Abstract
Urinary tract infections (UTIs) are caused by uropathogenic Escherichia coli (UPEC) strains. In contrast to many enteric E. coli pathogroups, no genetic signature has been identified for UPEC strains. We conducted a high-resolution comparative genomic study using E. coli isolates collected from the urine of women suffering from frequent recurrent UTIs. These isolates were genetically diverse and varied in urovirulence, or the ability to infect the bladder of a mouse model of cystitis. Importantly, we found no set of genes, including previously defined putative urovirulence factors (PUFs), that were predictive of urovirulence. In addition, in some patients, the E. coli strain causing a recurrent UTI had fewer PUFs than the supplanted strain. In competitive experimental infections in mice, the supplanting strain was more efficient at colonizing the mouse bladder than the supplanted strain. Despite the lack of a clear genomic signature for urovirulence, comparative transcriptomic and phenotypic analyses revealed that the expression of key conserved functions during culture, such as motility and sugar metabolism, could be used to predict subsequent mouse bladder colonization. Taken together, our findings suggest that UTI risk and outcome may be determined by complex interactions between host susceptibility and the urovirulence potential of diverse bacterial strains.
Introduction
Urinary tract infections (UTIs) are a health and financial burden, particularly affecting sexually active, premenopausal women (1). Approximately 50% of women will suffer a UTI in their lifetime with 10.5 million cases of UTIs occurring in 2007 in the United States alone (2). In placebo-controlled studies of UTI outcome, most women maintain bladder infections for weeks after an acute cystitis episode if not treated with antibiotics, whereas other women clear the infection without antibiotic treatment (3, 4). These two outcomes of infection, either self-limiting or chronic cystitis, have been modeled in mice (5–7). Effective treatment of UTIs is becoming more challenging as antibiotic resistance rates are increasing (8). In addition, even though antibiotic therapy effectively resolves the majority of UTIs, approximately 20–30% of women will experience a recurrent UTI (rUTI) within six months of initial infection (9, 10). This highlights the need to better understand the pathogenic mechanisms that facilitate acute bacterial colonization of the urinary tract and determine UTI outcome.
The most common cause of community-acquired UTIs is uropathogenic E. coli (UPEC) (2). E. coli strains are diverse and have been characterized into pathotypes based on their ability to cause particular diseases (11, 12). Pathotypes include intestinal pathogens such as Shiga toxin-producing E. coli as well as extra-intestinal pathogenic E. coli, strains such as UPEC. E. coli are also sub-divided into phylogenetic clades (e.g., A, B1, B2, D, E) based on their genetic similarity (13, 14). In the U.S. and Europe, the majority of UPEC are from the B2 clade though members of clades A, B1 and D cause 25–50% of cystitis (15–19). In East Asia, clade D strains predominate in community-acquired UTI followed by B2 strains (20, 21). All commonly studied UPEC isolates, including UTI89, CFT073, NU14, and 536 (22–25), are B2 members whereas non-B2 strains have not been extensively characterized. Thus, it is unknown whether non-B2 UPEC strains use the same virulence mechanisms as B2 UPEC strains.
Study of prototypical B2 UPEC isolates, such as the model UPEC strain UTI89, originally isolated from a woman with cystitis (27), has shown that the chaperone usher pathway (CUP) type 1 pilus is critical for bladder colonization in diverse mouse models (26). The fim operon, which encodes the proteins for assembly of type 1 pili, is part of the core E. coli genome, found in nearly every E. coli strain (28). Further, in research of the model strain UTI89, it has been shown that type 1 pili expression is tightly co-regulated with numerous factors including flagella, and S and P CUP pili (29). Type 1 pili are tipped by the adhesin FimH, which binds specifically to mannosylated uroplakins lining the superficial bladder epithelium (30) and mannosylated proteins expressed on underlying layers of epithelial cells (31), thus mediating critical steps in bladder colonization and UTI progression. Subsequent to FimH attachment, E. coli can invade the superficial umbrella bladder cells (32) and escape into the cytoplasm where they rapidly replicate to form clonal, biofilm-like intracellular bacterial communities (IBCs) of ~104 colony forming units (CFU) while protected from the host immune response (33, 34). Upon IBC maturation, UTI89 transform into long filaments and burst out of the superficial umbrella cells of the bladder into the bladder lumen where they invade other epithelial cells and form new IBCs (34). The majority of clinical UPEC isolates that have been tested are able to form IBCs in mice, and each of six different inbred mouse strains tested support IBC formation (35). Importantly, after the discovery of IBCs in mouse UTI models, IBCs and filamentous bacteria were documented in human urine collected from clinical UTI cases (36, 37).
Despite decades of research using model UPEC strains, universal bacterial features that enable E. coli uropathogenesis remain largely undefined. Carriage of putative urovirulence factors (PUFs) is thought to enhance E. coli uropathogenicity and is used to measure and categorize clinical UPEC strains isolated from different patient populations (16, 20, 38, 39). PUFs were identified through their enrichment in UTI isolates when compared to non-UTI associated E. coli (38–41). While numerous sets of PUFs have been used to assess urovirulence (16, 20, 38–41) and strains with more PUFs are considered more uropathogenic than strains with fewer PUFs, a direct role in pathogenicity remains to be defined for the majority of PUFs.
Here, we examined a set of 43 human urine-associated E. coli (UAEC) strains isolated at symptomatic and asymptomatic time points from the urine of 14 women with frequent rUTIs. We tested whether UAEC isolates shared common genes that classified strains with increased virulence from those with lesser virulence in defined experimental mouse models. Surprisingly, we observed no correlation between gene content, including PUFs, and the ability to cause cystitis in C3H/HeN mouse models. However, we found that environmentally-responsive phenotypes and specific transcriptional responses to in vitro conditions could discriminate whether strains would be more or less efficient at bladder colonization when tested in the C3H/HeN acute cystitis mouse model. We also determined that some UAEC were robust colonizers in multiple mouse models whereas others were only efficient at colonizing the bladder in some models.
Results
Phylogenetic origin and relatedness of human urine-associated E. coli strains
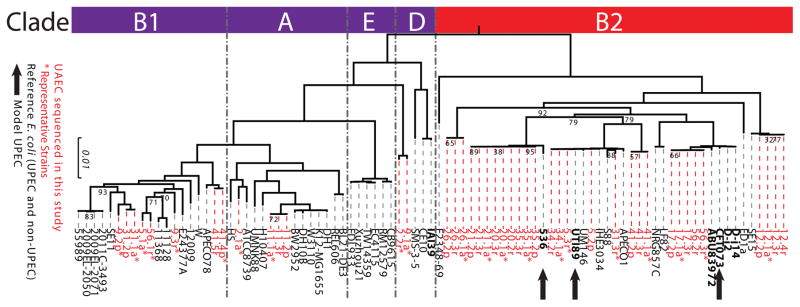
A total of 43 E. coli isolates was collected from 14 women with frequent rUTI (Fig. S1, Table S1, Table S2). Since all of these strains were isolated from the urine of women, but not necessarily collected during diagnosed UTIs, we refer to these strains as urine-associated E. coli (UAEC) rather than UPEC. Whole genome sequences of all 43 UAEC were determined, assembled, annotated and combined with genomic data from 46 previously sequenced representative E. coli from various clades and pathotypes to evaluate aspects of their relatedness and gene content (Table S3). We found that 60–75% of each strain’s genome consisted of core genes, defined as genes present in all strains being analyzed, while the variable genome comprised the remaining 25–40% (Fig. S2A). We constructed a maximum-likelihood phylogenetic tree using the 2,746 single-copy genes present in the core genome of all the E. coli isolates (Fig. 1). Consistent with previous studies of UPEC (15–19), more than two-thirds (67.4%, 29 of 43) of our UAEC strains fell within the B2 clade. The remaining UAEC fell within three other E. coli clades (4 in clade A, 8 in clade B1 and 2 in clade D).
Figure 1. Phylogenetic distribution of UAEC strains from rUTI patients.
The phylogenetic relatedness of the urine-associated E. coli (UAEC) strains (n=43, taxon labels in red) was contextualized within the broader phylogeny of reference E. coli strains (n=46, taxon labels in black) by comparing the single-copy core genes of the strains using the RAxML algorithm. Reference E. coli strains that were associated with urinary disease (e.g. cystitis, pyelonephritis, or asymptomatic bacteriuria) are in bold. Asterisks indicate UAEC strains chosen as representative isolates for their clonal clusters (Fig. S2B). Bootstrap supports are indicated at internal nodes, and bootstrap values >95 have been removed. UAEC strains were found in four out of five E. coli clades (indicated by red and purple bars on the left). Black arrows indicate model UPEC strains commonly used in UTI research.
In some patients, UAEC isolated at consecutive time points were tightly grouped within the phylogenetic tree indicating same-strain rUTI. In contrast, UAEC collected at consecutive time points in other patients were not closely related, which is consistent with different-strain rUTI. As has been done in previous studies (43), cluster analysis of pairwise SNP distances between all UAEC isolates revealed 13 different clonal groups comprised of two to four isolates each (Fig. S2B and Table S2). Eight UAEC did not group with any other isolate, and were each assigned to their own core clonal group. Based on clonal group membership, we identified 12 same-strain recurrences and 6 different-strain recurrences from a total of 18 rUTI events (Table S2). Eleven of 12 same-strain rUTIs were caused by B2 strains, which was a statistically significant enrichment relative to the phylogenetic distribution of the UAEC tested in our cohort (Hypergeometric test, p<0.05). For further analyses, we then selected a representative UAEC strain from each of the clonal groups, including core clonal groups with a single member, resulting in a representative collection of 21 UAEC isolates that encompassed the genetic, phylogenetic, and phenotypic diversity of our strain set, including 11 clade B2, 2 clade A, 6 clade B1 and 2 clade D strains (Table S2).
Carriage of PUFs correlates with B2 clade membership in human urine and non-urine isolates of E. coli
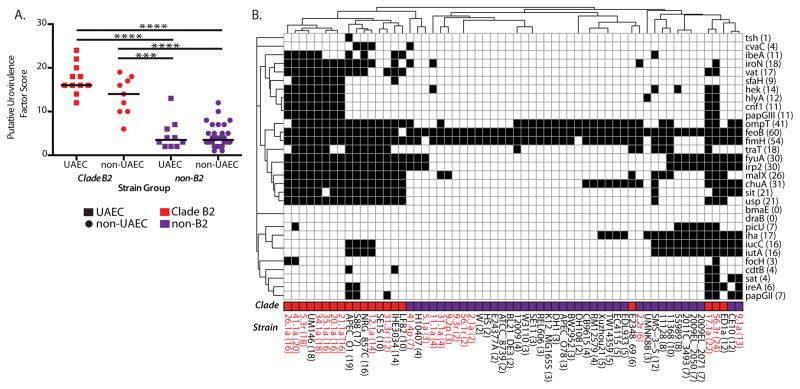
As done previously (38–40), we calculated a PUF score for each representative UAEC and non-UAEC control strain using an alignment-based analysis of 31 previously defined PUFs (20, 38–40) (Table S4). Carriage of PUFs varied considerably among strains, ranging from 2 to 24 PUFs per UAEC genome (median 13; the model B2 UPEC strain, UTI89, encodes 17 PUFs) to 2 to 19 PUFs per non-UAEC genome (median 5) (Fig. 2). Out of the 31 PUFs, 28 were found in at least one UAEC genome. We found that B2 E. coli strains carried more PUFs than did non-B2 E. coli strains whether or not they were associated with urinary disease in humans (Mann-Whitney U test, p<0.001) (Fig. 2A). Further, using unsupervised hierarchical clustering of PUF carriage, we found that strains generally clustered based on B2 clade membership, rather than on their association with urinary disease (Fig. 2B), indicating that PUF carriage was biased by phylogeny and was not specific to uropathogens.
