Abstract
Soil salinity, a major environmental stress, reduces agricultural productivity by restricting plant development and growth. Jute (Corchorus spp.), a commercially important bast fiber crop, includes two commercially cultivated species, Corchorus capsularis and Corchorus olitorius. We conducted high-throughput transcriptome sequencing of 24 C. capsularis and C. olitorius samples under salt stress and found 127 common differentially expressed genes (DEGs); additionally, 4489 and 492 common DEGs were identified in the root and leaf tissues, respectively, of both Corchorus species. Further, 32, 196, and 11 common differentially expressed transcription factors (DTFs) were detected in the leaf, root, or both tissues, respectively. Several Gene Ontology (GO) terms were enriched in NY and YY. A Kyoto Encyclopedia of Genes and Genomes analysis revealed numerous DEGs in both species. Abscisic acid and cytokinin signal pathways enriched respectively about 20 DEGs in leaves and roots of both NY and YY. The Ca2+, mitogen-activated protein kinase signaling and oxidative phosphorylation pathways were also found to be related to the plant response to salt stress, as evidenced by the DEGs in the roots of both species. These results provide insight into salt stress response mechanisms in plants as well as a basis for future breeding of salt-tolerant cultivars.
Introduction
Soil salinity is a major environmental stress imposed on plants that reduces agricultural productivity by restricting plant development and growth[1]. Salinity has primary effects including ion toxicity and osmotic stress as well as secondary effects such as oxidative stress[1]. Plants have a variety of salt tolerance mechanisms that depend on mitogen-activated protein kinase (MAPK/MPK) and hormone signaling as well as posttranslational modification of proteins.
Na+ influx into roots occurs via different transporters. Plants use the Na+/H+ salt overly sensitive (SOS)1 antiporter, high-efficiency potassium transporter (HKT), and the tonoplast-localized Na+, K+/H+ exchanger (NHX) for sodium transport and detoxification[2, 3]. Na+ influx triggers an increase in cytosolic Ca2+ level; this is sensed by SOS3, which activates the serine/threonine protein kinase SOS2. Activated SOS2 phosphorylates and activates SOS1[2], a plasma membrane Na+/H+ antiporter that plays an important role in maintaining a low concentration of Na+ in the cytoplasm of cortex cells by extruding Na+ into the soil and loading Na+ into the xylem for long-distance transport to leaves[4, 5]. HKT1 mediates the reverse flux and unloads Na+ from xylem vessels to prevent accumulation of Na+ in the transpirational stream.
Salt stress can lead to elevated levels of reactive oxygen species (ROS) such as superoxide anion and hydrogen peroxide (H2O2) that are toxic and can cause oxidative damage to proteins, DNA, and lipids in the cell membrane[6]. ROS are scavenged by antioxidant metabolites (e.g., ascorbate and glutathione) and by ROS-detoxifying enzymes (e.g., superoxide dismutase and ascorbate peroxidase). Despite their toxicity to cells, ROS also function as signal transduction molecules that mediate responses to stress[7] by activating various MAPK signaling cascades, including MAPK kinase kinase 1, MPK4, and MPK6[8, 9].
Hormone signaling is important for mediating salt stress responses in plants[2]. For example, when abscisic acid (ABA) accumulates and binds to the pyrabactin resistance/Pyr1-like (PYL)/regulatory components of ABA receptor, the resultant conformational change leads to interaction with protein phosphatase (PP)2C and the formation of the ternary ABA-ABAR-PP2C complex. This in turn relieves inhibition of sucrose non-fermenting-1-related protein kinase (SnRK)2 via PP2C suppression. SnRK2 then phosphorylates and activates ABA response element-binding factor (ABF/AREB)/ABA-INSENSITIVE 5 transcription factors (TFs), resulting in ABA-dependent gene expression. During the abiotic stress response, ABA signaling activates the MAPK cascade, which regulates ABA effector proteins[10]. ABA can also induce H2O2 generation via phosphorylation of the plasma membrane NADPH oxidase respiratory burst oxidase homolog protein F. H2O2 can then mediate various ABA responses by modulating Ca2+ signaling[1].
Jute (Corchorus spp.) is one of the most commercially important bast fiber crops in the world as it provides biodegradable and renewable lignocellulose fiber. Corchorus capsularis and Corchorus olitorius are two commercially cultivated species of jute[11]. The plant is mainly distributed in China, India, Bangladesh, and east-central Africa[12] but global demand has been increasing due to its broad-spectrum applications and environmentally friendly characteristics. Various studies have investigated jute salt tolerance[13]. Naik et al. [14] studied a few of Corchorus capsularis accessions under differential salt concentration, and the results showed that some varieties could be grown at 160 mM NaCl concentration. Taneenah et al. studied the germination of Corchorus olitorius L underlying the Dead Sea water, sea water and water with differential NaCl concentration, and the results indicated that Corchorus olitorius L. showed high tolerance to Dead Sea water, sea water and NaCl up to 6‰[15]. So far, which one is comparatively more tolerant between Corchorus capsularis and Corchorus olitorius has not been determined, and it maybe depend on which accessions or varieties[16]. However, the underlying mechanism has not been reported at the molecular level. To this end, the present study investigated the gene expression profiles of C. capsularis and C. olitorius under salt stress by high-throughput transcriptome sequencing. Our findings provide some insight into the mechanisms of salt tolerance in plants.
Materials and methods
Plant materials and salt stress treatment
Yueyuan NO.5, an important cultivar in China of C. capsularis L. (named YY) and NY/253C, a accession of C. olitorius L. (named NY) originated from China were hydroponically cultivated at 25°C–28°C in Yoshida nutrient solution in a greenhouse. And the salt tolerance of YY was only slightly better than that of NY. At the nine-leaf stage, six uniform seedlings of each species were selected; three were treated with 250 mM NaCl and the other three served as controls. After 12 h, the roots and leaves were collected for RNA extraction.
RNA isolation and transcriptome sequencing
Total RNA was extracted from tissue samples using TRIzol reagent (Invitrogen, Carlsbad, CA, US) according to the manufacturer’s protocol. RNA degradation and contamination was monitored in 1% agarose gels. RNA purity, integrity and the concentration was measured with a NanoPhotometer spectrophotometer (Implen, Westlake Village, CA, USA), RNA Nano 6000 Assay kit with the Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA), and Qubit RNA Assay kit with the Qubit 2.0 flurometer (Life Technologies, Carlsbad, CA, USA). A total of 24 RNA sequencing libraries (NYCR, NY control root; NYCL, NY control leaf; NYSR, NY salt-stressed root; NYSL, NY salt-stressed leaf; YYCR, YY control root; YYCL, YY control leaf; YYSR, YY salt-stressed root; YYSL, YY salt-stressed leaf. three biological repetition for above each sample) were generated using the NEBNext Ultra RNA Library Prep kit for Illumina (New England Biolabs, Ipswich, MA, USA) according to the manufacturers’ instructions. The quality of the libraries was assessed with the Agilent Bioanalyzer 2100 system. Libraries were sequenced on an Illumina HiSeq 4000 platform (San Diego, CA, USA) and paired-end reads were generated for transcriptome sequencing.
