Abstract
Pancreatic cancer (PC) is one of the most lethal malignances. Identification of biomarkers for early diagnosis of PC is a key imperative. MicroRNAs (miRNAs) have been shown to be valuable biomarkers in the context of several cancers. Exosomes refer to vesicles released by the tumor cells at the early stage of disease. Thus, detection of miRNA in exosomes can be used as a potential biomarker for PC. In this study, we profiled serum levels of miRNAs in patients with chronic pancreatitis (CP) and PC; the role of miR-23b-3p in PC progression was assessed in vitro. Additionally, we assessed, the expression of miR-23b-3p in exosomes isolated from serum samples and assessed the correlation between the expression of miR-23b-3p and carbohydrate antigen 19-9 (CA19-9). Three serum samples each were randomly selected from healthy controls (n=20), and patients with CP (n=18) and PC (n=16) for miRNA microarray profiling. The dysregulated miRNAs were confirmed using qRT-PCR. Four dysregulated miRNAs common to patients with CP and PC were identified on miRNA microarray analysis and confirmed by qRT-PCR. miR-23b-3p level was consistently higher in serum samples from PC patients as compared to those from healthy controls and CP patients (p<0.05). Overexpression of miR-23b-3p promoted proliferation, migration, and invasion capability of PC cells in vitro (p<0.05). Furthermore, miR-23b-3p was upregulated in exosomes of PC serum samples and the supernatant of pancreatic cancer cells (PANC-1), and the expression levels of miR-23b-3p were associated with those of serum CA19-9 levels. This study provides insights into the potential role of miR-23b-3p as a novel biomarker and target for treatment of PC.
Keywords: miRNA, exosome, pancreatic cancer, invasion
Introduction
Pancreatic cancer (PC) is a biologically and clinically challenging disease due to the limited treatment options. Thus, early diagnosis is critical to improve treatment outcomes (1). Circulating miRNAs have been shown to be non-invasive diagnostic biomarkers in many diseases (2). Earlier studies have demonstrated the association between aberrant miRNA expression and development and progression of human cancers (3). For example, circulating miRNAs were proposed as potential biomarkers of diagnostic and prognostic relevance in the context of pancreatic cancer (4).
Chronic pancreatitis (CP) is considered as a premalignant lesion (5); however, the relationship between CP and PC is not fully understood. Thus, in this study, we first profiled serum miRNA levels in patients with PC, those with CP and in normal controls. Then the overlap miRNAs, miR-23b-3p were selected for the following in vitro studies. We assessed the effects of miR-23b-3p knock-in on proliferation, migration, and invasive growth properties of PC cells.
Exosomes are small membranous vesicles (30–100 nm) that are produced by liver cells. They represent a powerful diagnostic tool owing to their relative stability, recoverability in all body fluids, and composition covering a wide range of cancer-related biomarkers including proteins, metabolites, DNA, DNA modifications, and coding and non-coding RNAs. Cancer cells secrete exosomes, and transfer exosomes from primary tumors to the circulation (6). Thus, detection of exosomic miRNA levels in body fluids may serve as novel biomarkers for early diagnosis of tumors, therapeutic monitoring, and for prognostic assessment. In this study, the exosomes in the serum of PC patients were extracted, and the expression of miR-23b-3p was determined; finally the correlation of exosomic miR-23b-3p with CA-19-9 was studied. We analyzed the role of altered miRNAs in the development and progression of PC. The objective was to provide novel insights into use of miR-23b-3p as a therapeutic target and predictor for prevention and treatment of PC in future.
Materials and methods
Ethics statement
The study was approved by the Institutional Review Board (IRB) of Nanjing Medical University. All patients provided written informed consent prior to their enrolment.
Collection of human body liquid samples, RNA extraction and miRNA microarray
The diagnosis of PC was based on the National Comprehensive Cancer Network (NCCN) clinical practice guidelines 2016 (7). CP was diagnosed based on the American Pancreatic Association's practice guidelines for chronic pancreatitis, 2014 (8). Blood samples from 20 healthy controls and serum samples of 18 CP and 16 PC patients were collected at the Wuxi People's Hospital. All body fluid specimens were flash-frozen upon collection and stored at −80°C until further use.
