Abstract
The stiffness and nanotopographical characteristics of the extracellular matrix (ECM) influence numerous developmental, physiological, and pathological processes in vivo. These biophysical cues have therefore been applied to modulate almost all aspects of cell behavior, from cell adhesion and spreading to proliferation and differentiation. Delineation of the biophysical modulation of cell behavior is critical to the rational design of new biomaterials, implants, and medical devices. The effects of stiffness and topographical cues on cell behavior have previously been reviewed, respectively; however, the interwoven effects of stiffness and nanotopographical cues on cell behavior have not been well described, despite similarities in phenotypic manifestations. Herein, we first review the effects of substrate stiffness and nanotopography on cell behavior, and then focus on intracellular transmission of the biophysical signals from integrins to nucleus. Attempts are made to connect extracellular regulation of cell behavior with the biophysical cues. We then discuss the challenges in dissecting the biophysical regulation of cell behavior and in translating the mechanistic understanding of these cues to tissue engineering and regenerative medicine.
Keywords: Extracellular matrix, Stiffness, Nanotopography, Adhesive ligands, Cell behavior
1. Introduction
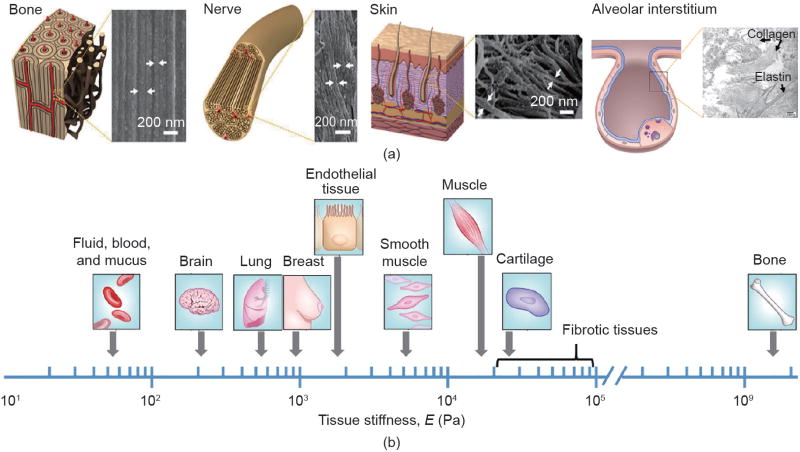
A growing body of literature shows that cell fate can be dictated by the stiffness and topographical characteristics of the extracellular matrix (ECM). The ECM, which is constructed from diverse, nanometer-sized biomacromolecules including collagen, elastin, and fibronectin [1], often displays topography at nanoscales, as shown in Fig. 1(a) [2–8]. For example, collagen fibers, being several microns in diameter, are hierarchically structured from collagen fibrils of 10–300 nm in diameter [9,10]. The lung interstitial matrix displays an interrelated framework of nanoscale fibrous collagen and elastin proteins [8,11]. Depending on the composition of the ECM as well as on interstitial fluids [12], the ECM exhibits various degrees of stiffness, as shown in Fig. 1(b) [13–15]. The biophysical (stiffness and nanotopographical) cues, in concert with the spatiotemporally arranged biochemical and biomechanical cues, regulate cell phenotype and function.
Fig. 1.
Biophysical characteristics of human tissues. (a) Nanoscale structures displayed in various tissues. The arrows indicate various nanostructures. (Reproduced with permission from Ref. [6] for the graphical illustrations and scanning electron microscope (SEM) micrographs of bone, nerve, and skin. The graphical illustration and SEM micrographs of the alveolar interstitium are reproduced from Refs. [7] and [8], respectively) (b) Stiffness of human tissues. The fibrotic tissues become stiffer than those in normal conditions. (Reproduced with permission from Ref. [15])
The stiffness and nanotopographical characteristics of the ECM influence numerous developmental, physiological, and pathological processes in vivo [16–20]. For example, tissue stiffness can be altered by the disease state. The stiffness of mammary tissue increases from ~1 kPa in its normal condition to ~4 kPa during breast cancer [21]. Lung stiffness is lower in emphysema [22], but higher in fibrotic tissues than in the normal condition [23,24]. Moreover, fibroblasts respond to increases in matrix stiffness with promoted proliferation and collagen synthesis; the induced ECM stiffening can further promote, amplify, and perpetuate fibrosis via a positive feedback loop [24,25].
Biophysical cues have therefore been applied to modulate almost all aspects of cell behavior [26]. Since the first report in 1997 [27], emerging compelling evidence has shown that substrate stiffness plays important roles in cell modulation and many biological processes [27–32]. For example, C2C12 mouse myoblasts exhibit definitive actomyosin striations only on polyacrylamide (PAAm) gels with a stiffness that is typical of normal muscle, but not on softer gel or stiffer glass substrate [33]. Furthermore, the neurogenic, myogenic, and osteogenic differentiation of human mesenchymal stem cells (hMSCs) can be facilitated by PAAm gels with stiffnesses matching those of brain, muscle, and collagenous bone, respectively [28]. Meanwhile, a large body of literature underscores the phenomenon that cellular responses are highly sensitive to nanotopography [34–39]. In addition to having a pronounced influence on cell morphology, nanotopographical cues could regulate cell proliferation and facilitate stem cell differentiation into certain lineages such as neuron [35,40,41], muscle [42], and bone [36,37].
Many excellent review articles discuss cellular responses to substrate stiffness [14,43,44] or topography [45–50]. However, despite similarities in phenotypic manifestations, the interwoven effects of stiffness and nanotopographical cues on cell behavior have not been well described [51]. Herein, we first review the effects of substrate stiffness and nanotopography on cell behavior, and then focus on intracellular transmission of the biophysical signals from integrins to nucleus. Attempts are made to connect extracellular regulation of cell behavior with the biophysical cues. We then discuss the challenges in dissecting the biophysical regulation of cell behavior and in translating the mechanistic understanding of these cues to tissue engineering and regenerative medicine.
2. Biophysical regulation of cell phenotype and function
2.1. Stiffness cues
A broad spectrum of materials has been adopted as substrates/matrices for cellular studies. These materials range from very hard metals such as titanium oxide (TiO2; Young’s modulus E ≈ 150 GPa) [52], to hard glass (65 GPa) [53], to thermoplastic polymers such as polystyrene (PS; 2.3 GPa) [54] and poly(lactic-co-glycolic acid) (PLGA; 1.31 GPa for PLGA 50/50) [55], to elastomeric polymers such as polydimethylsiloxane (PDMS; 3.4 MPa) [56], and to soft hydrogels (from several pascals to several kilopascals), as shown in Fig. 2(a). In the literature, different terms such as elasticity, stiffness, rigidity, and shear modulus have been used to characterize the mechanical property of substrates. Elasticity is an intensive property of the material, while stiffness is an extensive property, depending on the material and the shape and boundary conditions. Throughout this review, the value in the brackets gives the Young’s modulus of the substrate, unless otherwise specified.
Fig. 2.
Substrate stiffness affects cell differentiation. (a) Cell culture substrates with a variety of stiffnesses. (b) Relationship between stem cell differentiation and substrate stiffness; each symbol represents one cell type. PEG: poly(ethylene glycol); PCL: polycaprolactone; rMSC: rat mesenchymal stem cell; ESC: embryonic stem cell; SMC: smooth muscle cell; NSC: neural stem cell; rNSC: rat neural stem cell.
2.1.1. Stiffness effects
With an increase in substrate stiffness, cells usually exhibit enhanced cell adhesion [57–60], enlarged cell spreading with defined actin organization [60–67], increased cellular contractility [60–68], decreased migration speed [69,70], and promoted proliferation [57,61,67,71,72]. For example, when hMSCs adhere onto collagen I-modified PAAm gels, paxillin-labeled adhesions change from undetectable diffuse focal complexes on soft gels (1 kPa), to punctate adhesions on gels with intermediate stiffness (11 kPa), to long, thin, and more stable focal adhesions on the stiffest gels (34 kPa) [28]. The expression of the focal adhesion protein vinculin in MC3T3-E1 osteoblasts on alginate gels increases 1.5-fold as the gel stiffness increases from 20 kPa to 110 kPa [57]. It has also been shown that NIH 3T3 fibroblasts on the stiffer collagen I-coated PAAm gels (7.69 kPa) are more dispersed and have better attachment, with > 80% of cells remaining after a centrifugation assay, as compared with the softer gels (2.68 kPa), which only have about 30% of cells remaining [58].
Although many studies show monotonic dependence of cell behavior on substrate stiffness, biphasic relations between cell adhesion [73], migration [59,74–76], and proliferation [77–79] and substrate stiffness have also been observed. On the one hand, when primary adult human dermal fibroblasts are grown on poly(ethylene glycol) (PEG) hydrogels, the average cell migration speed decreases significantly from 0.81 µm·min−1 on soft gels (95 Pa) to 0.38 µm·min−1 on stiff gels (4.3 kPa) [70]. In addition, when the Young’s modulus of PAAm gels increases from 4.7 kPa to 14 kPa, NIH 3T3 fibroblasts show ~2- and ~4-fold higher cell proliferation after 24 h and 48 h, respectively [61]. On the other hand, the migration speed of MC3T3-E1 cells on collagen I-coated PAAm gels monotonically increases with increasing stiffness on low collagen densities, while at higher collagen densities, the cells exhibit a biphasic dependence of migration speed on substrate stiffness and reach a maximum on 21.6 kPa gels [59]. A peak proliferation rate is observed on the gels of an intermediate stiffness for rat neural stem cells cultured on PEG substrates (10 Pa–10 kPa) [78]. In addition, the proliferation of murine stem cells on alginate hydrogels does not show any dependence on gel stiffness [72].
