Abstract
Chemokines and their receptors function in the recruitment and activation of cells of the immune system to sites of inflammation. As such, chemokines play an important role in mediating pathophysiological events during microbial infection. In particular, CXCL9, CXCL10 and CXCL11 and their cognate receptor CXCR3 have been associated with the clinical course of several infectious diseases, including severe acute respiratory syndrome (SARS) and influenza. While CXCL9, CXCL10 and CXCL11 share the same receptor and have overlapping functions, each can also have unique activity in host defense. The lack of a preferred characterized animal model for SARS has brought our attention to ferrets, which have been used for years in influenza studies. The lack of immunological reagents for ferrets prompted us to clone CXCL9, CXCL10, CXCL11 and CXCR3 and, in the case of CXCL10, to express the gene as a recombinant protein. In this study we demonstrate that endogenous ferret CXCL10 exhibits similar mRNA expression patterns in the lungs of deceased SARS patients and ferrets experimentally infected with SARS coronavirus. This study therefore represents an important step towards development of the ferret as a model for the role of CXCL9, CXCL10 and CXCL11:CXCR3 axis in severe viral infections.
Keywords: Ferret, Chemokines, CXCL9, CXCL10, CXCL11, CXCR3, SARS, Animal model
1. Introduction
The CXCL9 (MIG), CXCL10 (IP-10), and CXCL11 (I-TAC) chemokines, along with their cognate receptor CXCR3 participate in the development and regulation of Th1 responses. The expression of CXCL9, CXCL10, and CXCL11 can be induced in virtually all cell types by a variety of stimulants including IFN-γ, TLR activation and viral infection (Cameron and Kelvin, 2003, Farber, 1997, Rossi and Zlotnik, 2000). There is evidence in the literature that strongly suggests that CXCL10 is an important regulator of Th1 polarized immune responses (Krathwohl and Anderson, 2006), and that, in some instances, the CXCL9, CXCL10 and CXCL11:CXCR3 axis can generate pathological inflammatory conditions (Liu et al., 2001, Qin et al., 1998, Ruth et al., 2001, Sorensen and Ransohoff, 1998). While several reports have identified an increased expression of CXCL9 or CXCL10 following viral infection in animal models or humans (Cameron et al., 2007a; Christen et al., 2004, Dufour et al., 2002, Liu et al., 2001, Ruth et al., 2001), the role of these chemokines and their receptor CXCR3 in the development of acute respiratory distress syndrome (ARDS)-like diseases associated with pathogenic pulmonary viral infections is unknown. Support for a role of this subfamily of chemokines in SARS comes from observations by ourselves and other groups showing a correlation between increased systemic levels of CXCL10 in SARS coronavirus (SARS CoV) (Cameron et al., 2007b; Cheung et al., 2005, Tang et al., 2005) and H5N1-infected individuals (Peiris et al., 2004), as well as chronically elevated levels of systemic expression of CXCL10 in patients with poor outcome post-infection (Cameron et al., 2007b, Tang et al., 2005).
It has been suggested that the ferret is a more appropriate animal model for SARS compared to murine and primate models (Martina et al., 2003). Cynomolgus macaques (Macaca fascicularis) fail to show clinical symptoms following infection with SARS CoV, despite viral replication in the species (Marshall and Enserink, 2004, Martina et al., 2003). In mice, the SARS CoV replicates well, but the value of the model as representative of the human condition has been debated due to rapid clearance of the virus and the absence of clinical symptoms (Glass et al., 2004). In contrast, SARS CoV replicates well in ferrets with infected animals demonstrating a number of clinical manifestations similar to those seen in humans, such as lethargy and elevated body temperature (Martina et al., 2003). Furthermore, SARS CoV-infected ferrets display histological evidence of pneumonitis, and a certain degree of mortality has been reported, which is also consistent with the human disease (Marshall and Enserink, 2004, Martina et al., 2003). Replication of SARS CoV occurs both in the upper and lower respiratory tract, and development of ARDS-like disease also occurs in some patients (Ng et al., 2006, Ye et al., 2007). Diarrhea has been reported in a portion of cases of human H5N1 infection, and was also noted in many infected ferrets (Sidwell and Smee, 2000, Sweet and Smith, 1980, Zitzow et al., 2002).
The ferret is the preferred animal model for influenza research, including that with avian strains such as the H5N1 sub-type, as it closely mimics the human disease in terms of susceptibility to infection, transmission, pathology and spectrum of symptoms observed upon infection. For example, ferrets infected with highly pathogenic human isolates of H5N1 virus develop rhinorrea and ocular discharge, anorexia, otologic manifestations, fever, weight loss, dyspnea, pulmonary infiltrates and lymphopenia (Govorkova et al., 2005, Maines et al., 2005, Zitzow et al., 2002).
The major drawback to the use of the ferret model for either SARS CoV or influenza virus research is the general lack of immunological reagents. To investigate the roles of CXCL9, CXCL10, CXCL11 and CXCR3 in SARS and avian influenza pathogenesis in the ferret model, there is a need to develop ferret-specific antibodies to accurately assess chemokine expression levels in infected animals, and to neutralize any effects these chemokines might have in pathogenesis progression and clearance of infection.
As a first step towards this goal, we have cloned and sequenced the genes encoding ferret CXCL9, CXCL10, CXCL11 and CXCR3 orthologues, and then expressed and purified recombinant ferret CXCL10 protein. Next we showed that expression of CXCL9, CXCL10, and CXCL11 were induced by LPS stimulation of ferret PBMC cultures. We have also demonstrated that levels of the mRNA expression of CXCR3 and CXCL10 are increased in the lungs of deceased patients infected with SARS CoV during the SARS outbreak in Toronto. Furthermore, we have confirmed that the increased CXCL10 expression observed in these patients is paralleled in ferrets infected with SARS CoV.