Three serum samples from each group were randomly selected based on the shortest sampling time for miRNA microarray analysis. Total RNA was isolated from these frozen body fluids using RNeasy Mini kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The quality and quantity of the isolated RNA was then assessed using agarose gel electrophoresis and a spectrophotometer (Sangon Biotech, Shanghai, China), and then labeled with biotin and hybridized to a GeneChip miRNA 3.0 Array from Affymetrix (Santa Clara, CA, USA), according to the manufacturer's protocol. Following hybridization, the images were digitized and analyzed using a laser scanner interfaced with Affymetrix GeneChip Command Console (AGCC). The most differentially expressed miRNAs in the CCA serum samples were identified and compared to those from normal control and PC samples.
Quantitative real-time RT-PCR (qRT-PCR)
miRNA microarray-identified miRNA expression was further confirmed with qRT-PCR using total RNA from serum samples and exosomes. In brief, RNA samples were converted into cDNA using 0.5 µg RNA sample and a reverse transcription kit (Takara, Ohtsu, Shiga, Japan). qRT-PCR was performed using the iQ™ SYBR® Green Supermix (Bio-Rad) in an ABI 7500 qPCR system (Applied Biosystems, Foster City, CA, USA). The optimal dilution and melting curves were utilized to quantify each of the amplified production using specific primer sets. Bulge-loop™ miRNA qRT-PCR primer sets (one RT primer and a pair of qRT-PCR primers for each miRNA) were designed and produced by RiboBio (Guangzhou, China) and ribosome RNA 5S was used as an internal control for miR-23b-3p level in sera and bile. The relative quantification of each gene was measured using the 2−∆∆CT method. We performed qRT-PCR in serum samples from 20 healthy controls, 18 patients with CP and 16 patients with PC; all measurements were performed in triplicate.
Cell culture and transfection
Human PC cell lines pancreatic cancer cells (PANC-1) cultured in DMEM containing 10% fetal bovine sera, 100 g/ml streptomycin, and 100 U/ml penicillin. The synthesized miR-23b-3p mimic (miR-23b-3pM) and non-specific mimic (NSM) were purchased from RiboBio. Here, 70% of confluent cells were transfected using 20 nM of miR-23b-3pM or NSM with Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA). Cells were allowed to incubate for 48 h after transfection and then harvested for further studies.
Cell growth curve, proliferation, invasion study, and wound healing assay
Ten thousand cells were transfected and placed in 24-well plates (Corning, USA). The number of cells was determined every other day with a hemocytometer under an inverted-light microscope.
We also performed the 5-ethynyl-20-deoxyuridine-Apollo 488 (EdU-Apollo 488) incorporation assay to assess the cell proliferation capacity. CCA cells were seeded into 96-well plates at a density of 3×104 cells/ml and 24 h later, cells were transfected with 20 nM of NSM or 23b-3pM for 6 h. The cells were stained with EdU and Hoechst 33342 separately from a kit from RiboBio, according to the manufacturer's instructions. The proliferating cells were checked under the fluorescence microscope and the cell proliferation rate calculated by the formula: (number of proliferating cells/total cells) × 100%.
For the invasion assay, 2.5×104 transfected cells were placed in 24-well plates with inserts
This established a two-chamber system divided by a cell-permeable membrane coated with Matrigel (R&D Systems, USA). Cells were allowed to incubate for 24 h, fixed, and stained with crystal violet. Then invasive cells were counted and images of 10 random fields under an inverted-light microscope were captured.
For the wound healing assay, 2.5×104 transfected cells were cultured in 24-well plates. A line was drawn when cells reached ~90% confluence. At 0, 24, and 48 h, images of the same 10 fields were recaptured.
