Abstract
Many taxa exhibit plastic immune responses initiated after primary microbial exposure that provide increased protection against disease-induced mortality and the fitness costs of infection. In several arthropod species, this protection can even be passed from parents to offspring through a phenomenon called trans-generational immune priming. Here, we first demonstrate that trans-generational priming is a repeatable phenomenon in flour beetles (Tribolium castaneum) primed and infected with Bacillus thuringiensis (Bt). We then quantify the within-host dynamics of microbes and host physiological responses in infected offspring from primed and unprimed mothers by monitoring bacterial density and using mRNA-seq to profile host gene expression, respectively, over the acute infection period. We find that priming increases inducible resistance against Bt around a critical temporal juncture where host septicemic trajectories, and consequently survival, may be determined in unprimed individuals. Our results identify a highly differentially expressed biomarker of priming, containing an EIF4-e domain, in uninfected individuals, as well as several other candidate genes. Moreover, the induction and decay dynamics of gene expression over time suggest a metabolic shift in primed individuals. The identified bacterial and gene expression dynamics are likely to influence patterns of bacterial fitness and disease transmission in natural populations.
Keywords: trans-generational immune priming, Tribolium castaneum, trained immunity, metabolism, EIF4-e, Bacillus thuringiensis, gene expression
Introduction
The ability to respond to fluctuating environments through phenotypic plasticity allows organisms to balance the trade-offs of expressing traits that might confer unequal fitness in different environments. Immunity is a trait that benefits from plastic patterns of expression because immune responses exert energetic (Povey et al. 2009), immunopathological (Sadd & Siva-Jothy 2006), and multiple-fronts (Sadd & Schmid-Hempel 2009; Tate & Graham 2015) costs that decrease organismal fitness in the absence of infection (Contreras-Garduño et al. 2014). Despite lacking a traditional adaptive immune system based on somatic recombination, arthropods from many taxa are able to plastically increase their immune response upon re-exposure to previously encountered microbes and parasites (Hernández López et al. 2014; Little et al. 2003; Milutinović et al. 2013; Pham et al. 2007; Roth & Kurtz 2009; Roth et al. 2009; Sadd & Schmid-Hempel 2007; Zanchi et al. 2012). This phenomenon, termed “immune priming,” increases the odds of surviving infection, although relatively little is known about the underlying temporal dynamics of the primed response. Immune priming can even be passed trans-generationally from parents to offspring (Roth et al. 2010).
Trans-generational immune priming can spring from both maternal and paternal sources, although the immunological and fitness consequences of priming for the offspring may differ based on parental sex (Roth et al. 2010; Zanchi et al. 2011). A recent study in the honeybee Apis millifera suggests that bacteria-challenged mothers import bacterial fragments bound to the ubiquitous egg yolk protein vitellogenin (Salmela et al. 2015) into eggs, providing a means for offspring to sample and respond to specific challenges in the maternal environment. Indeed, eggs from bacteria-challenged mothers exhibit increased antibacterial activity in the beetle Tenebrio molitor (Zanchi et al. 2012) and in bumblebees (Sadd & Schmid-Hempel 2007), although it is not clear whether these antimicrobial proteins were produced maternally or zygotically. Moreover, there is conflicting evidence as to the protective mechanism of priming once these eggs hatch and the larvae develop, with some studies showing a continued concentration of antimicrobial peptides (Barribeau et al. 2016; Sadd & Schmid-Hempel 2007), some suggesting a differentiated cellular response (Hernández López et al. 2014), and some implicating an inducible transcriptional program (Trauer-Kizilelma & Hilker 2015). However, studies on trans-generational priming, to date, have not explored the importance of feedbacks between bacterial load and immune responses, and most (but see (Barribeau et al. 2016)) only assay targets previously identified as important in the unprimed immune response against infection.
To understand how trans-generational priming gives rise to the post-infection survival advantage, we believe that it is important to consider the temporal dynamics of both bacterial growth and the full primed physiological response (Graham et al. 2011) in the offspring during acute infection. From a gene regulation perspective, priming may manifest as a change in constitutive levels of gene expression, wherein a primed individual may produce more or less, respectively, of a beneficial or detrimental transcript before infection (Barribeau et al. 2016). Priming may also manifest as an altered rate of change in gene expression over time, as we might expect if primed individuals are able to jump-start pathogen recognition, signaling and effector machinery more quickly than their unprimed counterparts upon detection of infection. Finally, priming may manifest as a change in the integral of gene expression over time, as we would expect if the overall magnitude of gene expression, or increase or decrease in negative regulation, produces an increase in resistance or tolerance (Ayres & Schneider 2012) to infection relative to unprimed insects.
To address these hypotheses, we performed multiple trans-generational immune priming experiments, differing in offspring disease-induced mortality rates, using an insect host (Tribolium castaneum) and a bacterial pathogen (Bacillus thuringiensis serovar Berliner) that induces rapid septicemia when introduced directly into the insect hemocoel. To characterize the within-host dynamics of bacterial growth preceding disease-induced mortality, we measured bacterial density across time in the infected larval offspring of primed and unprimed mothers. To characterize and quantify gene expression profiles across priming status and experiment and in correlation to bacterial load and time, we then used next-generation mRNA sequencing. Here, we present the results of these analyses, identify candidate genes and pathways that differentiate primed larvae from their unprimed cohorts, and provide evidence for the repeatability of priming in this system. We discuss the implications of our results for the temporal dynamics of host-pathogen interactions, host survival, and pathogen fitness.
Methods
Beetle and bacteria sources and rearing conditions
We obtained T. castaneum beetles from the Carolina Biological Supply (#144356). For these experiments we used three beetle lines that were confirmed uninfected with common stock parasites (mites and protozoa) or sources of unexplained mortality for at least five generations, and crossed 20 from each line into a single colony two generations before the beginning of the experiment. All beetle stocks were maintained in All-Purpose Gold Medal Flour supplemented with 5% baker’s yeast in a 30°C incubator in the dark.
To prepare B. thuringiensis (ATCC 10792/DSM 2046 as used in (Roth et al. 2010)) for experiments, we started a 30°C overnight culture in nutrient broth from a −80 °C Bt stock in glycerol. When the culture optical density reached 1.0 at 600nm we centrifuged the desired volume of culture at 16,000×g for 3 minutes and discarded the supernatant. We re-suspended the pellet in 1mL insect saline (pH 7.4) and then either heat-killed the bacteria at 95 °C for ten minutes (for maternal injections) (Roth et al. 2009) or else proceeded with live bacteria (for larval offspring infections).
Trans-generational immune priming
To create the parental generation for each trans-generational priming experiment, we collected fully mature adult beetles from the T. castaneum stock colony described above and divided them randomly into breeding groups, with 15–18 adults per group (Fig. S1). Adults were allowed to lay eggs for four days. When offspring reached the pupal stage, we divided them by sex and breeding group. We considered the adult population mature and ready to reproduce when 90% of adults were ten days post eclosion, and we excluded from the experiment any under-developed adults at that time. We then randomly assigned females to three maternal treatment groups (30/group): Naïve unprimed treatment, sterile saline wounding treatment (hereafter “saline control”), or heat-killed B. thuringensis priming treatment.
We placed female beetles from all treatment groups on ice. Females in the saline and primed treatments received a jab between the pronotum and occiput at an angle nearly parallel to the anteroposterior axis (to prevent puncturing the gut) with an ultra-thin needle (0.10×12mm) dipped in sterile saline or heat-killed bacteria solution (2×10^8 CFU/mL, or about 100–200 vegetative cells per challenge estimated by rinsing and plating needle contents from non-heat-killed aliquots). Females were allowed to recover in individual wells of 96-well plates overnight and then placed in individual glass vials with about 2mL of loosely packed flour/yeast mixture. Post-treatment maternal mortality in saline control and primed treatments did not differ significantly from naïve and was less than 5% for each experiment. We paired a naïve male from a different breeding group with each female and allowed the pair to lay eggs for five days.