2. Materials and methods
2.1. Animals
Six-month-old male Fitch ferrets (Mustela putorius furo) were purchased from Triple F Farms Inc. (Sayre, PA, USA). Animals used for non-infectious experimentation were housed at Toronto General Research Institute animal facility. The animal use protocol was approved by Animal Care Committee of the University Health Network, Toronto, Ontario. All experiments were conducted in accordance with committee recommendations. Animals were quarantined and monitored for 1 week prior to tissue and blood collection. The animals’ diet was based on a low fat/high protein regimen recommended by Triple F Farms for small carnivores. Animal experiments involving virus work were performed in the Animal Biohazard Safety Level 3 (ABSL3) facility at Southern Research Institute (Birmingham, AL, USA), in accordance with the approved protocols. Briefly, three male ferrets (castrated and descented), weighing approximately 800–1000 g, were infected intranasally with 103 TCID50 SARS CoV virus (TOR2 strain) in 1 mL PBS. An additional three animals were mock-infected using 1 mL PBS alone. Infected animals were deeply anesthetized and euthanized via exsanguination between days 5 and 7 post-infection, and lung tissue was obtained at necropsy and immediately homogenized in Tripure reagent (Roche Diagnostics, IN, USA).
2.2. Human lung biopsies and total RNA purification
Total RNA was purified using Tripure mRNA kits (Roche Diagnostics, Indianapolis, USA). Briefly, lung lower bilateral lobe biopsies collected from deceased SARS patients at autopsy were homogenized in 2 mL Tripure reagent in a 50 mL polypropylene tube at room temperature. Chloroform extraction of RNA was performed according to the manufacturer's recommended protocol. Informed consent was obtained from subjects or their substitute decision makers under the approval of the Research Ethics Boards of the University Health Network and participating Toronto area hospitals.
2.3. Ferret total RNA purification and cDNA synthesis
Ferret whole blood was diluted 1:1 with RPMI (Invitrogen, USA) and stimulated with mitogens, lipopolysaccharide (LPS) (1 μg/mL), phorbol myristate acetate (PMA) (50 ng/mL) and ionomycin (0.1 mM), or polyinosinic:polycytidylic acid (poly I:C) (25 μg/mL) by incubating at 37 °C in 5% CO2 for 1, 2, 4, 8, and 16 h prior to RNA purification. Paxgene RNA isolation method (Qiagen, Mississauga, Canada) was used according to manufacturer's protocols. Purified human or ferret total RNA was reverse transcribed to cDNA using reverse transcriptase II (Invitrogen, Carlsbad, USA) according to manufacturer's protocol.
2.4. Real-time RT-PCR
The following components were added to the reaction mixture plus cDNA to a total volume of 10 μL in distilled water: 0.25 μL cDNA, 250 nM forward gene-specific primer, 250 nM reverse gene-specific primer and 5 μL Cyber Green (Applied Biosystems, Foster City, CA, USA). In a single experiment, each reaction was performed in triplicate. An ABI 7900 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) was used for amplification. Initial denaturation was 15 min at 95 °C, followed by 40 cycles of amplification. Each cycle consisted of a denaturation step (15 s at 95 °C) and an annealing/extension step (1 min at 60 °C). Gene expression levels were normalized to β-actin and data was analyzed by SDS 2.1 software (Applied Biosystems, CA, USA).
2.5. Amplification, cloning and sequencing of ferret-specific genes
Purified RNA was reverse transcribed to cDNA using reverse transcriptase II according to the manufacturer's protocol (Invitrogen, Carlsbad, USA). Gene-specific degenerate primers were designed based on multiple gene sequence alignment analysis of several species using the ClustalW (1.83) web-based program from European Bioinformatics Institute (EBI) and then used to clone the cDNAs for each gene. Primers were tested in silico using Primer Express (Applied Biosystems, CA, USA). Standard PCR was performed using degenerate primers and template cDNA. Specific bands were gel purified according to manufacturer's instructions (Qiagen, Mississauga, Canada) and cloned into the pCR 2.1-TOPO vector (Invitrogen, Calsbad, USA). Sequences of positive clones were confirmed using ABI 3730XL DNA analyzers (Center for Applied Genomics, Toronto, Ontario). Identification of genes was carried out by basic local alignment search tool (BLAST) analyses against National Centre for Biotechnology Information (NCBI) data (http://www.ncbi.nlm.nih.gov/BLAST/).
2.6. Generation of an expression vector for ferret CXCL10
Kozak sequences were engineered at the 5′ end of the ferret CXCL10 cDNA and the 3′ end termination codon was removed prior to sub-cloning into the pcDNA3.1/His6.V5/TOPO expression vector (Invitrogen, Mississauga, Canada). The removal of the termination codon enabled the cloned gene to be expressed as a fusion protein that is tagged at the C-terminus with the His6 and V5 epitopes. Sequencing was repeated to ensure that no errors had been introduced and to verify that the gene had been inserted in the correct open reading frame.
2.7. Cell culture, plasmid transfections, and protein purification
COS-7 cells were maintained in Dulbecco's modified eagle's medium (DMEM), substituted with 10% fetal bovine serum (Invitrogen, Mississauga, Canada) at 37 °C, 5% CO2. COS-7 cells (1 × 107) were transiently transfected with Effectene (Qiagen, Canada) according to recommended protocols of the manufacturer. After 24–48 h of incubation, conditioned media was run through Ni–NTA metal immobilized affinity columns (0.5–1 mL). Bound protein was washed and eluted according to manufacturer's suggested protocols. Eluted fractions were screened via Western blot using antibodies that specifically recognize the epitope tags. Positive fractions were pooled and dialyzed against phosphate buffered saline (PBS) at 4 °C and concentrated by lyophilization.
2.8. Western blot analysis
SDS-polyacrylamide gel electrophoresis (SDS-PAGE) was performed with pre-cast gels (10–15%, Bio-Rad, USA) according to standard protocols. Protein blots were blocked with 5% milk protein/phosphate buffered saline-Tween 0.02% (TPBS) buffer for 1 h at room temperature followed by 16 h incubation at 4 °C with mouse-anti-His6 primary antibody (Invitrogen, Canada) at 508 ng/mL. The blots were washed with TPBS and incubated with goat-anti-mouse-HRP secondary antibody (Santa Cruz, USA) for 1 h at room temperature. Bands were visualized using enhanced chemiluminescent (ECL) reagents according to the manufacturer's protocol (GE Healthcare, Canada). Goat polyclonal anti-human CXCL10 antibody (R&D Systems, USA) was used to demonstrate cross-reactivity with ferret recombinant CXCL10.