Figure 2. Carriage of putative urovirulence factors (PUFs) is enriched in both UAEC and non-UAEC strains from the B2 clade.
(A) Comparisons between indicated groups were performed using the Mann-Whitney U test. Significant differences were calculated for each group and significant results are indicated as: ***, P<0.001; ****, P<0.0001. (B) Clinical UAEC strains and reference E. coli strains that were not associated with urinary disease (non-UAEC) (x-axis) were examined for the presence of 31 PUF sequences (y-axis) using BLAST and alignment-based searches. The binary presence of PUFs (indicated by black squares) was tallied for each strain and for each PUF (indicated in parentheses on both axes). Two-dimensional hierarchical clustering identified clusters of PUFs that tended to co-occur in UAEC strains (dendrogram along the y-axis) and showed that PUF carriage was associated with phylogeny (dendrogram along the x-axis, phylogeny indicated in column labeled ‘Clade’).
Bladder colonization in C3H/HeN mice by UAEC is variable and not restricted to B2 strains
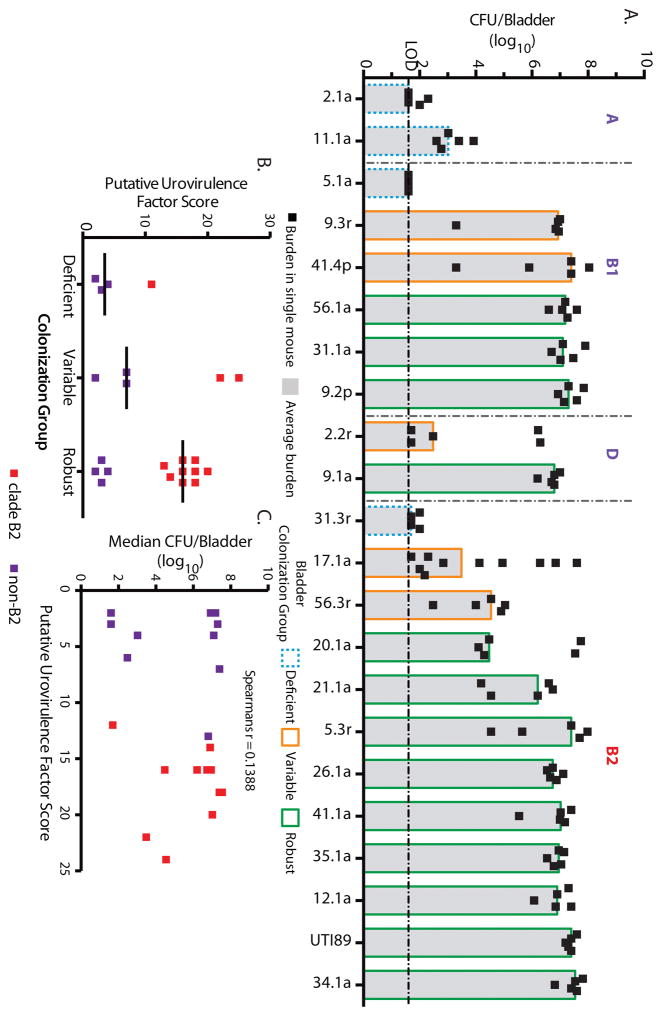
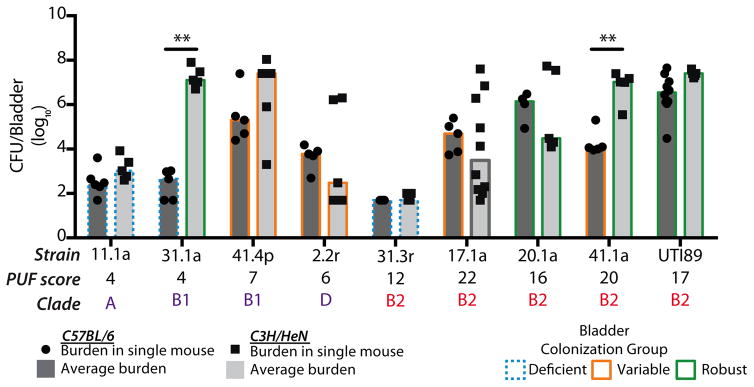
Although each of our UAEC strains was recovered from the urine of a patient and thus able to colonize a human bladder, differences in patient genetics, behavior, and infection history can influence susceptibility to UTI (9, 43–45). To control for host genetics and environmental factors that could confound comparisons of colonization efficiency among the UAEC strains, we characterized the ability of each of 21 representative strains to colonize the genetically homogenous C3H/HeN mouse model of cystitis. Each UAEC strain and a model UPEC strain, UTI89, was inoculated into the bladder of at least 5 C3H/HeN mice. Bacterial burdens, measured in colony forming units (CFU), were determined in both the bladder and kidneys at 24 hours post-infection (hpi) (Fig. 3A, Fig. S3). We observed a range of bladder bacterial burdens (200 to 108 CFU/bladder) at 24 hpi with three colonizer groups emerging among the tested UAEC strains. Twelve of 21 strains were “robust colonizers” where UAEC titers were greater than 104 CFU in every infected mouse bladder; 4 of 21 strains were “deficient colonizers” where UAEC titers were less than 104 in every infected bladder, and; 5 of 21 were “variable colonizers” where UAEC titers ranged both above and below 104 CFU/bladder of infected mice (Fig. 3A). Kidney colonization generally correlated with bladder colonization (Fig. S3). Notably, both B2 and non-B2 strains were among the robust colonizers, capable of colonizing the mouse bladder at rates similar to UTI89, indicating that efficient bladder colonization of C3H/HeN mice was achievable by strains outside of the B2 clade. Importantly, whereas C3H/HeN bladder colonization varied among UAEC at 24 hpi, the titers of these isolates were markedly higher than those of gut-associated E. coli strains collected from adult humans that were not associated with urinary tract disease. These strains included model E. coli strains and strains from the feces of healthy adults (63, 64) and were comparable to our UAEC collection in the distribution of B2 versus non-B2 strains (Table S5). We found that only 1 of 9 gut-associated E. coli, a clade A strain, was capable of robustly colonizing the bladders at 24 hpi, while 5 of 9 strains were deficient and the remaining 4 of 9 were variable (Fig. S4a). Thus, a significantly higher proportion of UAEC strains robustly colonized the C3H/HeN mouse model than gut-associated E. coli at 24 hpi (Fisher’s Exact test, p<0.05) (Table S5, Fig. S4).
Figure 3. Increased PUF carriage does not correlate with increased colonization efficiency.
(A) The colonization efficiencies of 21 representative UAEC strains and the model strain UTI89 from indicated clades were tested in C3H/HeN mice. Bacteria were enumerated from individual harvested bladders at 24 hours post-infection (hpi) (black boxes). Each UAEC strain was categorized as: “deficient” (n=4, blue stippled outline) at <104 CFU/bladder, “variable” (n=5, orange outline) above and below 104 CFU/bladder, or “robust” (n=12, green outline) at >104 CFU/bladder. The black broken horizontal line represents the limit of detection of bacteria. Data presented represent the median (gray bar) of mouse inoculations for each strain. (B) No enrichment of PUF carriage, as measured by PUF scores, was found in comparisons of robust, variable or deficient colonizer strains. Comparisons were performed with Mann-Whitney U test; horizontal solid bars indicate median values. (C) Using Spearman’s rank correlation (ρ statistic indicated at the top), there was no significant correlation between carriage of PUFs and bladder burden with UAEC strains at 24 hpi. Squares indicate the median bladder colonization of B2 (red) or non-B2 (purple) UAEC strains.
Given that early bladder colonization is enhanced by the formation of IBCs characterized using model B2 UPEC strains (33, 34, 46, 47), we sought to determine if diverse B2 and non-B2 UAEC strains from our collection followed the same pathogenic cascade as model B2 strains. As for the UPEC strain UTI89 (47), we found that 5 of 6 phylogenetically diverse UAEC were competent for IBC formation (Fig. S5, Table S6) in C3H/HeN mice at 6 hpi, suggesting that both B2 and non-B2 UAEC strains exhibited pathogenic mechanisms similar to that of the model B2 UPEC strain. Notably, the only deficient colonizer strain tested amongst the group clade A strain 11.1a was also the strain that did not form IBCs at this single time point supporting previous reports that IBC formation is linked to bladder colonization (47).
Carriage of PUFs is not required for bladder colonization in C3H/HeN mice, but may provide a competitive advantage in chronic cystitis
Although UAEC strains with both high and low PUF scores caused rUTI among the women in our cohort, we sought to determine whether PUF content could explain the observed differences in bladder colonization in the C3H/HeN mouse model (Figure 3A). We observed no significant differences in the carriage of PUFs when comparing UAEC strains that were robust (range 2 to 20, median 16), variable (range 2 to 24, median 7) or deficient (range 4 to 12, median 3.5) colonizers of the C3H/HeN mice (p>0.05, Kruskal-Wallis test and Mann-Whitney U tests) (Fig. 3B). Further, we found no significant correlation between PUF carriage and bladder bacterial burden in C3H/HeN mice (Spearman’s rank correlation, p>0.05) (Fig. 3C).
