Abstract
Better biomarkers to detect smoking are needed given the tremendous public health burden caused by smoking. Current biomarkers to detect smoking have significant limitations, notably a short half-life for detection and lack of sensitivity for light smokers. These limitations may be particularly problematic in populations with less accurate self-reporting. Prior epigenome-wide association studies indicate that methylation status at cg05575921, a CpG residue located in the aryl hydrocarbon receptor repressor (AHRR) gene, may be a robust indicator of smoking status in individuals with as little as half of a pack-year of smoking. In this study, we show that a novel droplet digital PCR assay for measuring methylation at cg05575921 can reliably detect smoking status, as confirmed by serum cotinine, in populations with different demographic characteristics, smoking histories, and rates of false-negative self-report of smoking behavior. Using logistic regression models, we show that obtaining maximum accuracy in predicting smoking status depends on appropriately weighting self-report and cg05575921 methylation according to the characteristics of the sample being tested. Furthermore, models using only cg05575921 methylation to predict smoking perform nearly as well as those also including self-report across populations. In conclusion, cg05575921 has significant potential as a clinical biomarker to detect smoking in populations with varying rates of accuracy in self-report of smoking behavior.
Keywords: Biomarkers, epigenetics, substance use disorders, addiction, tobacco
Introduction
Cigarette smoking is the leading cause of preventable morbidity and mortality in the United States, with the complications of smoking being responsible for nearly half a million deaths per year (DHHS 2014). In 2010, nearly 20 percent of all US adults were current cigarette smokers, with rates varying by age group, gender, and race (Centers for Disease Control and Prevention 2011). Smoking is more prevalent among those aged 25–44 (22%), men compared to women, and higher among African Americans and European Americans (both approximately 21%) than Hispanics (12.5%). A critical period in the trajectory of smoking behaviors is adolescence and early adulthood. Over the age interval of 14 to 25, the rate of smoking increases from 4.3% to 32.8% (SAMHSA 2011).
Accurate detection of smoking status is a high priority in multiple clinical scenarios. For adolescents, detection of intermittent smoking may allow targeting of preventative interventions in order to prevent progression to nicotine dependence and the long-term risks associated with regular smoking (Shadel and others 2000). In smokers who are already nicotine dependent, detection of current smoking status can allow clinicians to refer those individuals for smoking cessation treatments (Ranney and others 2006). Lastly, detection of smoking status is important in monitoring for relapse after smoking cessation treatment has been completed (Jatlow and others 2008; McClure 2001).
In current clinical practice, assessment of smoking is usually limited to self-report (Larzelere and Williams 2012). While self-report has been shown to be generally accurate in adults in epidemiologic studies (Caraballo and others 2001; Vartiainen and others 2002), there are important exceptions in some populations. For example, pregnant women (Britton and others 2004), adolescents (Caraballo and others 2004; Kandel and others 2006), and African Americans (Wagenknecht and others 1992) have all been reported to have elevated rates of disagreement between self-report of smoking behavior and objective measures of recent smoking, such as cotinine concentration. In addition, unreliable self-report is a well-established phenomenon in nicotine dependence treatment populations (Hilberink and others 2011; Philibert and others 2016).
Cotinine, the major metabolite of nicotine, is typically measured in body fluids such as saliva, urine, or serum by enzyme-linked immunoassay (ELISA) (Benowitz and others 2009b). Although cotinine is commonly cited as the gold standard biomarker for smoking detection, it has significant limitations, most notably a short half-life of approximately 20 hours (Benowitz and others 2009a). Determination of current smoking status by cotinine assay can also result in false-positives in individuals who have quit smoking but are using nicotine replacement therapies, which limits its usefulness in populations being treated for nicotine dependence (Benowitz and others 2009b; Florescu and others 2009; Jatlow and others 2008; Matsumoto and others 2010). Exhaled carbon monoxide (CO), an alternative biomarker for smoking, has an even shorter half-life of 4–5 hours and requires specialized equipment at the point of testing (Florescu and others 2009). Importantly, neither cotinine nor CO offer adequate sensitivity in the detection of the light or intermittent smoking patterns that characterize adolescent use, a time when preventative interventions may be able to interrupt progression to nicotine dependence (Flay 2009).
Epigenetic biomarkers have the potential to overcome many of the limitations of current biomarkers in detecting smoking. Epigenetic marks are mitotically heritable modifications to DNA that provide structural and regulatory functions for the genome without the presence of a change in base pair sequence (Goldberg and others 2007). These marks include methylation of the cytosine base at cytosine-phospho-guanine dinucleotide residues (CpGs), covalent modification of histones, and open versus closed chromatin states, all of which can affect the expression of genes (Jiang and others 2004). Critically, some epigenetic marks are responsive to environmental influences, such as tobacco smoke exposure (Bollati and Baccarelli 2010; Ladd-Acosta 2015). The development of array-based platforms such as Illumina’s Infinium HumanMethylation450 BeadChip system has allowed investigators to perform epigenome-wide association studies to uncover links between environmental exposures and epigenetic changes. As a result of these investigations, tobacco smoke has emerged as a paradigmatic environmental exposure with epigenetic effects (Mikeska and Craig 2014).
In particular, one locus, cg05575921, a CpG residue located in the gene for the aryl hydrocarbon receptor repressor (AHRR), is a leading candidate for translation as a clinical biomarker for smoking (Ladd-Acosta 2015; Mikeska and Craig 2014). In over thirty methylome-wide studies of smoking conducted to date using self-report and cotinine validation of smoking status, across a wide variety of age ranges, ethnicities, both sexes, and even in infants exposed prenatally, demethylation of cg05575921 has been consistently linked with tobacco smoke exposure (Andersen and others 2015). Studies by Fasanelli and Baglietto and colleagues have further demonstrated that demethylation of cg05575921 is an independent risk factor for lung cancer after adjusting for smoking status (Baglietto and others 2017; Fasanelli and others 2015).
The mechanism underlying this consistent epigenetic response to smoking is not fully understood, but is thought to involve activation of the xenobiotic pathway. The xenobiotic pathway, responsible for detoxifying harmful components of tobacco smoke including polyaromatic hydrocarbons and dioxins (Nguyen and Bradfield 2008), is induced by the binding of polyaromatic hydrocarbons to the aryl hydrocarbon receptor (AHR). Subsequently, AHRR expression increases to compete with AHR for binding to its nuclear receptor, preventing over-expression of downstream genes such as p450 enzymes that detoxify components of smoke. Increased AHRR expression is directly linked to demethylation at CpG sites in the AHRR gene, including cg05575921, and measurement of this change in methylation then serves as a robust indicator of both smoking and other exposures that upregulate the xenobiotic pathway (Philibert and others 2015).