2.9. Isolation of peripheral blood mononuclear cells (PBMCs) for chemotaxis assays
PBMCs were isolated from ferret whole blood by centrifuging over Histopaque density gradient (Sigma Chemicals, St. Louis, MO) according to manufacturer's protocol. Briefly, whole blood was diluted 1:1 with PBS (10 mL), layered over 5 mL histopaque solution, and centrifuged at 400 × g for 20 min. The enriched mononuclear cell layer at the histopaque–plasma interface was harvested, washed twice in PBS, and resuspended in chemotaxis binding buffer (RPMI 1640, 1% bovine serum albumin, 25 mM HEPES).
2.10. Chemotaxis assay
The Boyden chamber procedure was performed as described previously (Xu et al., 1995). Briefly, a 25 μL aliquot of recombinant ferret CXCL10 was diluted in chemotaxis medium was placed in the lower compartment and 50 μL of PBMC suspension (1 × 106 cells/mL) was placed in the upper compartment. The two compartments were separated by a polycarbonate filter (5 μm pore size; Neuroprobe, Cabin John, MD). The chamber was incubated for 3 h at 37 °C in humidified air with 5% CO2. At the end of the incubation period, filters were removed fixed and stained with Diff-Quik (Harlew, Gibbs-town, NJ). The number of migrating cells was counted in three high power fields (400×) by light microscopy. Results are expressed as mean ± S.D. for at least three independent experiments.
2.11. Gene accession numbers
Nucleotide sequences of the complete coding regions of CXCL9, CXCL10 and CXCL11 were submitted to the GeneBank. Partial nucleotide sequence of CXCR3 coding sequence was also submitted and the following accession numbers were obtained:
Ferret CXCL9 (EF492057), Ferret CXCL10 (EF492058), Ferret CXCL11 (EF492059) and Ferret CXCR3 (EF492060).
2.12. Statistical analysis
The Student's t-test or the Mann–Whitney rank sum test for two independent populations was used for statistical analysis via SPSS for Windows V13.0 software (SPSS Inc., Chicago, IL). P≤0.05 was considered significant.
3. Results
3.1. Cloning and sequence analysis of ferret CXCL9, CXCL10, CXCL11, and CXCR3
The cloning of ferret genes encoding CXCL9, CXCL10, CXCL11 and CXCR3 was performed using ferret cDNA derived from PBMC cultures stimulated with various mitogens to induce cytokines and chemokines expression (see Section 2). Each gene was amplified by primers designed from consensus sequences derived from multiple nucleotide sequence alignments among various species. In the case of CXCL10, the full-length coding region was not fully sequenced due to the lack of sequence conservation at the 5′ and 3′ end of the coding region. To address this issue, sequences upstream and downstream of the open reading frame were sequenced by RNA ligase mediated rapid amplification of cDNA ends (RLM–RACE) (data not shown). For CXCR3, we amplified and sequenced a partial cDNA within the coding region. As expected, the predicted full-length amino acid sequences for ferret CXCL9, CXCL10, CXCL11 (126, 98 and 94 amino acids, respectively) and the predicted partial CXCR3 sequence (229 out of 368 amino acids) show significant homology to corresponding sequences in various other species (Fig. 1A–D). Phylogenetic analysis was performed using ClustalW and phylogenetic trees constructed using protein parsimony with a bootstrap value of 100. The ferret nucleotide sequences were translated to amino acid sequences using “Translate” (ExPASy, proteomics). Amino acid sequences of other species were obtained from Genbank (http://www.ncbi.nlm.nih.gov/Genbank). The phylogenetic tree shown for genes indicated a higher similarity existed between ferret gene and those of humans and order carnivora than between the ferret and order rodentia (Fig. 2A and B, Table 1 ).
Fig. 1.
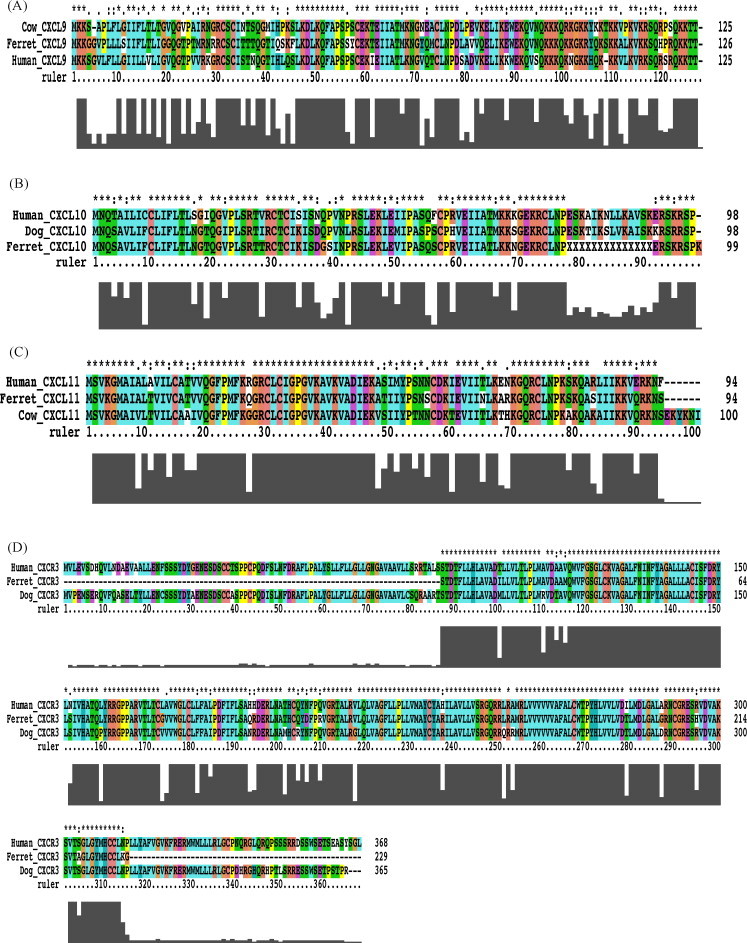
Multiple alignments of the primary amino acid sequences of CXCL9, CXCL10, CXCL11, and CXCR3. The putative translated full-length protein sequences of ferret CXCL9 (A), CXCL10 (B), CXCL11 (C) and the putative translated partial sequence of ferret CXCR3 (D) were compared with orthologous protein sequences from human, dog and or cow, were available. CXCL9: EF492057, XM_591770, BC063122; CXCL10: EF492058, AB183191, NM_001565; CXCL11: EF492059, XM_594243, NM_005409; CXCR3: EF492060, AB185149, NM_001504.
