SD). P<0.05 was considered to indicate a statistically significant difference.
Results
RRS1 is expressed in human HCC tissues and cell lines
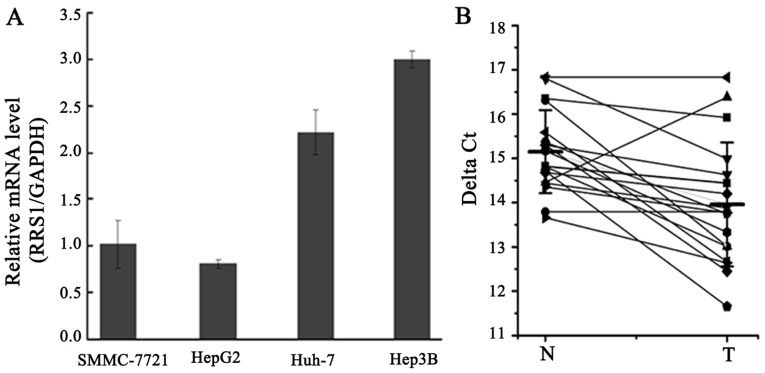
The expression of RRS1 mRNA was assessed in human liver cancer cell lines SMMC-7721, HepG2, Huh-7, and Hep3B. PCR analysis indicated that RRS1 mRNA was expressed in all four human liver cancer cell lines (Fig. 1A). Furthermore, the expression of RRS1 mRNA in 18 paired tissue samples (HCC and paracancerous tissues) was measured by qPCR. As shown in Fig. 1B, the level of RRS1 mRNA in HCC tissues was approximately 3–4-fold higher than that in adjacent tissues. Thus, elevated expression of RRS1 may play an important role in the pathogenesis of human HCC.
Figure 1.
RRS1 is frequently expressed in human HCC tissues and cell lines. (A) PCR analysis of RRS1 expression in human liver cancer cell lines SMMC-7721, HepG2, Huh-7, and Hep3B. (B) Expression of RRS1 mRNA in 18 cases of HCC and adjacent tissues. N, paracancerous tissue and T, cancerous tissue. Patient cancer tissues were directly compared to the matched adjacent tissues. The levels of RRS1 mRNA in the tissues were detected by RT-qPCR. The smaller the value of ∆Ct, the higher the expression level.
Lentiviral shRNA inhibits the mRNA and protein expression of RRS1 in SMMC-7721 cells
To investigate the role of RRS1 in HCC, the human HCC cell line SMMC-7721 was selected for in vitro study. A loss-of-function study was performed via lentivirus-mediated shRNA knockdown of RRS1. A lentiviral vector system expressing shRNA against RRS1 and GFP, as a reporter gene, was established. To determine the infection efficiency of lentivirus in SMMC-7721 cells at 30% cell density, cells infected with Lv-shRRS1 or Lv-shCon vector were observed under a fluorescence microscope at 3 days post-infection. As shown in Fig. 2A, >90% of SMMC-7721 cells were found to express GFP, which indicated a high infection efficiency of the lentivirus.
Figure 2.
Lentivirus-mediated shRNA knockdown of RRS1 in HMMC-7721 cells. (A) Infection efficiency of lentivirus in the human HCC cell line SMMC-7721. Representative fluorescence images of SMMC-7721 cells after 72 h of lentiviral infection are shown (magnification, ×100). (B) RT-qPCR analysis of RRS1 mRNA expression. (C) Western blot analysis of RRS1 protein expression. Lv-shCon: cells infected with Lv-shCon lentivirus. Lv-shRRS1, cells infected with Lv-shRRS1 lentivirus. The expression of GAPDH was used as an internal control for RT-qPCR and western blot analysis. *P<0.05; **P<0.01.
To verify the knockdown efficiency of Lv-shRRS1, the mRNA and protein expression levels of RRS1 in SMMC-7721 cells were detected by PCR and western blot analysis at 5 days post-infection. As shown in Fig. 2B, the mRNA expression levels of RRS1 in the Lv-shRRS1 infection group were downregulated by 68.0% when compared with the Lv-shCon infection group (P<0.01). As shown in Fig. 2C, the protein expression levels of RRS1 in the Lv-shRRS1 infection group were also significantly decreased (73.0% decrease) when compared with the Lv-shCon infection group.
