Abstract
As a consequence of recent discoveries of intimate involvement of mitochondria with key cellular processes, there has been a resurgence of interest in all aspects of mitochondrial biology, including the intricate mechanisms of mitochondrial DNA maintenance and expression. Despite four decades of research, there remains a lot to be learned about the processes that enable transcription of genetic information from mitochondrial DNA to RNA, as well as their regulation. These processes are vitally important, as evidenced by the lethality of inactivating the central components of mitochondrial transcription machinery. Here, we review the current understanding of mitochondrial transcription and its regulation in mammalian cells. We also discuss key theories in the field and highlight controversial subjects and future directions as we see them.
Keywords: Mitochondrial DNA, Mitochondrial Promoters, Processing, Regulation, TFAM, Transcription, Transcriptome, Review
2. INTRODUCTION
In mammalian cells, mitochondria generate the bulk of ATP required to sustain diverse cellular processes while also playing important roles in intracellular calcium signaling (1), apoptosis (2), reactive oxygen species (ROS) production (3), biosynthesis of heme and iron-sulfur clusters (4, 5), and other processes. In mammalian cells, mitochondrial DNA (mtDNA) is a covalently closed circular molecule that encodes 37 “formal” genes: 13 polypeptide components of the oxidative phosphorylation system (OXPHOS), two rRNAs and 22 tRNAs (Figure 1). The rRNAs and tRNAs, together with a number of nuclearly encoded and posttranslationally imported proteins, form a separate mitochondrial translational apparatus. The need for a separate translational apparatus in mitochondria is dictated by the fact that the 13 mitochondrial mRNAs are encoded using a genetic code distinct from that used by nuclear genes (6). In addition to 37 “formal” genes, mtDNA encodes at least two peptides, human in (7) and MOTS-c (8, 9), whose expression and function remain to be fully elucidated.
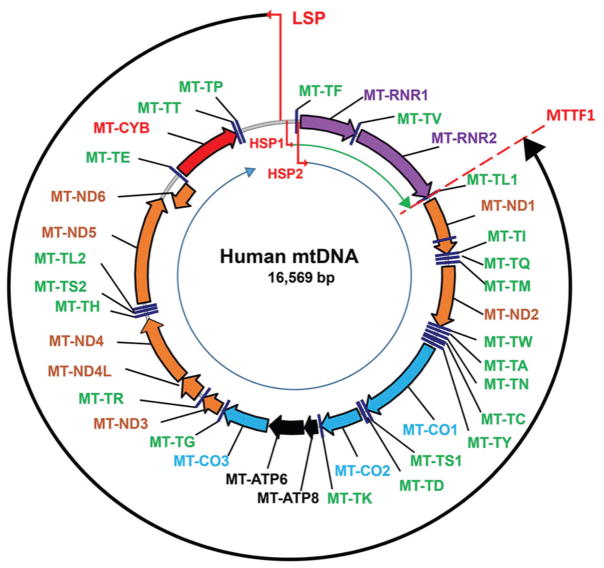
Figure 1.
The map of human mtDNA. The clockwise orientation is according to the revised Cambridge Reference Sequence (rCRS, GenBank NC_012920.1.). Thin red bent arrows designate mitochondrial promoters that transcribe either H-strand (HSP1 and HSP2) or L-strand (LSP). Thin black and blue circular arrows, LSP and HSP2 transcripts, respectively. Thin green arch arrow, HSP1 transcript. Brown arrows: MT-ND1, -2, -3, -4, -5, -6, NADH dehydrogenase subunits 1 through 6. Purple arrows: MT-RNR1 and MT-RNR2, 12S and 16S rRNA genes. Blue arrows: MT-CO1-MT-CO3, cytochrome oxidase subunits 1 through 3. Black arrows: MT-ATP6 and MT-ATP8, subunits 6 and 8 of the mitochondrial ATPase, Red arrow: MT-CYB, cytochrome b. tRNAs are designated by green labels as per HGNC. Red broken line through MT-TL1, position of the bidirectional transcription terminator within MT-TL1. The putative D-Term transcription terminator is not shown as it has been described in murine mtDNA only.
All mitochondrial functions are linked, directly or indirectly, to OXPHOS. Therefore, mitochondrial transcription is a key to proper mitochondrial function as it both represents the means for the expression of mtDNA-encoded OXPHOS subunits, and is required for mtDNA replication (10, 11). Despite considerable progress made in understanding mitochondrial transcription in mammalian cells, even today, more than 40 years since the pioneering studies on mtDNA transcription (12, 13), the field remains full of challenges and controversies, which we will attempt to outline in this review along with current state of the field.
3. THE MAIN mtDNA CODING STRAND: HEAVY OR LIGHT?
One long standing terminological confusion in the field of mtDNA transcription is that of the main coding mtDNA strand. In 1981, Anderson et al. (14) and Bibb et al. (15) published sequences of human and mouse mitochondrial genomes, respectively. Even though the organization of the two genomes is identical, the authors used different notations to designate coding strands. Anderson et al. used a definition of the coding strand identical to the contemporary one and stated that “The sequence shown is that of the L strand which we define as the main coding strand containing the sense sequence of the rRNAs and most of the tRNAs and mRNAs. Thus, these RNAs are transcribed from (and therefore hybridize to) the H strand” (14). In contrast, Bibb et al. used a definition of the coding strand that is identical to today’s definition of a template (noncoding) strand and stated “The major coding strand of mouse mitochondrial DNA is the heavy (H) strand; most transcripts are therefore of light (L)- strand sequence and are complementary to the H strand. The sequence shown is L-strand sequence”. Therefore, both studies agree in that sequences of most mitochondrial transcripts are identical to those found in the L-strand of mtDNA, and hence in today’s terms, the L-strand of mtDNA is the main coding strand. Also, Bibb et al. (15) depicted mtDNA organization in the form of a circle with a counterclockwise order of mtDNA-encoded transcripts. This notation has since become obsolete, and newly sequenced mitochondrial genomes are annotated on their light strand, with clockwise annotation of encoded genes. Therefore, in our review we used this latter notation (Figure 1). However, it should be noted that due to the enormous contributions of the labs of Clayton and Attardi (who used the same notation as Bibb (16)), Bibb’s notation currently dominates the fields of mammalian mtDNA transcription and replication (17).
4. MITOCHONDRIAL TRANSCRIPTION: TWO OR THREE PROMOTERS IN mtDNA?
Genetic information is encoded on both strands of mtDNA. Therefore, at least two promoters are needed to drive mitochondrial transcription: one for the H-strand, and one for the L-strand. However, while the existence of a single light strand promoter (LSP) is generally accepted, the exact number of heavy strand promoters (HSP) remains controversial. In their seminal study, Ojala et al. noted about 15- to 60-fold higher abundance and 50–100 fold higher rate of synthesis of the 12S mitochondrial rRNA transcript (MT-RNR1) as compared to the most abundant mRNA transcript encoded by the same mtDNA strand (16, 18). They outlined two possible explanations: a) the existence of two HSPs, and b) premature termination downstream of the mitochondrial 16S rRNA (MT-RNR2) (Figure 1). Strictly speaking, these two explanations are not necessarily mutually exclusive, and subsequent studies by the same group provided support for both models. Indeed, very soon two distinct H-strand transcription initiation sites were discovered: one at bp 561 of human mtDNA, 16 nucleotides upstream of the MT-TF gene, and a second at bp 646, just 2 nucleotides upstream of the MT-RNR1 gene and inside the MT-TF gene (Figure 1) (19–21). The existence of these two sites was confirmed by two techniques: 1) 5′ end labeling of in vivo HeLa cell mtDNA nascent transcripts with (α-32P)-GTP and guanylyltransferase, followed by an S1 nuclease protection assay and analysis of the capped products by polyacrylamide/urea gel electrophoresis, and by 2) primer extension by reverse transcriptase using a mitochondrial lysate. Promoters responsible for the synthesis of the two transcripts were designated HSP1 and HSP2, respectively. Importantly, in vitro, MTERF1 stimulates HSP1, but not HSP2 activity (21), further supporting the existence of two independent H-strand promoters. The activity of both HSP promoters can also be observed in an in vitro system, although in this system the major transcription start site of HSP2 maps to A644 instead C646 (22, 23). While the in vivo and in vitro transcription start sites of HSP2 are in reasonable agreement, this discrepancy raises an interesting possibility of regulation of the choice of HSP2 transcription start site by extrinsic factors like template topology, additional transacting factors, and/or nucleotide pools (22). This evidence notwithstanding, an argument has been made in favor of one HSP promoter model (17, 24). This argument is based on the observation that MTERF1 knockout mice are viable and do not manifest changes in the relative abundance of mitochondrial HSP1 and HSP2 transcripts, which would be expected if MTERF1 indeed selectively stimulated HSP1 transcription by DNA looping (21). However, it can be argued that since MTERF1 has not been implicated in the regulation of HSP2 transcription, results with MTERF1 knockouts cannot have any bearing on the question of whether HSP2 exists. These results can only challenge the model for stimulation of HSP1 activity by DNA looping with the help of MTERF1. Moreover, even though two major HSP transcripts are observed in both human and mouse cells, transcription start sites in murine mtDNA, unlike human mtDNA, are closely spaced (Figure 2), raising the possibility that they are a result of alternative choice of transcription initiation sites from a single HSP promoter. In other words, caution should be exercised when trying to extrapolate observations made in human mitochondrial transcription system into murine cells and vice versa, even though an assumption that these two systems have very similar or identical regulation appears plausible. In fact, some evidence suggests that regulation of at least LSP1 may be different in human and murine cells. Indeed, mLSP1 was reported to have more extensive requirements for maximal activity in vitro in the upstream sequences than hLSP (Figure 2 and (25)). To summarize, the available evidence does not allow us to rule out the existence of two separate HSP promoters in human cells in vivo.