Fig. 2.
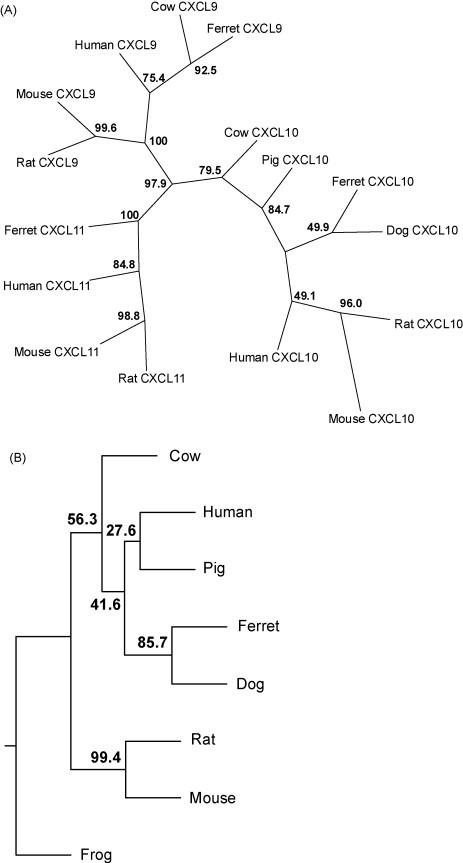
Phylogenetic analysis of ferret CXCL9, CXCL10, CXCL11, and CXCR3. The full-length putative protein sequences obtained for (A) ferret CXCL9, CXCL10 and CXCL11 and (B) partial putative protein sequence of ferret CXCR3 were aligned with all available orthologues from cow, dog, human, mouse, rat, pig and frog using ClustalW 1.83, and phylogenetic trees constructed using protein parsimony with a bootstrap value of 100. All chemokines cluster specifically within their orthologous groups. Accession numbers: CXCL9—NP_663705, NP_032625, NP_002407, XP_591770; CXCL10: NP_001008691, NP_620789, NP_067249, NP_001556, NP_001010949, NP_001040016; CXCL11: NP_891997, NP_062367, NP_005400; CXCR3: NP_001011067, CAH64842, NP_445867, NP_034040, NP_001011673, NP_0011495, NP_001011887.
Table 1.
Amino acid identity of ferret CXCL9, CXCL10, CXCL11 and CXCR3 with known species sequences
| Species | CXCL9 | CXCL10 | CXCL11 | CXCR3 |
|---|---|---|---|---|
| Human | 68 | 80 | 86 | 91 |
| Dog | – | 81 | – | 92 |
| Cow | 72 | 80 | 82 | 89 |
| Mouse | 58 | 67 | 63 | 86 |
Values shown represent % amino acid identity with highest % for each gene in bold.
3.2. Expression of recombinant ferret CXCL10 protein and detection by western blot
A C-terminal His6/V5-tagged expression construct was generated in order to produce recombinant ferret CXCL10 protein (predicted molecular weight of 18.5 kDa, including epitope tags). We transiently transfected COS-7 cells with this expression construct and purified recombinant ferret CXCL10 by affinity chromatography. Eluted fractions were subjected to western blot analysis using anti-His antibody. A single band was noted at the expected molecular weight of 18.5 kDa. COS-7 cells transfected with empty vector did not show reactivity with anti-His antibody, while recombinant ferret IFN-γ reacted with same antibody at 34 kDa as a positive control (Fig. 3A). As recombinant ferret CXCL10 shows a high degree of homology with human CXCL10, we tested whether our ferret recombinant CXCL10 would be recognized by an anti-human CXCL10 polyclonal antibody. As shown in Fig. 3B, ferret recombinant CXCL10 and human recombinant CXCL10 are reactive with a polyclonal goat anti-human CXCL10 antibody.
Fig. 3.
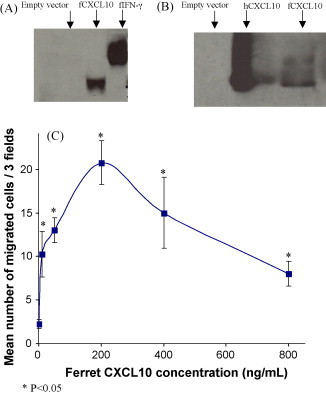
The recombinant ferret CXCL10 protein migrates to 18.5 kDa and is active in cell migration assays. (A) Anti-His6 western blot of purified recombinant ferret CXCL10 (fCXCL10) protein (from elution 2) shows a single band at 18.5 kDa compared with ferret IFN-γ (fIFN-γ) with a band at 34 KDa. (B) Goat anti-human CXCL10 (hCXCL10) polyclonal antibody is cross-reactive with ferret CXCL10 (fCXCL10) recombinant protein. (C) Migration of ferret PBMCs following the exposure to increasing doses of recombinant ferret CXCL10 protein. The results are expressed as mean cell count of three high-powered fields obtained in three experiments. *P < 0.05 compared to migration induced by medium alone using Student's t-test.
3.3. Chemotactic activity of recombinant ferret CXCL10 protein
The purified recombinant ferret CXCL10 protein was tested for chemotactic activity using ferret PBMC cultures in a Boyden chamber. Chemotactic migration of ferret PBMCs increased upon treatment with ferret CXCL10 protein in a dose-dependent manner. Maximal migration occurred at 200 ng/mL ferret CXCL10 protein, above which concentration a declining response was observed. Therefore our CXCL10-induced chemotaxis data conformed to a classical bell-shaped chemotaxis dose–response curve, and was consistent with previous reports for recombinant human CXCL9 (Liao et al., 1995) and recombinant human IL-8 (Xu et al., 1995). Recombinant ferret CXCL10 attracted a statistically significant greater number of PMBCs when compared to the negative control wells (media alone) at all concentrations used in the assay (P < 0.05) (Fig. 3C). These results confirmed that our recombinant ferret CXCL10 protein demonstrated biological activity.
