Abstract
Time-resolved analysis of resting-state functional magnetic resonance imaging (rs-fMRI) data allows researchers to extract more information about brain function than traditional functional connectivity analysis, yet a number of challenges in data analysis and interpretation remain. This article briefly summarizes common methods for time-resolved analysis and presents some of the pressing issues and opportunities in the field. From there, the discussion moves to interpretation of the network dynamics observed with rs-fMRI and the role that rs-fMRI can play in elucidating the large-scale organization of brain activity.
Keywords: : dynamic analysis, dynamic connectivity, network dynamics, time-resolved resting-state fMRI
Introduction
Resting-state functional magnetic resonance imaging (rs-fMRI), based on spontaneous fluctuations of the blood oxygenation level-dependent (BOLD) signal, has become a powerful and popular tool for the study of normal and dysfunctional brain activities. Traditional methods of analysis identify spatial patterns of BOLD signal coordination that are assumed to persist for the duration of the entire rs-fMRI scan (∼5–10 min or longer), which we will refer to as average functional connectivity. However, using the entire time series for a single connectivity calculation disregards the vast amount of dynamic information that is present in the rs-fMRI data. Researchers are increasingly turning to analyses that capture time dependence in the data as a way to extract more information about brain function, using methods ranging from windowed versions of standard seed-based correlation or independent component analysis (ICA) techniques to new methods that consider information from individual time points and/or identify change points in the rs-fMRI signal.
As the field of time-resolved rs-fMRI and functional connectivity analysis has grown, a number of challenges, opportunities, open questions, and new areas of inquiry have arisen. This article summarizes discussion of these topics from the Dynamic Connectivity Satellite Symposium at the Resting-State Functional Connectivity Workshop in Vienna in September 2016. We begin with a summary of current approaches to analysis of rs-fMRI data that incorporate time dependence and describe some of the existing technical challenges in the field, including the definition of null models for validation and statistical inference. Note, in this overview, we use the terms time-varying and dynamic interchangeably, although we recognize that one can distinguish between, for example, dynamic state models and static models, both of which can be used to characterize time-dependent signals (e.g., an oscillatory signal can be modeled with a static model). Our main focus is on approaches that move beyond querying parameters that represent averages over the entire experiment (e.g., a single set of nodes and edges) and instead capture information about changes over time in activity or connectivity. From there, we move to a more open-ended consideration of how to interpret large-scale patterns of time-varying activity and explore rs-fMRI's potential contribution to neuroscience.
Summary of Current Approaches
The vast majority of studies that use rs-fMRI to examine time-varying changes in the brain employ a windowed version of traditional analysis techniques, primarily correlation and/or ICA (Fig. 1). When used over the whole scan period, these techniques map the spatial distribution of the networks and provide a single measure of statistical dependence, for example, linear correlation, between the time courses for any pair of voxels, regions, or networks of interest. When applied in a windowed manner, in contrast, the results are maps of spatial extent and/or correlation values that vary over time. However, sliding windows are not the only available approach; there is a rich set of tools that have been proposed over the past few years that are unveiling the utility in characterizing the dynamic reconfiguration of brain activity and connectivity. For example, change points in functional connectivity can be identified based on the covariance matrix of partitioned (i.e., temporally windowed) data, or dynamic analyses can focus on the signal amplitude to identify individual events (Fig. 2). One can also extract spatiotemporal patterns of dynamic activity that repeat over the course of the scan (Fig. 3). In this section, we briefly summarize some of the most widely used techniques.
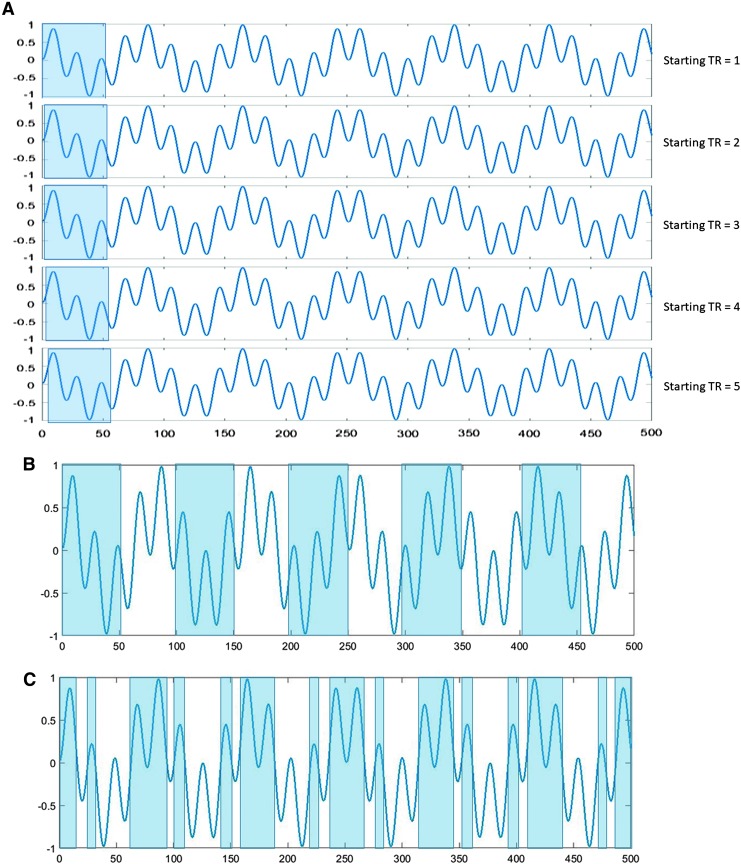
FIG. 1.
Windowed analysis of time series. (A) Sliding window, showing the initial window position and four subsequent window positions. Fifty time points are included in each window and the window position is incremented by one time point, resulting in 451 windows across the 500-point time series. (B) Segmented time course using constant nonoverlapping windows (shown in alternating blue/white). Each window contains fifty time points, resulting in 10 windows over the course of the 500-time point scan. (C) Data-driven segmentation or adaptive windowing, with nonoverlapping windows of variable size. For this demonstration, the windows are determined by the zero crossings of the signal and result in 29 windows over the course of the scan. Color images available online at www.liebertpub.com/brain
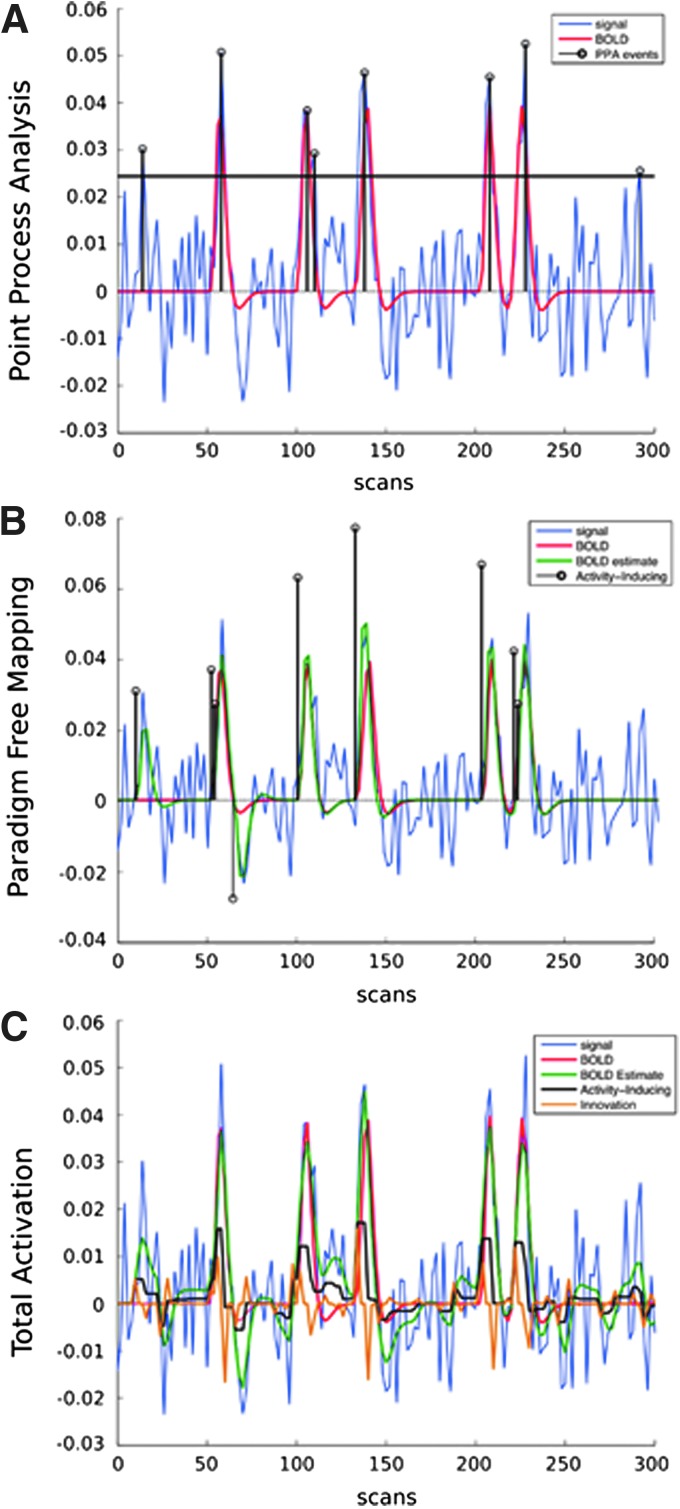
FIG. 2.