Transcriptome data analysis and annotation
Quality control of raw data was carried out with Perl scripts developed in house by removing reads containing adapter sequences, poly-N, and low-quality reads. Clean reads were assembled into a transcriptome using Trinity[17] with default settings for all parameters and mapped to transcripts; those with less than 5× coverage were removed. The transcripts which would be used as reference transcripts for analysis of DEGs were further assembled into non-redundant unigenes. Gene function was annotated using the following databases: National Center for Biotechnology Information (NCBI) non-redundant protein sequences (Nr); NCBI non-redundant nucleotide sequences (Nt); Protein Family (Pfam); Clusters of Orthologous Groups of Proteins (KOG); Swiss-Prot, Kyoto Encyclopedia of Genes and Genomes (KEGG) Ortholog (KO) database; and Gene Ontology (GO) using the Basic Local Alignment Search Tool with an E-value threshold of 10−5[18]
Analysis of differentially expressed genes (DEGs)
Gene expression levels for each sample were estimated using RSEM[19]. Clean data from each sample were respectively mapped back onto the assembled transcripts, and the mapping results for each gene were used for differential expression analysis of unigenes. Differential expression analysis of the two conditions was carried out using the DESeq R package (1.10.1)[20]. Genes with Padjusted < 0.05 assigned by DESeq were considered as differentially expressed. GO enrichment and KEGG pathways analysis of DEGs were implemented with the GOseq R package[21] and KOBAS[22] software.
Quantitative real-time (qRT)-PCR analysis
Eight DEGs randomly selected from the RNA-seq results and eight DEGs selected from the list of some differentially expressed genes (DEGs) playing important roles in salinity stress were analyzed by qRT-PCR to validate RNA-seq results in the same samples used for RNA-seq with two independent biological and three technological replicates. The jute elongation factor-alpha (ELF) gene was used as the endogenous control[23]. Primer sequences for amplifying DEGs and ELF were listed in S1 File. For qRT-PCR, total RNA was reverse transcribed into first-strand cDNA using the M-MuLV Reverse Transcriptase kit (Fermentas, Burlington, ON, Canada) according to the manufacturer’s instructions. The reaction was performed using the FastStart Universal SYBR Green Master kit (Roche Diagnostics, Basel, Switzerland) according to the manufacturer’s instructions in an optical 384-well plate on a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, USA). The cycle threshold method was used to calculate relative expression levels[24].
Results
Transcriptome assembly and functional annotation
A total of 24 NY and YY samples under salt stress and control conditions were used for Illumina paired-end transcriptome sequencing and analysis. We obtained 1,234,376,376 raw reads (Table 1). After quality control, 1,200,159,212 clean reads (97.23% of raw reads) were obtained with a high base quality score (average Q20 = 97.51%), accounting for approximately 180.06 Gb of sequencing data. A total of 161,760 transcripts were assembled with lengths varying from 201 to 15,953 bp and an average length of 1578 bp (Table 2). Transcripts were assembled into 72,278 non-redundant unigenes with an average length of 1240 bp. All sequencing data were deposited into the NCBI SRA database under accession numbers SRP116874 (included sequence data of 12 NY samples) and SRP119015 (included sequence data of 12 YY samples and unigenes annotation files).
Table 1. Details of the raw and clean data of 24 NY and YY sample transcriptomes.
| Genotypes | Tissues | Raw reads | Clean reads | Clean bases(Gb)(GC%) | Raw reads avg Q20 | |||
|---|---|---|---|---|---|---|---|---|
| Control | Salinity | Control | Salinity | Control | Salinity | (%) | ||
| NY | Leaf | 154,357,990 | 157,905,474 | 149,840,624 | 151,807,922 | 22.48(45.27%) | 22.78(45.12%) | 97.51 |
| Root | 170,983,540 | 150,144,930 | 165,732,832 | 145,221,480 | 24.86(44.06%) | 21.78(44.05%) | ||
| YY | Leaf | 160,566,334 | 148,384,870 | 157,631,384 | 145,816,236 | 23.64(43.61%) | 21.88(43.54%) | |
| Root | 156,290,594 | 135,742,644 | 151,681,828 | 132,426,906 | 22.76(43.47%) | 19.88(43.34%) | ||
| Total | 1,234,376,376 | 1,200,159,212 (97.23%) | 180.06 | |||||
Table 2. Assembly output statistics of all clean data using the Trinity assembler.
| Parameters | Value |
|---|---|
| Number of unigenes | 72,278 |
| Number of Transcripts | 161,760 |
| Average unigenes length (bp) | 1,240 |
| Average Transcripts length (bp) | 1,578 |
| Min length unigenes length (bp) | 201 |
| Min length Transcripts length (bp) | 201 |
| Max length unigenes length (bp) | 15,953 |
| Max length Transcripts length (bp) | 15,953 |
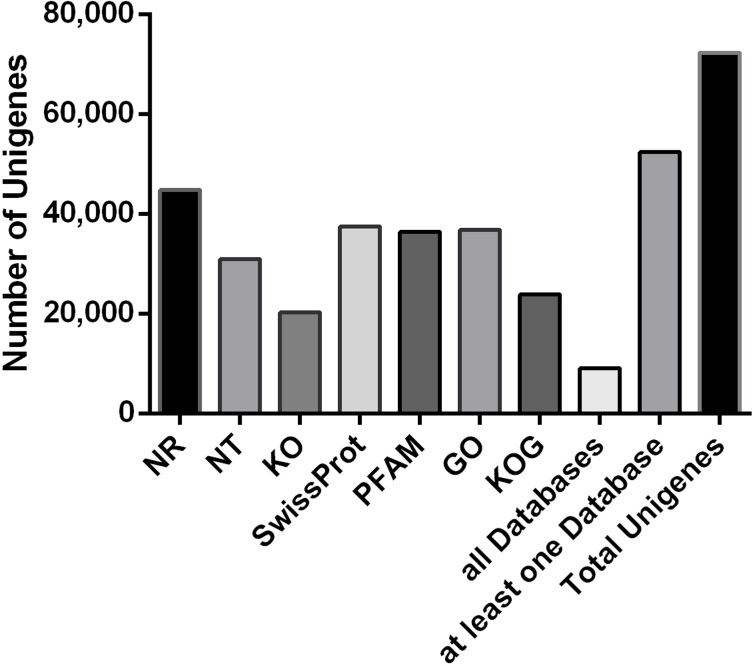
In total, 9108 (12.6%) unigenes were found to have homologs in the seven databases (Fig 1), with 52,417 (72.52%) annotated in at least one database. The largest number of unigenes (44,764, 61.93%) had hits to protein sequences in Nr, while only 20,283 (28.06%) were mapped in KO.
Fig 1. Unigenes annotated success across multiple databases.
Differential gene expression in response to salt treatment
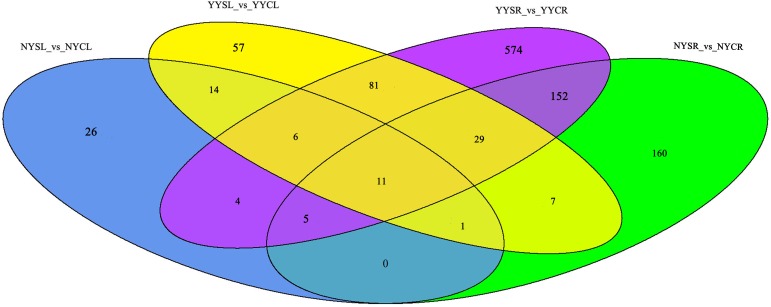
We analyzed genes that were differentially expressed in various Corchorus tissues in response to salt stress. In general, there were more DEGs in roots than in leaves in the two species. There were 1932 (792/1140) and 9608 (1904/7704) (S2 File, Table 3) DEGs (up-/downregulated) in NY and 3089 (1799/1290) (S2 File, Table 3) and 15,099 (6401/8698) (S2 File, Table 3) DEGs (up-/downregulated) in YY in the leaf and root tissues, respectively. A comparative analysis revealed that 4489 (948/3294) (S3 File, Table 3) and 492 (181/131) (S4 File, Table 3) root and leaf DEGs (upregulated/downregulated), respectively, were to both species, while 127 (107/12) (S5 File, Table 3) DEGs (upregulated/downregulated) were present in both tissues of both species.
