SUMMARY
Do all animals sleep? Sleep has been observed in many vertebrates, and there is a growing body of evidence for sleep-like states in arthropods and nematodes [1–5]. Here we show that sleep is also present in Cnidaria [6–8], an earlier branching metazoan lineage. Cnidaria, along with Ctenophora, are the first metazoan phyla to evolve tissue-level organization and differentiated cell types, such as neurons and muscle [9–15]. In Cnidaria, neurons are organized into a non-centralized radially symmetric nerve net [11,13,15–17] that nevertheless shares fundamental properties with the vertebrate nervous system: action potentials, synaptic transmission, neuropeptides, and neurotransmitters [15–20]. It was reported that cnidarian soft corals [21] and box jellyfish [22,23] exhibit periods of quiescence, a pre-requisite for sleep-like states, prompting us to ask if sleep is present in Cnidaria. Within Cnidaria, the upside-down jellyfish Cassiopea spp. displays a quantifiable pulsing behavior, allowing us to perform long-term behavioral tracking. Monitoring Cassiopea pulsing activity for consecutive days and nights revealed behavioral quiescence at night that is rapidly reversible, and a delayed response to stimulation in the quiescent state. When deprived of nighttime quiescence, Cassiopea exhibited decreased activity and reduced responsiveness to a sensory stimulus during the subsequent day, consistent with homeostatic regulation of the quiescent state. Together these results indicate that Cassiopea has a sleep-like state, supporting the hypothesis that sleep arose early in the metazoan lineage, prior to the emergence of a centralized nervous system.
eTOC Blurb
Understanding the phylogenetic roots of behaviors sheds light on evolutionary forces that shape them. Sleep has been observed in worms, flies, zebrafish, and mice. Nath et al. discover that jellyfish have a sleep-like state. This shifts the known root of sleep in the phylogenetic tree prior to the emergence of a centralized nervous system.
RESULTS AND DISCUSSION
Three behavioral characteristics define a sleep state [6,7,24]: (1) behavioral quiescence, a period of decreased activity; (2) reduced responsiveness to stimuli during the quiescent state; and (3) homeostatic regulation of the quiescent state. Both behavioral quiescence and reduced responsiveness must be rapidly reversible to differentiate sleep-like states from other immobile states (e.g. paralysis or coma) and reduced responsiveness distinguishes sleep from quiet wakefulness. Homeostatic regulation results in a rebound response, i.e. a compensatory period of increased sleep following sleep deprivation. Here we asked whether the cnidarian jellyfish Cassiopea exhibits these behavioral characteristics.
Cassiopea are found throughout the tropics in shallow ocean waters and mudflats (Figure 1; [25,26]). They rarely swim and rather remain stationary with their bell on a surface, hence their name, the upside-down jellyfish (Figure 1B; Figure S1A; Movie S1; [25,26]). Cassiopea, like coral and sea anemones, have a photosynthetic obligate endosymbiote, Symbiodinium (Figure 1C). Cassiopea continuously pulse by relaxing and contracting their bell at a rate of about 1 pulse/second (Figure 1D). This pulsing behavior generates fluid currents that facilitate vital processes such as filter feeding, circulation of metabolites, expulsion of byproducts, and gamete dispersion [25,27]. The pulsing behavior is controlled by light and gravity sensing organs called rhopalia (Figure 1C; [11,13]). This stationary pulsing behavior makes Cassiopea a suitable jellyfish for behavioral tracking.
Figure 1. The pulsing behavior of the upside-down jellyfish, Cassiopea spp., is trackable.
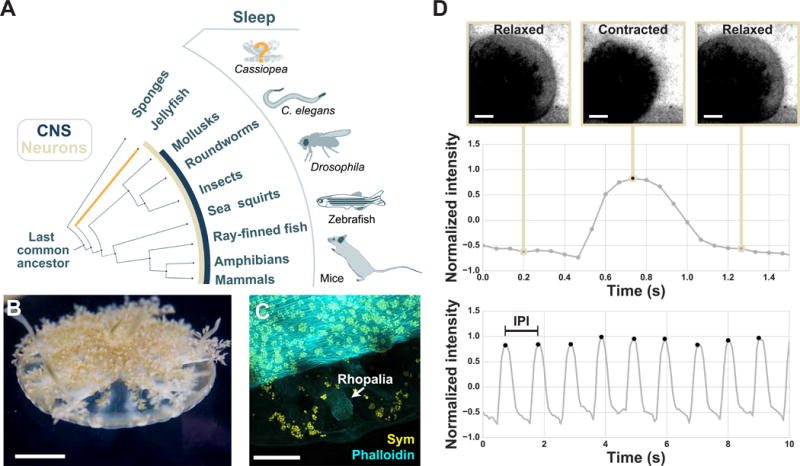
(A) Phylogenetic tree schematic highlighting animals in which sleep behavior has been described, the presence of neurons (tan), and the emergence of a centralized nervous system (dark blue). See boxed key. (B) An image of Cassiopea. (C) Higher magnification view of Cassiopea with labeled actin-rich muscle (phalloidin stain; cyan), autofluorescent Symbiodinium (yellow), and a rhopalia, the sensory organ that controls pulsing, which is free of Symbiodinium. (D) As Cassiopea pulse the relaxation and contraction of the bell causes a corresponding change in average pixel intensity. Pulsing behavior was tracked by measuring this change in pixel intensity within the region of interest. (top) Representative frames and corresponding normalized pixel intensities for one pulse event. The local maxima in the pulse-trace was used to count pulse events. (bottom) A 10-second recording of one jellyfish shows multiple pulsing events. The inter-pulse interval (IPI) was calculated as the time between the maxima. See Figure S1, Figure S2, Movie S1.
To track behavior in Cassiopea, we designed an imaging system (Figure S1C–F) for counting pulses of individual jellyfish over successive cycles of day and night, defined as a 12-hour period when the light is on or off, respectively. As Cassiopea pulse, the relaxation and contraction of the bell causes a corresponding change in average pixel intensity, which was measured for each frame of the recording, producing a pulse-trace (Figure 1D). Pulse events were counted using the peak of the pulse-trace, and the inter-pulse interval (IPI) was calculated as the time between the peaks (Figure 1D; Figure S2).