Simulation results for (A, top) PPA; (B, middle) PFM; and (C, bottom) TA. The simulated time series includes five BOLD events modeled as the convolution of 4-sec-long activations with the canonical HRF, plus uncorrelated Gaussian noise. PPA identifies time points that are local maxima as well as with amplitude larger than 1.5 of the standard deviation of the signal. PFM deconvolves voxelwise time series to estimate activity-inducing signal (events) based on the shape of the canonical HRF through a sparse temporal regularization estimator. Once events are identified, the corresponding BOLD signal can be estimated as the convolution of the identified events with the canonical HRF. Finally, the TA model is also able to detect sustained activity-inducing events (e.g., several seconds long). It also allows to compute innovation signals as the temporal derivative of activity-inducing signals. To allow comparisons between PFM and TA, TA was implemented using only the sparse temporal regularization term (and not spatial) for the sole purpose of this figure. BOLD, blood oxygenation level-dependent; PFM, paradigm free mapping; PPA, point process analysis; TA, total activation. Color images available online at www.liebertpub.com/brain
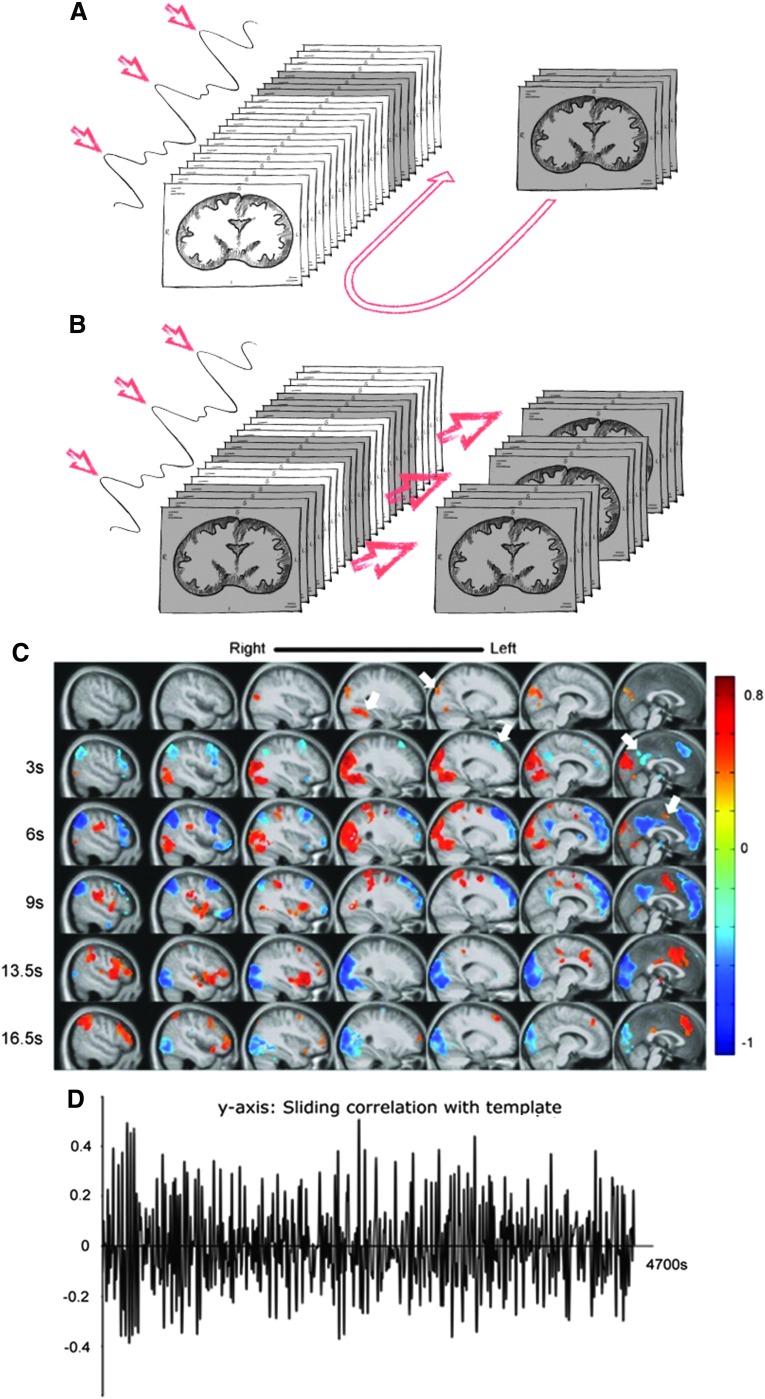
FIG. 3.
Detection of quasi-periodic patterns. (A) A spatiotemporal chunk of data with a predefined length (the template) is randomly chosen from the scan and correlation between it and the rest of the scan is calculated in a sliding window manner. (B) The time points with the highest correlation with the template are identified and the corresponding chunks of data are averaged together to create a new template. The process is repeated until the template converges, resulting in a single 4D template for the scan and a correlation time course that describes the strength of the template at any given time. (C) The final template calculated from a group of healthy human subjects (Majeed et al., 2011). The DMN and TPN exhibit alternating activity. (D) The time course of correlation between the scan and the template, showing the strength of the template at each time point for the concatenated subject data. DMN, default mode network; TPN, task positive network. Color images available online at www.liebertpub.com/brain
Windowed coherence, correlation, or covariance-based methods
As in standard studies of average functional connectivity, dynamic analysis is often used under the assumption that the relationships between areas are of greater interest than the relative signal amplitudes. Coherence, correlation, and covariance provide information about the similarity between signals from different areas. Sliding window correlation analysis is widely used to examine dynamic connectivity presumably because it is relatively straightforward and can be implemented using regions of interest (ROIs) or ICA-derived time courses (Allen et al., 2014; Chang and Glover, 2010; Handwerker et al., 2012; Hutchison et al., 2013; Keilholz et al., 2013; Petridou et al., 2013; Sakoǧlu et al., 2010). For this method, a window is moved along the scan from beginning to end and correlation between the areas or components of interest is calculated for each window, resulting in a plot of correlation as a function of time (Fig. 1A). Different studies use different window lengths, but the length is generally kept constant throughout the analysis. Consecutive windows may overlap maximally (all time points are the same except one), minimally (no time points are the same; Fig. 1B), or at some level in between. Window length, shape, and overlap for best performance are still not known, as discussed further in the section on technical challenges. For whole-brain studies, the brain is often first parcellated into a manageable number of ROIs or components. Coarse parcellations increase the signal-to-noise ratio (SNR) of the time courses through averaging, while finer parcellations improve the homogeneity of the time courses that are averaged. Sliding window correlation is then calculated pairwise between the time courses from all parcels for each window to create a series of correlation matrices that can be used for further analysis (Allen et al., 2014; Gonzalez-Castillo et al., 2015; Hutchison et al., 2013; Li et al., 2014; Ma et al., 2017). Sliding window correlation can also be computed on the first-order temporal derivative of the time series, an approach referred to as multiplication of temporal derivatives (Shine et al., 2015, 2016). This is equivalent to high-pass filtering with a cutoff frequency fcutoff = 0.25/repetition time (TR) (Oppenheim and Schafer, 2009).
The choice of window length is critical for sliding window approaches because it influences the ability to extract information from the data and the interpretation of the results (Leonardi and Van De Ville, 2015). To overcome this constraint, time–frequency analyses enable the exploration of time-varying connectivity at multiple frequencies, which is conceptually equivalent to adapting the analysis to different window lengths (Fig. 1C). In one of the earliest demonstrations, Chang and Glover (2010) focused on wavelet coherence between selected ROIs. They identified periods with significant levels of coherence. The statistical rigor and use of both frequency and time information taken by this approach are appealing, but difficult to transfer to whole-brain studies due to the explosion in the dimensionality of the data, perhaps explaining why this approach has not been widely utilized. More recently, approaches to obtain information about the multiple frequencies that mediate dynamic functional connectivity at the level of the whole brain have been introduced (Miller et al., 2016a; Yaesoubi et al., 2015, 2017).
Change point detection
Another approach that avoids the challenges involved in choosing an appropriate window length involves data-driven temporal segmentation of the rs-fMRI data (Chen et al., 2016a; Cribben et al., 2012, 2013; Lindquist et al., 2014; Ou et al., 2014; Xu and Lindquist, 2015; Zhang et al., 2014). The goal of these methods is to determine when a change in brain state occurs based on properties of the data itself. The segmentation can be accomplished using simple methods (clustering based on the amplitude of the signal) or with more sophisticated state–space models (hidden Markov models) that consider the covariance as well as the amplitude of the time series, although at the cost of additional computational complexity (Chen et al., 2016a; Eavani et al., 2013; Ryali et al., 2016; Suk et al., 2016; Taghia et al., 2017). Still other methods use the properties of the signal or the relationship between the signals from different areas to identify times when the large-scale organization of brain activity changes (Lindquist et al., 2014; Ou et al., 2014; Xu and Lindquist, 2015; Zhang et al., 2014). In some cases, these methods can be considered to be a version of windowed analysis techniques in which the window size is adaptively varied in response to data properties (Xu and Lindquist, 2015). The spatial patterns in each cluster and the timing of their occurrence through the scan can then be used in further analysis.
Event-based analysis
In contrast to methods based on relationships between brain areas, analysis techniques based on amplitude changes do not necessarily assume that changes occur at the network level. At the heart of many of these methods is the idea that activity in any given area primarily comprises distinct spontaneous events that each give rise to a hemodynamic response, similar to the response that occurs for a task or stimulus. This assumption is based on the hypothesis that these spontaneous BOLD events originate from neuronal events such as avalanching activity (Tagliazucchi et al., 2011, 2012a). The timing of the events can be deciphered from BOLD fluctuations using a variety of approaches. Straightforward detection of single events can be accomplished by thresholding the time courses of the voxel or ROI based on amplitude, where the threshold can be based on standard deviation of the time series (Tagliazucchi et al., 2011, 2012a; Wu et al., 2013) or the local maximum (or minimum) of the signal (Laumann et al., 2016; Tagliazucchi et al., 2016). This approach is known as point process analysis (PPA; Fig. 2A). Alternatively, events can be identified through deconvolution of a given hemodynamic model from the time series (Caballero Gaudes et al., 2013; Karahanoğlu et al., 2013; Petridou et al., 2013). Hemodynamic deconvolution to estimate the underlying neuronal signal is commonly applied to investigate psychophysiologic interactions (PPIs) in task-based functional connectivity studies (Gerchen et al., 2014; Gitelman et al., 2003) and in rs-fMRI (Di and Biswal, 2015). Contrary to the classical formulation of PPI analysis, recent deconvolution approaches employ sparsity-promoting estimators based on the assumption that the dynamics of spontaneous brain activity can be characterized by looking at sparse BOLD events (Karahanoğlu et al., 2013; Petridou et al., 2013), similar to the hypothesis underlying PPA-based approaches (Tagliazucchi et al., 2012a).
The dynamics of the brain activity generated by individual events can be visualized by watching the sequence of events. The complexity of these sequences (which explodes with large number of ROIs or in whole-brain analyses) and the inherent variability in the timing of events across datasets make drawing inferences a challenging task. Consequently, once the events are detected, a variety of postprocessing methods have been developed to summarize the spatial and temporal distribution of the events. As a first approximation, the timings of the events can be employed as onsets in a standard general model analysis (Caballero Gaudes et al., 2013; Petridou et al., 2013). On the other hand, the time volumes corresponding to the single events can be either averaged or clustered to generate the so-called coactivation patterns (CAPs), that is, patterns of regions that repeatedly activate and deactivate together (Chen and Glover, 2015; Chen et al., 2017; Liu and Duyn, 2013; Tagliazucchi et al., 2012a). Furthermore, estimating the shape of the hemodynamic response function at rest is feasible by averaging the signal (i.e., fitting a finite impulse response model) around the times of the identified events, for example, with a PPA-based approach (Tagliazucchi et al., 2012a; Wu and Marinazzo, 2016; Wu et al., 2013). The retrieved resting-state hemodynamic response function (HRF) exhibits a similar temporal pattern to the HRF that is obtained for task-related fMRI data, which partially validates the employment of deconvolution-based methods to identify these events.
In all these studies, the spatial distribution of maps obtained based on these brief spontaneous events closely resembles the resting-state networks obtained using static seed-based correlation or ICA, even though the number of data observations or events is substantially reduced (Liu and Duyn, 2013; Petridou et al., 2013; Tagliazucchi et al., 2012a). Furthermore, removal of the spontaneous events considerably diminishes the strength of correlation between the nodes of the network as computed with a sliding window approach (Petridou et al., 2013). These observations demonstrate that a substantial part of the brain's functional connectivity observed in rs-fMRI is driven by spontaneous BOLD events that sometimes occur simultaneously in all the nodes of the network or in a subset of nodes (Allan et al., 2015).
As an extension of the CAP technique, Karahanoğlu and Van de Ville (2015) proposed to identify innovation-driven CAPs (iCAPs) where k-means clustering is applied to the temporal derivatives of the deconvolved time courses, which encode changes in the original BOLD time courses. In contrast to conventional CAPs, iCAPs identify regions whose signal simultaneously increases or decreases, that is, regions with similar temporal dynamics (Preti et al., 2016). Using this framework, Karahanoğlu and Van de Ville (2015) found evidence that well-known resting-state networks, such as the default mode network, might divide into multiple subsystems with their own temporal dynamics and therefore possibly functionally heterogeneous subnetworks (see also Chen et al., 2017 for similar observations using PPA and CAPs). Moreover, backprojection of iCAPs to the deconvolved fMRI volumes allows reconstruction of iCAP time courses and therefore evaluation of temporal overlaps between different patterns. Interestingly, it has been found that (on average) between 3 and 4 iCAPs overlap in time and that the brain activity associated with these patterns is sustained for 5–10 sec, which might explain why a window length of at least 20 sec is required to obtain robust inferences with a sliding window approach (Karahanoğlu and Van de Ville, 2015; Preti et al., 2016).
