ABSTRACT
Primary hepatocellular carcinoma (PHC) is a major health problem worldwide and is one of the 10 most commonly diagnosed cancers in China. Heat shock protein 27 (HSP27) were found to be overexpressed in a wide range of malignancies including PHC, however, post-translational modification of HSP27 still needs exploration in PHC. Recently, SUMOylation, an important post-translational modification associating with the development of many kinds of cancers has been intensively studied. In the current study, mRNA and protein level of HSP27 in archived tumor samples representing various pathological characteristics of PHC were examined, and modification of HSP27 by SUMO2/3 was investigated. HSP27 were expressed abundantly in patients' tumor tissues, and found to be associated with pathological progression. Besides, HSP27 was also elevated significantly in liver cancer cell lines Huh7 and HepG2 compared with human hepatocyte cells L02. Furthermore, knockdown of HSP27 was found to be associated with the decreased proliferation and invasion ability in Huh7 and HepG2 cells. Immunofluorescence assay showed that HSP27 and SUMO2/3 were co-localized in the subcellular, and co-immunoprecipitation verified the interaction between HSP27 and SUMO2/3. Overexpression of SUMO2/3 upregulated the HSP27 protein level and promotes Huh7 and HepG2 cell proliferation and invasion, and vice versa when the SUMO2/3 was knockdown. Taken together, increased protein level of HSP27 through SUMO2/3-mediated SUMOylation plays crucial roles in the progression of PHC, and this finding may shed light on developing potential therapeutic targets for PHC.
KEYWORDS: HSP27, SUMOylation, SUMO2/3, liver cancer
Introduction
Primary hepatocellular carcinoma (PHC) is a common malignant tumor in clinical with the characteristic of high degree of malignancy, aggressive, poor prognosis and high mortality. The mortality of PHC was the second among all the malignant tumors in the world.1 The metastasis and recurrence of PHC is one of the difficult and hot spots in the present study. Heat shock proteins (HSP) have been found to be overexpressed in a variety of tumor cells, which play an important role in the development of tumor, tumor immunity, anti-tumor therapy and prognosis prediction.2 HSPs are ubiquitous and evolutionarily conserved proteins among species.3 Mammalian HSPs are classified according to their molecular mass into 5 families; Large HSPs, HSP90, HSP70, HSP60 and small HSPs including; HSP27.4 HSP27 is an important member of the HSP family, with the important biologic functions in protecting the cells from various stress factors of injury, in addition it also can be involved in cell proliferation, differentiation and signal transduction regulation.5 But the mechanism of HSP27 on the pathogenesis of PHC and its invasion and metastasis is not clear at present.
Small ubiquitin-related modifier (SUMO) proteins are a family of small proteins which resemble the 3 dimensional structure of ubiquitin.6 There are 4 SUMO paralogs in human genome, and they can be found in liver tissues except SUMO4.7 Human SUMO2 and SUMO3 share ∼97% sequence and presently cannot be distinguished by antibodies, collectively named as SUMO2/3.8 SUMO proteins participate in an important post-translational modification called SUMOylation, which promotes SUMO proteins binding to target proteins.9 A number of studies have shown that SUMOylation plays an important role in human pathogenesis, especially inflammation related cancer, such as hepatocellular carcinoma (HCC).10
In this study, we detect the HSP27 mRNA expression levels in peripheral blood of different stages PHC patients with the real-time quantitative PCR and the protein expression levels in tumor tissues by immunohistochemistry. We further revealed the inner link between HSP27 and the PHC occurrence, invasion metastasis to obtain potential PHC diagnosis markers and therapeutic targets. Furthermore, we confirmed whether there is an interaction between HSP27 and SUMO2/3 via immunofluorescence and co-immunoprecipitation assays, and the detailed molecular mechanism and biologic function of how SUMO2/3 modified HSP27 was explored.
