Abstract
‘Massilibacteroïdes vaginae’ sp. nov. strain MV12T is the type strain of ‘Massilibacteroïdes’ gen. nov., a new genus within the family of Porphyromonadaceae. This strain was isolated in Marseille from a patient with vaginosis. ‘Massilibacteroïdes vaginae’ is a Gram-negative, aero-anaerobic bipolar bacillus. Here we describe the phenotypic characteristics and complete genome sequence of this bacterium. The EMBL accession number is FXDJ00000000 (FXDJ01000001–FXDJ01000009), and its genome consists of nine scaffolds. The strain was deposited in two culture collections under the numbers CSUR P1477 and DSM 100704.
Keywords: Bacterial vaginosis, culturomics, genome, Massilibacteroïdes vaginae, taxonogenomics
Introduction
Bacterial vaginosis presents a pressing health concern, as it regularly affects young women of childbearing age and is involved in cases of prematurity, with a prevalence of 10% to 30% in developed countries [1], [2]. In eastern and southern Africa, the prevalence is even higher, at more than 50%, and is often associated as a risk factor for sexually transmitted diseases, such as herpes simplex virus type 2 and HIV [2].
In bacterial vaginosis, normal bacterial flora are modified. The main species present in a normal flora, Lactobacillus sp., are depleted and replaced by an abnormal population consisting of a higher diversity of bacterial species, including the anaerobic species Gardnerella vaginae and Atopobium vaginae, which are overrepresented compared to healthy patients [2].
Vaginal flora are now the subject of increasing interest, thanks to several advances in microbial investigation methods including molecular tools such as metagenomics, sequencing and phylogenetic analysis [3], [4], [5], as well as culture techniques like culturomics and bacterial identification using matrix-assisted desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) [6], [7].
As a result of these advances, during the microbiologic exploration of vaginal samples from women with bacterial vaginosis in a postpregnancy study, we were able to isolate a new bacterial strain, which represents the first species of a new genus, named ‘Massilibacteroïdes vaginae,’ strain MV12T (= CSUR P1477 = DSM 100704).
Here we describe this new bacterial genus, including a summary classification, set of phenotypical characteristics and description of the complete genome sequencing and annotation. Those features support the circumscription of the genus and species ‘Massilibacteroïdes vaginae.’
Organism information
Classification and features
A vaginal specimen was collected during a prospective postpregnancy study at the Conception University Hospital of Marseille (France). The vaginal swab was performed on 9 March 2013. The specimen was collected from a 27-year-old woman with bacterial vaginosis that had a molecular detection of 1010/mL A. vaginae [8]. After sample collection, the specimen was stored at −30°C. The ‘Massilibacteroïdes vaginae’ MV12T strain was isolated in October 2014 by anaerobic cultivation on 5% sheep's blood–enriched Columbia agar (bioMérieux, Marcy l’Etoile, France) after incubation in an anaerobic blood culture bottle (Becton Dickinson [BD], San Diego, CA, USA), supplemented with sheep's blood and clarified sterile sheep rumen.
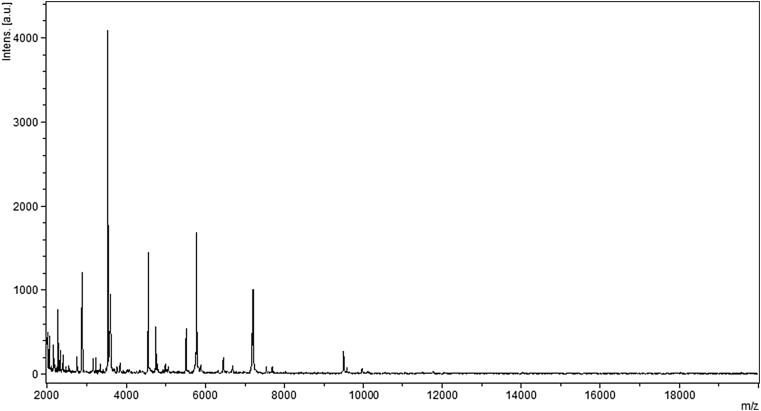
MALDI-TOF MS protein analysis was carried out using a Microflex spectrometer (Bruker Daltonics, Bremen, Germany) in order to identify the bacterium. Twenty separated colonies were deposited on a MTP 384 MALDI-TOF target plate (Bruker). Each spot was covered with 2 μL of matrix solution (saturated solution of α-cyano-4-hydroxycinnamic acid) in 50% acetonitrile and 2.5% trifluoroacetic acid, then left to dry for 5 minutes at room temperature. The spectra were imported into the MALDI BioTyper software (version 2.0, Bruker) and analysed using standard pattern matching (with default parameter settings) against the main spectra of 6335 bacteria in the BioTyper database. A score made it possible to identify or not identify the tested species: a score of >2 with a validated species enabled identification at the species level; a score of >1.7 but <2 enabled identification at the genus level; and a score of <1.7 did not enable any identification. For strain MV12T, the highest score reached was 1.34, which does not enable any identification. Therefore, this isolate could not be ranged within any known genus. The reference mass spectrum, generated by comparing 20 spectra obtained from 20 individual colonies of strain MV12T, was integrated into our database (Fig. 1).