Exosome isolation, characterization, uptake, and miRNA expression detection
Serum as well as supernatant of PANC-1 cells were transferred to ultracentrifuge tubes, and centrifuged at 16,500 × g for 20 min at 4°C to further remove cells and cell debris; the supernatant was filtered through a 0.2-µm filter to remove particles sized >200 nm. The filtered supernatant was transferred to new ultracentrifuge tubes and the tubes sealed prior to ultracentrifugation at 120,000 × g for 70 min at 4°C to obtain exosome pellets. The supernatant was discarded, and the exosomes were collected. For maximal exosome retrieval, the exosome enriched pellet was resuspended repeatedly in a small volume tube (9). Then total RNA was isolated from these exosomes using TRIzol as described elsewhere (10).
For the cell uptake experiment, the exosomes were stained with oil red, and washed using an ultra-filtration membrane (10 kDa, Merck Millipore, Darmstadt, Germany) to remove any free nucleotides. 1,1′-dioctadecyl-3,3,3′,3–tetramethylindocarbocyanineperchlorate (Dil) was used to label the membrane component of the exosomes. The exosomes were incubated with 10 µg/ml DiI (Beyotime, Nantong, China) for 15 min at 37°C and then washed twice with cold phosphate-buffered saline (PBS). The labeled exosomes were added to PANC-1 cells grown on chamber slides of a 35-mm dish with sterile small circular glass and incubated at 37°C. After 60 min, the cells were harvested and washed twice with PBS. Then, the cells were stained with 4′-6-diamidino-2-phenylindole (DAPI; Invitrogen) to visualize the nuclei and were examined using an UltraView VoX confocal fluorescence microscope (Perkin-Elmer, Waltham, MA, USA) (11).
Statistical analysis
All data are presented as mean ± SEM. One-way or two-way ANOVA was performed for statistical analysis, using GraphPad Prism 5. Differences between group means with p-values <0.05 were regarded as statistically significant. The correlations were assessed by calculation of Pearson correlation coefficients.
Results
Bodily fluid miRNA expression profiles are significantly altered in CP and PC
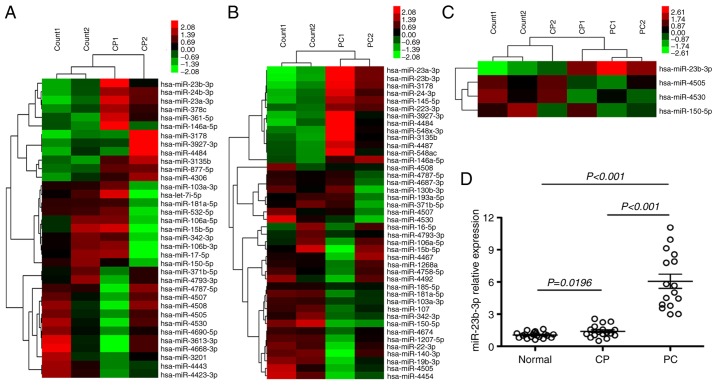
miRNA microarrays were used to assess the miRNA expression profiles of patients with CP and PC in order to assess changes in body fluid miRNAs from chronic inflammation to cancers. Thirty-five dysregulated serum miRNAs were found in CP, which were not observed in healthy controls (p<0.01). Of these, 12 were upregulated and 23 were downregulated (Fig. 1A). There were 42 dysregulated serum miRNAs in PC that were not found in healthy controls (p<0.01). Of these, 14 were upregulated and 28 were downregulated (Fig. 1B). Four miRNAs were verified altered in both CP and PC; among these, miR-23b-3p was upregulated, while 3 miRNAs, miR-4505, 4530, and 150-5p were downregulated (Fig. 1C). qRT-PCR was performed to determine the expression of miR-23b-3p in the serum of CP and PC patients. miR-23b-3p was found upregulated in sera of both CP and PC, and the expression level in PC was higher than that in CP (p<0.01) (data pertaining to 3 downregulated miRNAs is not shown). This finding was consistent with the microarray data (Fig. 1D).
Figure 1.