Stem cell differentiation is also profoundly affected by substrate stiffness. As previously mentioned, hMSCs exhibit up-regulated expression of neurogenic, myogenic, and osteogenic biomarkers on PAAm gels with stiffnesses matching those of brain (0.1–1 kPa), muscle (8–17 kPa), and collagenous bone (25–40 kPa), respectively [28]. Adult neural stem cells also exhibit peak levels of neurogenic biomarker β-tubulin III on substrates having the approximate stiffness of brain tissue. In addition, softer PAAm gels (100–500 Pa) promote neuronal differentiation, whereas stiffer substrates (1–10 kPa) lead to glial differentiation [78]. Because of the variation in cell sources, substrate preparation, and differentiation protocols, the optimal substrate stiffness is not the same across different studies. For example, another study showed that the myogenic and osteogenic differentiation peaks on PAAm gels with stiffnesses of 25 kPa and 80 kPa, respectively [67], are slightly different from those in the previous report [28]. Nonetheless, general trends have been observed, in that neural differentiation prefers soft substrates whereas osteogenesis favors stiff substrates, while myogenesis falls in the intermediate range (Fig. 2(b); see Supplementary Information for references on each data point). The remarkable consistency over a large number of studies involving diverse cell sources highlights the important role of mechanosensing in stem/progenitor cell differentiation.
Stiffness-dependent cell behavior has seen more applications. Substrate stiffness impacts the cellular uptake of nanoparticles: Soft PAAm gels (1.61 kPa) lower cell membrane tension, favoring bovine aortic endothelial cells uptaking PS nanoparticles compared with stiffer gels (3.81 kPa and 5.71 kPa) [80]. More intriguingly, recent studies have revealed that the cells can retain stiffness information from the past culture environment and that the previous mechanical history or mechanical dosing influences future cell fate decisions [32,81–84]. For example, skeletal muscle stem cells lose their in vivo regenerative potential rapidly on stiff plastic dishes, but sustain their self-renewal and regenerative capacity on soft hydrogels of physiologically relevant stiffness [32]. It is further demonstrated that hMSCs are increasingly differentiated toward osteogenesis after long-term culture on stiff PS, but remain plastic and can differentiate toward adipogenic and osteogenic lineages without previous mechanical dosing on a stiff PS surface [82].
2.1.2. Challenges in delineating stiffness regulation
Cellular responses to substrate stiffness cues are not always consistent, and are sometimes contradictory. One of the important reasons is that tuning the stiffness of hydrogels, the extensively used materials in stiffness studies, may affect the surface chemistry, backbone flexibility, and binding properties of adhesive ligands of the gel, in addition to its bulk stiffness and porosity [85–87]. It has been shown that hMSCs respond to the variation in stiffness of PAAm gels but not to that of PDMS; thus, it is speculated that it is the alteration of anchoring points of attached collagen I on the gels, rather than substrate stiffness per se, that regulates the cell behavior [85]. It is further suggested that hMSC differentiation is regulated by the fibronectin strain, which is not affected by the stiffness variation of smooth PDMS but is affected by that of hydrogels [88]. On the contrary, a recent study shows that hMSC differentiation is not affected by protein-substrate linker density up to 50 folds; thus, it is argued that substrate stiffness regulates stem cell differentiation independently of protein tethering and porosity [89]. Another important issue is that cells can sense the stiffness of underlying hydrogels, and even the stiffness of the supporting substrate when the gel is thin [90–92]. It is estimated that cells can sense the “hidden” substrate at a depth of approximately 5 µm [93], and even deform a substrate to a depth of 20 µm [94]. Collectively, the complexity of hydrogel structures in both lateral and vertical dimensions makes it challenging to dissect the role of substrate stiffness in cell regulation. Model systems in which the stiffness cues can be investigated independently of other environmental variables are highly desirable.
2.2. Nanotopographical cues
Cells can perceive variations of a few nanometers on the surface topography and actively respond to the nanotopography [38]. Cells exhibit diverse behavior on a wide variety of nanotopographies. Although nanoscale is defined as a length scale of 1–100 nm in the physical realm [95], the length scale of nanotopographies discussed here is extended beyond 100 nm and upward to the submicrometer range because cells can interact with the ECM exhibiting features with size up to several micrometers.
2.2.1. Nanotopographical effects
Shape (e.g., pillars, pits, and gratings), dimension (feature size, spacing, and height), and arrangement of nanoscale features all have pronounced effects on cell behavior, from cell adhesion and spreading to proliferation and differentiation, which is cell-type specific. Mesenchymal stem cells (MSCs) display different cell adhesion, proliferation, and differentiation on TiO2 nanotubes of 15–100 nm in diameter compared with a flat TiO2 surface [37,96]. On small (~30 nm diameter) TiO2 nanotubes, hMSCs exhibit enhanced adhesion. A ~10-fold increase in cell elongation occurs on larger (70–100 nm) nanotubes compared with the flat control, thus inducing cytoskeletal stress and biasing the osteogenic differentiation [37]. Other than feature size, nanotopography height can effectively regulate cell behavior [97]. On randomly distributed nanoislands produced by demixing, a variety of cell types exhibit more pronounced focal adhesions and actin stress fibers, highly spread morphology, and larger cell area on the shallow (11–13 nm height) nanoislands compared with the flat control surface [98–101]. When the height increases to ~90 nm, some cells—such as human fetal osteoblastic cells [98], human bone marrow cells [102], and human fibroblasts [103]—display a reduced cell-spreading morphology, with diffuse actin and fewer stress fibers. In contrast, human endothelial cells display larger lamellae and have increased numbers of stress fibers on 95 nm nanoislands [100]. Cell-type specific responses to nanotopography have also been observed in other systems [104,105]. For example, human embryonic stem cells (hESCs) exhibit enhanced proliferation and long-term self-renewal on smooth surfaces but tend to differentiate on nanorough glass surfaces, whereas nanorough surfaces promote the adhesion of NIH 3T3 fibroblasts compared with smooth surfaces [105].
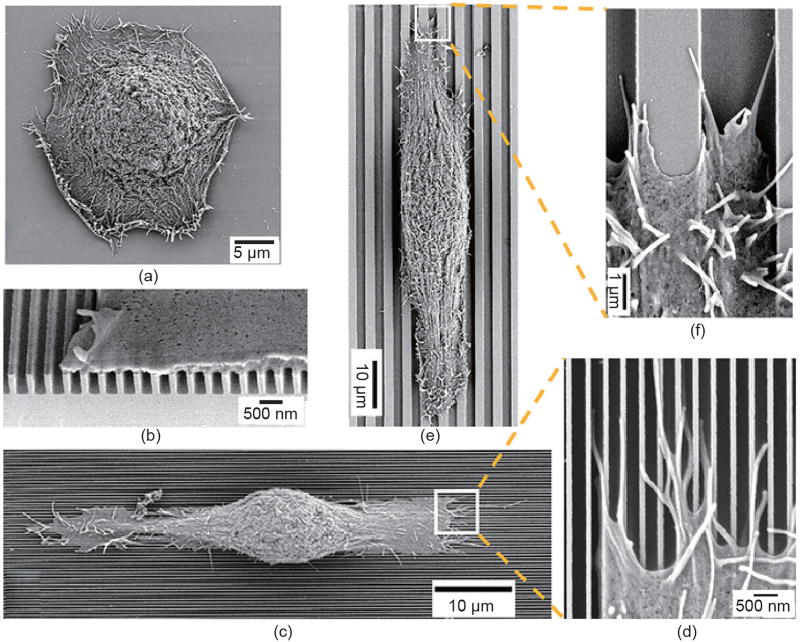
In contrast to isotropic nanotopographies, anisotropic nanotopographies such as nanogratings may result in smaller cell sizes and lower proliferation rates—even apoptosis—while promoting cell alignment, elongation, and migration [35,101,106–111]. Compared with random distribution on a smooth control, the focal adhesions and stress fibers of human corneal epithelial cells align along silicon nanogratings that are 70–1900 nm in ridge width, 400–4000 nm in pitch, and 150 nm and 600 nm in depth (Fig. 3) [112]. The focal adhesion size increases with the ridge width up to 400 nm and remains constant for ridge widths greater than 650 nm. Compared with the smooth control surfaces, the cells display smaller average cell areas on all nanogratings, yet significantly elongated morphology on all gratings that are 600 nm deep [112]. Nanograting-induced decrease in cell area results in lower proliferation rates. On PDMS nanogratings of 350 nm in width, 700 nm in pitch, and 350 nm in depth, hMSCs display elongated cytoskeletons and nuclei along the nanograting direction, and a significantly lower cell proliferation rate of (26.9 ± 3.1)% compared with (35.7 ± 7.6)% on smooth surfaces [35]. In addition to the observation that various human cell types exhibit enhanced motility on nanotopographies compared with smooth surfaces [113–117], directional cell migration can be realized on anisotropic nanotopographies, as the cell extends and retracts lamellipodia preferentially along the long axis of anisotropic nanotopographies, compared with random cell migration on isotropic nanotopographies [118]. It is suggested that directional cell migration can be regulated by the polarization of the microtubule organizing centers [109], and that migration speed is dependent on the width [48] and depth [119] of underlying nanogratings. Note that unidirectional cell migration can be achieved by using nano/microtopographical gradients such as sawteeth geometry on scales smaller than that of a single cell but comparable to those of collagen fibers [120].
Fig. 3.