For experiment one, conducted in Feb. 2012, we collected 210 approximately 16 day old larvae randomly from breeding pairs across each treatment (630 total larvae, Fig. S1). We infected 60 mid-instar larvae per maternal treatment with a needle dipped in live B. thuringiensis solution (5×10^7 CFU/mL, or an estimated 15 cells/larva (Fig. S2)) while another 40/treatment were left unchallenged. Offspring of unprimed mothers are hereafter referred to as unprimed hosts, while offspring of saline control and Bt-primed mothers are referred to as “saline” and “primed” hosts, respectively. All subjects were maintained in individual wells of 96 well plates. These groups were monitored for survival and development (time to pupation and time to adulthood) for 30 days, replacing flour periodically. After confirming a statistically significant difference in survival at 24 hours post infection among infected groups (to verify the primed phenotype), we divided the remaining 110 unmanipulated larvae into time point groups by treatment (N = 10/treatment for t = 0, 2, and 6 hours, and N = 25/treatment for t = 12 and 24 hours and an extra group in case of higher than anticipated mortality) and infected all but the t = 0 groups with a needle dipped in live B. thuringiensis solution (5×10^7 CFU/mL). We recorded the time of infection of each group, and the individual beetles from each group (N = 7–10/treatment/time point) that were still clearly alive (i.e., active, responsive to touch, and normally colored) were flash frozen in liquid nitrogen within 2 minutes of the desired sampling hour before storage at −80 °C. We froze the unchallenged t = 0 beetles at the same time as the challenged t = 6 beetles to control for circadian effects. During the t = 12 time point, we also froze three larvae from each treatment that were clearly moribund (unresponsive to touch and stiff but not yet systemically melanized).
Experiment two (July 2013) was set up the same way as experiment one (Fig. S1) and was performed with the larval cohorts of beetles that showed the primed survival phenotype in the adult stage (same group as in the lab T. castaneum experiment in (Tate & Graham 2015)). 85 larvae per treatment were challenged with around 15 CFUs of bacteria as in the previous experiment. 25 larvae per treatment were sacrificed at t = 0, 2, 6, and 8 hours post infection (four per time point), while 60 were monitored for survival.
Collection of mRNA and creation of cDNA
For both experiments, we homogenized whole frozen larvae with a mortar and pestle on ice before proceeding with RNA extraction as per manufacturer’s recommended protocol using RNeasy Microarray mini-kits and the Qiashredder (Qiagen) to remove excess chitinous tissue prior to the chloroform step. This method resulted in high yields (1.8–3.6µg total RNA/larva) of high quality RNA, as determined by the Agilent Bioanalyzer. For bacterial density estimation and qPCR validation, we eliminated remaining gDNA contamination using a heat-inactivated RNAse-free DNAse treatment (Qiagen) and reverse-transcribed the RNA into cDNA using the Quantitect Reverse Transcription kit (Qiagen) as per manufacturer’s instructions.
Measurement of bacterial load by qPCR
In order to design primers specific to B. thuringiensis rRNA, we followed a design protocol suggested by Alex Louie and David Schneider (pers. comm.), and then created a standard curve for the within-host quantification of Bt. Both protocols are outlined in the Supplementary Methods (Online Supporting Information).
To quantify Bt 16s with qPCR for both the standards and all time-series individuals, cDNA samples were first run with the Bt primers (Table S1) at an annealing temperature of 55°C and an extension temperature of 72°C as per manufacturer’s protocol (Sybr Select Master Mix, Applied Biosystems) and immediately followed by the quantification of reference gene RPL13a expression at the optimal 60°C annealing and extension temperature for those primers.
Statistical analysis of larval survival within experiments
To determine the impact of priming status on larval survival post infection for experiment one, we performed right-censored Cox proportional hazard regression analyses using the “survival” package in R (Therneau), with treatment as a main factor. We compared uninfected survival across treatments, infected survival across treatments, and survival of infected vs. uninfected individuals within treatments. The early-weighted distribution of mortality prompted us to perform a Chi-squared analysis comparing overall survival and death at 24 hours post infection across treatments, but as this yielded very similar results as the Cox regression, we dropped this analysis. While maternal ID was not retained for the larvae in experiment one, previous analysis (Tate and Graham 2015) suggests that maternal ID as a random effect explains little variance in survival and does not change inference on priming.
Statistical analysis of larval survival across experiments
Over the course of three years (2011 – 2013, Table S2), we had performed 22 other trans-generational priming offspring survival assays from nine maternal challenge experiments for various purposes (e.g. (Tate & Graham 2015)) where we had recorded survival at 24 hours post infection, using similar priming and offspring infection methods but often with different disease-induced mortality rates (Fig. 1A). We decided to take advantage of this multi-experiment data to investigate the average effect size of primed and saline wounding control offspring survival rates, relative to unprimed offspring. Therefore, we conducted a binomial generalized linear mixed model analysis of 24 hour mortality with treatment as a main effect and offspring survival experiment as a random effect nested within maternal challenge experiment, using the lme4 package in R, to derive estimates for any survival advantage provided by priming and control treatments. While some experiments infected larval offspring and others used adult offspring (Table S2), this factor was not significant and did not improve model fit and was therefore dropped from the analysis.
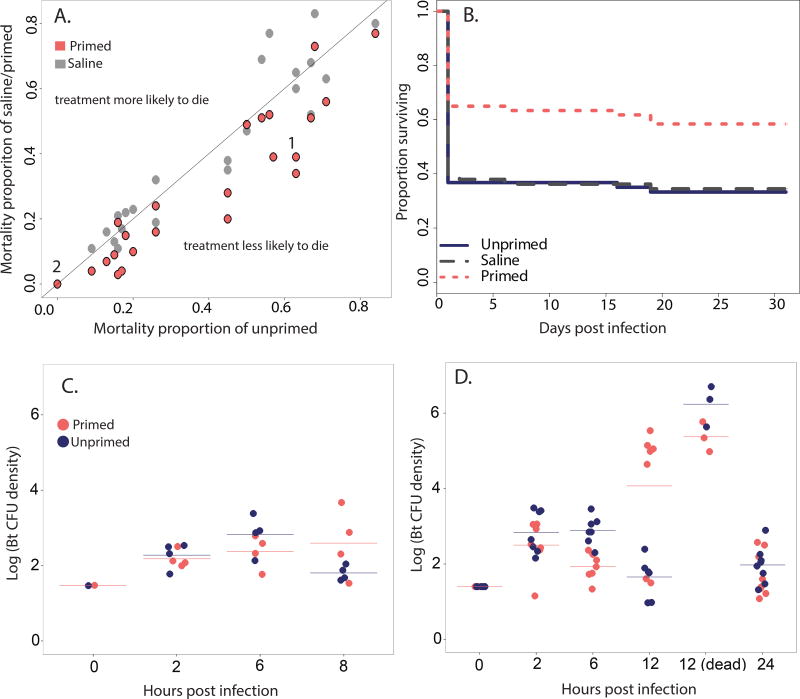
Figure 1.
Trans-generational priming increases offspring survival and alters bacterial density dynamics during the acute phase of Bt infection. Comparison of mortality rates for unprimed and primed or saline-control offspring infected with Bt for 24 offspring survival experiments, ordered by unprimed mortality rate (A). Unprimed versus primed mortality proportions are represented in pink, and unprimed versus saline control proportions are in gray. The black 1:1 line represents an equal mortality rate for offspring of unprimed and treated mothers. Numbers correspond to the experiments further explored in this paper (experiments 1 and 2). For experiment one, from which half of the mRNA-seq data is derived, nearly all disease-induced mortality occurred within the first 12 hours after infection with Bt (B), where primed individuals (dotted pink line) were significantly more likely to survive than unprimed (solid blue line) or saline-control (dashed gray line) individuals. For both experiment two (C) and experiment one (D), unprimed (blue) and primed (pink) larvae received an initial dose of around 15 vegetative Bt cells, which grew rapidly within the hemocoel in the first two hours but plateaued until after six hours. By 12 hours post infection the larval survival fates became clear, as larvae destined for death appeared clearly moribund (D, “12-dead”) and had high bacterial loads. Horizontal bars indicate the mean for each treatment at each time point.
Statistical analysis of differences in bacterial density across treatments and over time
Differences in log-transformed bacterial density across treatments, time, experiments, and their interactions were analyzed with factorial ANOVAs in R (formula = density ~ treatment*time*experiment, reduced for subsetted analyses as noted in text), followed by contrast tests (Tukey’s HSD) for a priori planned and post-hoc tests.