Microarray analysis of the serum miRNA expression profile of patients with CP and PC using qRT-PCR verification. Serum miRNA expression profile was determined using the LAN-based miRNA microarray. (A) There were 35 dysregulated serum miRNAs (p<0.01) in patients with CP and (B) 42 dysregulated serum miRNAs (p<0.01) in patients with PC, as compared to that in the healthy control group. (C) Among these dysregulated miRNAs, 4 miRNAs were verified to be common to both the groups. The top bar represents the signal levels of miRNA expression from 2.08 (green) to 2.73 (red). The individual identity of the significantly dysregulated miRNAs is shown on the right. (D) The expression of miR-23b-3p was verified on qRT-PCR. The x-axis represents the different groups and the y-axis represents the relative expressions. qRT-PCR was performed in sera obtained from 18 CP, 16 PC and 20 healthy controls. The horizontal bars represent mean (± SEM) miR-23b-3p expression level. qRT-PCR was performed in triplicate.
miR-23b-3p promotes the ability of growth, proliferation, invasion, and migration of PANC-1 cells in vitro
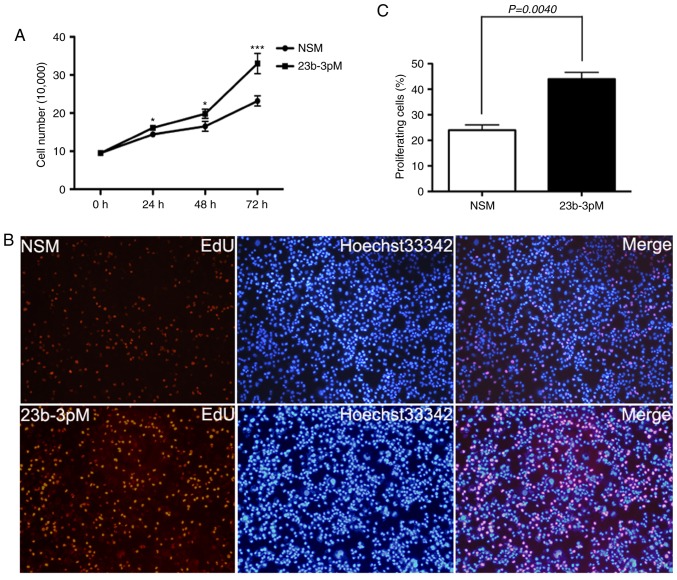
Cell counting and EdU incorporation assays showed that transfection of PANC-1 cells with 23b-3pM increase tumor cell growth and proliferation rate as compared to those of NSM-transfected cells (p<0.05; Fig. 2).
Figure 2.
Results of cell assay showing the effect of miR-23b-3p expression on regulation of growth and proliferation of PC. (A) Cell counting assay. PANC-1 cells (10,000) were transiently transfected with miR-23b-3p mimic (23b-3pM) and non-specific mimic (NSM), respectively, and the number of cells were counted at 0, 24, 48 and 72 h. Mean (± SEM) values of three independent experiments are presented; **p<0.01. (B) EdU incorporation assay. 3×104 cells/ml were seeded on 96-well plates and grown for 24 h and then transiently transfected with 23b-3pM and NSM for 48 h. Cells were stained with EdU-Apollo 488 and Hoechst 33342; Hoechst 33342 blue fluorescence represents all live cells and the Edu-Apollo488 green fluorescence represents the proliferating cells. (C) The number of cells in 10 randomly selected fields were counted under inverted microscope. The proliferation ratio was calculated as number of proliferating cells/total number of cells × 100%. Mean (± SEM) values of three independent experiments are presented; *p<0.05, **p<0.01, ***p<0.001.
Transwell and wound healing assays were used to analyze the effect of miR-23b-3p on in vitro invasion and metastasis of PANC-1 cells, respectively. The overexpression of miR-23b-3p in PANC-1 cells caused substantially greater cell invasion through the Transwell chamber at 48 h, as compared to that observed among cells transfected with NSM (p<0.05). The wound healing scratch test revealed that introduction of miR-23b-3p also accelerated the mobility of PANC-1 cells at 48 h after scratching, relative to cells transfected with NSM (p<0.05) (Fig. 3).
Figure 3.