Fig. 1.
Phylogenetic reconstruction based on 16S rRNA gene sequence, highlighting phylogenetic position of ‘Massilibacteroïdes vaginae’ relative to other bacteria belonging to other genera in Porphyromonadaceae family. Sequences were aligned using Muscle [9] and phylogenetic tree was built using MEGA6 software [10] and maximum likelihood method.
Because MALDI-TOF MS identification failed, we performed identification by amplifying and sequencing the 16S rRNA gene, as previously described [9]. The strain MV12T exhibited a 93.5% 16S rRNA gene sequence similarity with Parabacteroides goldsteinii, the closest related species with a validly published name [10]. This similarity percentage is lower than the threshold recommended to delineate a new genus without carrying out DNA-DNA hybridization [11], [12]. The genus Parabacteroides belongs to the family of Porphyromonadaceae [13], which contains 18 different genera. Of them, the genus found to be the most closely related to ‘Massilibacteroïdes vaginae’ strain MV12T, according to the comparison of the 16S rRNA gene sequences, is first Parabacteroides, followed by Tannerella, Porphyromonas and Dysgonomonas. Phylogenetic analysis was performed, and in the inferred phylogenetic tree obtained by comparing the 16S rRNA gene sequence obtained for strain MV12T with the closest related species, it forms a distinct lineage close to the genus Parabacteroides (Fig. 2). Sequences were aligned using CLUSTALW software, and the phylogenetic tree was built using MEGA software [14].
Fig. 2.
Reference mass spectrum obtained by concatenation of 20 spectra generated from 20 colonies of ‘Massilibacteroïdes vaginae’ strain MV12T.
The strain was incubated at several growth temperatures (25, 28, 32, 35, 37, 40 and 50°C). The bacterium grew between 25 and 40°C, with optimal growth at 37°C. No growth was evident at 50°C. Morphologically, the colonies were light beige, slightly bright and nonhaemolytic, approximately 0.5 mm in diameter. On Gram staining they appeared as bipolar Gram-negative bacilli, nonmotile and unable to form spores (Fig. 3). The morphology of the bacteria was observed by electron microscopy and the following method: detection-coated grids were deposited on 40 μL of bacterial suspension and incubated for 30 minutes at 37°C. The grids were then brought into contact for 1 second with a drop of ammonium molybdate 1%, dried on filter paper and observed with a Tecnai G20 Cryo electron microscope (FEI Company, Limeil-Brevannes, France) at an operating voltage of 60 kV. Using the electron microscopy, we determined that the cells had a mean length of 2.06 μm (range 1.27–3.21 μm) and a width of 0.73 μm (range 0.62–0.81 μm) (Fig. 4).
Fig. 3.
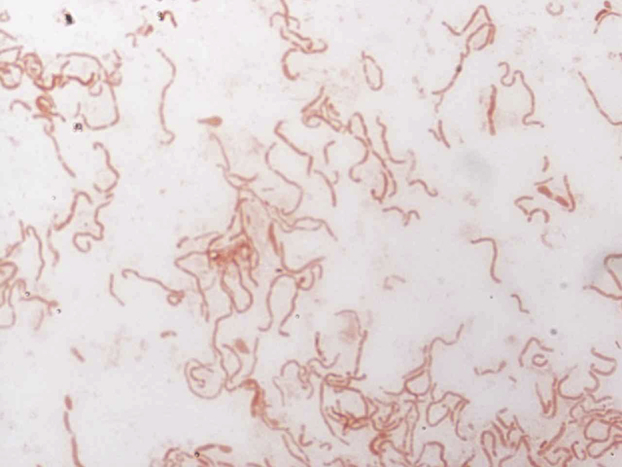
Gram staining of ‘Massilibacteroïdes vaginae’ strain MV12T.