The advantages of both PPA and deconvolution approaches for the study of dynamic functional connectivity are that they can potentially allow a reduction in data dimensionality since time points with nonsignificant amplitude, which are more likely to be corrupted by noise, are excluded. In addition, they easily enable whole-brain analyses at the voxel level (Petridou et al., 2013; Tagliazucchi et al., 2016) even though in practice, it is common to constrain the analysis to specific brain regions (Chen et al., 2017; Liu and Duyn, 2013) to ease interpretability of the results and reduce computational time. However, a critical issue in both approaches is the sensitivity of detection of events to the choice of amplitude thresholds or regularization parameters (Caballero Gaudes et al., 2013; Karahanoğlu et al., 2013; Tagliazucchi et al., 2012a; Tagliazucchi et al., 2016) (Fig. 2). Varying these parameters may considerably modulate the sensitivity and specificity of the algorithm to detect true neuronally related BOLD events and, in turn, subsequent analysis (e.g., the definition of [i]CAPs) and results. Often, to ensure the functional significance of the detected events or voxels, some type of additional spatial or temporal thresholding is applied. For example, one can select only those time points in a time course summarizing the activations or activation time series (Caballero Gaudes et al., 2013) where a minimum number of voxels exhibit an event (Karahanoğlu and Van de Ville, 2015; Petridou et al., 2013) or a subset of voxels of single volumes with a minimum signal change (Liu and Duyn, 2013).
Principal component analysis and independent component analysis
ICA is commonly used not only to identify functional networks that persist across the duration of an rs-fMRI scan but it has also been used to characterize dynamic connectivity by computing the correlation or coherence between the components' time courses in a sliding window approach (Allen et al., 2014; Calhoun and Adali, 2016; Sakoǧlu et al., 2010). ICA maximizes spatial independence among brain networks, which is effectively finding networks that are not systematically overlapping. As such, it provides a powerful and intuitive framework for analyzing resting fMRI data (Beckmann, 2012; Calhoun and Adali, 2012). This, however, does not necessarily imply that the brain is actually organized into spatially independent units, rather it represents a modeling framework for organizing and understanding high-dimensional data at a particular scale (Calhoun and deLacy, in press). Instead of examining the relationships between the windowed time courses of ICA components, ICA can also be applied independently to the rs-fMRI data from each temporal window to provide information about the spatial extent of the networks as a function of time (Kiviniemi et al., 2011), similar to a related approach called independent vector analysis (Ma et al., 2014).
In traditional rs-fMRI analysis, principal component analysis (PCA) is primarily used as a data cleaning/reduction step before ICA. For dynamic analysis, PCA can also be used in conjunction with sliding window correlation to identify patterns of connectivity termed eigen components that serve as the basis for the observed network dynamics (Leonardi et al., 2013, 2014). In contrast to hard clustering, PCA provides a weighted combination for the basis patterns of functional connectivity at each time point, rather than a discretized assignment to a single cluster. Such fuzzy clustering is useful and fuzzy membership in possibly overlapping states can be computed from a variety of approaches, including the hard clustering approaches mentioned earlier (e.g., k-means, PCA, spatial ICA, and temporal ICA) (Miller et al., 2016b).
Repetitive patterns
Most dynamic analysis methods do not assume a particular temporal sequence of events. However, a number of experimental observations of quasi-periodic sequences of activity have been reported (Chow et al., 2013; Majeed et al., 2009), and researchers have begun to explore analysis methods that explicitly search for repeated patterns. Mitra et al. (2015) demonstrated reproducible propagation across the cortex on the time scale of seconds, while other studies have observed repeated whole-brain patterns of activity that can be characterized with an autoregressive pattern-finding algorithm (Chow et al., 2013; Kiviniemi et al., 2016; Majeed et al., 2011) (Fig. 3). The patterns of activity typically involve sequential activation and deactivation of one or more of the large-scale functional networks detected with traditional rs-fMRI analysis. At least some of these repetitive patterns are linked to infraslow (<1 Hz) electrical activity (Grooms et al., 2017; Pan et al., 2013; Thompson et al., 2014b) and appear to arise from a different mechanism than the variability that reflects activity in typical electroencephalography (EEG) bands (Thompson et al., 2014a, 2015). These reproducible spatiotemporal patterns of activity contribute to both average functional connectivity and dynamics measured with other analysis methods unless they are explicitly accounted for.
Challenges in Dynamic Analysis
All of the approaches for obtaining dynamic information from rs-fMRI data face considerable challenges. Because no other imaging modality can map dynamic activity throughout the brain with spatial and temporal resolution at the finest scale, there is no gold standard for evaluating the accuracy of a particular analysis method. The SNR in rs-fMRI is low and it is known to be contaminated with non-neuronal components such as head motion and physiological respiratory- and cardiac-related fluctuations (Caballero-Gaudes and Reynolds, 2017; Laumann et al., 2016; Murphy et al., 2013; Power et al., 2012). Even the neuronal portion of the signal may be dominated by changes in vigilance levels over the course of the scan (Allen et al., 2014; Chang et al., 2016; Laumann et al., 2016; Wong et al., 2013). In this section, we highlight some of the most pressing issues in analysis; in the following section, we address how these issues affect the interpretation of dynamic rs-fMRI data.
The utility of sliding window correlation
To date, sliding window correlation has been the most widely used approach for the analysis of rs-fMRI data. Network dynamics measured with sliding window correlation have been linked to behavioral variability (Gonzalez-Castillo et al., 2015; Kucyi and Davis, 2014; Kucyi et al., 2017; Thompson et al., 2013a), can distinguish patient populations from healthy controls (Damaraju et al., 2014; Sakoǧlu et al., 2010), and are even shown to be more accurate than static connectivity for individual subject classification (Rashid et al., 2014). However, some recent modeling studies have shown that sliding window correlation is inherently highly variable for noisy autocorrelated signals and that it may not accurately represent the underlying correlation (Hindriks et al., 2015; Shakil et al., 2016). How, then, to reconcile its poor correspondence to the true correlation structure with its sensitivity to behaviorally relevant changes and its success in distinguishing patient groups? One key to understanding the robustness and clear results of the sliding window correlation approach may be that most of the successful studies have examined sliding window correlation from large arrays of segments from the whole brain, rather than bivariate correlation between small ROIs (Damaraju et al., 2014; Gonzalez-Castillo et al., 2015; Rashid et al., 2014; Sakoǧlu et al., 2010). This suggests that sliding window correlation may retain information about the underlying correlation structure despite any shortcomings in the analysis or noise in the data and that consideration of the changes in correlation across many areas improves sensitivity to the differences between groups. The few studies that have successfully used bivariate sliding window correlation to identify behaviorally relevant changes in connectivity typically apply it to large widespread networks (i.e., default mode network vs. task-positive network) rather than small ROI (Kucyi et al., 2016; Thompson et al., 2013a). This improves the SNR and allows more accurate estimation of the correlation values in each window. Along similar lines, the growing use of fast imaging sequences to obtain subsecond TRs (Feinberg et al., 2010; Moeller et al., 2010) should improve the performance of sliding window correlation for a given window size, assuming other sources of noise remain constant. Improvements in null models used for statistical analysis, as described in a later section, also hold the potential to increase the sensitivity of sliding window correlation with the neurally linked variability.
The characteristics of the window itself are important considerations for sliding window correlation and other windowed techniques. Long windows average the signal over longer periods and approach the traditional measures of average functional connectivity; short windows are more sensitive to transient changes, but provide much noisier estimates of correlation. Intuitively, the appropriate window size should approximate the amount of time that the brain spends in a single configuration, a hypothesis that has been confirmed by modeling (Shakil et al., 2016) (Fig. 4). Since the duration of a typical brain state is unknown, unfortunately, researchers have turned to other methods to identify appropriate window sizes. Sakoǧlu et al. (2010) showed that the first saddle point in the plot of time-windowed correlation occurs at ∼0.5/f, where the lowest and highest frequencies in the data provide bounds for the longest and shortest windows, respectively. Similarly, Leonardi and Van De Ville (2015) showed analytically that spurious fluctuations can arise when the window is shorter than the period of the lowest frequency present in the data, typically ∼50–100 sec. Expanding on this study, Zalesky and Breakspear (2015) showed that using the period of the lowest frequency maximizes statistical power, but may be overly conservative when the SNR is moderately high. Gonzalez-Castillo et al. (2015) have tested the efficacy of different sliding window lengths for identifying and differentiating several ongoing cognitive processes that were 3 min in duration. They found that while windows of 3 min (matching the duration of each imposed cognitive process) were optimal, windows as short as 20 sec in duration nearly matched the performance of the longer windows. From an experimental perspective, Thompson et al. (2013b) found that the correlation between sliding window BOLD correlation and band-limited power correlation reached a plateau at ∼50 sec, but that shorter windows could exhibit less error.
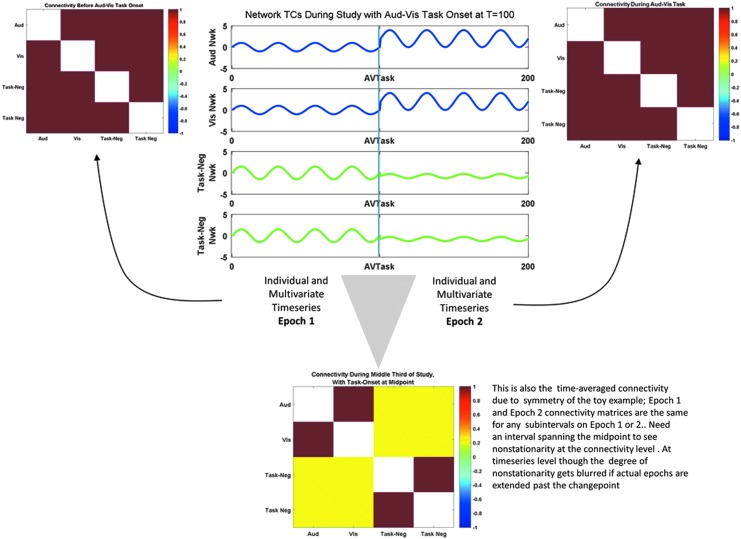
FIG. 4.
Changes in activity and connectivity are both relevant to rs-fMRI. Example of two epochs, in which the covariance is similar throughout, but there is a clear change point. In this case, the data show clearly bounded epochs for individual and multivariate time series that completely obscure the correlation nonstationarity that happens (exclusively) at the midpoint of the experiment. rs-fMRI, resting-state functional magnetic resonance imaging. Color images available online at www.liebertpub.com/brain
The type of the window is also debated. Many studies use simple square windows, but these windows can be extremely sensitive to outliers in the data since the inclusion or exclusion of outlier observations may cause a sudden apparent change in dynamic functional connectivity (Lindquist et al., 2014). Other groups have advocated the use of tapered windows, for example, Hamming windows, in which the weight of the points far from the center of the window is reduced. One modeling study found that the square window produced a more accurate estimation of the underlying correlation than the Hamming window, but with the caveat that changes in brain states were explicitly modeled as discontinuous jumps that occurred between one TR and the next (Shakil et al., 2016). Presumably, a model that employed slower transitions might be better served with a smoother window. Again, the ideal choice depends strongly on which model better describes the underlying brain activity.