Result
HSP27 was upregulated in PHC patient's samples and cell lines
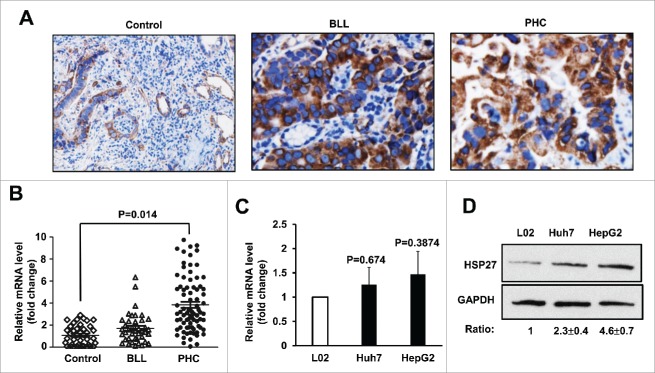
To test whether Hsp27 plays an important role in occurrence and development of PHC, we detected the Hsp27 protein level as well as mRNA expressions in tumor tissues from PHC patients, BLL patients and healthy control group (excluded all liver diseases when undergo physical examination) by immunohistochemistry or real-time RT-PCR assay, respectively. The protein level of HSP27 was verified by immunohistochemistry assay in all tissues from subjects of different groups, and the result showed that HSP27 protein was significantly upregulated in the PHC group and the BLL group than those in the healthy control group (Fig. 1A). In addition, the HSP27 mRNA levels were slightly higher in the PHC group than those in the Control or the BLL group (p < 0.05), while there was no significant difference in the expression of HSP27 mRNA between the BLL group and the control group (>0.05). (Fig. 1B). Furthermore, we examined HSP27 expressions of PHC patients in different clinical stages with real-time RT-PCR, PHC patients were divided into 4 phases (phase I, II, III, IV) according to the TNM Classification of Malignant Tumors (TNM) of International Union against cancer. The results showed that the differential expression of HSP27 mRNA was involved in the occurrence and progress of PHC, and the plasma HSP27 level in recurrent patients were significantly lower than non-recurrent ones (p < 0.05)(Table 1). To further determine the function of HSP27 in PHC development, we chose the human hepatoma cells Huh7 and the human hepatoblastoma cells HepG2 for further study, and used the human hepatocyte cells L02 as a normal hepatocyte control. Firstly, we detected the mRNA level of HSP27 in 3 cell lines, we did not observe the significant difference among these groups (p > 0.05, when either the Huh7 or HepG2 cells compared with the L02 cells) (Fig. 1C). In addition, western blot assay was used to determine the protein level of HSP27 in 2 carcinoma cells and the normal hepatocyte, and the result showed that the protein level of HSP27 was significantly upregulated in Huh7 and HepG2 cells than that in L02 cells, which indicated that some post-transcriptional modification affecting protein stability may have occurred in hepatocellular carcinoma (Fig. 1D).
Figure 1.
HSP27 was upregulated in PHC patient's samples and cell lines. (A) The expression of Hsp27 protein in healthy control group (Control) group, BLL group and PHC group via immunohistochemistry assay. Magnification, 200 ×. (B) The mRNA level of Hsp27 in healthy control group (Control), benign liver lesions patients group (BLL) and primary hepatocellular carcinoma patients group (PHC) via real time RT-PCR assay. Fold change of HSP27 mRNA in BLL and PHC group was compared with Control group. (C) The mRNA level of Hsp27 in L02 cells, Huh7 cells and HepG2 cells via real time RT-PCR assay. Fold change of HSP27 mRNA in Huh7 and HepG2 cells was compared with L02 cells. (D) The expression of HSP27 protein in L02 cells, Huh7 cells and HepG2 cells via western blot assay. The ratios of HSP27 to GAPDH for 3 independent experiments are shown as indicated below the blots.
Table 1.
Relationship between plasma HSP27 level and clinicopathological features in patients with HCC.
| Clinicopathological parameters | Cases (%) | plasma HSP27 level (Median) | P-value |
|---|---|---|---|
| Gender | |||
| male | 52 | 32.33 | 0.3212 |
| female | 48 | 30.61 | |
| Age(years) | |||
| ≤ 60 | 43 | 31.23 | 0.5622 |
| >60 | 57 | 30.69 | |
| Diameter(cm) | |||
| ≤ 5.0 | 61 | 15.65 | 0.0015 |
| >5.0 | 39 | 44.32 | |
| Viral infection (HBV or HCV) | |||
| Positive | 68 | 34.23 | 0.2314 |
| Negative | 32 | 29.01 | |
| Pathological differentiation | |||
| Well | 25 | 16.66 | 0.0023 |
| Moderate or poor | 75 | 42.45 | |
| Liver cirrhosis | |||
| Yes | 56 | 34.54 | 0.2115 |
| No | 44 | 28.23 | |
| pStage (TNM: AJCC/UICCg) | |||
| I | 35 | 14.45 | 0.0011 |
| II-IV | 65 | 45.21 | |
| Recurrence | |||
| Yes | 30 | 20.23 | 0.0055 |
| No | 70 | 39.88 |
HSP27 level associates proliferation and invasion of the HCC cells
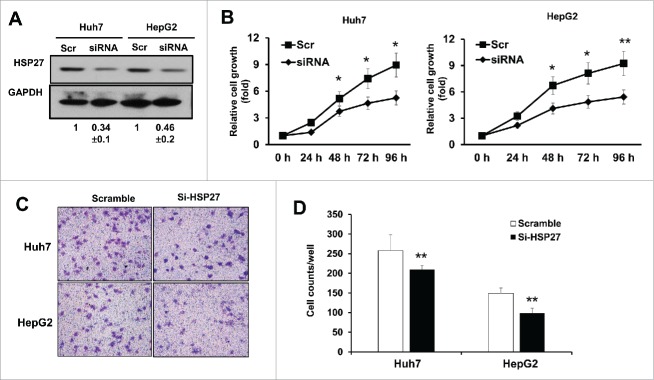
To determine the biologic functions of HSP27 in PHC carcinogenesis, we performed gene manipulation of HSP27 in Huh7 and HepG2 cells. After transfection of small interfering RNA (siRNA) specifically targeting HSP27 gene for 48 hours, protein level of HSP27 was analyzed using western blotting in both the Huh7 and HepG2 cells. Compared with the non-targeting scramble control group (Scr), the protein level of HSP27 in the siRNA group was significantly reduced in both the Huh7 and the HepG2 cells (Fig. 2A). Further, the Huh7 and HepG2 cells with HSP27 knockdown were selected for cancer proliferation and invasion ability assays. Our results showed that compared with the scramble control (Scr) group, cell proliferation in the presence of the siRNA began to decrease at 24 h, and presented significant difference after 48 hours to 96 hours (p < 0.05), indicating that the HSP27 expression was correlated with the proliferation of the HSP tumor cells (Fig. 2B). Moreover, transwell chambers coated with Matrigel were used to study the invasion ability of the Huh7 and HepG2 cells with HSP27 knockdown. Compared with the scramble control (Scr) group, the number of invading cells that crossed the membranes diminished after 48 h in both the HSP27 cells and HepG2 cells (Fig. 2C), and quantitative analysis of the invasion cell numbers have shown significant differences between the scramble and si-HSP27 groups, both in Huh7 and in HepG2 cells (Fig. 2D), indicating that HSP27 was correlated with the invasiveness of the hepatocellular carcinoma cells. Thus, the inhibition of HSP27 may reduce the proliferation and invasiveness of HPS cells.