3.4. Stimulation of ferret PBMCs with LPS induces mRNA expression of CXCL9, CXCL10 and CXCL11
We predict that stimulation of ferret PBMC cells with lipopolysaccharide (LPS) should promote the secretion of pro-inflammatory cytokines and chemokines in accordance to data from other species (Loos et al., 2006, Nau et al., 2003, Tassiulas et al., 2007). Sequence analysis of ferret CXCL9, CXCL10 and CXCL11 genes enabled us to design primers for use in real-time PCR experiments in order to detect gene expression of these chemokines. Initially, we set out to validate the specificity of the primers and then aimed to test whether CXCL9, CXCL10 and CXCL11 transcription is induced in ferret blood cells upon LPS treatment. Quantitative real-time PCR on cDNA derived from ferret whole blood stimulated with LPS was performed and demonstrated that CXCL9, CXCL10 and CXCL11 mRNA expression was induced at various times following LPS treatment relative to unstimulated control cDNA. The peak increase for CXCL9 (3-fold above unstimulated control, P < 0.05) and CXCL11 (greater than 8-fold above unstimulated control, P < 0.01) occurred at 4 h post-stimulation, while for CXCL10 the peak increase (60-fold above unstimulated control, P < 0.01) occurred at 2 h following LPS treatment (Fig. 4 ).
Fig. 4.
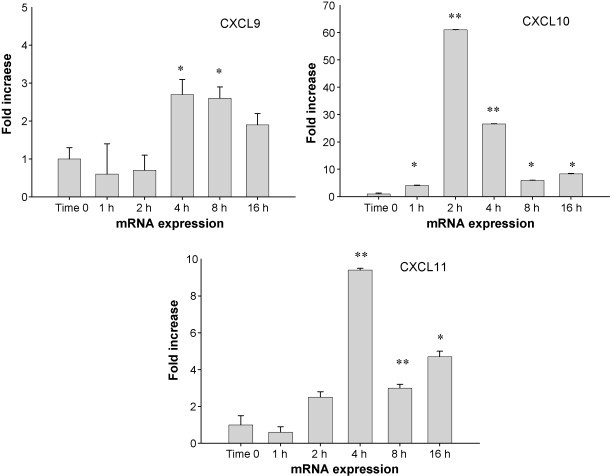
Induction of chemokine gene expression by lipopolysaccharide. Ferret peripheral blood was stimulated with 0.1 mg/mL LPS for the time points indicated. Utilizing primers specific for the ferret CXCL9, CXCL10, and CXCL11 genes, mRNA expression was quantified. The Y-axis indicates the abundance of mRNA after stimulation with LPS compared to untreated samples. Treatment time is indicated by X-axis. Error bars indicate the S.D. of the relative quantities of the triplicates. Quantity readings of triplicates at each time point are compared with quantity readings of unstimulated triplicates, using Student's t-test. The peak increases for CXCL9 and CXCL11 occurred at 4 h post-stimulation (P < 0.05 and P < 0.01, respectively), while for CXCL10 was at 2 h following LPS treatment (P < 0.01) (*P < 0.05, **P < 0.01).
3.5. Increased CXCL10 and CXCR3 mRNA expression in lung biopsies from fatal outcome SARS patients at autopsy
We and others have demonstrated increased levels of CXCL10 in the plasma of SARS patients during acute and progressive illness (Cameron et al., 2007b, Chien et al., 2006, Ng et al., 2005, Tang et al., 2005). In order to compare the expression of CXCL10 in lung biopsies of ferrets infected with SARS CoV, we evaluated the expression of CXCL10 and its receptor CXCR3 in the lungs of fatal outcome SARS patients as well. Real-time PCR analysis was performed on cDNA purified from the lung biopsies of three deceased SARS patients and three control patients who died from non-respiratory illnesses. The results show high expression of CXCL10 and CXCR3 mRNA in SARS patients (P < 0.05) and no expression in control cadaveric lung biopsies (Fig. 5A and B).
Fig. 5.
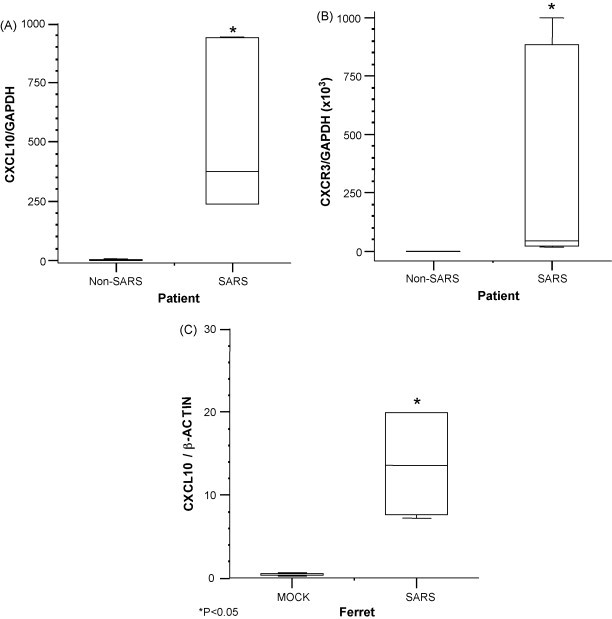
CXCL10 and CXCR3 mRNA expression in lung tissues of humans and ferrets infected with SARS CoV. (A) CXCL10 and (B) CXCR3 are significantly increased in the lungs of fatal outcome SARS (P < 0.05). RNA was isolated from bilateral lower lung biopsies from three deceased SARS patients and from lower lung biopsies from three control cadaveric lungs. (C) CXCL10 mRNA levels in the lungs of mock vs. SARS CoV-infected ferrets. Data represent averages from three animals per group, performed in triplicate wells. Asterisks indicate statistical significance using Mann–Whitney rank sum test (P < 0.05).