Fig. 4.
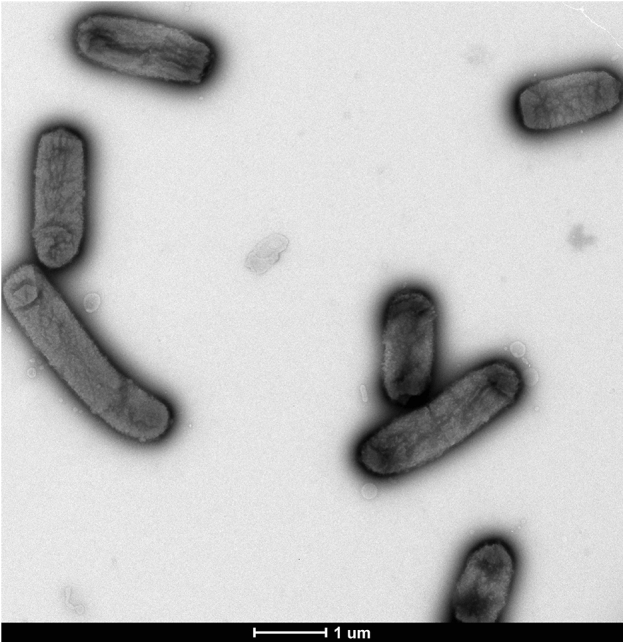
Transmission electron microscopy of ‘Massilibacteroïdes vaginae’ strain MV12T, obtained with Tecnai G20Cryo electron microscope, at operating voltage of 60 kV. Scale bar represents 1 μm.
The growth of the bacterium was tested in different atmospheric conditions using GENbag anaer and GENbag microaer systems (bioMérieux). The bacteria multiplied in aerobic and anaerobic atmospheres. It also multiplied in microaerophilic atmosphere supplemented with 5% CO2. However, optimal growth was shown in anaerobic conditions. Colonies also grew on brain–heart infusion (BHI) agar and on BHI agar supplemented with 1% NaCl. This strain exhibited oxidase and catalase activities.
Using an API ZYM (bioMérieux), positive reactions were observed for phosphatase alkaline, leucine arylamidase, trypsin, phosphatase acid, naphtol phosphohydrolase, β-galactosidase, α-glucosidase, β-glucuronidase and N-acetyl β-glucosaminidase. Negative reactions were observed for esterase, esterase lipase, lipase, valine arylamidase, cystine arylamidase, α-chymotrypsin, α-galactosidase, β-glucuronidase, α-mannosidase and α-fucosidase.
A rapid ID 32 A API strip (bioMérieux) was conducted, and positive reactions were obtained with arginine dihydrolase, β-galactosidase, β-galactosidase 6 phosphate, α-glucosidase, β-glucosidase, α-arabinosidase, N-acetyl-β-glucosaminidase, fermentation of mannose, fermentation of raffinose, glutamic decarboxylase acid, α-fucosidase, reduction of nitrates, phosphatase alkaline, arginine arylamidase, leucyl glycine arylamidase, leucine arylamidase, alanine arylamidase, glutamyl acid and glutamic arylamidase. The reaction was negative for the following enzymes: urease, α-galactosidase, β-glucuronidase, production of indole, proline arylamidase, phenylalanine arylamidase, pyroglutamic acid arylamidase, tyrosine arylamidase, glycine arylamidase, histidine arylamidase and serine arylamidase.
The susceptibility of M. vaginae strain MV12T was tested in vitro using the Etest method. We observed susceptibility to benzylpenicillin (MIC = 0.094 μg/mL), cefepim (MIC = 6 μg/mL), imipenem (MIC = 0.5 μg/mL), metronidazole (MIC = 0.094 μg/mL), vancomycin (MIC = 3 μg/mL) and amoxicillin/clavulanic acid (MIC = 125 μg/mL).
Phenotypic features and comparison with those of the closest related genus are summarized in Table 1.
Table 1.