We observed that Cassiopea pulse less at night than during the day (Figure 2; Data S1). To quantify this difference in pulsing frequency, we tracked the pulsing behavior of 23 jellyfish over 6 consecutive days and nights (Figure 2C). We define activity as the total number of pulses in the first 20 minutes of each hour. While individual jellyfish showed different basal activity levels (Figure 2C), all showed a large decrease in mean activity (~32%) at night (781 ± 199 pulses/20 min, mean ± s.d.) compared to the day (1155 ± 315 pulses/20 min, mean ± s.d.; Figure 2C,E). To determine if fast and slow pulsing jellyfish change their activity to a similar degree, we normalized activity of individual jellyfish by their mean day activity. Despite variations in basal activity, the relative change from day to night was similar between jellyfish (Figure 2D). Jellyfish activity decreased throughout the first 3–6 hours of the night, with the lowest activity occurring 6–12 hours after the day to night transition. Pulsing activity peaked upon feeding, occurring on the 4th hour of each day (Figure 2C,D). To ensure that day feeding does not cause the day-night behavioral difference, we tracked the activity of 16 jellyfish over three consecutive days and nights without feeding and observed results consistent with those including feeding (Figure 2F,G; Figure S3D). These results demonstrate that Cassiopea have a quiescent state during the night. To test the reversibility of this nighttime quiescent state we introduced a food stimulus at night, which transiently increased activity to daytime levels (Figure S3E). The nighttime quiescent state in Cassiopea is thus rapidly reversible, consistent with a sleep-like behavior.
Figure 2. Continuous tracking of Cassiopea reveals pulsing quiescence at night.
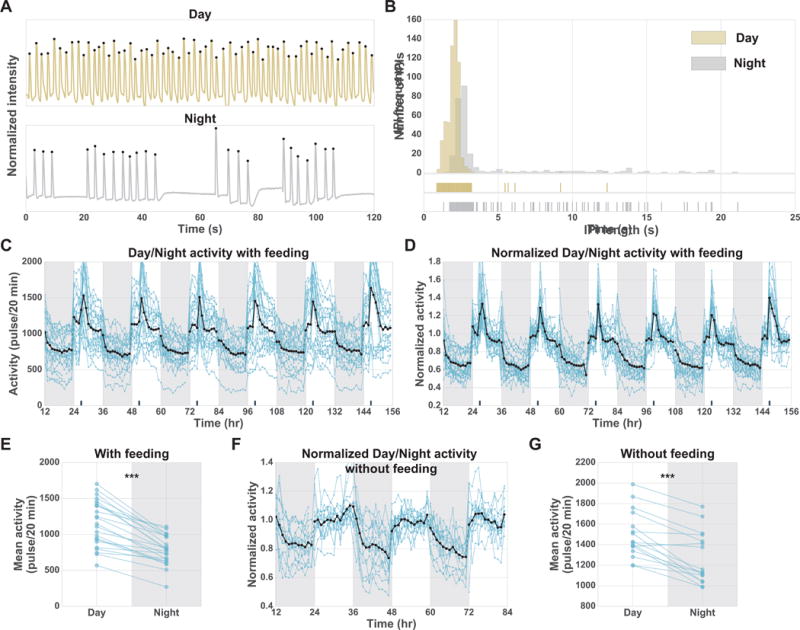
(A) Pulsing-traces for individual jellyfish during day and night over 120 s. (B) The distribution of IPI length for a 12-hour day and a 12-hour night for the same jellyfish shown in A. Tick marks below the distribution show each IPI length during the day and night. This highlights the long-pause events, which are more common at night (Figure S3A; Data S1). (C-G) Each blue line corresponds to a single jellyfish. The black line indicates the mean activity of all jellyfish. Dark gray shading indicates night periods. Dark tick marks on the x-axis indicate time of feeding. (C) Baseline activity (pulses/20 min) of 23 jellyfish tracked for six days from four laboratory replicates. (D) Normalized baseline activity for jellyfish shown in C, where each jellyfish is normalized by their mean day activity. (E) Mean day activity versus mean night activity for each jellyfish over the six-day experiment shown in C. Two-sided paired t-test, day versus night, P = 6×10−9. (F) Normalized baseline activity without feeding of 16 jellyfish tracked over three days from two laboratory replicates, where each jellyfish is normalized by its mean day activity. (G) Mean day activity versus mean night activity for each jellyfish over the three-day experiment shown in F. Two-sided paired t-test, day versus night, P =10−5. ***P<10−3. See Figure S3.
To better understand the nighttime quiescence, we compared day and night pulse-traces of individual jellyfish. The day and night pulse-traces of one representative jellyfish are shown in Figure 2A. During the night, the IPI is typically longer than during the day (Figure 2A,B; Data S1; Figure S3A). Two features contribute to this lengthening of the IPI: (1) the mode of the IPI distribution is longer at night than during the day, and (2) night pulsing is more often interrupted by pauses of variable length. These pauses are seen as a tail in the IPI frequency distribution (Figure 2B: 95th percentile of night IPI frequency distribution (gray) is 13.9 s). Such long pauses are rarely seen during the day (Figure 2B: 95th percentile of day IPI frequency distribution (yellow) is 2.5 s). This pause behavior may be analogous to long rest bouts observed in Drosophila and zebrafish, which are suggested to be periods of deep quiescence with reduced responsiveness to stimuli [1,28].
To test whether Cassiopea exhibit reduced responsiveness to stimuli during their nighttime-quiescent state, we designed an experiment to deliver a consistent arousing stimulus to the jellyfish. We observed in our nursery that Cassiopea prefer staying on solid surfaces as is found in nature. If Cassiopea are released into the water column, they quickly reorient and move to the bottom of the tank. We used placement into the water column as a stimulus to compare responsiveness during the night versus the day. Cassiopea were put inside a short PVC pipe with a screen bottom (Figure 3A). This was lifted to a fixed height, held for 5 min to allow the jellyfish to acclimate, and then rapidly lowered, which placed the jellyfish free-floating into the water column. We then scored the time it took for the jellyfish to first pulse and the time to reach the screen bottom (Figure 3A; Star Methods). At night, the jellyfish showed an increase in the time to first pulse and the time to reach bottom, compared to day (time to first pulse day: 2.1 ± 0.9 s versus night: 5.9 ± 4.0 s, and the time to reach bottom day: 8.6 ± 2.9 s versus night: 12.0 ± 3.2 s, mean ± s.d.; n = 23 animals) (Figure 3B,C). This increased latency in response to stimulus indicates that Cassiopea have reduced responsiveness to stimulus during the night.