3.6. The expression of CXCL10 in lung biopsies of ferrets infected with SARS CoV
In order to further dissect the ferret model of SARS CoV infection and determine if it is representative of the disease in humans on a molecular basis, we endeavored to determine whether if the mRNA expression of the chemokine CXCL10 is increased in SARS-infected ferrets. Real-time PCR analysis of CXCL10 gene expression in lung tissue from both SARS-infected and uninfected ferrets was carried out. Consistent with our findings in human SARS patients, ferrets experimentally infected with SARS CoV demonstrated a significant increase in CXCL10 mRNA expression in the lower respiratory tract compared to uninfected control animals (P < 0.05) (Fig. 5C).
4. Discussion
The CXCL9, CXCL10 and CXCL11–CXCR3 axis has been reported to play a role in the immune response following viral infection (Christensen et al., 2006, Luster, 1998, Tang et al., 2005, Tschen et al., 2006, Xie et al., 2003). The ferret represents an important animal model for a number of human viral infectious diseases (Martina et al., 2003, Suguitan et al., 2006, Vos et al., 2004). We report here for the first time cloning and sequencing of the ferret genes for CXCL9, CXCL10, CXCL11 and CXCR3. The ferret sequences of these chemokines and their cognate receptor were highly conserved with human, dog and cow sequences.
Purified recombinant ferret CXCL10 was detected as a single band on anti-His6 western blots that ran close to the predicted molecular weight of 18.5 kDa. Since recombinant CXCL10 was expressed in a mammalian cell line (COS-7), it is reasonable to assume that the recombinant protein is properly processed at the post-translational level. Furthermore, we have demonstrated that recombinant ferret CXCL10 protein exhibited biological activity in terms of stimulating chemotaxis in peripheral blood mononuclear cells.
CXCL9, CXCL10 and CXCL11 are non-ELR chemokines found at increased levels in a variety of viral diseases (Luster, 1998). Previous work has shown that CXCL10 mRNA expression is strongly up-regulated in conjunction with CXCR3 in mice infected with lymphocytic choriomeningitis virus (LCMV), particularly in the pancreas and at early stages of infection (Christen et al., 2004). More recently, it has demonstrated that LCMV infection of the murine central nervous system (CNS) is associated with CXCL9 and CXCL11 up-regulation (Christensen et al., 2006). Several animal models have shown that inflammation-based pathology induced by viral infections can be reduced by either neutralization of chemokines or blockade of their receptor (Carr et al., 2003, Dawson et al., 2000, Xie et al., 2003). Treatment of spontaneously diabetic NOD mice with CXCL10 neutralizing antibodies suppressed the incidence of diabetes (Shigihara et al., 2005). In the MHV mouse model of encephalitis, neutralization of CXCL9 or CXCL10 (or in CXCL10-null mice) resulted in reduced neuropathology despite increased viral load (Dufour et al., 2002, Liu et al., 2001).
The absence of immunological reagents for the ferret model of SARS and H5N1 has hindered a direct investigation into the role played by the CXCL9, CXCL10 and CXCL11–CXCR3 axis in the immmunopathogenesis of viral disease. The availability of the cloned genes for ferret CXCL9, CXCL11 and CXCR3, will enable the expression and purification of recombinant proteins. Biologically active recombinant purified CXCL10 protein has been prepared and will be utilized immediately in a number of in vitro and in vivo assays. Using these proteins we intend to generate specific monoclonal and polyclonal neutralizing antibodies to investigate the involvement of these chemokines in the pathogenesis of SARS and H5N1 diseases.
We and others have previously reported the elevation of CXCL10 in the serum of SARS patients (Cameron et al., 2007b, Chien et al., 2006; Ng et al., 2005; Tang et al., 2005). The chemoattractant CXCL10, which recruits mainly Th1 cells and NK cells to the site of infection, was markedly increased in SARS (Cameron et al., 2007b). Furthermore, the depletion of lymphocyte subsets from peripheral blood (unpublished data) might be partly due to the influence of CXCL10 in that up-regulation of CXCL10 in the lung might be influential in the recruitment and sequestration of lymphocyte subsets to the site of infection. This in turn may lead to severe pathological consequences as described in a number of viral infections including SARS and bird flu (Ng et al., 2006).
In conclusion, we have demonstrated increased gene expression of CXCL10 and its receptor CXCR3 in lung biopsies of deceased SARS patients. We observed a similar pattern of CXCL10 mRNA expression in the lungs of ferrets infected with SARS CoV, lending further support to the argument that ferrets are a highly representative model of the human disease. We have identified a subfamily of chemokines and their cognate receptor in ferrets and carried out an initial characterization of one of these chemokines. This study is a fundamental step towards the development of a more readily available array of immunological tools for use in investigating the mechanism of disease in this natural model of viral infection.
Acknowledgments
We thank Feseha Abebe-Akele for technical assistance. This project was supported by funding from NIAID through NIH/NIAID Contract No. N01-AI-30063 Task Order No. 03.