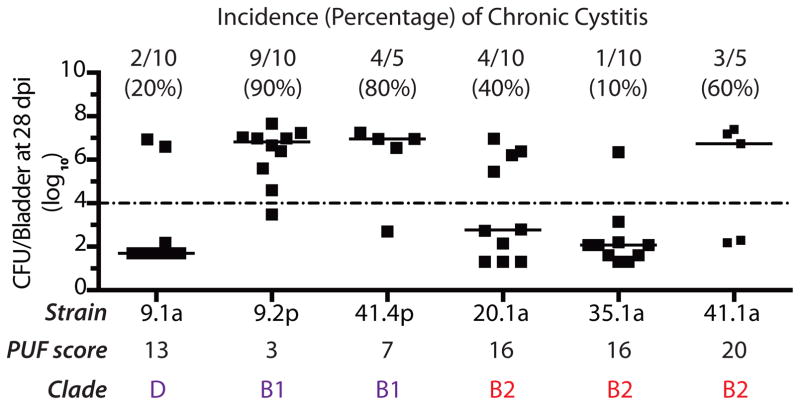
In mice, chronic cystitis is defined as the development of persistent high titer (>104 CFU/ml) bacteriuria, high titer bladder bacterial burdens and chronic inflammation with persistent lymphoid aggregates at sacrifice >4 weeks post-infection (wpi). Chronic cystitis and bladder inflammation result in large-scale remodeling of the bladder mucosa, which can last for weeks/months after antibiotic treatment to clear the infection (6, 7). This remodeling is evident in changes to the bladder itself and in host susceptibility to recurrent infection, exemplified by: (i) an altered transcriptional profile; (ii) an altered urothelial membrane proteome; (iii) defects in superficial cell maturation; (iv) changes to bacterial occupation of different habitats and; (v) differential host responses upon subsequent bacterial exposure (6, 7). To determine the effects of PUF carriage on the ability of UAEC to cause chronic cystitis, we examined three non-B2 strains: 41.4p (clade B1, PUF score=7), 9.1a (clade D, PUF score=13), and 9.2p (clade B1, PUF score=3) and three B2 UAEC strains: 20.1a (PUF score=16), 35.1a (PUF score=16), and 41.1a (PUF score=20) for their ability to cause persistent, high-titer bacteriuria (>104 CFU/mL urine), high-titer bladder colonization (>104 CFU/bladder) and chronic inflammation at 28 days post-infection in juvenile C3H/HeN mice. We found that all of the tested UAEC strains were capable of causing chronic cystitis at varying frequencies, including the non-B2 UAEC strains (range 20–90%) (Fig. 4). These rates were comparable to the rates of chronic cystitis caused by B2 UAEC strains and UTI89 (range 10–60%) (5). Taken together, our results indicate that several non-B2 strains carrying few PUFs were capable of causing both acute and chronic UTI in C3H/HeN mice in addition to causing clinical infection in women. While the carriage of many PUFs was not essential for non-B2 UAEC strains to cause chronic cystitis in mice, B2 UAEC strains that carried many PUF genes were enriched in isolates causing same-strain rUTI (11 of 12 same-strain UTIs were caused by B2 UAEC). Indeed, we found that specific deletion from UTI89 of a large pathogenicity-associated island, PAI IIUTI89, carrying 124 genes including 4 PUFs (papgIII, hek, cnf1 and hlyA) did not result in a competitive defect against the WT strain during acute time points (1–3 dpi) in urine; however, the mutant strain was severely outcompeted in the urine during chronic time points (≥7 dpi)(Fig. S6a), and in the bladder and kidney tissue at sacrifice 28 dpi (Fig. S6b).
Figure 4. Both B2 and non-B2 UAEC strains cause chronic cystitis in mice.
Incidence of chronic cystitis, defined as persistent high-titer bacteriuria (>104 CFU/mL urine), high-titer bladder colonization (>104 CFU/bladder) and chronic inflammation at 28 days post-infection (dpi), was measured for a subset of B2 and non-B2 UAEC strains infecting C3H/HeN mice. Both B2 and non-B2 UAEC strains could cause chronic cystitis in mice. PUF scores and incidence of chronic cystitis are indicated for each strain. Horizontal bars indicate median values.
Later time point rUTI isolates outcompete those collected at enrollment regardless of PUF carriage
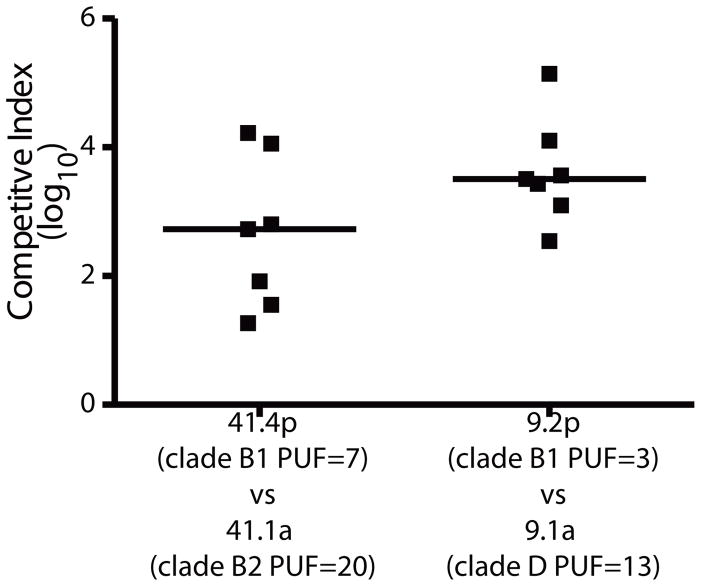
Previous research has shown that different-strain rUTIs can occur when the recurrent strain outcompetes the initial UTI strain in the gastrointestinal tract and bladder (48). In two patients in our cohort, patient 9 and patient 41, a UAEC strain with a higher PUF score (strain 9.1a, PUF score=13; strain 41.1a, PUF score=20) was supplanted by a UAEC strain with a lower PUF score (strain 9.2p, PUF score=3; strain 41.4p, PUF score=7). We hypothesized that the supplanting strains would be fitter than the initial UTI strains collected from the same patient in colonizing the murine bladder. Thus, in two separate experiments, we inoculated equal numbers of the initial and supplanting strains from patients 9 and 14 into the bladders of C3H/HeN mice, respectively, and the relative bacterial titers were determined from urine at 24hpi using strain-specific antibiotic resistance markers (Fig. 5). In both experiments, we found that the supplanting strains outcompeted the initial UTI strains at 24 hpi despite the fact that strains 41.4p and 9.2p carried considerably fewer PUF genes. Taken together, these data suggest that bladder colonization was not limited to B2 UAEC strains or strains with high carriage of PUFs.
Figure 5. PUF carriage does not increase competitive advantage during co-infection.
UAEC strains isolated from two patients (9 and 41) with different strain-induced rUTIs were differentially marked with antibiotic resistance markers. C3H/HeN mice were then coinfected with the supplanting strain (9.2p or 41.4p) at an equal dose with their enrollment strain from the same patient (9.1a or 41.1a, respectively). A competitive index (CI) was calculated from the ratio of the supplanting strains over their enrollment strains in the urine from each mouse at 24 hpi. Clades and PUF scores are indicated for each strain. Horizontal bars indicate median values.
Variations in UAEC gene expression under defined conditions predicts the outcome of acute cystitis in mice
It is possible that UAEC strains share some genomic feature that is not currently identified as a PUF. Thus, we also performed an in-depth, unbiased comparative genomic analysis between robust and deficient colonizer strains of UAEC to find any possible cryptic PUF genes or genomic signature that would clearly delineate these two groups. Our analysis revealed that no orthologous genes were exclusive to either robust or deficient colonizers and that there was no significant enrichment of specific functional domains or enzymatic pathways in either colonizer group after correcting for variance between strains. Using a relaxed definition of enrichment (i.e., present in >80% of one group and <20% of the other group), we found that two orthogroups were enriched in the deficient colonizer UAEC relative to robust colonizer strains, namely the brnTA genes encoding a type II toxin-antitoxin (80) (Table S7). The brnTA genes, whose function in pathogenesis and regulation in E. coli are unknown, were identified in all four deficient colonizer strains; however, they were not specific to deficient colonizers, as they were also found in 2 of the 12 robust colonizers (12.1a and 26.1a). Further, the number of hits identified here fell below our threshold for Type I errors, which was determined by extensive permutations of the enrichment analysis (see Supplemental Methods). Thus, we find that the enrichment of these two genes in deficient colonizers is likely due to random chance.
Whereas differences in gene carriage could not clearly discriminate between these two groups of UAEC strains, we hypothesized that differences in how core bacterial functions are regulated, such as pili production, might be able to explain the variation that we observed in mouse bladder colonization and UTI outcome. We found significant variability in pili function amongst our UAEC strains with mannose-sensitive hemagglutination titers, denoting the expression of type 1 pili, ranged from zero to 210 (Fig. S7a) despite the presence of an intact fim operon in 20 of the 21 UAEC strains, as determined using custom BLAST and alignment-based searches (see Supplemental Methods). Notably, the closely related B2 strains 20.1a, 21.1a and 35.1a had high mannose-resistant hemagluttination titers (>24), denoting expression of a non-type 1 pilus, under culture conditions that typically induce mannose-sensitive hemagglutination phenotypes in model B2 strains such as UTI89, including S pili (Fig. S7b) (76, 83). Importantly, we found that robust colonizer strains had significantly higher hemagglutination titers (median 27.25) than deficient colonizers (median 22.25) (Mann-Whitney U test, p< 0.01) or variable colonizers (median 26) (Mann-Whitney U test, p<0.01) (Fig. S7c), and that hemagglutination titer was significantly correlated to bladder bacterial burden in C3H/HeN mice at 24 hpi (Spearman’s rank correlation, p<0.001)(Fig. S7d). Taken together, these data suggest that the regulatory networks that drive the expression of CUP pili genes differ significantly between UAEC strains and that hemagglutination titer is correlated with the ability to cause acute cystitis in C3H/HeN mice.
To systematically compare core gene expression between phylogenetically and phenotypically diverse UAEC strains, we generated RNA-Seq data from triplicate cultures of a subset of 11 strains after growth in static LB broth (bladder infection inoculum). These strains included: (i) representative UAEC from four phylogenetic clades; (ii) robust, variable, and deficient colonizers; (iii) strains with disparate hemagglutination titers and; (iv) the model B2 strain, UTI89 (Table S8). Pairwise comparisons of transcript abundance for the 10 UAEC strains relative to the UTI89 strain revealed 753 genes (of 3,516 core genes or ~21.4%) that were differentially expressed in at least 1 of the 10 UAEC strains under these culture conditions (Padj < 0.05, fold change > 3) (Table S9) with B2 strains having fewer differentially expressed genes relative to UTI89 than non-B2 strains (Fig. S8A). However, the expression of a large number of core genes differed among the clinical UAEC strains, including both the B2 and non-B2 strains, even though they were grown under identical culture conditions. Differentially expressed genes were scattered throughout the genome, and many were clustered in operons encoding flagella, pilus, and chemotaxis machinery (Fig. S8B). Notably, under this culture condition, the average expression of genes in the fim operon encoding type 1 pili was lower in deficient colonizers than robust colonizers and corresponded with the results of our hemagglutination assay. We also found that most PUFs present in each strain were expressed under these conditions, although at different levels when comparing across strains (Table S10).