Figure 3. Cassiopea show reduced responsiveness to a sensory stimulus at night.
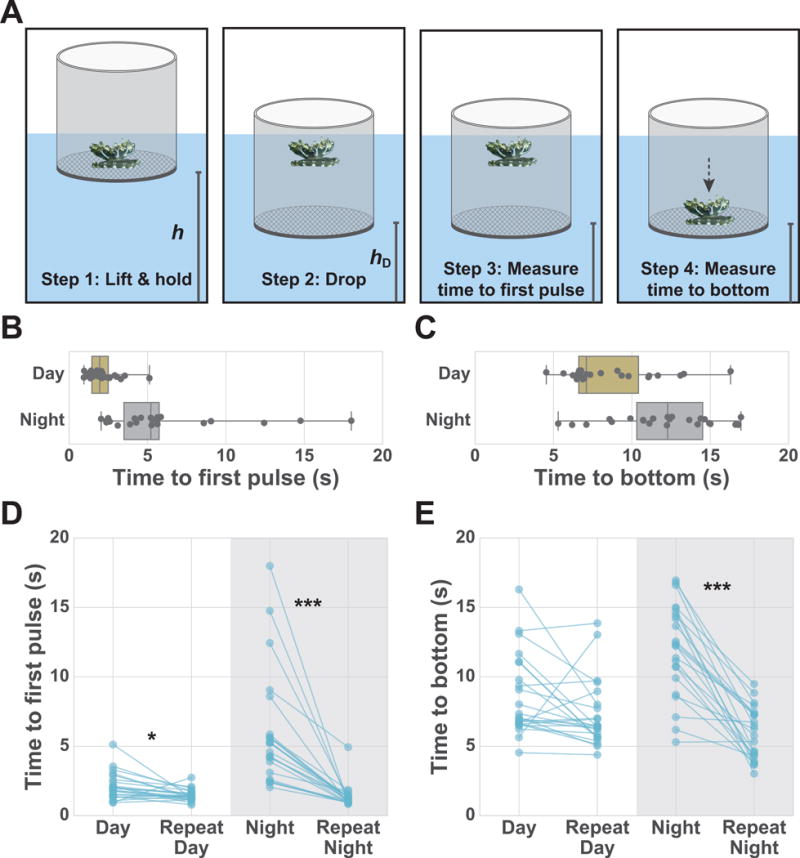
(A) Schematic of experiment to test sensory responsiveness. Jellyfish were lifted and held at a fixed height (hL) and then dropped to a fixed height (hD). hL and hD were kept constant throughout experiments. Boxplots of time to first pulse after drop (B) for 23 jellyfish and time to reach bottom after drop (C) for 23 jellyfish during the day and night. Dots represent individual jellyfish collected from two laboratory replicates. Two-sided unpaired t-test, day versus night, (B) P < 10−4 and (C) P = 5×10−4. (D) Time to first pulse after initial drop and after perturbation for both day and night for 23 jellyfish. (E) Time to reach bottom after initial drop and after perturbation for both day and night for 23 jellyfish. Two-way analysis of variance (ANOVA) for data shown in D and E, followed by post-hoc comparisons between experimental groups using Bonferroni posttest (*P<5×10−2, ***P<10−3). For the time to first pulse, two-sided unpaired t-test (B) and two-way ANOVA (D) were performed after log-transformation (Star Methods).
To determine if the increased latency at night is rapidly reversible, a second drop was initiated within 30 s of the first drop, that is, after the jellyfish have been aroused. Reversibility was tested during both the day and night for 23 jellyfish. During the night, there is a large decrease in the time to first pulse and time to reach the bottom, after the second drop when compared to the first drop (Figure 3D,E). During the day and night, the time to first pulse and time to bottom after the second drop were indistinguishable, demonstrating that after perturbation, animals have similar arousal levels during the day and night. These results indicate that Cassiopea have rapidly reversible reduced responsiveness to a stimulus during the night.
To test whether Cassiopea nighttime quiescence is homeostatically regulated, we deprived jellyfish of behavioral quiescence for either 6 or 12 hours using a mechanical stimulus (Figure 4). The stimulus consisted of a brief (10 s) pulse of water every 20 min, which caused a transient increase in pulsing activity (Movie S2). This increase in pulsing activity lasts for approximately 5 min after the 10 s pulse of water. Thus, the perturbation disrupts quiescence for approximately 25% of the perturbation period (either 6 hours or 12 hours). When the perturbation was performed during the last 6 hours of the night (Figure 4A), we observed a significant decrease in activity (~12%) during the first 4 hours of the following day relative to the pre-perturbation day (mean of first 4 hours of pre-perturbation day: 1146 ± 232 pulses/20 min compared to post-perturbation day: 1008 ± 210 pulses/20 min, mean ± s.d.; n = 30 animals; Figure 4C). This period of decreased activity is due to both decreased pulsing frequency (increased mode of IPI-length) and increased pause length (increase in the IPI-length 95th percentile) (Figure S4B,C). This result is consistent with an increased sleep-drive after sleep deprivation. After a single day of decreased activity, the jellyfish return to baseline levels of day and night activity. Similar results were observed after an entire night of perturbation (12 hours; Figure 4D), with a large decrease in activity (~17%) throughout the following day (mean of 12 hours of pre-perturbation day: 1361 ± 254 pulses/20 min compared to post-perturbation day: 1132 ± 263 pulses/20 min, mean ± s.d.; n = 16 animals; Figure 4F). The decrease in activity caused by the 12-hour perturbation was larger than that of the 6-hour perturbation, indicating that the amount of sleep rebound is dependent on the level of sleep deprivation. During periods of decreased activity after either the 6-hour or 12-hour perturbation, we also observed increased response latency to a sensory stimulus (Figure S4A), indicating a sleep-like state.
Figure 4. Homeostatic rebound in Cassiopea.