RNA-seq library preparation and sequencing
We selected three individual larval samples per treatment (primed and unprimed), time point (0, 2, 6, 8 (exp. 1.) or 12 (exp. 2) hours), and experiment (one and two, N = 48 total) that captured the range of bacterial loads within each treatment at each time point. For the 12 hour time point, we chose to use samples from primed larvae that had high bacterial loads (as quantified by qPCR on the cDNA), but had survived to 12 hours, to investigate the apparent tolerance effect (see results). The RNA from these samples was inspected on the Agilent Bioanalyzer to ensure high quality, un-degraded RNA, and then mRNA-selected libraries were created from approximately 0.75 – 1ug total RNA using the Illumina Tru-Seq Sample Prep Kit v2 as per manufacturer’s instructions, except all reactions were scaled down by half. Library adapters contained one of 24 unique barcodes to facilitate multiplexing after library construction. After 13 PCR cycles and AMPure bead cleanup, cDNA libraries were quantified on the Qubit and analyzed on the Agilent Bioanalyzer for quality and consistency and then reduced into five pools based on non-overlapping barcodes. Each pool was run on a single lane of the Illumina HiSeq 2500 sequencer at the Princeton University Lewis Sigler Institute Genomics Core Facility to obtain approximately 130 million reads (single end 65bp) per pool, and a range of 9–25 million reads per sample (Table S3).
RNA-seq data analysis
We processed raw reads using the Galaxy interface (Blankenberg et al. 2001; Giardine et al. 2005; Goecks et al. 2010) through the Princeton Galaxy server. Reads were first subjected to FASTQC (Andrews 2010) quality filtering and trimming to exclude low quality reads and to determine sample barcodes for each read. The Galaxy Fastx-Toolkit (Gordon & Hannon 2010) barcode splitter algorithm was used to sort reads by sample. Aided by the Tribolium castaneum gene annotation file (Richards & al. 2008) downloaded from the Ensembl Metazoa database, TopHat2 (Kim et al. 2013) was used to map raw reads to the genome (alignment statistics are presented in Table S3 and program settings are outlined in the Supplementary Methods (Online Supporting Information)). The python program HTseq-count (Anders et al. 2014) was then used to assign and count mapped reads associated with gene features. Raw reads and processed count data files are available under accession GSE95387 of the NCBI GEO database (Edgar et al. 2002).
HT-seq count data for each sample was imported into R (v. 3.0.1) (Team 2012) and transformed into a count table for use by the DESeq2 package (Love et al. 2014) (further details in Supplementary Methods (Online Supporting Information)). The linear models used to determine relationships between DESeq2 results and factors are included in the notes for Tables 2, S7, S8, and S9. Models include terms to capture variation in gene expression due to bacterial load and experiment, as preliminary analysis revealed that many genes were expressed proportionally to bacterial load rather than time. Our goal in implementing these models is to avoid confounding treatment and bacterial load effects, given that primed and unprimed individuals display different bacterial density dynamics. Because the t = 12 bacterial density was especially high for some samples in experiment one, we also repeated a subset of analyses on experiment two data only to ensure that the results were not being driven by this time point even after statistically controlling for bacterial density. Gene lists were functionally annotated using the BioMART package (Durinck et al. 2005) and KEGG pathway analysis was performed on them using the GAGE (Luo et al. 2009) package. Only a subset of KEGG pathways are annotated for T. castaneum, however, and as disease pathways are among those excluded, this approach was used mostly for metabolic pathway analyses.
Table 2.
KEGG pathway enrichment of genes differentially expressed between primed and unprimed individuals before and during the acute infection period
| Treatment, prior to infection (t=0) | |||||
|---|---|---|---|---|---|
| Ensembl ID | KEGG Pathway | stat.mean | p val | adj. p val | set.size |
| tca03010 | Ribosome | 3.53 | 0.0003 | 0.0190 | 111 |
| tca00190 | Oxidative phosphorylation | 3.30 | 0.0006 | 0.0190 | 93 |
| tca04141 | Protein processing in endoplasmic reticulum | 3.28 | 0.0006 | 0.0190 | 119 |
| tca04120 | Ubiquitin mediated proteolysis | 3.13 | 0.0011 | 0.0250 | 75 |
| tca00380 | Tryptophan metabolism | 2.93 | 0.0027 | 0.0511 | 23 |
| tca00280 | Valine, leucine and isoleucine degradation | 2.59 | 0.0059 | 0.0932 | 35 |
| tca00071 | Fatty acid degradation | 2.49 | 0.0080 | 0.1077 | 27 |
| tca00051 | Fructose and mannose metabolism | 2.40 | 0.0103 | 0.1077 | 25 |
|
| |||||
| Treatment × Time2, full time series | |||||
| Ensembl ID | KEGG Pathway | stat.mean | p val | adj. p val | set.size |
|
| |||||
| tca00190 | Oxidative phosphorylation | 11.03 | 1.30E-21 | 1.23E-19 | 93 |
| tca00280 | Valine, leucine and isoleucine degradation | 5.11 | 1.49E-06 | 7.05E-05 | 35 |
| tca00020 | Citrate cycle (TCA cycle) | 3.74 | 0.0002 | 0.0076 | 34 |
| tca00071 | Fatty acid degradation | 2.95 | 0.0024 | 0.0448 | 27 |
| tca04146 | Peroxisome | 2.87 | 0.0024 | 0.0448 | 73 |
| tca00630 | Glyoxylate and dicarboxylate metabolism | 2.85 | 0.0031 | 0.0448 | 30 |
| tca00310 | Lysine degradation | 2.83 | 0.0033 | 0.0448 | 29 |
| tca00650 | Butanoate metabolism | 2.88 | 0.0039 | 0.0461 | 15 |
| tca03013 | RNA transport | 2.52 | 0.0062 | 0.0602 | 96 |
| tca00620 | Pyruvate metabolism | 2.58 | 0.0063 | 0.0602 | 30 |
| tca00260 | Glycine, serine and threonine metabolism | 2.52 | 0.0074 | 0.0642 | 31 |
| tca00062 | Fatty acid elongation | 2.41 | 0.0113 | 0.0833 | 16 |
| tca00640 | Propanoate metabolism | 2.41 | 0.0114 | 0.0833 | 17 |
| tca00010 | Glycolysis / Gluconeogenesis | 2.13 | 0.0187 | 0.1267 | 33 |
Notes:Few genes were returned as significant or marginally significant for the treatment and treatment × time models and so the results for those modes were not enriched for KEGG pathways. Full model for DESeq2: gene expression ~ series:btload + poly(time, 2):condition, which controls for the major effects of experimental series and bacterial load to isolate genes that show treatment-based temporal patterns that are largely independent of these confounding variables. KEGG pathway analysis was performed with the full gene results list using the GAGE package in R, and employing SE-adjusted log2foldchange to reduce false positives driven by large variances or differences in amplitude across temporal patterns. The stat.mean statistic indicates the mean of the absolute vale of the magnitude of gene set changes. P values were adjusted for false discovery rate using the Benjamini & Hochberg method. Adjusted p values < 0.05 are emboldened.
Validating RNA-seq with qPCR for a subset of immune genes
To confirm that the RNA-seq data correctly reflect gene expression patterns in the high-mortality samples, we compared the temporal expression profiles to previous qPCR results from the same samples on several genes (Table S1) associated with immunological function, including the AMPs Coleoptericin-1 and Attacin-2 (Shrestha & Kim 2010). A description of qPCR methods and analysis can be found in the Supplementary Methods. The comparison of seven qPCR samples/time point and the three RNA-seq samples per time point for Coleoptericin-1 and Attacin −1 are shown in Fig. S3 and demonstrate the concordance between RNA-seq and qPCR results as well as the concordance between the subset of samples selected for RNA-seq and the entire set of samples.
For a gene of particular interest from the RNA-seq results (TC009881), we also used qPCR to assay expression from t = 0 and t = 2 post infection samples from experiment one, experiment two larvae, and experiment two adults (see (Tate & Graham 2015)), from unprimed, saline control, and primed individuals (N = 21/primed and unprimed treatments, 11/saline treatment). In separate experiments using two different constructs, we attempted to knock down the expression of TC009881 using RNAi (Posnien et al. 2009)) but met with a fecundity loss phenotype and inefficient knockdown, respectively. We have included the details in the Supplementary Methods.