Phenotypic features differencing Massilibacteroïdes vaginae strain MV12T to closest related genera Parabacteroides goldsteinii strain DSM19448 and Parabacteroïdes gordonii strain DSM 23371
| Property | M. vaginae | P. goldsteinii | P. gordonii |
|---|---|---|---|
| Cell width (μm) | 0.2–0.5 | NA | 0.5–1.7 |
| Oxygen requirement | Aero-anaerobic | Anaerobic | Anaerobic |
| Gram stain | Negative | Negative | Negative |
| Optimal growth temperature | 37°C | 37°C | 37°C |
| Habitat | Vagina | Specimens from humans | Blood culture |
| Use of: | |||
| α-Glucosidase | + | + | + |
| α-Galactosidase | − | + | + |
| β-Galactosidase | + | + | + |
| N-acetyl-β-glucosaminidase | + | + | NA |
| Acid phosphatase | + | + | NA |
| Alkaline phosphatase | + | + | + |
| Leucine arylamidase | + | + | + |
| Arginine arylamidase | + | + | + |
| Leucyl glycine arylamidase | + | + | + |
| Phenylalanine arylamidase | − | + | − |
| Alanine arylamidase | + | + | + |
| Glycine arylamidase | − | + | − |
| Histidine arylamidase | − | + | − |
| Glutamyl glutamic acid arylamidase | + | + | − |
| Production of: | |||
| Indole | − | − | − |
+, positive result; −, negative result; NA, data not available.
Genome sequencing information
Growth conditions and genomic DNA (gDNA) preparation
Strain MV12T was grown under anaerobic conditions on 5% sheep's blood–enriched Columbia agar (bioMérieux) at 37°C. Colonies from five plates were collected and resuspended in 4 × 100 μL of Tris-EDTA (TE) buffer. An aliquot of 200 μL of this suspension was then diluted in 1 mL of TE buffer and incubated with 2.5 μg/μL of lysozyme at 37°C for 30 minutes, for lysis treatment. This was followed by overnight incubation with 20 μg/μL proteinase K at 37°C. DNA was then extracted and purified with three successive phenol–chloroform extractions, then ethanol precipitation at −20°C overnight. After a final centrifugation, the DNA was resuspended in 160 μL of TE buffer.
Genome sequencing and assembly
The gDNA of M. vaginae was sequenced on MiSeq Technology (Illumina, San Diego, CA, USA) using the mate-pair strategy. The gDNA was barcoded in order to be mixed with 11 other projects with the Nextera mate-pair strategy. The gDNA was quantified using a Qubit assay with a high-sensitivity kit (Life Technologies, Carlsbad, CA, USA) to 333.7 ng/μL. The mate pair library was prepared with 1.5 μg of gDNA using the Nextera mate pair Illumina guide. The gDNA sample was simultaneously fragmented and tagged using a mate-pair junction adapter. The pattern of fragmentation was validated on an Agilent 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA, USA) with a DNA 7500 labchip. The DNA fragments ranged in size from 1.5 to 11 kb, with an optimal size of 6.08 kb. No size selection was performed, and 716.8 ng of tagmented fragments were circularized. The circularized DNA was mechanically sheared to small fragments with an optimal size of 1049 bp on the Covaris device S2 in T6 tubes (Covaris, Woburn, MA, USA). The library profile was visualized using a High Sensitivity Bioanalyzer LabChip (Agilent Technologies), and the final concentration library was measured at 14.65 nmol/L. The libraries were normalized at 2 nM and pooled. After a denaturation step and dilution at 15 pM, the pool of libraries was loaded onto the reagent cartridge and then onto the instrument with the flow cell. Automated cluster generation and sequencing runs were performed in a single 39-hour run at a 2 × 251 bp read length. A total of 5.5 Gb of information was obtained from a 572 k/mm2 cluster density with a cluster passing quality control filters of 96.3% (10 764 000 passed filter clusters). Within this run, the index representation for M. vaginae MV12T was identified as 5.81%. The 625 022 paired reads were filtered according to the read qualities. These reads were trimmed then assembled.