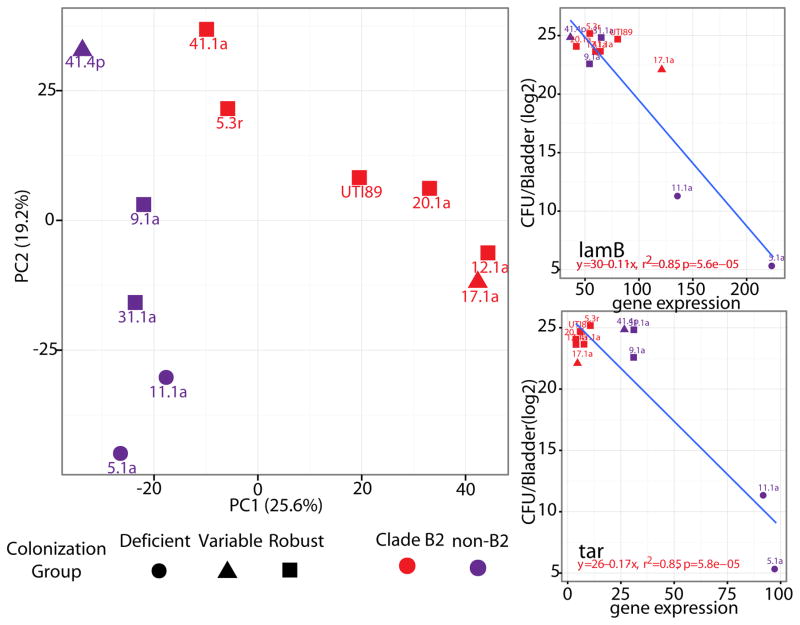
To explore whether transcriptional profiles under inoculum growth conditions were able to predict bladder colonization efficiency, we performed principal component analysis (PCA) on the complete set of core genome transcriptional profiles relative to UTI89 (Fig. 6A). We found a distinct pattern that separated the profiles of deficient and robust colonizers along the principal component 2 (PC2) axis, which explained 19.2% of the variance between the UAEC strains. Variable colonizers grouped with robust colonizers and away from deficient colonizers in this analysis. Further, PCA showed a separation between UAEC strains along PC1 (25.6% of variance) that correlated with clade membership (i.e. B2 vs. non-B2) suggesting that differences in gene expression associated with phylogeny are distinct from those that separate deficient and robust colonizers. Using a linear regression analysis, we systematically evaluated how expression of each gene (using normalized read counts per gene) correlated with bacterial burden at 24 hpi in the mouse bladder. After correcting for false discovery, we identified a total of 42 core genes whose expression in static LB broth correlated with subsequent bladder colonization efficiency in C3H/HeN mice at 24 hpi (Fig. S9 and Table S11). These included genes encoding sugar-transport proteins, such as the LamB maltoporin (Fig. 6B), and other genes associated with maltose transport (49, 50), as well as the Tar protein (Fig. 6C), a chemotactic sensor of aspartate and maltose (51, 52). The majority of the linear regression curves had negative beta coefficients indicating that deficient colonizers had higher expression of these genes than robust colonizers. We grouped these 42 core genes into functional pathways (53, 54), which showed a total of six functions that were enriched including motility, chemotaxis and carbohydrate transport (Table S12). These results indicate that under culture conditions used for growth of bacteria prior to infecting mice, core bacterial behaviors, such as swimming motility and nutrient transport, were regulated differently between robust and deficient colonizers in UAEC strains. Interestingly, despite differences in how genes mediating motility and chemotaxis were expressed, the motility of the clinical UAEC strains did not correlate with bladder colonization in C3H/HeN mice at 24 hpi (Fig S10), suggesting that transcriptional control of motility, rather than the in vitro phenotype itself, was a predictor of subsequent colonization efficiency.
Figure 6. Differential expression of core genes by UAEC strains distinguishes robust from deficient colonizers.
(A) Principle component analysis (PCA) was used to cluster UAEC strains based on their expression of 3,340 core genes under type 1 pili-inducing culture conditions used to culture bacteria before inoculation into mouse bladders. B2 strains separated from non-B2 strains along PC1, whereas robust colonizers separated from deficient colonizers along PC2. Linear regression identified the expression of 42 core genes in type 1-inducing conditions as correlated to bladder bacterial burden at 24hpi in C3H/HeN mice after correction for false discovery (P-adjusted<0.1). These 42 genes included pathways genes mediating nutrient uptake, such as lamB (B) and chemotaxis, such as and tar (C), whose correlations to bladder burden are visualized here as representative examples. Data presented represent averages derived from three independent experiments that were corrected for variance in read depth between samples and gene length.
UAEC strains vary in their ability to colonize the bladders of mice of different genetic backgrounds
Our data suggest that dynamic features (e.g., transcriptional control of core E. coli genes) rather than static characteristics (e.g., carriage of PUFs) could predict the relative efficiency of UAEC strain colonization of mouse bladder. Thus, differential regulation of transcriptional networks during in vitro growth in the colonization inoculum may have served to prime UAEC bladder colonization. To determine if this “priming” transcriptional response was universal in predicting colonization among different mouse strains, we inoculated a subset of our UAEC strains into C57BL/6 mice, which was a different host environment with a different immune response to infection. As with the C3H/HeN mouse model, the UAEC strains showed a variable ability to colonize the bladders of C57BL/6 mice, with bladder bacterial burdens ranging from undetectable to >107 CFU/bladder at 24 hpi (Fig. 7). Overall, we observed less variation in bladder colonization for each UAEC strain in C57BL/6 mice than seen in C3H/HeN mice e.g., the highly variable strains in C3H/HeN colonization experiments, 2.2r and 17.1a, showed a more narrow distribution of bladder burden at 24hpi in the C57BL/6 mice. Interestingly, whereas most UAEC strains colonized the C57BL/6 mouse bladder as well as the C3H/HeN bladder at 24 hpi, two strains, 31.1a (clade B1, PUF score=4) and 41.1a (clade B2, PUF score=20), showed significantly lower bladder bacterial burdens in C57BL/6 mice (Mann-Whitney U test, p<0.01) (Fig. 7). Notably, the reduction in colonization ability did not appear to be based on genetic background or phylogeny of the bacterial strains, as other UAEC strains carrying similar numbers of PUFs and strains from the same clades as 31.1a and 41.1a colonized C3H/HeN and C57BL/6 mouse bladders at similar levels.
Figure 7. Colonization efficiency by UAEC strains varies between mouse models.
Select UAEC strains were inoculated into C3H/HeN mice (black squares and light gray bars) and C57BL/6 mice (black circles and dark gray bars). Bacteria were enumerated from individual harvested bladders at 24 hpi (black squares and circles). Median values for each infection are given (gray bars). Each UAEC infection of the two mouse strains was categorized separately as “deficient” (blue stippled outline), “variable” (orange outline), or “robust” (green outline). Bladder colonization between C3H/HeN and C57BL/6 mice was significantly different for strains 31.1a and 41.1a by Mann-Whitney U test with **, P<0.01.
Quiescent intracellular reservoirs are small collections of dormant UPEC cells that remain in LAMP-1-positive vesicles in the underlying uroepithelium and represent another mode of bladder colonization within a different host environment. Quiescent intracellular reservoirs persist in the bladder even after apparent clearance of bacteria from the kidneys and urine and may be able to seed same-strain rUTI (55). In addition to acute cystitis, C57BL/6 mice have been used to model the formation of quiescent intracellular reservoirs. In general, quiescent intracellular reservoirs can be detected through plating bladder homogenates 14dpi after the clearance of bacteriuria and kidney infection, as has been done previously (55). We tested 4 robust colonizer UAEC strains, including 9.1a (clade D, PUF score=13), 9.2p (clade B1, PUF score =3), 35.1a (clade B2, PUF score=16) and 41.1a (clade B2, PUF score=20), and the model B2 UTI89 (PUF score=17) and found that all 5 strains, resulted in detectable bladder CFUs at 14dpi (Fig. S11), despite having sterile urines and kidneys, indicating the existence of quiescent intracellular reservoirs. However, the non-B2 strain, 9.1a, produced significantly lower bladder burdens at this time point than other tested strains, including the non-B2 strain, 9.2p, which was isolated from the same patient. Strain 9.1a also produced fewer IBCs than other strains tested (Table S6). Thus, diverse UAEC strains could colonize multiple host backgrounds. However, the relative efficiency of mouse bladder colonization varied between different UAEC strains in the same host background, and individual UAEC strains had different relative abilities to infect the bladders of mice with different genetic backgrounds.
UAEC strains that robustly colonized mice were correlated with markers of increased UTI severity in patients
An overly exuberant inflammatory response to an initial UTI is associated with severity of subsequent infections in mice (5) and susceptibility to recurrent infection in women (6). This inflammatory response is cyclooxygenase-2 (cox-2) dependent and is associated with increased neutrophil infiltration across the urothelium during infection, which results in increased levels of white blood cells in the urine (5,6). In addition to neutrophils, the host protects itself from UTI through secretion of lipocalin-2, which is induced upon infection of the bladder and helps to sequester iron away from the infecting organism (61). Importantly, levels of lipocalin 2 in the urine correlate to increasing levels of bacteria in the urine, or bacteriuria, in women (61). To determine if UAEC strains having increased virulence in the mouse model caused more severe UTI in women, we examined levels of white blood cells and lipocalin 2 in our cohort of patients infected with robust, variable, and deficient colonizer UAEC. We found that strains defined as robust and variable colonizer in the C3H/HeN mouse model were associated with markedly higher levels of white blood cells in the urine than deficient colonizers during UTI episodes (Table S13). Further, we found that robust colonizers were associated with higher levels of lipocalin 2 in the urine than either variable or deficient colonizers (Table S13).
Discussion
Although virulence in other E. coli pathotypes has been tightly linked to carriage of specific genes, UPEC are distinct in their lack of a signature set of genes that universally distinguishes them from non-UPEC strains. This likely reflects the broad definition of UPEC – any E. coli strain that is recovered from the urine of a symptomatic UTI patient – a classification that fails to account for differences in host susceptibility, or the possibility that multiple evolutionary and mechanistic paths can lead to urovirulence. To better define the determinants of urovirulence, we focused our study on a collection of 43 diverse UAEC strains isolated from a cohort of 14 women with frequent rUTI. Here, we have integrated genomic and transcriptomic approaches with in vitro and in vivo characterization of isolate phenotypes, including their colonization and pathogenic potential in defined experimental mouse models of cystitis. Our findings expand our understanding of the genomic diversity of UPEC, show that differential transcriptional regulation of core genes contributes to pathogenic potential and that the relative virulence of different E. coli strains varies depending on host background. We conclude that the range of clinical UTI outcomes reflects a range of complex host-pathogen interactions driven, in part, by variation in how uropathogens regulate core functions, impacting their pathogenic potential in hosts with different genetics and health status.
Phylogenomic analysis of this collection of UAEC strains revealed patterns of phylogenetic diversity previously reported in other clinical studies (15–20), including an enrichment of clade B2 strains (67% of total) and a mixture of same-strain and different-strain rUTIs, with B2 strains dominating same-strain rUTIs (11 of 12 same-strain rUTI isolates fell within the B2 clade) (20). We observed variation in the genomic content of these isolates, up to 40% in pairwise comparisons between strains, similar to the level of genomic diversity observed in E. coli overall (13, 14). Genomic diversity among E. coli strains is thought to be key to the species’ ability to thrive in a multitude of environments (14). Thus, it is possible that the genetic diversity observed in our cohort of UAEC strains, where two strains may share only 60% of the genes in their genome, reflects the sum of selection pressures across multiple habitats including but not limited to reservoirs in the gut, intracellular and extracellular locales in the bladder and diverse host backgrounds. In addition to other genes, we found that UAEC were variable in their carriage of PUF genes, which have previously been identified as being enriched in UTI isolates relative to other E. coli strains (20, 38–40). In contrast to this paradigm, we found that, if phylogeny was taken into account, there was no enrichment of PUFs in UAEC strains relative to non-UAEC strains. Thus, the enrichment of PUFs in urinary isolates of E. coli reported previously may be due to a combination of the enrichment of clade B2 in urinary isolates and the phylogenetic bias in PUF carriage in clade B2 strains.