Results
Trans-generationally primed larvae are significantly more likely to survive than unprimed larvae
There are currently no descriptions or tests of repeatability within systems for invertebrate primed immunity, fueling controversy about the robustness and even existence of immune priming in invertebrates. Over the 24 survival experiments (including experiments one and two analyzed in depth in this study) performed on Bt-infected offspring from unprimed, saline-injected, and Bt-primed mothers in our system, priming did not always result in a survival advantage for primed larvae (Fig. 1A). However, in a binomial generalized linear mixed model analysis with treatment as a main effect and offspring infection experiment as a random effect nested within maternal challenge experiment (Table S2), trans-generationally primed cohorts were significantly more likely to survive infection than unprimed cohorts, overall (Table S4, primed estimate relative to unprimed = −0.301, p < 0.001, while saline control cohorts showed no significant survival advantage (estimate relative to unprimed = −0.013, p = 0.85). Relative to saline control individuals, primed were also significantly more likely to survive (estimate = −0.288, p < 0.001).
Experiment one: Following infection with Bt (Figure 1B) (N = 175), primed larvae had a 58% survival rate into adulthood while 33% of unprimed and 34% of offspring from saline-injected mothers survived into adulthood. Primed larvae were significantly more likely to survive than unprimed larvae (Table S5, Cox proportional hazard regression, hazard ratio = 0.49, p = 0.0052), but infected larvae from saline control mothers showed no significant survival advantage over unprimed larvae (hazard ratio = .96, p = 0.86). Given the equality of unprimed and saline control survival phenotypes, coupled with the heightened degree of immunogenic variability introduced by sterile wounding relative to both naïve and Bt injection treatments, we chose to focus subsequent comparisons on the difference between unprimed and primed treatments. There was no significant difference in uninfected offspring survival by maternal treatment (Table S5, Cox regression, primed:unprimed hazard ratio = 1.28, p = 0.71; saline:unprimed hazard ratio = 1.026, p = 0.97), indicating that priming is not significantly costly to survival into adulthood in the absence of infection. All treatments were significantly more likely to die when infected, relative to their uninfected treatment cohorts (Table S5).
Experiment two: there was no (0%) larval mortality following Bt infection and thus differences between treatment groups could not be assessed. However, an uninfected cohort of these larvae subsequently challenged with Bt as adults showed a significant effect of maternal treatment (Tate & Graham 2015) (indicated in Fig. 3 in that study), with primed individuals once again significantly more likely to survive than unprimed individuals.
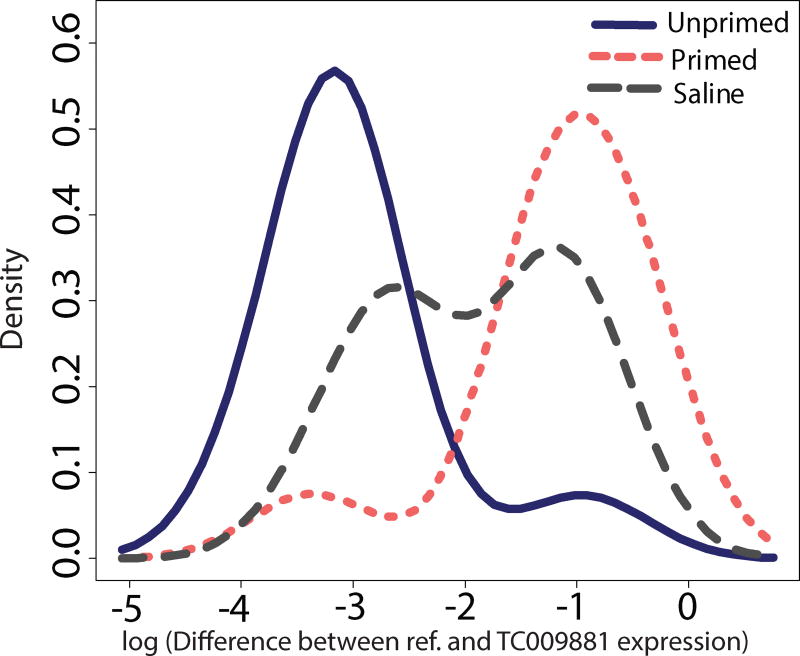
Figure 3.
A histogram of TC009881 expression by treatment showed nearly bimodal patterns of differential expression between primed (pink/light dotted line) and unprimed (solid dark line) individuals. All available cDNA samples from t = 0 and t = 2 hours post-infection experiment one larvae, experiment two larvae, and adult experiments were assayed for relative TC009881 expression via qPCR. Offspring from saline challenged mothers (dashed gray line) showed intermediate and bimodal expression of the gene. A log(relative expression) value closer to or greater than 0 indicates higher expression of TC009881. The y axis represents sample density.
Temporal dynamics of microbe load in infected larvae differ between treatments
There was no significant main effect of experiment on bacterial density for the time points in common (t = 2 and 6, Table 1). In both experimental series, bacterial load increased rapidly from the initial 15 CFU dose so that by two hours post infection, larvae from all treatment groups averaged around 400 CFU/individual (Figure 1C, D). We have previously observed that log phase Bt doubles every 30–35 minutes in LB at 30°C (unpublished obs.), so this suggests the bacteria grow exponentially, and with no appreciable lag time, in the first two hours after infection. This bacterial load remained relatively stable through six hours for unprimed larvae, but decreased significantly at six hours post infection for primed larvae in comparison to both the previous primed time point and the unprimed time point cohort (Table S6).
Table 1.
Comparison of bacterial density across time, treatment, and experiment
| Global Model: Log (Bacterial Density) ~ Time × Treatment × Exp., all data | |||||
|---|---|---|---|---|---|
| D.f. | Sum. Sq | Mean Sq. | F value | p value | |
| Time | 5 | 126.87 | 25.374 | 54.324 | 2.00E-16 |
| Treatment | 1 | 0.21 | 0.213 | 0.456 | 0.501 |
| Experiment | 1 | 0.01 | 0.013 | 0.027 | 0.87 |
| Time × Treatment | 5 | 26.11 | 5.222 | 11.181 | 2.96E-08 |
| Time × Exp. | 2 | 1.56 | 0.78 | 1.669 | 0.195 |
| Treatment × Exp. | 1 | 0.3 | 0.298 | 0.638 | 0.427 |
| Full interaction | 2 | 0.1 | 0.051 | 0.11 | 0.896 |
|
| |||||
| Residuals | 83 | 38.77 | 0.467 | ||
| Early time points (t = 2 and t = 6): Log (Bact. Density) ~ Time × Treatment × Exp | |||||
| D.f. | Sum. Sq | Mean Sq. | F value | p value | |
|
| |||||
| Time | 1 | 0.022 | 0.022 | 0.072 | 0.789 |
| Treatment | 2 | 2.966 | 1.483 | 4.966 | 0.011 |
| Experiment | 1 | 0.124 | 0.124 | 0.417 | 0.522 |
| Time × Treatment | 2 | 0.872 | 0.436 | 1.461 | 0.242 |
| Time × Exp. | 1 | 0.913 | 0.913 | 3.059 | 0.087 |
| Treatment × Exp. | 1 | 0.350 | 0.350 | 1.173 | 0.284 |
| Full interaction | 1 | 0.051 | 0.051 | 0.170 | 0.682 |
|
| |||||
| Residuals | 48 | 14.335 | 0.299 | ||
Notes: Due to the tri-modality introduced by t = 0 and primed t = 12, data in the global model do not have a gaussian distribution. Data in the early infection analysis are normally distributed. Contrast tests are presented in Table S3.