Figure 2.
HSP27 siRNA reduces the proliferation and invasion of the HCC cells. (A) The expression of HSP27 protein in Huh7 cells and HepG2 cells transfected with Scramble siRNA (Scr) or siRNA targeting Hsp27 (siRNA) for 48 hours via western blot assay, respectively. GAPDH was used as the loading control for each group. The ratios of HSP27 to GAPDH for 3 independent experiments are shown as indicated below the blots. (B) The relative cell growth rate (normalized with the cell numbers at day 0) in Huh7 cells (left panel) and HepG2 cells (right panel) transfected with Scr or siRNA for 0, 24, 48, 72 and 96 hours after transfection by adding 10 μl of CCK8 reagent. *, p < 0.05 and **, p < 0.01 in siRNA group compared with Scr group. (C) The invasion capability in Huh7 cells and HepG2 cells transfected with Scr or siRNA via transwell assay after 48 hours. (D) Quantitative analysis of the invasion cell numbers per well of a 24-well plate in the Huh7 and HepG2 cells transfected with scramble control or si-HSP27. Data presents Mean±SD for 12 independent visions per group in the Huh7 and HepG2 cells, respectively. *, p < 0.05, **, p < 0.01, the scramble groups vs. si-HSP27 group.
HSP27 was post-transcriptionally modified by protein degradation
As shown above, we found that HSP27 mRNA expression was only slightly upregulated in Huh7 and HepG2 cells compared with L02 cells, whereas the protein level of Huh7 and HepG2 cells showed significantly upregulated. As we all known, the post-transcriptional modification includes the microRNA regulation, histone modification and protein degradation. To illustrate the detailed molecular mechanism concerning how HSP27 expression was different between mRNA level and protein level, we determined from the 4 aspect. Firstly, we found that there was no conserved binding site of microRNAs on the mRNA of HSP27 (http://feb2014.archive.ensembl.org/index.html, Transcript: ENST00000248553) 3′UTR (Untranslated Regions) based on the Targetscan website (http://www.targetscan.org/vert_71/). And we detected some poorly conserved microRNAs, the miR-214 and miR-539, targeting HSP27 (S Fig. 1A), and found these microRNA levels had no significant difference among L02 and Huh7 or HepG2 cells (S Fig. 1B). In addition, the histone modification of H3K4 trimethylation (H3K4me3) H3K27me3 on the HSP27 gene promoter was detected using chromatin immunoprecipitation (ChIP) assay, however, we did not observe any significant difference (S Fig. 1C).
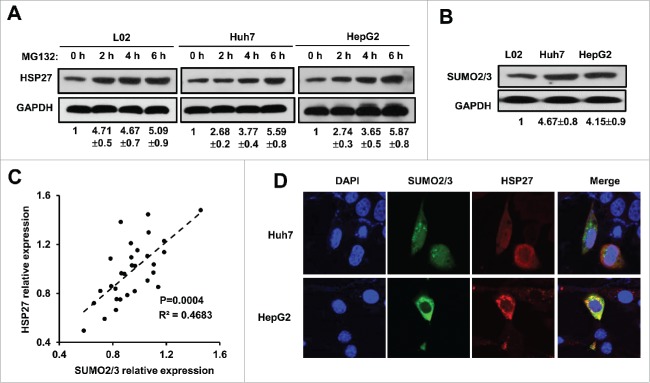
To explore the mechanism by which post-translational modification participated in regulating the cytoplasmic HSP27 level, the cells were treated with a reversible proteasome inhibitor MG132 (20 μM) in 3 cell lines for up to 6 hours, we found that HSP27 protein level was significantly upregulated in L02 cells as well as the Huh7 and HepG2 cells, which indicated that post-translational modification indeed existed in HSP27 expression (Fig. 3A). To confirm whether SUMO modification exerts its function in regulating HSP27 expression, we detected the expression of SUMO1, SUMO2/3 protein in 3 cell lines. We found that there was no significant difference for SUMO1 expression in L02 cells and the Huh7 and HepG2 cancer cells (data not shown), however, L02 cells showed lower SUMO2/3 expression compared with Huh7 and HepG2 cells (Fig. 3B). The protein expression of SUMO2/3 and HSP27 were positively correlated in patients' tissue samples, which indicated that HSP27 might be involved in SUMO2/3 related modification (Fig. 3C). To indicate whether SUMO2/3 participated in the regulation of HSP27 degradation, immunofluorescence assay was used to detect the subcellular localization of HSP27 and SUMO2/3, the result showed that both SUMO 2/3 and HSP27 located and merged in the cytoplasm (Fig. 3D). These observations suggest a relationship between HSP27 and SUMO 2/3.