Effect of miR-23b-3p expression on regulation of PANC-1 cell migration and invasion capacity in vitro. (A) Wound healing study. After 48 h of transfection, 2.5×104 cells were trypsinized and seeded in 24-well plates. A line was scratched using a sterile 50-µl pipette tip after the cells reached 90% confluence. The cells were observed in same 10 views under inverted microscope and images captured at 0 and 48 h after scratching. Results showed the migration distance of cells transfected with miR-23b-3p mimic (23b-3pM) relative to that of cells transfected with non-specific mimic (NSM). Data represents the mean value of three independent experiments. Bars represent the mean ± SEM (n=3) of area % of migrated cells per field. (B and C) Invasion of PANC-1 cells. After 48 h of transfection, 2.5×104 cells were trypsinized and seeded in 24-well plates with Matrigel-coated membranes for the invasion assays. Cells that invaded through the Matrigel and reached the bottom of the insert were fixed with formalin, and stained with crystal violet. The number of cells in 10 randomly selected fields was counted under inverted microscope at 48 h after incubation. Data represents the mean value of three independent experiments. Bars represent the mean (± SEM) number of invaded cells per field.
miR-23b-3p was translocated into human PC cells in an exosome-mediated manner
The exosomes in supernatant of PC patients sera as well as in pancreatic cell lines were isolated; the expression of exosomic miR-23b-3p was checked by RT-PCR. The results showed overexpression of exosomic miR-23b-3p in sera of patients with PC when compared with that in sera of healthy controls (Fig. 4A). Moreover, the exosomic miR-23b-3p expression in the supernatant of PANC-1 was higher than that in the pancreatic ductal epithelial cells (HPDE6c-7) (Fig. 4B). Furthermore, exosomes were labeled with the membrane dye Dil and the labeled exosomes were added to the culture medium of the PANC-1 cells. After co-culture for 60 min with unlabeled recipient cells, the labeled exosomes as well as PANC-1 cells were observed using a confocal microscope; it was detected that PANC-1 cells can take up exosomes (Fig. 4C). Our results demonstrated that the pancreatic cancer cells can release exosomes into the circulation and that the serum exosomes containing miRNAs can in turn be taken up by pancreatic cancer cells and may subsequently mediate cellular functions.
Figure 4.
Characterization of serum exosomic miR-23b-3p translocation into PANC-1 cells. Exosomes were isolated using centrifugation, filtration, and ultracentrifugation as described in Materials and methods. (A) qRT-PCR analysis of exosomic miR-23b-3p expression in 16 serum samples of PC patients and 20 serum samples of healthy controls (normal). (B) qRT-PCR analysis of exosomic miR-23b-3p contained in the supernatant of pancreatic ductal epithelial cell line (HPDE6c-7) and pancreatic cancer cell line (PANC-1 cells). Mean (± SEM) values of three independent experiments are presented. (C) Microscopic analysis of the translocation of the pre-labeled exosomes; the DiI-labeled exosomes display red color; the cell nuclei are stained with DAPI (blue).
Upregulated sera as well as exosomic miR-23b-3p was closely related to CA-19-9
In order to investigate the relationship between miR-23b-3p and CA-19-9, we examined the correlation between the two. miR-23b-3p in sera of PC patients showed a significant positive correlation with CA-19-9 (R=0.6511; Fig. 5A). Moreover, the exosomic miR-23b-3p expression in the sera of PC patients also showed a positive correlation with that of CA-19-9 (R=0.7142; Fig. 5B).
Figure 5.
Correlation analysis of miR-23b-3p with CA-19-9. (A) The correlation between expression of miR-23b-3p and CA-19-9 in serum samples of PC patients was examined. (B) The correlation between the expression of exosomic miR-23b-3p and CA-19-9 from the serum samples of PC patients was examined. r represents a correlation coefficient; a positive r-value implies a positive correlation; mean ± SEM.