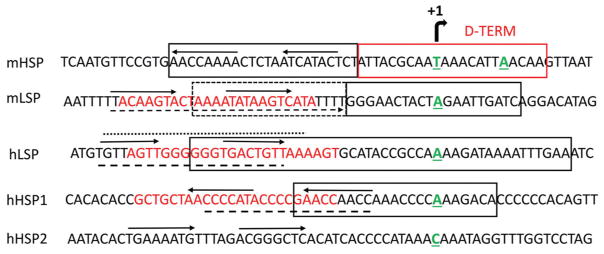
Figure 2.
mtDNA organization around mitochondrial promoters in mouse and human cells. Nontemplate strand is shown. Transcription start sites are aligned and designated by a bent arrow at the top. 5′ nucleotides of mitochondrial transcripts are green, bold and underlined (human and mouse HSP1 and LSP initiating nucleotides as per (96) and (63), respectively. hHSP2 as per (21)). Promoter sequences are enclosed into solid black boxes (hHSP1 and hLSP as per (96), mLSP and mHSP as per (25)). The sequence for the putative murine transcription termination site in the D-loop, D-TERM, is enclosed into a solid red box (46). Horizontal arrows, putative HMG binding sequences and their direction as per (59). Broken box, sequences that are important for preinitiation complex formation at mLSP as per (25). Red font, TFAM footprints as per (29). Broken lines underline TFAM footprints as per (58). Dotted line over the sequence, TFAM footprint as per (60). Broken arrow under the sequence, TFAM footprint as per (62).
5. THE CORE MITOCHONDRIAL TRANSCRIPTION MACHINERY CLUB: IS TFAM A MEMBER?
In vitro, transcription at mitochondrial promoters can be reconstituted using only three proteins: mitochondrial transcription factor A (TFAM), mitochondrial transcription factor B2 (TFB2M) and mitochondrial RNA polymerase (POLRMT). However, specific transcription initiation at HSP1, HSP2 and LSP promoters has been demonstrated in vitro in the absence of TFAM (22, 23, 26–29), which led to a two-component model in which TFAM is ascribed the role of transcriptional activator instead of being an obligate core component of the transcription initiation complex (27). This model, nevertheless, was challenged on the grounds that the experimental conditions used in those assays may not faithfully recapitulate those found in vivo because they favor promoter “breathing” (i.e. spontaneous separation of DNA strands) (28). It should be noted, however, that every in vitro system is a mere approximation of in vivo conditions and therefore none of them faithfully recapitulate the full complexity of the intracellular or intramitochondrial milieu in terms of the exact roster and concentrations of players. Therefore, the ultimate test of any model is in vivo. In vivo, homozygous TFAM knockout is embryonically lethal (30), which can be interpreted either in support of the notion that TFAM is an essential component of the mitochondrial transcription apparatus, or may be viewed as a mere reflection of the fact that altered mitochondrial transcription in the absence of TFAM disrupts the developmental program in a manner that is incompatible with embryo’s survival. Tissue-specific TFAM knockouts also produced inconclusive results: in one study, mice with TFAM ablation in the heart and skeletal muscle died from dilated cardiomyopathy 2–4 weeks after birth (31). This lethality was accompanied by reduced TFAM protein and mitochondrial transcript levels in the heart and skeletal muscle and by reduced activity of the respiratory complexes in the heart, but not in the skeletal muscle (31). The reduced, rather than absent, levels of TFAM and mitochondrial transcripts in hearts appears to be in good agreement with the presence of nonmuscle cells. However, the wild type activities of respiratory complexes I and IV in the skeletal muscle of TFAM KO mice are difficult to reconcile with the proposed essential role of TFAM in mitochondrial transcription and with the fact that the activities of both complexes critically depend on mitochondrially-encoded subunits. This may be a result of the mitochondrial transcription in the absence of TFAM, which agrees with a two-component model of mitochondrial transcription, which leaves some room for a two-component model of mitochondrial transcription. In summary, both proponents and critics of the two-component model agree that under certain conditions, POLRMT and TFB2M can initiate transcription at mitochondrial promoters in the absence of TFAM, and the only disagreement is in whether such initiation is biologically relevant (i.e. whether it contributes to mitochondrial transcription in vivo).
6. MITOCHONDRIAL TRANSCRIPTION: STEP BY STEP
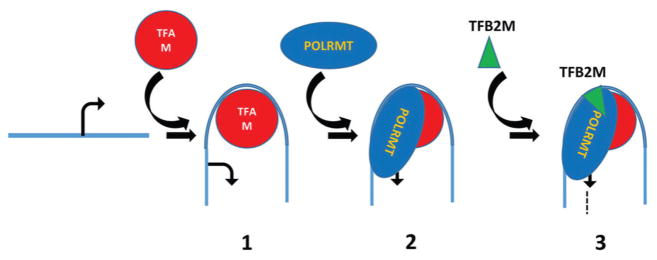
Mitochondrial transcription consists of the three stages: initiation, elongation, and termination. These stages remain incompletely understood, with transcription initiation being the most controversial due to the debated role of TFAM (see above). According to the prevailing model, transcription initiation in mitochondria proceeds through three steps (Figure 3):
Figure 3.
A model for mitochondrial transcription initiation. 1. TFAM binds to a high affinity binding site upstream of a mitochondrial promoter (designated by the bent arrow at the transcription start site) and induces a sharp bend in mtDNA; 2. POLRMT is recruited to the mtDNA/TFAM complex presumably through interaction with TFAM CTD; 3. Recruitment of the TFB2M to mtDNA/TFAM/POLRMT ternary complex facilitates promoter melting, recruitment of the initiating nucleotide, and initiates RNA synthesis (broken line).
TFAM first binds to high-affinity binding sites upstream of HSP1 and LSP promoters (32) and induces a sharp bend in mtDNA (33–35). These sites have been mapped to position -12-35 relative to the LSP and HSP1 transcription start sites by DNAse I footprinting (29). A 180° bend in the LSP promoter region may be important for the proper positioning of mitochondrial transcription machinery relative to the transcription start site (36).
The TFAM-mtDNA complex recruits POLRMT through interactions localized to the C-terminal domain (CTD) of TFAM, thus forming a pre-initiation complex (pre-IC) (37).
TFB2M recruitment to the pre-IC facilitates promoter melting and recruitment of the priming substrate to the catalytic site of POLRMT (38).
Once transcription has started, TFB2M is believed to dissociate from POLRMT, initiating the elongation step (38, 39). This step is conducted by the elongation complex, which likely includes, along with POLRMT, a mitochondrial transcription elongation factor (TEFM, (40)) and possibly a helicase. TEFM increases the processivity of POLRMT and, in the case of LSP-mediated transcription, TEFM facilitates bypass by POLRMT of a G-quadruplex structure in the human conserved sequence block II (CSBII). It has been proposed that this bypass regulates the switch between mtDNA replication and transcription (41). When TEFM is unavailable, transcription termination around CSBII creates a primer for mtDNA replication. When TEFM is present, POLRMT bypasses CSBII and continues synthesis of a near-genomic length polycistronic transcript.
Transcription termination in mitochondria remains poorly understood. Precise 3′ ends of near-genomic length transcripts from HSP2 and LSP have not been mapped, although it is likely that the LSP transcript terminates within MT-TL1 (see below). For the HSP1 transcript, a termination site has been established within MT-TL1. One factor responsible for transcription termination at this position is MTERF1 (42, 43). Recombinant MTERF1 acts bidirectionally (i.e. mediates transcription termination on both strands) and promotes almost complete transcription termination at a molar ratio slightly above 1:1 of protein to DNA template (44). To effect transcription termination, MTERF1 binds along the major groove of mtDNA, imposes a 25° bend, partially unwinds the mtDNA, and induces eversion of the three nucleotides within its binding site (45). This eversion stabilizes protein binding to mtDNA and is critical for efficient termination. Therefore, MTERF1 is likely to terminate transcription through interference with the transcription elongation machinery. Interestingly, MTERF1 is a more efficient terminator in the orientation that promotes LSP transcript termination than it is in the orientation that terminates the HSP1 transcript (44, 45), suggesting that both HSP1 and LSP transcripts in human mtDNA may terminate around the same transcription termination site within MT-TL1, at least in human cells. In murine cells, a second putative transcription termination site for H-strand transcription has been described within the D-loop region, immediately upstream of MT-TF. This site, D-TERM, maps to nucleotides 16274–16295 of the mouse genome (GenBank J01420), lies immediately downstream of the HSP promoter and overlaps with both HSP transcription initiation sites (Figure 2). Two major proteins of 45 kDa and 70 kDa can be purified by DNA affinity chromatography using the 22 bp D-TERM sequence. Mouse D-TERM can mediate transcription termination in a unidirectional manner in a human mitochondrial transcription system, only in the presence of D-TERM DNA binding proteins purified from mouse liver (46).
Apart from MTERF1, three other members of the MTERF family, MTERF2, MTERF3 and MTERF4, have been identified (47). These proteins share similar structure but do not appear to contribute to mitochondrial transcription termination in vivo. Instead, MTERF3 appears to be a negative regulator of mitochondrial transcription (48) and MTERF4 may be involved in the assembly of mitoribosomes (49)
7. COMPONENTS OF THE MITOCHONDRIAL TRANSCRIPTION MACHINERY
While the full set of mitochondrial proteins involved in transcription in vivo remains to be determined, some components have already been identified, and an in vitro transcription system has been reconstituted. While not attempting to provide a comprehensive description of all known mitochondrial transcription factors, we will emphasize some of the salient features of the major players in mitochondrial transcription here.
7.1. TFAM
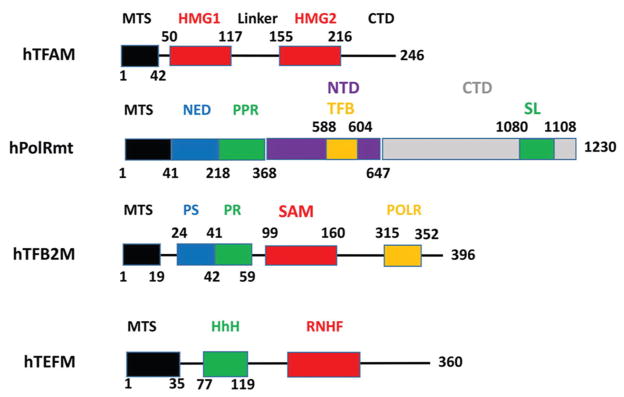
TFAM is the first described and arguably the best studied component of the mitochondrial transcription and replication machineries (50). TFAM is a member of the high mobility group (HMG) superfamily of DNA binding proteins (51) defined by the HMG DNA binding domain (known as the HMG box). The HMG box is an L-shaped three-helix domain that binds to DNA in the minor groove and dramatically bends and unwinds DNA with the help of DNA intercalating residues. TFAM consists of a 42 amino acid matrix targeting sequence (MTS), one canonical (HMG1) and one noncanonical (HMG2) HMG domain, a 37 amino acid linker between HMG domains, and a 30 amino acid CTD (Figure 4). Note that this domain annotation is somewhat different from the original Clayton annotation and is based on the most recent definition of HMG domains by NCBI’s Conserved Domain Database (52).