References
- Cameron, M.J., Bermejo-Martin, J.F., Danesh, A., Muller, M.P., Kelvin, D.J., 2007a. Human immunopathogenesis of severe acute respiratory syndrome (SARS). Virus Res. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- Cameron M.J., Kelvin D.J. Cytokines and chemokines—their receptors and their genes: an overview. Adv. Exp. Med. Biol. 2003;520:8–32. doi: 10.1007/978-1-4615-0171-8_2. [DOI] [PubMed] [Google Scholar]
- Cameron M.J., Ran L., Xu L., Danesh A., Bermejo-Martin J.F., Cameron C.M., Muller M.P., Gold W.L., Richardson S.E., Poutanen S.M., Willey B.M., Devries M.E., Fang Y., Seneviratne C., Bosinger S.E., Persad D., Wilkinson P., Greller L.D., Somogyi R., Humar A., Keshavjee S., Louie M., Loeb M.B., Brunton J., McGeer A.J., Kelvin D.J. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in severe acute respiratory syndrome (SARS) patients. J. Virol. 2007;81(16):8692–8706. doi: 10.1128/JVI.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr D.J., Chodosh J., Ash J., Lane T.E. Effect of anti-CXCL10 monoclonal antibody on herpes simplex virus type 1 keratitis and retinal infection. J. Virol. 2003;77:10037–10046. doi: 10.1128/JVI.77.18.10037-10046.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung C.Y., Poon L.L., Ng I.H., Luk W., Sia S.F., Wu M.H., Chan K.H., Yuen K.Y., Gordon S., Guan Y., Peiris J.S. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J. Virol. 2005;79:7819–7826. doi: 10.1128/JVI.79.12.7819-7826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien J.Y., Hsueh P.R., Cheng W.C., Yu C.J., Yang P.C. Temporal changes in cytokine/chemokine profiles and pulmonary involvement in severe acute respiratory syndrome. Respirology. 2006;11:715–722. doi: 10.1111/j.1440-1843.2006.00942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen U., Benke D., Wolfe T., Rodrigo E., Rhode A., Hughes A.C., Oldstone M.B., von Herrath M.G. Cure of prediabetic mice by viral infections involves lymphocyte recruitment along an IP-10 gradient. J. Clin. Invest. 2004;113:74–84. doi: 10.1172/JCI200417005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J.E., de Lemos C., Moos T., Christensen J.P., Thomsen A.R. CXCL10 is the key ligand for CXCR3 on CD8+ effector T cells involved in immune surveillance of the lymphocytic choriomeningitis virus-infected central nervous system. J. Immunol. 2006;176:4235–4243. doi: 10.4049/jimmunol.176.7.4235. [DOI] [PubMed] [Google Scholar]
- Dawson T.C., Beck M.A., Kuziel W.A., Henderson F., Maeda N. Contrasting effects of CCR5 and CCR2 deficiency in the pulmonary inflammatory response to influenza A virus. Am. J. Pathol. 2000;156:1951–1959. doi: 10.1016/S0002-9440(10)65068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour J.H., Dziejman M., Liu M.T., Leung J.H., Lane T.E., Luster A.D. IFN-gamma-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. J. Immunol. 2002;168:3195–3204. doi: 10.4049/jimmunol.168.7.3195. [DOI] [PubMed] [Google Scholar]
- Farber J.M. Mig and IP-10: CXC chemokines that target lymphocytes. J. Leukoc. Biol. 1997;61:246–257. [PubMed] [Google Scholar]
- Glass W.G., Subbarao K., Murphy B., Murphy P.M. Mechanisms of host defense following severe acute respiratory syndrome-coronavirus (SARS-CoV) pulmonary infection of mice. J. Immunol. 2004;173:4030–4039. doi: 10.4049/jimmunol.173.6.4030. [DOI] [PubMed] [Google Scholar]
- Govorkova E.A., Rehg J.E., Krauss S., Yen H.L., Guan Y., Peiris M., Nguyen T.D., Hanh T.H., Puthavathana P., Long H.T., Buranathai C., Lim W., Webster R.G., Hoffmann E. Lethality to ferrets of H5N1 influenza viruses isolated from humans and poultry in 2004. J. Virol. 2005;79:2191–2198. doi: 10.1128/JVI.79.4.2191-2198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krathwohl M.D., Anderson J.L. Chemokine CXCL10 (IP-10) is sufficient to trigger an immune response to injected antigens in a mouse model. Vaccine. 2006;24:2987–2993. doi: 10.1016/j.vaccine.2005.11.032. [DOI] [PubMed] [Google Scholar]
- Liao F., Rabin R.L., Yannelli J.R., Koniaris L.G., Vanguri P., Farber J.M. Human Mig chemokine: biochemical and functional characterization. J. Exp. Med. 1995;182:1301–1314. doi: 10.1084/jem.182.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.T., Keirstead H.S., Lane T.E. Neutralization of the chemokine CXCL10 reduces inflammatory cell invasion and demyelination and improves neurological function in a viral model of multiple sclerosis. J. Immunol. 2001;167:4091–4097. doi: 10.4049/jimmunol.167.7.4091. [DOI] [PubMed] [Google Scholar]
- Loos T., Dekeyzer L., Struyf S., Schutyser E., Gijsbers K., Gouwy M., Fraeyman A., Put W., Ronsse I., Grillet B., Opdenakker G., Van Damme J., Proost P. TLR ligands and cytokines induce CXCR3 ligands in endothelial cells: enhanced CXCL9 in autoimmune arthritis. Lab. Invest. 2006;86:902–916. doi: 10.1038/labinvest.3700453. [DOI] [PubMed] [Google Scholar]
- Luster A.D. Chemokines—chemotactic cytokines that mediate inflammation. N. Engl. J. Med. 1998;338:436–445. doi: 10.1056/NEJM199802123380706. [DOI] [PubMed] [Google Scholar]
- Maines T.R., Lu X.H., Erb S.M., Edwards L., Guarner J., Greer P.W., Nguyen D.C., Szretter K.J., Chen L.M., Thawatsupha P., Chittaganpitch M., Waicharoen S., Nguyen D.T., Nguyen T., Nguyen H.H., Kim J.H., Hoang L.T., Kang C., Phuong L.S., Lim W., Zaki S., Donis R.O., Cox N.J., Katz J.M., Tumpey T.M. Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. J. Virol. 2005;79:11788–11800. doi: 10.1128/JVI.79.18.11788-11800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall E., Enserink M. Medicine. Caution urged on SARS vaccines. Science. 2004;303:944–946. doi: 10.1126/science.303.5660.944. [DOI] [PubMed] [Google Scholar]