Genome annotation
Open reading frames (ORFs) were predicted using Prodigal [15] with default parameters. Functional annotation was performed using comparative genomics by BLASTp search against the GenBank nonredundant (nr) and Clusters of Orthologous Groups (COGs) databases [16], with an E-value threshold of <1e-3 for amino acid sequences >80 aa and <1e-5 for sequences of <80 aa. The transfer RNAs were searched for using tRNAScanSE tool [17], ribosomal RNAs by using RNAmmer [18] and BLASTn against GenBank. The ORFans were identified as sequences which were larger than 80 aa without a significant hit, which was defined as an E value of >1e-03 against the nr database. Artemis was used to visualize genomic features [19]. Pairwise genome comparisons were performed using Proteinortho software [20] to detect best BLASTp reciprocal hits between M. vaginae MV12T and nine other bacteria (Dysgonomonas capnocytophagoides, Dysgonomonas mossii, Dysgonomonas gadei, Porphyromonas cangingivalis, Proteiniphilum acetatigenes, Tannerella forsythia, Bacteroïdes massiliensis, Parabacteroïdes gordonii and Parabacteroïdes distasonis), using 1e-5, 30% and 70% as thresholds for the E value, identity amino acid percentage and coverage.
Genome properties
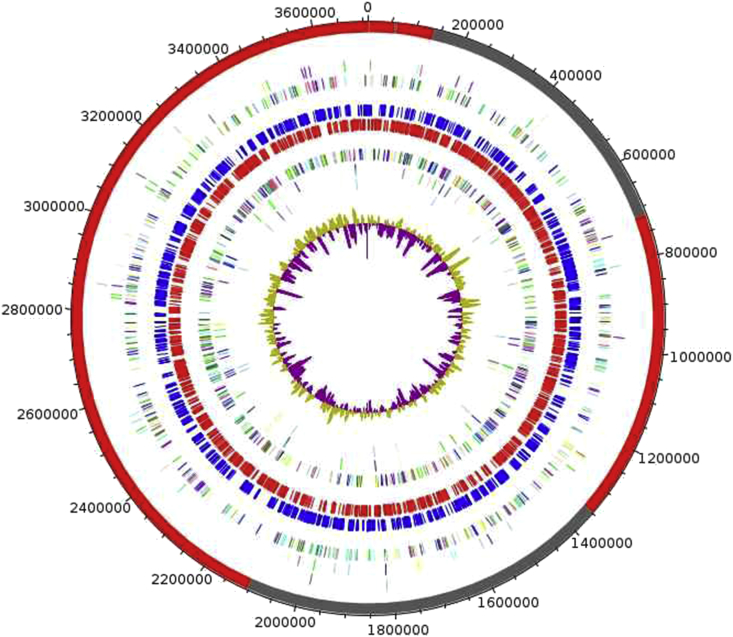
The genome of M. vaginae MV12T is 3 710 320 bp long, composed of one chromosome and no plasmid, and harbouring 40.11% G+C content (Fig. 5). Of the 3062 predicted genes, 3001 were protein-coding genes and 61 were RNAs (10 rRNA and 51 tRNA genes). A total of 1969 genes (65.61%) were assigned a putative function. A total of 172 genes were identified as ORFans (5.73%). The remaining genes were annotated as hypothetical proteins (774 genes = 25.79%). The distribution of genes into COGs functional categories is provided in Table 2, and the properties of the genome are summarized in Table 3.
Fig. 5.
Graphical circular map of ‘Massilibacteroïdes vaginae’ genome. From outside to center: contigs (red/gray), COGs category of genes on forward strand (three circles), genes on forward strand (blue circle), genes on reverse strand (red circle), COGs category on reverse strand (three circles), GC content. COGs, Clusters of Orthologous Groups database.
Table 2.