Primed and unprimed individuals continued to show divergent Bt density dynamics later in the acute infection phase. By 8 hours (assayed in experiment two), unprimed individuals had largely and significantly suppressed their bacterial load by an order of magnitude compared to two and six hours post infection, while primed larvae showed signs of continued increase in bacterial load, although not significantly different from two and six hours. By 12 hours (assayed in experiment one), all surviving unprimed larvae as well as a third of surviving primed larvae had suppressed their bacterial load to less than 100 CFU/individual. However, two thirds of the surviving primed larvae exhibited bacterial loads of 105 CFU/individual, and the primed group overall had a significantly higher bacterial load than any other living group at any other time point (Tukey’s HSD, all adjusted p values <= 0.001). The moribund larvae sampled at 12 hours had a significantly higher bacterial load (around 106 CFU/individual) than any other treatment at any other time point except the 12 hour primed larvae (Tukey’s HSD, all p values <= 0.000001 except: primed moribund vs. primed 12 hours: log10 diff = 1.33, p = 0.33; unprimed moribund vs. primed 12 hours: log10 diff. = 2.17, p = 0.0013). By 24 hours post infection, all but two surviving larvae still had detectable levels of Bt averaging around 102 CFU/individual, but there was no longer a significant difference among treatments (Tukey’s HSD, p = 0.91). Bacterial density patterns for offspring of saline-control mothers did not significantly differ from completely unprimed larvae and are not presented here.
Constitutive differences in gene expression between primed and unprimed individuals
15 genes were significantly differentially expressed (after FDR adjustment, p < 0.05) in uninfected primed individuals compared to unprimed individuals (Table S7). One gene in particular, TC009881, was over-expressed almost 7-fold in all primed individuals relative to unprimed individuals before infection, and an investigation across the entire time series revealed nearly non-overlapping expression distributions between primed and unprimed individuals (Figure 2A). We evaluated differences in expression using qPCR for t = 0 and t = 2 larvae from experiments one and two and adults from experiment two, as well as saline control offspring from experiment one, to demonstrate that this bimodal expression pattern was robust across experiments and life stages (Fig. 3). Intriguingly, the saline group showed intermediate values for the expression of this gene. KEGG pathway enrichment of t = 0 data (Table 2) revealed significant differential expression between primed and unprimed individuals in pathways related to protein production and degradation, and oxidative phosphorylation.
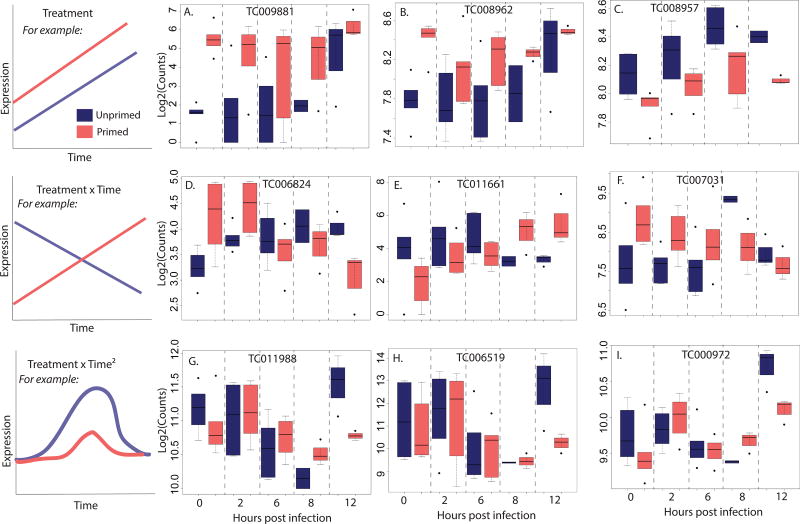
Figure 2.
Modes of differential expression between primed (light/pink) and unprimed (dark/blue) individuals across time, and examples of genes that show significant differential expression conforming to each mode. Gene expression patterns with different intercepts but monotonic slopes may be significant under the main effect of treatment in the generalized linear model fit of mRNA-seq count data by the DESeq2 program in R. For example, A. is TC009881, with an EIF4-e domain, B. is TC008962, an acyltransferase, and C. is TC008957, a protein kinase. Genes may have the same intercept but different rates of induction or decay, or have different intercepts but crossing slopes, and will be best fit by the interaction of treatment and time. The gene TC006824 (D., a POU transcription factor) demonstrates the interaction of time and treatment through nearly orthogonal expression patterns for each treatment over time, and the genes TC011661 (E., a GNBP-like osiris family gene) and TC007031 (F., saposin B domain) also show a treatment × time interaction pattern. Finally, genes that experience induction and decay will be best fit by a quadratic expression for time, such as TC011988 (G., mitochondrial succinate dehydrogenase) TC006519 (H., cytochrome C oxidase) and TC000972 (I., mitochondrial function). Experimental series have been combined for this analysis, and full gene lists for each mode appear in the supplementary tables (Supporting Information).
Differential dynamics of gene expression across time for primed and unprimed individuals
TC009881, a transcript encoding an EIF4-e-like protein, is significantly upregulated in primed individuals prior to infection and is among a handful of genes (Table S8) that remained significant after FDR adjustment for the main effect of treatment across the entire dataset (Fig. 2A–C). Two of the other three genes that were significantly differentially expressed over all time points (TC008962 (Fig. 2B) and TC008957 (Fig. 2C)) co-localize with TC009881 on a 39 kb region of chromosome LG7. Other annotated genes in this region were not differentially expressed.
For the interaction of treatment and time, a gram-negative binding protein (GNBP) (TC011661) that is probably involved in microbial recognition and upstream signaling was significantly upregulated in primed individuals (Fig. 2D), but declined over time compared to unprimed individuals. A handful of additional genes carried marginally significant p-values (Fig. 2E & F; Table S8). Due to the small number of differentially expressed genes for treatment and treatment by time analyses, we did not pursue these modes with KEGG pathway analysis.
The interaction of treatment and quadratic time should capture differences in the integral of gene expression over time between primed and unprimed individuals. An analysis of differential gene expression for this term (Table S9) reveals high representation of components of the TCA cycle, oxidative phosphorylation, peroxisome, and protein and lipid metabolic processes. This pattern was formalized by KEGG pathway functional enrichment analysis in GAGE using log fold change values, corrected for standard error, from the DESeq2 output (Table 2). Oxidative phosphorylation and the TCA cycle are downregulated over time for both treatments, but less so for primed larvae. To ensure that these results were not being driven solely by the t = 12 high primed bacterial load (despite controlling for bacterial load in the statistical model), we visualized the most differentially expressed genes using experiment two data only (Fig. S4). These results supported the larger analysis, showing similar qualitative patterns over time.
Discussion
The T. castaneum response to microbial challenge is characterized by the dynamic regulation of physiological, immunological, and metabolic pathways. Previous studies have annotated putative immune genes in this species (Zou et al. 2007), identified a subset of these genes that are differentially expressed in response to microbial challenge (Altincicek et al. 2008; Yokoi et al. 2012), and even characterized differences in gene expression between oral and septic challenge with Bt (Behrens et al. 2014). However, relatively little is known about the finer scale temporal dynamics of gene expression in response to live infection in this or almost any other insect species, especially as it relates to the manifestation of immune priming and patterns of microbial density at the individual level. In this paper, we describe the differential dynamics of unprimed and primed T. castaneum larval gene expression during the acute phase of infection over two separate experimental series, one that produced no larval mortality and one that killed almost 65% of unprimed larvae. To our knowledge, this is the first study to describe systemic temporal gene expression patterns and identify a potential bio-marker transcript associated with trans-generational immune priming in beetles. Moreover, we provide a large-scale description of the repeatability of trans-generational priming within a study system (Fig. 1A), assuaging concerns (Hauton & Smith 2007; Reber & Chapuisat 2012) surrounding the existence, robustness, and experimental tractability of priming.
The temporal dynamics of bacterial density and gene expression distinguish primed larvae from unprimed larvae
How do primed larvae gain a survival advantage from the observed host-pathogen dynamics? If primed individuals were better able to resist infection primarily through an increased concentration of standing antimicrobial peptides (Barribeau et al. 2016; Sadd & Schmid-Hempel 2007; Zanchi et al. 2012) or more efficient nodule formation at the wound site (Dunn & Drake 1983), then the bacterial load at two hours should be lower for primed larvae. Instead, the differences in pathogen load emerge at 6 hours (Fig. 1D) suggesting that primed individuals may mount a more efficient inducible immune response early (4–6 hours) in the acute infection phase. This dynamic is consistent with observations of enhanced cellular immune responses in within-generation primed mosquitoes (Ramirez et al. 2015; Rodrigues et al. 2010), woodlice (Roth & Kurtz 2009), and fruit flies (Pham et al. 2007), as well as trans-generationally primed honeybees (Hernández López et al. 2014). In many of the within-generation priming studies, this early increase in primed resistance carries over to later time points, where primed individuals continue to sustain lower microbe loads at 24 hours (Pham et al. 2007) and even two weeks (Rodrigues et al. 2010) post challenge.