Knockdown of RRS1 significantly inhibits SMMC-7721 cell proliferation
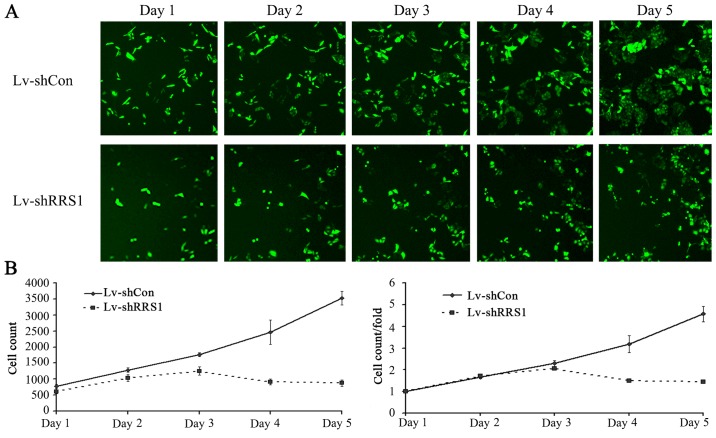
To investigate the effect of RRS1 knockdown on cell proliferation, the two cell groups were seeded into 96-well plates and subjected to daily cellomics analysis for 5 days (Fig. 3A). As shown in Fig. 3B, lentivirus-mediated shRNA knockdown of RRS1 significantly inhibited the proliferation of SMMC-7721 cells when compared with cells infected with control lentivirus (P<0.05). This result indicated that RRS1 played an important role in the proliferation of SMMC-7721 cells.
Figure 3.
In vitro proliferation assay of SMMC-7721 cells. Lv-shCon, cells infected with Lv-shCon lentivirus. Lv-RRS1, cells infected with Lv-shRRS1 lentivirus. Cell proliferation in the Lv-shRRS1 group was significantly inhibited compared with the Lv-shCon group. (A) Representative cell fluorescence images on days 1–5. (B) Cell counting statistics curve representative of 5 consecutive days.
Knockdown of RRS1 significantly inhibits the colony-forming ability of SMMC-7721 cells
To study the long-term effect of RRS1 shRNA lentivirus on cell growth, a colony forming assay was conducted in SMMC-7721 cells. As shown in Fig. 4A, the number of colonies in the Lv-shRRS1 group was lower than that in the Lv-shCon group. Specifically, the colony number was 9±5 in the Lv-shRRS1 group compared with 104±9 in the Lv-shCon group (P<0.01; Fig. 4B). These results indicate that downregulation of RRS1 significantly decreases the colony formation capacity of HCC cells in vitro.
Figure 4.
Colony formation capacity of SMMC-7721 cells. Cells were grown into natural colonies in 6-well plates. After staining with Giemsa, the number of colonies was counted in each group. (A, upper panel) Three representative colony formation wells of each group are shown. (A, lower panel) Three representative fluorescence images are shown. (B) Colony numbers. Lv-shCon: cells infected with Lv-shCon lentivirus. Lv-shRRS1: cells infected with Lv-shRRS1 lentivirus. **P<0.01.
Knockdown of RRS1 significantly increases apoptosis of SMMC-7721 cells
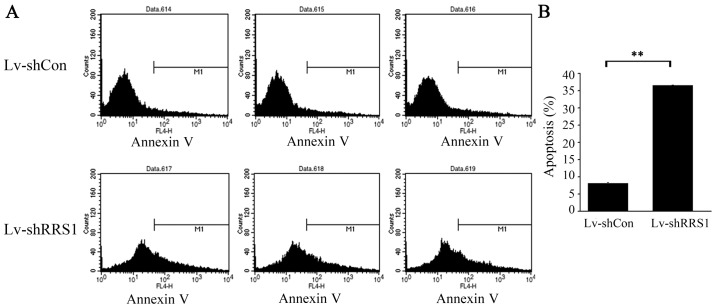
To determine the effect of RRS1 knockdown in HCC cells, Annexin V staining and flow cytometric analysis were performed on SMMC-7721 cells 5–6 days after lentiviral infection, when cell confluence reached 85%. As shown in Fig. 5, the apoptotic rate of SMMC-7721 cells in the Lv-shRRS1 group was significantly higher than that in the Lv-shCon group (36.52±0.19% vs. 8.15±0.30%; P<0.001).