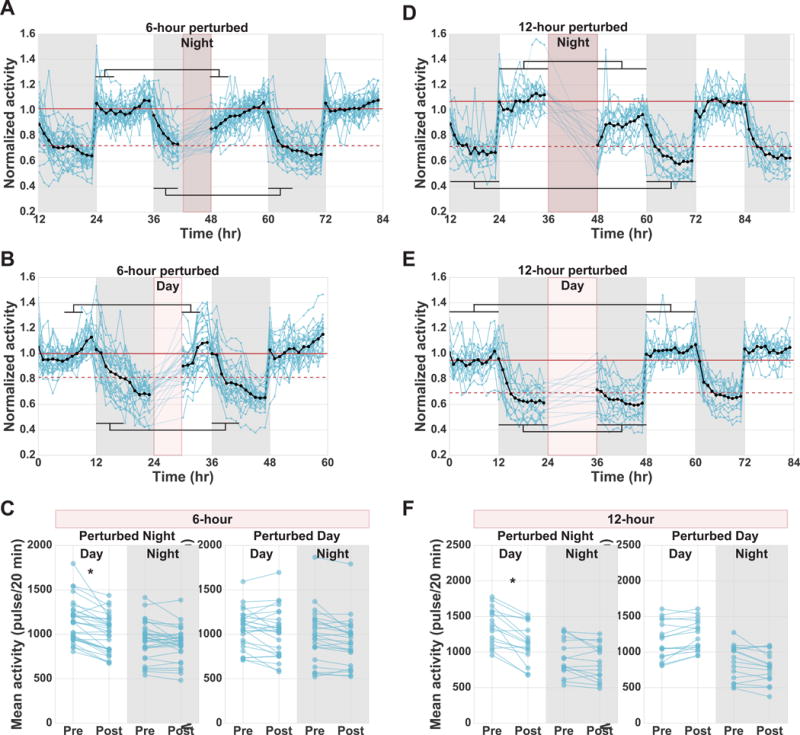
Each blue line corresponds to a single jellyfish. The black line indicates the mean activity of all jellyfish. Dark gray shading indicates night periods. Maroon shading indicates perturbation periods with 10 s water pulses every 20 min. Jellyfish were exposed to different perturbation lengths (6 or 12 hours) at different times (day or night). The normalized activity of all jellyfish tracked over multiple days is plotted. Maroon horizontal lines show the mean activity of pre-perturbation day (solid) and pre-perturbation night (dashed). (A) Perturbation of 30 jellyfish for the last 6 hours of the night. (B) Perturbation of 26 jellyfish for the first 6 hours of the day. (C) Mean day and night activity pre- and post-perturbation for experiments shown in A and B. (D) Perturbation of 16 jellyfish for an entire 12-hour night. (E) Perturbation of 16 jellyfish for an entire 12-hour day. (F) Mean day and night activity pre- and post-perturbation for experiments shown in D and E. Black-horizontal lines in A, B, D, and E indicate the windows of time used for calculating pre- and post-perturbation means shown in C and F for both the night (bottom lines) and day (top lines). For the 6-hour experiments we compared the first 4 hours of the post-perturbation day to the equivalent time pre-perturbation, and also compared the first 6 hours of post-perturbation night to the equivalent time pre-perturbation. For the 12-hour experiments we compared the full 12-hour days and nights pre- and post-perturbation. Two-way ANOVA followed by post-hoc comparisons between experimental groups using Bonferroni posttest (*P<5×10−2). Both day and night 6-hour perturbation experiments include data from four laboratory replicates. Both day and night 12-hour perturbation experiments include data from two laboratory replicates. See Figure S4, Movie S2.
If the reduced activity following nighttime perturbation is due to sleep deprivation rather than muscle fatigue, applying the perturbation during the day, when Cassiopea are much less quiescent, should not result in reduced activity. To distinguish between sleep deprivation and muscle fatigue, we performed the 6- or 12-hour mechanical stimulus experiments during the day (Figure 4B,E). We observed no significant difference between pre- and post-perturbation activity levels (Figure 4C,F), indicating that the rebound response is specific to deprivation of nighttime quiescence. Taken together, these results demonstrate that Cassiopea have a nighttime-quiescent state that is homeostatically controlled.
In many animals sleep is regulated by both homeostatic and circadian systems [29], but this is not always the case [4–7,30]. For instance, the nematode C. elegans exhibits a developmentally regulated sleep state, and adult C. elegans show a non-circadian stress-induced-sleep state [4,5,31]. A fully functioning circadian system is also not essential for sleep to occur; animals with null mutations of circadian rhythm genes still sleep, though sleep timing is altered [30]. To test if nighttime quiescence in Cassiopea is regulated by a circadian rhythm, we first entrained the jellyfish for one week in a normal 12:12-hour light/dark cycle, and then shifted them to constant lighting conditions for 36 hours. We tested low- (~0.5 Photosynthetic Photon Flux [PPF]), mid- (~100 PPF), and full-intensity (~200 PPF) light, as well as dark (Figure S4D,E). If jellyfish activity is regulated by a circadian rhythm, cycling activity should persist in the absence of entraining stimuli, such as light. We observed no circadian oscillation of jellyfish activity under any of the constant light conditions (Figure S4D). However, we do observe circadian oscillation of activity in constant dark conditions (Figure S4E). This result suggests that the quiescent state may be under circadian regulation.
Cassiopea display the key behavioral characteristics of a sleep-like state: a reversible quiescent state with reduced responsiveness to stimuli and both homeostatic and possibly circadian regulation. To our knowledge, our finding is the first example of a sleep-like state in an organism with a diffuse nerve net [7,8], suggesting that this behavioral state arose prior to the evolution of a centralized nervous system. Though at least 600 million years of evolution separate cnidarians from bilaterians [9–16], many aspects of the nervous system are conserved, including neuropeptides and neurotransmitters [15–20]. One such conserved molecule, melatonin [32], promotes sleep in diurnal vertebrates, including zebrafish [33] and humans [34], and induces quiescence in invertebrates [35]. We observed that melatonin induces a reversible decrease in activity in Cassiopea during the day in a concentration-dependent manner (Figure S4F–H), suggesting that melatonin has a conserved quiescence-inducing effect in Cassiopea. Pyrilamine, a histamine H1 receptor antagonist that induces sleep in vertebrates [36], also induces concentration-dependent quiescence in Cassiopea (Figure S4F). These results suggest that at least some mechanisms involved in vertebrate sleep may be conserved in Cassiopea.