Discussion
There is a need for more sensitive diagnostic markers for PC as it is a highly aggressive malignancy with no evident symptoms at early stages and a low overall 5-year survival rate (<5%). miRNA is a desirable tool and biomarker because of its high stability in many types of body fluids. Several blood-circulating miRNAs have been shown to be associated with cancer development and progression and can serve as novel potential diagnostic and prognosis biomarkers in the context of PC (4). Earlier studies have shown that miR-224 and miR-150-5p regulate tumor invasion and migration of cholangiocarcinoma cells and can be biomarkers for tumor diagnosis (12,13). In this study, microarray was applied to check the deregulation of circulating miRNAs in PC. miR-23b-3p was verified to be the only upregulated miRNA in both PC and CP groups, as compared to that in normal controls. Furthermore, the extent of upregulation in PC group was higher than that in the CP group. Previous studies have found upregulated expression of miR-23b-3p in renal (14), gastric (15), and non-small cell lung cancers (16). miR-23b-3p acts as an oncogene in these cancers; however, this was not found to be the case in PC, which may be attributable to racial differences in the study population (17–19). This study verified the overexpression of serum miR-23b-3p in patients with PC and CP; the level of upregulation in PC patients was higher than that in CP, which was consistent with the microarray data described above. Overexpression of miR-23b-3p in vitro promoted the PANC-1 cells proliferation, invasion, and migration ability. It is speculated that miR-23b-3p may serve as a novel potential diagnostic and prognostic biomarker in the context of PC. However, the function and role of miR-23b-3p alterations in PC development and progression needs to be further studied.
Exosomes are extracellular vesicles 30–100 nm in size and play a key role in intercellular communications and in regulating diverse biological processes. We sought to build on recently reported findings and sought to establish the potential diagnostic value of serum exosomes in the context of PC (20). In this study, PANC-1 cells were shown to produce exosomes in the supernatant and take them up as well; furthermore, miR-23b-3p was found to be overexpressed in the exosomes in sera of patients with PC. Currently, CA-19-9 is a traditional clinical biomarker for PC diagnosis (21), but several more novel biomarkers, such as miRNAs and proteomics need to be further studied (22). Many studies have suggested the utility of combined use of miRNAs and CA19-9 as biomarkers for diagnosis of PC (23,24). Exosomic miRNAs have also been proposed as potential biomarkers for the diagnosis of PC (25–27); however, the relationship between exosomic miRNA and CA19-9 is not clear. In this study, miR-23b-3p expression in sera or that in the exosomes isolated from sera showed a close relationship with CA-19-9 expression. However, larger studies on higher number of patients are required to assess the superiority of use of miR-23b-3p over that of CA-19-9 for diagnostic purposes, and to determine if combined use of miR-23b-3p and CA-19-9 is more sensitive and specific for diagnosis of early PC, as compared to use of either of these parameters individually.
In conclusion, we determined differential miRNA expression profiles in sera of patients with CP and PC. The expression of miR-23b-3p was upregulated in the sera specimens, and in vitro overexpression of miR-23b-3p promoted proliferation, migration, and invasive growth of PC cells. Moreover, upregulated miR-23b-3p was also detected in the exosomes from the sera of PC patients; miR-23b-3p in both serum and exosomes was positively associated with CA19-9 in PC patients. However, this study is just a proof-of-principle investigation; more studies are needed to fully disclose the function and role of exosomic miRNA alterations in the development and progression of PC, and for the establishment of miR-23b-3p as a novel target and diagnostic and prognostic marker for PC.
Acknowledgements
The authors would like to thank our staff in both Departments for their contributions to this study. This study was supported in part by grants from the National Natural Science Foundation of China (no. 81502038 to F.A., no. 81302104 to J.D. and no. 81302382 to Y.J.), the Natural Science Foundation of Jiangsu Province (no. BK2012098 to F.A.), the Youth Medical Talent of Jiangsu Province (no. QNRC2016187 to F.A.) and Wuxi Medical Innovation Team (no. CXTD005 to Q.Z.).