The performance characteristics of cg05575921 in detecting smoking are excellent, with an AUC of 0.99 reported in one study which used serum cotinine to confirm smoking status (Philibert and others 2015). This excellent predictive ability appears to be driven by two key factors. First, cg05575921 appears to have a fairly narrow biological “set-point”, with non-smokers almost always having a value of 80% to 90% methylation. Second, the magnitude of the change in percent methylation or “delta beta” in this locus in response to smoking is large compared to other smoking-associated loci (Andersen and others 2015). While delta beta values for methylation biomarkers in non-communicable diseases are typically less than 5% (Mikeska and Craig 2014), multiple studies have reported a delta beta of 15–20% at cg05575921 in response to smoking (Dogan and others 2014; Elliott and others 2014; Tsaprouni and others 2014; Zeilinger and others 2013). A recent meta-analysis of epigenetic signatures of smoking by Joehanes and colleagues reported a delta beta of 18% for cg05575921, the largest of 2623 CpGs associated with smoking, although it ranked only 36th in statistical significance (Joehanes and others 2016) whereas a 2015 review of 14 epigenome wide studies by Brenner and colleagues showed that cg05575921 showed both that cg05575921 had both the largest delta beta and the smallest p-value in the epigenome.(Gao and others 2015).
A third critical factor is that this locus is responsive even to light or intermittent smoking. In one study of 19 year-old smokers versus controls, smokers with less than ½ pack-year smoking history had a measurable delta beta at cg05575921 of 4.9% (Philibert and others 2012).
The extent of reversion of demethylation of cg05575921 with smoking cessation is uncertain and in need of further study. Several studies show demethylation at this locus in subjects who reported quitting smoking decades earlier (Baglietto and others 2017; Fasanelli and others 2015; Guida and others 2015; Shenker and others 2013). However, using biochemically confirmed measures, we have shown average reversion of nearly 5% after one month of cessation whereas Bauer and colleagues show complete erasure of the demethylation response present in the cord blood in offspring of smoking mothers after only 2 years.
While the epigenome-wide studies cited above used chip-based assays for discovery, the cost of arrays and their long turnaround time (1 week) prevent their use in routine clinical practice. In contrast, digital polymerase chain reaction (dPCR) methods are emerging as an attractive alternative for precise measurement of methylation that may be more suitable for translation of epigenetic biomarkers to clinical settings. Digital PCR methylation assays require genomic DNA that has been treated with bisulfite, a process that converts unmethylated cytosines to uracils, while sparing methylated cytosines (Frommer and others 1992). Bisulfite-converted DNA is then amplified using primers specific for both the methylated and unmethylated alleles. The relative abundance of each allele is then measured, allowing for calculation of the relative abundance of methylated and unmethylated alleles in the original sample. Digital PCR methods differ from quantitative PCR (qPCR) techniques by allowing highly precise quantification of the original sample without an external reference (Hindson and others 2011). This is accomplished by fractionating the sample into a large number of separate PCR reactions, typically 15,000–20,000. The fractional abundance of each allele in the original sample is calculated based on the number of positive and negative reactions, assuming a Poisson distribution. One application of this method is to use hydrophobic droplets to encapsulate and fractionate the samples, a technique known as droplet digital PCR (ddPCR) (Hindson and others 2011). In one study, measurement of methylation at a promoter CpG in the CD3Z gene was used to estimate T cell numbers in peripheral blood using both ddPCR and qPCR, and ddPCR was reported to offer superior precision, accuracy, and technical simplicity (Wiencke and others 2014).
For the detection of smoking, a clear application of methylation-specific ddPCR assays is the measurement of cg05575921 methylation in an individual in order to determine if it is lower than the population “set-point” of 80–90%. Values progressively lower than this population average signal an increasing probability that the person is a smoker. Indeed, Zhang and colleagues have demonstrated a strong linear relationship between progressive demethylation of cg05575921 and both cotinine and the number of cigarettes smoked per day (Zhang and others 2016).
In clinical practice, however, this information must also be integrated with self-report data and knowledge of the smoking habits characteristic of the individual’s population, given that neither laboratory testing nor self-report have perfect sensitivity or specificity. Thus when integrating information from an epigenetic biomarker such as cg05575921 with self-report, it is important to develop models that weight each source of information appropriately to guide clinical decision in different populations.
Here, we employ ddPCR to compare methylation at cg05575921 in two cotinine-validated samples of current smokers versus non-smokers with differing rates of false-negative self-reporting and cumulative smoking histories. We show that cg05575921 is a robust predictor of cotinine positivity in both samples. We further show that logistic models trained in different samples vary in predictive accuracy depending on the level of false negative self-reporting of smoking status in both training and test samples.
Materials and Methods
Samples
The Iowa Adoption Studies (IAS) is a long-running study of adoptees from the State of Iowa whose focus is substance use disorders and related psychopathology. The methods used in this study have been described previously (Philibert 2006). Briefly, 475 adoptees with a biological family history of substance use or antisocial personality disorder were selected from among 11,700 adoptees statewide, along with a matched sample of 475 controls. These adoptees and their families then were followed over 30 years with repeated assessments including semi-structured interviews including a modified version of the Semi Structured Assessment for the Genetics of Alcoholism, Version 2 (SSAGA-II) (Bucholz and others 1994) administered by a trained research assistant during the most recent wave (2005–2009). The clinical data used in this study was extracted from the tobacco use module contained within the SSAGA. The biomaterials for the current study were obtained via phlebotomy conducted during the last wave of the study.
The Family and Community Health Study (FACHS) is a multi-site investigation of neighborhood and family effects on health and development, consisting of 867 African American families living in Georgia and Iowa. A more detailed description of the sample and methods for the study is available elsewhere (Kogan and others 2013; Simons and others 2012). Of particular relevance, each family who participated in the FACHS study included at least one child between the ages of 10–12 years old at the time of recruitment (1995–97). During the last wave of the study, wave seven, (2014–2016), each subject was interviewed by a trained research assistant using a custom designed structured interview, then phlebotomized to provide materials for the current study. The structured interview included assessment of subject smoking status and total number of cigarettes smoked in their lifetime. Of note, data to calculate the number of pack-years smoked were not available for this sample but participants did report on whether they had smoked 500 or more cigarettes in their lifetime.
All procedures and protocols used in the IAS sample were approved by the University of Iowa Institutional Review Board (IRB). Similarly, all procedures and protocols for the FACHS sample were approved by IRB panels at Iowa State University, University of Iowa or University of Georgia.
Biomaterials
Following phlebotomy of the FACHS subjects, whole blood DNA and sera were prepared according to previously published protocols (Philibert and others 2012; Philibert and others 2013b). Similarly, for IAS subjects, whole blood DNA was obtained via the same method (Lahiri and Schnabel 1993) while plasma was obtained via ultrafiltration of blood samples frozen at −80°C.
Cotinine levels were assayed using enzyme-linked immunoassay (ELISA) with kits supplied by Abnova (Taiwan) as previously described (Philibert and others 2013a). To minimize the effects of unfiltered hemoglobin, washes conducted during processing of the plasma samples were conducted with phosphate buffered saline, pH 8.0, per the manufacturer’s suggestion.