SEM micrographs of human corneal epithelial cells cultured on (a) a smooth silicon oxide substrate and (b–f) nanogratings. On nanogratings that are 70 nm in width, 400 nm in pitch, and 600 nm in depth (b–d), the cell adheres to the top of the nanogratings (b), and aligns along the nanograting direction (c), with filopodia extending along the top of ridges and bottom of grooves (d). In contrast, the cell elongates along nanogratings that are 1900 nm in width, 4000 nm in pitch, and 600 nm in depth (e), with lamellipodia reaching the bottom of the grooves (f). (Reproduced with permission from Ref. [112])
Anisotropic nanotopography is crucial to neuron growth and differentiation, in addition to facilitating myogenic differentiation [42,121]. The neurites of dorsal root ganglion neurons elongate and exhibit little to no branching on aligned nanofibers; however, they have noticeably more branching on random nanofibers, which is detrimental to nerve regeneration. Furthermore, the neurites exhibit bipolar extension parallel to nanofibers that are 500 nm in diameter, in an identical manner to the organization in in vivo neurite outgrowth [122]. Interestingly, neural stem cells elongate and their neurites outgrow along with the aligned fibers independent of their diameter; however, nanofibers that are 250 nm in diameter promote cell differentiation compared with microfibers (1.25 µm) [123]. The influence of nanogratings on neuronal differentiation is significant. On the aforementioned 350 nm PDMS nanogratings, hMSCs exhibit significant up-regulation of the expression of neuronal markers such as β-tubulin III and microtubule-associated protein 2 (MAP2), compared with microgratings and flat controls. Although the combination of nanotopographical cues with biochemical cues such as retinoic acid (RA) further enhances the up-regulation of the neuronal markers, nanogratings demonstrate a stronger effect than RA alone on a smooth surface [35]. Even in the absence of RA, hESCs grown on equally spaced gratings that are 350 nm in width and 500 nm in height are differentiated into neuronal lineage, but not into glial cells [40]. Interestingly, anisotropic topographies are shown to enhance neuronal differentiation, while isotropic topographies enhance glial differentiation under the same conditions [41]. While cell polarity is critical to cell regulation and organ development, and loss of cell polarity is associated with many human diseases [124,125], anisotropic nanotopographies provide a powerful tool to establish and maintain cell polarity.
Intriguing findings show that the arrangement of nanoscale features can have a profound influence on cell phenotype and function. On arrays of nanopits (120 nm in diameter, 300 nm center-to-center spacing, and 100 nm in depth) in three different arrangements—square, hexagonal, and near-square (i.e., a square pattern with ±50 nm disorder)—primary human osteoblasts display a mean fibrillar adhesion length of approximately 11 µm on near-square nanopits, which is significantly larger than those on hexagonal and square nanopits (~6.6 µm) and on the flat control (~7.2 µm) [126]. Moreover, the near-square nanopits alone stimulate osteogenic differentiation of hMSCs at levels similar to the differentiation induced by osteogenic supplements, whereas highly ordered or completely randomly positioned nanopits and the flat control only induce limited osteogenic differentiation [36]. On the other hand, highly ordered, square nanopits permit the retention of multipotency of hMSCs for up to 8 weeks [39].
Being a potent regulator of cell behavior, topography can alter the cell-substrate interactions in order to strengthen or weaken cell adhesion, consequently affecting cellular processes. Nanotopography has therefore been utilized in various applications, from capturing circulating tumor cells (CTCs) [127–131] and optimizing the fibroblast-to-neuron reprogramming process [132] to modulating the fibrogenic responses of fibroblasts to nanoparticles [133]. Inspired by the nanostructured surface (e.g., microvilli, microridges, and cilia) of tumor cells [134] and by enhanced tumor cell-nanotopography interactions [135], a variety of nanotopographies such as nanowires [127,128], nanotubes [129], and nanorough surfaces [130] have been fabricated to improve the sensitivity and efficiency of CTC capturing. Compared with isotropic discrete nanopillars, nanogratings are shown to favor tumor cell adhesion, leading to more effective tumor cell capturing [131]. On the other hand, nanocrater pitch is designed to disrupt the formation of mature focal adhesions, thus favoring NIH 3T3 fibroblast migration toward higher-pitched regions [136]. Because of weakened cell-substrate interaction, the bovine corneal endothelial cell monolayer on nanopillars demonstrates a higher density of microvilli than the flat control, as well as enhanced formation and function that are similar to those of the native corneal endothelium [137].
2.2.2. Cell sensing of nanotopography
Discrepancies in the literature confound current understanding of the nanotopographical regulation of cell behavior. For example, one group shows that nanogratings significantly increase the expression of osteogenic markers of hMSCs [138,139]. In contrast, another group reports that nanogratings do not strongly influence the osteogenic phenotype of hMSCs [140]. Moreover, some groups conclude that biochemical cues exert a stronger influence on cell behavior as compared with nanotopography [107,141,142]. For example, on a silicon substrate with a pore-size gradient ranging from 19 nm to 920 nm and an orthogonal cyclic arginine-glycine-aspartic acid (Arg-Gly-Asp or RGD) ligand gradient, rat MSCs respond to both nanotopographical and biochemical cues; however, they respond more strongly to the change in RGD density than to the change in pore size [141]. It is also shown that MC3T3-E1 cells predominantly align along the nanogratings (100 nm in width, spacing, and depth) that are uniformly coated with fibronectin. However, when the nanogratings are orthogonally contact-printed with fibronectin lanes that are 10 µm wide and separated by non-adhesive lanes, the cells elongate along the fibronectin lanes rather than along the nanogratings [107]. It is unclear whether the aforementioned discrepancy results from the difference in nanogratings per se or from the nanograting-altered ligand presentation.
It is generally thought that nanotopography can increase surface area, thus enhancing cell adhesion. However, the apparent surface that cells can perceive is determined by the shape and dimension of nanoscale features. Whether the cell membrane will bridge over the top or reach the bottom of nanofeatures is dependent on the stiffness of the cell membrane at nanoscales [143]. On equally spaced nanogratings that are 500 nm in height, it is shown that neonatal rat ventricular myocytes extend toward but do not reach the bottom of gratings that are 400 nm wide; this action is accompanied by limited cell-substrate adhesion. In contrast, the cells can completely fill gratings that are 800 nm wide, and show increased cell-substrate adhesion [144].
When the nanotopography reduces the apparent surface area that cells can perceive, the nanotopography restricts focal adhesions, thus weakening cell adhesion and facilitating cell migration [102]. On nanopillars that are 700 nm in diameter and 1.2–5.6 µm in pillar-to-pillar distances, the hMSCs are stretched and favor osteogenesis on the nanopillars with longer pillar-to-pillar distance (5.6 µm), but are rounded and favor adipogenesis on the nanopillars with shorter distances (1.2 µm) [145]. The relation between cell spreading and spacing can be biphasic. For example, on nanodot arrays with diameters ranging between 10 nm and 200 nm and spacings between 20 nm and 200 nm, cardiomyoblasts exhibit maximal surface area and proliferation on 50 nm nanodot arrays [146]. In addition, among nanodot arrays with diameters of 150 nm, 400 nm, and 600 nm, osteogenic differentiation of hMSCs peaks on the 400 nm dot array [147]. It is thus speculated that effective nanotopographical cell modulation is, first, determined by whether the nanotopography increases the substrate surface area that cells can perceive and, second, determined by how significant the increase in apparent surface area is. Small spacing can limit the apparent surface area, while large spacing may alleviate the increase in apparent surface area. The aspect ratio of height to spacing of nanotopography is thus suggested to provide more comprehensive characterization of nanotopography than a single dimensional parameter [41,50,148–151]. It has been shown that, on gratings of 1–10 µm in width and spacing and 0.35–10 µm in height, hMSCs are mostly elongated on the gratings with an aspect ratio of 1.04, whereas cell elongation is not significant on gratings with the smallest width or the greatest height [152].
While nanotopography provides a potent regulator of cell growth and differentiation by modulating the cell shape [153,154], the underlying mechanisms remain unclear. Will nanotopography affect cell behavior via contact guidance when nanotopography does not affect the presentation of adhesive ligands? Will nanotopography affect cell behavior when nanotopography affects the cell sensing of the substrate surface or adhesive ligands?
2.3. Interwoven substrate nanotopographical and stiffness cues
Cells constantly exert force on the ECM, remodel the ECM, and affect physiological and pathological processes [155–157]. When a flat, pliant substrate is used, cells may detect a difference in substrate stiffness and respond to the stiffness cues [158]. Furthermore, when topography is fabricated on stiff substrates that cells cannot deform, cells will only respond to topographical cues, whereas cells will sense and respond to both topographical and stiffness cues if cells can deform the topography. Note that the substrate stiffness sensing is also cell-type specific. For example, bovine pulmonary artery smooth muscle cells behave similarly on both poly(methyl methacrylate) (PMMA) and PDMS nanogratings that are 350 nm in width, spacing, and depth [109]. However, hMSCs are observed to deform PDMS nanogratings but not PS nanogratings, which have a similar stiffness to the PMMA nanogratings (E = 2.3 GPa and 3.7 GPa for PS and PMMA, respectively) [54], as shown in Fig. 4(a, b) [159]. In addition, on PS substrates, hMSCs exhibit lower mechanical properties when attached to the 350 nm gratings as compared with flat controls. On the other hand, hMSCs cultured on PDMS substrates show lower mechanical properties than those on PS substrates, regardless of topography [160]. Evidently, when the building material for the topography is soft enough for the cell to deform the substrate, topographical and stiffness cues are interwoven to exert influence on cell phenotypes and functions [161]. It is worth mentioning that the surface of thermoplastic polymers, usually at a length scale of less than 100 nm, has different properties from the bulk [162–165]. Because nanofeatures make up a significant portion of surface areas, thus providing different mechanical properties, the nanotopography may provide cells with stiffness in addition to nanotopographical cues.
Fig. 4.