In our study, however, the increase in resistance is transient in primed larvae, where surviving primed larvae sustain increased bacterial loads from 8 to 12 hours while surviving unprimed larvae largely repress microbial burdens over the same period (Fig. 1C and D). Despite the average increase in bacterial loads, these primed larvae are very unlikely to be destined for death, as no larvae in any treatment died of infection in experiment two, and in experiment one, 95% of larvae that eventually died were clearly moribund by the 12 hour sampling time. These bacterial density patterns suggest that an early resistance advantage may subsequently promote the appearance of tolerance (Medzhitov et al. 2012) in primed larvae, whereas only sufficiently resistant unprimed larvae will survive infection with Bt.
There were surprisingly few differentially expressed genes with known immunological functions between treatments, either before or during infection, after controlling for differences in bacterial load that could confound the main effect of treatment (Tables S7–9). Possible exceptions include TC011661 (Fig. 2E), a probable GNBP gene (Gobert et al. 2003) involved in microbial recognition, and TC007031 (Fig. 2F), a gene with a saposin B domain. In humans, saposin B facilitates the recognition of lipid antigens by NKT cells for lysosomal degradation (Yuan et al. 2007). Both these genes were differentially expressed between treatments interacting with time (Table S8). Notably, no antimicrobial peptides were differentially expressed between treatments before or during infection after controlling for microbe load (the two are positively correlated). This suggests that if AMPs play a role in immune priming in flour beetles as they might in bumblebees (Barribeau et al. 2016), they are not being differentially regulated at the transcriptional level. Since immunological pathways have not been annotated for T. castaneum in the KEGG database, and GO annotation is similarly under-annotated, we could not determine whether the lack of differential expression for individual immune genes might be hiding any subtle shift in aggregate pathway expression.
However, a new hypothesis arises from the KEGG enrichment analysis of uninfected differential expression between trans-generationally primed and unprimed larvae. These results contained a high prevalence of amino-acid and protein processing machinery and raise the question of whether more efficient translation and post-translational modifications may underlie a more robust and effective inducible response.
TC009881, the most differentially expressed gene in primed individuals both before and during infection (Tables S7–8 and Figs. 2A and 3), contains an EIF4-e domain. Eukaryotic translation initiation factor 4E proteins are involved in stabilizing, shuttling, and selectively enhancing the translation of capped mRNAs (Rhoads 2009). Interestingly, they play a crucial role in regulating immune responses in Drosophila (Bernal & Kimbrell 2000) and mammals (Piccirillo et al. 2014). Moreover, an EIF4-e binding protein is significantly differentially expressed in murine macrophages trained (Cheng et al. 2014) against β-glucan, resulting in a post-infection survival advantage similar to invertebrate immune priming. While it is often assumed by transcriptomics studies that expressed transcripts are likely to become expressed proteins in proportion to their abundance, increasing evidence suggests that translational control is at least as important as transcriptional control in modulating the abundance of specific proteins (Khan et al. 2013; Piccirillo et al. 2014). EIF4-e is an important player in producing the immunological “translatome” by selectively favoring the translation of highly structured transcripts that would normally be inefficiently translated. Previous studies have implicated EIF4-e proteins in cell proliferation, macrophage phenotype differentiation, and the mTOR-mediated cellular response to hypoxia (Piccirillo et al. 2014; Rhoads 2009). It is possible that T. castaneum TC009881 plays a similar role in priming hemocyte polarization toward activated, pro-inflammatory phenotypes, and would be consistent with previous observations on the importance of cellular differentiation and increased activity for producing the primed resistance advantage (Pham et al. 2007; Rodrigues et al. 2010).
We attempted to knock down TC009881 in primed mothers to ascertain the functional significance of the differential expression, but the treatment resulted in a dearth of viable offspring, possibly due to pleiotropic effects of this gene on fecundity (Dinkova et al. 2005) or off-target effects (Online Supplementary Methods), and the knockdown effect wore off in the offspring by the time they became old enough to infect. Therefore, a more permanent manipulation like CRISPR-Cas9 may provide a way forward for future investigation into the function of this gene in the context of trans-generational priming. However, two other genes significantly differentially expressed between primed and unprimed individuals over the entire time series (Table S8) lie within a small region of ChLG7, suggesting that this region could be groomed for enhanced transcription through chromatin modification or some other epigenetic mechanism. Interestingly, offspring from saline-control mothers showed intermediate expression of TC009881 (Fig. 3), possibly reflecting a partial signal of environmental stress or contamination of the maternal wound site with bacteria from the cuticle. We would like to emphasize that while we consider TC009881 to be an interesting biomarker for distinguishing primed from unprimed individuals, this gene is still just a candidate for the causality between expression and primed protection since our attempts to functionally validate it were met with biological difficulty. However, these genes and chromosomal region do represent salient targets for future studies investigating the cellular processes underlying trans-generational immune priming.
Trans-generational immune priming and metabolism
A large proportion of differentially expressed genes from uninfected individuals and for the integral of expression over time (quadratic time by treatment analysis) after infection are associated with metabolic functions. When we pursued this observation through KEGG pathway analysis, we found that differences between treatments were enriched for oxidative phosphorylation both before and during infection. In addition, components of the TCA cycle also showed differential induction and decay patterns between treatments over time. After infection, both primed and unprimed larvae show a quadratic down-regulation of oxidative phosphorylation and the TCA cycle (e.g. Fig. 2G–I and Table 2) over time. These expression patterns are in firm agreement with major patterns observed in Anopheles gambiae after sterile and gram-positive septic injury (Dimopoulos et al. 2002). In mitochondria, the TCA cycle processes nutrients to supply NADH and succinate to the oxidative phosphorylation cascade, which in turn produces ATP and oxygen free radicals. The down-regulation of oxidative phosphorylation is common in mammalian models of sepsis (Lee & Hüttemann 2014), and is associated with poor outcomes due to the death of energy-starved cells (Bauerfeld et al. 2012) in key organs. It is not entirely clear why organisms suppress oxidative phosphorylation during infection, but it may be to prevent excessive pathology induced by free radicals, or to prevent microbes from acquiring the energy produced in this cascade (Lee & Hüttemann 2014). The insect phenoloxidase and melanization cascade responses generate high levels of reactive oxygen species (Kumar et al. 2003), and so downregulating oxidative phosphorylation may be a general counter-response to high ROS levels induced by infection. Finally, hypoxic conditions within the cell may also prevent the use of oxidative phosphorylation and force the cell to use glycolysis to fuel its metabolic needs (Pearce & Pearce 2013).
Our results suggest that primed larvae may be able to mitigate the metabolic dysfunction commonly observed during acute infection and provide more energy to fuel important physiological and immunological processes. Whether the shift in energy production comes at a cost of potentially harmful reactive oxygen species, increased microbial growth, or trade-offs with other life history traits like longevity is an interesting question that requires further study. However, it is worth noting that similar metabolic shifts are observed in β-glucan-trained macrophages in mice (Cheng et al. 2014). These shifts are thought to increase the activation and metabolic endurance of the macrophages during the acute infection phase, increasing overall antimicrobial resistance. Our results invite further investigation into a potential metabolic homology between invertebrate immune priming and innate trained immunity in mammals.
Priming increases resistance, but could it also increase infection tolerance?
Our bacterial density data suggest that primed individuals could transiently tolerate elevated bacterial loads and still survive to a time point by which most others have died, while unprimed individuals succumbed to infection unless they succeeded in quickly resisting it. If priming increases the metabolic endurance or energy management strategies of host cells, then primed individuals may be able to avoid the catastrophic organ failure that precedes mortality during septic infections. This could lead to increased survival despite high bacterial burdens until later stage antimicrobial peptide responses can take effect (Johnston et al. 2013) and ultimately knock back microbial density to 24 hour post infection levels. Alternately, microbial burden measured at a single time point may give a misleading impression of infection tolerance. Surviving primed individuals at twelve hours may have had to deal with high microbe burdens for less time than moribund individuals due to the resistance-induced delay in exponential microbial growth. Thus, the integral of microbe load over time may influence infection virulence in this system (Regoes et al. 2002). Potential causal links between infection resistance and point tolerance during the acute infection phase represent an interesting avenue for future investigation.