References
- 1.Karanikas M, Esempidis A, Chasan ZT, Deftereou T, Antonopoulou M, Bozali F, Amarantidis K, Man YG. Pancreatic cancer from molecular pathways to treatment opinion. J Cancer. 2016;7:1328–1339. doi: 10.7150/jca.15419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghai V, Wang K. Recent progress toward the use of circulating microRNAs as clinical biomarkers. Arch Toxicol. 2016;90:2959–2978. doi: 10.1007/s00204-016-1828-2. [DOI] [PubMed] [Google Scholar]
- 3.Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell. 2013;153:516–519. doi: 10.1016/j.cell.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Ebrahimi S, Hosseini M, Ghasemi F, Shahidsales S, Maftouh M, Akbarzade H, Parizadeh SA, Hassanian SM, Avan A. Circulating microRNAs as novel potential diagnostic and prognosis biomarkers in pancreatic cancer. Curr Pharm Des. 2016;22:6444–6450. doi: 10.2174/1381612822666160817095047. [DOI] [PubMed] [Google Scholar]
- 5.Midha S, Sreenivas V, Kabra M, Chattopadhyay TK, Joshi YK, Garg PK. Genetically determined chronic pancreatitis but not alcoholic pancreatitis is a strong risk factor for pancreatic cancer. Pancreas. 2016;45:1478–1484. doi: 10.1097/MPA.0000000000000684. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z, Chen JQ, Liu JL, Tian L. Exosomes in tumor microenvironment: Novel transporters and biomarkers. J Transl Med. 2016;14:297. doi: 10.1186/s12967-016-1056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Hawary M. Role of imaging in diagnosing and staging pancreatic cancer. J Natl Compr Canc Netw. 2016;14(Suppl):678–680. doi: 10.6004/jnccn.2016.0191. [DOI] [PubMed] [Google Scholar]
- 8.Conwell DL, Lee LS, Yadav D, Longnecker DS, Miller FH, Mortele KJ, Levy MJ, Kwon R, Lieb JG, Stevens T, et al. American Pancreatic Association Practice Guidelines in Chronic Pancreatitis: Evidence-based report on diagnostic guidelines. Pancreas. 2014;43:1143–1162. doi: 10.1097/MPA.0000000000000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lässer C, Eldh M, Lötvall J. Isolation and characterization of RNA-containing exosomes. J Vis Exp. 2012;59:e3037. doi: 10.3791/3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.An F, Gong B, Wang H, Yu D, Zhao G, Lin L, Tang W, Yu H, Bao S, Xie Q. miR-15b and miR-16 regulate TNF mediated hepatocyte apoptosis via BCL2 in acute liver failure. Apoptosis. 2012;17:702–716. doi: 10.1007/s10495-012-0704-7. [DOI] [PubMed] [Google Scholar]
- 11.Tang Y, Cui Y, Li Z, Jiao Z, Zhang Y, He Y, Chen G, Zhou Q, Wang W, Zhou X, et al. Radiation-induced miR-208a increases the proliferation and radioresistance by targeting p21 in human lung cancer cells. J Exp Clin Cancer Res. 2016;35:7. doi: 10.1186/s13046-016-0285-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang M, Wu X, Cao H, Zhan Q, Xia M, Zhou Q, Cai X, An F. Regulatory role of serum miR-224 in invasiveness and metastasis of cholangiocarcinoma. Zhonghua Gan Zang Bing Za Zhi. 2015;23:748–753. doi: 10.3760/cma.j.issn.1007-3418.2015.10.008. (In Chinese) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu X, Xia M, Chen D, Wu F, Lv Z, Zhan Q, Jiao Y, Wang W, Chen G, An F. Profiling of downregulated blood-circulating miR-150-5p as a novel tumor marker for cholangiocarcinoma. Tumour Biol. 2016;37:15019–15029. doi: 10.1007/s13277-016-5313-6. [DOI] [PubMed] [Google Scholar]