Figure 4.
Structures of major proteins involved in the mitochondrial transcription. CTD, C-terminal domain; HhH, helix-hairpin-helix motif; HMG1 and 2, high mobility group-like domain; MTS, matrix targeting sequence; NED, N-terminal extension domain; NTD, N-terminal domain; POLR, POLRMT binding domain of the TFB2M; PR, Promoter-interacting domain; PS, priming substrate-interacting domain; RNHF, RNaseH fold, RuvC type; SAM, S-adenosylmethionine-dependent methyltransferase-like domain; SL, specificity loop; TFB, TFB2M-binding domain.
Available experimental support for the notion of indinspensability of TFAM to proper cellular function remains somewhat equivocal. On one hand, disruption of the TFAM gene in murine cardiomyocytes (31, 53), skeletal muscle cells (54), pancreatic β-cells (55) and pyramidal neurons (56) leads to tissue-specific depletion of mtDNA, depletion of mitochondrial transcripts (mtRNA) and severe respiratory chain deficiency. On the other hand, TFAM knockout in stem cells of the basal layer of the epidermis, which is responsible for the production of skin keratinocytes, does not disrupt normal epidermal development and skin barrier function despite a profound depletion of mtDNA and complete absence of respiratory chain complexes. Also, it does not affect differentiation of epidermal layers, and no proliferation defect or major increase in apoptosis could be observed, suggesting that the absence of TFAM is not biologically relevant in these cells (57).
TFAM binds DNA both specifically and nonspecifically. Specific binding has been mapped to positions -12 to -35 upstream of major mitochondrial promoters HSP1 and LSP (29, 32) by DNase I protection assays. Importantly, a high-affinity site has not yet been precisely defined upstream of the HSP2 promoter (however, see (22)), which can be interpreted either in support of a two-promoter model of mitochondrial transcription (if absence of a high-affinity site is interpreted as suggesting the artefactual nature of HSP2), or as a weakness of a two-promoter model for transcription initiation (which requires a high affinity site upstream of the promoter). It is worth mentioning that different studies mapped high-affinity TFAM binding sites to different positions upstream of the transcription start site in human HSP and LSP (e.g., they were mapped to positions -18-39 and -8-29 relative to the transcription start site in LSP and HSP1 respectively in (58), see Figure 2). This is particularly puzzling considering that precise spacing between TFAM-binding sites and transcription initiation sites appears to be important for efficient transcription initiation (59). Also, functional interchangeability of human and mouse TFAMs in vitro and in vivo (60, 61) and similar footprints of mouse and human TFAMs on both human and mouse LSPs (62), together with the lack of considerable sequence similarity in upstream regions of human and mouse promoters (63), collectively suggest that correct TFAM positioning upstream of the transcription start site may be sequence-independent. Finally, some studies failed to detect substantial differences in TFAM binding to specific and non-specific sequences as reported by dissociation constants (64) or by genome-scale mapping of TFAM-mtDNA interactions (65).
The role of TFAM in mitochondrial transcription is complex. Low-level transcription from all three mitochondrial promoters can be observed in vitro without TFAM (22, 23, 26–29). Varying TFAM concentrations in transcription reactions at all three mitochondrial promoters in vitro has a biphasic effect: both stimulation and repression were observed at different concentrations, but the principal effect at higher concentrations was to suppress transcription (26, 66). This finding has given rise to the suggestion that TFAM acts to modulate mitochondrial transcription, increasing the availability of rRNAs at low concentrations, increasing mRNA and mtDNA replication at moderate concentrations, and shutting down both transcription and replication at high concentrations (22, 27).
There is an ongoing controversy regarding the amount of TFAM present in mitochondria. Some studies suggest that TFAM is present in saturating quantities sufficient to cover mtDNA (67–69), whereas other authors report much lower in vivo TFAM: mtDNA ratios of 50:1 (70) and 35:1 (71, 72) and argue that, based on in vitro observations, the saturating TFAM: mtDNA ratios that have been reported would render transcription impossible (70). Also, in vivo methylation protection and crosslinking protection assays suggest existence of “naked” regions in mtDNA (73–76). Finally, even though TFAM is a believed to be a major component of mitochondrial nucleoids (77), it has been observed that not all nucleoids contain TFAM (65, 78), and that some replicating nucleoids do not stain for TFAM (78).
7.2. POLRMT
The mitochondrial RNA polymerase, POLRMT, is a 139 kDa protein structurally related to the single-subunit RNA polymerase encoded by bacteriophage T7 (79). It consists of a 41-aa MTS, an N-terminal extension (NED, aa 42-368), an N-terminal domain (NTD), and a CTD which is responsible for the enzyme’s catalytic activity and which contains the specificity loop (Figure 4). The specificity loop interacts with DNA at positions -5 and -10 relative to the transcription start site (80), and is likely responsible for promoter recognition (81). Within the NED, there is a PPR domain, which is often found in proteins interacting with RNA (82–85). NED is not found in T7 RNA polymerase and appears to be dispensable for POLRMT activity: its deletion results in a hyperactive enzyme that does not require TFAM for transcription initiation in vitro and can initiate transcription both at mitochondrial promoters and at non-specific sequences (86). During transcription initiation, residues within NED interact with aa 271 and 227 of TFAM, and residues within NTD interact with aa 228 and 237 within TFAM CTD. Also, NTD interacts with TFB2M CTD and with DNA at positions around −50 relative to the transcription start site. Finally, CTD of POLRMT interacts with DNA at positions -5 to -10 (80).
In the absence of TFAM and TFB2M, POLRMT is unable to initiate transcription on double-stranded DNA, but can initiate transcription on relaxed circular single-stranded DNA (87). In the absence of DNA, POLRMT forms a weak complex with TFAM, but not with TFB2M (88). POLRMT, unlike TFAM and TFB2M, is unable to initiate transcription at heterologous mitochondrial promoters, and therefore it was suggested to be a factor that determines species specificity of mitochondrial transcription (22, 60) and a primary factor responsible for promoter recognition (22). Apart from its role in mitochondrial transcription, POLRMT forms a separate complex with TFB1M and thereby has a transcription-independent role in the biogenesis and/or function of mitochondrial ribosomes through modulating methylation of the small ribosomal subunit (89).
7.3. TFB2M
TFB2M was initially identified in a search for homologs of Mtf1, a yeast protein which is essential for mitochondrial transcription (90). This 396 aa, 45 kDa protein is composed of a putative 19 aa MTS, a priming substrate-interacting domain, a promoter-interacting domain, an S-adenosylmethionine-dependent methyltransferase-like domain, and a POLRMT-interacting domain (Figure 4 and (38, 91)). Interestingly, both the priming substrate interacting and promoter-interacting domains are dispensable for in vitro run-off transcription (38). Despite the presence of the S-adenosylmethionine-dependent methyltransferase-like domain, TFB2M possesses minimal methyltransferase activity as compared to its homolog, TFB1M, likely due to a combination of suboptimal architecture of the methyltransferase catalytic site combined with an insertion that is predicted to impede access of the nucleic acid substrate to the active site (91–93).
TFB2M is not required for transcription on pre-melted templates (38) or for promoter-independent transcription on relaxed circular single-stranded DNA templates (87), suggesting its role in DNA melting; however, it is required for TFAM-independent transcription on supercoiled plasmids (87). While murine TFB2M is capable of activating transcription in the human system, it is about 3 fold less active than its human cognate (91).
7.4. TEFM
TEFM (c17orf42) has been discovered only recently and remains poorly understood. Three domains have been identified in TEFM so far: a 35 aa MTS, a helix-hairpin-helix domain, and an RNaseH-like fold (Figure 4). TEFM knockdown by RNA interference leads to severe respiratory chain dysfunction due to reduced levels of promoter-distal H- and L-strand transcripts (40). In mitochondria, TEFM interacts with mitochondrial transcripts, POLRMT, pentatricopeptide repeat domain 3 protein (PTCD3), and a putative DEAD-box RNA helicase known as DHX30 to form a large-molecular weight complex. After the treatment of this complex with RNase, the only component that remains associated with TEFM is POLRMT, suggesting that TEFM is an accessory subunit of POLRMT (40). In vitro, TEFM strongly promotes POLRMT processivity and dramatically stimulates the formation of longer transcripts. TEFM also abolishes premature transcription termination at various sites in the template, including conserved sequence block II and 8-Oxo-2′-deoxyguanosine. TEFM also substantially increases POLRMT’s affinity for an elongation-like DNA: RNA template (94).
8. PRIMARY TRANSCRIPTS
mtDNA transcription results in three primary transcripts. The first transcript is HSP1 originating at A561 and terminating within MT-TL1, downstream of MT-RNR2 (21, 95). This is the shortest of mitochondrial transcripts. It encodes MT-TF, MT-TV and both mitochondrial rRNA genes MT-TRNR1 and MT-RNR2. The second transcript is HSP2, which originates at C646 inside MT-TF just upstream of MT-RNR1 and runs almost the full circumference of mtDNA (21). The final transcript, LSP, is a near genomic length primary transcript originating at a single LSP promoter (29, 95, 96).
The total rate of mtDNA transcription is between 1 and 4 × 106 nucleotides per cell per minute (18). The rate of L-strand transcription in vivo has been estimated to be 2–3 fold higher than the rate of H-strand transcription (97). The estimated rates of synthesis of mitochondrial mRNAs are fairly similar to each other and fall in the range between 1 to 2 molecules per cell per minute, which is equivalent to about one transcript per mtDNA molecule per generation. These rates are 50–100 times lower than rates of rRNA synthesis (18).