Determination of methylation status at cg05575921 was conducted using ddPCR implementation of the previously described quantitative PCR approach (Dogan and others 2014). First, 1 μg of DNA from each subject was bisulfite converted using an EpiTect Fast 96 DNA Bisulfite kit (Qiagen, Germany) according to the manufacturer’s direction. The methylation ratio at cg05575921 (C/(C+T)) in each bisulfite treated sample was then determined using the Smoke Signature™ Assay (IBI Scientific, Peosta, IA) and a QX200 Droplet Digital PCR System™ (Bio-Rad, Hercules, CA) according to the manufacturer’s protocols. In brief, an aliquot of the bisulfite converted DNA was pre-amplified with the Smoke Signature™ Pre-Amp Master Mix under high stringency conditions per the manufacturer’s protocol, then diluted between 1:1000 and 1:5000. Then, 5 μl of the resulting solution was mixed with 1.1 ul of 20X Smoke Signature Assay, 4.9 ul of water, and 11 ul of BioRad 2X ddPCR Supermix (no dUTP), and vortexed. The resulting mixture was then processed with a Bio Rad Automated Droplet Generator, which generated approximately 20,000 micelles each containing approximately 1 nanoliter of PCR mixture, and quickly PCR amplified (95°C × 10′, then 40 cycles of 95°C × 15″ and 55°C × 60″, and finally 98°C × 10′). After amplification was complete, the post-amplification allele content status (either C, T, C+T, or blank) of each micelle by the QX200 Droplet Reader and the percent methylation status of each sample calculated using BioRad’s proprietary QuantaSoft software (v1.7).
Statistical analysis
All subsequent analyses were conducted with the R (Team 2014) statistical software, using the pROC (Robin and others 2011) and the scoring (Merkle and Steyvers 2013) packages Cotinine positivity was coded as 0 for individuals with 3 ng/mL or less and 1 for those above 3 ng/mL (Benowitz and others 2009a). Positive self-report of smoking was coded as a 1 and negative self-report as a 0. Subjects failing quality control, defined as greater than a 5% confidence interval for the ddPCR assay, were excluded from further analysis (N=31 in IAS and N=17 in FACHS).
For the primary analysis, logistic models were fit for each sample using cotinine positivity as the outcome and, cg05575921 methylation only (model 1), self-report only (model 2), and both self-report and cg05575921 methylation (model 3) as predictors. An intercept-only model (model 0) was also fit with no predictors. For between-sample predictions the full IAS or FACHS sample was used for training and subsequent testing. For within-sample model fitting and prediction, 70%–30% random splits within each sample were made using the R package caret (Kuhn 2015). Each model was then trained on the 70% split and cotinine status predicted in the corresponding 30% split within each sample.
Regression coefficients were estimated via maximum likelihood methods and Akaike’s information criterion (AIC) was used to assess relative model fit in each of the models above. Each of the four logistic models trained in each sample were then used to predict the probability of cotinine positivity within and across samples, using the full or split samples for training and testing as applicable.
Predicted probabilities were used to construct receiver operating characteristic curves (Zou and others 2007) and calculate the area under the curve (AUC) with 95% confidence interval for each set of predictions. The Brier score (Redelmeier and others 1991) is a quadratic scoring rule that measures the accuracy of probabilistic predictions in tasks with discrete outcomes, in this case testing positive or negative for cotinine. Brier scores closer to 0 indicate better accuracy. Brier scores scaled against a null model according to the equation (BSnull-BSfull)/BSnull are an estimate of the proportion of variation (R2) explained by the model, with scores closer to 1 indicating better accuracy (Gerds and others 2008; Steyerberg and others 2010). Brier scores for each set of predictions were calculated as the sum of squared distance between a participant’s smoking status as classified by cotinine and their predicted status based on the estimated regression coefficients (i.e., based on the linear predictor). Scaled Brier scores were then calculated to estimate the corresponding R2 for each set of predictions.
We performed an additional adjusted analysis of model 3 in the IAS sample to examine the effect of including pack-years and age as predictors of cg05575921 methylation and cotinine positivity. First, a linear model was fit using cg05575921 methylation as the outcome and age, pack-years, self-report and cotinine positivity as predictors, and P-values for each parameter were inxpected. Next, we included age and pack-years as additional predictors in our full logistic model (model 3) in the IAS 70% training split. Predictions from this adjusted model were then made in the IAS 30% testing split and used to calculate an AUC and Brier score using the same methods as above. The function roc.test() in the pROC package was then used to compare the resulting AUC with that of the unadjusted model.
A secondary analysis was performed to examine differences in the regression coefficients among between samples. The goal was to test the null hypothesis of equal population regression coefficients for self-report and cg05575921 among the population counterparts of the FACHS and IAS samples. Additional logistic regression models were estimated using the combined data with a dummy code for sample membership (FACHS = 0, IAS = 1), which was included as another predictor. The interaction of the dummy code and self-report, and the dummy code and cg05575921 was included in the full model (Model 4) in order to test for invariance of the regression coefficients among FACHS and IAS. We refer to this secondary analysis as the sample-as-group analysis.
Results
Clinical characteristics of the two samples are given in Table 1. In brief, the IAS sample (n=209) was older, predominantly European American, and had reported greater cumulative smoke exposure, with a mean lifetime consumption of 17.9 pack years among the self-reported current smokers. In contrast, subjects in FACHS (n=592) were younger, almost entirely African American, more likely to be female, and had significantly less self-reported smoking. Although data to calculate self-reported pack-years were not available for the FACHS subjects, only 15.2% of FACHS subjects reported having smoked more than 500 cigarettes (roughly 0.07 pack-years) or more in their lifetime.
Table I.
Demographics.
| Sample | FACHS | IAS |
|---|---|---|
| N | 592 | 209 |
| Median Age | 29 | 44 |
| Male (%) | 254 (42.9) | 101 (48.3) |
| Race/Ethnicity | ||
| African-American (%) | 526 (88.9) | 4 (1.9) |
| European-American (%) | 33 (5.6) | 192 (91.9) |
| Hispanic (%) | 4 (0.7) | 4 (1.9) |
| Other (%) | 4 (0.7) | 2 (0.9) |
| Smoking History | ||
| Never Smoker (%) | 346 (58.4) | 137 (65.6) |
| Former Smoker (%) | 101 (17.1) | 16 (7.7) |
| Current Smoker (%) | 145 (24.5) | 56 (26.8) |
Comparison of self-reported current smoking status with biochemical evidence of current smoking via serum cotinine revealed distinct patterns across the two samples, as shown in Table 2. Self-reported rates of smoking in IAS and FACHS were 26.8% and 31.8%, respectively, while serum cotinine positivity was seen in 33.5% of IAS subjects and 51.7% of FACHS subjects. Among IAS subjects, the proportion denying current smoking but positive for serum cotinine was 7%, while in FACHS the corresponding proportion was 22.6%. Only one (0.5%) of the IAS subjects self-reported current smoking but was not positive for serum cotinine, while 16 (2.7%) in FACHS did, and the remainder in each sample were negative for cotinine as well as self-report.
Table II.