Interwoven substrate topographical and stiffness effects on cells. (a, b) SEM micrographs of hMSCs on (a) stiff PS and (b) pliant PDMS nanogratings. (c–e) SEM micrographs of hMSCs on PDMS micropillars with heights of (c) 0.97 µm, (d) 6.1 µm, and (e) 12.9 µm. On micropillars that are 0.97 µm in height, hMSCs are well spread in (c), but they display a rounded morphology with prominent microvilli on 12.9 µm pillars in (e). (f) Immunofluorescent images of Chinese hamster ovary (CHO) cells grown on nanogratings with different stiffnesses. Cells are immunostained for actin (red), vinculin (green), and nuclear material (blue). (Parts (a) and (b) are reproduced with permission from Ref. [159], parts (c–e) are reproduced with permission from Ref. [166], and part (f) is reproduced with permission from Ref. [167])
Recently developed microscale PDMS pillar arrays provide convincing evidence regarding cellular responses to combined topographical and stiffness cues [166,168–175]. The spring constant of the pillar is proportional to the fourth power of the diameter and inversely proportional to the cube of the height [166]. The dimensions of the pillars have been designed to generate stiffness varying from about 1 kPa to 1.2 MPa, thus affecting focal adhesions, cell morphology, contractility, and differentiation [174,176]. For example, hMSCs demonstrate different cell spreading on micropillar arrays of various stiffnesses, as shown in Fig. 4(c–e) [166], which biases hMSC differentiation: Stiff arrays favor osteogenesis, whereas soft ones promote adipogenesis [166]. Cellular studies using an anisotropic micropillar array further highlight the importance of substrate stiffness cues. On anisotropic PDMS microarrays with an oval cross-section (major axis/minor axis: 0.95 µm/0.55 µm, leading to the pillars being about three times stiffer along their major axis than along their minor axis), epithelial cells align and migrate preferentially along the long axis direction or the stiffest direction [173]. The preferential orientation of focal adhesions and actin stress fibers, cell migration, and tissue growth along the stiffest direction of such substrates is correlated to a greater traction force concentrated at the edges of cellular assemblies [44]. In comparison, no preferential orientation of cell alignment or assemblies is observed on a cylindrical pillar array [170]. Moreover, on polyurethane nanogratings, which are 800 nm in width, spacing, and depth, and which have different Young’s moduluses from 1.8 MPa to 1.1 GPa, Chinese hamster ovary (CHO) cells exhibit increasing cell spreading and elongation, and cellular and nuclear areas with increasing substrate stiffness, as shown in Fig. 4(f) [167].
While the roles of substrate stiffness and nanotopographical cues in cell regulation are elusive, the interweaving of the biophysical cues escalates the complexity. To facilitate the mechanical understanding of the biophysical regulation, we next discuss some common elements in intracellular and extracellular transduction of the biophysical signals in terms of cell regulation.
3. Intracellular transduction of biophysical signals
Biophysical signals can be transmitted from integrins, through focal adhesions and the actin cytoskeleton to the nucleus, and regulate cell phenotype and function. We therefore focus on how the biophysical cues affect focal adhesions, the cytoskeleton, and the nucleus.
Before discussing how biophysical cues affect cells, we will describe how cells sense and respond to the substrate. The first step in cellular response to a substrate is to form focal adhesions via the binding and clustering of integrins onto the adhesive ligands on the substrate. As illustrated in Fig. 5, heterodimeric integrin receptors, containing one α- and one β-subunit, bind to the RGD peptide of ECM proteins with their extracellular domain and link to cytoskeletal adaptor proteins with their cytoplasmic tail, subsequently recruiting scaffolding proteins that connect the integrins to the actin cytoskeleton [177]. The earliest forms of integrin-mediated contacts are focal complexes. These small (~500 nm) but highly dynamic focal complexes are located at the leading edge of lamellipodia and membrane protrusions [178]. When the lamellipodia retract or stop protruding, focal contacts are replaced by focal adhesions, and cytoplasmic anchor proteins including paxillin, vinculin, and talin are recruited to the adhesion sites [179]. Maturation of nascent focal complexes to stable streak-like focal adhesions and fibrillar adhesions is induced by cytoskeletal tension driven by cross-bridging interactions of actin and myosin filaments (actomyosin) [180]. The highly anisotropic growth of focal adhesions is in the direction of the force exerted by the cytoskeleton [181]. Downstream signaling of proteins in the Rho family of small GTPases subsequently occurs [43,182], regulating nanoscale sensing (Cdc42), stress fiber formation (RhoA), and cell spreading (Rac) [183]. These processes then control the elongation and contraction of filamentous actin fibers through proteins such as myosin [184]. Increase in RhoA activity decreases the activities of Cdc42 and Rac, driving the formation of focal adhesions and actin stress fibers [178]. Substrate stiffness and nanotopographical cues can mediate the size and distribution of focal adhesions and, subsequently, cytoskeletal organization and tension, which regulate cell morphology and, ultimately, cell function.
Fig. 5.
Transmission of biophysical signals from integrin through focal adhesions and the cytoskeleton to the nucleus. ARP: actin-related protein; FAK: focal adhesion kinase; ROCK: Rho-associated protein kinase; TAZ: transcriptional co-activator with PDZ-binding motif; VASP: vasodilator-stimulated phosphoprotein; YAP: yes-associated protein.
A recent study indicates that nascent focal complexes, which have a smallest size of 0.19 µm2 [185], are critical to mechanosensing [186]. In line with focal complexes, stable integrin-fibronectin clusters are found to be assembled above an area threshold (0.11 µm2); below the threshold, no stable integrin-fibronectin clusters are assembled or appreciable adhesive forces are generated [187]. The focal adhesion size is not a predictor of the local tension exerted at the adhesion. It has been shown that the force exerted at the focal adhesions continues to increase while the elongated focal adhesion protein paxillin remains within 8 µm of the cell periphery without further size change [188]. As a primary regulator of focal adhesion signaling, focal adhesion kinase (FAK) regulates cell proliferation [189] and differentiation [190,191], and its activation increases upon mechanical strain [192]. By means of FAK and the Src-mediated phosphorylation of paxillin, vinculin can be recruited to focal adhesions through myosin-dependent tension, and will further stabilize adhesions [193]. Indeed, tyrosine phosphorylation and dephosphorylation of FAK play a key role in cellular responses to substrate stiffness [194] and nanotopographical cues [160]. On a compliant ECM, FAK signaling is suppressed and intracellular tension is decreased [195]. With an increase in substrate stiffness, the expression of the mature focal adhesion protein zyxin is up-regulated [56]. It has also been observed that FAK phosphorylation increases on equally spaced nanogratings that are 250 nm [196] and 500 nm [160] in width. The increased expression of phosphorylated FAK (pFAK) facilitates the neuronal differentiation of hMSCs, suggesting that the phosphorylation of FAK may act as a signal transducer between integrins and the cytoskeleton in order to relay nanotopographical stimuli to the nucleus via intracellular contractility [196]. In addition, the expression of zyxin is down-regulated on 350 nm nanogratings, correlating with smaller ((3.2 ± 0.26) µm2 versus (5.3 ± 0.55) µm2 on flat controls) and more dynamic focal adhesions, indicating that the traction force in focal adhesions on the nanogratings is decreased. As a result, hMSCs migrate along the nanograting direction at a speed of 15.6 µm·h−1, which is significantly faster than their speed of 8.3 µm·h−1 on a flat surface [197].
The assembly of focal adhesions depends on, and can be regulated by, intracellular tension through the actin cytoskeleton [198], and the members of the small Rho family of GTPases are master regulators of actin cytoskeleton remodeling [199]. Activating Rho and its downstream effector Rho-associated protein kinase (ROCK), and hence inhibiting myosin-light-chain phosphatase, promotes the contraction of actin stress fibers [200]. Through ROCK-dependent contractility, the actin cytoskeleton plays a dominant role in mediating cell shape, which is a proven regulator of cell growth and differentiation [201–203]. Cells that are restricted on micropatterned proteins have been shown to switch from growth to apoptosis when the micropattern size decreases [153]. Well-spread and flattened hMSCs favor osteogenesis, while unspread, round cells undergo adipogenesis [57,59,203]. Investigation of shape-dependent differentiation of hMSCs indicates that focal adhesions and myosin-generated intracellular tension during differentiation play crucial roles in stem cell lineage commitment [204]. Well-spread, polarized shapes are associated with high RhoA/ROCK activity, while cells with small, rounded shapes have low RhoA/ROCK activity [154]. Pharmacological drug studies further confirm that increasing the intracellular tension drives the majority of hMSCs toward osteoblasts despite the variation in shape; conversely, inhibiting ROCK activity biases adipogenesis [204]. Moreover, hMSCs undergoing osteogenic differentiation in osteogenic medium demonstrate higher intracellular tension than the non-differentiating cells, whereas hMSCs that do not differentiate into adipocytes in adipogenic medium are more contractile than the cells undergoing adipogenesis or the cells maintained in the growth medium [166]. Increasing substrate stiffness promotes actin polymerization and actomyosin force generation, leading to increased intracellular tension [205–207] and Rho activity, which is attenuated by decreasing substrate stiffness [21,208,209]. For example, a stiff substrate increases the activation of RhoA and Cdc42 and thus inhibits the neurogenesis of neural stem cells, while inhibition of RhoA/ROCK signaling modestly increases neuronal differentiation. Inhibition of RhoA/ROCK signaling also blocks the osteogenesis of hMSCs on stiff substrates [210]. In the case of nanotopography, the dimension (height in particular) and shape of nanotopography affect intracellular tension. When examined on a variety of nanotopographies, human lung fibroblasts show a significantly stiffer cytoskeleton on shallow (150 nm height) nanotopographies than on their 560 nm counterparts. The nanogratings also increase cytoskeletal stiffness compared with nanopillars featuring the same size and height and similar spacing. The stiffer cytoskeleton is associated with increased synthesis of collagen I [211]. Nanogratings are also found to induce high actomyosin contractility, which is crucial for the neural differentiation of hESCs [212].