Both mature mitochondrial rRNAs and the putative H-strand encoded mRNA species are metabolically unstable. Their half-lives, as determined in the kinetics of labeling experiments, were found to be 2.5. to 3.5. hours for the rRNA components and between 25 and 90 min for the different mRNAs (18). In contrast to mitochondrial mRNAs, rRNAs within polysomes appear to be more stable than free rRNAs.
It should be noted here that rates cited in this section have been determined using rapidly proliferating cancer cells, and therefore may or may not accurately approximate corresponding rates in post-mitotic cells.
9. PROCESSING OF PRIMARY TRANSCRIPTS
Processing of mitochondrial primary transcripts involves cleavage, polyadenylation, addition of CCA tails to tRNAs, and tRNA and rRNA base modification. The tRNA punctuation hypothesis initially proposed by Ojala et al. (16) suggests that processing of primary transcripts occurs at tRNAs, which therefore represent “punctuation marks”. Even though tRNA punctuation is unable to fully account for all cleavage sites necessary to generate the full complement of mature mitochondrial RNAs (98), it accounts for most of them, and the processing sites predicted by this hypothesis are in good agreement with those observed experimentally (99). In addition to canonical processing and posttranscriptional modification of primary transcripts, RNAseq analysis has revealed numerous types of aberrant processing, including polyadenylation of some tRNAs (99). The biological significance of these modifications remains to be determined.
Several enzymatic activities are known to be involved in the cleavage of primary transcripts in mammalian mitochondria: cleavage of tRNAs at their 5′ ends is mediated by a special protein-only RNAseP, which consists of three subunits (MRPP1-3), and 3′terminal cleavage is mediated by ELAC2 and PTCD1 (100). tRNAs released by the activity of RNAseP and ELAC2/PTCD1 are further modified by nontemplate addition of the CCA tail by activity of tRNA nucleotidyltransferase 1 (TRNT1) (101). Furthermore, up to 7% of tRNA bases in mammalian mitochondria are modified by various enzymatic activities to yield mature tRNAs (reviewed in (102)). Recently, SUV3 helicase has also been demonstrated to be essential for the proper processing of mitochondrial transcripts: SUV3 knockout in Drosophila leads to a severe decrease in mitochondrial tRNAs accompanied by an accumulation of unprocessed precursor transcripts. These processing defects lead to reduced mitochondrial translation and a severe respiratory chain complex deficiency, resulting in a pupal-lethal phenotype (103).
Polyadenylation of mitochondrial mRNAs is common, and involves the addition of stretches of adenosines to the 3′ ends of mRNAs. However, human and mouse mitochondria mRNA for MT-ND6 is not polyadenylated (104, 105), which is in contrast to Drosophila, in which the MT-ND6 message is polyadenylated (106). Similarly, 3′ ends of mammalian rRNAs contain few or no adenylates, whereas in Drosophila rRNAs are extensively polyadenylated. Intriguingly, in yeast mitochondria, mRNAs are not polyadenylated, but rather are modified by the addition of a conserved dodecamer sequence (107).
In Drosophila (108) and human cells (109), a single poly(A) polymerase (MTPAP) is responsible for the polyadenylation of mitochondrial transcripts, with no other enzyme capable of completing stop signals. MTPAP knockout in Drosophila demonstrated that polyadenylation does not regulate transcript stability, nor is it required for translation, but that it might be involved in the maturation of certain mitochondrial tRNAs (108). Similarly, in human cells, MTPAP is involved in tRNA repair prior to polyadenylation (110).
In different systems, polyadenylation of mitochondrial RNAs has different effects on mRNA stability. In plants, polyadenylation generally leads to degradation, while in mammals and trypanosomes polyadenylation may have opposite effects on RNA stability for different transcripts (reviewed in (111)).
10. HUMAN MITOCHONDRIAL TRANSCRIPTOME
The advent of RNAseq enabled accurate evaluation of whole mitochondrial transcriptomes. In human osteosarcoma 143B cells, transcripts detected by RNAseq analysis aligned to 99.9.% of the H-strand and to 97.6.% of the L-strand suggesting the existence of D-loop transcripts, which is consistent with previous observations made in murine and rat cells (112, 113). Eighty percent of mitochondrial transcripts can be attributed to activity of the HSP1 promoter, 9% to the HSP2 promoter, and 11% to the activity of the LSP. The least abundant mRNA is that for MT-ND6, which is about 10-fold less abundant than mRNAs for MT-CO1 and MT-CO2 (99). These observations do not agree with previous studies in HeLa cells that found that steady-state levels of mRNAs encoded by H- and L-strands are comparable to each other (18). Also, the ratio of steady-state levels of rRNAs to mRNAs determined by RNAseq (10:1) differs dramatically from estimates previously obtained by other techniques (which are in the 50:1 to 300:1 range) (18, 99). It is unclear whether these differences are cell-line specific, or whether they are a consequence of the different techniques used. Mitochondrial mRNAs constitute between 5% (lung and leucocytes) and 30% (heart) of the total cellular mRNA pool (99). Interestingly, low steady-state levels of abutted MT-ND5 and MT-ND6 transcripts inversely correlate with relatively high levels of the antisense transcript. Of note, several nuclear tRNAs are enriched in mitoplasts as compared to whole mitochondrial preparations, suggesting that they may be imported. However, the exact contribution of these tRNAs to mitochondrial translation, if any, remains to be determined (99). In the mitochondrial transcriptome the MT-ND6 mRNA stands out as the only mRNA generated from the LSP transcript, the least abundant mRNA, and the only non-polyadenylated mRNA. Congruent with previous studies, the most abundant polyadenylated RNA species appears to be the 160–185 nucleotide (nt) LSP transcript previously called 7S RNA (16, 18) whose functional significance remains unclear.
RNAseq analysis also revealed an unexpected complexity of the mitochondrial transcriptome, which was influenced by the existence of non-canonical primary transcript processing sites and by the discovery of novel antisense RNAs and mitochondrial small RNAs. The latter were most frequently associated with tRNAs (84%) and formed two prominent classes, 21 nt and 26 nt (99).
11. TRANSCRIPTION REGULATION
Since mtDNA encodes key components of the OXPHOS machinery, regulation of mitochondrial gene expression can be paramount for energy homeostasis in the cell. However, since the OXPHOS apparatus is composed of subunits encoded by both nuclear DNA and mtDNA, it is possible, in principle, to regulate OXPHOS by modulating the supply of nuclearly encoded subunits without altering mitochondrial transcription. Indeed, the abundance of mitochondrial transcripts correlates well with mtDNA copy number (114) and therefore may be regulated by nuclear genes controlling mtDNA replication. Furthermore, the supply of mtDNA-encoded OXPHOS subunits can be regulated at the post-transcriptional level, at the level of translation, and post-translationally. Finally, OXPHOS can be regulated at the level of assembly of both respiratory complexes and supercomplexes. The relative contributions, if any, of these potential mechanisms of regulation of mitochondrial energy production remain to be established. Here, we review mechanisms that may regulate mitochondrial transcription in vivo.
11.1. Regulation by ATP
Transcription from human HSP1 and LSP is affected by ATP concentration (58, 115), an effect that has also been observed at mitochondrial promoters in Saccharomyces cerevisiae (116). Increasing ATP concentration up to 100 μM stimulates both HSP1 and HSP2. Further increases in ATP concentration suppress HSP1 transcription but have no effect on HSP2. These observations are likely to be physiologically relevant, since the levels at which a decline in HSP1 transcription is seen are within the range of expected matrix ATP concentrations (117). In HeLa mitochondrial extracts, HSP1 transcription is lost at ATP levels above 1 mM (115). Therefore, it has been suggested that suppression of HSP1 transcription at high mitochondrial ATP levels could represent a mechanism for the downregulation of mitochondrial translation when there is no need to increase activity of the electron transport chain (23). Since HSP2 transcription is not affected by ATP concentration, there is the potential for differential regulation of the two HSP promoters by ATP availability. A variant of this model proposed by Sologub et al. suggests that mitochondrial transcription initiation operates through the regulating ability of TFB2M to bind priming nucleotides (38).
11.2. Regulation by MTERF proteins
Above, we mentioned a model for positive regulation of HSP1 transcription in human cells by MTERF1. It has been shown that MTERF1 can bind not only its cognate transcription terminator within MT-TL1, but also to a site upstream of HSP1, thus inducing mtDNA looping and activating HSP1 transcription (21). However, this model has been challenged on the grounds that MTERF knockout mice do not show any gross aberrations in mitochondrial transcription or any reduction in the relative abundance of MT-RNR1 and MT-RNR2 encoded within looped region in human mtDNA (24). It should be noted, though, that unlike in human cells, the existence of two independent promoters for heavy strand transcription in murine cells has not yet been demonstrated. Therefore, evidence from MTERF1 knockout mice does not conclusively disprove the existence of this regulatory mechanism in human cells.
Another model for the regulation of mitochondrial transcription by a member of the MTERF family involves MTERF3. This protein binds to the mitochondrial promoter region and represses transcription. Whole-organism MTERF3 knockout is embryonically lethal, and tissue-specific cardiac knockout of this gene is accompanied by increased transcription initiation at both LSP and HSP but decreased levels of promoter-distal LSP transcripts in MTERF3-deficient mouse hearts (48). Transcriptional abnormalities in the heart were accompanied by severe respiratory chain deficiency and decreased lifespan (48).
The role of MTERF proteins in the regulation of mitochondrial transcription in other metazoan systems has been reviewed elsewhere (118).
11.3. Regulation by TFAM
In human mitochondria, HSP1, HSP2 and LSP promoters demonstrate differential requirements for TFAM for transcription activation. Therefore, a model for mitochondrial transcription regulation has been proposed in which HSP2 is the most active promoter at low intramitochondrial TFAM levels, followed by HSP1 and LSP (HSP2 > HSP1 ⋙ LSP). At intermediate TFAM levels, relative activity of the promoters changes to LSP ≫ HSP1 ≫HSP2. Finally, at high TFAM levels, relative activity of the promoters changes once again to HSP1 ⋙ LSP1 ⋙ HSP2 (22). Thus, according to this model, at low TFAM levels, mitochondria execute a “maintenance program” supplying subunits for OXPHOS through the activity of HSP2, whereas at higher TFAM levels, a “mitochondrial biogenesis” program turns on, which is wired to supply ribosomal RNAs (HSP1) and primers for mtDNA replication (LSP).