The molecular mechanisms of pathogenicity for many PUFs remain poorly defined and may have functions outside of bladder pathogenicity that still enhance the overall virulence of UPEC strains, such as increasing successful migration from the gut reservoir to the bladder or enabling persistence in host niches outside of the bladder, such as the vagina. One limitation of our study is that it is solely focused on bladder virulence and thus, functions outside of the bladder were not tested. Further, given the fact that many of these factors are co-localized on pathogenicity-associated islands common to the B2 clade and absent from other E. coli (56), it is possible that the enrichment of PUFs results from a type of “genetic hitchhiking” due to some selection for B2 strains, such as their transcriptional profiles or persistence in gut reservoirs, rather than selection for the PUF genes themselves. Strikingly, we found that PUF carriage did not correlate with urovirulence in C3H/HeN mice. Indeed, non-B2 UAEC that carried few PUFs were as proficient as B2 UAEC carrying many PUFs in causing cystitis in mice. We also observed that in two patients, a UAEC strain with a higher number of PUFs was supplanted by a UAEC strain with a lower PUF score. In addition, we found in a mouse model, that the supplanting strains outcompeted the initial UTI strains in co-infection experiments. However, we found that same-strain rUTI isolates were much more likely to be from clade B2 than from other clades, which suggests that B2-specific genomic features, such as carriage of PUFs, other genes and/or transcriptional regulatory machinery, may provide an advantage for persisting within hosts between symptomatic episodes of UTI. Further, while PUF carriage was neither necessary nor sufficient for bladder colonization at acute time points, we did find that deletion of a large pathogenicity island from UTI89, which carried 124 genes including 4 PUFs, resulted in a loss of fitness during competition in chronic cystitis. While this result does not directly implicate these four PUFs in bladder colonization, it does show that carriage of at least some of the genes in this island may provide competitive advantages during different aspects of disease. All of our studied isolates came from the urine of women either during or between episodes of rUTI and thus, all of them, by definition, were uropathogens able to colonize at least one human bladder. However, since host genetics, environment and behavior are important for UTI susceptibility, it is likely that many UAEC strains isolated from a single patient may be unable to cause infection in all or even most human bladders. To control for these inter-host variations, we assessed the colonization potential of UAEC and non-UAEC strains in genetically defined animal models under identical environmental conditions. While mouse bladder colonization by UAEC strains was markedly higher than by gut-associated E. coli, we observed that bladder colonization varied among UAEC isolates under these conditions. However, even after comparing the gene content of robust and deficient colonizer UAEC isolates, we found no significant differences in specific gene content or overall functional potential that could account for the variability of these strains in colonizing mouse bladders. Another important limitation of our study is our focus on gene carriage among strains. Since our comparative analysis focused solely on gene content, as determined by orthology, we would not have been able to identify more subtle genomic differences that could influence uropathogenicity including variation in promoter sequences or in coding sequences that might impact gene expression or protein structure and function.
While we found no clear genomic signatures that could be identified to discriminate between robust and deficient colonizers, we found that differences in the expression of core genes shared among all UAEC strains were predictive of urovirulence in C3H/HeN mouse bladders. These included genes involved in maltose transport, chemotaxis and flagellar assembly that mediate core bacterial behaviors such as nutrient utilization and motility. The variance in expression of core functions contrasts with studies showing that shared genes are expressed similarly in different E. coli strains (57), possibly reflecting different definitions of the “core genome/transcriptome” between the studies. Our definition was based on comparisons of genomic sequences while previous reports have identified genes that were “commonly transcribed” to identify a core transcriptome. Importantly, this variance in transcriptional regulation of core bacterial behaviors has a direct impact on the ability of UAEC strains to colonize host bladders. This is exemplified by the differential regulation of the conserved fim operon, which encodes type 1 pili. While all but one of our UAEC strains carried an intact fim operon, the strains varied considerably in their hemagglutination titers, a measure of piliation. Further, hemagglutination titers correlated well with colonization of the C3H/HeN mouse bladder. We posit that that the regulatory networks that control type 1 pili expression within the different UAEC strains respond differently to environmental cues. For example, a set of highly related strains (from patients 20, 21, and 35), all carrying the fim gene cluster, expressed S pili instead of type 1 pili, when grown under standard type 1 pilus-inducing conditions (24). These strains were robust colonizers of C3H/HeN mice, indicating that their increased expression of S pili under inoculum conditions did not prevent their urovirulence. In the UTI89 strain, expression of S and type 1 pili is inversely controlled so that blocking expression of one pilus type induces the expression of the other (28). Further, differences in type 1 pili regulation are seen in the ways that UAEC regulate their motility and pili expression relative to model B2 UPEC strains. For example, increased expression of type 1 pili is inversely correlated with expression of flagellar genes in UTI89 (58). Here, we found that the non-B2 strains exhibiting high expression of type 1 pili also had increased motility, indicating that the networks mediating coordinated regulation of flagella and type 1 pilus expression may differ between B2 and non-B2 UAEC strains. Taken together, our findings implicate the divergence of transcriptional and regulatory networks as a key driver of UPEC pathogenesis and underscore the importance of examining transcriptional regulation of genes in addition to their patterns of carriage to more fully understand the factors underlying bacterial virulence and host-pathogen interactions. Moreover, our findings suggest that transcriptional responses to the host environment likely also diverge among UAEC strains, and that to gain richer insights into the determinants of UTI risk and outcome, we need a better understanding of how gene expression differences during infection influence bacterial physiology and interactions with the host.
The progression and outcome of a UTI is determined not only by the virulence potential of the infecting bacteria, but by myriad factors in the host environment. Mouse models of UTI recapitulate the histological markers of human UTIs and multiple mouse models have been developed that each reflect a portion of the diversity of UTI pathology seen in the clinic [reviewed in (59, 60)]. Consistent with their ability to cause UTIs in humans, many of our UAEC strains were able to elicit key hallmarks of human pathogenesis in mouse models of UTI, including acute cystitis, the formation of IBCs, the development of chronic cystitis, and persistence in quiescent intracellular reservoirs. Importantly, we found that the ability of certain UAEC strains to cause acute cystitis in the C3H/HeN mice correlated with markers of increased UTI severity in patients. Specifically, UAEC strains that exhibited an increased ability to cause acute cystitis in the C3H/HeN mouse model were associated with an increase in leukocytes and lipocalin 2 in the urine of the patients from which the strains were derived. Lipocalin 2 is known to be associated with increased activation of the immune response in human UTIs (61). Additionally, transmigration of neutrophils across the bladder epithelium, which results in an increase in leukocytes in the urine, has been associated with UTI severity and recurrence (6).
Our study suggests that each infection by a single UAEC strain in a single mouse background captures only a small part of the complex landscape of factors that govern UTI progression and outcome. Specifically, we found that, while most UAEC strains had similar success colonizing the bladder of C57BL/6 and C3H/HeN mice, two phylogenetically and genetically diverse strains were able to colonize the C3H/HeN bladder much better than the C57BL/6 bladder, indicating that some feature of the C57BL/6 bladder environment presents a specific barrier to colonization that could be overcome by some strains but not others. These findings suggest that the barriers to infection vary among mouse backgrounds as do the capacities of different UAEC strains to overcome these barriers. Thus, the outcome of each UTI is determined by the compatibility of bacterial virulence and host susceptibility factors involved in that infection. Gaining a more comprehensive understanding of the factors that determine UTI risk and outcome will require studies of urovirulence using more combinations of UAEC isolates and mouse backgrounds that better represent the diversity of bacterial urovirulence potentials and host susceptibilities.
Materials and Methods
Study design
This study was conducted to identify conserved bacterial features that enabled E. coli bladder colonization and virulence and determine if these features enabled virulence in all host backgrounds. In a previous study that was approved by the Human Subjects Review Committee at the University of Washington, 104 women aged 18–49 years with a self-reported history of UTI and a current diagnosis of acute cystitis were enrolled in the analysis cohort and self-collected mid-stream urine samples daily for 90 days (d), as described previously (62). Women were recruited from University of Washington Health Centers in Seattle, Washington. Exclusion criteria for this cohort included known anatomic of functional abnormalities of the urinary tract, chronic illness, pregnancy, and development of acute pyelonephritis (62). From this cohort, a total of 29 women experienced a rUTI within the 90d study window and provided urine samples containing urine-associated E. coli (UAEC). We examined strains isolated from the first 14 of these 29 women, resulting in a total of 43 UAEC isolates collected throughout the study (Fig. S1, Table S2; See Supplemental Methods). These isolates 14 isolates collected at enrollment, 18 isolates collected during rUTI, and 11 isolates from daily urine samples collected at home by the patients in the days preceding the diagnosis of UTI (Table 1). A single E. coli isolate was collected from each urine sample using selective plating techniques (62), and all isolates were stored at −80°C. When possible, levels of white blood cells and lipocalin 2 were measured in the urine samples, as described previously (62). E. coli rUTI was defined based on: i) the presence of E. coli in the urine at levels ≥102 CFU/mL and ii) medical diagnosis of a UTI based on symptoms of acute cystitis, such as foul-smelling urine, increased frequency of urination, and/or urgency to urinate.
Table 1.
Summary of 43 UAEC isolates from women with recurrent UTIs.
Diagnosed UTI
No UTI diagnosis
Additional strains used in this project
The model strains MG1655, Sakai and Nissle 1917 were derived from pure cultures and stored in glycerol at −80°C. As described in a previous study, gut-associated E. coli were isolated from human feces collected from healthy adults (63, 64). Briefly, single colonies were identified as E. coli with isolation plating on differential media and glycerol stocks were stored at −80°C.
Genome dataset construction
We constructed draft genomes of the 43 UAEC strains by sequencing extracted DNA on the Illumina HiSeq 2000 platform and assembling the reads into ordered contigs using Velvet v1.2 (65) and Mauve v2.3.1 (66)(Table S14; see Supplemental Methods). In order to contextualize the UAEC genomes, we constructed a database containing the 43 draft UAEC genomes as well as 46 closed E. coli genomes from the National Center for Biotechnological Information (NCBI) (Table S3). All genomes were re-annotated using the Broad Institute’s prokaryote annotation pipeline (67) and genes were clustered into orthogroups using the reciprocal best BLAST hits method (68).
Phylogenetic and gene content analysis
Phylogenetic trees were generated using RAxML (69) with bootstrapping on a concatenated alignment of 2746 single-copy core orthogroups across 89 organisms aligned using MUSCLE (70). SNPs were calculated in this single-copy core genome and used to identify clusters of highly-related strains (See Supplemental Methods). Targeted analysis of PUF genes (Table S4) and the fim operon was performed using a combination of BLAST-based interrogation and confirmed by mapping DNA reads to a database of the PUF and fim genes (see Supplemental Methods). We compared global gene content differences between deficient colonizing UAEC and robust colonizers (with UTI89) using a stringent set of criteria as well as more relaxed definition of enrichment (see Supplemental Methods). In addition to gene content, we also investigated differences in pathways or functional domains between groups of strains using Pfam (71) and KEGG (72). To provide context for our analysis and estimate the chance of “false positives” in our results, we calculated the rate Type I errors arising from random chance, as described previously (73)(see Supplemental Methods).