Current smoking and epigenetic characteristics of samples: cotinine positivity, self-report of current smoking, smoking history, cg05575921 methylation.
| Sample | Cotinine | Self-Report | N (%) | N 500 or more lifetime cigarettes smoked (%) | Cg05575921 % methylation (SD) |
|---|---|---|---|---|---|
| FACHS | 0 | 0 | 270 (45.6) | 9 (5.9) | 84.6 (6.6) |
| 0 | 1 | 16 (2.7) | 1 (10.9) | 76.4 (17.1) | |
| 1 | 0 | 134 (22.6) | 9 (11.7) | 62.1 (18.1) | |
| 1 | 1 | 172 (29.1) | 71 (71.7) | 58.5 (16.9) | |
| Sample | Cotinine | Self-Report | N (%) | Pack Years smoked (SD) | Cg05575921 % methylation (SD) |
| IAS | 0 | 0 | 138 (66) | 1.5 (6.6) | 81.7 (11.4) |
| 0 | 1 | 1 (0.5) | 3 (NA) | 90.5 (NA) | |
| 1 | 0 | 15 (7) | 9.5 (16.3) | 66.4 (21.5) | |
| 1 | 1 | 55 (26.3) | 18.7 (17.9) | 47.1 (17.4) |
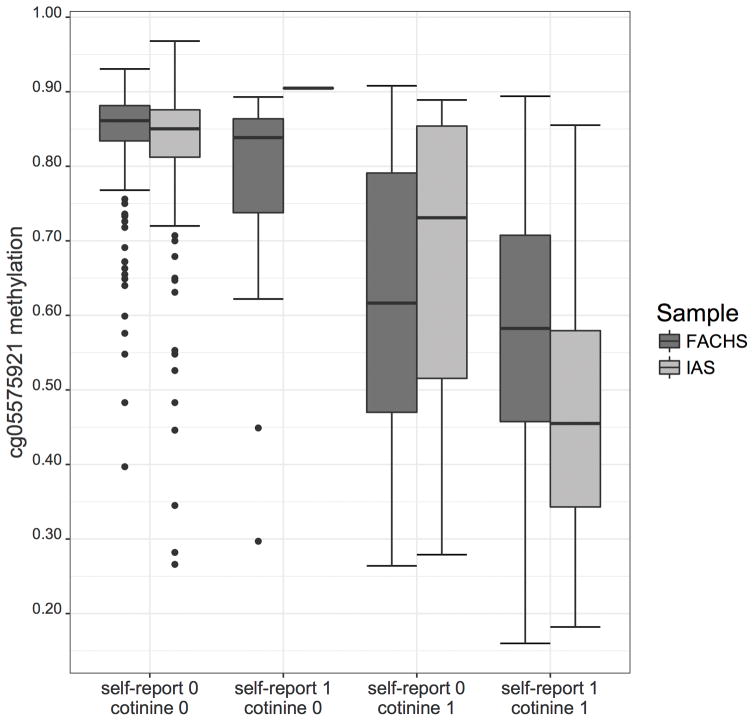
Mean methylation values at cg05575921 for the sample groups, broken down by self-report of current smoking and serum cotinine positivity, are also given in Table II and depicted in Figure I. In general, those with either self-reported current smoking and/or serum cotinine positivity had markedly lower cg05575921 methylation values than those without either positive self-report or cotinine. Those in IAS with both positive self-report and positive serum cotinine were the most demethylated, followed by the equivalent group in FACHS, and then those denying smoking but with serum positivity in FACHS and IAS, respectively. For both FACHS and IAS, those without positive self-report or cotinine positivity had cg05575921 methylation values clustered around 80–85%.
Figure I.
Boxplot showing cg05575921 methylation by cotinine status and self-report of current smoking (0 indicating negative, 1 indicating positive) in IAS and FACHS.
Results of the primary analysis are shown in Table III, which lists the regression coefficient estimates for the intercept (B0), self-report of current smoking (BSR), and cg05575921 methylation (BM) predicting cotinine smoking for each sample. In summary, those models trained in the FACHS sample weighted self-report less heavily and cg05575921 methylation more heavily, consistent with the lower accuracy of self-report in the FACHS population as compared to the IAS population. Models including both self-report and cg05575921 methylation had the lowest AIC values, indicating the best fit, in both samples. In contrast, the best fitting single-predictor models differed among the samples. In FACHS (full and split samples), model 1 (cg05575921 only) had a lower AIC than model 2 (self-report only), while the reverse was true in IAS.
Table III.
Model goodness of fit statistics (AIC), estimated regression coefficients, standard errors (SEs), Z statistics, and P-values of four models by sample.
| IAS (full sample) | Predictor | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Intercept | Cg05575921 Methylation | Self-Report | |||||||||||
| Model | AIC | B0 | SE | Z | P-value | BM | SE | Z | P-value | BSR | SE | Z | P-value |
| 0 (Null) | 268.5 | −0.686 | 0.147 | −4.681 | 2.86e-06 | ||||||||
| 1 | 158.4 | 6.186 | 0.947 | 6.535 | 6.36e-11 | −9.811 | 1.290 | −7.605 | 2.84e-14 | ||||
| 2 | 112.2 | −2.219 | 0.272 | −8.162 | 3.27e-16 | 6.227 | 1.045 | 5.959 | 2.54e-09 | ||||
| 3 | 98.4 | 2.189 | 1.088 | 2.011 | 4.42e-02 | −5.844 | 1.469 | −3.978 | 6.94e-05 | 5.177 | 1.067 | 5.850 | 1.23e-06 |
| FACHS (full sample) | Predictor | ||||||||||||
| Intercept | Cg05575921 Methylation | Self-Report | |||||||||||
| Model | AIC | B0 | SE | Z | P-value | BM | SE | Z | P-value | BSR | SE | Z | P-value |
| 0 (Null) | 822.0 | 0.068 | 0.082 | 0.82 | 0.411 | ||||||||
| 1 | 476.3 | 11.10 | 1.001 | 11.1 | 1.46e-28 | −14.52 | 1.245 | −11.66 | 2.10e-31 | ||||
| 2 | 626.8 | −0.701 | 0.106 | −6.63 | 3.36e-11 | 3.075 | 0.282 | 10.91 | 1.04e-27 | ||||
| 3 | 411.2 | 8.867 | 0.977 | 9.08 | 1.12e-19 | −12.42 | 1.219 | −10.18 | 2.40e-24 | 2.417 | 0.323 | 7.47 | 7.93e-14 |
| IAS (70% training sample) | Predictor | ||||||||||||
| Intercept | Cg05575921 Methylation | Self-Report | |||||||||||
| Model | AIC | B0 | SE | Z | P-value | BM | SE | Z | P-value | BSR | SE | Z | P-value |
| 0 (Null) | 119.3 | −0.507 | 0.169 | −2.999 | 2.71E-03 | ||||||||
| 1 | 118.9 | 6.281 | 1.099 | 5.718 | 1.08E-08 | −9.790 | 1.505 | −6.506 | 7.72E-11 | ||||
| 2 | 88.0 | −2.037 | 0.307 | −6.636 | 3.21E-11 | 5.821 | 1.057 | 5.508 | 3.63E-08 | ||||
| 3 | 76.7 | 2.633 | 1.294 | 2.034 | 4.19E-02 | −6.245 | 1.759 | −3.550 | 3.86E-04 | 4.811 | 1.087 | 4.424 | 9.68E-06 |
| FACHS (70% training sample) | Predictor | ||||||||||||
| Intercept | Cg05575921 Methylation | Self-Report | |||||||||||
| Model | AIC | B0 | SE | Z | P-value | BM | SE | Z | P-value | BSR | SE | Z | P-value |
| 0 (Null) | 578.2 | 0.067 | 0.098 | 0.686 | 4.93E-01 | ||||||||
| 1 | 320.7 | 11.825 | 1.260 | 9.384 | 6.32E-21 | −15.475 | 1.567 | −9.872 | 5.49E-23 | ||||
| 2 | 454.3 | −0.662 | 0.125 | −5.293 | 1.20E-07 | 2.867 | 0.318 | 9.022 | 1.84E-19 | ||||
| 3 | 285.1 | 9.915 | 1.255 | 7.899 | 2.81E-15 | −13.708 | 1.566 | −8.755 | 2.04E-18 | 2.183 | 0.379 | 5.760 | 8.41E-09 |
Comparisons of AUCs and Brier scores for each model, in both within-sample and across-sample testing, are given in Table IV. In general, results indicate a high level of agreement across samples in the utility of cg05575921 alone as a predictor of cotinine positivity, with AUCs of ranging from 0.87 to 0.90. Self-report alone performed equally well or better in models tested in IAS, while in contrast, in models tested in FACHS, AUCs were markedly worse at 0.75 (external validation) and 0.78 (internal validation). Models including both self-report and cg05575921 were the best-performing in both internal and external validation for both samples, with AUCs ranging from 0.90 to 0.93.