11.4. Regulation by MRPL12
MRPL12 has been shown to form a distinct, ribosome-free pool inside mitochondria and to associate with PolRmt in complexes distinct from those containing TFB2M (119). In vitro, MRPL12 directly stimulates transcription at LSP and HSP1 promoters. MRPL12 overexpression increases steady-state levels of mitochondrial transcripts (120). Conversely, knockdown of the MRPL12 leads to reduced steady-state levels of mitochondrial transcripts (119). MRPL12 exists in mitochondria as two distinct mature forms: the short and the long one. The long form is associated with ribosomes, while the short one is likely to be associated with PolRmt (121).
11.5. Regulation by methylation
Emerging evidence suggests that mtDNA is methylated and that mitochondrial transcription may be regulated by mtDNA methylation (122–124). However, the evidence at this point is correlative, and mechanistic studies are needed in order to establish mtDNA methylation as a mechanism for regulation of mitochondrial transcription (125).
12. RNA TURNOVER
RNA degradation plays a key role in controlling the abundance of mitochondrial transcripts. Also, mitochondrial homeostasis requires prompt removal of aberrant RNA molecules and maturation products. In Saccharomyces cerevisiae, the mitochondrial degradosome consists of two subunits: the NTP-dependent RNA helicase SUV3 (126) and the 3′- to 5′-exoribonuclease DSS1 (127). Inactivation of either the SUV3 or DSS1 gene results in respiratory deficiency and instability of the mitochondrial genome. Inactivation of the degradosome results in the accumulation of transcripts with abnormally processed 5′ and 3′ termini, excised introns and unprocessed RNA precursors (10–12). Interestingly, in Schizosaccharomyces pombe, inactivation of SUV3 and DSS1 orthologs has only a mild effect on RNA turnover (128). In contrast, in vivo knockdown of SUV3 in Drosophila mitochondria resulted in a severe mitochondrial dysfunction and pupal lethality accompanied by the accumulation of mitochondrial RNA processing intermediates, mRNA and anti-sense RNA (103).
Even though mammalian mitochondria contain no introns, the LSP transcript contains large noncoding regions that have to be degraded after excision. In human cells, silencing of SUV3 or the expression of a dominant-negative variant increases mRNA steady-state levels and results in the accumulation of mRNA decay intermediates, truncated ND2 transcripts, processing byproducts and antisense RNAs, all of which implicate SUV3 as a component of the human mitochondrial degradosome (129, 130). Even though the search for a DSS1 ortholog in human cells proved futile, human polynucleotide phosphorylase (PNPT1), which has 3′–5′ exoribonuclease activity, interacts with SUV3 in vitro to form a functional degradosome (131) and copurifies with SUV3 during isolation from cultured cells (129).
13. CONCLUDING REMARKS
Due to the prominent role played by mitochondria in health and disease, recent years have seen a renewed interest in different aspects of mitochondrial biology, including transcription. However, despite considerable progress, our understanding of mitochondrial transcription remains incomplete. It will be important to resolve the issue of the number of HSP promoters in human and mouse cells and to see whether MTERF1-mediated transcription termination within MT-TL1 can fully account for the differences in the abundance of HSP-encoded rRNAs and mRNAs. In this respect, the differential regulation of HSP1 and HSP2 transcripts, if detected in vivo, may further support the notion of the existence of two H-strand promoters. It has been observed that while LSP promoter is the strongest promoter in vitro, HSP1 appears to dominate in vivo (22). Also, mutation T642G potently activates the HSP2 promoter in vitro, but has no effect on its activity in mitochondrial extracts (23). These observations suggest that existing in vitro transcription systems can be further improved by identifying additional transcription factors to more closely reflect the situation in vivo. Furthermore, it will be interesting to determine how TFB2M recruits different priming nucleotides for transcription initiation at alternative transcription start sites. The substantial disagreements between different studies about the exact position(s) of TFAM footprints upstream of mitochondrial promoters require an explanation. Past and emerging reports that the whole mitochondrial genome is transcribed (including the D-loop region) deserve attention because they can inform our models of mitochondrial transcription termination in a profound way. Processing of mitochondrial transcripts at sites not punctuated by tRNAs needs to be studies in more depth. It will be important to gain new insights into the regulation of mammalian mitochondrial transcription in vivo. Finally, the field can benefit from standardization of terminology and nomenclature. These and many other issues are likely to keep the field busy in the coming years.
Acknowledgments
The authors were supported by the National Institutes of Health grants OD010944 and HL66299 and by the Department of Defense grant PR150220 (to M.A.).
Abbreviations
- HSP
heavy strand promoter
- KO
knockout
- LSP
light strand promoter
- mtDNA
mitochondrial DNA
- MTERF
mitochondrial transcription termination factor
- MT-RNR1
mitochondrial 12S rRNA
- MT-RNR2
mitochondrial 16S RNA
- MT-TF
mitochondrial phenylalanine tRNA
- OXPHOS
Oxidative Phosphorylation system
- POLRMT
mitochondrial RNA polymerase
- rRNA
ribosomal RNA
- ROS
reactive oxygen species
- TEFM
mitochondrial transcription elongation factor
- TFAM
mitochondrial transcription factor A
- TFB2M
mitochondrial transcription factor B2
- tRNA
transfer RNA
- TRNT1
tRNA nucleotidyltransferase 1
References
- 1.Blackstone NW. The impact of mitochondrial endosymbiosis on the evolution of calcium signaling. Cell Calcium. 2015;57(3):133–139. doi: 10.1016/j.ceca.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Elkholi R, Renault TT, Serasinghe MN, Chipuk JE. Putting the pieces together: How is the mitochondrial pathway of apoptosis regulated in cancer and chemotherapy? Cancer Metab. 2014;2:16. doi: 10.1186/2049-3002-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zorov DB, Juhaszova M, Sollott SJ. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol Rev. 2014;94(3):909–50. doi: 10.1152/physrev.00026.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shah DI, Takahashi-Makise N, Cooney JD, Li L, Schultz IJ, Pierce EL, Narla A, Seguin A, Hattangadi SM, Medlock AE, Langer NB, Dailey TA, Hurst SN, Faccenda D, Wiwczar JM, Heggers SK, Vogin G, Chen W, Chen C, Campagna DR, Brugnara C, Zhou Y, Ebert BL, Danial NN, Fleming MD, Ward DM, Campanella M, Dailey HA, Kaplan J, Paw BH. Mitochondrial Atpif1 regulates haem synthesis in developing erythroblasts. Nature. 2012;491(7425):608–12. doi: 10.1038/nature11536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lill R, Srinivasan V, Muhlenhoff U. The role of mitochondria in cytosolic-nuclear iron-sulfur protein biogenesis and in cellular iron regulation. Curr Opin Microbiol. 2014;22C:111–119. doi: 10.1016/j.mib.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 6.Barrell BG, Bankier AT, Drouin J. A different genetic code in human mitochondria. Nature. 1979;282(5735):189–94. doi: 10.1038/282189a0. [DOI] [PubMed] [Google Scholar]
- 7.Hashimoto Y, Niikura T, Tajima H, Yasukawa T, Sudo H, Ito Y, Kita Y, Kawasumi M, Kouyama K, Doyu M, Sobue G, Koide T, Tsuji S, Lang J, Kurokawa K, Nishimoto I. A rescue factor abolishing neuronal cell death by a wide spectrum of familial Alzheimer’s disease genes and Abeta. Proc Natl Acad Sci U S A. 2001;98(11):6336–41. doi: 10.1073/pnas.101133498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee C, Kim KH, Cohen P. MOTS-c. A novel mitochondrial-derived peptide regulating muscle and fat metabolism. Free Radical Biology & Medicine. 2016 doi: 10.1016/j.freeradbiomed.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee C, Zeng J, Drew BG, Sallam T, Martin-Montalvo A, Wan J, Kim SJ, Mehta H, Hevener AL, de Cabo R, Cohen P. The Mitochondrial-Derived Peptide MOTS-c Promotes Metabolic Homeostasis and Reduces Obesity and Insulin Resistance. Cell Metab. 2015;21(3):443–54. doi: 10.1016/j.cmet.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wanrooij S, Fuste JM, Farge G, Shi Y, Gustafsson CM, Falkenberg M. Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro. Proc Natl Acad Sci U S A. 2008;105(32):11122–7. doi: 10.1073/pnas.0805399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76:679–99. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 12.Aloni Y, Attardi G. Expression of the mitochondrial genome in HeLa cells. II. Evidence for complete transcription of mitochondrial DNA. J Mol Biol. 1971;55(2):251–67. doi: 10.1016/0022-28367190197-5. doi: 10.1016/0022-28367190197-5. [DOI] [PubMed] [Google Scholar]
- 13.Aloni Y, Attardi G. Expression of the mitochondria genome in HeLa cells. IV. Titration of mitochondrial genes for 16 s, 12 s and 4 s RNA. J Mol Biol. 1971;55(2):271–6. doi: 10.1016/0022-28367190195-1. doi: 10.1016/0022-28367190195-1. [DOI] [PubMed] [Google Scholar]
- 14.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290(5806):457–65. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 15.Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26(2 Pt 2):167–80. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- 16.Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290(5806):470–4. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 17.Gustafsson CM, Falkenberg M, Larsson NG. Maintenance and Expression of Mammalian Mitochondrial DNA. Annu Rev Biochem. 2016 doi: 10.1146/annurev-biochem-060815-014402. [DOI] [PubMed] [Google Scholar]