Table 3. Statistics of differentially expressed genes (DEGs) in multiple NY and YY comparison groups under salt stress and control conditions.
| Comparison groups | No. DEG | Upregulated | Downregulated |
|---|---|---|---|
| NYSL_vs_NYCL | 1932 | 792 | 1140 |
| NYSR_vs_NYCR | 9608 | 1904 | 7704 |
| YYSL_vs_YYCL | 3089 | 1799 | 1290 |
| YYSR_vs_YYCR | 15099 | 6401 | 8698 |
| NYSR_vs_NYCR & YYSR_vs_YYCR | 4489 | 948 | 3294 |
| NYSL_vs_NYCL & YYSL_vs_YYCL | 492 | 181 | 131 |
| NYSL_vs_NYCL&YYSL_vs_YYCL&NYSR_vs_NYCR&YYSR_vs_YYCR | 127 | 107 | 12 |
NYSR (NY salt-stressed root); NYCR (NY control root); NYSL (NY salt-stressed leaf); NYCL (NY control leaf); YYSR (YY salt-stressed root); YYCR (YY control root); YYSL (YY salt-stressed leaf); TCCL (TC control leaf)
Differentially expressed (D)TFs
In the study, a total of 2303 TFs were discovered (S6 File). There were more DTFs in YY than in NY (206/862 and 67/365 DTFs in leaves/roots of YY and NY, respectively) (Fig 2). In the leaves, there were 32 DTFs common to the two species (S7 File and Fig 2). These DTFs belonged to 17 TF families, with homeobox (HB) and heat shock factor (HSF) being the most highly represented. In the roots, there were 196 DTFs (S8 File and Fig 2) from 43 TF families, of which six had more than 10 unigenes including myeloblastosis (MYB) (25), WRKY (12), CCAAT (12), Apetala (AP)2/ethylene response element binding protein (EREBP) (11), Orphans (10), and forkhead-associated domain (FHA) (10). There were 11 DTFs (S9 File and Fig 2) common to both tissues and both species, including four HSF, three HB, and one each of NAM/ATAF/CUC (NAC), basic leucine zipper (bZIP), CCAAT, and tafazin.
Fig 2. DTFs in the roots and leaves of C. capsularis and C. olitorius cultivated under high salt conditions.
DTFs, Differentially expressed (D)TFs; NYCL, NY control leaf; NYCR, NY control root; NYSL, NY salt-stressed leaf; NYSR, NY salt-stressed root; YYCL, YY control leaf; YYCR, YY control root; YYSL, YY salt-stressed leaf; YYSR, YY salt-stressed root.
GO and KEGG analyses of DEG function
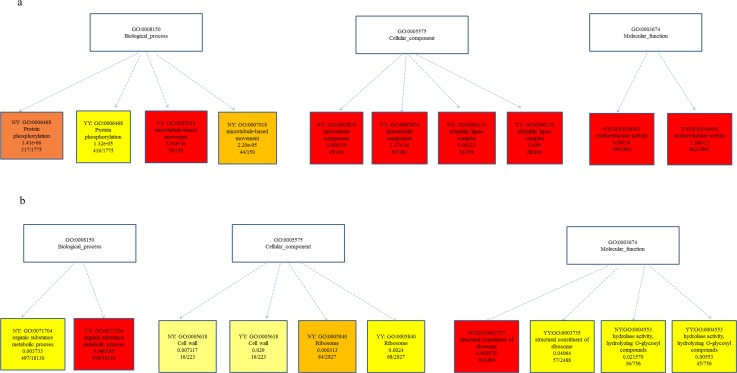
To gain insight into the functions of DEGs induced by salt stress in root and leaf tissues of Corchorus, we analyzed GO term annotations. In the roots, massive downregulated unigenes were enriched in the GO terms ‘protein phosphorylation’ (GO:0006468), ‘microtubule-based movement’ (GO:0007018), and ‘microtubule component’ (GO:0005874), whereas numerous upregulated unigenes were enriched in ‘ubiquitin ligase complex’ (GO:0000151) and ‘oxidoreductase activity’ (GO:0016491) (Fig 3A). In leaves, the GO terms ‘organic substance metabolic process’ (GO:0071704), ‘cell wall’ (GO:0005618), ‘ribosome’ (GO:0005840), ‘structural constituent of ribosome’ (GO:0003735), and ‘hydrolase activity/hydrolyzing O-glycosyl compounds’ (GO:0004553) were significantly enriched in both NY and YY (Fig 3B).
Fig 3.
Differentially expressed genes (DEGs) induced by salt stress in both NY and YY root (a) and leaf (b) tissues of Corchorus according to GO term annotation analysis.
In order to identify pathways involved in the salt stress response, we carried out a KEGG pathways analysis of DEGs and found that pathways associated with metabolism and signaling were highly represented. There were 13 pathways enriched in both tissues in both species (Table 4) such as those related to plant hormone signal transduction and the peroxisome; and six pathways (Table 4) were enriched in roots in both NY and YY, including MAPK and Ca2+ signaling. On the other hand, methane metabolism and cutin, suberin, and wax biosynthesis were only enriched in leaves of both species, although methane metabolism accounted for 10 and nine DEGs in NYL and YYL, respectively, and cutin, suberin, and wax biosynthesis accounted for four DEGs in the leaves of NY and YY (here, we only list the number of DEGs within enriched pathways in root and leaf of NY or YY in Table 4, while a few of DEGs (or no DEGs) without obviously enrichment in root and leaf of NY or YY were not showed (such as TCA cycle, only in root enrichment). Of the 127 DEGs common to both tissues and species, 13 were enriched in plant hormone signal transduction.
Table 4. The differentially expressed genes (DEGs) number and pathways associated with metabolism and signaling highly represented under salt stress.
| Pathway ID | The number of differentially expressed genes | The number of backgroud genes |
Pathway terms | |||
|---|---|---|---|---|---|---|
| NYSL vs. NYCL | YYSL vs. YYCL | NYSR vs. NYCR | YYSR vs. YYCR | |||
| ko01210 | 9 | 11 | 23 | 42 | 164 | 2-Oxocarboxylic acid metabolism |
| ko00520 | 19 | 21 | 29 | 60 | 213 | Amino sugar and nucleotide sugar metabolism |
| ko00330 | 11 | 14 | 34 | 51 | 179 | Arginine and proline metabolism |
| ko01230 | 50 | 39 | 67 | 130 | 570 | Biosynthesis of amino acids |
| ko00710 | 18 | 19 | 31 | 42 | 236 | Carbon fixation in photosynthetic organisms |
| ko01200 | 45 | 39 | 83 | 136 | 676 | Carbon metabolism |
| ko00260 | 16 | 15 | 31 | 44 | 168 | Glycine, serine and threonine metabolism |
| ko00040 | 10 | 13 | 24 | 45 | 156 | Pentose and glucuronate interconversions |
| ko00030 | 8 | 9 | 17 | 37 | 135 | Pentose phosphate pathway |
| ko00360 | 15 | 22 | 33 | 92 | 175 | Phenylalanine metabolism |
| ko00940 | 24 | 33 | 40 | 125 | 240 | Phenylpropanoid biosynthesis |
| ko04075 | 28 | 57 | 49 | 125 | 288 | Plant hormone signal transduction |
| ko04146 | 14 | 15 | 63 | 68 | 262 | Peroxisome |
| ko00020 | - | - | 44 | 62 | 267 | Citrate cycle (TCA cycle) |
| ko00190 | - | - | 59 | 57 | 738 | Oxidative phosphorylation |
| ko04010 | - | - | 47 | 49 | 292 | MAPK signaling pathway |
| ko04020 | - | - | 45 | 38 | 213 | Calcium signaling pathway |
| ko04120 | - | - | 43 | 50 | 322 | Ubiquitin mediated proteolysis |
| ko00250 | - | - | 14 | 25 | 98 | Alanine, aspartate and glutamate metabolism |
| ko00680 | 10 | 9 | - | - | 115 | Methane metabolism |
| ko00073 | 4 | 4 | - | - | 26 | Cutin, suberine and wax biosynthesis |
Some DEGs playing important roles in salinity stress
Here, numerous DEGs that played important roles in salinity stress were discovered. In Table 5, we list DEGs encoding HKT1, CBL-interacting protein kinases (CIPK), late embryogenesis abundant protein gene (LAE) etc. In the DEGs, the most of DEGs were the unigenes encoding CIPK (27), and 17 were up-regulated, following by the DEGs encoding MKK (9) with 6 down-regulated, 2 up-regulated and one up and down-regulated in NY or YY. The two DEGs (c122990_g1 and c122990_g2) were both up-regulated in the two tissues of both species. The two LAE DEGs and two SOS1 DEGs were all up-regulated in roots of YY. And one HKT1 was upregulated only in NY root.