Null models and validation
Several early studies showed that apparent variations in connectivity can arise in signals that share no temporal information (Handwerker et al., 2012; Keilholz et al., 2013). In other words, networks, each oscillating at their own unique set of frequencies, can show transient correlations due to the beat frequency correlation (from the difference in the frequency content) that inevitably would arise between different networks (Handwerker et al., 2012). It is extremely difficult to dissociate this beat frequency effect from true transient correlation. Perhaps one approach would be to surmise that if the transient correlations are periodic, then the correlations are spurious, and if they occur in a nonperiodic behaviorally correlated manner, then they represent true correlations. Multivariate approaches may serve an important role here as well as they take into account the full structure of the data and, as such, can be more robust than pairwise approaches (Damaraju et al., 2014; Kudela et al., 2017; Lindquist et al., 2014). Because apparent changes in the network configuration of the brain can arise from properties of the signal itself rather than neural activity, methods for validating the findings and statistical analysis are a critical need in dynamic rs-fMRI.
Statistical analysis relies on the appropriate definition of a null model. One common process is to construct an empirical model of a null distribution by permuting the data (i.e., matching time courses from different scans or different subjects), creating surrogate data [e.g., by shuffling the phase of the voxels' time course (Karahanoğlu and Van de Ville, 2015), or using a spatiotemporal wavelet resampling approach (Patel et al., 2006)] so that shared temporal information is destroyed. This empirical approach has the advantage of preserving features that arise from properties of the signal itself, which for rs-fMRI, is typically heavily processed and strongly autocorrelated in space and time. However, depending on the type of procedure, permutation does not usually preserve other properties of the signal (such as the average correlation value) and may not be the appropriate control (Hindriks et al., 2015). The design of a good null distribution for time-resolved rs-fMRI is challenging and requires careful consideration (Shi et al., 2016), particularly since it is not understood exactly what type of dynamic activity is expected. There are a large number of ways a signal can vary in time, and most existing simulation approaches have made rather strong assumptions about the form of the dynamic activity. In this case, it is quite easy to create a scenario where a certain dynamic behavior (not anticipated by a given null model) has a distribution that is indistinguishable from the aforementioned null model, rendering it essentially useless (Miller et al., “Resting-State fMRI Dynamics and Null Models: Perspectives, Sampling Variability, and Simulations,” bioRxiv, preprint).
Validation with multimodal imaging studies
Particularly because the identification of statistically significant network dynamics is still under development, it is crucial to validate the findings against other modalities whenever possible. Simultaneous acquisition of EEG and rs-fMRI has shown that the changes observed in the BOLD signal are linked to changes in the pattern of neural activity (Allen et al., 2017; Chang et al., 2013a; Grooms et al., 2017; Tagliazucchi et al., 2012b), increasing confidence that rs-fMRI is sensitive to dynamic reconfigurations of brain networks. In animal models where more localized invasive recordings can be obtained, spontaneous BOLD fluctuations are correlated with local field potentials from the same site (Pan et al., 2011; Shmuel and Leopold, 2008), and BOLD sliding window correlation between the left and right somatosensory cortex is significantly correlated with simultaneously acquired local field potential, particularly in the theta, beta, and gamma bands (Thompson et al., 2013b). In contrast, the repetitive quasi-periodic patterns of whole-brain activity are more closely linked to infraslow (<1 Hz) electrical activity (Pan et al., 2013; Thompson et al., 2014b). The relative independence of the quasi-periodic patterns and the time-varying interactions observed with sliding window correlation (Thompson et al., 2014a, 2015) raises the intriguing possibility that it may prove possible to selectively sensitize dynamic rs-fMRI to particular types of activities based on their spatial, spectral, and temporal signatures (Keilholz et al., 2017). Simultaneous monitoring of neuronal calcium signals and whole-brain hemodynamic signals with optical imaging in mice has also provided evidence of two apparently independent types of fluctuations in large-scale functional connectivity, one related to global waves propagating across the neocortex and transient coactivations among cortical areas sharing high functional connectivity (Matsui et al., 2016). These findings not only help to validate ongoing efforts with rs-fMRI but may also aid in the development of better models of brain dynamics.
Validation by correlation with behavior
In human studies, a growing number of researchers are taking a different approach to validation, using differences in behavioral outcomes as a proxy for differences in neural activity (Kucyi et al., 2016; Thompson et al., 2013a). These studies differ by necessity from standard rs-fMRI, in that brain activity is no longer truly spontaneous, although the task may have low cognitive requirements. An early example incorporated a psychomotor vigilance task in which subjects pressed a button as rapidly as they could whenever the fixation dot changed colors. Correlation between the default mode network and task-positive network within a short window before the color change predicted reaction time (Thompson et al., 2013a). A more recent study showed that patterns of functional connectivity predicted whether an auditory stimulus would be perceived (Sadaghiani et al., 2015). Other groups have looked at reproducible changes across subjects listening to the same narratives or watching the same movies (Simony et al., 2016). This work builds on existing literature linking activity in areas or networks before a task to the task response (Boly et al., 2007; Hesselmann et al., 2008) (for review, see Sadaghiani and Kleinschmidt, 2013). Note, however, these approaches tell nothing about the dynamics of areas that are not involved in the task and can be confounded by other factors that vary with task performance, such as head motion (Siegel et al., 2016). Still, the use of behavioral variability as a surrogate for measures of underlying neural variability may prove to be a powerful tool for interpreting rs-fMRI data.
Replication, reliability, and sensitivity to individual differences
One very basic criterion for the validation of dynamic rs-fMRI analysis is that the metrics measured must be reproducible. It is less natural to think of reproducibility for dynamic rs-fMRI than for average functional connectivity or task-based fMRI since dynamic analysis attempts to capture the time-varying unconstrained activity of the brain. Nevertheless, properties such as the number of states, the primary networks contributing to each state, and the relative number of transitions between states should be reproducible at least at the level of a population average. A recent study by Abrol et al. used sliding window correlation followed by clustering on 28 groups of 250 age-matched subjects and identified five distinct connectivity states for each group. The patterns of correlation in each state were very reproducible across the groups (Abrol et al., 2016). Choe et al. (2017) examined test-retest data and found that summary statistics for dynamic analysis (mean and variance) could be reliably detected across sessions. Indeed, recent work shows that even at the individual level, patterns of dynamic activity provide important information such that the inclusion of dynamic connectivity improves classification of individuals compared with average connectivity alone (Rashid et al., 2016).
Sensitivity to changes related to brain disorders
Another indication that time-varying rs-fMRI analysis is sensitive to vital aspects of dynamic brain activity comes from the growing body of studies showing that it can successfully differentiate between patients with psychiatric or neurological disorders and healthy control subjects. One of the earliest reports from patients with schizophrenia (Sakoǧlu et al., 2010) showed that dynamic analysis has the potential to provide information that is different from the information obtained from average functional connectivity. Differences in the dynamic connectivity of the brain were subsequently observed in mild cognitive impairment (Chen et al., 2016b), schizophrenia (Damaraju et al., 2014; Ma et al., 2014; Miller et al., 2016b; Yaesoubi et al., in press, Yu et al., 2015), post-traumatic stress disorder (Li et al., 2014), attention-deficit/hyperactivity disorder (Ou et al., 2014), major depression (Demirtaş et al., 2016), and autism/autism spectrum (de Lacy et al., 2017; Falahpour et al., 2016). In a study of particular note, Rashid et al. (2014) found differences between schizophrenic and bipolar patients, groups that can be very challenging to distinguish. A follow-up study showed that dynamic connectivity was able to predict individual diagnoses within these groups (Rashid et al., 2016).
The differences between healthy subjects and patients with brain disorders provide support for dynamic rs-fMRI sensitivity to altered brain activity. However, physiological variables such as cerebral perfusion and levels of motion can also vary across groups and may influence the results of dynamic analysis (although many of the studies mentioned above do careful correction and evaluation to true to rule out motion). In the sense that metrics from dynamic rs-fMRI can serve as specific biomarkers for different disorders, it may not matter whether the differences reflect brain activity or other physiological processes. For studies that make inferences about how the brain's organization is affected by a particular disorder, however, caution should be used and other potential sources of differences should be examined.
Validating and constraining whole-brain computational models
The nature of the neuronal mechanisms that generate whole-brain temporal dynamics is still elusive. Whole-brain computational models aim to balance complexity and realism to describe the most important features of brain activity in vivo. This balance is extremely difficult to achieve because of the astronomical number of neurons and the underspecified connectivity at the neural level. Thus, the most successful whole-brain computational models have taken their lead from statistical physics, where it has been shown that macroscopic physical systems obey laws that are independent of their mesoscopic constituents. The emerging collective macroscopic behavior of brain models depends only weakly on individual neuron behavior (Breakspear and Jirsa, 2007). Thus, these models typically use mesoscopic top-down approximations of brain complexity with dynamical networks of local brain area attractor networks.
Indeed, whole-brain models can provide a detailed understanding of the causal dynamics of the human brain by linking anatomical structure with functional dynamics. The structural connectivity of the brain forms the framework that patterns of coordinated activity play across (although it should be noted that activity can also influence structure, something that is ignored in most modeling approaches). To better understand how network structure constrains and informs large-scale patterns of activity, researchers have created models based on diffusion-weighted MR tractography or other tractographic techniques that result in a matrix of pairwise connectivity for all ROIs in the brain. The global dynamics of the whole-brain model result from the mutual interactions of local node dynamics coupled through the underlying empirical structural connectivity matrix. Typically, the temporal dynamics of local brain areas in these models are taken to be either asynchronous (spiking models or their respective mean-field reduction) or oscillatory (Cabral et al., 2011; Deco et al., 2009; Deco and Jirsa, 2012; for review, see Deco and Kringelbach, 2014).
Whole-brain computational models have shown that the structural connectivity of the brain is a major determinant of the patterns of functional connectivity that it can support. However, major functional networks can be identified using numerous models for activity at individual nodes and a wide range of parameters that describe the coupling between nodes. Most of the models (especially those that are linear) perform poorly when asked to reproduce the network dynamics observed with rs-fMRI (Messé et al., 2014). This suggests that incorporating information obtained with dynamic analysis into the modeling process can serve as a constraint on the types of models and parameters that are appropriate. If the dynamic connectivity of the brain can be successfully modeled, the features of the model itself may provide insight into the organization and coordination of the neural processes that produce these dynamics.
Consider a study by Hansen et al. (2014) that demonstrated that average functional connectivity is closely linked to the underlying structural connectivity and proposed to characterize the time-dependent structure of resting fluctuations with the functional connectivity dynamics (FCD) matrix, which is based on the sliding window approach. For each window, centered at time t, one calculates a separate FC matrix, FC(t). The FCD matrix is an MxM symmetric matrix whose (t1, t2) entry is defined by the Pearson correlation between the upper triangular parts of the two matrices FC(t1) and FC(t2). Epochs of stable FC(t) configurations are reflected around the diagonal of the FCD matrix in blocks of elevated inter-FC(t) correlations. When nonlinearities are considered in the network models, the spatiotemporally dynamic repertoire of the network is significantly enhanced and the resting-state dynamics show nonstationary FCD. While Hansen et al. proposed FCD as a novel biomarker and demonstrated that all known resting state networks can be derived from the nonlinear network dynamics of FCD, they did not fit the model to the empirical functional time series data. The patterns in the FCD matrix arise from what is essentially a random process and are thus different for different measurements. This renders the fitting process for brain network models more complex than fitting with average functional connectivity, for which a Pearson correlation across empirical and simulated FC matrices is sufficient.