Number of genes associated with 25 general COGs functional categories
| Code | Description | Value | Percentage |
|---|---|---|---|
| [J] | Translation | 144.0 | 4.7 |
| [A] | RNA processing and modification | 0.0 | 0.0 |
| [K] | Transcription | 88.0 | 2.9 |
| [L] | Replication, recombination and repair | 119.0 | 3.9 |
| [B] | Chromatin structure and dynamics | 0.0 | 0.0 |
| [D] | Cell cycle control, mitosis and meiosis | 13.0 | 0.4 |
| [Y] | Nuclear structure | 0.0 | 0.0 |
| [V] | Defense mechanisms | 53.0 | 1.7 |
| [T] | Signal transduction mechanisms | 42.0 | 1.4 |
| [M] | Cell wall/membrane biogenesis | 119.0 | 3.9 |
| [N] | Cell motility | 0.0 | 0.0 |
| [Z] | Cytoskeleton | 0.0 | 0.0 |
| [W] | Extracellular structures | 0.0 | 0.0 |
| [U] | Intracellular trafficking and secretion | 15.0 | 0.5 |
| [O] | Posttranslational modification, protein turnover, chaperones | 55.0 | 1.8 |
| [C] | Energy production and conversion | 109.0 | 3.6 |
| [G] | Carbohydrate transport and metabolism | 139.0 | 4.5 |
| [E] | Amino acid transport and metabolism | 139.0 | 4.5 |
| [F] | Nucleotide transport and metabolism | 56.0 | 1.8 |
| [H] | Coenzyme transport and metabolism | 82.0 | 2.7 |
| [I] | Lipid transport and metabolism | 44.0 | 1.4 |
| [P] | Inorganic ion transport and metabolism | 68.0 | 2.2 |
| [Q] | Secondary metabolites biosynthesis, transport and catabolism | 18.0 | 0.6 |
| [R] | General function prediction only | 189.0 | 6.2 |
| [S] | Function unknown | 72.0 | 2.4 |
COGs, Clusters of Orthologous Groups database.
Table 3.
Nucleotide content and gene count level of genome
| Attribute | Value | Percent |
|---|---|---|
| Genome size (bp) | 3 710 320 | 100 |
| DNA G+C content | 1 488 269 | 40.1 |
| No. of genes | 3062 | 100 |
| Protein-encoding genes | 3001 | 98.0 |
| RNA genes | 61 | 2.0 |
| DNA coding sequence size | 3 361 204 | 90.6 |
| Proteins associated to COGs | 1324 | 44.1 |
| ORFans | 172 | 5.7 |
| Hypothetical proteins | 774 | 25.8 |
| Paralogous genes (E value = 1e-25) | 441 | 14.7 |
| Proteins assigned with a predicted function | 731 | 24.4 |
| Protein with transmembrane helices | 608 | 20.3 |
| Genes with atypical GC and codon bias usage | 19 | 0.6 |
| Genes associated with antimicrobial resistance | 0 | 0 |
| Genes associated with bacteriocin | 9 | 0.3 |
| Genes associated with mobilome | 1315 | 43.8 |
| Genes associated with toxin/antitoxin | 78 | 2.6 |
COGs, Clusters of Orthologous Groups database; ORF, open reading frame.
Insights from the genome sequence
The M. vaginae strain MV12T genome (3.71 Mbp) is smaller than those of D. mossii (3.92 Mbp), B. massiliensis (4.51 Mbp), D. gadei (5.17 Mbp), P. gordonii (6.67 Mbp), P. distasonis (4.81 Mbp), P. acetatigenes (4.78 Mbp) and D. capnocytophagoides (4.38 Mbp). It is larger than those of T. forsythia (3.41 Mbp) and P. cangingivalis (2.36 Mbp).
The G+C content of M. vaginae strain MV12T (40.11%) is in the same range as those observed in the Porphyromonadaceae family, which vary between 37.47% for D. mossii and 47.60% for P. cangingivalis.
The M. vaginae strain MV12T gene content is the same size as T. forsythia ATCC43037 (3001 Kb), larger than P. cangingivalis (2459 Kb) and smaller than the other bacteria of the family: D. mossii (3386 Kb), B. massiliensis (3935 Kb), D. gadei (4156 Kb), P. gordonii (5329 Kb), P. distasonis (3850 Kb), P. acetatigenes (3875 Kb) and D. capnocytophagoides (3578 Kb).
The comparison of M. vaginae strain MV12T to the other related genera, regarding the number of orthologous proteins shared between genomes, is represented in Table 4.
Table 4.