Table IV.
ROC area under the curve (AUC) statistics, Brier scores, and scaled Brier scores for logistic models predicting cotinine positivity, trained and tested in IAS and FACHS samples.
| Validation Type | Training Sample | Testing Sample | Model | AUC (CI) | Brier | R2 |
|---|---|---|---|---|---|---|
| Internal | IAS (70%) | IAS (30%) | 0 (Null) | 0.5 (0.5, 0.5) | 0.199 | 0.000 |
| 1 | 0.873 (0.735, 1.000) | 0.090 | 0.546 | |||
| 2 | 0.893 (0.781, 1.000) | 0.049 | 0.752 | |||
| 3 | 0.900 (0.76, 1.000) | 0.045 | 0.773 | |||
| Internal | FACHS (70%) | FACHS (30%) | 0 (Null) | 0.5 (0.5, 0.5) | 0.250 | 0.000 |
| 1 | 0.889 (0.839, 0.940) | 0.134 | 0.464 | |||
| 2 | 0.779 (0.727, 0.883) | 0.162 | 0.350 | |||
| 3 | 0.926 (0.886, 0.965) | 0.110 | 0.561 | |||
| External | IAS (full) | FACHS (full) | 0 (Null) | 0.5 (0.5, 0.5) | 0.283 | 0.000 |
| 1 | 0.898 (0.971, 0.924) | 0.171 | 0.395 | |||
| 2 | 0.753 (0.722, 0.784) | 0.215 | 0.241 | |||
| 3 | 0.917 (0.894, 0.941) | 0.165 | 0.417 | |||
| External | FACHS (full) | IAS (full) | 0 (Null) | 0.5 (0.5, 0.5) | 0.256 | 0.000 |
| 1 | 0.897 (0.848, 0.947) | 0.130 | 0.493 | |||
| 2 | 0.889 (0.840, 0.938) | 0.111 | 0.568 | |||
| 3 | 0.929 (0.884, 0.973) | 0.096 | 0.625 |
Differences in scaled Brier scores, a measure of the variation in the outcome (R2) explained by the predictors, were seen in within-sample and across-sample testing depending on the model used. For model 1 (cg05575921 only), the R2 was highest in IAS internal testing at 0.55 and worst in IAS to FACHS testing at 0.40. For model 2 (self-report only), the R2 was again best in IAS internal testing at 0.75, and markedly worse in IAS to FACHS testing at 0.24. Finally, in each set of testing, model 3 (cg05575921 + self-report) performed best, with the worst accuracy seen in IAS to FACHS testing, with an R2 of 0.42, whereas IAS to IAS gave an R2 of 0.77. For the equivalent model trained in FACHS, internal testing gave an R2 of 0.56, while external testing gave an R2 of 0.63.
In adjusted analyses in the IAS sample, self-report and cotinine positivity were both significant predictors (P<0.001) of cg05575921 methylation, while pack-years was an even more significant (4.34e-07). Although age was not a significant predictor in this model, we chose to include both age and pack-years as additional covariates in an adjusted full logistic model (model 3) predicting cotinine positivity. Compared with predictions generated by the unadjusted model 3, the addition of age and pack-years as covariates did not significantly improve the AUC (0.925 vs 0.900, P=0.450). Additionally, the scaled Brier score for the adjusted model (R2=0.646) was not better than the unadjusted model than the unadjusted model (R2=0.773).
Results of the secondary samples-as-groups analyses showed that for model 1 (cg05575921 only), the difference in regression coefficients for cg05575921 was not statistically significant (p > 0.05). For model 2 (self-report only), the coefficient for self-report was significantly different among FACHS and IAS (p < 0.05), and for model 3 (cg05575921 methylation and self-report), only the cg05575921 regression coefficient was statistically different between FACHS and IAS (p < .001).
Discussion
In this study, we show that cg05575921 methylation status is an excellent predictor of current smoking status, as ascertained by serum cotinine, in two samples with very different demographics, smoking histories, and rates of false self-reporting of smoking behavior. Out-of-sample prediction was similar to in-sample prediction illustrating that cg05575921 methylation is robust to differences in sample characteristics.
This ddPCR assay is a different measurement approach than used in prior studies investigating epigenetic signatures of smoking, the majority of which used the Illumina Infinium HumanMethylation450 BeadChip array and found methylation at cg05575921 in cotinine confirmed non-smokers to be between 86 and 93% (Andersen and others 2015). In the current study, the range for was largely between 80 and 88%. Although each of these approaches have their biases (Olson and others 2016; Soto and others 2016) and the results between the assays are highly correlated (Dogan and others 2014), we note that the normal range observed herein is very similar to that obtained by Novakovic and colleagues who used a mass spectroscopy approach (Novakovic and others 2014). Therefore, when comparing results obtained from the two platforms, it is important to keep in mind that while both of these techniques appear quite reliable, subtle differences between studies may arise from both sample and assay characteristics.
We focused our analyses on two complementary measures of model performance to assess the relative contributions of cg05575921 methylation and self-report in predicting current smoking status: the AUC and the Brier score. Despite some limitations, the calculation of overall AUCs remains the de facto method of comparing classifiers such as biomarkers (Zou and others 2007). The AUC values associated with cg05575921 in our study constitute a “good” to “excellent” level of prediction. The effect was consistent in within-sample and across-sample testing, and consistent with prior reports (Zhang and others 2016). Self-report performed equally well or better than cg05575921 methylation in testing in the IAS sample, whereas in FACHS the AUC of self-report would be only be classified as “fair”. The combination of self-report and cg05575921 methylation provided improved model fit and the best AUC in each train-test combination, although the magnitude of improvement over cg05575921 alone was modest.