Although future studies are required to test whether other cnidarians sleep, field studies showing behavioral quiescence, diel vertical migration, and swimming speeds that vary with diel period [21,22,23,37] suggest that a sleep-like state may not be specific to Cassiopea. A cnidarian sleep-like state could result from either divergent or convergent evolution. The observation of behaviorally and mechanistically conserved sleep-like states across the animal kingdom [6,7] strongly supports the possibility for an early rooted sleep state rather than many instances of convergent evolution. It has been hypothesized that sleep has multiple functions, including synaptic homeostasis, regulation of neurotransmitters, repair of cellular damage, removal of toxins, memory consolidation and energy conservation [7], although the ancestral role and selective advantage of sleep remains elusive. Our discovery of a sleep-like state in an ancient metazoan phylum suggests that the ancestral role of sleep is rooted in basic requirements that are conserved across the animal kingdom. The ancestral function of sleep may be revealed by further study of early branching metazoa.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Lea Goentoro (goentoro@caltech.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cassiopea spp. medusae used in this study were originally collected from the Florida Keys. For the majority of the experiments, a collection of multiple Cassiopea species were used (Figure S1A,B). For the experiments shown in Figure S4A,E,F a young (2–4 months old) clonal population of medusa were used (Cassiopea xamachana). This clonal polyp line was generated in Monica Medina’s lab at Pennsylvania State University.
Cassiopea were reared in artificial seawater (ASW, Instant Ocean, 30–34 ppt) at pH 8.1–8.3, 26–28°C with a 12-hour day/night cycle. During the day, 450 and 250 W light sources were used to generate 200–300 PPF (Photosynthetic Photon Flux, a measurement of light power between 400 and 700 nm). To limit waste buildup, the Cassiopea aquarium was equipped with a refugium (Chaetomorpha algae aquaculture), a protein skimmer (Vertex Omega Skimmer), carbon dosing bio-pellets (Bulk Reef Supply), activated carbon in a media reactor (Bulk Reef Supply), and a UV sterilizer (Emperor Aquatics 25 W). Waste products were kept at or below the following levels: 0.1 ppm ammonia, 5 ppb phosphorus, 0 ppm nitrite, and 0 ppm nitrate.
Cassiopea were fed daily with brine shrimp (Artremia nauplii, Brine Shrimp Direct) enriched with Nannochloropsis algae (Reed Mariculture), and they were fed oyster roe once per week (Reed Mariculture). Cassiopea were group housed in a 60 gallon holding tank. Animals were randomly assigned to experimental groups. Medusae between 3–6 cm in diameter were used for experiments.
Cassiopea Genotyping
Cassiopea is a genus with many species that have not been classified. All of our experiments were performed with Cassiopea spp. of a range of sizes, ages, sex and morphologies (Figure S1A,B). To assess the diversity of Cassiopea spp. within our population we genotyped several animals by amplification and sequencing of the Mitochondrial cytochrome c oxidase I (COI). Genomic DNA extractions were performed as described [38]. Jellyfish fragments, about 2 mm of tissue from the tentacles, were placed in 400 μL DNA extraction buffer (50% w/v guanidinium isothiocyanate; 50 mM Tris pH 7.6; 10 μM EDTA; 4.2% w/v sarkosyl; 2.1% v/v β-mercaptoethanol). Samples were incubated at 72°C for 10 min, centrifuged at 16,000 g for 5 min, and the resulting supernatant mixed with an equal volume of isopropanol and incubated at −20°C overnight. The DNA was precipitated by centrifugation at 16,000 g for 15 min and the DNA pellet washed in 70% ethanol and resuspended and stored in water.
Amplification of COI was performed using primers designed by Folmer et al. [39], which amplify a ~710 base pair fragment of COI across the broadest array of invertebrates. COI primers:
LCO1490 forward primer: 5′-ggtcaacaaatcataaagatattgg-3′
HC02198 reverse primer: 5′-taaacttcagggtgaccaaaaaatca-3′
Amplifications were performed under the following PCR conditions: 2 min at 92°C, 30 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 45 s, with a final 72°C extension for 7 min. Amplification products were then TOPO-cloned using OneTaq (NEB) and sequenced.
Multiple sequence alignment of Cassiopea spp. COI sequences were generated using Clustal Omega software. Sequences were aligned with each other (see Figure S1B), and to the previously identified cryptic species Cassiopea ornata, Cassiopea andromeda, and Cassiopea frondosa [26]. The level of identity between these sequences is presented in Figure S1B. Of the 15 Cassiopea spp. sequenced there were 8 identical COI sequences and 7 COI sequences with 45–90% identity.
METHODS DETAILS
Cassiopea behavioral tracking
Individual jellyfish were placed into 700 mL square clear plastic containers (cubbies), with white sand bottoms, in 35 L (10 gallon) glass tanks (Figure S1C–F). Eight containers can fit in each tank, so eight jellyfish can be simultaneously recorded per tank. Tanks were housed inside Sterilite utility cabinets (65 cm W × 48 cm L × 176 cm H) with a door to eliminate ambient light in the recording setup. During the 12-hour day (lights on) tanks were illuminated with 24-inch florescent lamps, each containing four florescent bulbs that provide a combination of wavelengths optimized for photosynthesis in water: two 24 W, 6000 K Mid-day lights, and two 24 W Actinic lights (Giesemann), which combined provided 200–300 PPF. During the 12-hour night (lights off) low-intensity red-LEDs were used to illuminate jellyfish to enable visualization. For all jellyfish recordings we used Unibrain 501b cameras above the tank running Firei software capturing at 15 frames per second. Camera aperture and Firei settings were adjusted to increase the contrast between jellyfish and background. Recordings were saved directly onto hard drives.
Jellyfish were acclimated in the recording tank in their cubbies for 2–3 days before starting recordings. 24-hour recordings were taken for successive days (7 am – 7 pm) and nights (7 pm – 7 am), unless otherwise indicated. Cassiopea were fed each day at 10:30 am, 3.5 hours after the lights turn on. Each jellyfish received 5 mL of 16 g/L brine shrimp. For each circadian rhythm experiment a different light condition was left on for 36-hours: dark conditions, low-intensity light conditions (an array of white-LED lights, 0–0.5 PPF), mid-intensity light conditions (two 24 W, 6000 K Mid-day lights, 75–150 PPF), or full light conditions (two 24 W, 6000 K Mid-day lights, and two 24 W Actinic lights, 200–300 PPF). For 6-hour and 12-hour rebound experiments the mechanical stimulus (Movie S2) was applied for 10 s every 20 min.