Numbers of orthologous proteins shared between genomes
| Dysgonomonas capnocytophagoides | Dysgonomonas mossii strain CCUG43457 | Dysgonomonas gadei | Porphyromonas cangingivalis | Proteiniphilum acetatigenes | Tannerella forsythia | Bacteroides massiliensis | Parabacteroides gordonii | Massilibacteroides vaginae | Parabacteroides distasonis | |
|---|---|---|---|---|---|---|---|---|---|---|
| D. capnocytophagoides | — | 1592 | 1702 | 715 | 1318 | 1023 | 1164 | 1372 | 1248 | 1284 |
| D. mossii strain CCUG43457 | 0.70 | — | 1771 | 721 | 1320 | 1049 | 1178 | 1404 | 1267 | 1331 |
| D. gadei | 0.70 | 0.74 | — | 720 | 1475 | 1112 | 1209 | 1530 | 1392 | 1404 |
| P. cangingivalis | 0.61 | 0.61 | 0.61 | — | 729 | 708 | 760 | 803 | 741 | 791 |
| P. acetatigenes | 0.65 | 0.65 | 0.65 | 0.61 | — | 1114 | 1182 | 1479 | 1316 | 1358 |
| T. forsythia | 0.63 | 0.63 | 0.63 | 0.62 | 0.64 | — | 1081 | 1281 | 1197 | 1225 |
| B. massiliensis | 0.65 | 0.65 | 0.65 | 0.61 | 0.64 | 0.64 | — | 1542 | 1227 | 1488 |
| P. gordonii | 0.65 | 0.65 | 0.65 | 0.62 | 0.65 | 0.68 | 0.67 | — | 1627 | 1910 |
| M. vaginae | 0.66 | 0.66 | 0.66 | 0.62 | 0.64 | 0.66 | 0.66 | 0.70 | — | 1538 |
| P. distasonis | 0.64 | 0.64 | 0.64 | 0.62 | 0.64 | 0.67 | 0.66 | 0.74 | 0.69 | — |
Conclusion
Regarding the analysis of the phenotypic, phylogenic and genomic features of strain MV12T, we propose the creation of the new genus ‘Massilibacteroïdes,’ for which the type strain is ‘Massilibacteroïdes vaginae’ strain MV12T. This strain was isolated from the vaginal flora of a 27-year-old woman with bacterial vaginosis.
Description of Massilibacteroïdes gen. nov.
‘Massilibacteroïdes’ (Ma.ssi.li'bac.te.ro.i.des, from Massilia, the Roman name for Marseille, the city in which the strain was isolated, and bacteroides, as being phylogenetically close to the genus bacteroides) was chosen because the strain was isolated in Marseille, France. Gram-negative rods. Facultative anaerobic. Mesophilic. Nonmotile. Exhibits catalase and oxidase activity. Positive for phosphatase alkaline, leucine arylamidase, trypsin, phosphatase acid, naphtol phosphohydrolase, β-galactosidase, α-glucosidase, β-glucuronidase, N-acetyl β-glucosaminidase, arginine dihydrolase, β-galactosidase, β-galactosidase 6 phosphate, α-glucosidase, β-glucosidase, α-arabinosidase, N-acetyl-β-glucosaminidase, fermentation of mannose, fermentation of raffinose, glutamic decarboxylase acid, α-fucosidase, reduction of nitrates, phosphatase alkaline, arginine arylamidase, leucyl glycine arylamidase, leucine arylamidase, alanine arylamidase, glutamyl acid and glutamic arylamidase. Habitat: human vaginal flora. Type species: ‘Massilibacteroïdes vaginae.’
Description of ‘Massilibacteroïdes vaginae’ sp. nov.
‘Massilibacteroïdes vaginae’ (va.gi'nae, L. n., from vagina, ‘vagina’; L. gen. n. vaginae, ‘of the vagina’). Gram-negative bacilli. Facultative anaerobic. Mesophilic. Nonmotile. Optimal growth at 37°C. Nonsporulating. Catalase and oxidase activity. Colonies are light beige, slightly bright, nonhaemolytic and approximately 0.5 mm in diameter. Cells are bacilli with a mean length of 2.06 μm and a mean width of 0.73 μm. Positive for phosphatase alkaline, leucine arylamidase, trypsin, phosphatase acid, naphtol phosphohydrolase, β-galactosidase, α-glucosidase, β-glucuronidase, N-acetyl β-glucosaminidase, arginine dihydrolase, β-galactosidase, β-galactosidase 6 phosphate, α-glucosidase, β-glucosidase, α-arabinosidase, N-acetyl-β-glucosaminidase, fermentation of mannose, fermentation of raffinose, glutamic decarboxylase acid, α-fucosidase, reduction of nitrates, phosphatase alkaline, arginine arylamidase, leucyl glycine arylamidase, leucine arylamidase, alanine arylamidase, glutamyl acid and glutamic arylamidase. Strain MV12T has susceptibility to penicillin G, cefotetan, imipenem, metronidazole, vancomycin and amoxicillin/clavulanic acid.