Table 5. the list of some differentially expressed genes (DEGs) playing important roles in salinity stress.
| Gene ID | treatment vs. contral |
Log2(FC) | q_value | code protein |
gene_ length |
function |
|---|---|---|---|---|---|---|
| c127423_g2 | NYSR vs. NYCR | 4.879 | 2.20E-02 | HKT1 | 1983 | transmembrane transport//cation transport |
| c108681_g1 | YYSL vs. YYCL | 2.927 | 1.70E-02 | CIPK6 | 771 | signal transduction//protein phosphorylation |
| c119693_g1 | NYSR vs. NYCR | -4.625 | 8.15E-03 | CIPK23 | 1762 | protein phosphorylation |
| c120042_g2 | YYSL vs. YYCL | 4.023 | 1.87E-03 | CIPK6 | 1880 | protein phosphorylation//signal transduction |
| c122990_g1 | NYSL vs. NYCL | 1.752 | 2.91E-03 | CIPK25 | 812 | protein phosphorylation |
| c122990_g1 | NYSR vs. NYCR | 2.197 | 1.19E-08 | CIPK25 | 812 | protein phosphorylation |
| c122990_g1 | YYSL vs. YYCL | 1.874 | 1.00E-10 | CIPK25 | 812 | protein phosphorylation |
| c122990_g1 | YYSR vs. YYCR | 3.238 | 1.38E-84 | CIPK25 | 812 | protein phosphorylation |
| c122990_g2 | NYSL vs. NYCL | 1.802 | 5.54E-04 | CIPK25 | 1877 | signal transduction//protein phosphorylation |
| c122990_g2 | NYSR vs. NYCR | 2.079 | 1.39E-08 | CIPK25 | 1877 | signal transduction//protein phosphorylation |
| c122990_g2 | YYSL vs. YYCL | 1.850 | 1.49E-11 | CIPK25 | 1877 | signal transduction//protein phosphorylation |
| c122990_g2 | YYSR vs. YYCR | 3.182 | 4.45E-51 | CIPK25 | 1877 | signal transduction//protein phosphorylation |
| c125842_g1 | NYSR vs. NYCR | 1.844 | 4.46E-04 | CIPK10 | 2412 | protein phosphorylation//signal transduction |
| c125842_g1 | YYSL vs. YYCL | 0.777 | 3.18E-02 | CIPK10 | 2412 | protein phosphorylation//signal transduction |
| c125842_g1 | YYSR vs. YYCR | 1.939 | 2.10E-33 | CIPK10 | 2412 | protein phosphorylation//signal transduction |
| c126096_g1 | YYSR vs. YYCR | -1.437 | 6.13E-18 | CIPK7 | 1840 | protein phosphorylation//signal transduction |
| c127863_g1 | NYSR vs. NYCR | 2.126 | 1.56E-06 | CIPK2 | 3180 | signal transduction//protein phosphorylation |
| c127863_g1 | YYSL vs. YYCL | 0.720 | 2.63E-02 | CIPK2 | 3180 | signal transduction//protein phosphorylation |
| c127863_g1 | YYSR vs. YYCR | 2.506 | 2.17E-35 | CIPK2 | 3180 | signal transduction//protein phosphorylation |
| c128981_g1 | YYSL vs. YYCL | 2.357 | 7.29E-08 | CIPK25 | 2389 | signal transduction//protein phosphorylation |
| c128981_g1 | YYSR vs. YYCR | 0.514 | 3.68E-03 | CIPK25 | 2389 | signal transduction//protein phosphorylation |
| c129602_g4 | YYSR vs. YYCR | 1.752 | 1.16E-31 | CIPK6 | 1063 | protein phosphorylation |
| c129602_g5 | YYSR vs. YYCR | 1.870 | 2.16E-36 | CIPK6 | 922 | protein phosphorylation//signal transduction |
| c130525_g2 | NYSR vs. NYCR | 1.148 | 2.41E-03 | CIPK10 | 4968 | protein phosphorylation//signal transduction |
| c130525_g2 | YYSL vs. YYCL | 0.945 | 1.72E-03 | CIPK10 | 4968 | protein phosphorylation//signal transduction |
| c130525_g2 | YYSR vs. YYCR | 0.782 | 1.46E-06 | CIPK10 | 4968 | protein phosphorylation//signal transduction |
| c131066_g1 | NYSR vs. NYCR | 2.018 | 7.03E-06 | CIPK8 | 2721 | signal transduction//protein phosphorylation |
| c131066_g1 | YYSR vs. YYCR | 1.402 | 2.44E-17 | CIPK8 | 2721 | signal transduction//protein phosphorylation |
| c132458_g2 | YYSL vs. YYCL | 1.034 | 1.31E-02 | CIPK9 | 4385 | signal transduction//protein phosphorylation |
| c132458_g2 | YYSR vs. YYCR | 0.889 | 1.69E-03 | CIPK9 | 4385 | signal transduction//protein phosphorylation |
| c133247_g1 | NYSR vs. NYCR | 1.526 | 6.50E-04 | CIPK9 | 2301 | protein phosphorylation |
| c133247_g1 | YYSR vs. YYCR | 2.933 | 9.96E-05 | CIPK9 | 2301 | protein phosphorylation |
| c133300_g1 | NYSR vs. NYCR | 3.610 | 7.49E-13 | CIPK21 | 2694 | protein phosphorylation//signal transduction |
| c133300_g1 | YYSR vs. YYCR | 1.167 | 3.19E-06 | CIPK21 | 2694 | protein phosphorylation//signal transduction |
| c133961_g3 | NYSR vs. NYCR | 1.918 | 4.84E-03 | CIPK12 | 1865 | protein phosphorylation//signal transduction |
| c133961_g3 | YYSR vs. YYCR | 1.073 | 1.15E-09 | CIPK12 | 1865 | protein phosphorylation//signal transduction |
| c134486_g1 | YYSR vs. YYCR | 0.946 | 2.35E-05 | CIPK8 | 3079 | protein phosphorylation//signal transduction |
| c135536_g3 | YYSR vs. YYCR | -0.956 | 2.59E-10 | CIPK32 | 1922 | signal transduction//protein phosphorylation |
| c135536_g4 | YYSR vs. YYCR | -1.240 | 3.51E-03 | CIPK32 | 1084 | signal transduction |
| c1961_g1 | NYSR vs. NYCR | -7.549 | 3.43E-09 | CIPK23 | 1477 | protein phosphorylation//defense response to bacterium |
| c1961_g1 | YYSR vs. YYCR | -5.827 | 4.89E-05 | CIPK23 | 1477 | protein phosphorylation//defense response to bacterium |