- Martina B.E., Haagmans B.L., Kuiken T., Fouchier R.A., Rimmelzwaan G.F., Van Amerongen G., Peiris J.S., Lim W., Osterhaus A.D. Virology: SARS virus infection of cats and ferrets. Nature. 2003;425:915. doi: 10.1038/425915a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nau G.J., Schlesinger A., Richmond J.F., Young R.A. Cumulative toll-like receptor activation in human macrophages treated with whole bacteria. J. Immunol. 2003;170:5203–5209. doi: 10.4049/jimmunol.170.10.5203. [DOI] [PubMed] [Google Scholar]
- Ng P.C., Lam C.W., Li A.M., Wong C.K., Leung T.F., Cheng F.W., Hon K.L., Chan I.H., Wong E., Fok T.F. Chemokine response in children with SARS. Arch. Dis. Child. 2005;90:422–423. doi: 10.1136/adc.2004.053660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng W.F., To K.F., Lam W.W., Ng T.K., Lee K.C. The comparative pathology of severe acute respiratory syndrome and avian influenza A subtype H5N1—a review. Hum. Pathol. 2006;37:381–390. doi: 10.1016/j.humpath.2006.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Yu W.C., Leung C.W., Cheung C.Y., Ng W.F., Nicholls J.M., Ng T.K., Chan K.H., Lai S.T., Lim W.L., Yuen K.Y., Guan Y. Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet. 2004;363:617–619. doi: 10.1016/S0140-6736(04)15595-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S., Rottman J.B., Myers P., Kassam N., Weinblatt M., Loetscher M., Koch A.E., Moser B., Mackay C.R. The chemokine receptors CXCR3 and CCR5 mark subsets of T cells associated with certain inflammatory reactions. J. Clin. Invest. 1998;101:746–754. doi: 10.1172/JCI1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D., Zlotnik A. The biology of chemokines and their receptors. Annu. Rev. Immunol. 2000;18:217–242. doi: 10.1146/annurev.immunol.18.1.217. [DOI] [PubMed] [Google Scholar]
- Ruth J.H., Rottman J.B., Katschke K.J., Jr., Qin S., Wu L., LaRosa G., Ponath P., Pope R.M., Koch A.E. Selective lymphocyte chemokine receptor expression in the rheumatoid joint. Arthritis Rheum. 2001;44:2750–2760. doi: 10.1002/1529-0131(200112)44:12<2750::aid-art462>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Shigihara T., Shimada A., Oikawa Y., Yoneyama H., Kanazawa Y., Okubo Y., Matsushima K., Yamato E., Miyazaki J., Kasuga A., Saruta T., Narumi S. CXCL10 DNA vaccination prevents spontaneous diabetes through enhanced beta cell proliferation in NOD mice. J. Immunol. 2005;175:8401–8408. doi: 10.4049/jimmunol.175.12.8401. [DOI] [PubMed] [Google Scholar]
- Sidwell R.W., Smee D.F. In vitro and in vivo assay systems for study of influenza virus inhibitors. Antiviral Res. 2000;48:1–16. doi: 10.1016/s0166-3542(00)00125-x. [DOI] [PubMed] [Google Scholar]
- Sorensen T.L., Ransohoff R.M. Etiology and pathogenesis of multiple sclerosis. Semin. Neurol. 1998;18:287–294. doi: 10.1055/s-2008-1040879. [DOI] [PubMed] [Google Scholar]
- Suguitan A.L., Jr., McAuliffe J., Mills K.L., Jin H., Duke G., Lu B., Luke C.J., Murphy B., Swayne D.E., Kemble G., Subbarao K. Live, attenuated influenza A H5N1 candidate vaccines provide broad cross-protection in mice and ferrets. PLoS Med. 2006;3:e360. doi: 10.1371/journal.pmed.0030360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet C., Smith H. Pathogencity of influenza viruses. Microbiol. Rev. 1980;44:303–330. doi: 10.1128/mr.44.2.303-330.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N.L., Chan P.K., Wong C.K., To K.F., Wu A.K., Sung Y.M., Hui D.S., Sung J.J., Lam C.W. Early enhanced expression of interferon-inducible protein-10 (CXCL-10) and other chemokines predicts adverse outcome in severe acute respiratory syndrome. Clin. Chem. 2005;51:2333–2340. doi: 10.1373/clinchem.2005.054460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassiulas I., Park-Min K.H., Hu Y., Kellerman L., Mevorach D., Ivashkiv L.B. Apoptotic cells inhibit LPS-induced cytokine and chemokine production and IFN responses in macrophages. Hum. Immunol. 2007;68:156–164. doi: 10.1016/j.humimm.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschen S.I., Stohlman S.A., Ramakrishna C., Hinton D.R., Atkinson R.D., Bergmann C.C. CNS viral infection diverts homing of antibody-secreting cells from lymphoid organs to the CNS. Eur. J. Immunol. 2006;36:603–612. doi: 10.1002/eji.200535123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos A., Muller T., Cox J., Neubert L., Fooks A.R. Susceptibility of ferrets (Mustela putorius furo) to experimentally induced rabies with European Bat Lyssaviruses (EBLV) J. Vet. Med. B: Infect. Dis. Vet. Public Health. 2004;51:55–60. doi: 10.1111/j.1439-0450.2004.00730.x. [DOI] [PubMed] [Google Scholar]
- Xie J.H., Nomura N., Lu M., Chen S.L., Koch G.E., Weng Y., Rosa R., Di Salvo J., Mudgett J., Peterson L.B., Wicker L.S., DeMartino J.A. Antibody-mediated blockade of the CXCR3 chemokine receptor results in diminished recruitment of T helper 1 cells into sites of inflammation. J. Leukoc. Biol. 2003;73:771–780. doi: 10.1189/jlb.1102573. [DOI] [PubMed] [Google Scholar]
- Xu L., Kelvin D.J., Ye G.Q., Taub D.D., Ben Baruch A., Oppenheim J.J., Wang J.M. Modulation of IL-8 receptor expression on purified human T lymphocytes is associated with changed chemotactic responses to IL-8. J. Leukoc. Biol. 1995;57:335–342. doi: 10.1002/jlb.57.2.335. [DOI] [PubMed] [Google Scholar]
- Ye J., Zhang B., Xu J., Chang Q., McNutt M.A., Korteweg C., Gong E., Gu J. Molecular pathology in the lungs of severe acute respiratory syndrome patients. Am. J. Pathol. 2007;170:538–545. doi: 10.2353/ajpath.2007.060469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitzow L.A., Rowe T., Morken T., Shieh W.J., Zaki S., Katz J.M. Pathogenesis of avian influenza A (H5N1) viruses in ferrets. J. Virol. 2002;76:4420–4429. doi: 10.1128/JVI.76.9.4420-4429.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]