All analysis was done using open-source packages in the SciPy ecosystem [40–42]. To monitor jellyfish activity, pulsing information was extracted from the individual frames of each recording. Approximately 648,000 frames were collected every 12 hours. To quantify pulsing activity, we processed the first 18,000 frames of every hour (20 min). As Cassiopea pulse, the relaxation and contraction of the bell causes a corresponding change in average pixel intensity. To measure this change in average pixel intensity we drew a rectangular region of interest (ROI) around each jellyfish (Figure 1D; Figure S1F). A user manually selected a ROI around each of the eight jellyfish in the first and last of the 18,000 frames. This was done so that the selected ROI accounts for any movement of the jellyfish. To control for noise from oscillations in ambient lighting, we perform background subtraction using a similarly sized ROI containing no jellyfish.
We analyzed pixel intensity data, and identified pulse events and inter-pulse intervals (IPI) in a four-step process. Step 1: Gaussian smoothing of the mean intensity over time to eliminate high frequency oscillations (Figure S2A). This smoothed trace was used to account for large movements in the mean intensity due to jellyfish translational movement within the selected ROI. Step 2: Normalization of the mean intensity values with the max mean intensity and the smoothed mean intensity:
Where Traw is the raw intensity trace, Tsmooth is the smoothed trace generated in Step 1, Tmax is maximum intensity across the raw trace, and n is the index of each frame of the recording. Step 3: find the indices (time) of local maxima and minima in the normalized trace. Because of noise in the pulsing trace there is a high rate of false positives when finding local maxima and minima (Figure S2B). We have used a set of criteria to identify a true pulse event from the local maxima and local minima. Step 4: identifying pulses from local maxima and minima (Figure S2C). A local maximum can be defined as a pulse peak if it meets two criteria. First, it must be above a set threshold (to eliminate local maxima due to noise in pause regions of the pulse trace). Second, it must be above a set distance from the next local maxima (to prevent double counting of a single pulse). The standard deviation of the Gaussian smoothing, the threshold level, and the minimum distance between pulses can all be changed from one jellyfish to another. For all data analysis these parameter values were optimized to quantify pulsing events for each animal.
We calculated the total number of pulses and the IPI for each 20-min time bin. With some jellyfish the difference in pixel intensity from the contracted to non-contracted state was not big enough to easily identify pulsing above the noise. These jellyfish were excluded from analysis. During the 20-min recordings jellyfish would occasionally move out of the selected ROI. We would then exclude that 20-min recording for that jellyfish from the analysis. In compiling data to generate activity versus time plots we excluded jellyfish that we could not analyze for more than three 20-min recordings during a 12-hour day or night period.
For the arousal assay we designed an experiment to systematically test this sensory responsiveness. Cassiopea respond to being placed in the water column by rapidly orienting themselves and moving towards a stable surface. For the experimental system, Cassiopea were placed inside a 20 cm tall, 12 cm diameter, PVC pipe with a 53 m filter screen bottom, called a Cassiopea dropper (CD). The experiment consists of four steps, as seen in the four panels in Figure 3A. Step 1, the jellyfish were placed on the screen bottom of the CD, which was positioned two cm below the water surface (hL) and were acclimated for five min. At night jellyfish took less than five min to return to quiescence after being placed in the CD. Step 2, the CD was then “dropped” to a set depth (18 cm from the surface, hD). This action leaves the jellyfish free-floating, two cm below the water surface. Step 3, the time to first pulse was measured. Step 4, the time to reach bottom was measured. To determine if the nighttime arousal latency is reversible, a second drop experiment was performed within 30 s of the initial drop. The CD was returned to two cm below the water surface, but instead of waiting for five min, steps 2 and 3 were performed immediately. Time to first pulse and time to bottom are not completely independent measures, though there is also not a perfect correlation. A jellyfish could pulse quickly but be delayed in reaching the bottom due to, for example, inactivity after the first pulse.
Cassiopea staining and imaging
Actin was stained using Alexa Flour 488-Phalloidin (ThermoFisher A12379). Jellyfish were anesthetized in ice-cold 0.8 mM menthol/ASW, and then fixed in 4% formaldehyde on ice for 45 min. Fixed jellyfish were permeabilized in 0.5% Triton/PBS for 2 hours and blocked using 3% BSA for 1 hour. They were then incubated in 1:100 Phalloidin solution in 0.5% Triton/PBS, for 18–24 hours in the dark at 4°C [43]. Stained jellyfish were mounted in refractive index matching solution [44] and imaged using a LSM 780 confocal microscope (Zeiss).
QUANTIFICATION AND STATISTICAL ANALYSIS
The following statistical tests were used: two-sided paired Student’s t-tests, two-sided unpaired Student’s t-tests, and two-way ANOVA with Bonferroni posttest. We performed D’Agostino’s omnibus K2 normality test on all data sets to assess whether or not to reject the null hypothesis that all values were sampled from a population that follows a Gaussian distribution. For paired values, we tested if the pairs were sampled from a population where the difference between pairs follows a Gaussian distribution. Experimental groups that were statistically compared were tested for equal variance. The normality tests showed that all datasets were approximately Gaussian distributed with the exception of the time to first pulse arousal data. The time to first pulse data also showed grounds for rejecting the null hypothesis that there was equal variance between experimental groups. Tests of the log transformed time to first pulse data showed that the transformed data was approximately Gaussian distributed with equal variance between experimental groups, validating the use of standard two-way ANOVA and unpaired t-tests on the transformed data. Statistical tests were performed using either statistical functions from the SciPy ecosystem or GraphPad Prism (version 6.04 for Windows, GraphPad Software, San Diego California USA, www.graphpad.com). No statistical methods were used to predetermine sample size. For these experiments we performed at least two laboratory replicates within our recording setup, which is limited to 8 jellyfish. Investigators were not blinded to allocation during experiments and outcome assessment. No specific method for randomization was used.
DATA AND SOFTWARE AVAILABILITY
Code used for tracking jellyfish activity and analysis are available at https://github.com/GradinaruLab/Jellyfish.
Supplementary Material
Movie S1. Cassiopea pulsing behavior, Related to Figure 1. Two jellyfish pulsing during the day with ruler for scale.
Movie S2. Cassiopea exposed to brief water pulse perturbation, Related to Figure 4. A mechanical stimulation perturbs a single jellyfish.