Figure 3.
HSP27 was post-transcriptionally modified by protein degradation. (A) The expression of HSP27 protein in L02 cells, Huh7 cells and HepG2 cells treated with MG132 (20 μM) at 0, 2, 4, 6 hours via western blot assay. The ratios of HSP27 to GAPDH for 3 independent experiments are shown as indicated below the blots. (B) The expression of SUMO2/3 protein in L02 cells, Huh7 cells and HepG2 cells via western blot assay. The ratios of SUMO2/3 to GAPDH for 3 independent experiments are shown as indicated below the blots. (C) The correlation analysis of the protein expression between HSP27 and SUMO2/3. (D) Immunofluorescence of nucleus (DAPI, blue), SUMO2/3 (green) and HSP27 (red) in Huh7 cells and HepG2 cells.
SUMO 2/3 protected HSP27 from degradation
It was reported that SUMOylation of the majority of substrate proteins occur through the covalently attachment of SUMO to a lysine (K) in a consensus sequence KxD/E, although SUMO conjugation may also occur in non-consensus sites.11 Then the amino acid sequence was analyzed, and we found that there was a putative SUMO2/3 binding sequence GKHE in HSP27 protein (Fig. 4A). To determine whether SUMO 2/3 contributed in HSP27 degradation, immunoprecipitation assay was used in both Huh7 and HepG2 cells, in which we used the anti-HSP27 antibodies to precipitate the HSP protein and then used the anti-SUMO2/3 antibody detect the possible SUMOylation, and the result showed that there was amount of HSP27 binds to SUMO 2/3 in Huh7 (Fig. 4B) and HepG2 cells (Fig. 4C).
Figure 4.

SUMO 2/3 directly binds HSP27 protein. (A) Shown is the amino acid sequence of HSP27 protein and the binding site of SUMO2/3 (bold and underlined fonts). Immunoprecipitation assay showed the interaction of SUMO2/3 with HSP27 in (B) Huh7 cells and (C) HepG2 cells. 20 μg of the whole cell lysate was served as the input control, and 2 μg of IgG control or α-HSP27 monoclonal antibody was used to precipitate the target protein HSP27, and after blotting, the SUMO2/3 antibody was incubated to detect the binding on HSP27 protein at dilution of 1:500. The ratios of SUMO2/3 in the IgG or α-HSP27 group to that in the input group for 3 independent experiments are shown as indicated below the blots.
Manipulation of SUMO2/3 affected the hepatocellular carcinoma cells proliferation and invasion
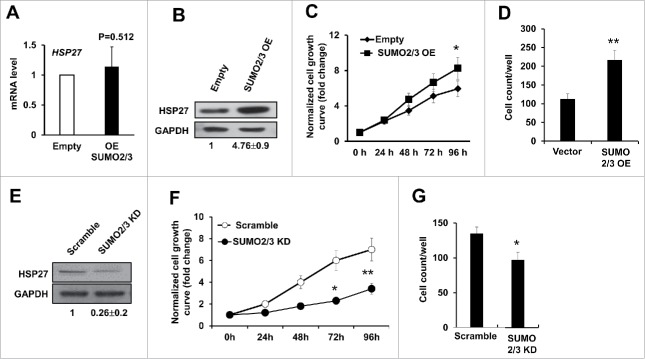
Since our data suggested a positive evidence for HSP27 SUMOylation modification, we hypothesize that manipulation of SUMO2/3 level would affect hepatocellular carcinoma cells proliferation and invasion as well as affecting the HSP27 protein level. To examine this possibility, we manipulated the SUMO2/3 level by overexpression and knockdown experiments in HepG2 cells by transfected with SUMO2/3 expressing vector (SUMO2/3 EO) or the empty vector control (Empty) for 48 hours, and we found that although overexpression of SUMO2/3 did not affect the mRNA level of HSP27 (Fig. 5A), the HSP27 protein level was significantly upregulated in the cells overexpressing SUMO2/3, whereas did not occur in the cells transfecting empty vector (Fig. 5B). These above data indicated that SUMO2/3 contributed in protecting HSP27 from degradation. Functionally, we found that overexpression of SUMO2/3 could increase the cell growth rate in Huh7 cells compared with the empty vector control (p < 0.05 after 96 hours after transfection) (Fig. 5C). The invasion capability was detected via transwell assay, and the result showed that overexpressing SUMO2/3 could significantly increase the invasion capability of Huh7 cells (Fig. 5D). On the contrary, when we knockdown the SUMO2/3 using siRNA, the HSP27 protein level was downregulated obviously (Fig. 5E), and the Huh7 cell proliferation and invasion abilities were all significantly decreased compared with the scramble control (Fig. 5F, 5G). We also tested the whether HSP27 knockdown affected the growth of L02 cells in vitro. Although the mRNA level of HSP27 in the L02 cells was similar with that in the Huh7 and HepG2 cells, protein level of HSP27 was very low in the L02 cells (as shown above in Fig. 1D), therefore knockdown efficacy of HSP27 in the L02 cells was limited and we did not find obvious change in cell growth (data not shown). Hence, we believe the function of HSP27 is more specific for cancer cells than for the normal cells. These data strongly supported our hypothesis that SUMO2/3 modifies HSP27 protein level to affect hepatocellular carcinoma cells proliferation and invasion.