The molecular connections between focal adhesions, the cytoskeleton, and the nucleus are associated with cellular and nuclear structure [197,213,214], enabling biophysical regulation of cell behavior [215,216]. For example, chondrogenesis of murine MSCs requires a rounded cell shape, and a more rounded nuclear shape is shown to be associated with the greatest expression of chondrogenic biomarkers in MSCs, through the comparison of cellular and nuclear shapes [217]. The plasticity of nuclei has also been shown to be strongly linked to the lineage commitment of stem/progenitor cells [218]. Nuclear deformation, regulated by cell shape through not only the content but also the organization of the actin cytoskeleton [219], can result in conformational adaptation in chromatin structure and organization, which affects transcriptional regulation [220], gene expression, and protein synthesis [216,221], eventually leading to changes in proliferation, differentiation, or cell death [159,218]. Increasing the spreading area of circular cells from 300 µm2 to 2500 µm2 results in a 36% increase in nuclear volume of cells in the G1 phase, a 50-fold cell stiffening, and a 10-fold rise in proliferation rates [222]. It is envisioned that changes in substrate stiffness and/or nanotopographical configuration can alter the size and distribution of focal adhesions and the cytoskeleton, leading to nuclear deformation and changes in cell phenotype and function [223]. On equally spaced PDMS nanogratings that are 350 nm in width, hMSCs exhibit preferential nuclear (62% nuclei) alignment along the nanograting direction and more elongated nuclei (elongation aspect ratio: 1–5) compared with a random nuclear orientation and an elongation ratio of 1–3 on the flat control. The average nuclear area decreases to (145.1 ± 4.1) µm2 on the nanogratings, from (194.8 ± 4.8) µm2 on the flat control [224]. Compared with the flat control, the nanogratings also significantly down-regulate the expression of A-type lamin nuclear protein and retinoblastoma protein in hMSCs, thus decreasing cell proliferation and changing the differentiation potential [213].
The nuclear factors yes-associated protein (YAP) and transcriptional co-activator with PDZ-binding motif (TAZ) have been revealed to play important roles in developmental and pathological processes and to mediate cellular mechanosensing [82,225–229]. For example, YAP is excluded from the nucleus in the pluripotent cells of the inner cell mass in the early mouse embryo [230]. Knockdown of YAP in mouse ESCs leads to loss of pluripotency, whereas ectopic expression of YAP prevents ESC differentiation [231]. In addition, YAP and TAZ are prominently expressed in fibrotic but not normal lung tissue [229]. Transfer of the fibroblasts overexpressing YAP and TAZ in mice results in profound ECM remodeling and fibrosis in the lung [229]. YAP/TAZ intracellular localization and activity are primarily regulated by cell spreading and cytoskeletal tension [225,232]. When cells are spread, thick and abundant stress fibers form, leading to YAP/TAZ dephosphorylation and nuclear translocation, accompanied by promoted cell proliferation. In contrast, limited cell spreading (compact and round morphology) results in thin and less-evident stress fibers, leading to YAP/TAZ phosphorylation and cytoplasmic translocation, accompanied by suppressed cell proliferation [226,228,233]. Inhibiting myosin in cells reduces stress fibers and nuclear YAP [234]. YAP dephosphorylation can be completely blocked by Rho but not Rac or Cdc42 inhibitors [233].
YAP nucleocytoplasmic localization and activity can be mediated by cell-cell contacts or cell-substrate adhesion [226,228,233], and are sensitive to substrate stiffness [82,174,225,232,235] and nanotopography [196,232,234]. On microfabricated adhesive squares, YAP nucleocytoplasmic distribution gradually changes with square size. Cells mostly express cytoplasmic YAP on small squares, whereas they predominantly show nuclear YAP on squares that are larger than a threshold area between (30 × 30) µm2 and (40 × 40) µm2 [226]. YAP/TAZ nucleocytoplasmic localization is shown to be dependent on substrate stiffness in a physiologically relevant range (0.5–40 kPa) [232]. Soft substrates induce cytoplasmic YAP expression, inhibit cell proliferation [228], and promote the differentiation of human pluripotent stem cells (hPSCs) into motor neurons or GABAergic interneurons [174,235]. Knockdown of YAP/TAZ in hMSCs grown on stiff substrates or large adhesive areas enables adipogenic differentiation, which is commonly observed in cells grown on soft substrates or small adhesive areas; in addition, overexpression of YAP/TAZ causes cells grown on soft substrates to behave like those grown on stiff substrates [225]. Moreover, YAP/TAZ is suggested to act as an intracellular mechanical rheostat, mediating the influence of mechanical dosing on stem cell plasticity. Mechanically priming the cells for short periods of time leads to the reversible activation of YAP; however, a mechanical dosing beyond a threshold dose leads to the constitutive activation of YAP, which biases hMSC differentiation toward osteogenesis even after the mechanical dosing is removed [82]. Compared with well-studied stiffness cues, the effects of nanotopographical cues on YAP intracellular localization have not been well investigated [212,232]. Cytoplasmic YAP is suggested to be necessary yet insufficient for neural differentiation from human induced pluripotent stem cells, while nanograting-induced cell polarity is crucial to induced neural differentiation [234].
4. Comparison between biophysical regulations
4.1. Similarity between substrate stiffness and nanotopographical modulation
In response to a broad spectrum of substrate stiffnesses and a wide variety of nanotopographies, cells exhibit diverse phenotypes and function. Yet a striking similarity is observed in cellular responses to biophysical cues. For example, MC3T3-E1 osteoblasts exhibit different focal adhesions and cytoskeleton on PAAm gels of different stiffnesses, showing diffuse focal adhesions and poorly organized actin cytoskeleton on the softest gels (11.8 kPa), but distinct focal adhesions and mature actin stress fibers on stiffer gels (39 kPa), which are comparable to those in cells grown on glass surfaces [59]. Similarly, stress fiber development is perturbed in human osteoblasts grown on highly ordered nanopit arrays, but cells on randomly arranged nanopits are observed to be well-spread with organized stress fibers; the latter cells are similar to those on the smooth control substrates [236].
The similarity in cell spreading and migration is evident on substrates with stepwise changes in stiffness and topography. As shown in Fig. 6(a) [207], on PAAm substrates with soft (14 kPa) and stiff (30 kPa) regions, individual NIH 3T3 fibroblasts easily migrate from the soft side to the stiff side, with a concurrent increase in cell area and traction force, yet a decrease in cell migration speed: (0.44 ± 0.23) µm·min−1 on the soft side and (0.26 ± 0.13) µm·min−1 on the stiff side. In contrast, when the cells migrate from the stiff side toward the soft side, they turn around or retract at the boundary, as shown in Fig. 6(b) [207]. Even in the presence of many cell-cell contacts, NIH 3T3 fibroblasts and bovine pulmonary arterial endothelial cells accumulate preferentially on stiffer regions (34 kPa) compared with softer regions (1.8 kPa) of PAAm substrates [237]. Similarly, on consecutive arrays of PDMS micropillars that are 1 µm and 2 µm in diameter (resulting in a stiffness ratio of ~10 between the arrays), fibroblast cells from the 1 µm (soft) array probe the boundary and exert larger forces on the array, thus inducing polarization of the actin cytoskeleton and promoting cell migration toward the 2 µm (stiff) array, as shown in Fig. 6(c) [238]. Conversely, most of the cells on the 2 µm array do not migrate toward the 1 µm array, as shown in Fig. 6(d) [238]. Moreover, cell migration toward the 2 µm array is reduced when the stiffnesses of both arrays are greater than 50 nN·µm−1, as shown in Fig. 6(e), indicating that the stiffness effects on cellular response appear within a narrow stiffness range [238].
Fig. 6.
Cell migration on substrates with a step difference in (a, b) stiffness and (c–e) topography. (a) An NIH 3T3 cell migrates from the soft side toward the stiff side of the PAAm gel. (b) An NIH 3T3 cell migrates from the stiff side toward the soft side of the gel. The scale bar is 40 µ m. (c) SEM micrograph of a fibroblast cell migrating from the 1 µ m (top region) to the 2 µ m (bottom region) pillar array. The micropillar densities of the arrays are kept constant. (d) Statistics of cells migrating from a 1 µ m array toward a 2 µ m array as a function of the spring constant of the 1 µ m pillars. Red bars: percentage of cells migrating from the 1 µ m array to the 2 µ m array. Blue bars: percentage of non-migrated cells. Gray bars: percentage of cells with undefined movement. (e) Statistics of cells migrating from the 2 µ m array toward the 1 µ m array as a function of the spring constant of the 2 µ m pillars. Red bars: percentage of migrated cells on the 2 µ m array. Blue bars: percentage of cells migrated toward the 1 µ m pillars. Gray bars: percentage of cells with undefined movement. (Parts (a) and (b) are reproduced with permission from Ref. [207] and parts (c–e) are reproduced with permission from Ref. [238])
Cellular studies on the gradients of stiffness [239,240] and topography [118,241,242] further manifest the similarity of the biophysical modulation. On a PAAm substrate with a gradient of stiffness that is linearly varied from ~1 kPa to 240 kPa across 2 mm, NIH 3T3 fibroblastic and neuroblastoma cells display a rounded morphology with diffuse focal adhesions on the softer region, but are well spread with defined focal adhesions on the stiffer region, as shown in Fig. 7(a) [239]. The cells also migrate from the softer region to the stiffer region [240]. The density of topographies can also provide guidance for cell spreading and migration [118,241–243]. As shown in Fig. 7(b) [241], on an array of nanopillars that are 600 nm in diameter with a constant spacing of 600 nm in the y direction but a spacing varying between 0.3 µm and 4.2 µm in the x direction, 1205Lu melanoma cells exhibit long and parallel filopodia on the sparser pillar density region but short, thick, and randomly oriented protrusions on the denser pillar density region. The cell migration direction is dependent on both pillar density and fibronectin density [241]. On a rectangular lattice array, NIH 3T3 fibroblasts preferentially migrate toward the topographically denser areas and away from sparser ones [242].
Fig. 7.