Figure 5.
Knockdown of RRS1 increases apoptosis of SMMC-7721 cells in vitro. RRS1 knockdown with Lv-shRRS1 caused a significant increase in the rate of apoptosis when compared with Lv-shCon infection. **P<0.01, significant difference vs. Lv-shCon group.
Effects of RRS1 knockdown on the cell cycle distribution of SMMC-7721 cells
To elucidate the impact of RRS1 knockdown on the cell cycle progression of HCC cells, SMMC-7721 cells were subjected to a PI staining flow cytometry assay 6 days after lentiviral infection. The Lv-shRRS1 infection group exhibited an increased proportion of cells in G1 phase (Lv-shRRS1 62.49±0.59% vs. Lv-shCon 59.41±0.41%, P=0.003) and reduced proportions of cells in S phase (Lv-shRRS1 34.77±0.55% vs. Lv-shCon 36.21±0.51%, P=0.03) and G2/M phase (Lv-shRRS1 2.74±0.50% vs. Lv-shCon 4.39±0.12%, P=0.024) when compared with the Lv-shCon group (Fig. 6). These results indicated that knockdown of RRS1 could induce G1 phase arrest in HCC cells.
Figure 6.
Knockdown of RRS1 leads to cell cycle arrest in SMMC-7721 cells. Cell populations in the different cell cycle phases were quantified by PI staining followed by flow cytometry analyses. Lv-shCon, cells infected with Lv-shCon lentivirus. Lv-shRRS1, cells infected with Lv-shRRS1 lentivirus. *P<0.05, **P<0.01.
Discussion
The incidence and mortality rates of HCC have increased over recent years. An estimated 782,500 new liver cancer cases and 745,500 liver cancer-related deaths occurred worldwide during 2012, with China alone accounting for approximately 50% of the total cases and deaths (2).
RRS1 is a conserved eukaryotic nuclear protein. RRS1 can be regulated to control the speed of ribosome biogenesis according to the energy state of the cell, thereby maintaining cellular homeostasis. Additionally, RRS1 is an important protein in signal transduction pathways related to protein secretion and ribosome synthesis. It has been reported that the RRS1 gene may be involved in the pathogenesis of Huntington's disease, although the specific underlying mechanism is unclear (26). A study by Gambe et al (24) showed that RRS1 was involved in chromosome aggregation. Notably, their experiments identified abnormalities in the chromosome alignment and spindle organization of RRS1-depleted cells, which resulted in mitotic delay. Furthermore, it has been suggested that RRS1 may play a role in the development of cervical cancer through regulation of the cell cycle. However, the expression and function of RRS1 in other tumors, including HCC, have not been reported.
In the present study, we successfully constructed an shRNA lentiviral vector that targeted RRS1 mRNA, which may provide an experimental basis for further research. To evaluate the function of RRS1 in HCC cells, we efficiently silenced the RRS1 gene, and found that the proportion of SMMC-7721 cells in G1 phase was increased, while the proportions of cells in S and G2/M phases were decreased, relative to the control group. Further assays verified that silencing of the RRS1 gene decreased cell proliferation and colony formation ability, and promoted apoptosis. These results suggest that RRS1 may act as an oncogenic factor in liver cancer. However, the specific signaling pathway underlying the involvement of RRS1 in the biological behavior of HCC cells remains unclear, and further study is ongoing.
In conclusion, this is the first study to demonstrate that the downregulation of RRS1 in SMMC-7721 cells by RNAi could decrease cell proliferation, inhibit colony formation ability and induce cell apoptosis. Therefore, RRS1 may be involved in the regulation of hepatocellular proliferative activity, cell cycling and apoptotic ability. These findings may aid in developing gene targeting therapies for patients with liver cancer, and provide an experimental basis for further research into the functions of RRS1 in the progression, invasion, metastasis and tumor recurrence of HCC.