Figure 5.
SUMO2/3 promotes PHC cell proliferation and invasion. (A) The mRNA level of HSP27 in Huh7 cells transfected with empty vector (Empty) or SUMO2/3 expressing plasmid for 48 hours was detected using real time PCR. (B) Shown are western blot assays to determine the expression of HSP27 protein in Huh7 cells transfected with empty vector control or SUMO2/3 overexpression plasmids for 48 hours. The ratios of HSP27 to GAPDH for 3 independent experiments are shown as indicated below the blots. (C) The relative cell growth rate in Huh7 cells transfected with SUMO2/3 overexpression plasmid or Empty vector via cck-8 assay. Cell growth has been normalized to the day 0 and shown as fold change. (D) The invasion cell numbers per well of a 24-well plate of Huh7 cells transfected with SUMO2/3 overexpression plasmid or Empty vector for 48 hours via transwell assay. (E) Shown are western blot assays to determine the expression of HSP27 protein in Huh7 cells transfected with scramble control or siRNA specially targeting SUMO2/3 for 48 hours. The ratios of HSP27 to GAPDH for 3 independent experiments are shown as indicated below the blots. (F) The relative cell growth rate in Huh7 cells transfected with scramble control or siRNA specially targeting SUMO2/3 via cck-8 assay. Cell growth has been normalized to the day 0 and shown as fold change. (G) The invasion cell numbers per well of a 24-well plate of Huh7 cells transfected with scramble control or siRNA specially targeting SUMO2/3 for 48 hours via transwell assay. *, p < 0.05 and **, p < 0.01; vector control vs. OE or KD vectors.
Discussion
Different functions of HSPs have been described to explain their cytoprotective functions, including their most basic role as molecular chaperones, that is to regulate protein folding, transport, translocation and assembly, especially helping in the refolding of misfolded proteins, as well as their anti-apoptotic properties.12 Firstly, HSP27 was identified as a protein chaperone that assisted proper refolding of disordered proteins in response to heat shock.13 Recent evidences suggested that HSP27 was a multifunction protein, of which the deregulation had been implicated in neuro-degenerative diseases, cardiovascular diseases and cancers.14,15 Notably, HSP27 was expressed at basal level in normal liver cells, whereas hepatocellular carcinoma cells expressed very high levels of HSP27.16 High levels of HSP27 have been observed in many cancer types, and the tumorigenic potential of HSP27 has been observed in experimental models.17 Many clinical trials have also shown its association with promoting drug resistance, aggressive cancers, metastasis, and poor patient outcomes.18,19 In the present study, we found that the mRNA and protein level of HSP27 in patients with PHC was positively correlated with the clinical phases, which indicated that HSP27 might be involved in the progression of PHC. Though the function of HSP27 was illustrated in the present study, the detailed molecular mechanism was still not clear.
SUMOylation, an important post-translational modification, associates with the development of liver cancer. Since SUMOylation-deSUMOylation cycles can be highly dynamic and as protein SUMOylation is being attributed as an important event in controlling many aspects of cell physiology, including cell cycle regulation, transcription, nucleo-cytoplasmic transport, DNA replication and repair, chromosome dynamics, apoptosis and ribosome biogenesis.20,21 In the present study, the amino acid sequence has been analyzed first, and we found the binding site of SUMO2/3 on HSP27 protein at a consensus sequence KxD/E. Immunofluorescence assay showed that HSP27 and SUMO2/3 located in the cytoplasm, and both protein merge perfectly. Furthermore, co-immunoprecipitation assays showed that HSP27 and SUMO2/3 indeed existed in the same protein complex. Biologically, when SUMO2/3 was overexpressed in liver cancer cells, in which HSP27 showed low protein level though the HSP27 mRNA level is much higher, Huh7 cells showed more proliferation rate and migration capability.