Mouse infections
Mouse infections were done using previously established models (5, 74, 75). In preparation for inoculation into mice, bacterial strains were grown statically in LB at 37°C overnight and then subcultured 1:1,000 into 10 ml of fresh LB for a second static outgrowth at 37°C for 18 to 24 h to induce expression of type 1 pili (76). Bacteria from these cultures were harvested and resuspended in 1x PBS for inoculation. Mice used in these experiments were purchased from Charles River and Harlan Labs. Urine collections and harvested bladders and kidneys were serially plated on LB agar plates with or without appropriate antibiotics for enumeration of bacterial burdens. Competitive indexes were calculated based on the ratios of marked strains in each mouse replicate.
Detection of intracellular bacterial communities and persistent bladder reservoirs
The formation of intracellular bacterial communities (IBCs) was measured in 7–8 week old female C3H/HeN mice that were inoculated with the indicated strain as described (see Mouse infections above). IBCs were visualized at 6 hours post-infection (hpi) with α-E. coli antibodies and bladder cells were counter-stained with wheat germ agglutinin. Persistent bladder reservoirs were measured in C57BL/6 mice that were inoculated with the indicated strain. Mice were sacrificed after 2 weeks for enumeration of bacterial burdens in the bladders and kidneys. Mice that developed chronic infections, as determined by urine titers collected during the 2-week infection, or mice that had visible kidney abscesses were removed from this analysis.
Phenotypic analyses
For all phenotypic analyses, clinical UAEC were first grown under type 1 pili-inducing conditions (see Mouse infections above). Hemagglutination assays were performed with guinea pig erythrocytes in PBS with or without 2% methyl-α-D-mannopyranoside incubated at 4°C overnight. Swimming motility assays were performed in 0.25% agar LB and the diameter of the spread of the bacteria was measured after 6h incubation at 37°C.
Generation of RNA-Seq data
After growth in type 1 pili-inducing conditions (see Mouse Infections above), RNA was extracted from select UAEC strains grown in triplicate and Illumina cDNA libraries were constructed following the RNAtag-seq protocol (77) (see Supplemental Methods). Barcoded cDNA was then sequenced on the Illumina HiSeq 2000 platform. Triplicate RNA-seq libraries were de-multiplexed and mapped to their cognate strain using BWA (78). Targeting a set of 3,516 “core transcriptome genes” found in UTI89 and 9/10 UAEC strains tested, we calculated differences in expression using DESeq (79). We normalized for read abundance and gene length, averaged expression levels across the triplicate samples and then removed genes whose average expression level across the 11 UAEC fell into the lowest quartile of genes. The remaining genes were then used in principle component analysis and linear regression with appropriate controls for multiple hypothesis testing.
Statistical analysis
Colonization groups were defined based on the median value of mouse bladder colonization by UAEC strains at 24 hpi. Repeated non-parametric Mann-Whitney U tests were used to identify statistically significant differences in PUF gene carriage, hemagglutination assay titers, and swimming motility between colonization groups. Further, repeated Mann-Whitney U tests were used to identify differences in PUF gene carriage between clades, colonization efficiency of different inbred mouse strains by UAEC isolates, and persistence of quiescent intracellular reservoirs by UAEC in C57BL/6 mice. Correlations between median bladder burden in C3H/HeN mice and PUF gene carriage, hemagglutination assay titer, or motility in UAEC strains was determined using the Spearman rank correlation test in separate analyses. UAEC were clustered into related groups using unsupervised hierarchical clustering of SNP distances defined through pairwise alignments of the core genome. Further, unsupervised clustering was used to identify UAEC that shared similar carriage of PUF genes. A Hypergeometric Distribution test was used to compare enrichment of clade B2 strains causing same-strain rUTI relative to the distribution of B2 strains with our collection of clinical isolates. Differences in expression of core genes were measured using the DESeq algorithm. Principle component analysis (PCA) was used to visualize similarities in core gene expression and linear regression analysis was used to correlate these differences in gene expression to median bladder burden at 24 hpi in C3H/HeN mice. Type I error modeling was established with repeated permutation analysis with 1000 replicates and counts of gene presence/absence and is described in detail in the Supplementary Materials. Where p-values could be assigned, a threshold of significance was established at P<0.05 after multiple hypothesis testing correction (Padj<0.05), except for the linear regression analysis of gene expression to bladder burden, which used a significance threshold of Padj<0.1.
Supplementary Material
Figure S1. Sample collection and patient time line.
Figure S2. Measurement of UAEC gene carriage and nucleotide diversity in core genome.
Figure S3. Kidney colonization by B2 and non-B2 UAEC.
Figure S4. Gut-associated E. coli are poor colonizers in the C3H/HeN mouse models of UTI.
Figure S5. IBC formation by B2 and non-B2 UAEC.
Figure S6. Carriage of PAI IIUTI89 enhances competitive fitness in chronic UTI in C3H/HeN mice.
Figure S7. Hemagglutination is correlated to colonization efficiency in both B2 and non-B2 UAEC.
Figure S8. Differentially expressed genes in UAEC relative to UTI89.
Figure S9. Expression of core genes in type 1-inducing culture conditions correlates to bladder colonization in UAEC.
Figure S10. Motility is not correlated to colonization efficiency in either B2 or non-B2 UAEC.
Figure S11. Both B2 and non-B2 UAEC are capable of forming persistent bladder reservoirs in C57BL/6 mice.
Table S1. Clinical information of Enrolled Patients.
Table S2. Characteristics of UAEC Isolates.
Table S3. Reference Escherichia coli Strains.
Table S4. List of putative urovirulence factors.
Table S5. Gut associated E. coli Strain Characteristics.
Table S6. IBC Formation in Select UAEC at 6 hpi in C3H/HeN mice.
Table S7. Presence of brnAT genes in Robust and Deficient colonizer UAEC strains.
Table S8. Reads mapped to core genome of UAEC.
Table S10. Expression of PUF genes in select UAEC.
Table S13. UAEC clinical data.
Table S14. UAEC sequencing and genome assembly results.
Table S11. Linear correlation of core gene expression to bladder colonization.
Table S12. Functions enriched in genes associated with bladder colonization.
Table S9. DEseq Analysis of UAEC transcription of core genome.
Accessible Summary.
Urinary tract infections (UTIs) are most commonly caused by uropathogenic E. coli (UPEC). An interdisciplinary analysis of an extensive panel of UTI-associated E. coli revealed strain dependent variation in the colonization of distinct mouse models of UTI. The expression of conserved bacterial behaviors, such as metabolism or motility, diverged markedly among these strains and was a better predictor of bladder colonization than carriage of any set of genes, including previously identified putative urovirulence factors. Our findings suggest that UTI risk is determined by the pairing of variable host susceptibilities with bacterial virulence phenotypes governed by both gene content and expression.
Acknowledgments
We thank members of the S.J.H. laboratory for their helpful suggestions and Daniel Stoebel for the generous gift of the gut-associated E. coli strains.
Funding: H.L.S.IV. was supported in part by a 2013 Monsanto Excellence Fund Graduate Fellowship and a 2012 Lucille P. Markey Pathway award. This project was funded by the National Institutes of Health (NIH), specifically National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant number RO1 DK051406 and Office of Research on Women’s Health (ORWH) Specialized Center of Research (SCOR) grant number P50 DK064540 to S.J.H. and National Institute of Allergy and Infectious Disease (NIAID) grant number U19AI110818 to the Broad Institute. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
Author contributions H.L.S.IV., K.W.D., and S.J.H. conceived the project and H.L.S.IV., J.L., A.M.E., and S.J.H. designed the experiments. H.L.S.IV, M.S.C., M.E.H and T.J.H performed mouse infections under supervision of S.J.H. H.L.S.IV performed in vitro assays under supervision of S.J.H. H.L.S.IV and A.L.M. performed genomic comparisons and measured gene carriage under supervision of A.LM. W-C.C. and J.L. performed and analyzed RNAseq experiments. P.L.R., A.E.S., and T.M.H. analyzed clinical metadata. H.L.S.IV wrote the paper with all co-authors.
Competing interests: T.J.H. is a part-time employee at Fimbrion Therapeutics (St. Louis, MO), a company co-founded by T.M.H. and S.J.H. that is developing small molecule therapeutics to treat UTIs caused by UPEC. T.M.H. is an advisory board member with GlaxoSmithKlein (Brentford, United Kingdom), Paratek Pharmaceuticals (Boston, MA), Achaogen Pharmaceuticals (San Francisco, CA), and Ocean Spray (Middleborough, MA). S.J.H. is an advisory board member of Genentech (San Francisco, CA) and Roche Pharmaceuticals (Basel, Switzerland), a consultant for Regeneron (Tarrytown, NY) and Wellspect Healthcare (El Segundo, CA) and a founding member of QureTech Bio (Umea, Sweden).