The 16S rRNA gene sequence was deposited in GenBank database under the accession number LN866992. The European Molecular Biology Laboratory (EMBL) accession number for the complete genome sequence is FXDJ00000000 (FXDJ01000001–FXDJ01000009), and it consists of nine scaffolds. The strain was deposited in two culture collections: the Collection de Souches de l’Unité des Rickettsies (CSUR) under number CSUR P1477, and the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) under number DSM 100704.
Acknowledgements
The authors thank the Xegen Company (www.xegen.fr) for automating the genomic annotation process. This study was funded by the Fondation Méditerranée Infection.
Conflict of interest
None declared.
References
- 1.Madhivanan P., Krupp K., Chandrasekaran V., Karat C., Arun A., Cohen C.R. Prevalence and correlates of bacterial vaginosis among young women of reproductive age in Mysore, India. Indian J Med Microbiol. 2008;26:132–137. doi: 10.4103/0255-0857.40526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradshaw C.S., Brotman R.M. Making inroads into improving treatment of bacterial vaginosis—striving for long-term cure. BMC Infect Dis. 2015;15:292. doi: 10.1186/s12879-015-1027-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fredricks D.N., Fiedler T.L., Marrazzo J.M. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med. 2005;353:1899–1911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- 4.Fredricks D.N., Fiedler T.L., Thomas K.K., Oakley B.B., Marrazzo J.M. Targeted PCR for detection of vaginal bacteria associated with bacterial vaginosis. J Clin Microbiol. 2007;45:3270–3276. doi: 10.1128/JCM.01272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fredricks D.N., Fiedler T.L., Thomas K.K., Mitchell C.M., Marrazzo J.M. Changes in vaginal bacterial concentrations with intravaginal metronidazole therapy for bacterial vaginosis as assessed by quantitative PCR. J Clin Microbiol. 2009;47:721–726. doi: 10.1128/JCM.01384-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lagier J.C., Armougom F., Million M., Hugon P., Pagnier I., Robert C. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 7.Seng P., Drancourt M., Gouriet F., La Scola B., Fournier P.E., Rolain J.M. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 8.Menard J.P., Fenollar F., Henry M., Bretelle F., Raoult D. Molecular quantification of Gardnerella vaginalis and Atopobium vaginae loads to predict bacterial vaginosis. Clin Infect Dis. 2008;47:33–43. doi: 10.1086/588661. [DOI] [PubMed] [Google Scholar]
- 9.Rossi-Tamisier M., Benamar S., Raoult D., Fournier P.E. Cautionary tale of using 16S rRNA gene sequence similarity values in identification of human-associated bacterial species. Int J Syst Evol Microbiol. 2015;65:1929–1934. doi: 10.1099/ijs.0.000161. [DOI] [PubMed] [Google Scholar]
- 10.Sakamoto M., Benno Y. Reclassification of Bacteroides distasonis, Bacteroides goldsteinii and Bacteroides merdae as Parabacteroides distasonis gen. nov., comb. nov., Parabacteroides goldsteinii comb. nov. and Parabacteroides merdae comb. nov. Int J Syst Evol Microbiol. 2006;56:1599–1605. doi: 10.1099/ijs.0.64192-0. [DOI] [PubMed] [Google Scholar]
- 11.Stackebrandt E. Molecular taxonomic parameters. Microbiol Aust. 2011;32:59–61. [Google Scholar]
- 12.Rosselló-Mora R. DNA-DNA reassociation methods applied to microbial taxonomy and their critical evaluation. In: Stackebrandt E., editor. Molecular identification, systematics, and population structure of prokaryotes. Springer; Berlin: 2006. pp. 23–50. [Google Scholar]
- 13.Validation of the publication of new names and new combinations previously effectively published outside the IJSB: list no. 7. Int J Syst Bacteriol. 1981;31:382–383. doi: 10.1099/00207713-47-4-1274. List Editor. [DOI] [PubMed] [Google Scholar]
- 14.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyatt D., Chen G.L., LoCascio P.F., Land M.L., Larimer F.W., Hauser L.J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tatusov R.L., Galperin M.Y., Natale D.A., Koonin E.V. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lagesen K., Hallin P., Rødland E.A., Staerfeldt H.H., Rognes T., Ussery D.W. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M.A. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 20.Lechner M., Findeiss S., Steiner L., Marz M., Stadler P.F., Prohaska S.J. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinform. 2011;12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]