It was known that SUMOylation and ubiquitination exhibit similar biologic processes of post-translational modification,Therefore, SUMO proteins compete with ubiquitin for the same lysine and inhibit proteasome- mediated degradation of target proteins. Liu et al. showed that SUMO proteins may mediate p65 SUMOylation and then recruit an inhibitor of p65 in nucleus, in which SUMO2/3 played a tumor suppressor function. The data showed some contradiction with our observation. They showed that SUMO2/3 level was downregulated in the tumor tissues as compared with the adjacent non-tumor tissues. In their study, double-labeled immunofluorescent staining was performed using the antibodies against SUMO2/3 in the liver tissues, HepG2 cells and Huh7 cells. However, SUMO2/3 presented high expression in both liver tissues and liver cancer cell lines. The data indicated that SUMO2/3 might maintain relative high expression level in both normal liver cells and cancer cells. In our data, we found SUMO2/3 was slightly higher in liver cancer cells. On the other hand, as an extensive modification regulator, SUMO2/3 might interact with dozens of target proteins, so we hypothesis that P65 was not the only target genes.
Taken together, we found that HSP27 was significantly upregulated in PHC blood samples and tissues, indicating HSP27 might potentially contributes to the tumorigenesis and development of PHC. In addition, knocking down HSP27 resulted in a decrease in cellular growth and invasion in cancer cells. Furthermore, we reported a novel observation that SUMO2/3 interacts with HSP27 and stabilizes HSP27 in the cytoplasm. Additionally, SUMO2/3 overexpression in normal liver cells slightly increase the proliferation and migration of hepatoma cells.
Method and material
Patient sample
All the samples were obtained from the inpatients of Tianjin Third Central Hospital from January 1, 2014 to December 31, 2015, including 80 cases of PHC (53 males and 27 females, aging 34 to 82 with median age 53); 40 cases of benign liver lesions (BLL) (28 males and 12 females, aging 21 to 62 with median age 42) and 40 cases of healthy control group (21 males and 19 females, aging 25 to 51 with median age 37). The samples were rapidly frozen into liquid nitrogen, and stored at −80C until use. Informed consents were signed and obtained from all patients enrolled in this study. The use of clinical samples was in accordance with the Declaration of Helsinki and was approved by the Ethics Committee of Tianjin Third Central Hospital.
Immunohistochemistry
Tumor tissues were embedded into paraffin sections, treated for 2 h at 65°C and then deparaffinized. Slides were applied to the following procedures: deparaffine, antigen retrieval and endogenous peroxidase blockage before incubation of the primary antibodies at 4°C overnight (1:100, Santa Cruz Biotechnology, Dallas, TX, USA). The slides were incubated with a HRP-conjugated secondary antibody (1:100; ZSGB Biotech, Beijing, China) for 1 h at 25°C and then applied to liquid DAB+ Substrate (ZSGB Biotech, Beijing, China), and a final hymatoxylin staining was performed to localize cell nucleus.
Cell culture and transfections
L02 (human hepatocyte cells), Huh7 (human hepatoma cells) and HepG2 (human hepatoblastoma cells) were all purchased from the American Type Culture Collection (ATCC, Shanghai, China), and cultured in Dulbecco's modified Eagle's medium (DMEM, Sigma-Aldrich, St. Louis, MO, USA) with 10% fetal bovine serum (Biological Industries, Shanghai, China). For siRNA experiments, the cells were transfected with HSP27 siRNA (F: 5′-AAAUGUAUCAAAAGAACACAC-3′, R: 5′-GUGUUCUUUUGAUACAUUUAU-3′) or Scramble (Scr) siRNA (F: 5′-AAAUAAAGAGUAUCAACACAC-3′, R: 5′-GUGUUGAUAUCUUUCAUUUAU-3′) (Sigma-Aldrich, St. Louis, MO, USA) using Oligofectamine reagents (Invitrogen, Carlsbad, CA, USA). Protein level was analyzed by immunoblotting 48 h after transfection to evaluate the knockdown efficacy. For SUMO2/3 expression, Huh7 cells were transfected with empty vector control (Empty) or SUMO2/3-overexpressing vector (SUMO2/3-OE) (kind gifts from Prof. Zhiqiang Liu laboratory, Tianjin Medical University) respectively using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA), and 48 hr after the transfection, cells were collected for qPCR and Western blot assay or cell growth and invasion assay, respectively.
Real-time RT-PCR
Total RNA was extracted from the collected white blood cell precipitation using the Trizol reagent (Invitrogen, Shanghai, China) according to the manufacturer's instructions. cDNA was synthesized using 1 μg of RNA with avian myeloblastosis virus reverse transcriptase (Promega, Madison, WI, USA) and oligo (dT) primers. Transcript levels were assessed by quantitative real-time PCR (ABI 7300; Applied Biosystems), and all experiments were normalized to GAPDH. The primers used for HSP27: F: 5′-ACGGTCAAGACCAAGGATGG-3′, R: 5′-AGCGTGTATTTCCGCGTGA-3′; SUMO 2/3 F: 5′-CTGCCGCCTCCTTCTTCTGC-3′, R: 5′-ATCCTCCATTTCCAACTGTCGTTC-3′; GAPDH: F: 5′-AACGGATTTGGTCGTATTGGG-3′, R: 5′-CGCTCCTGGAAGATGGTGAT-3′.