- 18.Gelfand R, Attardi G. Synthesis and turnover of mitochondrial ribonucleic acid in HeLa cells: the mature ribosomal and messenger ribonucleic acid species are metabolically unstable. Molecular & Cellular Biology. 1981;1(6):497–511. doi: 10.1128/MCB.1.6.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montoya J, Christianson T, Levens D, Rabinowitz M, Attardi G. Identification of initiation sites for heavy-strand and light-strand transcription in human mitochondrial DNA. Proc Natl Acad Sci U S A. 1982;79(23):7195–9. doi: 10.1073/pnas.79.23.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montoya J, Gaines GL, Attardi G. The pattern of transcription of the human mitochondrial rRNA genes reveals two overlapping transcription units. Cell. 1983;34(1):151–9. doi: 10.1016/0092-8674(83)90145-9. [DOI] [PubMed] [Google Scholar]
- 21.Martin M, Cho J, Cesare AJ, Griffith JD, Attardi G. Termination factor-mediated DNA loop between termination and initiation sites drives mitochondrial rRNA synthesis. Cell. 2005;123(7):1227–40. doi: 10.1016/j.cell.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 22.Lodeiro MF, Uchida A, Bestwick M, Moustafa IM, Arnold JJ, Shadel GS, Cameron CE. Transcription from the second heavy-strand promoter of human mtDNA is repressed by transcription factor A in vitro. Proc Natl Acad Sci U S A. 2012;109(17):6513–8. doi: 10.1073/pnas.1118710109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zollo O, Tiranti V, Sondheimer N. Transcriptional requirements of the distal heavy-strand promoter of mtDNA. Proc Natl Acad Sci U S A. 2012;109(17):6508–12. doi: 10.1073/pnas.1118594109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Terzioglu M, Ruzzenente B, Harmel J, Mourier A, Jemt E, Lopez MD, Kukat C, Stewart JB, Wibom R, Meharg C, Habermann B, Falkenberg M, Gustafsson CM, Park CB, Larsson NG. MTERF1 binds mtDNA to prevent transcriptional interference at the light-strand promoter but is dispensable for rRNA gene transcription regulation. Cell Metab. 2013;17(4):618–26. doi: 10.1016/j.cmet.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 25.Chang DD, Clayton DA. Precise assignment of the light-strand promoter of mouse mitochondrial DNA: a functional promoter consists of multiple upstream domains. Molecular & Cellular Biology. 1986;6(9):3253–61. doi: 10.1128/MCB.6.9.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shutt TE, Lodeiro MF, Cotney J, Cameron CE, Shadel GS. Core human mitochondrial transcription apparatus is a regulated two-component system in vitro. Proc Natl Acad Sci U S A. 2010;107(27):12133–8. doi: 10.1073/pnas.0910581107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shutt TE, Bestwick M, Shadel GS. The core human mitochondrial transcription initiation complex: It only takes two to tango. Transcription. 2011;2(2):55–59. doi: 10.4161/trns.2.2.14296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi Y, Dierckx A, Wanrooij PH, Wanrooij S, Larsson NG, Wilhelmsson LM, Falkenberg M, Gustafsson CM. Mammalian transcription factor A is a core component of the mitochondrial transcription machinery. Proc Natl Acad Sci U S A. 2012;109(41):16510–5. doi: 10.1073/pnas.1119738109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fisher RP, Topper JN, Clayton DA. Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell. 1987;50(2):247–58. doi: 10.1016/0092-8674(87)90220-0. [DOI] [PubMed] [Google Scholar]
- 30.Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18(3):231–6. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 31.Wang J, Wilhelmsson H, Graff C, Li H, Oldfors A, Rustin P, Bruning JC, Kahn CR, Clayton DA, Barsh GS, Thoren P, Larsson NG. Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nat Genet. 1999;21(1):133–7. doi: 10.1038/5089. [DOI] [PubMed] [Google Scholar]
- 32.Dairaghi DJ, Shadel GS, Clayton DA. Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J Mol Biol. 1995;249(1):11–28. doi: 10.1006/jmbi.1995.9889. [DOI] [PubMed] [Google Scholar]
- 33.Fisher RP, Lisowsky T, Parisi MA, Clayton DA. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J Biol Chem. 1992;267(5):3358–67. [PubMed] [Google Scholar]
- 34.Ngo HB, Kaiser JT, Chan DC. The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA. Nat Struct Mol Biol. 2011;18(11):1290–6. doi: 10.1038/nsmb.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rubio-Cosials A, Sidow JF, Jimenez-Menendez N, Fernandez-Millan P, Montoya J, Jacobs HT, Coll M, Bernado P, Sola M. Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Nat Struct Mol Biol. 2011;18(11):1281–9. doi: 10.1038/nsmb.2160. [DOI] [PubMed] [Google Scholar]
- 36.Ngo HB, Lovely GA, Phillips R, Chan DC. Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation. Nat Commun. 2014;5:3077. doi: 10.1038/ncomms4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morozov YI, Agaronyan K, Cheung AC, Anikin M, Cramer P, Temiakov D. A novel intermediate in transcription initiation by human mitochondrial RNA polymerase. Nucleic Acids Res. 2014 doi: 10.1093/nar/gkt1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sologub M, Litonin D, Anikin M, Mustaev A, Temiakov D. TFB2 is a transient component of the catalytic site of the human mitochondrial RNA polymerase. Cell. 2009;139(5):934–44. doi: 10.1016/j.cell.2009.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mangus DA, Jang SH, Jaehning JA. Release of the yeast mitochondrial RNA polymerase specificity factor from transcription complexes. J Biol Chem. 1994;269(42):26568–74. [PubMed] [Google Scholar]
- 40.Minczuk M, He J, Duch AM, Ettema TJ, Chlebowski A, Dzionek K, Nijtmans LG, Huynen MA, Holt IJ. TEFM (c17orf42) is necessary for transcription of human mtDNA. Nucleic Acids Res. 2011;39(10):4284–99. doi: 10.1093/nar/gkq1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agaronyan K, Morozov YI, Anikin M, Temiakov D. Mitochondrial biology. Replication-transcription switch in human mitochondria. Science. 2015;347(6221):548–51. doi: 10.1126/science.aaa0986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kruse B, Narasimhan N, Attardi G. Termination of transcription in human mitochondria: identification and purification of a DNA binding protein factor that promotes termination. Cell. 1989;58(2):391–7. doi: 10.1016/0092-8674(89)90853-2. [DOI] [PubMed] [Google Scholar]
- 43.Asin-Cayuela J, Helm M, Attardi G. A monomer-to-trimer transition of the human mitochondrial transcription termination factor (mTERF) is associated with a loss of in vitro activity. J Biol Chem. 2004;279(15):15670–7. doi: 10.1074/jbc.M312537200. [DOI] [PubMed] [Google Scholar]
- 44.Asin-Cayuela J, Schwend T, Farge G, Gustafsson CM. The human mitochondrial transcription termination factor (mTERF) is fully active in vitro in the non-phosphorylated form. J Biol Chem. 2005;280(27):25499–505. doi: 10.1074/jbc.M501145200. [DOI] [PubMed] [Google Scholar]
- 45.Yakubovskaya E, Mejia E, Byrnes J, Hambardjieva E, Garcia-Diaz M. Helix unwinding and base flipping enable human MTERF1 to terminate mitochondrial transcription. Cell. 2010;141(6):982–93. doi: 10.1016/j.cell.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Camasamudram V, Fang JK, Avadhani NG. Transcription termination at the mouse mitochondrial H-strand promoter distal site requires an A/T rich sequence motif and sequence specific DNA binding proteins. Eur J Biochem. 2003;270(6):1128–40. doi: 10.1046/j.1432-1033.2003.03461.x. [DOI] [PubMed] [Google Scholar]
- 47.Linder T, Park CB, Asin-Cayuela J, Pellegrini M, Larsson NG, Falkenberg M, Samuelsson T, Gustafsson CM. A family of putative transcription termination factors shared amongst metazoans and plants. Curr Genet. 2005;48(4):265–9. doi: 10.1007/s00294-005-0022-5. [DOI] [PubMed] [Google Scholar]
- 48.Park CB, Asin-Cayuela J, Camara Y, Shi Y, Pellegrini M, Gaspari M, Wibom R, Hultenby K, Erdjument-Bromage H, Tempst P, Falkenberg M, Gustafsson CM, Larsson NG. MTERF3 is a negative regulator of mammalian mtDNA transcription. Cell. 2007;130(2):273–85. doi: 10.1016/j.cell.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 49.Metodiev MD, Spahr H, Loguercio Polosa P, Meharg C, Becker C, Altmueller J, Habermann B, Larsson NG, Ruzzenente B. NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet. 2014;10(2):e1004110. doi: 10.1371/journal.pgen.1004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fisher RP, Clayton DA. Purification and characterization of human mitochondrial transcription factor 1. Molecular & Cellular Biology. 1988;8(8):3496–509. doi: 10.1128/MCB.8.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parisi MA, Clayton DA. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991;252(5008):965–9. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- 52.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43(Database issue):D222–6. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, Wang J, Wilhelmsson H, Hansson A, Thoren P, Duffy J, Rustin P, Larsson NG. Genetic modification of survival in tissue-specific knockout mice with mitochondrial cardiomyopathy. Proc Natl Acad Sci U S A. 2000;97(7):3467–72. doi: 10.1073/pnas.97.7.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wredenberg A, Wibom R, Wilhelmsson H, Graff C, Wiener HH, Burden SJ, Oldfors A, Westerblad H, Larsson NG. Increased mitochondrial mass in mitochondrial myopathy mice. Proc Natl Acad Sci U S A. 2002;99(23):15066–71. doi: 10.1073/pnas.232591499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silva JP, Kohler M, Graff C, Oldfors A, Magnuson MA, Berggren PO, Larsson NG. Impaired insulin secretion and beta-cell loss in tissue-specific knockout mice with mitochondrial diabetes. Nat Genet. 2000;26(3):336–40. doi: 10.1038/81649. [DOI] [PubMed] [Google Scholar]