Recently, a powerful, novel whole-brain model emerged that uses, for each brain area, a local dynamical model given by a normal form of bifurcations (e.g., a supercritical Hopf bifurcation) (Deco et al., 2017; Kringelbach et al., 2015). The normal form of Hopf bifurcation can describe the transition from asynchronous noisy behavior to full oscillations and thus unify previous asynchronous and full oscillatory scenarios. One key finding of the Hopf whole-brain model is that previous findings using the optimal operating point based on average functional connectivity hold true if we take into account the temporal dynamics of FC, that is, FCD. Importantly, this model also demonstrated that fitting the temporal structure of fluctuations using the FCD provided a better way of constraining the model than simply using the average functional connectivity. Another remarkable and important finding is that high metastability is only present in a narrow range of parameters. In dynamical systems, metastability refers to a nonequilibrium state that persists for an extended period of time; in computational modeling of the brain, it is a measure of the variability of the whole-brain synchronization level, that is, global temporal fluctuation. In other words, the FCD of the spontaneous resting state, in conjunction with brain network modeling, provides evidence that the brain at rest is maximally metastable, refining and demonstrating the hypothesis of Tognoli and Kelso (2014)—an excellent demonstration of how empirical network dynamics can inform theoretical neuroscience.
Taming the data explosion
Dynamic analysis of rs-fMRI inherently multiplies the size of an already large data set by two or three orders of magnitude. Managing this data explosion and summarizing the relevant features are ongoing challenges. Many studies to date have defined a small number of states that describe the current configuration of the brain. These states are usually obtained through clustering or dimensionality reduction algorithms and can be based on features of the data such as the amplitude in each voxel, patterns of correlation between all brain parcels (Allen et al., 2014; Chen et al., 2016b; Hudetz et al., 2015; Ma et al., 2017; Suk et al., 2016; Taghia et al., 2017), or patterns of regions with similar temporal dynamics as with the (innovative) coactivation patterns (Karahanoğlu and Van de Ville, 2015; Liu and Duyn, 2013). In the simplest approach, each time point is ascribed to one of these states so that each N time point scan can be described by a single string of N numbers. If temporal overlap between states is modeled or allowed, each time point can also be assigned a set of states, resulting in more complex trajectories (Karahanoğlu and Van de Ville, 2015). Using these time series, the number of transitions between states, the number of occurrences of a given state, and the average dwell time in each state can be calculated and compared across individuals or groups [e.g., see Chen and Glover, 2015; Karahanoğlu and Van de Ville, 2015 for specific examples of (i)CAP approaches and Taghia et al. 2017 for an example based on hidden Markov models]. Additional summarization can be obtained by tracking the transitions between multiple (possibly overlapping) states through a metastate that characterizes each fMRI time point as an N-element metastate vector representing the contribution of each time point to each of the N states. These metastates can then be quantified in terms of multiple metrics such as total distance traveled, number of transitions, or more complex, but interesting, quantities such as hub states (Miller et al., 2016b).
Alternatively, graph theory has proven successful at summarizing average measures of functional connectivity across the brain and is increasingly being applied to dynamic rs-fMRI as well. The appeal of graph theory lies in its ability to condense connectivity matrices into measures that can be reported by node, by network, or even globally across the brain. Brain imaging researchers have adopted concepts of centrality, efficiency, modularity, and community structure from the well-developed field of complex networks (Betzel et al., 2016; van den Heuvel and Sporns, 2011; van den Heuvel et al., 2012). In the realm of dynamic analysis, graph theory is particularly applicable to techniques such as sliding window correlation that provide an estimate of the changing relationship between areas over time (Bassett et al., 2011; Yu et al., 2015). Graph metrics such as efficiency and modularity can then be calculated on a time-varying basis, allowing an examination of how the changes in brain network configuration influence communication between areas (Betzel et al., 2016; Fukushima et al., 2017; Yu et al., 2015; Zalesky et al., 2014). These studies are paving new ground in basic neuroscience by elucidating the dynamic balance of integration and segregation in the brain.
Interpretation of Network Dynamics
rs-fMRI can provide a unique view of dynamic activity throughout the whole brain. This leaves us in the challenging and somewhat circular position of attempting to interpret the rs-fMRI findings from analysis methods that rely on key assumptions about unknown processes in the brain. For example, when using sliding window correlation followed by clustering, one is likely to find that clusters persist for approximately the length of the window (Shakil et al., 2016). Ideally, the window length would approximate the length of a brain state, but since the length of a typical brain state is unknown, instead we obtain brain states that approximate the length of the window that was chosen. In this section, we discuss key underlying parameters that affect the design of experiments and interpretation of results.
What is a brain state? Inherent to many types of dynamic analyses is the concept of a brain state, which we will define as a spatial pattern of activity that remains relatively stable for some minimum amount of time. Even in the definition, ambiguities are apparent. How much must a spatial pattern change between states? How long should a state persist? The answers depend, in part, on the types of activities that are reflected in the rs-fMRI signal. The fast, brief brain states observed in electrophysiological data, either with EEG (Khanna et al., 2015) or magnetoencephalography (MEG) (Baker et al., 2014), involve large-scale patterns that cover most of the cortex, but persist for less than a second, and studies show that they partially contribute to the BOLD fluctuations in patterns that resemble resting-state networks (Britz et al., 2010; Musso et al., 2010; Yuan et al., 2012). Many cognitive processes occur on a scale of seconds and should be reflected in the BOLD fluctuations as well, as demonstrated by the marked similarity between activation maps and resting-state networks (Smith et al., 2009). At the slower end of the scale are changes in the level of arousal and vigilance that tend to occur over minutes or even longer and which appear to be one of the driving factors in identifying brain states in most rs-fMRI studies (Allen et al., 2017; Chang et al., 2016; Laumann et al., 2016; Tagliazucchi and Laufs, 2014). All of these processes have been shown to influence the rs-fMRI signal, and it is plausible that sensitivity to a particular contributor varies depending on analysis factors such as window length.
Interestingly, most studies that use states to summarize the brain's activity find a relatively small number of distinct states, whether using EEG/MEG or rs-fMRI (Allen et al., 2014; Britz et al., 2010; Chen et al., 2016a; Musso et al., 2010). These states seem to comprise a rather limited set of building blocks compared with the brain's rich dynamical repertoire, particularly given the wide range of temporal scales involved. Still, it is possible that these states represent some fundamental property of brain activity at each scale that is yet to be understood. The replicability of states and their various metrics suggest there may be some canonical aspect to these states (Abrol et al., 2016). However, this is certainly not the end of the story. It is possible and even probable that better acquisition and analysis methods might lead to the separation of some clusters into distinct subclusters. Higher temporal resolution to improve the estimation of correlation, higher spatial resolution to better localize the signal, improved registration methods to reduce blurring and averaging across subjects, and better noise removal could all increase sensitivity to distinct brain states. This is a similar position to that of average functional connectivity studies a few years ago in modeling the brain with a specific number of networks and/or components. As acquisition and analytic approaches evolved and data sets increased in size, many more interesting aspects of the resting-state networks have emerged.
Another fundamental issue in identifying brain states is choosing the right metric. One might cluster the data, for example, based on the amplitude of all voxels or ROIs at each time point, or one might instead choose to cluster based on the result of sliding window correlation between areas. In amplitude-based approaches, it is assumed that the activity in each voxel defines a brain state, whereas in correlation or coherence-based approaches, changes in the relationships between areas define states. At this time, it is unknown which provides a better picture of the changes that occur in the brain. One modeling study suggests that using amplitude produces a better representation of the true changes in network configuration (Shakil et al., 2016), but several experimental studies have found improved sensitivity with correlation (Gonzalez-Castillo et al., 2015; Thompson et al., 2013a). Indeed, these different measures may well be complementary, in which case a combined approach might be more comprehensive.
While the use of brain states to summarize dynamic rs-fMRI provides a practical simplification of the data that allows for easy comparisons across groups and conditions, it is far from certain that the brain state model accurately describes activity in the brain. One can also imagine a continuous evolution of activity over time, with certain spatial patterns evolving together, a view that has been studied with both PCA and ICA approaches (Leonardi et al., 2013; Miller et al., 2016b; Yaesoubi et al., 2014), as well as with PPA and deconvolution approaches and (i)CAPs (Karahanoğlu and Van de Ville, 2015). The presence of repeated quasi-periodic spatiotemporal patterns in the brain's activity is also somewhat at odds with the brain state model, where it may appear as alternating between two or three states without capturing the propagation of activity from one state to the next. As we learn more about the macroscale organization of the brain, we must update and revise our analytical models to best capture its features.
Even for theoretical models linking brain structure and function, a single comprehensive model for dynamic activity in the brain has proven elusive. There are many reasons for this failure, but the main reason comes from the realization that whole-brain dynamics are much more complex than previously thought. Traditional attractor states do not appear to adequately describe them (Amit and Treves, 1989). One can perhaps define a given brain state by its dynamical complexity, which must arise from the interplay between anatomy and functional dynamics. For a given brain state, a balance has to be found between the integration and segregation of information (Deco et al., 2015). The dynamical repertoire of a brain state depends on the underlying anatomical structural connectivity and local dynamics. A number of different methods have tried to describe the spatiotemporal unfolding of activity (Allen et al., 2014; Hansen et al., 2014). These methods are able to describe the evolution of global whole-brain activity, but they are less good at describing the interaction of how activity in a local region shapes global activity. A possible way to escape this problem, perhaps, is by generalizing the definition of brain states as an ensemble or cloud of possible steady states (attractors). This cloud of attractors can be defined by the underlying time-varying brain generators, which are the parameters of a generative whole-brain model describing each possible attractor contributing to the system dynamics. Thus, a given brain state could be characterized by the statistics and dynamical complexity of these intrinsic brain generators over time. Another approach is to focus on multiple scales, for example, interaction between networks versus domains (sets of networks) (Vergara et al., 2017).
What processes contribute to dynamic rs-fMRI? The conception of dynamic rs-fMRI as reflecting moment-to-moment changes related to cognition and information processing is appealing, but overly simplistic. Like traditional rs-fMRI functional connectivity measurements, dynamic rs-fMRI is sensitive to physiological cycles and small motions of the head (Caballero-Gaudes and Reynolds, 2017), and even changes related to neuronal activity are multiplexed, encompassing different processes that occur at different time scales. Moreover, neural and physiological processes can be intertwined. For example, heart rate variability is an important marker of autonomic function and can be affected by emotionally salient stimuli. Using sliding window correlation, Chang et al. (2013b) identified a network of areas that become more strongly connected during periods of high heart rate variability. While it is difficult to say whether autonomic fluctuations drive changes in functional connectivity or changes in the brain drive autonomic variability, it is clear that some portion of the dynamic activity in the brain is linked to autonomic processes (Nikolaou et al., 2016).
Motion can be a particularly tricky confound for dynamic rs-fMRI. Realignment and regression of motion parameters reduce, but do not eliminate, its effects on scans (Power et al., 2012), and recent work suggests that the residual effects of motion may account for a sizable portion of the variability in the BOLD correlation between areas (Laumann et al., 2016). Furthermore, head motion is linked to a number of behavioral and physiological traits, suggesting that some of the relationships observed between network dynamics and behavior may actually arise from head motion during the scan (Siegel et al., 2016). The development of better ways to characterize and correct for small motions during scans is critical for improved analysis and interpretation of dynamic rs-fMRI.