Cellular responses to gradients of (a) substrate stiffness, (b) nanotopography, and (c) gold nanoparticle arrays. (a) Phase contrast image of NIH 3T3 fibroblasts on a hydrogel with a gradient of stiffness. Substrate stiffnesses are given on the top, and the boxed areas are enlarged in panels (i–iii). Panel (iv) shows cell spreading on glass. (b) Upper panels: graphical illustration and SEM micrograph of 1205Lu melanoma cell on the nanopillar gradient. Lower panels: magnified boxed areas showing filopodia structure in the region of denser (blue box) and sparser (red box) pillars. (c) (i) Scheme of the gold nanoparticle array. (ii, iii) SEM micrographs of MC3T3-EI osteoblasts on the gold nanoparticle array of ~60 nm in spacing. The inset in (iii) shows a close-up of cellular protrusions interacting selectively with the gold nanoparticles. (iv) A 40° tilted view of cellular protrusions interacting with the gold nanoparticles. (v) Cells grown on the gold nanoparticle array with patch spacing from ~50 nm to ~80 nm. The stitched phase-contrast images (top) show cell spreading on the array, and the enlarged boxed areas (bottom) show the cells on areas having ~50 nm, ~60 nm, ~70 nm, and ~80 nm patch spacing. Scale bars: (ii) 500 nm, (iii) 200 nm (inset: 100 nm), (iv) 100 nm, (v) 100 mm. (Part (a) is reproduced with permission from Ref. [239], Part (b) is reproduced with permission from Ref. [241], and Part (c) is reproduced with permission from Ref. [244])
Recent studies of cellular responses to gold nanoparticle arrays advance our understanding of how substrate stiffness and nanotopographical cues regulate cell behavior [244–246]. The cyclic RGDfK peptides-conjugated gold nanoparticle (~8 nm) can bind only one integrin molecule of approximately 10 nm [247]. Inter-particle spacing is controlled using block-copolymer micelle nanolithography, and the inter-particle areas are passivated by PEG in order to eliminate topographical effects on cell adhesion [245]. On isotropic ligand patches with spacing between 28 nm and 85 nm, a variety of cell types spread in all directions, and display optimal integrin clustering, adhesion, actin stress fiber formation, and cell spreading for 58–73 nm spacings [245]. On a ligand patch with a gradient of spacings from 60 nm to 110 nm, the cells clearly sense the gradient and migrate toward the smaller spacing. As shown in Fig. 7(c[i–iv]) [244], the spacing between the adhesive patches increases from ~50 nm to ~80 nm, leading to a spacing gradient with a strength of ~Δ15 nm·mm−1. The cell morphology varies, from well spread on a ligand patch with a spacing of ~50 nm, to strongly elongated on a patch with a spacing of ~80 nm, as shown in Fig. 7(c[v]) [244]. In addition, the cells polarize and exhibit directional migration along the direction of the spacing gradient [244]. Regardless of the cell type, cell spreading and migration on the ligand spacing gradient [244] show striking similarity to those on the substrates with the stiffness gradient [239,240] and topographical gradient [241,242], suggesting that substrate stiffness and topographical cues share some common ground regarding cell modulation.
We thus hypothesize that biophysical regulation occurs mainly through the modulation of adhesive sites on the substrate. The level of stiffness of the substrates, usually hydrogels, is adjusted by changing the crosslinking density; increasing hydrogel stiffness reduces mesh size. When the stiffness of PAAm gels increases from 2 kPa to 20 kPa, the average pore size is measured as decreasing from 15 nm to 5.8 nm [85]. If adhesive ligands such as RGD are incorporated into hydrogel chains, stiffer substrates may provide more adhesive sites while softer substrates form fewer sites. When adhesive proteins are covalently grafted onto the hydrogel, the increase in mesh size results in increasing length of anchored protein fiber, which rapidly decreases the adhesion strength and mechanical feedback that cells sense on integrin ligation [85]. In the case of nanotopography, the shape, dimension, and arrangement of the nanoscale features determine the distribution and even conformation of adhesive proteins and thus restrict adhesive sites’ access to the cell [101,115]. Therefore, substrate stiffness and topography can be used to regulate the assembly and organization of focal adhesions via mediation of binding and clustering of integrins in a similar manner. The crucial role of adhesive site organization is further supported by the observation that modulation of focal adhesion geometry with ECM nanopatterns on stiff substrates can mimic soft matrices, thus controlling cell spreading and differentiation [248]. It is envisioned that on a pliant, nanostructured substrate/matrix, cell behavior can be guided physically by nanotopography and further modulated by stiffness via mechanical feedback upon cells deforming the surrounding nanostructures.
4.2. Theoretical modeling
Biophysical regulation of cell spreading and migration can be described by the model that Bischofs et al. proposed, which uses isotropic linear elasticity theory with the Young’s modulus, E, and the Poisson ratio, ν, of substrates [249,250]. The cell actively pulls on its surrounding matrix, and it is assumed that the amount of work that the cell invests on the matrix will be minimized. The cell-matrix contacts are coupled throughout the actin cytoskeleton such that the forces are balanced; only pairs of opposing forces need to be considered. The work ΔW required to build up the anisotropic force contraction dipole Pij (represented by a tensor Pij = Pninj, where P is the dipole strength and n is its orientation) at cell position r⃑c is proportional to the strain of the environment, :
| (1) |
The optimal cellular organization will minimize ΔW. Assume the matrix acts like a linear spring with spring constant K, and the cell pulls on the matrix through a single cell-matrix contact. The cell-matrix contact can be a single micropillar or part of a micropillar. The cell needs to invest energy W = F2/(2K) into the spring to generate sufficiently large force F. It is more efficient to generate the force by using a stiffer spring (a larger K), even with less work (a smaller W). The cell probes the matrix by pulling at many cell-matrix contacts, each having a different K (Fig. 8) [250]. In an isotropic substrate, as shown in Fig. 8(a), all Ks are equal and all cell-matrix contacts perform similarly; the cell does not orient preferentially and adopts a round or stellate morphology, as observed in cell spreading on isotropically arranged micro-/nano-sized features [98,166,170,251] and homogeneous hydrogels [59]. In an anisotropic matrix, as shown in Fig. 8(b), the force generation is more efficient in one specific direction and the corresponding contacts will eventually outgrow the others. Hence, the anisotropic elastic properties of the matrix can orient the cell along the direction of maximal effective stiffness followed by possible directional cell locomotion, as observed on the oval micropillars [173] and nanogratings [197], as well as on substrates with stiffness gradients [240]. It is notable that ΔW is inversely proportional to the substrate stiffness E: The stiffness effects will only work in a soft environment, as the difference in ΔW for different contacts of a stiffer matrix might become too small to induce oriented cellular responses [250]. This explains the previous observation that the motility of fibroblasts becomes insensitive to a step difference in the substrate once the stiffness is above a certain threshold [238].
Fig. 8.
A proposed mechanism for mechanosensing-induced cell organization. The local elastic property of the matrix is represented by linear springs with different spring constants K. (a) In an isotropic matrix, all spring constants are the same, the forces generated at different contacts are similar, and the cell does not orient in a specific direction. (b) In an anisotropic matrix, the force generation is favored at the contacts with larger spring constants, leading to cell orientation in the direction of maximal stiffness. (Adapted with permission from Ref. [250])
In summary, experimental observations and theoretical analysis imply that substrate stiffness and topographical cues share some common ground; while both types of cues regulate cell behavior, they do so by taking different approaches.
5. Perspectives
Biophysical cues provide significant opportunities for regulating cell fates. To realize this potential and thus advance cell engineering and regenerative medicine, it is crucial to dissect underlying mechanisms and to accurately translate physical cues from a 2D into a 3D environment.
5.1. Mechanistic understanding of biophysical regulation
Adhesive proteins are essential in order for adherent cells to sense and respond to the underlying substrate. It is therefore critical to determine how the substrate stiffness and topography affect protein adsorption, conformation, and distribution, and then to identify the roles of the biophysical cues in cell regulation.
The chemical activities of the adhesive ligands and substrate surface are paramount. A higher affinity of adhesive ligands for the integrin adhesion receptors favors cell spreading with increased traction forces. For example, the cyclic peptide RGDfC, which has approximately two orders of magnitude higher affinity than the linear peptide GRGDSC for ανβ3 integrin, leads to a more contractile cytoskeleton and promotes osteogenesis of hMSCs [252]. Substrate surface hydrophilicity also affects protein conformation. Collagen I proteins fold and clump to form aggregates on hydrophobic PDMS surfaces, thus delaying osteogenesis of hMSCs; conversely, the proteins adopt a more extended conformation on hydrophilic PDMS surfaces, which is associated with an increase in activation of α1β1 integrin and enhanced osteogenic behavior [253].
Moreover, evidence suggests that, at nanoscales, the substrate surface can have different surface energy, and thus affect adhesive protein deposition [254–256]. Nano-sized metals have a higher energy at the surface than in the bulk due to a larger proportion of atomic defects and delocalized surface electrons at the surface [254,256]. The macromolecular chains at the polymer surface also display higher mobility [257,258]. The alteration in surface energy may result in variation in protein deposition on the substrate and may influence the composition and conformation of adsorbed proteins, thus inducing different cellular responses [37,112,255,259–261]. Human fibrinogen exhibits different conformational structures on PLGA pillars that are 250 nm in diameter than on a flat surface [262]. Interestingly, the later-arriving adhesive protein molecules are suggested to have the highest biological activity, compared with the initially adsorbed proteins [263]. Along with the organization of adhesive sites altered by substrate stiffness and nanotopography, it is important to distinguish the biophysical regulation from biochemical regulation of cell behavior. In this regard, enabling technologies that are capable of probing the distribution and conformation of proteins at the nanoscale is desirable [264], such as real-time fluorescence imaging tools that can monitor the cellular responses to these external cues at a resolution comparable to electronic microscopy [265], and new biosensors that allow the detection of protein activities in subcellular regions with high spatial resolution [266].