Although the consistently high AUC values for cg05575921 methylation seen in this study help validate it as an epigenetic biomarker for current smoking status, this same consistency also highlights a known limitation of the AUC as a measure of model performance. Specifically, models with very similar AUCs may be associated with predictions with large differences in accuracy (Steyerberg and others 2010). For a biological relationship such as the one seen in the current study, a monotonic decrease in methylation at cg05575921 with increasing exposure to smoke, logistic models with very different parameter estimates may produce identical or nearly identical rankings of smoking probability. Meanwhile, the actual predictions derived from those parameter estimates may vary widely in terms of accuracy. This weakness was addressed by including the Brier score as an additional validation index in our analysis.
Unlike the AUC, a rank-based non-parametric method, the Brier scores reported in our study are indicators of absolute differences in the accuracy of predictions based on self-report and/or cg05575921 methylation. Once again, a high degree of consistency was seen in within-sample and between-sample testing, with scaled Brier scores (R2) for the full model ranging from 0.56 to 0.77. The exception was a lower R2 of 0.42 seen in IAS to FACHS testing.
Inspection of the parameter estimates for the model trained on the full IAS sample helps illuminate this discrepancy. In short, the models trained in IAS weighted cg05575921 less strongly than those in FACHS, resulting in weaker accuracy in IAS to FACHS prediction. In the full model trained in IAS, the parameter estimate for cg05575921 methylation was −5.8, while in FACHS the corresponding value was −12.4.
As shown by our secondary, samples-as-groups analysis, this effect was driven by the large differences in the accuracy of self-report between our samples, rather than differences in the performance of cg05575921. The difference in regression coefficients between samples for cg05575921 methylation alone was not significant. In contrast, there was a significant difference in the regression coefficient for cg05575921 in the full model that including both cg05575921 and self-report.
Smoking history and age may both influence the accuracy of cg05575921 methylation in predicting current smoking status. In addition, there may be subtle age-related changes in cg05575921 methylation due to unmeasured environmental exposures or intrinsic biological processes. Therefore, we performed additional analyses to assess the impact of age and smoking history on cg05575921 and self-report as predictors of current smoking status. Because pack-year smoking history data were not available in our FACHS sample, these analyses were restricted to IAS.
As shown above, a logistic model relating cg05575921 methylation to age, pack-years, cotinine positivity, and self-report of current smoking showed significant effects for each predictor except for age. Further modeling did not support any improvement in predictive utility or accuracy with the inclusion of age and pack-years as predictors, as measured by AUC and Brier scores, respectively. Although these results suggest that cg05575921 alone is a robust predictor of current smoking status even in populations of varying ages and cumulative smoking histories, some limitations of this analysis are notable. First, data were not able to allow us to examine the effect of time since quitting on cg0557521 methylation as a predictor of current smoking. Additionally, these analyses were restricted to the smaller of our two samples, as cumulative smoking exposure data in FACHS was limited to self-report of having smoked 500 or more cigarettes.
The lack of additional predictive utility or accuracy when including age and pack-years as predictors of current smoking is a surprising finding. Incomplete reversion of cg05575921 methylation after prolonged smoking cessation has been reported in prior work and would be expected to influence prediction of current smoking status (Baglietto and others 2017; Fasanelli and others 2015; Guida and others 2015; Shenker and others 2013). One potential explanation of this finding is diminished recall. Diminished recall is a well-established phenomenon in other forms of psychopathology including other forms of substance use (Giuffra and Risch 1994). Because few studies are designed to confirm longitudinal tobacco cessation by with validated biomarker ssuch as cotinine, some reports of incomplete reversion of demethylation at smoking-associated loci may actually underestimate the extent of reversion by including subjects who continued to smoke intermittently or have dimished recall. Using the multiwave data of the IAS, we are actively examining this phenomenon.
In summary, cg05575921 methylation performed well alone and in combination with self-report as a predictor of current smoking as measured by AUC and the Brier score, two measures of predictive performance. Differences in logistic model parameter estimates between samples were largely driven by differences in the accuracy of self-report rather than cg05575921 methylation in predicting smoking status. Inclusion of age and smoking history (pack-years) did not significantly influence the utility or accuracy of predictions. Our results support cg05575921 methylation as an effective biomarker for current smoking in a variety of potential clinical scenarios, including populations of differing ages, smoking histories, and in which prior smoking history may or may not be known. Because cg05575921 methylation is sensitive to even early smoking, and the extent of reversion after sustained heavy smoking has not been fully characterized, translational efforts investigating the use of cg05575921 as a clinical biomarker for smoking focusing on adolescent and young adult smokers may be most promising.
Acknowledgments
This work was supported by NIH grants R01DA037648 (Philibert), T32MH019113 (Andersen), and K12DA000357 (Andersen). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Disclosures:
The use of DNA methylation to assess alcohol use status is covered by pending property claims. The use of DNA methylation to assess smoking status is covered by US patent 8,637,652, 9,273,358 and other pending claims. Dr. Philibert is a potential royalty recipient on those intellectual right claims. Dr. Philibert is also an officers and stockholder of Behavioral Diagnostics. (www.bdmethylation.com).