- 56.Sorensen L, Ekstrand M, Silva JP, Lindqvist E, Xu B, Rustin P, Olson L, Larsson NG. Late-onset corticohippocampal neurodepletion attributable to catastrophic failure of oxidative phosphorylation in MILON mice. J Neurosci. 2001;21(20):8082–90. doi: 10.1523/JNEUROSCI.21-20-08082.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baris OR, Klose A, Kloepper JE, Weiland D, Neuhaus JF, Schauen M, Wille A, Muller A, Merkwirth C, Langer T, Larsson NG, Krieg T, Tobin DJ, Paus R, Wiesner RJ. The Mitochondrial Electron Transport Chain is Dispensable for Proliferation and Differentiation of Epidermal Progenitor Cells. Stem Cells. 2011 doi: 10.1002/stem.695. [DOI] [PubMed] [Google Scholar]
- 58.Lodeiro MF, Uchida AU, Arnold JJ, Reynolds SL, Moustafa IM, Cameron CE. Identification of multiple rate-limiting steps during the human mitochondrial transcription cycle in vitro. J Biol Chem. 2010;285(21):16387–402. doi: 10.1074/jbc.M109.092676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dairaghi DJ, Shadel GS, Clayton DA. Human mitochondrial transcription factor A and promoter spacing integrity are required for transcription initiation. Biochim Biophys Acta. 1995;1271(1):127–34. doi: 10.1016/0925-4439(95)00019-Z. [DOI] [PubMed] [Google Scholar]
- 60.Gaspari M, Falkenberg M, Larsson NG, Gustafsson CM. The mitochondrial RNA polymerase contributes critically to promoter specificity in mammalian cells. EMBO J. 2004;23(23):4606–14. doi: 10.1038/sj.emboj.7600465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Freyer C, Park CB, Ekstrand MI, Shi Y, Khvorostova J, Wibom R, Falkenberg M, Gustafsson CM, Larsson NG. Maintenance of respiratory chain function in mouse hearts with severely impaired mtDNA transcription. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fisher RP, Parisi MA, Clayton DA. Flexible recognition of rapidly evolving promoter sequences by mitochondrial transcription factor 1. Genes & Development. 1989;3(12B):2202–17. doi: 10.1101/gad.3.12b.2202. [DOI] [PubMed] [Google Scholar]
- 63.Chang DD, Clayton DA. Identification of primary transcriptional start sites of mouse mitochondrial DNA: accurate in vitro initiation of both heavy- and light-strand transcripts. Molecular & Cellular Biology. 1986;6(5):1446–53. doi: 10.1128/MCB.6.5.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wong TS, Rajagopalan S, Freund SM, Rutherford TJ, Andreeva A, Townsley FM, Petrovich M, Fersht AR. Biophysical characterizations of human mitochondrial transcription factor A and its binding to tumor suppressor p53. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang YE, Marinov GK, Wold BJ, Chan DC. Genome-wide analysis reveals coating of the mitochondrial genome by TFAM. PLoS One. 2013;8(8):e74513. doi: 10.1371/journal.pone.0074513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Litonin D, Sologub M, Shi Y, Savkina M, Anikin M, Falkenberg M, Gustafsson C, Temiakov D. Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genesin vitro. J Biol Chem. 2010 doi: 10.1074/jbc.C110.128918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alam TI, Kanki T, Muta T, Ukaji K, Abe Y, Nakayama H, Takio K, Hamasaki N, Kang D. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 2003;31(6):1640–5. doi: 10.1093/nar/gkg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takamatsu C, Umeda S, Ohsato T, Ohno T, Abe Y, Fukuoh A, Shinagawa H, Hamasaki N, Kang D. Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA-binding protein. EMBO Rep. 2002;3(5):451–6. doi: 10.1093/embo-reports/kvf099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kukat C, Wurm CA, Spahr H, Falkenberg M, Larsson NG, Jakobs S. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc Natl Acad Sci U S A. 2011;108(33):13534–9. doi: 10.1073/pnas.1109263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cotney J, Wang Z, Shadel GS. Relative abundance of the human mitochondrial transcription system and distinct roles for h-mtTFB1 and h-mtTFB2 in mitochondrial biogenesis and gene expression. Nucleic Acids Res. 2007;35(12):4042–54. doi: 10.1093/nar/gkm424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wiesner RJ, Zsurka G, Kunz WS. Mitochondrial DNA damage and the aging process: facts and imaginations. Free Radic Res. 2006;40(12):1284–94. doi: 10.1080/10715760600913168. [DOI] [PubMed] [Google Scholar]
- 72.Maniura-Weber K, Goffart S, Garstka HL, Montoya J, Wiesner RJ. Transient overexpression of mitochondrial transcription factor A (TFAM) is sufficient to stimulate mitochondrial DNA transcription, but not sufficient to increase mtDNA copy number in cultured cells. Nucleic Acids Res. 2004;32(20):6015–27. doi: 10.1093/nar/gkh921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.http://www.mitomap.org
- 74.Pardue ML, Fostel JM, Cech TR. DNA-protein interactions in the Drosophila virilis mitochondrial chromosome. Nucleic Acids Res. 1984;12(4):1991–9. doi: 10.1093/nar/12.4.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Potter DA, Fostel JM, Berninger M, Pardue ML, Cech TR. DNA-protein interactions in the Drosophila melanogaster mitochondrial genome as deduced from trimethylpsoralen crosslinking patterns. Proc Natl Acad Sci U S A. 1980;77(7):4118–22. doi: 10.1073/pnas.77.7.4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ghivizzani SC, Madsen CS, Nelen MR, Ammini CV, Hauswirth WW. In organello footprint analysis of human mitochondrial DNA: human mitochondrial transcription factor A interactions at the origin of replication. Molecular & Cellular Biology. 1994;14(12):7717–30. doi: 10.1128/MCB.14.12.7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bogenhagen DF. Mitochondrial DNA nucleoid structure. Biochim Biophys Acta. 2012;1819(9–10):914–20. doi: 10.1016/j.bbagrm.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 78.Wai T, Teoli D, Shoubridge EA. The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat Genet. 2008 doi: 10.1038/ng.258. [DOI] [PubMed] [Google Scholar]
- 79.Ringel R, Sologub M, Morozov YI, Litonin D, Cramer P, Temiakov D. Structure of human mitochondrial RNA polymerase. Nature. 2011;478(7368):269–73. doi: 10.1038/nature10435. [DOI] [PubMed] [Google Scholar]
- 80.Morozov YI, Parshin AV, Agaronyan K, Cheung AC, Anikin M, Cramer P, Temiakov D. A model for transcription initiation in human mitochondria. Nucleic Acids Res. 2015;43(7):3726–35. doi: 10.1093/nar/gkv235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nayak D, Guo Q, Sousa R. A promoter recognition mechanism common to yeast mitochondrial and phage t7 RNA polymerases. J Biol Chem. 2009;284(20):13641–7. doi: 10.1074/jbc.M900718200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aphasizheva I, Maslov D, Wang X, Huang L, Aphasizhev R. Pentatricopeptide repeat proteins stimulate mRNA adenylation/uridylation to activate mitochondrial translation in trypanosomes. Mol Cell. 2011;42(1):106–17. doi: 10.1016/j.molcel.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aphasizheva I, Maslov DA, Qian Y, Huang L, Wang Q, Costello CE, Aphasizhev R. Ribosome-associated pentatricopeptide repeat proteins function as translational activators in mitochondria of trypanosomes. Mol Microbiol. 2016;99(6):1043–58. doi: 10.1111/mmi.13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Binder S, Stoll K, Stoll B. P-class pentatricopeptide repeat proteins are required for efficient 5′ end formation of plant mitochondrial transcripts. RNA Biol. 2013;10(9):1511–9. doi: 10.4161/rna.26129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nakamura T, Yagi Y, Kobayashi K. Mechanistic insight into pentatricopeptide repeat proteins as sequence-specific RNA-binding proteins for organellar RNAs in plants. Plant & Cell Physiology. 2012;53(7):1171–9. doi: 10.1093/pcp/pcs069. [DOI] [PubMed] [Google Scholar]
- 86.Posse V, Hoberg E, Dierckx A, Shahzad S, Koolmeister C, Larsson NG, Wilhelmsson LM, Hallberg BM, Gustafsson CM. The amino terminal extension of mammalian mitochondrial RNA polymerase ensures promoter specific transcription initiation. Nucleic Acids Res. 2014;42(6):3638–47. doi: 10.1093/nar/gkt1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fukuoh A, Ohgaki K, Hatae H, Kuraoka I, Aoki Y, Uchiumi T, Jacobs HT, Kang D. DNA conformation-dependent activities of human mitochondrial RNA polymerase. Genes Cells. 2009;14(8):1029–42. doi: 10.1111/j.1365-2443.2009.01328.x. [DOI] [PubMed] [Google Scholar]
- 88.Yakubovskaya E, Guja KE, Eng ET, Choi WS, Mejia E, Beglov D, Lukin M, Kozakov D, Garcia-Diaz M. Organization of the human mitochondrial transcription initiation complex. Nucleic Acids Res. 2014;42(6):4100–12. doi: 10.1093/nar/gkt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Surovtseva YV, Shadel GS. Transcription-independent role for human mitochondrial RNA polymerase in mitochondrial ribosome biogenesis. Nucleic Acids Res. 2013;41(4):2479–88. doi: 10.1093/nar/gks1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat Genet. 2002;31(3):289–94. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 91.Moustafa IM, Uchida A, Wang Y, Yennawar N, Cameron CE. Structural models of mammalian mitochondrial transcription factor B2. Biochim Biophys Acta. 2015;1849(8):987–1002. doi: 10.1016/j.bbagrm.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 92.Cotney J, Shadel GS. Evidence for an early gene duplication event in the evolution of the mitochondrial transcription factor B family and maintenance of rRNA methyltransferase activity in human mtTFB1 and mtTFB2. J Mol Evol. 2006;63(5):707–17. doi: 10.1007/s00239-006-0075-1. [DOI] [PubMed] [Google Scholar]
- 93.Cotney J, McKay SE, Shadel GS. Elucidation of separate, but collaborative functions of the rRNA methyltransferase-related human mitochondrial transcription factors B1 and B2 in mitochondrial biogenesis reveals new insight into maternally inherited deafness. Hum Mol Genet. 2009;18(14):2670–82. doi: 10.1093/hmg/ddp208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Posse V, Shahzad S, Falkenberg M, Hallberg BM, Gustafsson CM. TEFM is a potent stimulator of mitochondrial transcription elongation in vitro. Nucleic Acids Res. 2015;43(5):2615–24. doi: 10.1093/nar/gkv105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chang DD, Clayton DA. Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc Natl Acad Sci U S A. 1985;82(2):351–5. doi: 10.1073/pnas.82.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chang DD, Clayton DA. Precise identification of individual promoters for transcription of each strand of human mitochondrial DNA. Cell. 1984;36(3):635–43. doi: 10.1016/0092-8674(84)90343-X. [DOI] [PubMed] [Google Scholar]