Data S1. Day versus night pulsing behavior, Related to Figure 2. Pulsing-traces for individual jellyfish during day (yellow) and night (gray) over 300 s. These recordings were taken from the 7th hour of the day or night. Each tab is a different jellyfish (jf1-jf22). The activity of the same jellyfish are presented in Figure 2C,D.
Highlights.
-
-
Cassiopea jellyfish exhibit reversible behavioral quiescence during the night
-
-
Cassiopea show reduced responsiveness to stimuli during their quiescent state
-
-
The nighttime quiescence is regulated by both homeostatic and circadian systems
-
-
Pharmacological studies show evidence of molecular conservation of sleep regulation
Acknowledgments
We thank Chris Blair from the National Aquarium, MD, Monica Medina and Aki Ohdera from Pennsylvania State University, PA, and Wyatt Patry from the Monterey Bay Aquarium, CA for generously supplying Cassiopea medusa and polyps; Dr. John Bedbrook for critical reading of the manuscript. We thank Kiersten Darrow and Michael Schaadt of the Cabrillo Marine Aquarium, CA for input on husbandry. This work was supported by the NIH Director’s New Innovator Award/PECASE (IDP20D017782-01, to V.G.), James S. McDonnell Foundation for Complex Systems Science (220020365, to L.G.), the NIMH under Ruth L. Kirschstein National Research Service Award (F31MH102913, to C.N.B.), the NINDS under Ruth L. Kirschstein National Research Service Award (F31NS100519, to R.D.N.), the National Science Foundation Graduate Research Fellowship (1144469, to M.J.A.), the NIH training grant (T32GM007616, to C.N.B. and R.D.N.). V.G. is a Heritage Principal Investigator supported by the Heritage Medical Research Institute; P.W.S. is an investigator with the HHMI, which supported this research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
R.D.N., C.N.B., and M.J.A. conceived the project. V.G., P.W.S., and L.G. oversaw the project. R.D.N., C.N.B., and M.J.A. performed experiments and data analysis. R.D.N., C.N.B., M.J.A., and T.B. conceptualized, designed, and built experimental setups. C.N.B. and R.D.N. wrote image processing and data analysis scripts with J.S.B.’s oversight. D.A.P. provided input on experimental design. R.D.N., C.N.B., and M.J.A. wrote the paper with input from J.S.B., D.A.P., V.G., P.W.S., and L.G.
References
- 1.Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, Pack AI. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–138. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 2.Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–1837. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- 3.Raizen DM, Zimmerman JE, Maycock MH, Ta UD, You YJ, Sundaram MV, Pack AI. Lethargus is a Caenorhabditis elegans sleep-like state. Nature. 2008;451:569–572. doi: 10.1038/nature06535. [DOI] [PubMed] [Google Scholar]
- 4.Hill AJ, Mansfield R, Lopez JMNG, Raizen DM, Van Buskirk C. Cellular stress induces a protective sleep-like state in C. elegans. Curr Biol. 2014;24:2399–2405. doi: 10.1016/j.cub.2014.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trojanowski NF, Raizen DM. Call it worm sleep. Trends in Neurosciences. 2016;39:54–62. doi: 10.1016/j.tins.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allada R, Siegel JM. Unearthing the phylogenetic roots of sleep. Curr Biol. 2008;18:R670–R679. doi: 10.1016/j.cub.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joiner WJ. Unraveling the evolutionary determinants of sleep. Curr Biol. 2016;26:R1073–R1087. doi: 10.1016/j.cub.2016.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirszenblat L, van Swinderen B. The yin and yang of sleep and attention. Trends in Neurosciences. 2015;38:776–786. doi: 10.1016/j.tins.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunn CW, Giribet G, Edgecombe GD, Hejnol A. Animal phylogeny and its evolutionary implications. Annu Rev Ecol Evol Syst. 2014;45:371–395. [Google Scholar]
- 10.Arendt D, Tosches MA, Marlow H. From nerve net to nerve ring, nerve cord and brain–evolution of the nervous system. Nat Rev Neuro. 2016;17:61–72. doi: 10.1038/nrn.2015.15. [DOI] [PubMed] [Google Scholar]
- 11.Katsuki T, Greenspan RJ. Jellyfish nervous systems. Curr Biol. 2013;23:R592–R594. doi: 10.1016/j.cub.2013.03.057. [DOI] [PubMed] [Google Scholar]
- 12.Erwin DH, Davidson EH. The last common bilaterian ancestor. Development. 2002;129:3021–3032. doi: 10.1242/dev.129.13.3021. [DOI] [PubMed] [Google Scholar]
- 13.Hejnol A, Rentzsch F. Neural nets. Curr Biol. 2015;25:R782–R786. doi: 10.1016/j.cub.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 14.Kelava I, Rentzsch F, Technau U. Evolution of eumetazoan nervous systems: insights from cnidarians. Philos Trans R Soc Lond, B, Biol Sci. 2015;370 doi: 10.1098/rstb.2015.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosch TCG, Klimovich A, Domazet-Lošo T, Gründer S, Holstein TW, Jékely G, Miller DJ, Murillo-Rincon AP, Rentzsch F, Gemma RS, et al. Back to the basics: cnidarians start to fire. Trends in Neurosciences. 2017;40:92–105. doi: 10.1016/j.tins.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grimmelikhuijzen CJ, Westfall JA. The nervous systems of cnidarians. EXS. 1995;72:7–24. doi: 10.1007/978-3-0348-9219-3_2. [DOI] [PubMed] [Google Scholar]
- 17.Satterlie RA. Do jellyfish have central nervous systems? J Exp Biol. 2011;214:1215–1223. doi: 10.1242/jeb.043687. [DOI] [PubMed] [Google Scholar]
- 18.Watanabe H, Fujisawa T, Holstein TW. Cnidarians and the evolutionary origin of the nervous system. Development, Growth & Differentiation. 2009;51:167–183. doi: 10.1111/j.1440-169X.2009.01103.x. [DOI] [PubMed] [Google Scholar]
- 19.Dupre C, Yuste R. Non-overlapping neural networks in Hydra vulgaris. Curr Biol. 2017;27:1–14. doi: 10.1016/j.cub.2017.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimmelikhuijzen CJ, Williamson M, Hansen GN. Neuropeptides in cnidarians. Can J Zool. 2002;80:1690–1702. [Google Scholar]