For a microbe like Bt that must kill its host in order to be transmitted (Garbutt et al. 2011; Raymond et al. 2010), priming could reduce microbial fitness through multiple avenues. By increasing survival, priming reduces the chance of any transmission, and even in primed individuals who do succumb, the microbe burden may be lower and reduce the force of infection. Thus, priming could interfere with the epidemic trajectories of entomopathogens and potentially place pressure on the evolution of microbial growth and virulence characteristics. The co-evolutionary implications of immune priming demand further theoretical and empirical study.
Supplementary Material
Table S1: Primers used for qPCR
Table S2: Summary of priming experiments used in aggregate survival analysis of priming
Table S3: RNA-seq converage depth, quality control, and alignment statistics
Table S4: Generalized linear mixed model statistics for 24 experiment survival analysis
Table S5: Hazard ratios for survival within infection and treatment factors
Table S6: Bacterial density contrast tests for time, treatment, and their interaction
Table S7: Genes differentially expressed in uninfected primed and unprimed larvae
Table S8: Genes differentially expressed for the main effect of treatment and time × treatment interaction
Table S9: Genes differentially expressed for Treatment × Time^2, full time series
Table S10: Primers used to construct dsRNA for RNAi experiments as describe in the online supporting information
Acknowledgments
We would like to thank Lance Parsons, Molly Schumer, and Ying Zhen for help with library prep and bioinformatics, Catherine Rodgers and Heath Blackmon for laboratory assistance for the RNAi experiment, Tina Hansen for general laboratory assistance, and Tim Cooper and two anonymous reviewers for comments that improved the manuscript. A.T.T. was supported by Agriculture and Food Research Initiative Competitive Grants, No. 2012-67011-19893 and No. 2014-67012-22278, from the USDA National Institute of Food and Agriculture. P.A. was partially supported by NIH R01 5R01GM083228.
Footnotes
Data Accessibility
The mRNA-seq experiment data have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al. 2002) and are accessible through GEO Series accession number GSE95387 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE95387). The qPCR data for microbe load and gene expression are deposited in Data Dryad under DOI: http://dx.doi.org/10.5061/dryad.hq6c7.
Author Contributions
A.T.T. conceived the experiments, A.T.T., A.L.G., P.A., and J.D. designed the experiments, P.A., A.L.G., and J.D. contributed reagents and resources, A.T.T. performed the experiments, A.T.T. analyzed the data., and A.T.T., A.L.G., J.D., and P.A. wrote the manuscript.
References
- Altincicek B, Knorr E, Vilcinskas A. Beetle immunity: Identification of immune-inducible genes from the model insect Tribolium castaneum. Developmental & Comparative Immunology. 2008;32:585–595. doi: 10.1016/j.dci.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq–A Python framework to work with high-throughput sequencing data. bioRxiv. 2014 doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. FastQC: A quality control tool for high throughput sequence data. Reference Source 2010 [Google Scholar]
- Ayres JS, Schneider DS. Tolerance of Infections. Annual Review of Immunology. 2012;30:271–294. doi: 10.1146/annurev-immunol-020711-075030. [DOI] [PubMed] [Google Scholar]
- Barribeau SM, Schmid-Hempel P, Sadd BM. Royal Decree: Gene Expression in Trans-Generationally Immune Primed Bumblebee Workers Mimics a Primary Immune Response. PloS One. 2016;11:e0159635. doi: 10.1371/journal.pone.0159635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerfeld CP, Rastogi R, Pirockinaite G, et al. TLR4-Mediated AKT Activation Is MyD88/TRIF Dependent and Critical for Induction of Oxidative Phosphorylation and Mitochondrial Transcription Factor A in Murine Macrophages. The Journal of Immunology. 2012;188:2847–2857. doi: 10.4049/jimmunol.1102157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens S, Peuss R, Milutinovi B, et al. Infection routes matter in population-specific responses of the red flour beetle to the entomopathogen Bacillus thuringiensis. BMC Genomics. 2014;15:445. doi: 10.1186/1471-2164-15-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal A, Kimbrell DA. Drosophila Thor participates in host immune defense and connects a translational regulator with innate immunity. Proceedings of the National Academy of Sciences. 2000;97:6019–6024. doi: 10.1073/pnas.100391597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Kuster GV, Coraor N, et al. Current Protocols in Molecular Biology. John Wiley & Sons, Inc; 2001. Galaxy: A Web-Based Genome Analysis Tool for Experimentalists. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S-C, Quintin J, Cramer RA, et al. mTOR-and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345:1250684. doi: 10.1126/science.1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras-Garduño J, Rodríguez MC, Rodríguez MH, Alvarado-Delgado A, Lanz-Mendoza H. Cost of immune priming within generations: trade-off between infection and reproduction. Microbes and Infection. 2014;16:261–267. doi: 10.1016/j.micinf.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Dimopoulos G, Christophides GK, Meister S, et al. Genome expression analysis of Anopheles gambiae: Responses to injury, bacterial challenge, and malaria infection. Proceedings of the National Academy of Sciences. 2002;99:8814–8819. doi: 10.1073/pnas.092274999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova TD, Keiper BD, Korneeva NL, Aamodt EJ, Rhoads RE. Translation of a small subset of Caenorhabditis elegans mRNAs is dependent on a specific eukaryotic translation initiation factor 4E isoform. Molecular and Cellular Biology. 2005;25:100–113. doi: 10.1128/MCB.25.1.100-113.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn PE, Drake DR. Fate of bacteria injected into naive and immunized larvae of the tobacco hornworm Manduca sexta. Journal of Invertebrate Pathology. 1983;41:77–85. [Google Scholar]
- Durinck S, Moreau Y, Kasprzyk A, et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbutt J, Bonsall MB, Wright DJ, Raymond B. Antagonistic competition moderates virulence in Bacillus thuringiensis. Ecology Letters. 2011;14:765–772. doi: 10.1111/j.1461-0248.2011.01638.x. [DOI] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, et al. Galaxy: A platform for interactive large-scale genome analysis. Genome Research. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert V, Gottar M, Matskevich AA, et al. Dual Activation of the Drosophila Toll Pathway by Two Pattern Recognition Receptors. Science. 2003;302:2126–2130. doi: 10.1126/science.1085432. [DOI] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J, Team TG. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biology. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon A, Hannon G. Fastx-toolkit. FASTQ/A short-reads preprocessing tools (unpublished) 2010 http://hannonlab.cshl.edu/fastx_toolkit.