Glossary
Abbreviations
- HCC
hepatocellular carcinoma
- RSS1
regulator of ribosome synthesis 1
- RNAi
RNA interference
- shRNA
short hairpin RNA
References
- 1.World Health Organization, corp-author. World Cancer Report 2014. In: Stewart BW, Wild CP, editors. International Agency for Research on Cancer. World Health Organization; WHO Press, Geneva: 2015. [DOI] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Zuo TT, Zheng RS, Zhang SW, Zeng HM, Chen WQ. Incidence and mortality of liver cancer in China in 2011. Chin J Cancer. 2015;34:508–513. doi: 10.1186/s40880-015-0056-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei KR, Yu X, Zheng RS, Peng XB, Zhang SW, Ji MF, Liang ZH, Ou ZX, Chen WQ. Incidence and mortality of liver cancer in China, 2010. Chin J Cancer. 2014;33:388–394. doi: 10.5732/cjc.014.10088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petrick JL, Braunlin M, Laversanne M, Valery PC, Bray F, McGlynn KA. International trends in liver cancer incidence, overall and by histologic subtype, 1978–2007. Int J Cancer. 2016;139:1534–1545. doi: 10.1002/ijc.30211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fitzmorris P, Shoreibah M, Anand BS, Singal AK. Management of hepatocellular carcinoma. J Cancer Res Clin Oncol. 2015;141:861–876. doi: 10.1007/s00432-014-1806-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen MH, Keeffe EB. Screening for hepatocellular carcinoma. J Clin Gastroenterol. 2002;35(Suppl 2):S86–S91. doi: 10.1097/00004836-200211002-00004. [DOI] [PubMed] [Google Scholar]
- 8.Shimizu D, Inokawa Y, Sonohara F, Inaoka K, Nomoto S. Search for useful biomarkers in hepatocellular carcinoma, tumor factors and background liver factors (Review) Oncol Rep. 2017;37:2527–2542. doi: 10.3892/or.2017.5541. [DOI] [PubMed] [Google Scholar]
- 9.Pecot CV, Calin GA, Coleman RL, Lopez-Berestein G, Sood AK. RNA interference in the clinic: challenges and future directions. Nat Rev Cancer. 2011;11:59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sioud M. RNA interference: Mechanisms, technical challenges, and therapeutic opportunities. Methods Mol Biol. 2015;1218:1–15. doi: 10.1007/978-1-4939-1538-5_1. [DOI] [PubMed] [Google Scholar]
- 11.Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet. 2015;16:71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu H, Liang S, Yang X, Ji Z, Zhao W, Ye X, Rui J. RNAi-mediated RPL34 knockdown suppresses the growth of human gastric cancer cells. Oncol Rep. 2015;34:2267–2272. doi: 10.3892/or.2015.4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu K, Li X, Cao Y, Ge Y, Wang J, Shi B. MiR-132 inhibits cell proliferation, invasion and migration of hepatocellular carcinoma by targeting PIK3R3. Int J Oncol. 2015;47:1585–1593. doi: 10.3892/ijo.2015.3112. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Q, Hu H, Shi X, Tang W. Knockdown of S100P by lentiviral-mediated RNAi promotes apoptosis and suppresses the colony-formation ability of gastric cancer cells. Oncol Rep. 2014;31:2344–2350. doi: 10.3892/or.2014.3104. [DOI] [PubMed] [Google Scholar]
- 15.Du P, Ye L, Yang Y, Jiang WG. Candidate of metastasis 1 regulates in vitro growth and invasion of bladder cancer cells. Int J Oncol. 2013;42:1249–1256. doi: 10.3892/ijo.2013.1802. [DOI] [PubMed] [Google Scholar]
- 16.Roop RP, Ma CX. Endocrine resistance in breast cancer: Molecular pathways and rational development of targeted therapies. Future Oncol. 2012;8:273–292. doi: 10.2217/fon.12.8. [DOI] [PubMed] [Google Scholar]
- 17.Li J, Huang L. Targeted delivery of RNAi therapeutics for cancer therapy. Nanomedicine (Lond) 2010;5:1483–1486. doi: 10.2217/nnm.10.124. [DOI] [PubMed] [Google Scholar]