References and Notes
- 1.Foxman B. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am J Med. 2002;113(Suppl 1A):5S–13S. doi: 10.1016/s0002-9343(02)01054-9. [DOI] [PubMed] [Google Scholar]
- 2.Foxman B. Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect Dis Clin North Am. 2014;28:1–13. doi: 10.1016/j.idc.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Mabeck CE. Treatment of uncomplicated urinary tract infection in non-pregnant women. Postgrad Med J. 1972;48:69–75. [PMC free article] [PubMed] [Google Scholar]
- 4.Ferry SA, Holm SE, Stenlund H, Lundholm R, Monsen TJ. The natural course of uncomplicated lower urinary tract infection in women illustrated by a randomized placebo controlled study. Scand J Infect Dis. 2004;36:296–301. doi: 10.1080/00365540410019642. [DOI] [PubMed] [Google Scholar]
- 5.Hannan TJ, Mysorekar IU, Hung CS, Isaacson-Schmid ML, Hultgren SJ. Early Severe Inflammatory Responses to Uropathogenic E. coli Predispose to Chronic and Recurrent Urinary Tract Infection. PLoS Pathog. 2010;6:e1001042. doi: 10.1371/journal.ppat.1001042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hannan TJ, Roberts PL, Riehl TE, van der Post S, Binkley JM, Schwartz DJ, Miyoshi H, Mack M, Schwendener RA, Hooton TM, Stappenbeck TS, Hansson GC, Stenson WF, Colonna M, Stapleton AE, Hultgren SJ. Inhibition of Cyclooxygenase-2 Prevents Chronic and Recurrent Cystitis. EBioMedicine. 2014;1:46–57. doi: 10.1016/j.ebiom.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Brien VP, Hannan TJ, Yu L, Livny J, Roberson EDO, Schwartz DJ, Souza S, Mendelsohn CL, Colonna M, Lewis AL, Hultgren SJ. A mucosal imprint left by prior Escherichia coli bladder infection sensitizes to recurrent disease. Nat Microbiol. 2016;2:16196EP. doi: 10.1038/nmicrobiol.2016.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta K, Bhadelia N. Management of urinary tract infections from multidrug-resistant organisms. Infect Dis Clin North Am. 2014;28:49–59. doi: 10.1016/j.idc.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Foxman B, Gillespie B, Koopman J, Zhang L, Palin K, Tallman P, Marsh JV, Spear S, Sobel JD, Marty MJ, Marrs CF. Risk factors for second urinary tract infection among college women. Am J Epidemiol. 2000;151:1194–1205. doi: 10.1093/oxfordjournals.aje.a010170. [DOI] [PubMed] [Google Scholar]
- 10.Gupta K, Hooton TM, Naber KG, Wullt B, Colgan R, Miller LG, Moran GJ, Nicolle LE, Raz R, Schaeffer AJ, Soper DE. International Clinical Practice Guidelines for the Treatment of Acute Uncomplicated Cystitis and Pyelonephritis in Women: A 2010 Update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin Infect Dis. 2011;52:e103–e120. doi: 10.1093/cid/ciq257. [DOI] [PubMed] [Google Scholar]
- 11.Russo TA, Johnson JR. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J Infect Dis. 2000;181:1753–1754. doi: 10.1086/315418. [DOI] [PubMed] [Google Scholar]
- 12.Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 13.Rasko DA, Rosovitz MJ, Myers GSA, Mongodin EF, Fricke WF, Gajer P, Crabtree J, Sebaihia M, Thomson NR, Chaudhuri R, Henderson IR, Sperandio V, Ravel J. The Pangenome Structure of Escherichia coli: Comparative Genomic Analysis of E. coli Commensal and Pathogenic Isolates. J Bacteriol. 2008;190:6881–6893. doi: 10.1128/JB.00619-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, Calteau A, Chiapello H, Clermont O, Cruveiller S, Danchin A, Diard M, Dossat C, Karoui ME, Frapy E, Garry L, Ghigo JM, Gilles AM, Johnson J, Le Bouguénec C, Lescat M, Mangenot S, Martinez-Jéhanne V, Matic I, Nassif X, Oztas S, Petit MA, Pichon C, Rouy Z, Ruf CS, Schneider D, Tourret J, Vacherie B, Vallenet D, Médigue C, Rocha EPC, Denamur E. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rijavec M, Erjavec MS, Avguštin JA, Reissbrodt R, Fruth A, Križan-Hergouth V, Žgur-Bertok D. High Prevalence of Multidrug Resistance and Random Distribution of Mobile Genetic Elements Among Uropathogenic Escherichia coli (UPEC) of the Four Major Phylogenetic Groups. Curr Microbiol. 2006;53:158–162. doi: 10.1007/s00284-005-0501-4. [DOI] [PubMed] [Google Scholar]
- 16.Erjavec MS, Rijavec M, Križan-Hergouth V. Chloramphenicol- and tetracycline-resistant uropathogenic Escherichia coli (UPEC) exhibit reduced virulence potential. Int J Antimicrob Agents. 2007;30:436–442. doi: 10.1016/j.ijantimicag.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 17.Piatti G, Mannini A, Balistreri M, Schito AM. Virulence factors in urinary Escherichia coli strains: phylogenetic background and quinolone and fluoroquinolone resistance. J Clin Microbiol. 2008;46:480–487. doi: 10.1128/JCM.01488-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skjøt-Rasmussen L, Hammerum AM, Jakobsen L, Lester CH, Larsen P, Frimodt-Moller N. Persisting clones of Escherichia coli isolates from recurrent urinary tract infection in men and women. J Med Microbiol. 2011;60:550–554. doi: 10.1099/jmm.0.026963-0. [DOI] [PubMed] [Google Scholar]
- 19.Ejrnaes K, Stegger M, Reisner A, Ferry S, Monsen T, Holm SE, Lundgren B, Frimodt-Moller N. Characteristics of Escherichia coli causing persistence or relapse of urinary tract infections: Phylogenetic groups, virulence factors and biofilm formation. Virulence. 2011;2:528–537. doi: 10.4161/viru.2.6.18189. [DOI] [PubMed] [Google Scholar]
- 20.Luo Y, Ma Y, Zhao Q, Wang L, Guo L, Ye L, Zhang Y, Yang J. Similarity and Divergence of Phylogenies, Antimicrobial Susceptibilities, and Virulence Factor Profiles of Escherichia coli Isolates Causing Recurrent Urinary Tract Infections That Persist or Result from Reinfection. J Clin Microbiol. 2012;50:4002–4007. doi: 10.1128/JCM.02086-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Zhao S, Han L, Guo X, Chen M, Ni Y, Zhang Y, Cui Z, He P. Drug resistance and virulence of uropathogenic Escherichia coli from Shanghai, China. J Antibiot. 2014;67:799–805. doi: 10.1038/ja.2014.72. [DOI] [PubMed] [Google Scholar]
- 22.Chen SL, Hung CS, Xu J, Reigstad CS, Magrini V, Sabo A, Blasiar D, Bieri T, Meyer RR, Ozersky P. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc Natl Acad Sci USA. 2006;103:5977–5982. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welch RA, Burland V, Plunkett G, Redford P, Roesch P, Rasko D, Buckles EL, Liou SR, Boutin A, Hackett J, Stroud D, Mayhew GF, Rose DJ, Zhou S, Schwartz DC, Perna NT, Mobley HLT, Donnenberg MS, Blattner FR. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci USA. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hultgren SJ, Schwan WR, Schaeffer AJ, Duncan JL. Regulation of production of type 1 pili among urinary tract isolates of Escherichia coli. Infect Immun. 1986;54:613–620. doi: 10.1128/iai.54.3.613-620.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brzuszkiewicz E, Brüggemann H, Liesegang H, Emmerth M, Olschläger T, Nagy G, Albermann K, Wagner C, Buchrieser C, Emődy L, Gottschalk G, Hacker J, Dobrindt U. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc Natl Acad Sci USA. 2006;103:12879–12884. doi: 10.1073/pnas.0603038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwartz DJ, Kalas V, Pinkner JS, Chen SL, Spaulding CN, Dodson KW, Hultgren SJ. Positively selected FimH residues enhance virulence during urinary tract infection by altering FimH conformation. Proc Natl Acad Sci USA. 2013;110:15530–15537. doi: 10.1073/pnas.1315203110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mulvey MA, Schilling JD, Hultgren SJ. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect Immun. 2001;69:4572–4579. doi: 10.1128/IAI.69.7.4572-4579.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wurpel DJ, Beatson SA, Totsika M, Petty NK, Schembri MA. Chaperone-usher fimbriae of Escherichia coli. PLoS ONE. 2013;8:e52835. doi: 10.1371/journal.pone.0052835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greene SE, Pinkner JS, Chorell E, Dodson KW, Shaffer CL, Conover MS, Livny J, Hadjifrangiskou M, Almqvist F, Hultgren SJ. Pilicide ec240 disrupts virulence circuits in uropathogenic Escherichia coli. mBio. 2014;5:e02038. doi: 10.1128/mBio.02038-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu XR, Sun TT, Medina JJ. In vitro binding of type 1-fimbriated Escherichia coli to uroplakins Ia and Ib: relation to urinary tract infections. Proc Natl Acad Sci USA. 1996;93:9630–9635. doi: 10.1073/pnas.93.18.9630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eto DS, Jones TA, Sundsbak JL, Mulvey MA. Integrin-mediated host cell invasion by type 1-piliated uropathogenic Escherichia coli. PLoS Pathog. 2007;3:e100. doi: 10.1371/journal.ppat.0030100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez JJ, Mulvey MA, Schilling JD, Pinkner JS, Hultgren SJ. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO J. 2000;19:2803–2812. doi: 10.1093/emboj/19.12.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson GG, Palermo JJ, Schilling JD, Roth R, Heuser J, Hultgren SJ. Intracellular bacterial biofilm-like pods in urinary tract infections. Science. 2003;301:105–107. doi: 10.1126/science.1084550. [DOI] [PubMed] [Google Scholar]
- 34.Justice SS, Hung C, Theriot JA, Fletcher DA, Anderson GG, Footer MJ, Hultgren SJ. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc Natl Acad Sci USA. 2004;101:1333–1338. doi: 10.1073/pnas.0308125100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garofalo CK, Hooton TM, Martin SM, Stamm WE, Palermo JJ, Gordon JI, Hultgren SJ. Escherichia coli from urine of female patients with urinary tract infections is competent for intracellular bacterial community formation. Infect Immun. 2007;75:52–60. doi: 10.1128/IAI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosen DA, Hooton TM, Stamm WE, Humphrey PA, Hultgren SJ. Detection of intracellular bacterial communities in human urinary tract infection. PLoS Med. 2007;4:e329. doi: 10.1371/journal.pmed.0040329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robino L, Scavone P, Araujo L, Algorta G, Zunino P, Vignoli R. Detection of intracellular bacterial communities in a child with Escherichia coli recurrent urinary tract infections. Pathog Dis. 2013;68:78–81. doi: 10.1111/2049-632X.12047. [DOI] [PubMed] [Google Scholar]
- 38.Johnson JR, O’Bryan TT, Delavari P, Kuskowski M, Stapleton A, Carlino U, Russo TA. Clonal Relationships and Extended Virulence Genotypes among Escherichia coliIsolates from Women with a First or Recurrent Episode of Cystitis. J Infect Dis. 2001;183:1508–1517. doi: 10.1086/320198. [DOI] [PubMed] [Google Scholar]
- 39.Johnson JR, Porter S, Johnston B, Kuskowski MA, Spurbeck RR, Mobley HLT, Williamson DA. Host Characteristics and Bacterial Traits Predict Experimental Virulence for Escherichia coli Bloodstream Isolates From Patients With Urosepsis. Open Forum Infect Dis. 2015;2:ofv083. doi: 10.1093/ofid/ofv083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis. 2000;181:261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- 41.Johnson JR, Delavari P, Kuskowski M, Stell AL. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J Infect Dis. 2001;183:78–88. doi: 10.1086/317656. [DOI] [PubMed] [Google Scholar]
- 42.Sahl JW, Sistrunk JR, Fraser CM, Hine E, Baby N, Begum Y, Luo Q, Sheikh A, Qadri F, Fleckenstein JM, Rasko DA. Examination of the Enterotoxigenic Escherichia coli Population Structure during Human Infection. mBio. 2015;6:e00501. doi: 10.1128/mBio.00501-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stapleton A, Hooton TM, Fennell C, Roberts PL, Stamm WE. Effect of secretor status on vaginal and rectal colonization with fimbriated Escherichia coli in women with and without recurrent urinary tract infection. J Infect Dis. 1995;171:717–720. doi: 10.1093/infdis/171.3.717. [DOI] [PubMed] [Google Scholar]