| c246105_g1 | NYSR vs. NYCR | -4.464 | 6.58E-05 | CIPK10 | 1435 | protein phosphorylation |
| c248820_g1 | NYSR vs. NYCR | -7.544 | 7.33E-09 | CIPK30 | 1480 | protein phosphorylation |
| c248820_g1 | YYSR vs. YYCR | -5.389 | 2.20E-04 | CIPK30 | 1480 | protein phosphorylation |
| c253056_g1 | NYSR vs. NYCR | -3.329 | 3.06E-02 | CIPK11 | 1378 | — |
| c68433_g1 | NYSR vs. NYCR | -6.042 | 6.32E-09 | CIPK28 | 1701 | protein phosphorylation |
| c68433_g1 | YYSR vs. YYCR | -4.270 | 2.94E-04 | CIPK28 | 1701 | protein phosphorylation |
| c72352_g1 | NYSR vs. NYCR | -3.222 | 4.25E-02 | CIPK30 | 891 | protein phosphorylation |
| c90168_g1 | NYSL vs. NYCL | 5.569 | 9.11E-05 | CIPK13 | 1424 | protein phosphorylation |
| c129602_g4 | YYSR vs. YYCR | 1.752 | 1.16E-31 | CIPK6 | 1063 | protein phosphorylation |
| c111482_g1 | YYSR vs. YYCR | 1.401 | 1.70E-03 | LEA | 832 | response to stress//response to water stimulus |
| c135934_g2 | YYSL vs. YYCL | 3.109 | 4.97E-07 | LEA | 1218 | response to stress |
| c105006_g2 | NYSR vs. NYCR | -3.547 | 3.62E-02 | MKK5 | 1081 | protein phosphorylation |
| c122526_g1 | YYSR vs. YYCR | -1.317 | 8.13E-06 | MKK6 | 1589 | protein phosphorylation |
| c124453_g1 | NYSR vs. NYCR | 2.407 | 5.36E-06 | MKK1 | 1461 | protein phosphorylation |
| c124453_g1 | YYSL vs. YYCL | -0.738 | 2.39E-02 | MKK1 | 1461 | protein phosphorylation |
| c124453_g1 | YYSR vs. YYCR | 3.907 | 5.36E-06 | MKK1 | 1461 | protein phosphorylation |
| c126229_g1 | NYSR vs. NYCR | 2.013 | 1.44E-07 | MKK13 | 1486 | protein phosphorylation//photosynthesis |
| c126229_g1 | YYSR vs. YYCR | 0.602 | 2.14E-02 | MKK13 | 1486 | protein phosphorylation//photosynthesis |
| c132388_g1 | YYSR vs. YYCR | -0.419 | 2.31E-02 | MKK5 | 3164 | protein phosphorylation |
| c132606_g1 | YYSR vs. YYCR | 3.298 | 4.40E-51 | MKK18 | 1537 | thiamine biosynthetic process//protein phosphorylation |
| c207571_g1 | NYSR vs. NYCR | -3.932 | 2.15E-03 | MKK5 | 2599 | protein phosphorylation//oxidation-reduction process |
| c207571_g1 | YYSR vs. YYCR | -2.974 | 4.99E-02 | MKK5 | 2599 | protein phosphorylation//oxidation-reduction process |
| c226247_g1 | NYSR vs. NYCR | -3.518 | 1.49E-02 | MKK1 | 1057 | protein phosphorylation |
| c81745_g2 | NYSR vs. NYCR | -4.174 | 6.90E-03 | MKK9 | 1669 | protein phosphorylation |
| c121728_g1 | YYSR vs. YYCR | -6.603 | 7.38E-22 | NHX3 | 1516 | cation transport//regulation of pH |
| c121728_g2 | YYSL vs. YYCL | 1.810 | 2.12E-02 | NHX3 | 980 | cation transport//regulation of pH |
| c181442_g1 | NYSR vs. NYCR | -3.497 | 3.07E-02 | NHX1 | 1290 | cation transport//regulation of pH |
| c135331_g1 | YYSR vs. YYCR | 0.548 | 1.30E-03 | SOS1 | 4448 | transmembrane transport//regulation of pH |
| c135331_g1 | YYSR vs. YYCR | 0.548 | 1.30E-03 | SOS1 | 4448 | transmembrane transport//regulation of pH |
Differential expression of plant hormone signal transduction genes
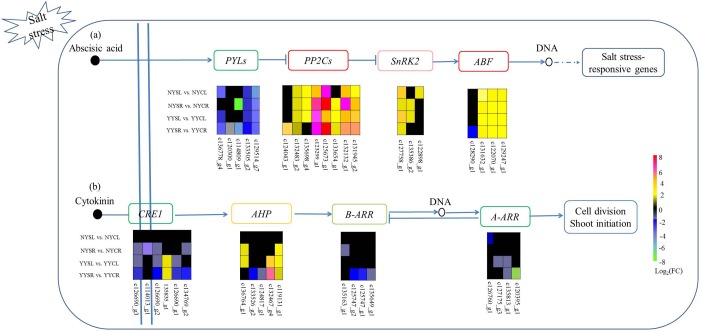
Hormones play an important role in the response to different environmental stressors (e.g., drought and salt stress) in plants. In this study, we identified numerous DEGs associated with plant hormone (ABA, auxin, cytokinin, gibberellin, ethylene, jasmonic acid, and salicylic acid) signaling in leaves and roots of both NY and YY, with ABA and cytokinin signaling being the most highly represented. In the ABA pathway, two genes encoding PYL were downregulated in all tissues and species whereas three other genes were downregulated in a subset of tissues and in one of the species. Five, one, and three genes encoding PP2C, SnRK2, and ABF were upregulated in all tissues and both species, respectively; and five genes encoding the three proteins were upregulated while one was downregulated in subset of tissues and in one of the species (Fig 4A). In the cytokinin signaling pathway, 13 genes encoding cytokinin response (CRE)1 (5), B-APR (4), and A-type Arabidopsis response regulator (A-ARR) (4) were downregulated in at least one of four groups under salt treatment as compared to the control condition. Most genes encoding Arabidopsis histidine-containing phosphotransmitter (AHP) were upregulated in NYR, YYL, and YYR (Fig 4B).
Fig 4. Differentially expressed genes (DEGs) belonging to the plant hormone pathway in root and leaf tissues of C. capsularis and C. olitorius cultivated under high salt conditions.
(a) ABA and (b) cytokinin signaling pathways are shown. NYCR, NY control root; NYCL, NY control leaf; NYSR, NY salt-stressed root; NYSL, NY salt-stressed leaf; YYCR, YY control root; YYCL, YY control leaf; YYSR, YY salt-stressed root; YYSL, YY salt-stressed leaf.