In terms of brain activity, the evidence is growing that changes in arousal level are major contributors to the variability in connectivity over the course of a scan (Allen et al., 2014, 2017; Chang et al., 2016; Laumann et al., 2016; Tagliazucchi and Laufs, 2014). Tagliazucchi and Laufs (2014) found that about one-third of their subjects fell asleep within the first 3 min of the scan. The patterns of changes associated with lowered arousal involve large portions of the cortex and are highly stereotyped such that Chang et al. (2016) suggest that a template might be derived and regressed from the signal to minimize this type of variability (or allow it to be specifically examined, depending on the researcher's interest).
The changes that occur as subjects relax and become drowsy within the scanner may also impact the global signal (Wong et al., 2013). Global signal regression is still widely debated in the neuroimaging community. It improves the spatial localization of networks, but can introduce artificial anticorrelations into the data (Murphy et al., 2009). In terms of network dynamics, greater network connectivity is observed during periods of high global signal (Scheinost et al., 2016). Recent work with positron emission tomography and MRI has shown that the global signal tends to follow the baseline level of [18F]-2-fluoro-2-deoxy-D-glucose, while the variance of the BOLD signal is mostly unaffected (Thompson et al., 2016). In support of the idea that the global signal represents a separable baseline level of brain activity, regression of the global signal improves the concordance between BOLD correlations and simultaneously measured local field potentials from the same areas (Thompson et al., 2013b).
Repeated spatiotemporal patterns of brain activity may also be linked to levels of vigilance or arousal. Work in animals has shown that the quasi-periodic patterns (QPPs) described by Majeed et al. (2009, 2011) are linked to infraslow electrical activity (Pan et al., 2013; Thompson et al., 2014b) and influence reaction time on a simple vigilance task (Abbas et al., 2016). Like the global signal, the QPPs appear to be separable from time-varying activity (Thompson et al., 2014a, 2015). The similarities between the global signal, templates associated with arousal level, and QPPs raise the question of whether they might represent a single neurophysiological process viewed through different lenses.
Despite the widespread contribution of changes related to vigilance levels, substantial variability in both BOLD correlation and local field potential correlation is observed in anesthetized rats, which are carefully maintained at a constant anesthetic depth and should theoretically not exhibit fluctuations in arousal level (Thompson et al., 2013b). These animals also exhibit minimal motion due to the use of a stereotaxic head holder, indicating that motion is not the primary source of network dynamics in this type of experiment (Keilholz et al., 2017).
What, then, of the time-varying patterns of activity or connectivity reflecting cognitive changes? After rs-fMRI scans, subjects report a variety of mental activities (daydreaming, counting, planning, thinking of music, remembering events, and dreaming, etc), and it has been shown that the tendency to daydream, for instance, correlates with variability in connectivity between certain brain areas (Kucyi and Davis, 2014). One can in some ways consider these to be tasks (although undirected and unknown tasks) that produce a response that we wish to detect. In this scenario, it seems clear that these varying mental activities over the course of the scan should result in variations in activity and/or connectivity in the rs-fMRI data. However, the relevant changes may be limited in spatial extent and difficult to detect with current analysis techniques, particularly because the timing of the tasks is unknown. Studies that analyze tasks with known timings or tie network dynamics to behavioral outcomes provide evidence that detection of time-varying activity with rs-fMRI is feasible (Gonzalez-Castillo et al., 2015; Kucyi et al., 2016; Thompson et al., 2013a). Gonzalez-Castillo et al. (2015) have found that specific cognitive states and steady-state tasks induced over several minutes were readily detectable by windowed connectivity analysis alone. In addition, it appears that the connectivity changes that occur are more extensive than the measured magnitude changes. Within this context, it is also interesting to consider individual vs. task-level contributions to time-varying connectivity (Xie et al., 2017). Further improvements in methods for minimizing noise from motion and physiological cycles, better data analysis methods for deconvolving the neuronal component of the fMRI signal (Caballero Gaudes et al., 2013; Karahanoğlu et al., 2013), and dynamic generative models of brain activity that do not assume a fixed window length or a priori number of brain states (e.g., using hidden Markov model formulations as in the study by Taghia et al., 2017) will definitely improve analysis of dynamic neural activity measured with rs-fMRI. It may also be possible to minimize contributions from unwanted types of variability (such as arousal) using template regression or other measures of alertness.
Conclusions
The ability of rs-fMRI to provide a noninvasive glimpse of dynamic activity throughout the brain has paved the way for a better understanding of how the brain is organized across spatial and temporal scales and how this organization is altered in neurological and psychiatric disorders. The combination of whole-brain coverage and reasonable spatial and temporal resolution with the noninvasive nature of the data acquisition can paint a picture of brain dynamics currently unobtainable with any other imaging modality. Further improvements in image acquisition, minimization of physiological noise, and better image registration and analysis techniques will continue to improve sensitivity to the activity of interest. In combination with whole-brain network models, dynamic rs-fMRI has the potential to give new insight into fundamental problems in basic neuroscience and may eventually enable the field to move beyond group analyses to characterize network dynamics within any given individual—possibly even to identify interventions that can restore normal function. While numerous challenges remain, particularly in the analysis and interpretation of data, dynamic rs-fMRI is poised to play a key role in fields ranging from basic neuroscience to clinical neurology.
Acknowledgments
The authors would like to thank all of the participants at the Dynamic Connectivity symposium for the invigorating discussion. The authors would also like to thank Anzar Abbas for help with figures.
Author Disclosure Statement
No competing financial interests exist.
References
- Abbas A, Majeed W, Thompson G, Keilholz SD. 2016. Phase of quasi-periodic patterns in the brain predicts performance on psychomotor vigilance task in humans. In Proceedings of the International Society of Magnetic Resonance in Medicine, Singapore, p. 1192 [Google Scholar]
- Abrol A, Chaze C, Damaraju E, Calhoun V. 2016. The chronnectome: replicability of dynamic connectivity patterns in 7500 resting FMRI datasets. In 38th Annual International Conference on IEEE Engineering in Medicine and Biology Society, Orlando, FL, pp. 5571–5574 [DOI] [PubMed] [Google Scholar]
- Allan TW, Francis ST, Caballero-Gaudes C, Morris PG, Liddle EB, Liddle PF, et al. 2015. Functional connectivity in MRI is driven by spontaneous BOLD events. PLoS One 10:e0124577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen EA, Damaraju E, Eichele T, Wu L, Calhoun VD. 2017. EEG Signatures of Dynamic Functional Network Connectivity States. Brain Topogr. [Epub ahead of print]; DOI: 10.1007/s10548-017-0546-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen EA, Damaraju E, Plis SM, Erhardt EB, Eichele T, Calhoun VD. 2014. Tracking whole-brain connectivity dynamics in the resting state. Cereb Cortex 24:663–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit DJ, Treves A. 1989. Associative memory neural network with low temporal spiking rates. Biophysics 86:7871–7815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker AP, Brookes MJ, Rezek IA, Smith SM, Behrens T, Probert Smith PJ, Woolrich M. 2014. Fast transient networks in spontaneous human brain activity. Elife 3:e01867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Wymbs NF, Porter MA, Mucha PJ, Carlson JM, Grafton ST. 2011. Dynamic reconfiguration of human brain networks during learning. Proc Natl Acad Sci U S A 108:7641–7646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann CF, 2012. Modelling with independent components. Neuroimage 62:891–901 [DOI] [PubMed] [Google Scholar]
- Betzel RF, Fukushima M, He Y, Zuo X-N, Sporns O. 2016. Dynamic fluctuations coincide with periods of high and low modularity in resting-state functional brain networks. Neuroimage 127:287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boly M, Balteau E, Schnakers C, Degueldre C, Moonen G, Luxen A, et al. 2007. Baseline brain activity fluctuations predict somatosensory perception in humans. Proc Natl Acad Sci U S A. [Epub ahead of print]; DOI: 10.1073/pnas.0611404104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breakspear M, Jirsa VK. 2007. Neuronal dynamics and brain connectivity. Underst Complex Syst 2007:3–64 [Google Scholar]
- Britz J, Van De Ville D, Michel CM. 2010. BOLD correlates of EEG topography reveal rapid resting-state network dynamics. Neuroimage 52:1162–1170 [DOI] [PubMed] [Google Scholar]
- Caballero Gaudes C, Petridou N, Francis ST, Dryden IL, Gowland PA. 2013. Paradigm free mapping with sparse regression automatically detects single-trial functional magnetic resonance imaging blood oxygenation level dependent responses. Hum Brain Mapp 34:501–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero-Gaudes C, Reynolds RC. 2017. Methods for cleaning the BOLD fMRI signal. Neuroimage. 154:128–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral J, Hugues E, Sporns O, Deco G. 2011. Role of local network oscillations in resting-state functional connectivity. Neuroimage 57:130–139 [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Adali T. 2012. Multisubject independent component analysis of fMRI: a decade of intrinsic networks, default mode, and neurodiagnostic discovery. IEEE Rev Biomed Eng 5:60–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T. 2016. Time-Varying Brain Connectivity in fMRI Data: whole-brain data-driven approaches for capturing and characterizing dynamic states. IEEE Signal Process Mag 33:52–66 [Google Scholar]
- Calhoun VD, deLacy N. 2017. 10 key observations on the analysis of resting-state fMRI data using independent component analysis. Neuroimag Clin North Am [Epub ahead of print]; DOI: 10.1016/j.nic.2017.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Glover GHH. 2010. Time-frequency dynamics of resting-state brain connectivity measured with fMRI. Neuroimage 50:81–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Leopold DA, Schölvinck ML, Mandelkow H, Picchioni D, Liu X, et al. 2016. Tracking brain arousal fluctuations with fMRI. Proc Natl Acad Sci U S A 113:4518–4523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Liu Z, Chen MCC, Liu X, Duyn JHH. 2013a. EEG correlates of time-varying BOLD functional connectivity. Neuroimage 72:227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Metzger CD, Glover GH, Duyn JH, Heinze H-J, Walter M. 2013b. Association between heart rate variability and fluctuations in resting-state functional connectivity. Neuroimage 68:93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JE, Glover GH. 2015. BOLD fractional contribution to resting-state functional connectivity above 0.1 Hz. Neuroimage 107:207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JE, Glover GH, Greicius MD, Chang C. 2017. Dissociated patterns of anti-correlations with dorsal and ventral default-mode networks at rest. Hum Brain Mapp 38:2454–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Langley J, Chen X, Hu X. 2016a. Spatiotemporal modeling of brain dynamics using resting-state functional magnetic resonance imaging with Gaussian Hidden Markov model. Brain Connect 6:326–334 [DOI] [PubMed] [Google Scholar]