- 44.Hawn TR, Scholes D, Li SS, Wang H, Yang Y, Roberts PL, Stapleton AE, Janer M, Aderem A, Stamm WE, Zhao LP, Hooton TM. Toll-like receptor polymorphisms and susceptibility to urinary tract infections in adult women. PLoS ONE. 2009;4:e5990. doi: 10.1371/journal.pone.0005990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scholes D, Hooton TM, Roberts PL, Stapleton AE, Gupta K, Stamm WE. Risk Factors for Recurrent Urinary Tract Infection in Young Women. J Infect Dis. 2000;182:1177–1182. doi: 10.1086/315827. [DOI] [PubMed] [Google Scholar]
- 46.Mulvey MA, Lopez-Boado YS, Wilson CL, Roth R, Parks WC, Heuser J, Hultgren SJ. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science. 1998;282:1494–1497. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- 47.Schwartz DJ, Chen SL, Hultgren SJ, Seed PC. Population Dynamics and Niche Distribution of Uropathogenic Escherichia coli during Acute and Chronic Urinary Tract Infection. Infect Immun. 2011;79:4250–4259. doi: 10.1128/IAI.05339-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen SL, Wu M, Henderson JP, Hooton TM, Hibbing ME, Hultgren SJ, Gordon JI. Genomic diversity and fitness of E. coli strains recovered from the intestinal and urinary tracts of women with recurrent urinary tract infection. Sci Transl Med. 2013;5:184ra60. doi: 10.1126/scitranslmed.3005497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szmelcman S, Hofnung M. Maltose transport in Escherichia coli K-12: involvement of the bacteriophage lambda receptor. J Bacteriol. 1975;124:112–118. doi: 10.1128/jb.124.1.112-118.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beg QK, Vazquez A, Ernst J, de Menezes MA, Bar-Joseph Z, Barabási AL, Oltvai ZN. Intracellular crowding defines the mode and sequence of substrate uptake by Escherichia coli and constrains its metabolic activity. Proc Natl Acad Sci USA. 2007;104:12663–12668. doi: 10.1073/pnas.0609845104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Springer MS, Goy MF, Adler J. Sensory transduction in Escherichia coli: two complementary pathways of information processing that involve methylated proteins. Proc Natl Acad Sci USA. 1977;74:3312–3316. doi: 10.1073/pnas.74.8.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mowbray SL, Koshland DE. Additive and independent responses in a single receptor: aspartate and maltose stimuli on the tar protein. Cell. 1987;50:171–180. doi: 10.1016/0092-8674(87)90213-3. [DOI] [PubMed] [Google Scholar]
- 53.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 55.Mysorekar IU, Hultgren SJ. Mechanisms of uropathogenic Escherichia coli persistence and eradication from the urinary tract. Proc Natl Acad Sci USA. 2006;103:14170–14175. doi: 10.1073/pnas.0602136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dobrindt U, Chowdary MG, Krumbholz G, Hacker J. Genome dynamics and its impact on evolution of Escherichia coli. Med Microbiol Immunol. 2010;199:145–154. doi: 10.1007/s00430-010-0161-2. [DOI] [PubMed] [Google Scholar]
- 57.Bielecki P, Muthukumarasamy U, Eckweiler D, Bielecka A, Pohl S, Schanz A, Niemeyer U, Oumeraci T, von Neuhoff N, Ghigo JM, Häussler S. In vivo mRNA profiling of uropathogenic Escherichia coli from diverse phylogroups reveals common and group-specific gene expression profiles. mBio. 2014;5:e01075–14. doi: 10.1128/mBio.01075-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lane MC, Simms AN, Mobley HLT. Complex interplay between type 1 fimbrial expression and flagellum-mediated motility of uropathogenic Escherichia coli. J Bacteriol. 2007;189:5523–5533. doi: 10.1128/JB.00434-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carey AJ, Tan CK, Ipe DS, Sullivan MJ, Cripps AW, Schembri MA, Ulett GC. Urinary tract infection of mice to model human disease: Practicalities, implications and limitations. Crit Rev Microbiol. 2016;42:780–799. doi: 10.3109/1040841X.2015.1028885. [DOI] [PubMed] [Google Scholar]
- 60.Barber AE, Norton JP, Wiles TJ, Mulvey MA. Strengths and Limitations of Model Systems for the Study of Urinary Tract Infections and Related Pathologies. Microbiol Mol Biol Rev. 2016;80:351–367. doi: 10.1128/MMBR.00067-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Steigedal M, Marstad A, Haug M, Damås JK, Strong RK, Roberts PL, Himpsl SD, Stapleton A, Hooton TM, Mobley HLT, Hawn TR, Flo TH. Lipocalin 2 imparts selective pressure on bacterial growth in the bladder and is elevated in women with urinary tract infection. J Immunol. 2014;193:6081–6089. doi: 10.4049/jimmunol.1401528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Czaja CA, Stamm WE, Stapleton AE, Roberts PL, Hawn TR, Scholes D, Samadpour M, Hultgren SJ, Hooton TM. Prospective Cohort Study of Microbial and Inflammatory Events Immediately Preceding Escherichia coli Recurrent Urinary Tract Infection in Women. J Infect Dis. 2009;200:528–536. doi: 10.1086/600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gordon DM, Stern SE, Collignon PJ. Influence of the age and sex of human hosts on the distribution of Escherichia coli ECOR groups and virulence traits. Microbiology (London) 2005;151:15–23. doi: 10.1099/mic.0.27425-0. [DOI] [PubMed] [Google Scholar]
- 64.Snyder E, Gordon DM, Stoebel DM. Escherichia coli lacking RpoS are rare in natural populations of non-pathogens. G3: Genes, Genomes, Genet. 2012;2:1341–1344. doi: 10.1534/g3.112.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rissman AI, Mau B, Biehl BS, Darling AE, Glasner JD, Perna NT. Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics. 2009;25:2071–2073. doi: 10.1093/bioinformatics/btp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lebreton F, van Schaik W, Manson McGuire A, Godfrey P, Griggs A, Mazumdar V, Corander J, Cheng L, Saif S, Young S, Zeng Q, Wortman J, Birren B, Willems RJL, Earl AM, Gilmore MS. Emergence of Epidemic Multidrug-Resistant Enterococcus faecium from Animal and Commensal Strains. mBio. 2013;4:e00534–13–e00534–13. doi: 10.1128/mBio.00534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Salichos L, Rokas A. Evaluating ortholog prediction algorithms in a yeast model clade. PLoS ONE. 2011;6:e18755. doi: 10.1371/journal.pone.0018755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stamatakis A, Ludwig T, Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 70.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer ELL, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–8. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, Burzyn D, Ortiz-Lopez A, Lobera M, Yang J, Ghosh S, Earl A, Snapper SB, Jupp R, Kasper D, Mathis D, Benoist C. Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science. 2015;349:993–997. doi: 10.1126/science.aaa9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hung CS, Dodson KW, Hultgren SJ. A murine model of urinary tract infection. Nat Protoc. 2009;4:1230–1243. doi: 10.1038/nprot.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Conover MS, Flores-Mireles AL, Hibbing ME, Dodson K, Hultgren SJ. Establishment and Characterization of UTI and CAUTI in a Mouse Model. J Visualized Exp. 2015;100:e52892. doi: 10.3791/52892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen SL, Hung CS, Pinkner JS, Walker JN, Cusumano CK, Li Z, Bouckaert J, Gordon JI, Hultgren SJ. Positive selection identifies an in vivo role for FimH during urinary tract infection in addition to mannose binding. Proc Natl Acad Sci USA. 2009;106:22439–22444. doi: 10.1073/pnas.0902179106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shishkin AA, Giannoukos G, Kucukural A, Ciulla D, Busby M, Surka C, Chen J, Bhattacharyya RP, Rudy RF, Patel MM, Novod N, Hung DT, Gnirke A, Garber M, Guttman M, Livny J. Simultaneous generation of many RNA-seq libraries in a single reaction. Nat Methods. 2015;12:323–325. doi: 10.1038/nmeth.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Heaton BE, Herrou J, Blackwell AE, Wysocki VH, Crosson S. Molecular structure and function of the novel BrnT/BrnA toxin-antitoxin system of Brucella abortus. J Biol Chem. 2012;287:12098–12110. doi: 10.1074/jbc.M111.332163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hannan TJ, Mysorekar IU, Chen SL, Walker JN, Jones JM, Pinkner JS, Hultgren SJ, Seed PC. LeuX tRNA-dependent and -independent mechanisms of Escherichia coli pathogenesis in acute cystitis. Mol Microbiol. 2008;67:116–128. doi: 10.1111/j.1365-2958.2007.06025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Connell I, Agace W, Klemm P, Schembri M, Mărild S, Svanborg C. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc Natl Acad Sci USA. 1996;93:9827–9832. doi: 10.1073/pnas.93.18.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Parkkinen J, Rogers GN, Korhonen T, Dahr W, Finne J. Identification of the O-linked sialyloligosaccharides of glycophorin A as the erythrocyte receptors for S-fimbriated Escherichia coli. Infect Immun. 1986;54:37–42. doi: 10.1128/iai.54.1.37-42.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wright KJ, Seed PC, Hultgren SJ. Uropathogenic Escherichia coli flagella aid in efficient urinary tract colonization. Infect Immun. 2005;73:7657–7668. doi: 10.1128/IAI.73.11.7657-7668.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Enright MC, Spratt BG. Multilocus sequence typing. Trends Microbiol. 1999;7:482–487. doi: 10.1016/s0966-842x(99)01609-1. [DOI] [PubMed] [Google Scholar]
- 86.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Sample collection and patient time line.
Figure S2. Measurement of UAEC gene carriage and nucleotide diversity in core genome.
Figure S3. Kidney colonization by B2 and non-B2 UAEC.
Figure S4. Gut-associated E. coli are poor colonizers in the C3H/HeN mouse models of UTI.
Figure S5. IBC formation by B2 and non-B2 UAEC.
Figure S6. Carriage of PAI IIUTI89 enhances competitive fitness in chronic UTI in C3H/HeN mice.
Figure S7. Hemagglutination is correlated to colonization efficiency in both B2 and non-B2 UAEC.
Figure S8. Differentially expressed genes in UAEC relative to UTI89.
Figure S9. Expression of core genes in type 1-inducing culture conditions correlates to bladder colonization in UAEC.
Figure S10. Motility is not correlated to colonization efficiency in either B2 or non-B2 UAEC.
Figure S11. Both B2 and non-B2 UAEC are capable of forming persistent bladder reservoirs in C57BL/6 mice.
Table S1. Clinical information of Enrolled Patients.
Table S2. Characteristics of UAEC Isolates.
Table S3. Reference Escherichia coli Strains.
Table S4. List of putative urovirulence factors.
Table S5. Gut associated E. coli Strain Characteristics.
Table S6. IBC Formation in Select UAEC at 6 hpi in C3H/HeN mice.
Table S7. Presence of brnAT genes in Robust and Deficient colonizer UAEC strains.
Table S8. Reads mapped to core genome of UAEC.
Table S10. Expression of PUF genes in select UAEC.
Table S13. UAEC clinical data.
Table S14. UAEC sequencing and genome assembly results.
Table S11. Linear correlation of core gene expression to bladder colonization.
Table S12. Functions enriched in genes associated with bladder colonization.
Table S9. DEseq Analysis of UAEC transcription of core genome.