Pathways represented only in the roots of Corchorus
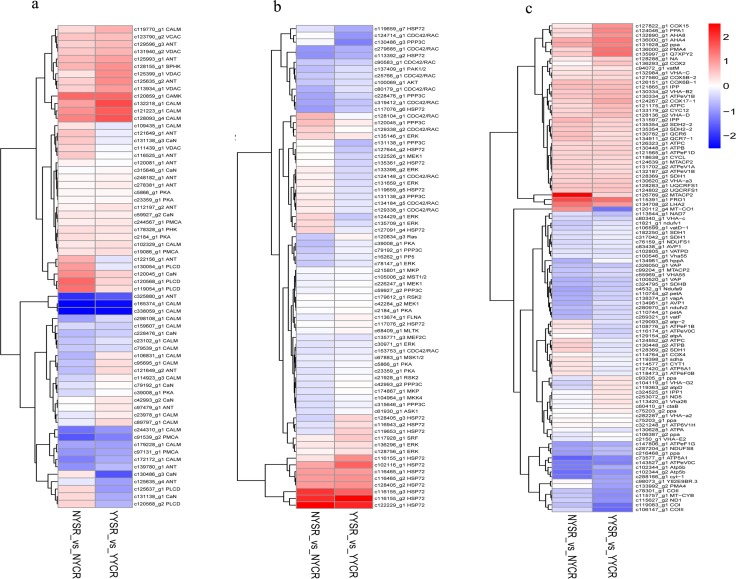
The Ca2+, MAPK signaling and oxidative phosphorylation pathways play major roles in the plant response to salt stress. Numerous DEGs identified in our study were associated with these pathways. Components of the Ca2+ signaling pathway included calmodulin (CaM)/CaM-like (CML) proteins and Ca2+-dependent protein kinase (CAMK) (Fig 5A). A total of 20 genes encoding CML (4/16 up-/downregulated) were found in YY or NY roots, with most exhibiting similar expression in the two species. One upregulated gene encoding CAMK was common to both YY and NY roots. In addition, nine DEGs encoding serine/threonine-protein phosphatase 2B catalytic subunit, four encoding Ca2+-transporting ATPase, and four encoding protein kinase (PK)A were detected in YY or NY roots. In the MAPK pathway, DEGs encoding HSP72 (n = 19), Ras-related C3 botulinum toxin substrate 1 (n = 12), MAPK1/3 (n = 9), and PP3C (n = 8) were detected in YY and NY roots (Fig 5B). In the oxidative phosphorylation pathway, 19 DEGs encoding H+-transporting ATPase subunits (V and F types) and six encoding cytochrome c oxidase subunit were identified (Fig 5C). The expression profiles of DEGs in these three pathways were similar in the roots of the two species.
Fig 5. Differentially expressed genes (DEGs) belonging to the following signaling pathways in root tissue of C. capsularis and C. olitorius cultivated under high salt conditions.
(a) Ca2+ signaling pathway; (b) MAPK signaling; and (c) oxidative phosphorylation pathways are shown. NYCR, NY control root; NYSR, NY salt-stressed root; YYCR, YY control root; YYSR, YY salt-stressed root.
Validation of DEGs
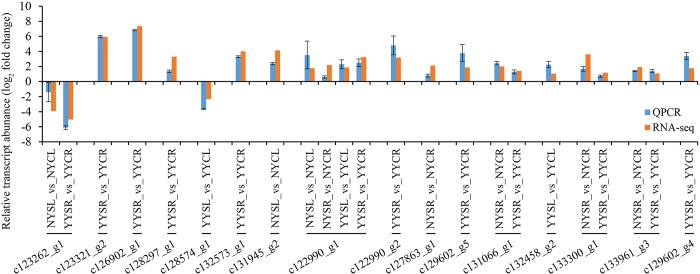
In order to validate the RNA-seq results, we carried out qRT-PCR analysis for eight randomly selected DEGs and eight DEGs selected from Table 5 in YY and NY under salt stress. All of the DEGs showed differential expression under salt stress as compared to normal conditions, and the expression profiles were in accordance with the results obtained by RNA-seq (correlation coefficient = 0.85). Log2 (fold change) values for each DEG are shown in Fig 6.
Fig 6. qRT-PCR validation of sixteen DEGs detected by RNA-seq.
Data represent log2 (fold changes) of the relative transcript abundance of each DEG in NYSR (NY salt-stressed root) vs. NYCR (NY control root); NYSL (NY salt-stressed leaf) vs. NYCL (NY control leaf); YYSR (YY salt-stressed root) vs. YYCR (YY control root); and YYSL (YY salt-stressed leaf) vs. YYCL (YY control leaf). Red and blue represent expression levels determined by RNA-seq and qRT-PCR, respectively.
Discussion
In this study, we carried out de novo transcriptome sequencing to investigate salt tolerance mechanisms in two jute species under salt stress. In previous studies, some reports showed that the number of DEGs was greater in roots than in leaves underlying salt stress. For example, under salt treatment, 4,884/3,692 up-/downregulated unigenes were discovered in cotton leaf samples; and much more up-/downregulated unigenes (6,303/11,068) were discovered in cotton roots DEGs[25]. while Yong et al. reported that[26] the number of DEGs was slightly greater in the roots (8,665 DEGs) than those in the leaves (7,795 DEGs) in Brassica napus. The number of DEGs in different tissues (e.g. roots and leaves) under salt stress might be related to crops. In the current study, much more DEGs common to both species in the roots than in leaves were discovered in jute, more similarly with in cotton; probably because that they were both classified as the Malvaceae[27]. Results of these studies might imply that roots were the main tissue responding to salt stress, especially in Malvaceae crops. In total, 127 DEGs were common to both tissues and both species, and the positive regulation DEGs (107) indwelled in all tissues and species were more than passive regulation DEGs (12) suggesting that the DEGs might play important roles in response to environmental stress in all of tissues and species.
Most of the identified DEGs common to both tissues and both species were directly related to the response to environmental stress. For example, 13 DEGs were enriched in plant hormone signal transduction, and most of these were upregulated; DEGs involved in oxidation-reduction and protein phosphorylation were also highly represented. Some DEGs, played important roles in salinity stress, were discovered. For example, there was evidence supports that HKT participated in the pathways of Na+ entry into the roots subjected to high salinity; and then Na+ was perceived and changed concentration of cytosolic Ca2+ decoded by Ca2+-sensing proteins, such as CIPK. Oscillations of Ca2+ induced by salt stress were sensed by SOS3, a myristoylated Ca2+-binding protein; finally, the SOS1 was phosphorylated and activated through SOS pathway which extruded sodium in cells[28]. In addition, NHXs could compartmentalize Na+ in vacuoles to prevent toxic effects in the cytosol. In the study, numerous DEGs encoding HKT (1, up-regulated), CIPK (27, 17 up-regulated,), SOS1 (2, up-regulated) and NHX (3, up and down-regulated) were found[29]. The DEGs might be play crucial role in Na+ homeostasis. previous studies have shown that TF families such as AP2/EREBP, HB, MYB, NAC, bZIP, HSF, Orphans, FHA, and WRKY are linked to salt stress[30, 31]. In the present study, DTFs expressed in leaves and roots differed; 17 DTF families including HB and HSF were detected in the former, while in the latter, 43 families including MYB, WRKY, and CCAAT were represented. Interestingly, 11 DTFs were common to both tissues in both species that are likely critical for the response to salt stress.
Pathways obviously involved in the salt stress response were identified in the study. The pathways should be important for plants in response to abiotic stress. However, some Pathways were only enriched in roots or leaves in NY and YY, such as TCA cycle and Cutin, suberine and wax biosynthesis. For example, TCA cycle was obviously enriched in root of both species in this study, consistent with previous some studies. Kreuzwieser et al. proposed that hypoxia stress led to the inhibition of the TCA cycle and differential expression of some genes involved in TCA cycle only in roots of poplar trees[32]. And cutin, suberine and wax biosynthesis pathway was only enriched in leaves in NY and YY, in previous studies, the pathway was enriched in leaves[33] or in roots[34], but Lauter et al. reported cutin, suberine and wax biosynthesis term was enriched only in leaves of soybean under iron deficiency stress. suggesting in which tissues cutin, suberine and wax biosynthesis pathway was enriched might be influenced by numerous factors, such as species, stress conditions etc. Cutin, suberine and wax played important roles at plant environment interfaces by serving as a barrier controlling the movements of water and solutes. Which was thus important for the abilities of plants to withstand various abiotic stresses, such as drought and salinity [35]. So, which cutin, suberine and wax biosynthesis pathway was not enriched in root in NY and YY might result in uptaking more ion inside the root tissues from the rhizosphere and finally causes jute susceptibility under salinity stress.