- Chen X, Zhang H, Gao Y, Wee CY, Li G, Shen D. 2016b. High-order resting-state functional connectivity network for MCI classification. Hum Brain Mapp 37:3282–3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe AS, Nebel MB, Barber AD, Cohen JR, Xu Y, Pekar JJ, et al. 2017. Comparing test-retest reliability of dynamic functional connectivity methods. Neuroimage 158:155–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow HM, Horovitz SG, Carr WS, Picchioni D, Coddington N, Fukunaga M, et al. 2013. Rhythmic alternating patterns of brain activity distinguish rapid eye movement sleep from other states of consciousness. Proc Natl Acad Sci U S A 110:10300–10305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribben I, Haraldsdottir R, Atlas LY, Wager TD, Lindquist MA. 2012. Dynamic connectivity regression: determining state-related changes in brain connectivity. Neuroimage 61:907–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribben I, Wager TD, Lindquist MA. 2013. Detecting functional connectivity change points for single-subject fMRI data. Front Comput Neurosci 7:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damaraju E, Allen EA, Belger A, Ford JM, McEwen S, Mathalon DH, et al. 2014. Dynamic functional connectivity analysis reveals transient states of dysconnectivity in schizophrenia. Neuroimage Clin 5:298–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lacy N, Doherty D, King BH, Rachakonda S, Calhoun VD. 2017. Disruption to control network function correlates with altered dynamic connectivity in the wider autism spectrum. Neuroimage Clin 15:513–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deco G, Jirsa V, McIntosh AR, Sporns O, Kotter R. 2009. Key role of coupling, delay, and noise in resting brain fluctuations. Proc Natl Acad Sci U S A 106:10302–10307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deco G, Jirsa VK. 2012. Ongoing cortical activity at rest: criticality, multistability, and ghost attractors. J Neurosci 32:3366–3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deco G, Kringelbach ML. 2014. Great expectations: using whole-brain computational connectomics for understanding neuropsychiatric disorders. Neuron 84:892–905 [DOI] [PubMed] [Google Scholar]
- Deco G, Kringelbach ML, Jirsa V, Ritter P. 2017. The dynamics of resting fluctuations in the brain: metastability and its dynamical cortical core. Sci Rep 7:3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deco G, Tononi G, Boly M, Kringelbach ML. 2015. Rethinking segregation and integration: contributions of whole-brain modelling. Nat Rev Neurosci 16:430–439 [DOI] [PubMed] [Google Scholar]
- Demirtaş M, Tornador C, Falcón C, López-Solà M, Hernández-Ribas R, Pujol J, et al. 2016. Dynamic functional connectivity reveals altered variability in functional connectivity among patients with major depressive disorder. Hum Brain Mapp 37:2918–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di X, Biswal BB. 2015. Dynamic brain functional connectivity modulated by resting-state networks. Brain Struct Funct 220:37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eavani H, Satterthwaite TD, Gur RE, Gur RC, Davatzikos C. 2013. Unsupervised learning of functional network dynamics in resting state fMRI. Inf Process Med. Imaging 23:426–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falahpour M, Thompson WK, Abbott AE, Jahedi A, Mulvey ME, Datko M, et al. 2016. “Underconnected,” but not broken? Dynamic fcMRI shows underconnectivity in autism is linked to increased intra-individual variability across time. Brain Connect 6:403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg DA, Moeller S, Smith SM, Auerbach E, Ramanna S, Gunther M, et al. 2010. Multiplexed echo planar imaging for sub-second whole brain FMRI and fast diffusion imaging. PLoS One 5:e15710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima M, Betzel RF, He Y, de Reus MA, van den Heuvel MP, Zuo X-N, Sporns O. 2017. Fluctuations between high- and low-modularity topology in time-resolved functional connectivity. Neuroimage. [Epub ahead of print]; DOI: 10.1016/j.neuroimage.2017.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerchen MF, Bernal-Casas D, Kirsch P. 2014. Analyzing task-dependent brain network changes by whole-brain psychophysiological interactions: a comparison to conventional analysis. Hum Brain Mapp 35:5071–5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitelman DR, Penny WD, Ashburner J, Friston KJ. 2003. Modeling regional and psychophysiologic interactions in fMRI: the importance of hemodynamic deconvolution. Neuroimage 19:200–207 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Castillo J, Hoy CW, Handwerker DA, Robinson ME, Buchanan LC, Saad ZS, Bandettini PA. 2015. Tracking ongoing cognition in individuals using brief, whole-brain functional connectivity patterns. Proc Natl Acad Sci U S A 112:8762–8767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grooms JK, Thompson GJ, Pan W-J, Billings J, Schumacher EH, Epstein CM, Keilholz SD. 2017. Infraslow EEG and dynamic resting state network activity. Brain Connect 7:265–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerker DAA, Roopchansingh V, Gonzalez-Castillo J, Bandettini PAA. 2012. Periodic changes in fMRI connectivity. Neuroimage 63:1712–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen ECA, Battaglia D, Spiegler A, Deco G, Jirsa VK. 2014. Functional connectivity dynamics: modeling the switching behavior of the resting state. Neuroimage 105:525–535 [DOI] [PubMed] [Google Scholar]
- Hesselmann G, Kell CA, Eger E, Kleinschmidt A. 2008. Spontaneous local variations in ongoing neural activity bias perceptual decisions. Proc Natl Acad Sci U S A 105:10984–10989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindriks R, Adhikari MH, Murayama Y, Ganzetti M, Mantini D, Logothetis NK, Deco G. 2015. Can sliding-window correlations reveal dynamic functional connectivity in resting-state fMRI? Neuroimage 127:242–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudetz AG, Liu X, Pillay S. 2015. Dynamic repertoire of intrinsic brain states is reduced in propofol-induced unconsciousness. Brain Connect 5:10–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Womelsdorf T, Gati JS, Everling S, Menon RS. 2013. Resting-state networks show dynamic functional connectivity in awake humans and anesthetized macaques. Hum Brain Mapp 34:2154–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karahanoğlu FI, Caballero-Gaudes C, Lazeyras F, Van De Ville D. 2013. Total activation: fMRI deconvolution through spatio-temporal regularization. Neuroimage 73:121–134 [DOI] [PubMed] [Google Scholar]
- Karahanoğlu FI, Van De Ville D. 2015. Transient brain activity disentangles fMRI resting-state dynamics in terms of spatially and temporally overlapping networks. Nat Commun 6:7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilholz SD, Magnuson ME, Pan WJ, Willis M, Thompson GJ. 2013. Dynamic properties of functional connectivity in the rodent. Brain Connect 3:31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilholz SD, Pan W-J, Billings J, Nezafati M, Shakil S. 2017. Noise and non-neuronal contributions to the BOLD signal: applications to and insights from animal studies. Neuroimage 154:267–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna A, Pascual-Leone A, Michel CM, Farzan F. 2015. Microstates in resting-state EEG: current status and future directions. Neurosci Biobehav Rev 49:105–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiviniemi V, Vire T, Remes J, Elseoud AA, Starck T, Tervonen O, Nikkinen J. 2011. A sliding time-window ICA reveals spatial variability of the default mode network in time. Brain Connect 1:339–347 [DOI] [PubMed] [Google Scholar]
- Kiviniemi V, Wang X, Korhonen V, Keinänen T, Tuovinen T, Autio J, et al. 2016. Ultra-fast magnetic resonance encephalography of physiological brain activity—glymphatic pulsation mechanisms?. J Cereb Blood Flow Metab 36:1033–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kringelbach ML, McIntosh AR, Ritter P, Jirsa VK, Deco G. 2015. The rediscovery of slowness: exploring the timing of cognition. Trends Cogn Sci 19:616–628 [DOI] [PubMed] [Google Scholar]
- Kucyi A, Davis KD. 2014. Dynamic functional connectivity of the default mode network tracks daydreaming. Neuroimage 100:471–480 [DOI] [PubMed] [Google Scholar]
- Kucyi A, Esterman M, Riley CS, Valera EM. 2016. Spontaneous default network activity reflects behavioral variability independent of mind-wandering. Proc Natl Acad Sci U S A 113:13899–13904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucyi A, Hove MJ, Esterman M, Hutchison RM, Valera EM. 2017. Dynamic brain network correlates of spontaneous fluctuations in attention. Cereb Cortex 27:1831–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudela M, Harezlak J, Lindquist MA. 2017. Assessing uncertainty in dynamic functional connectivity. Neuroimage 149:165–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumann TO, Snyder AZ, Mitra A, Gordon EM, Gratton C, Adeyemo B, et al. 2016. On the stability of BOLD fMRI correlations. Cereb Cortex [Epub ahead of print]; DOI: 10.1093/cercor/bhw265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi N, Richiardi J, Gschwind M, Simioni S, Annoni J-M, Schluep M, et al. 2013. Principal components of functional connectivity: a new approach to study dynamic brain connectivity during rest. Neuroimage 83:937–950 [DOI] [PubMed] [Google Scholar]
- Leonardi N, Shirer WR, Greicius MD, Van De Ville D. 2014. Disentangling dynamic networks: separated and joint expressions of functional connectivity patterns in time. Hum Brain Mapp 35:5984–5995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi N, Van De Ville D. 2015. On spurious and real fluctuations of dynamic functional connectivity during rest. Neuroimage 104:430–436 [DOI] [PubMed] [Google Scholar]
- Li X, Zhu D, Jiang X, Jin C, Zhang X, Guo L, et al. 2014. Dynamic functional connectomics signatures for characterization and differentiation of PTSD patients. Hum Brain Mapp 35:1761–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist MA, Xu Y, Nebel MB, Caffo BS. 2014. Evaluating dynamic bivariate correlations in resting-state fMRI: a comparison study and a new approach. Neuroimage 101:531–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Duyn JHH. 2013. Time-varying functional network information extracted from brief instances of spontaneous brain activity. Proc Natl Acad Sci U S A 110:4392–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Calhoun VD, Phlypo R, Adalı T. 2014. Dynamic changes of spatial functional network connectivity in healthy individuals and schizophrenia patients using independent vector analysis. Neuroimage 90:196–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Hamilton C, Zhang N. 2017. Dynamic connectivity patterns in conscious and unconscious brain. Brain Connect 7:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeed W, Magnuson M, Hasenkamp W, Schwarb H, Schumacher EHH, Barsalou L, Keilholz SDD. 2011. Spatiotemporal dynamics of low frequency BOLD fluctuations in rats and humans. Neuroimage 54:1140–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeed W, Magnuson M, Keilholz SD, 2009. Spatiotemporal dynamics of low frequency fluctuations in BOLD fMRI of the rat. J Magn Reson Imaging 30:384–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Murakami T, Ohki K. 2016. Transient neuronal coactivations embedded in globally propagating waves underlie resting-state functional connectivity. Proc Natl Acad Sci U S A 113:6556–6561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messé A, Rudrauf D, Benali H, Marrelec G. 2014. Relating structure and function in the human brain: relative contributions of anatomy, stationary dynamics, and non-stationarities. PLoS Comput Biol 10:e1003530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RL, Yaesoubi M, Calhoun VD. 2016a. Cross-frequency rs-fMRI network connectivity patterns manifest differently for schizophrenia patients and healthy controls. IEEE Signal Process Lett 23:1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RL, Yaesoubi M, Turner JA, Mathalon D, Preda A, Pearlson G, et al. 2016b. Higher dimensional meta-state analysis reveals reduced resting fMRI connectivity dynamism in schizophrenia Patients. PLoS One 11:e0149849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra A, Snyder AZ, Blazey T, Raichle ME. 2015. Lag threads organize the brain's intrinsic activity. Proc Natl Acad Sci U S A 112:E2235–E2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller S, Yacoub E, Olman CA, Auerbach E, Strupp J, Harel N, Ugurbil K. 2010. Multiband multislice GE-EPI at 7 tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain fMRI. Magn Reson Med 63:1144–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Birn RM, Bandettini PA. 2013. Resting-state fMRI confounds and cleanup. Neuroimage 80:349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Birn RM, Handwerker DA, Jones TB, Bandettini PA. 2009. The impact of global signal regression on resting state correlations: are anti-correlated networks introduced? Neuroimage 44:893–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso F, Brinkmeyer J, Mobascher A, Warbrick T, Winterer G. 2010. Spontaneous brain activity and EEG microstates. A novel EEG/fMRI analysis approach to explore resting-state networks. Neuroimage 52:1149–1161 [DOI] [PubMed] [Google Scholar]