Western blot analysis
Add 1 mL ice-cold protein lysis solution to the collected white blood cell precipitation and then homogenized in ice for 30 min. Equal amounts of cytosolic protein were loaded and electrophoresed on SDS polyacrylamide gel and then transferred to a PVDF membrane. Membranes were blocked with nonfat milk, and then incubated with the primary antibodies (1:1000) at 4°C, overnight. The primary antibody binding was detected with a secondary anti-rabbit antibody (1:2000) and visualized by the ECL method. HSP27 antibody (sc-1048) and SUMO2/3 antibody (sc-32873) were both purchased from Santa Cruz Biotech (Dallas, Texas, USA). Relative quantification for western blot bands were analyzed using ImageJ software (Wayne Rasband, NIH, USA), 3 protein band scans from 3 independent experiments were analyzed, and the results were shown as ratios of TARGET GENES/GAPDH.
Cell proliferation, invasion and wound healing assay
Cell proliferation assay was performed by the CCK8 method (DOJINDO, Japan). Briefly, approximately 5 × 103 cells were transfected and then seeded into 96-well plates for cell culture. Proliferation rates were determined at 0, 24, 48, 72 and 96 h after transfection by adding 10 μl of CCK8 reagent. For cell invasion assay, cells were starved for serum in DMEM media for 6 hours, then seeded onto the upper compartment of 8 μm trasnwell chamber (Corning Inc., Corning, NY, USA) and cultured for 1.5 hour to let the cells to adhere. The upper surface of transwell membrane was scraped by cotton swab to remove non-invasion cells. After fixing with 4% paraformaldehyde, migrated cells on the lower surface were stained with crystal violet and quantified by counting. For wound healing assay, cells were seeded at 5 × 104 per well in 6-well plates, and scratched one day after seeding, and then added fresh medium containing 0.1% FBS. Pictures were taken immediately and 24 h after scratching. Five separate experiments were performed. For quantification purposes, scratch widths were measured.
Immunoprecipitation assay
Immunoprecipitation was performed using ImmunoCruz™ IP/WB Optima B System (Santa Cruz Biotechnology, Dallas, TX, USA) based on the manufacturer's guideline. 2 μg of primary antibody, or immunoglobulin G (IgG) was used for immunoprecipitation and control respectively. Blots were incubated overnight at 4°C with designated primary antibodies at 1:500 dilutions. Proteins were visualized using the Odyssey system (Li-Cor Biosciences, Lincoln, Nebraska, USA).
Statistical analysis
All of the experiments were performed at least in triplicate. Each experiment was independently performed at least 3 times. Data were presented as the mean ± standard deviation (SD) and analyzed using GraphPad Prism 5 software. Statistical significance was assessed using a 2-tailed unpaired Student's t-test. When a p value was less than 0.05, the differences were considered as statistically significant.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This study was supported by Grant from Hebei Education Department (QN20131072).
References
- 1.Duan YF, Li XD, Zhu F, Zhang F. Expression and clinical significance of angiotensin II type 1 receptor in human hepatocellular carcinoma. Exp Ther Med 2014; 7:323-8; PMID:24396398; https://doi.org/ 10.3892/etm.2013.1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karademir B, Bozaykut P, Kartal Ozer N. Heat shock proteins and proteasomal degradation in normal and tumor cells. Free Radic Biol Med 2014; 75(Suppl 1):S35; PMID:26461349; https://doi.org/ 10.1016/j.freeradbiomed.2014.10.774 [DOI] [PubMed] [Google Scholar]
- 3.Seigneuric R, Mjahed H, Gobbo J, Joly AL, Berthenet K, Shirley S, Garrido C. Heat shock proteins as danger signals for cancer detection. Front Oncol 2011; 1:37; PMID:22649762; https://doi.org/ 10.3389/fonc.2011.00037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iwai LK, Luczynski MT, Huang PH. Discoidin domain receptors: A proteomic portrait. Cell Mol Life Sci 2014; 71:3269-79; PMID:24705941; https://doi.org/ 10.1007/s00018-014-1616-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciocca DR, Calderwood SK. Heat shock proteins in cancer: Diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones 2005; 10:86-103; PMID:16038406; https://doi.org/ 10.1379/CSC-99r.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin S, Wilkinson KA, Nishimune A, Henley JM. Emerging extranuclear roles of protein SUMOylation in neuronal function and dysfunction. Nat Rev Neurosci 2007; 8:948-59; PMID:17987030; https://doi.org/ 10.1038/nrn2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo D, Li M, Zhang Y, Yang P, Eckenrode S, Hopkins D, Zheng W, Purohit S, Podolsky RH, Muir A, et al.. A functional variant of SUMO4, a new I kappa B alpha modifier, is associated with type 1 diabetes. Nat Genet 2004; 36:837-41; PMID:15247916; https://doi.org/ 10.1038/ng1391 [DOI] [PubMed] [Google Scholar]