- 21.Kremien M, Shavit U, Mass T, Genin A. Benefit of pulsation in soft corals. Proc Natl Acad Sci USA. 2013;110:8978–8983. doi: 10.1073/pnas.1301826110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garm A, Bielecki J, Petie R, Nilsson DE. Opposite patterns of diurnal activity in the box jellyfish Tripedalia cystophora and Copula sivickisi. Biol Bull. 2012;222:35–45. doi: 10.1086/BBLv222n1p35. [DOI] [PubMed] [Google Scholar]
- 23.Seymour JE, Carrette TJ, Sutherland PA. Do box jellyfish sleep at night? Med J Aust. 2004;181:707. doi: 10.5694/j.1326-5377.2004.tb06529.x. [DOI] [PubMed] [Google Scholar]
- 24.Campbell SS, Tobler I. Animal sleep: a review of sleep duration across phylogeny. Neurosci Biobehav Rev. 1984;8:269–300. doi: 10.1016/0149-7634(84)90054-x. [DOI] [PubMed] [Google Scholar]
- 25.Jantzen C, Wild C, Rasheed M, El-Zibdah M. Enhanced pore water nutrient fluxes by the upside-down jellyfish Cassiopea sp in a Red Sea coral reef. Mar Ecol Prog Ser. 2010;411:117–125. [Google Scholar]
- 26.Holland BS, Dawson MN, Crow GL, Hofmann DK. Global phylogeography of Cassiopea (Scyphozoa: Rhizostomeae): molecular evidence for cryptic species and multiple invasions of the Hawaiian Islands. Marine Biology. 2004;145:1119–1128. [Google Scholar]
- 27.Santhanakrishnan A, Dollinger M, Hamlet CL, Colin SP, Miller LA. Flow structure and transport characteristics of feeding and exchange currents generated by upside-down Cassiopea jellyfish. J Exp Biol. 2012;215:2369–2381. doi: 10.1242/jeb.053744. [DOI] [PubMed] [Google Scholar]
- 28.Zhdanova IV. Sleep and its regulation in zebrafish. Rev Neurosci. 2011;22:27–36. doi: 10.1515/RNS.2011.005. [DOI] [PubMed] [Google Scholar]
- 29.Borbely AA, Achermann P. Sleep homeostasis and models of sleep regulation. J Biol Rhythms. 1999;14:557–568. doi: 10.1177/074873099129000894. [DOI] [PubMed] [Google Scholar]
- 30.Zimmerman JE, Naidoo N, Raizen DM, Pack AI. Conservation of sleep: insights from non-mammalian model systems. Trends in Neurosciences. 2008;31:371–376. doi: 10.1016/j.tins.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nath RD, Chow ES, Wang H, Schwarz EM, Sternberg PW. C. elegans stress-induced sleep emerges from the collective action of multiple neuropeptides. Curr Biol. 2016;26:2446–2455. doi: 10.1016/j.cub.2016.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peres R, Reitzel AM, Passamaneck Y, Afeche SC, Cipolla-Neto J, Marques AC, Martindale MQ. Developmental and light-entrained expression of melatonin and its relationship to the circadian clock in the sea anemone Nematostella vectensis. EvoDevo. 2014;5:26–18. doi: 10.1186/2041-9139-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhdanova IV, Wang SY, Leclair OU, Danilova NP. Melatonin promotes sleep-like state in zebrafish. Brain Research. 2001;903:263–268. doi: 10.1016/s0006-8993(01)02444-1. [DOI] [PubMed] [Google Scholar]
- 34.Brzezinski A, Vangel MG, Wurtman RJ, Norrie G, Zhdanova I, Ben-Shushan A, Ford I. Effects of exogenous melatonin on sleep: a meta-analysis. Sleep Medicine Reviews. 2005;9:41–50. doi: 10.1016/j.smrv.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 35.Tosches MA, Bucher D, Vopalensky P, Arendt D. Melatonin signaling controls circadian swimming behavior in marine zooplankton. Cell. 2014;159:46–57. doi: 10.1016/j.cell.2014.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gandhi AV, Mosser E, Oikonomou G, Prober DA. Melatonin is required for the circadian regulation of sleep. Neuron. 2015;85:1193–1199. doi: 10.1016/j.neuron.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moriarty PE, Andrews KS, Harvey CJ, Kawase M. Vertical and horizontal movement patterns of scyphozoan jellyfish in a fjord-like estuary. Mar Ecol Prog Ser. 2012;455:1–12. [Google Scholar]
- 38.Stat M, Pochon X, Cowie R, Gates RD. Specificity in communities of Symbiodinium in corals from Johnston Atoll. Mar Ecol Prog Ser. 2009;386:83–96. [Google Scholar]
- 39.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Marine Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 40.Walt S, Colbert SC, Varoquaux G. The NumPy array: a structure for efficient numerical computation. Computing in Science and Eng. 2011;13:22–30. [Google Scholar]
- 41.Hunter JD. Matplotlib: A 2D Graphics Environment. Computing in Science and Eng. 2007;9:90–95. [Google Scholar]
- 42.Oliphant TE. Python for Scientific Computing. Computing in Science and Eng. 2007;9:10–20. [Google Scholar]
- 43.Abrams MJ, Basinger T, Yuan W, Guo CL, Goentoro L. Self-repairing symmetry in jellyfish through mechanically driven reorganization. Proc Natl Acad Sci USA. 2015;112:E3365–E3373. doi: 10.1073/pnas.1502497112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Treweek JB, Chan KY, Flytzanis NC, Yang B, Deverman BE, Greenbaum A, Lignell A, Xiao C, Cai L, Ladinsky MS, et al. Whole-body tissue stabilization and selective extractions via tissue-hydrogel hybrids for high-resolution intact circuit mapping and phenotyping. Nat Protoc. 2015;10:1860–1896. doi: 10.1038/nprot.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Cassiopea pulsing behavior, Related to Figure 1. Two jellyfish pulsing during the day with ruler for scale.
Movie S2. Cassiopea exposed to brief water pulse perturbation, Related to Figure 4. A mechanical stimulation perturbs a single jellyfish.
Data S1. Day versus night pulsing behavior, Related to Figure 2. Pulsing-traces for individual jellyfish during day (yellow) and night (gray) over 300 s. These recordings were taken from the 7th hour of the day or night. Each tab is a different jellyfish (jf1-jf22). The activity of the same jellyfish are presented in Figure 2C,D.