- 97.Cantatore P, Attardi G. Mapping of nascent light and heavy strand transcripts on the physical map of HeLa cell mitochondrial DNA. Nucleic Acids Res. 1980;8(12):2605–25. doi: 10.1093/nar/8.12.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Temperley RJ, Wydro M, Lightowlers RN, Chrzanowska-Lightowlers ZM. Human mitochondrial mRNAs--like members of all families, similar but different. Biochim Biophys Acta. 2010;1797(6–7):1081–5. doi: 10.1016/j.bbabio.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, Filipovska A, Mattick JS. The human mitochondrial transcriptome. Cell. 2011;146(4):645–58. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lopez Sanchez MI, Mercer TR, Davies SM, Shearwood AM, Nygard KK, Richman TR, Mattick JS, Rackham O, Filipovska A. RNA processing in human mitochondria. Cell Cycle. 2011;10(17):2904–16. doi: 10.4161/cc.10.17.17060. [DOI] [PubMed] [Google Scholar]
- 101.Nagaike T, Suzuki T, Tomari Y, Takemoto-Hori C, Negayama F, Watanabe K, Ueda T. Identification and characterization of mammalian mitochondrial tRNA nucleotidyltransferases. J Biol Chem. 2001;276(43):40041–9. doi: 10.1074/jbc.M106202200. [DOI] [PubMed] [Google Scholar]
- 102.Powell CA, Nicholls TJ, Minczuk M. Nuclear-encoded factors involved in post-transcriptional processing and modification of mitochondrial tRNAs in human disease. Front Genet. 2015;6:79. doi: 10.3389/fgene.2015.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Clemente P, Pajak A, Laine I, Wibom R, Wedell A, Freyer C, Wredenberg A. SUV3 helicase is required for correct processing of mitochondrial transcripts. Nucleic Acids Res. 2015;43(15):7398–413. doi: 10.1093/nar/gkv692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Slomovic S, Laufer D, Geiger D, Schuster G. Polyadenylation and degradation of human mitochondrial RNA: the prokaryotic past leaves its mark. Molecular & Cellular Biology. 2005;25(15):6427–35. doi: 10.1128/MCB.25.15.6427-6435.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ruzzenente B, Metodiev MD, Wredenberg A, Bratic A, Park CB, Camara Y, Milenkovic D, Zickermann V, Wibom R, Hultenby K, Erdjument-Bromage H, Tempst P, Brandt U, Stewart JB, Gustafsson CM, Larsson NG. LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 2012;31(2):443–56. doi: 10.1038/emboj.2011.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stewart JB, Beckenbach AT. Characterization of mature mitochondrial transcripts in Drosophila, and the implications for the tRNA punctuation model in arthropods. Gene. 2009;445(1–2):49–57. doi: 10.1016/j.gene.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 107.Butow RA, Zhu H, Perlman P, Conrad-Webb H. The role of a conserved dodecamer sequence in yeast mitochondrial gene expression. Genome. 1989;31(2):757–60. doi: 10.1139/g89-134. [DOI] [PubMed] [Google Scholar]
- 108.Bratic A, Clemente P, Calvo-Garrido J, Maffezzini C, Felser A, Wibom R, Wedell A, Freyer C, Wredenberg A. Mitochondrial Polyadenylation Is a One-Step Process Required for mRNA Integrity and tRNA Maturation. PLoS Genet. 2016;12(5):e1006028. doi: 10.1371/journal.pgen.1006028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tomecki R, Dmochowska A, Gewartowski K, Dziembowski A, Stepien PP. Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucleic Acids Res. 2004;32(20):6001–14. doi: 10.1093/nar/gkh923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fiedler M, Rossmanith W, Wahle E, Rammelt C. Mitochondrial poly(A) polymerase is involved in tRNA repair. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chang JH, Tong L. Mitochondrial poly(A) polymerase and polyadenylation. Biochim Biophys Acta. 2012;1819(9–10):992–7. doi: 10.1016/j.bbagrm.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sbisa E, Nardelli M, Tanzariello F, Tullo A, Saccone C. The complete and symmetric transcription of the main non coding region of rat mitochondrial genome: in vivo mapping of heavy and light transcripts. Curr Genet. 1990;17(3):247–53. doi: 10.1007/BF00312616. [DOI] [PubMed] [Google Scholar]
- 113.Vijayasarathy C, Zheng YM, Mullick J, Basu A, Avadhani NG. Identification of a stable RNA encoded by the H-strand of the mouse mitochondrial D-loop region and a conserved sequence motif immediately upstream of its polyadenylation site. Gene Expr. 1995;4(3):125–41. [PMC free article] [PubMed] [Google Scholar]
- 114.D’Erchia AM, Atlante A, Gadaleta G, Pavesi G, Chiara M, De Virgilio C, Manzari C, Mastropasqua F, Prazzoli GM, Picardi E, Gissi C, Horner D, Reyes A, Sbisa E, Tullo A, Pesole G. Tissue-specific mtDNA abundance from exome data and its correlation with mitochondrial transcription, mass and respiratory activity. Mitochondrion. 2015;20:13–21. doi: 10.1016/j.mito.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 115.Enriquez JA, Fernandez-Silva P, Perez-Martos A, Lopez-Perez MJ, Montoya J. The synthesis of mRNA in isolated mitochondria can be maintained for several hours and is inhibited by high levels of ATP. Eur J Biochem. 1996;237(3):601–10. doi: 10.1111/j.1432-1033.1996.0601p.x. [DOI] [PubMed] [Google Scholar]
- 116.Amiott EA, Jaehning JA. Mitochondrial transcription is regulated via an ATP “sensing” mechanism that couples RNA abundance to respiration. Mol Cell. 2006;22(3):329–38. doi: 10.1016/j.molcel.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 117.Metelkin E, Demin O, Kovacs Z, Chinopoulos C. Modeling of ATP-ADP steady-state exchange rate mediated by the adenine nucleotide translocase in isolated mitochondria. FEBS J. 2009;276(23):6942–55. doi: 10.1111/j.1742-4658.2009.07394.x. [DOI] [PubMed] [Google Scholar]
- 118.Roberti M, Polosa PL, Bruni F, Manzari C, Deceglie S, Gadaleta MN, Cantatore P. The MTERF family proteins: mitochondrial transcription regulators and beyond. Biochim Biophys Acta. 2009;1787(5):303–11. doi: 10.1016/j.bbabio.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 119.Surovtseva YV, Shutt TE, Cotney J, Cimen H, Chen SY, Koc EC, Shadel GS. Mitochondrial Ribosomal Protein L12 selectively associates with human mitochondrial RNA polymerase to activate transcription. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1108852108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang Z, Cotney J, Shadel GS. Human mitochondrial ribosomal protein MRPL12 interacts directly with mitochondrial RNA polymerase to modulate mitochondrial gene expression. J Biol Chem. 2007;282(17):12610–8. doi: 10.1074/jbc.M700461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nouws J, Goswami AV, Bestwick M, McCann BJ, Surovtseva YV, Shadel GS. Mitochondrial Ribosomal Protein L12 Is Required for POLRMT Stability and Exists as Two Forms Generated by Alternative Proteolysis during Import. J Biol Chem. 2016;291(2):989–97. doi: 10.1074/jbc.M115.689299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bellizzi D, D’Aquila P, Scafone T, Giordano M, Riso V, Riccio A, Passarino G. The control region of mitochondrial DNA shows an unusual CpG and non-CpG methylation pattern. DNA Res. 2013;20(6):537–47. doi: 10.1093/dnares/dst029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ghosh S, Sengupta S, Scaria V. Comparative analysis of human mitochondrial methylomes shows distinct patterns of epigenetic regulation in mitochondria. Mitochondrion. 2014;18:58–62. doi: 10.1016/j.mito.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 124.Infantino V, Castegna A, Iacobazzi F, Spera I, Scala I, Andria G, Iacobazzi V. Impairment of methyl cycle affects mitochondrial methyl availability and glutathione level in Down’s syndrome. Molecular Genetics & Metabolism. 2011;102(3):378–82. doi: 10.1016/j.ymgme.2010.11.166. [DOI] [PubMed] [Google Scholar]
- 125.van der Wijst MG, Rots MG. Mitochondrial epigenetics: an overlooked layer of regulation? Trends Genet. 2015 doi: 10.1016/j.tig.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 126.Stepien PP, Margossian SP, Landsman D, Butow RA. The yeast nuclear gene suv3 affecting mitochondrial post-transcriptional processes encodes a putative ATP-dependent RNA helicase. Proc Natl Acad Sci U S A. 1992;89(15):6813–7. doi: 10.1073/pnas.89.15.6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dmochowska A, Golik P, Stepien PP. The novel nuclear gene DSS-1 of Saccharomyces cerevisiae is necessary for mitochondrial biogenesis. Curr Genet. 1995;28(2):108–12. doi: 10.1007/BF00315775. [DOI] [PubMed] [Google Scholar]
- 128.Hoffmann B, Nickel J, Speer F, Schafer B. The 3′ ends of mature transcripts are generated by a processosome complex in fission yeast mitochondria. J Mol Biol. 2008;377(4):1024–37. doi: 10.1016/j.jmb.2008.01.038. [DOI] [PubMed] [Google Scholar]
- 129.Szczesny RJ, Borowski LS, Brzezniak LK, Dmochowska A, Gewartowski K, Bartnik E, Stepien PP. Human mitochondrial RNA turnover caught in flagranti: involvement of hSuv3p helicase in RNA surveillance. Nucleic Acids Res. 2010;38(1):279–98. doi: 10.1093/nar/gkp903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Khidr L, Wu G, Davila A, Procaccio V, Wallace D, Lee WH. Role of SUV3 helicase in maintaining mitochondrial homeostasis in human cells. J Biol Chem. 2008;283(40):27064–73. doi: 10.1074/jbc.M802991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang DD, Shu Z, Lieser SA, Chen PL, Lee WH. Human mitochondrial SUV3 and polynucleotide phosphorylase form a 330-kDa heteropentamer to cooperatively degrade double-stranded RNA with a 3′-to-5′ directionality. J Biol Chem. 2009;284(31):20812–21. doi: 10.1074/jbc.M109.009605. [DOI] [PMC free article] [PubMed] [Google Scholar]