- 8.Liu J, Sha M, Wang Q, Ma Y, Geng X, Gao Y, Feng L, Shen Y, Shen Y. Small ubiquitin-related modifier 2/3 interacts with p65 and stabilizes it in the cytoplasm in HBV-associated hepatocellular carcinoma. BMC Cancer 2015; 15:675; PMID:26458400; https://doi.org/ 10.1186/s12885-015-1665-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dasso M. Biochemistry: Rear view of an enzyme. Nature 2013; 497:576-7; PMID:23698365; https://doi.org/ 10.1038/nature12249 [DOI] [PubMed] [Google Scholar]
- 10.Ning BF, Ding J, Liu J, Yin C, Xu WP, Cong WM, Zhang Q, Chen F, Han T, Deng X, et al.. Hepatocyte nuclear factor 4alpha-nuclear factor-kappaB feedback circuit modulates liver cancer progression. Hepatology 2014; 60:1607-19; PMID:24752868; https://doi.org/ 10.1002/hep.27177 [DOI] [PubMed] [Google Scholar]
- 11.Xu J, He Y, Qiang B, Yuan J, Peng X, Pan XM. A novel method for high accuracy SUMOylation site prediction from protein sequences. BMC Bioinformatics 2008; 9:8; PMID:18179724; https://doi.org/ 10.1186/1471-2105-9-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Chen M, Zhou J, Zhang X. HSP27, 70 and 90, anti-apoptotic proteins, in clinical cancer therapy (Review). Int J Oncol 2014; 45:18-30; PMID:24789222; https://doi.org/ 10.3892/ijo.2014.2399 [DOI] [PubMed] [Google Scholar]
- 13.Rogalla T, Ehrnsperger M, Preville X, Kotlyarov A, Lutsch G, Ducasse C, Paul C, Wieske M, Arrigo AP, Buchner J, et al.. Regulation of Hsp27 oligomerization, chaperone function, and protective activity against oxidative stress/tumor necrosis factor alpha by phosphorylation. J Biol Chem 1999; 274:18947-56; PMID:10383393; https://doi.org/ 10.1074/jbc.274.27.18947 [DOI] [PubMed] [Google Scholar]
- 14.Brophy CM. Stress and vascular disease at the cellular and molecular levels. World J Surg 2002; 26:779-82; PMID:11948370; https://doi.org/ 10.1007/s00268-002-4052-6 [DOI] [PubMed] [Google Scholar]
- 15.Jaattela M. Escaping cell death: Survival proteins in cancer. Exp Cell Res 1999; 248:30-43; PMID:10094811; https://doi.org/ 10.1006/excr.1999.4455 [DOI] [PubMed] [Google Scholar]
- 16.Matsushima-Nishiwaki R, Toyoda H, Nagasawa T, Yasuda E, Chiba N, Okuda S, Maeda A, Kaneoka Y, Kumada T, Kozawa O. Phosphorylated heat shock protein 20 (HSPB6) regulates transforming growth factor-alpha-induced migration and invasion of hepatocellular carcinoma cells. PLoS One 2016; 11:e0151907; PMID:27046040; https://doi.org/ 10.1371/journal.pone.0151907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Straume O, Shimamura T, Lampa MJ, Carretero J, Oyan AM, Jia D, Borgman CL, Soucheray M, Downing SR, Short SM, et al.. Suppression of heat shock protein 27 induces long-term dormancy in human breast cancer. Proc Natl Acad Sci U S A 2012; 109:8699-704; PMID:22589302; https://doi.org/ 10.1073/pnas.1017909109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciocca DR, Arrigo AP, Calderwood SK. Heat shock proteins and heat shock factor 1 in carcinogenesis and tumor development: An update. Arch Toxicol 2013; 87:19-48; PMID:22885793; https://doi.org/ 10.1007/s00204-012-0918-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pavan S, Musiani D, Torchiaro E, Migliardi G, Gai M, Di Cunto F, Erriquez J, Olivero M, Di Renzo MF. HSP27 is required for invasion and metastasis triggered by hepatocyte growth factor. Int J Cancer 2014; 134:1289-99; PMID:23996744; https://doi.org/ 10.1002/ijc.28464 [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Dasso M. SUMOylation and deSUMOylation at a glance. J Cell Sci 2009; 122:4249-52; PMID:19923268; https://doi.org/ 10.1242/jcs.050542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bigarella CL, Ferro KP, Barcellos KS, Martins-de-Souza D, Traina F, Novello JC, Saad ST, Archangelo LF. Post-translational modification of the RhoGTPase activating protein 21, ARHGAP21, by SUMO2/3. FEBS Lett 2012; 586:3522-8; PMID:22922005; https://doi.org/ 10.1016/j.febslet.2012.08.012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.