- 18.Montanaro L, Treré D, Derenzini M. Changes in ribosome biogenesis may induce cancer by down-regulating the cell tumor suppressor potential. Biochim Biophys Acta. 2012;1825:101–110. doi: 10.1016/j.bbcan.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 19.Donati G, Montanaro L, Derenzini M. Ribosome biogenesis and control of cell proliferation: p53 is not alone. Cancer Res. 2012;72:1602–1607. doi: 10.1158/0008-5472.CAN-11-3992. [DOI] [PubMed] [Google Scholar]
- 20.Montanaro L, Treré D, Derenzini M. Nucleolus, ribosomes, and cancer. Am J Pathol. 2008;173:301–310. doi: 10.2353/ajpath.2008.070752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsuno A, Miyoshi K, Tsujii R, Miyakawa T, Mizuta K. RRS1, a conserved essential gene, encodes a novel regulatory protein required for ribosome biogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:2066–2074. doi: 10.1128/MCB.20.6.2066-2074.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Harnpicharnchai P, Jakovljevic J, Tang L, Guo Y, Oeffinger M, Rout MP, Hiley SL, Hughes T, Woolford JL., Jr Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 2007;21:2580–2592. doi: 10.1101/gad.1569307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fossale E, Wheeler VC, Vrbanac V, Lebel LA, Teed A, Mysore JS, Gusella JF, MacDonald ME, Persichetti F. Identification of a presymptomatic molecular phenotype in Hdh CAG knock-in mice. Hum Mol Genet. 2002;11:2233–2241. doi: 10.1093/hmg/11.19.2233. [DOI] [PubMed] [Google Scholar]
- 24.Gambe AE, Matsunaga S, Takata H, Ono-Maniwa R, Baba A, Uchiyama S, Fukui K. A nucleolar protein RRS1 contributes to chromosome congression. FEBS Lett. 2009;583:1951–1956. doi: 10.1016/j.febslet.2009.05.033. [DOI] [PubMed] [Google Scholar]
- 25.Wan K, Kawara H, Yamamoto T, Kume K, Yabuki Y, Goshima T, Kitamura K, Ueno M, Kanai M, Hirata D, et al. The essential function of Rrs1 in ribosome biogenesis is conserved in budding and fission yeasts. Yeast. 2015;32:607–614. doi: 10.1002/yea.3083. [DOI] [PubMed] [Google Scholar]
- 26.Carnemolla A, Fossale E, Agostoni E, Michelazzi S, Calligaris R, De Maso L, Del Sal G, MacDonald ME, Persichetti F. Rrs1 is involved in endoplasmic reticulum stress response in Huntington disease. J Biol Chem. 2009;284:18167–18173. doi: 10.1074/jbc.M109.018325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyoshi K, Tsujii R, Yoshida H, Maki Y, Wada A, Matsui Y, Toh-E A, Mizuta K. Normal assembly of 60 S ribosomal subunits is required for the signaling in response to a secretory defect in Saccharomyces cerevisiae. J Biol Chem. 2002;277:18334–18339. doi: 10.1074/jbc.M201667200. [DOI] [PubMed] [Google Scholar]
- 28.Morita D, Miyoshi K, Matsui Y, Toh-E A, Shinkawa H, Miyakawa T, Mizuta K. Rpf2p, an evolutionarily conserved protein, interacts with ribosomal protein L11 and is essential for the processing of 27 SB Pre-rRNA to 25 S rRNA and the 60 S ribosomal subunit assembly in Saccharomyces cerevisiae. J Biol Chem. 2002;277:28780–28786. doi: 10.1074/jbc.M203399200. [DOI] [PubMed] [Google Scholar]
- 29.Nariai M, Tanaka T, Okada T, Shirai C, Horigome C, Mizuta K. Synergistic defect in 60S ribosomal subunit assembly caused by a mutation of Rrs1p, a ribosomal protein L11-binding protein, and 3′-extension of 5S rRNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2005;33:4553–4562. doi: 10.1093/nar/gki772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyoshi K, Shirai C, Horigome C, Takenami K, Kawasaki J, Mizuta K. Rrs1p, a ribosomal protein L11-binding protein, is required for nuclear export of the 60S pre-ribosomal subunit in Saccharomyces cerevisiae. FEBS Lett. 2004;565:106–110. doi: 10.1016/j.febslet.2004.03.087. [DOI] [PubMed] [Google Scholar]
- 31.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.Zhang C, Liu K, Li T, Fang J, Ding Y, Sun L, Tu T, Jiang X, Du S, Hu J, et al. miR-21: A gene of dual regulation in breast cancer. Int J Oncol. 2016;48:161–172. doi: 10.3892/ijo.2015.3232. [DOI] [PubMed] [Google Scholar]