The ABA signaling pathway is central to the salt stress responses in plants[1]. In this study, 20 DEGs were associated with this pathway, including genes encoding PP2C, SnRK2, and ABF (upregulated) and PYL (downregulated). Although PYL and PP2C expression is expected to be positively correlated based on what is known of ABA signaling, our observations are supported by recent studies[36, 37]. The cytokinin signaling pathway includes cytokinin receptors; membrane-bound histidine kinases (e.g., CRE1) with a cytokinin-binding domain; AHP (which transmits signals to the nucleus); and B- and A-type ARR, which are phosphorylated by AHP in the nucleus[38, 39]. Plants lacking or harboring mutant CRE1 show increased tolerance to high salt and drought conditions[40]; furthermore, B- and A-ARR mutations also increase salt tolerance[41], implying that cytokinin signaling negatively regulates the response to salt stress. In the present study, CRE1 and B- and A-ARR were downregulated in roots and leaves of both Corchorus species in response to high salinity. However, most AHP genes were upregulated, suggesting that they negatively regulate cytokinin signaling[42] to increase salt tolerance in plants.
CaM and CML are Ca2+ sensors that transmit Ca2+ signals to downstream effectors under conditions of environmental stress. Ca2+-loaded CaM/CML interacts with and regulates a broad spectrum of proteins including TFs, protein kinases and phosphatases, and metabolic enzymes[43]. Overexpression of CaM in Arabidopsis as a result of high salinity conferred salt stress tolerance via upregulation of the DNA-binding activity of MYB[44]. Interestingly, 20 DEGs corresponding to CML proteins were detected while MYB was the most highly represented TF family in YY and NY roots. CAMK upregulated in both YY and NY roots may enhance tolerance to salt stress[45] and interact with serine/threonine kinases that activate a Na+/H+ antiporter, resulting in the extrusion of Na+ into soil or xylem for transporting to leaves[46]. CAMK and the serine/threonine protein kinase PP3C may activate Ca2+ and MAPK signaling pathways in plants in response to high salt conditions or drought[10]. H+-ATPases were highly expressed in roots under salt stress, likely providing the proton-motive force required for maintaining membrane potential[47]. In the roots of both Corchorus species, there were 19 DEGs encoding H+-ATPase subunits under salt stress. This is consistent with previous studies[47] and demonstrates the importance of H+-ATPases for salt tolerance in plants.
In conclusion, our results provide a comprehensive resource for investigations into the mechanisms of salt response in plants, and can serve as a basis for breeding salt-tolerant cultivars of Corchorus species in the future.
Supporting information
(XLSX)
(XLSX)
(XLS)
(XLS)
(XLS)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
The authors like to express their gratitude to Dr. Dongwei Xie; Hong Gao; Haibing Wen; Dongbing Fang of Institute of Bast Fiber Crops, Chinese Academy of Agricultural Sciences. The expert technical assistance of Jian Zhao is gratefully acknowledged. The study was supported by grants of the National Natural Science Foundation of China (grant number 31601351) to Zemao Yang, the Science and Technology Innovation Project of Chinese Academy of Agricultural Sciences (grant number ASTIP-IBFC01) to Jianguang Su, the National Key Technology Support Program (grant number 2013BAD01B03-13) to Jianguang Su, and the Germplasm Resources Protection Project (grant number 2016NWB044) to Jianguang Su.
Data Availability
All sequencing data were deposited into the NCBI SRA database under accession numbers SRP116874 (including sequencing data for12 NY samples) and SRP119015 (including sequencing data for 12 YY samples and unigenes annotation files).
Funding Statement
This study was financially supported by the National Natural Science Foundation of China (grant number 31601351) to Zemao Yang, the Science and Technology Innovation Project of Chinese Academy of Agricultural Sciences (grant number ASTIP-IBFC01) to Jianguang Su, the National Key Technology Support Program (grant number 2013BAD01B03-13) to Jianguang Su, and the Germplasm Resources Protection Project (grant number 2016NWB044) to Jianguang Su.
References
- 1.Zhu JK. Abiotic stress signaling and responses in plants. Cell. 2016;167(2):313–24. doi: 10.1016/j.cell.2016.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu JK. Salt and drought stress signal transduction in plants. Annual review of plant biology. 2002;53(1):247–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanin M, Ebel C, Ngom M, Laplaze L, Masmoudi K. New Insights on plant salt tolerance mechanisms and their potential use for breeding. Frontiers in Plant Science. 2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olías R, Eljakaoui Z, Li J, DE MORALES PA, MARÍN-MANZANO MC, Pardo JM, et al. The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant, Cell & Environment. 2009;32(7):904–16. [DOI] [PubMed] [Google Scholar]
- 5.Shi H, Quintero FJ, Pardo JM, Zhu J-K. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. The Plant Cell. 2002;14(2):465–77. doi: 10.1105/tpc.010371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–99. doi: 10.1146/annurev.arplant.55.031903.141701 [DOI] [PubMed] [Google Scholar]
- 7.Miller G, Suzuki N, CIFTCI-YILMAZ S, Mittler R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant, cell & environment. 2010;33(4):453–67. [DOI] [PubMed] [Google Scholar]
- 8.Xing Y, Jia W, Zhang J. AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. The Plant Journal. 2008;54(3):440–51. doi: 10.1111/j.1365-313X.2008.03433.x [DOI] [PubMed] [Google Scholar]
- 9.Jammes F, Song C, Shin D, Munemasa S, Takeda K, Gu D, et al. MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proceedings of the National Academy of sciences. 2009;106(48):20520–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Zelicourt A, Colcombet J, Hirt H. The role of MAPK modules and ABA during abiotic stress signaling. Trends in plant science. 2016;21(8):677–85. doi: 10.1016/j.tplants.2016.04.004 [DOI] [PubMed] [Google Scholar]
- 11.Kundu A, Topdar N, Sarkar D, Sinha MK, Ghosh A, Banerjee S, et al. Origins of white (Corchorus capsularis L.) and dark (C. olitorius L.) jute: a reevaluation based on nuclear and chloroplast microsatellites. Journal of plant biochemistry and biotechnology. 2013;22(4):372–81. [Google Scholar]
- 12.Xiong H. Breeding sciences of bast and leaf fiber crops Chinese Agricultural Science and Technology Press, Beijing: 2008:319–41. [Google Scholar]
- 13.Ma H, Yang R, Song L, Yang Y, Wang Q, Wang Z, et al. Differential proteomic analysis of salt stress response in jute (Corchorus capsularis & olitorius L.) seedling roots. Pak J Bot. 2015;47(2):385–96. [Google Scholar]
- 14.Naik MR, Kumar M, Barman D, Meena P, Kumar AA, Kundu D. In vitro screening of white Jute (Corchorus capsularis L) against salinity stress. Journal of Applied and Natural Science. 2015;7(1):344–7. [Google Scholar]
- 15.Taneenah A, Nulit R, Yusof UK, Janaydeh M. Tolerance of Molokhia (Corchorus olitorius L.) seed with dead sea water, sea water, and NaCl: germination and anatomical approach. Advances in Environmental Biology. 2015;9(27):106–16. [Google Scholar]
- 16.Ma H, Yang R, Wang Z, Yu T, Jia Y, Gu H, et al. Screening of salinity tolerant jute (Corchorus capsularis & C. olitorius) genotypes via phenotypic and phsiology-assisted procedures. Pakistan Journal of Botany. 2011;43(6):2655–60. [Google Scholar]