To identify the roles of biophysical cues in cell regulation, precisely defined model systems with a single variable—substrate stiffness, nanotopography, or biochemical cues—are essential. Currently used substrates such as hydrogels [28] and elastomeric micropillar arrays [166] are not ideal models. As discussed in Section 2, when the stiffness of hydrogels is tuned by changing the crosslinker concentration, not only bulk stiffness but also molecular-scale material properties are altered [85–87]. Most micropillar array studies adopt microscale pillars and thus overlook early-stage focal complexes [186,238]. As a matter of fact, recent studies reveal that the mechanosensing for focal adhesion formation and the coupling of focal adhesions and actin on nanopillars are fundamentally different from the processes on micropillars [171], and that traction force increases with size for focal adhesions larger than 1 µm2, whereas no such correlation exists for smaller adhesions [169]. These studies collectively suggest that nanoscale topographies can closely mimic continuous substrates of a specified stiffness. In addition, elastomeric micropillar arrays are usually oxidized to facilitate protein adsorption and then promote cell adhesion [166,267]. The oxidization process may alter the substrate stiffness as well [56]. The peptide-tethering process can also alter the mechanical properties of the substrate, making it difficult to tune biophysical and biochemical cues independently [268]. Therefore, model systems containing predefined adhesive ligands are advantageous. Adhesive ligands of less than 10 nm should be precisely positioned onto the topography or embedded in the compliant substrate, allowing the uncoupling of these cues. This demands advances in nanotechnology, material synthesis, and surface chemistry. Innovative nanotechnology is needed in order to pattern a large area with a single-nanometer resolution. Block-copolymer micelle nanolithography has been used to control the position and spacing of adhesive ligands [244,245]. Dippen nanolithography (DPN) has been used to directly write integrin ανβ3 nanoarrays to investigate the integrin ανβ3-vitronectin interaction, thus providing a powerful tool to investigate cell-substrate interactions at the single biomolecule level [269]. It is also highly relevant to fabricate nanoscale structures onto compliant substrates with stiffnesses matching those of their in vivo counterparts [144]. Moreover, because the mechanical properties of polymeric nanostructures may differ significantly from those in the bulk, advanced measurement tools are required to map the nanoscale property of the substrate [54,270], not only laterally but also vertically.
5.2. Translation of biophysical regulation from 2D to 3D
Substrate stiffness and nanotopography need to be optimized for a specific cell type, ideally by using high-throughput screening techniques. Gradients of stiffness ranging from ~1 kPa to 240 kPa have been created on single substrates [239,240], covering the stiffness of soft tissues (0.1–100 kPa) [11]. Libraries of topographies of various shapes and dimensions can be directly fabricated onto or stitched into a single platform [41,97,271–273], enabling the investigation of relations between the topographical configuration and the cell-fate decision under the same conditions.
Compared with simple nanotopographies, bioinspired matrices are more attractive because the matrices may provide a more suitable environment for cell functions, as they better mimic the architecture of the natural ECM [274–276]. Hierarchical structures composed of both microscale and nanoscale components are of interest [272,276–285]. Microstructured titanium surfaces are shown to promote osteogenesis of osteoblasts yet inhibit their proliferation, whereas micro-nano-hybrid structures promote both osteogenesis and proliferation [276–278]. Another study shows that microgratings (2 µm in width, height, and spacing) overlaid with 250 nm gratings are effective in the production of dopaminergic neurons. Production of astrocytes is suppressed on microgratings with perpendicularly arranged nanogratings but promoted on those with parallel nanogratings [272]. To determine the exact roles played by microtopographies and nanotopographies, novel techniques such as multiple nanoimprint lithography (NIL) have been developed, in order to fabricate defined nanotopography on micropatterns [149,286,287]. However, NIL requires expensive equipment and expertise, and thus reliable yet cost-effective techniques remain highly desirable, especially techniques that can produce hierarchical structures for direct clinical use [276].
Although 2D in vitro studies help to elucidate fundamental principles of cell regulation with substrate stiffness and topographical cues, they do not recapitulate the complexity found in a 3D environment. Therefore, the biophysical cues have to be translated into a 3D milieu in order to provide the most meaningful answers [30,288–290]. Indeed, cells have demonstrated distinct cell adhesion [288], morphology [291], proliferation, and differentiation [30,292] in a 3D environment compared with those in a 2D environment. For example, hMSCs show the most efficient osteogenesis when cultured on top of stiff gels, while switching to predominantly terminal chondrogenesis when encapsulated within the same stiff gels [292]. In addition, compared with 2D conditions, cancer cells display different gene expression patterns and distinct sensitivities to chemotherapy drugs in 3D environments [293–296]. Moreover, many physiological (e.g., morphogenesis and organogenesis) and pathological (e.g., tumorigenesis) processes are exclusively observed in a 3D milieu [297]. Therefore, efforts have been made to incorporate the biophysical cues into a 3D context. For example, a conformal nanopatterning technique has been developed to pattern adhesive proteins onto topographically complex surfaces [298]. Gold nanoparticles conjugated with adhesion ligands are coordinated on a hydrogel matrix with tunable stiffness, including the surface of 2D gels [299–301] and the inner surface of micro-sized, circular channels [302]. Taking advantage of the fact that hydrogels can be processed under biologically permissive conditions, which allows the cells to be encapsulated inside, novel strategies have been developed to synthesize hydrogels with independently tuned stiffness, adhesive ligands, and architecture [292,303,304]. Still, it has been a long-standing challenge to preserve the merits of the 2D matrix and to control topographical and biochemical cues in all three dimensions with a physiologically relevant stiffness.
The fact that biophysical regulation is a dynamic process has often been overlooked. Biophysical characteristics of the cell microenvironment may change during developmental, physiological, and pathological processes. Changes in ECM stiffness may occur due to the production of ECM proteins, enzymatic degradation of the ECM, alterations in the remodeling process, and the extent of mineralization [305]. One example is tissue stiffening during fibrogenesis [306]. Programmable materials have recently been developed; as a result, cellular and biological processes in response to temporal alterations in stiffness [307–311], nanotopography [232,312], and adhesiveness [313] of the matrices have been investigated. For example, studies using light-induced softening (degradable) [82] and stiffening (crosslinkable) [309] hydrogels show that stem cell differentiation is sensitive to the dynamic environment.
A comprehensive understanding of cell-substrate interactions, particularly in a 3D milieu, is not only crucial to the elucidation of many fundamental biological processes, but also beneficial for stem cell bioprocessing, endogenous tissue engineering, and the design of next-generation biomaterials and biomedical devices. Although significant challenges abound, so do the rewards.
Supplementary Material
Acknowledgments
The authors would like to acknowledge funding support for Yong Yang from the National Science Foundation (CBET 1511759) and the National Institute of Health (NIH) (R15GM122953), and for Kam W. Leong from NIH (HL109442, AI096305, GM110494, and UH3 TR000505), Guangdong Innovative and Entrepreneurial Research Team Program (2013S086), and the Global Research Laboratory Program (Korean NSF GRL; 2015032163).
Footnotes
Compliance with ethics guidelines
Yong Yang, Kai Wang, Xiaosong Gu, and Kam W. Leong declare that they have no conflict of interest or financial conflicts to disclose.