References
- Andersen AM, Dogan MV, Beach SRH, Philibert RA. Current and Future Prospects for Epigenetic Biomarkers of Substance Use Disorders. Genes. 2015;6(4):991–1022. doi: 10.3390/genes6040991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baglietto L, Ponzi E, Haycock P, Hodge A, Bianca Assumma M, Jung CH, Chung J, Fasanelli F, Guida F, Campanella G. DNA methylation changes measured in pre-diagnostic peripheral blood samples are associated with smoking and lung cancer risk. International Journal of Cancer. 2017;140(1):50–61. doi: 10.1002/ijc.30431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benowitz NL, Bernert JT, Caraballo RS, Holiday DB, Wang JT. Optimal Serum Cotinine Levels for Distinguishing Cigarette Smokers and Nonsmokers Within Different Racial/Ethnic Groups in the United States Between 1999 and 2004. American Journal of Epidemiology. 2009a;169(2):236–248. doi: 10.1093/aje/kwn301. [DOI] [PubMed] [Google Scholar]
- Benowitz NL, Hukkanen J, Jacob P., III . Nicotine psychopharmacology. Springer; 2009b. Nicotine chemistry, metabolism, kinetics and biomarkers; pp. 29–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati V, Baccarelli A. Environmental epigenetics. Heredity (Edinb) 2010;105(1):105–12. doi: 10.1038/hdy.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton GR, Brinthaupt J, Stehle JM, James GD. Comparison of self-reported smoking and urinary cotinine levels in a rural pregnant population. J Obstet Gynecol Neonatal Nurs. 2004;33(3):306–11. doi: 10.1177/0884217504264866. [DOI] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock V, Nurnberger J, Jr, Reich T, Schmidt I, Schuckit M. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. Journal of studies on alcohol. 1994;55(2):149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Caraballo RS, Giovino GA, Pechacek TF. Self-reported cigarette smoking vs. serum cotinine among US adolescents. Nicotine & Tobacco Research. 2004;6(1):19-+. doi: 10.1080/14622200310001656821. [DOI] [PubMed] [Google Scholar]
- Caraballo RS, Giovino GA, Pechacek TF, Mowery PD. Factors associated with discrepancies between self-reports on cigarette smoking and measured serum cotinine levels among persons aged 17 years or older - Third National Health and Nutrition Examination Survey, 1988–1994. American Journal of Epidemiology. 2001;153(8):807–814. doi: 10.1093/aje/153.8.807. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Vital signs: current cigarette smoking among adults aged >/=18 years--United States, 2005–2010. MMWR. 2011;60:1207–12. [PubMed] [Google Scholar]
- DHHS. The Health Consequences of Smoking - 50 Years of Progress: A Report of the Surgeon General. Office of the Surgeon General; 2014. [Google Scholar]
- Dogan MV, Shields B, Cutrona C, Gao L, Gibbons FX, Simons R, Monick M, Brody GH, Tan K, Beach SR, et al. The effect of smoking on DNA methylation of peripheral blood mononuclear cells from African American women. BMC Genomics. 2014;15:151. doi: 10.1186/1471-2164-15-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott HR, Tillin T, McArdle WL, Ho K, Duggirala A, Frayling TM, Davey Smith G, Hughes AD, Chaturvedi N, Relton CL. Differences in smoking associated DNA methylation patterns in South Asians and Europeans. Clin Epigenetics. 2014;6(1):4. doi: 10.1186/1868-7083-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasanelli F, Baglietto L, Ponzi E, Guida F, Campanella G, Johansson M, Grankvist K, Johansson M, Assumma MB, Naccarati A. Hypomethylation of smoking-related genes is associated with future lung cancer in four prospective cohorts. Nature communications. 2015:6. doi: 10.1038/ncomms10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flay BR. School-based smoking prevention programs with the promise of long-term effects. Tobacco Induced Diseases. 2009;5(1):1. doi: 10.1186/1617-9625-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florescu A, Ferrence R, Einarson T, Selby P, Soldin O, Koren G. Methods for quantification of exposure to cigarette smoking and environmental tobacco smoke: focus on developmental toxicology. Ther Drug Monit. 2009;31(1):14–30. doi: 10.1097/FTD.0b013e3181957a3b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer M, Mcdonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A Genomic Sequencing Protocol That Yields a Positive Display of 5-Methylcytosine Residues in Individual DNA Strands. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(5):1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Jia M, Zhang Y, Breitling LP, Brenner H. DNA methylation changes of whole blood cells in response to active smoking exposure in adults: a systematic review of DNA methylation studies. Clinical Epigenetics. 2015;7(1):113. doi: 10.1186/s13148-015-0148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerds TA, Cai T, Schumacher M. The performance of risk prediction models. Biom J. 2008;50(4):457–79. doi: 10.1002/bimj.200810443. [DOI] [PubMed] [Google Scholar]
- Giuffra LA, Risch N. Diminished recall and the cohort effect of major depression: a simulation study. Psychol Med. 1994;24(2):375–83. doi: 10.1017/s0033291700027355. [DOI] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128(4):635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Guida F, Sandanger TM, Castagné R, Campanella G, Polidoro S, Palli D, Krogh V, Tumino R, Sacerdote C, Panico S. Dynamics of smoking-induced genome-wide methylation changes with time since smoking cessation. Human molecular genetics:ddu751. 2015 doi: 10.1093/hmg/ddu751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilberink SR, Jacobs JE, van Opstal S, van der Weijden T, Keegstra J, Kempers P, Muris JW, Grol RP, de Vries H. Validation of smoking cessation self-reported by patients with chronic obstructive pulmonary disease. Int J Gen Med. 2011;4:85–90. doi: 10.2147/IJGM.S15231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83(22):8604–10. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jatlow P, Toll BA, Leary V, Krishnan-Sarin S, O’Malley SS. Comparison of expired carbon monoxide and plasma cotinine as markers of cigarette abstinence. Drug Alcohol Depend. 2008;98(3):203–9. doi: 10.1016/j.drugalcdep.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YH, Bressler J, Beaudet AL. Epigenetics and human disease. Annu Rev Genomics Hum Genet. 2004;5:479–510. doi: 10.1146/annurev.genom.5.061903.180014. [DOI] [PubMed] [Google Scholar]
- Joehanes R, Just AC, Marioni RE, Pilling LC, Reynolds LM, Mandaviya PR, Guan W, Xu T, Elks CE, Aslibekyan S. Epigenetic Signatures of Cigarette SmokingCLINICAL PERSPECTIVE. Circulation: Cardiovascular Genetics. 2016;9(5):436–447. doi: 10.1161/CIRCGENETICS.116.001506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel DB, Schaffran C, Griesler PC, Hu MC, Davies M, Benowitz N. Salivary cotinine concentration versus self-reported cigarette smoking: Three patterns of inconsistency in adolescence. Nicotine Tob Res. 2006;8(4):525–37. doi: 10.1080/14622200600672732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogan SM, Lei MK, Grange CR, Simons RL, Brody GH, Gibbons FX, Chen YF. The contribution of community and family contexts to African American young adults’ romantic relationship health: a prospective analysis. J Youth Adolesc. 2013;42(6):878–90. doi: 10.1007/s10964-013-9935-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn M. caret: Classification and regression training. Astrophysics Source Code Library. 2015;1:05003. [Google Scholar]