- Nikolaou F, Orphanidou C, Papakyriakou P, Murphy K, Wise RG, Mitsis GD. 2016. Spontaneous physiological variability modulates dynamic functional connectivity in resting-state functional magnetic resonance imaging. Philos Trans A Math Phys Eng Sci 374 pii: [DOI] [PubMed] [Google Scholar]
- Oppenheim AV, Schafer RW. 2009. Discrete-Time Signal Processing. Upper Saddle River, NJ: Prentice Hall Press [Google Scholar]
- Ou J, Lian Z, Xie L, Li X, Wang P, Hao Y, et al. 2014. Atomic dynamic functional interaction patterns for characterization of ADHD. Hum Brain Mapp 35:5262–5278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W-J, Thompson G, Magnuson M, Majeed W, Jaeger D, Keilholz S. 2011. Broadband local field potentials correlate with spontaneous fluctuations in functional magnetic resonance imaging signals in the rat somatosensory cortex under isoflurane anesthesia. Brain Connect 1:119–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W-JJ, Thompson GJ, Magnuson ME, Jaeger D, Keilholz S. 2013. Infraslow LFP correlates to resting-state fMRI BOLD signals. Neuroimage 74:288–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel RS, Van De Ville D, Bowman FD. 2006. Determining significant connectivity by 4D spatiotemporal wavelet packet resampling of functional neuroimaging data. Neuroimage 31:1142–1155 [DOI] [PubMed] [Google Scholar]
- Petridou N, Gaudes CC, Dryden IL, Francis ST, Gowland PA. 2013. Periods of rest in fMRI contain individual spontaneous events which are related to slowly fluctuating spontaneous activity. Hum Brain Mapp 34:1319–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. 2012. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage 59:2142–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preti MG, Bolton TA, Van De Ville D. 2016. The dynamic functional connectome: state-of-the-art and perspectives. Neuroimage [Epub ahead of print]; DOI: 10.1016/j.neuroimage.2016.12.061 [DOI] [PubMed] [Google Scholar]
- Rashid B, Arbabshirani MR, Damaraju E, Cetin MS, Miller R, Pearlson GD, Calhoun VD. 2016. Classification of schizophrenia and bipolar patients using static and dynamic resting-state fMRI brain connectivity. Neuroimage 134:645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid B, Damaraju E, Pearlson GD, Calhoun VD. 2014. Dynamic connectivity states estimated from resting fMRI Identify differences among Schizophrenia, bipolar disorder, and healthy control subjects. Front Hum Neurosci 8:897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryali S, Shih Y-YI, Chen T, Kochalka J, Albaugh D, Fang Z, et al. 2016. Combining optogenetic stimulation and fMRI to validate a multivariate dynamical systems model for estimating causal brain interactions. Neuroimage 132:398–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaghiani S, Kleinschmidt A. 2013. Functional interactions between intrinsic brain activity and behavior. Neuroimage 80:379–386 [DOI] [PubMed] [Google Scholar]
- Sadaghiani S, Poline J-B, Kleinschmidt A, D'Esposito M. 2015. Ongoing dynamics in large-scale functional connectivity predict perception. Proc Natl Acad Sci U S A 112:8463–8468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoǧlu U, Pearlson GD, Kiehl KA, Wang YM, Michael AM, Calhoun VD. 2010. A method for evaluating dynamic functional network connectivity and task-modulation: application to schizophrenia. Magma 23:351–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinost D, Tokoglu F, Shen X, Finn ES, Noble S, Papademetris X, Constable RT. 2016. Fluctuations in global brain activity are associated with changes in whole-brain connectivity of functional networks. IEEE Trans Biomed Eng 63:2540–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakil S, Lee CH, Keilholz SD. 2016. Evaluation of sliding window correlation performance for characterizing dynamic functional connectivity and brain states. Neuroimage 133:111–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Rogers BP, Chen LM, Morgan VL, Mishra A, Wilkes DM, Gore JC. 2016. Realistic models of apparent dynamic changes in resting-state connectivity in somatosensory cortex. Hum Brain Mapp 37:3897–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine JM, Koyejo O, Poldrack RA. 2016. Temporal meta-states are associated with differential patterns of dynamic connectivity, network topology and attention. Proc Natl Acad Sci USA 113:1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine JM, Oluwasanmi K, Bell PT, Gorgolewski KJ, Gilat M, Poldrack RA. 2015. Estimation of dynamic functional connectivity using Multiplicative Analytical Coupling. Neuroimage [Epub ahead of print]; DOI: 10.1016/j.neuroimage.2015.07.064 [DOI] [PubMed] [Google Scholar]
- Shmuel A, Leopold DA. 2008. Neuronal correlates of spontaneous fluctuations in fMRI signals in monkey visual cortex: implications for functional connectivity at rest. Hum Brain Mapp 29:751–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel JS, Mitra A, Laumann TO, Seitzman BA, Raichle M, Corbetta M, Snyder AZ. 2016. Data quality influences observed links between functional connectivity and behavior. Cereb Cortex [Epub ahead of print]; DOI: 10.1093/cercor/bhw253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simony E, Honey CJ, Chen J, Lositsky O, Yeshurun Y, Wiesel A, Hasson U. 2016. Dynamical reconfiguration of the default mode network during narrative comprehension. Nat Commun 7:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, et al. 2009. Correspondence of the brain's functional architecture during activation and rest. Proc Natl Acad Sci 106:13040–13045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suk H-I, Wee C-Y, Lee S-W, Shen D. 2016. State-space model with deep learning for functional dynamics estimation in resting-state fMRI. Neuroimage 129:292–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghia J, Ryali S, Chen T, Supekar K, Cai W, Menon V. 2017. Bayesian switching factor analysis for estimating time-varying functional connectivity in fMRI. Neuroimage 155:271–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliazucchi E, Balenzuela P, Fraiman D, Chialvo DR. 2012. Criticality in large-scale brain FMRI dynamics unveiled by a novel point process analysis. Front Physiol 3:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliazucchi E, Balenzuela P, Fraiman D, Montoya P, Chialvo DR. 2011. Spontaneous BOLD event triggered averages for estimating functional connectivity at resting state. Neurosci Lett 488:158–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliazucchi E, Crossley N, Bullmore ET, Laufs H. 2016. Deep sleep divides the cortex into opposite modes of anatomical–functional coupling. Brain Struct Funct 221:4221–4234 [DOI] [PubMed] [Google Scholar]
- Tagliazucchi E, Laufs H. 2014. Decoding wakefulness levels from typical fMRI resting-state data reveals reliable drifts between wakefulness and sleep. Neuron 82:695–708 [DOI] [PubMed] [Google Scholar]
- Tagliazucchi E, von Wegner F, Morzelewski A, Brodbeck V, Laufs H. 2012c. Dynamic BOLD functional connectivity in humans and its electrophysiological correlates. Front Hum Neurosci 6:339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson GJ, Magnuson ME, Merritt MD, Schwarb H, Pan WJ, Mckinley A, et al. 2013a. Short-time windows of correlation between large-scale functional brain networks predict vigilance intraindividually and interindividually. Hum Brain Mapp 34:3280–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson GJ, Merritt MD, Pan WJ, Magnuson ME, Grooms JK, Jaeger D, Keilholz SD. 2013b. Neural correlates of time-varying functional connectivity in the rat. Neuroimage 83:826–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson GJ, Pan W-J, Billings JCW, Grooms JKK, Shakil S, Jaeger D, Keilholz SD. 2014a. Phase-amplitude coupling and infraslow (<1 Hz) frequencies in the rat brain: relationship to resting state fMRI. Front Integr Neurosci 8:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson GJ, Pan W-J, Keilholz SD. 2015. Different dynamic resting state fMRI patterns are linked to different frequencies of neural activity. J Neurophysiol 114:114–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson GJ, Pan WJ, Magnuson ME, Jaeger D, Keilholz SD. 2014b. Quasi-periodic patterns (QPP): large-scale dynamics in resting state fMRI that correlate with local infraslow electrical activity. Neuroimage 84:1018–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson GJ, Riedl V, Grimmer T, Drzezga A, Herman P, Hyder F. 2016. The whole-brain “global” signal from resting state fMRI as a potential biomarker of quantitative state changes in glucose metabolism. Brain Connect 6:435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognoli E, Kelso JAS. 2014. The metastable brain. Neuron 81:35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Kahn RS, Goni J, Sporns O. 2012. High-cost, high-capacity backbone for global brain communication. Proc Natl Acad Sci U S A 109:11372–11377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Sporns O. 2011. Rich-club organization of the human connectome. J Neurosci 31:15775–15786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara VM, Miller R, Calhoun V. 2017. An information theory framework for dynamic functional domain connectivity. J Neurosci Methods 284:103–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CW, Olafsson V, Tal O, Liu TT. 2013. The amplitude of the resting-state fMRI global signal is related to EEG vigilance measures. Neuroimage 83:983–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G-R, Liao W, Stramaglia S, Ding J-R, Chen H, Marinazzo D. 2013. A blind deconvolution approach to recover effective connectivity brain networks from resting state fMRI data. Med Image Anal 17:365–374 [DOI] [PubMed] [Google Scholar]
- Wu G-R, Marinazzo D. 2016. Sensitivity of the resting-state haemodynamic response function estimation to autonomic nervous system fluctuations. Philos Trans A Math Phys Eng Sci 374 pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Calhoun VD, Gonzalez-Castillo J, Damaraju E, Miller R, Bandettini PA, Mitra S. 2017. Whole-brain connectivity dynamics reflect both task-specific and individual-specific modulation: a multitask study. Neuroimage [Epub ahead of print]; DOI: 10.1016/j.neuroimage.2017.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Lindquist MA. 2015. Dynamic connectivity detection: an algorithm for determining functional connectivity change points in fMRI data. Front Neurosci 9:285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaesoubi M, Allen EA, Miller RL, Calhoun VD. 2015. Dynamic coherence analysis of resting fMRI data to jointly capture state-based phase, frequency, and time-domain information. Neuroimage 120:133–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaesoubi M, Miller R, Bustillo J, Lim KO, Vaidya J, Calhoun VD. 2017. A joint time-frequency analysis of resting-state functional connectivity reveals novel patterns of connectivity shared between or unique to schizophrenia patients and healthy controls. Neuroimage Clin 15:761–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaesoubi M, Miller RL, Calhoun VD. 2014. Mutually temporally independent connectivity patterns: a new framework to study resting state brain dynamics with application to explain group difference based on gender. Neuroimage 107:85–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaesoubi M, Miller RL, Calhoun VD. 2017. Time-varying spectral power of resting-state fMRI networks reveal cross-frequency dependence in dynamic connectivity. PLoS One 12:e0171647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Erhardt EB, Sui J, Du Y, He H, Hjelm D, et al. 2015. Assessing dynamic brain graphs of time-varying connectivity in fMRI data: application to healthy controls and patients with schizophrenia. Neuroimage 107:345–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Zotev V, Phillips R, Drevets WC, Bodurka J. 2012. Spatiotemporal dynamics of the brain at rest—exploring EEG microstates as electrophysiological signatures of BOLD resting state networks. Neuroimage 60:2062–2072 [DOI] [PubMed] [Google Scholar]
- Zalesky A, Breakspear M. 2015. Towards a statistical test for functional connectivity dynamics. Neuroimage 114:466–470 [DOI] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Cocchi L, Gollo LL, Breakspear M. 2014. Time-resolved resting-state brain networks. Proc Natl Acad Sci U S A 111:10341–10346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li X, Li C, Lian Z, Huang X, Zhong G, et al. 2014. Inferring functional interaction and transition patterns via dynamic Bayesian variable partition models. Hum Brain Mapp 35:3314–3331 [DOI] [PMC free article] [PubMed] [Google Scholar]