- 17.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature biotechnology. 2011;29(7):644–52. doi: 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic acids research. 1997;25(17):3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics. 2011;12(1):323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26(1):136–8. doi: 10.1093/bioinformatics/btp612 [DOI] [PubMed] [Google Scholar]
- 21.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome biology. 2010;11(2):R14 doi: 10.1186/gb-2010-11-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mao X, Cai T, Olyarchuk JG, Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21(19):3787–93. doi: 10.1093/bioinformatics/bti430 [DOI] [PubMed] [Google Scholar]
- 23.Ferdous AS, Islam MT, Alam SS, Khan H. Identification of stable reference genes for quantitative PCR in jute under different experimental conditions: An essential assessment for gene expression analysis. Australian Journal of Crop Science. 2015;9(7):646. [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, Yao D, Wang Q, Xu W, Wei Q, Wang C, et al. mRNA-seq analysis of the Gossypium arboreum transcriptome reveals tissue selective signaling in response to water stress during seedling stage. PloS one. 2013;8(1):e54762 doi: 10.1371/journal.pone.0054762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yong H-Y, Zou Z, Kok E-P, Kwan B-H, Chow K, Nasu S, et al. Comparative transcriptome analysis of leaves and roots in response to sudden increase in salinity in Brassica napus by RNA-seq. BioMed research international. 2014;2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bremer B, Bremer K, Chase M, Fay M, Reveal J, Soltis D, et al. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Botanical Journal of the Linnean Society. 2009. [Google Scholar]
- 28.Miranda RdS Mesquita RO, Costa JH, Alvarez-Pizarro JC, Prisco JT, Gomes-Filho E. Integrative Control Between Proton Pumps and SOS1 Antiporters in Roots is Crucial for Maintaining Low Na+ Accumulation and Salt Tolerance in Ammonium-Supplied Sorghum bicolor. Plant and Cell Physiology. 2017;58(3):522–36. doi: 10.1093/pcp/pcw231 [DOI] [PubMed] [Google Scholar]
- 29.Xia K, Wang B, Zhang J, Li Y, Yang H, Ren D. Arabidopsis phosphoinositide-specific phospholipase C4 negatively regulates seedling salt tolerance. Plant, cell & environment. 2017;40(8):1317–31. [DOI] [PubMed] [Google Scholar]
- 30.Du X, Wang G, Ji J, Shi L, Guan C, Jin C. Comparative transcriptome analysis of transcription factors in different maize varieties under salt stress conditions. Plant Growth Regulation. 2016;81(1):183–95. doi: doi: 10.1007/s10725-016-0192-9 [Google Scholar]
- 31.Shankar R, Bhattacharjee A, Jain M. Transcriptome analysis in different rice cultivars provides novel insights into desiccation and salinity stress responses. Scientific reports. 2016;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kreuzwieser J, Hauberg J, Howell KA, Carroll A, Rennenberg H, Millar AH, et al. Differential response of gray poplar leaves and roots underpins stress adaptation during hypoxia. Plant physiology. 2009;149(1):461–73. doi: 10.1104/pp.108.125989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li P, Cao W, Fang H, Xu S, Yin S, Zhang Y, et al. Transcriptomic Profiling of the Maize (Zea mays L.) Leaf Response to Abiotic Stresses at the Seedling Stage. Frontiers in Plant Science. 2017;8:290 doi: 10.3389/fpls.2017.00290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krishnamurthy P, Ranathunge K, Franke R, Prakash H, Schreiber L, Mathew M. The role of root apoplastic transport barriers in salt tolerance of rice (Oryza sativa L.). Planta. 2009;230(1):119–34.36. doi: 10.1007/s00425-009-0930-6 [DOI] [PubMed] [Google Scholar]
- 35.Vishwanath SJ, Delude C, Domergue F, Rowland O. Suberin: biosynthesis, regulation, and polymer assembly of a protective extracellular barrier. Plant Cell Reports. 2015;34(4):573–586. doi: 10.1007/s00299-014-1727-z [DOI] [PubMed] [Google Scholar]
- 36.Chan Z. Expression profiling of ABA pathway transcripts indicates crosstalk between abiotic and biotic stress responses in Arabidopsis. Genomics. 2012;100(2):110–5. doi: 10.1016/j.ygeno.2012.06.004 [DOI] [PubMed] [Google Scholar]
- 37.Zhang F, Zhu G, Du L, Shang X, Cheng C, Yang B, et al. Genetic regulation of salt stress tolerance revealed by RNA-Seq in cotton diploid wild species, Gossypium davidsonii. Scientific reports. 2016;6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Zwack PJ, Rashotte AM. Interactions between cytokinin signalling and abiotic stress responses. Journal of experimental botany. 2015;66(16):4863–71. doi: 10.1093/jxb/erv172 [DOI] [PubMed] [Google Scholar]
- 39.Arnaud D, Lee S, Takebayashi Y, Choi D, Choi J, Sakakibara H, et al. Cytokinin-Mediated Regulation of Reactive Oxygen Species Homeostasis Modulates Stomatal Immunity in Arabidopsis. The Plant Cell. 2017;29(3):543–59. doi: 10.1105/tpc.16.00583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tran L- SP, Urao T, Qin F, Maruyama K, Kakimoto T, Shinozaki K, et al. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proceedings of the National Academy of Sciences. 2007;104(51):20623–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mason MG, Jha D, Salt DE, Tester M, Hill K, Kieber JJ, et al. Type-B response regulators ARR1 and ARR12 regulate expression of AtHKT1; 1 and accumulation of sodium in Arabidopsis shoots. The Plant Journal. 2010;64(5):753–63. doi: 10.1111/j.1365-313X.2010.04366.x [DOI] [PubMed] [Google Scholar]
- 42.Kushwah S, Laxmi A. The interaction between glucose and cytokinin signal transduction pathway in Arabidopsis thaliana. Plant Cell Environ. 2014;37(1):235–53. doi: doi: 10.1111/pce.12149. . [DOI] [PubMed] [Google Scholar]
- 43.Zeng H, Xu L, Singh A, Wang H, Du L, Poovaiah BW. Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front Plant Sci. 2015;6:600. doi: doi: 10.3389/fpls.2015.00600. ; PubMed Central PMCID: PMC4532166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoo JH, Park CY, Kim JC, Do Heo W, Cheong MS, Park HC, et al. Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in Arabidopsis. Journal of Biological Chemistry. 2005;280(5):3697–706. doi: 10.1074/jbc.M408237200 [DOI] [PubMed] [Google Scholar]
- 45.Campo S, Baldrich P, Messeguer J, Lalanne E, Coca M, San Segundo B. Overexpression of a calcium-dependent protein kinase confers salt and drought tolerance in rice by preventing membrane lipid peroxidation. Plant physiology. 2014;165(2):688–704. doi: 10.1104/pp.113.230268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu M, Shabala L, Cuin TA, Huang X, Zhou M, Munns R, et al. Nax loci affect SOS1-like Na+/H+ exchanger expression and activity in wheat. Journal of experimental botany. 2016;67(3):835–44. doi: 10.1093/jxb/erv493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bose J, Rodrigo-Moreno A, Lai D, Xie Y, Shen W, Shabala S. Rapid regulation of the plasma membrane H+-ATPase activity is essential to salinity tolerance in two halophyte species, Atriplex lentiformis and Chenopodium quinoa. Annals of botany. 2015;115(3):481–94. doi: 10.1093/aob/mcu219 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLS)
(XLS)
(XLS)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All sequencing data were deposited into the NCBI SRA database under accession numbers SRP116874 (including sequencing data for12 NY samples) and SRP119015 (including sequencing data for 12 YY samples and unigenes annotation files).