- Graham AL, Shuker DM, Pollitt LC, et al. Fitness consequences of immune responses: strengthening the empirical framework for ecoimmunology. Functional Ecology. 2011;25:5–17. [Google Scholar]
- Hauton C, Smith VJ. Adaptive immunity in invertebrates: A straw house without a mechanistic foundation. BioEssays. 2007;29:1138–1146. doi: 10.1002/bies.20650. [DOI] [PubMed] [Google Scholar]
- Hernández López J, Schuehly W, Crailsheim K, Riessberger-Gallé U. Trans-generational immune priming in honeybees. Proceedings of the Royal Society B: Biological Sciences. 2014;281 doi: 10.1098/rspb.2014.0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston PR, Makarova O, Rolff J. Inducible Defenses Stay Up Late: Temporal Patterns of Immune Gene Expression in Tenebrio molitor. G3: Genes|Genomes|Genetics. 2013 doi: 10.1534/g3.113.008516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan Z, Ford MJ, Cusanovich DA, et al. Primate Transcript and Protein Expression Levels Evolve under Compensatory Selection Pressures. Science (New York, N.Y.) 2013;342:1100–1104. doi: 10.1126/science.1242379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Christophides GK, Cantera R, et al. The role of reactive oxygen species on Plasmodium melanotic encapsulation in Anopheles gambiae. Proceedings of the National Academy of Sciences. 2003;100:14139–14144. doi: 10.1073/pnas.2036262100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Hüttemann M. Energy crisis: The role of oxidative phosphorylation in acute inflammation and sepsis. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2014;1842:1579–1586. doi: 10.1016/j.bbadis.2014.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little TJ, O’Connor B, Colegrave N, Watt K, Read AF. Maternal transfer of strain-specific immunity in an invertebrate. Current Biology. 2003;13:489–492. doi: 10.1016/s0960-9822(03)00163-5. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. bioRxiv. 2014 doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Schneider DS, Soares MP. Disease Tolerance as a Defense Strategy. Science. 2012;335:936–941. doi: 10.1126/science.1214935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milutinović B, Fritzlar S, Kurtz J. Increased survival in the red flour beetle after oral priming with bacteria-conditioned media. Journal of Innate Immunity. 2013;6:306–314. doi: 10.1159/000355211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce Erika L, Pearce Edward J. Metabolic Pathways in Immune Cell Activation and Quiescence. Immunity. 2013;38:633–643. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham LN, Dionne MS, Shirasu-Hiza M, Schneider DS. A specific primed immune response in Drosophila Is dependent on phagocytes. PLoS Pathog. 2007;3:e26. doi: 10.1371/journal.ppat.0030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo CA, Bjur E, Topisirovic I, Sonenberg N, Larsson O. Translational control of immune responses: from transcripts to translatomes. Nat Immunol. 2014;15:503–511. doi: 10.1038/ni.2891. [DOI] [PubMed] [Google Scholar]
- Posnien N, Schinko J, Grossmann D, et al. RNAi in the Red Flour Beetle (Tribolium) Cold Spring Harb Protoc. 2009;2009:pdb.prot5256-. doi: 10.1101/pdb.prot5256. [DOI] [PubMed] [Google Scholar]
- Povey S, Cotter SC, Simpson SJ, Lee KP, Wilson K. Can the protein costs of bacterial resistance be offset by altered feeding behaviour? Journal of Animal Ecology. 2009;78:437–446. doi: 10.1111/j.1365-2656.2008.01499.x. [DOI] [PubMed] [Google Scholar]
- Ramirez JL, de Almeida Oliveira G, Calvo E, et al. A mosquito lipoxin/lipocalin complex mediates innate immune priming in Anopheles gambiae. Nat Commun. 2015;6 doi: 10.1038/ncomms8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond B, Johnston PR, Nielsen-LeRoux C, Lereclus D, Crickmore N. Bacillus thuringiensis: an impotent pathogen? Trends in Microbiology. 2010;18:189–194. doi: 10.1016/j.tim.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Reber A, Chapuisat M. No Evidence for Immune Priming in Ants Exposed to a Fungal Pathogen. PloS One. 2012;7:e35372. doi: 10.1371/journal.pone.0035372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regoes RR, Staprans SI, Feinberg MB, Bonhoeffer S. Contribution of Peaks of Virus Load to Simian Immunodeficiency Virus Pathogenesis. Journal of Virology. 2002;76:2573–2578. doi: 10.1128/jvi.76.5.2573-2578.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads RE. eIF4E: new family members, new binding partners, new roles. Journal of Biological Chemistry. 2009;284:16711–16715. doi: 10.1074/jbc.R900002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards Sale. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- Rodrigues J, Brayner FA, Alves LC, Dixit R, Barillas-Mury C. Hemocyte Differentiation Mediates Innate Immune Memory in Anopheles gambiae Mosquitoes. Science. 2010;329:1353–1355. doi: 10.1126/science.1190689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth O, Joop G, Eggert H, et al. Paternally derived immune priming for offspring in the red flour beetle, Tribolium castaneum. Journal of Animal Ecology. 2010;79:403–413. doi: 10.1111/j.1365-2656.2009.01617.x. [DOI] [PubMed] [Google Scholar]
- Roth O, Kurtz J. Phagocytosis mediates specificity in the immune defence of an invertebrate, the woodlouse Porcellio scaber (Crustacea: Isopoda) Developmental & Comparative Immunology. 2009;33:1151–1155. doi: 10.1016/j.dci.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Roth O, Sadd BM, Schmid-Hempel P, Kurtz J. Strain-specific priming of resistance in the red flour beetle, Tribolium castaneum. Proceedings of the Royal Society B: Biological Sciences. 2009;276:145–151. doi: 10.1098/rspb.2008.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadd BM, Schmid-Hempel P. Facultative but persistent trans-generational immunity via the mother’s eggs in bumblebees. Current Biology. 2007;17:R1046–R1047. doi: 10.1016/j.cub.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Sadd BM, Schmid-Hempel P. A distinct infection cost associated with trans-generational priming of antibacterial immunity in bumble-bees. Biology Letters. 2009;5:798–801. doi: 10.1098/rsbl.2009.0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadd BM, Siva-Jothy MT. Self-harm caused by an insect’s innate immunity. Proceedings of the Royal Society B: Biological Sciences. 2006;273:2571–2574. doi: 10.1098/rspb.2006.3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmela H, Amdam GV, Freitak D. Transfer of Immunity from Mother to Offspring Is Mediated via Egg-Yolk Protein Vitellogenin. PLoS Pathog. 2015;11:e1005015. doi: 10.1371/journal.ppat.1005015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha S, Kim Y. Activation of immune-associated phospholipase A2 is functionally linked to Toll/Imd signal pathways in the red flour beetle, Tribolium castaneum. Developmental & Comparative Immunology. 2010;34:530–537. doi: 10.1016/j.dci.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Tate AT, Graham AL. Trans-generational priming of resistance in wild flour beetles reflects the primed phenotypes of laboratory populations and is inhibited by co-infection with a common parasite. Functional Ecology. 2015;29:1059–1069. [Google Scholar]
- Team RC. R: A language and environment for statistical computing 2012 [Google Scholar]
- Therneau T. A Package for Survival Analysis in S. R package version 2.37-7 2014 [Google Scholar]
- Trauer-Kizilelma U, Hilker M. Insect parents improve the anti-parasitic and anti-bacterial defence of their offspring by priming the expression of immune-relevant genes. Insect Biochemistry and Molecular Biology. 2015;64:91–99. doi: 10.1016/j.ibmb.2015.08.003. [DOI] [PubMed] [Google Scholar]
- Yokoi K, Koyama H, Ito W, et al. Involvement of NF-κB transcription factors in antimicrobial peptide gene induction in the red flour beetle, Tribolium castaneum. Developmental & Comparative Immunology. 2012;38:342–351. doi: 10.1016/j.dci.2012.06.008. [DOI] [PubMed] [Google Scholar]
- Yuan W, Qi X, Tsang P, et al. Saposin B is the dominant saposin that facilitates lipid binding to human CD1d molecules. Proceedings of the National Academy of Sciences. 2007;104:5551–5556. doi: 10.1073/pnas.0700617104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanchi C, Troussard J-P, Martinaud G, Moreau J, Moret Y. Differential expression and costs between maternally and paternally derived immune priming for offspring in an insect. Journal of Animal Ecology. 2011;80:1174–1183. doi: 10.1111/j.1365-2656.2011.01872.x. [DOI] [PubMed] [Google Scholar]
- Zanchi C, Troussard J-P, Moreau J, Moret Y. Relationship between maternal transfer of immunity and mother fecundity in an insect. Proceedings of the Royal Society B: Biological Sciences. 2012:rspb20120493. doi: 10.1098/rspb.2012.0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Evans J, Lu Z, et al. Comparative genomic analysis of the Tribolium immune system. Genome Biology. 2007;8:R177. doi: 10.1186/gb-2007-8-8-r177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Primers used for qPCR
Table S2: Summary of priming experiments used in aggregate survival analysis of priming
Table S3: RNA-seq converage depth, quality control, and alignment statistics
Table S4: Generalized linear mixed model statistics for 24 experiment survival analysis
Table S5: Hazard ratios for survival within infection and treatment factors
Table S6: Bacterial density contrast tests for time, treatment, and their interaction
Table S7: Genes differentially expressed in uninfected primed and unprimed larvae
Table S8: Genes differentially expressed for the main effect of treatment and time × treatment interaction
Table S9: Genes differentially expressed for Treatment × Time^2, full time series
Table S10: Primers used to construct dsRNA for RNAi experiments as describe in the online supporting information