- 14.Zaman MS, Thamminana S, Shahryari V, Chiyomaru T, Deng G, Saini S, Majid S, Fukuhara S, Chang I, Arora S, et al. Inhibition of PTEN gene expression by oncogenic miR-23b-3p in renal cancer. PLoS One. 2012;7:e50203. doi: 10.1371/journal.pone.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.An Y, Zhang Z, Shang Y, Jiang X, Dong J, Yu P, Nie Y, Zhao Q. miR-23b-3p regulates the chemoresistance of gastric cancer cells by targeting ATG12 and HMGB2. Cell Death Dis. 2015;6:e1766. doi: 10.1038/cddis.2015.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Begum S, Hayashi M, Ogawa T, Jabboure FJ, Brait M, Izumchenko E, Tabak S, Ahrendt SA, Westra WH, Koch W, et al. An integrated genome-wide approach to discover deregulated microRNAs in non-small cell lung cancer: Clinical significance of miR-23b-3p deregulation. Sci Rep. 2015;5:13236. doi: 10.1038/srep13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kojima M, Sudo H, Kawauchi J, Takizawa S, Kondou S, Nobumasa H, Ochiai A. MicroRNA markers for the diagnosis of pancreatic and biliary-tract cancers. PLoS One. 2015;10:e0118220. doi: 10.1371/journal.pone.0118220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin MS, Chen WC, Huang JX, Gao HJ, Sheng HH. Aberrant expression of microRNAs in serum may identify individuals with pancreatic cancer. Int J Clin Exp Med. 2014;7:5226–5234. [PMC free article] [PubMed] [Google Scholar]
- 19.Cao Z, Liu C, Xu J, You L, Wang C, Lou W, Sun B, Miao Y, Liu X, Wang X, et al. Plasma microRNA panels to diagnose pancreatic cancer: Results from a multicenter study. Oncotarget. 2016;7:41575–41583. doi: 10.18632/oncotarget.9491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu L, Risch HA. Exosomes: Potential for early detection in pancreatic cancer. Future Oncol. 2016;12:1081–1090. doi: 10.2217/fon-2015-0005. [DOI] [PubMed] [Google Scholar]
- 21.Cao S, Hu Y, Gao X, Liao Q, Zhao Y. Serum carbohydrate antigen 19-9 in differential diagnosis of benign and malignant pancreatic cystic neoplasms: A meta-analysis. PLoS One. 2016;11:e0166406. doi: 10.1371/journal.pone.0166406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ballehaninna UK, Chamberlain RS. Biomarkers for pancreatic cancer: Promising new markers and options beyond CA 19-9. Tumour Biol. 2013;34:3279–3292. doi: 10.1007/s13277-013-1033-3. [DOI] [PubMed] [Google Scholar]
- 23.Gao L, He SB, Li DC. Effects of miR-16 plus CA19-9 detections on pancreatic cancer diagnostic performance. Clin Lab. 2014;60:73–77. doi: 10.7754/Clin.Lab.2013.121210. [DOI] [PubMed] [Google Scholar]
- 24.Wang WS, Liu LX, Li GP, Chen Y, Li CY, Jin DY, Wang XL. Combined serum CA19-9 and miR-27a-3p in peripheral blood mononuclear cells to diagnose pancreatic cancer. Cancer Prev Res (Phila) 2013;6:331–338. doi: 10.1158/1940-6207.CAPR-12-0307. [DOI] [PubMed] [Google Scholar]
- 25.Machida T, Tomofuji T, Maruyama T, Yoneda T, Ekuni D, Azuma T, Miyai H, Mizuno H, Kato H, Tsutsumi K, et al. miR-1246 and miR-4644 in salivary exosome as potential biomarkers for pancreatobiliary tract cancer. Oncol Rep. 2016;36:2375–2381. doi: 10.3892/or.2016.5021. [DOI] [PubMed] [Google Scholar]
- 26.Madhavan B, Yue S, Galli U, Rana S, Gross W, Müller M, Giese NA, Kalthoff H, Becker T, Büchler MW, et al. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int J Cancer. 2015;136:2616–2627. doi: 10.1002/ijc.29324. [DOI] [PubMed] [Google Scholar]
- 27.Zöller M. Pancreatic cancer diagnosis by free and exosomal miRNA. World J Gastrointest Pathophysiol. 2013;4:74–90. doi: 10.4291/wjgp.v4.i4.74. [DOI] [PMC free article] [PubMed] [Google Scholar]