- Ladd-Acosta C. Epigenetic Signatures as Biomarkers of Exposure. Curr Environ Health Rep. 2015;2(2):117–25. doi: 10.1007/s40572-015-0051-2. [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Schnabel B. DNA isolation by a rapid method from human blood samples: effects of MgCl2, EDTA, storage time, and temperature on DNA yield and quality. Biochem Genet. 1993;31(7–8):321–8. doi: 10.1007/BF02401826. [DOI] [PubMed] [Google Scholar]
- Larzelere MM, Williams DE. Promoting smoking cessation. Am Fam Physician. 2012;85(6):591–8. [PubMed] [Google Scholar]
- Matsumoto A, Ino T, Ohta M, Otani T, Hanada S, Sakuraoka A, Matsumoto A, Ichiba M, Hara M. Enzyme-linked immunosorbent assay of nicotine metabolites. Environ Health Prev Med. 2010;15(4):211–6. doi: 10.1007/s12199-009-0129-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure JB. Are biomarkers a useful aid in smoking cessation? A review and analysis of the literature. Behav Med. 2001;27(1):37–47. doi: 10.1080/08964280109595770. [DOI] [PubMed] [Google Scholar]
- Merkle EC, Steyvers M. Choosing a Strictly Proper Scoring Rule. Decision Analysis. 2013;10(4):292–304. [Google Scholar]
- Mikeska T, Craig JM. DNA methylation biomarkers: cancer and beyond. Genes (Basel) 2014;5(3):821–64. doi: 10.3390/genes5030821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LP, Bradfield CA. The search for endogenous activators of the aryl hydrocarbon receptor. Chem Res Toxicol. 2008;21(1):102–16. doi: 10.1021/tx7001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakovic B, Ryan J, Pereira N, Boughton B, Craig JM, Saffery R. Postnatal stability, tissue, and time specific effects of AHRR methylation change in response to maternal smoking in pregnancy. Epigenetics. 2014;9(3):377–86. doi: 10.4161/epi.27248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson C, Lo FY, Deutsch K, Austin S, Howard K, Leonti A, Maassel L, Subia C, Saloranta T, Christopherson N. Synergistic effects of promoter associated DNA methylation and genetic alterations to better understand oncogenic gene expression profiles. AACR 2016 [Google Scholar]
- Philibert R. Merging genetic and environmental effects in the Iowa Adoption Studies: focus on depression. Ann Clin Psychiatry. 2006;18(4):219–22. doi: 10.1080/10401230600948399. [DOI] [PubMed] [Google Scholar]
- Philibert R, Hollenbeck N, Andersen E, McElroy S, Wilson S, Vercande K, Beach SRH, Osbom T, Gerrard M, Gibbons FX, et al. Reversion of AHRR Demethylation Is a Quantitative Biomarker of Smoking Cessation. Frontiers in Psychiatry. 2016:7. doi: 10.3389/fpsyt.2016.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert R, Hollenbeck N, Andersen E, Osborn T, Gerrard M, Gibbons FX, Wang K. A quantitative epigenetic approach for the assessment of cigarette consumption. Front Psychol. 2015;6:656. doi: 10.3389/fpsyg.2015.00656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Beach SR, Brody GH. Demethylation of the aryl hydrocarbon receptor repressor as a biomarker for nascent smokers. Epigenetics. 2012;7(11):1331–1338. doi: 10.4161/epi.22520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Beach SR, Lei MK, Brody GH. Changes in DNA methylation at the aryl hydrocarbon receptor repressor may be a new biomarker for smoking. Clin Epigenetics. 2013a;5(1):19. doi: 10.1186/1868-7083-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Beach SRH, Lei MK, Brody GH. Changes in DNA methylation at the aryl hydrocarbon receptor repressor may be a new biomarker for smoking. Clinical Epigenetics. 2013b;5(19) doi: 10.1186/1868-7083-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranney L, Melvin C, Lux L, McClain E, Lohr KN. Systematic review: smoking cessation intervention strategies for adults and adults in special populations. Ann Intern Med. 2006;145(11):845–56. doi: 10.7326/0003-4819-145-11-200612050-00142. [DOI] [PubMed] [Google Scholar]
- Redelmeier DA, Bloch DA, Hickam DH. Assessing Predictive Accuracy - How to Compare Brier Scores. Journal of Clinical Epidemiology. 1991;44(11):1141–1146. doi: 10.1016/0895-4356(91)90146-z. [DOI] [PubMed] [Google Scholar]
- Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Muller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12(1):77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAMHSA. Results from the 2010 National Survey on Drug Use and Health: Summary of National Findings. 2011. [Google Scholar]
- Shadel WG, Shiffman S, Niaura R, Nichter M, Abrams DB. Current models of nicotine dependence: what is known and what is needed to advance understanding of tobacco etiology among youth. Drug and Alcohol Dependence. 2000;59:S9–S22. doi: 10.1016/s0376-8716(99)00162-3. [DOI] [PubMed] [Google Scholar]
- Shenker NS, Ueland PM, Polidoro S, van Veldhoven K, Ricceri F, Brown R, Flanagan JM, Vineis P. DNA methylation as a long-term biomarker of exposure to tobacco smoke. Epidemiology. 2013;24(5):712–716. doi: 10.1097/EDE.0b013e31829d5cb3. [DOI] [PubMed] [Google Scholar]
- Simons RL, Simons LG, Lei MK, Landor AM. Relational schemas, hostile romantic relationships, and beliefs about marriage among young African American adults. Journal of Social and Personal Relationships. 2012;29(1):77–101. doi: 10.1177/0265407511406897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto J, Rodriguez-Antolin C, Vallespin E, de Castro Carpeno J, Ibanez de Caceres I. The impact of next-generation sequencing on the DNA methylation-based translational cancer research. Transl Res. 2016;169:1–18 e1. doi: 10.1016/j.trsl.2015.11.003. [DOI] [PubMed] [Google Scholar]
- Steyerberg EW, Vickers AJ, Cook NR, Gerds T, Gonen M, Obuchowski N, Pencina MJ, Kattan MW. Assessing the performance of prediction models: a framework for some traditional and novel measures. Epidemiology (Cambridge, Mass) 2010;21(1):128. doi: 10.1097/EDE.0b013e3181c30fb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2014. 2012. [Google Scholar]
- Tsaprouni LG, Yang TP, Bell J, Dick KJ, Kanoni S, Nisbet J, Vinuela A, Grundberg E, Nelson CP, Meduri E, et al. Cigarette smoking reduces DNA methylation levels at multiple genomic loci but the effect is partially reversible upon cessation. Epigenetics. 2014;9(10):1382–96. doi: 10.4161/15592294.2014.969637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartiainen E, Seppala T, Lillsunde P, Puska P. Validation of self reported smoking by serum cotinine measurement in a community-based study. J Epidemiol Community Health. 2002;56(3):167–70. doi: 10.1136/jech.56.3.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenknecht LE, Burke GL, Perkins LL, Haley NJ, Friedman GD. Misclassification of smoking status in the CARDIA study: a comparison of self-report with serum cotinine levels. Am J Public Health. 1992;82(1):33–6. doi: 10.2105/ajph.82.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiencke JK, Bracci PM, Hsuang G, Zheng S, Hansen H, Wrensch MR, Rice T, Eliot M, Kelsey KT. A comparison of DNA methylation specific droplet digital PCR (ddPCR) and real time qPCR with flow cytometry in characterizing human T cells in peripheral blood. Epigenetics. 2014;9(10):1360–5. doi: 10.4161/15592294.2014.967589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeilinger S, Kuhnel B, Klopp N, Baurecht H, Kleinschmidt A, Gieger C, Weidinger S, Lattka E, Adamski J, Peters A, et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS One. 2013;8(5):e63812. doi: 10.1371/journal.pone.0063812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Florath I, Saum K-U, Brenner H. Self-reported smoking, serum cotinine, and blood DNA methylation. Environmental research. 2016;146:395–403. doi: 10.1016/j.envres.2016.01.026. [DOI] [PubMed] [Google Scholar]
- Zou KH, O’Malley AJ, Mauri L. Receiver-operating characteristic analysis for evaluating diagnostic tests and predictive models. Circulation. 2007;115(5):654–7. doi: 10.1161/CIRCULATIONAHA.105.594929. [DOI] [PubMed] [Google Scholar]