References
- 1.Klein G. The extracellular matrix of the hematopoietic microenvironment. Experientia. 1995;51(9):914–26. doi: 10.1007/BF01921741. [DOI] [PubMed] [Google Scholar]
- 2.Shirato I, Tomino Y, Koide H, Sakai T. Fine structure of the glomerular basement membrane of the rat kidney visualized by high-resolution scanning electron microscopy. Cell Tissue Res. 1991;266(1):1–10. doi: 10.1007/BF00678705. [DOI] [PubMed] [Google Scholar]
- 3.Hironaka K, Makino H, Yamasaki Y, Ota Z. Renal basement membranes by ultrahigh resolution scanning electron microscopy. Kidney Int. 1993;43(2):334–45. doi: 10.1038/ki.1993.51. [DOI] [PubMed] [Google Scholar]
- 4.Abrams GA, Schaus SS, Goodman SL, Nealey PF, Murphy CJ. Nanoscale topography of the corneal epithelial basement membrane and Descemet’s membrane of the human. Cornea. 2000;19(1):57–64. doi: 10.1097/00003226-200001000-00012. [DOI] [PubMed] [Google Scholar]
- 5.Liliensiek SJ, Nealey P, Murphy CJ. Characterization of endothelial basement membrane nanotopography in rhesus macaque as a guide for vessel tissue engineering. Tissue Eng Part A. 2009;15(9):2643–51. doi: 10.1089/ten.tea.2008.0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim J, Kim HN, Lim KT, Kim Y, Seonwoo H, Park SH, et al. Designing nano-topographical density of extracellular matrix for controlled morphology and function of human mesenchymal stem cells. Sci Rep. 2013;3:3552. doi: 10.1038/srep03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suki B, Sato S, Parameswaran H, Szabari MV, Takahashi A, Bartolák-Suki E. Emphysema and mechanical stress-induced lung remodeling. Physiology. 2013;28(6):404–13. doi: 10.1152/physiol.00041.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shvedova AA, Kisin ER, Mercer R, Murray AR, Johnson VJ, Potapovich AI, et al. Unusual inflammatory and fibrogenic pulmonary responses to single-walled carbon nanotubes in mice. Am J Physiol Lung Cell Mol Physiol. 2005;289(5):L698–708. doi: 10.1152/ajplung.00084.2005. [DOI] [PubMed] [Google Scholar]
- 9.Mwenifumbo S, Stevens MM. ECM interactions with cells from the macroto nanoscale. In: Gonsalves KE, Halberstadt CR, Laurencin CT, Nair LS, editors. Biomedical nanostructures. New York: John Wiley & Sons, Inc.; 2008. pp. 225–60. [Google Scholar]
- 10.Silver FH, Freeman JW, Seehra GP. Collagen self-assembly and the development of tendon mechanical properties. J Biomech. 2003;36(10):1529–53. doi: 10.1016/s0021-9290(03)00135-0. [DOI] [PubMed] [Google Scholar]
- 11.Gonçalves CA, Figueiredo MH, Bairos VA. Three-dimensional organization of the elastic fibres in the rat lung. Anat Rec. 1995;243(1):63–70. doi: 10.1002/ar.1092430108. [DOI] [PubMed] [Google Scholar]
- 12.Ma Z, Ramakrishna S. Nanostructured extracellular matrix. Enc Nanosci Nanotechnol. 2004;7:641–55. [Google Scholar]
- 13.Discher DE, Mooney DJ, Zandstra PW. Growth factors, matrices, and forces combine and control stem cells. Science. 2009;324(5935):1673–7. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nemir S, West JL. Synthetic materials in the study of cell response to substrate rigidity. Ann Biomed Eng. 2010;38(1):2–20. doi: 10.1007/s10439-009-9811-1. [DOI] [PubMed] [Google Scholar]
- 15.Cox TR, Erler JT. Remodeling and homeostasis of the extracellular matrix: implications for fibrotic diseases and cancer. Dis Model Mech. 2011;4(2):165–78. doi: 10.1242/dmm.004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gong H, Freddo TF, Johnson M. Age-related changes of sulfated proteoglycans in the normal human trabecular meshwork. Exp Eye Res. 1992;55(5):691–709. doi: 10.1016/0014-4835(92)90174-q. [DOI] [PubMed] [Google Scholar]
- 17.Orr AW, Helmke BP, Blackman BR, Schwartz MA. Mechanisms of mechanotransduction. Dev Cell. 2006;10(1):11–20. doi: 10.1016/j.devcel.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 18.Wozniak MA, Chen CS. Mechanotransduction in development: a growing role for contractility. Nat Rev Mol Cell Biol. 2009;10(1):34–43. doi: 10.1038/nrm2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore SW, Sheetz MP. Biophysics of substrate interaction: influence on neural motility, differentiation, and repair. Dev Neurobiol. 2011;71(11):1090–101. doi: 10.1002/dneu.20947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Tan Y, Zhang H, Zhang Y, Xu P, Chen J, et al. Soft fibrin gels promote selection and growth of tumorigenic cells. Nat Mater. 2012;11(8):734–41. doi: 10.1038/nmat3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8(3):241–54. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 22.Parameswaran H, Majumdar A, Suki B. Linking microscopic spatial patterns of tissue destruction in emphysema to macroscopic decline in stiffness using a 3D computational model. PLOS Comput Biol. 2011;7(4):e1001125. doi: 10.1371/journal.pcbi.1001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Booth AJ, Hadley R, Cornett AM, Dreffs AA, Matthes SA, Tsui JL, et al. Acellular normal and fibrotic human lung matrices as a culture system for in vitro investigation. Am J Resp Crit Care. 2012;186(9):866–76. doi: 10.1164/rccm.201204-0754OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu F, Mih JD, Shea BS, Kho AT, Sharif AS, Tager AM, et al. Feedback amplification of fibrosis through matrix stiffening and COX-2 suppression. J Cell Biol. 2010;190(4):693–706. doi: 10.1083/jcb.201004082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marinković A, Mih JD, Park JA, Liu F, Tschumperlin DJ. Improved throughput traction microscopy reveals pivotal role for matrix stiffness in fibroblast contractility and TGF-β responsiveness. Am J Physiol Lung Cell Mol Physiol. 2012;303(3):169–80. doi: 10.1152/ajplung.00108.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alenghat FJ, Ingber DE. Mechanotransduction: all signals point to cytoskeleton, matrix, and integrins. Sci STKE. 2002;2002(119):pe6. doi: 10.1126/stke.2002.119.pe6. [DOI] [PubMed] [Google Scholar]
- 27.Pelham RJ, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA. 1997;94(25):13661–5. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 29.Chowdhury F, Na S, Li D, Poh Y, Tanaka T, Wang F, et al. Material properties of the cell dictate stress-induced spreading and differentiation in embryonic stem cells. Nat Mater. 2010;9(1):82–8. doi: 10.1038/nmat2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huebsch N, Arany PR, Mao AS, Shvartsman D, Ali OA, Bencherif SA, et al. Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat Mater. 2010;9(6):518–26. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holst J, Watson S, Lord MS, Eamegdool SS, Bax DV, Nivison-Smith LB, et al. Substrate elasticity provides mechanical signals for the expansion of hemopoietic stem and progenitor cells. Nat Biotechnol. 2010;28(10):1123–8. doi: 10.1038/nbt.1687. [DOI] [PubMed] [Google Scholar]
- 32.Gilbert PM, Havenstrite KL, Magnusson KE, Sacco A, Leonardi NA, Kraft P, et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329(5995):1078–81. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Engler AJ, Griffin MA, Sen S, Boennemann CG, Sweeney HL, Discher DE. Myotubes differentiate optimally on substrates with tissue-like stiffness: pathological implications for soft or stiff microenvironments. J Cell Biol. 2004;166(6):877–87. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, et al. Selective differentiation of neural progenitor cells by high-epitope density nanofibers. Science. 2004;303(5662):1352–5. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 35.Yim EKF, Pang SW, Leong KW. Synthetic nanostructures inducing differentiation of human mesenchymal stem cells into neuronal lineage. Exp Cell Res. 2007;313(9):1820–9. doi: 10.1016/j.yexcr.2007.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dalby MJ, Gadegaard N, Tare R, Andar A, Riehle MO, Herzyk P, et al. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat Mater. 2007;6(12):997–1003. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- 37.Oh S, Brammer KS, Li YSJ, Teng D, Engler AJ, Chien S, et al. Stem cell fate dictated solely by altered nanotube dimension. Proc Natl Acad Sci USA. 2009;106(7):2130–5. doi: 10.1073/pnas.0813200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brunetti V, Maiorano G, Rizzello L, Sorce B, Sabella S, Cingolani R, et al. Neurons sense nanoscale roughness with nanometer sensitivity. Proc Natl Acad Sci USA. 2010;107(14):6264–9. doi: 10.1073/pnas.0914456107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McMurray R, Gadegaard N, Tsimbouri P, Burgess K, McNamara L, Tare R, et al. Nanoscale surfaces for the long-term maintenance of mesenchymal stem cell phenotype and multipotency. Nat Mater. 2011;10(8):637–44. doi: 10.1038/nmat3058. [DOI] [PubMed] [Google Scholar]
- 40.Lee MR, Kwon KW, Jung H, Kim HN, Suh KY, Kim K, et al. Direct differentiation of human embryonic stem cells into selective neurons on nanoscale ridge/groove pattern arrays. Biomaterials. 2010;31(15):4360–6. doi: 10.1016/j.biomaterials.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 41.Moe AAK, Suryana M, Marcy G, Lim SK, Ankam S, Goh JZW, et al. Microarray with micro- and nano-topographies enables identification of the optimal topography for directing the differentiation of primary murine neural progenitor cells. Small. 2012;8(19):3050–61. doi: 10.1002/smll.201200490. [DOI] [PubMed] [Google Scholar]
- 42.Dang JM, Leong KW. Myogenic induction of aligned mesenchymal stem cell sheets by culture on thermally responsive electrospun nanofibers. Adv Mater. 2007;19(19):2775–9. doi: 10.1002/adma.200602159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Discher DE, Janmey P, Wang Y. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310(5751):1139–43. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 44.Guilak F, Cohen DM, Estes BT, Gimble JM, Liedtke W, Chen CS. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell. 2009;5(1):17–26. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flemming RG, Murphy CJ, Abrams GA, Goodman SL, Nealey PF. Effects of synthetic micro- and nano-structured surfaces on cell behavior. Biomaterials. 1999;20(6):573–88. doi: 10.1016/s0142-9612(98)00209-9. [DOI] [PubMed] [Google Scholar]
- 46.Stevens MM, George JH. Exploring and engineering the cell surface interface. Science. 2005;310(5751):1135–8. doi: 10.1126/science.1106587. [DOI] [PubMed] [Google Scholar]
- 47.Yang Y, Leong KW. Nanoscale surfacing for regenerative medicine. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2010;2(5):478–95. doi: 10.1002/wnan.74. [DOI] [PubMed] [Google Scholar]
- 48.Kim DH, Provenzano PP, Smith CL, Levchenko A. Matrix nanotopography as a regulator of cell function. J Cell Biol. 2012;197(3):351–60. doi: 10.1083/jcb.201108062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dalby MJ, Gadegaard N, Oreffo RO. Harnessing nanotopography and integrin-matrix interactions to influence stem cell fate. Nat Mater. 2014;13(6):558–69. doi: 10.1038/nmat3980. [DOI] [PubMed] [Google Scholar]
- 50.Nguyen AT, Sathe SR, Yim EK. From nano to micro: topographical scale and its impact on cell adhesion, morphology and contact guidance. J Phys Condens Matter. 2016;28(18):183001. doi: 10.1088/0953-8984/28/18/183001. [DOI] [PubMed] [Google Scholar]
- 51.Janson IA, Putnam AJ. Extracellular matrix elasticity and topography: material-based cues that affect cell function via conserved mechanisms. J Biomed Mater Res A. 2015;103(3):1246–58. doi: 10.1002/jbm.a.35254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elastic moduli data for polycrystalline ceramics [Internet] Gaithersburg: National Institute of Standards and Technology; [cited 2017 Jan 8]. c2017 Available from: https://srdata.nist.gov/CeramicDataPortal/elasticity/TiO2. [Google Scholar]
- 53.Halliday D, Resnick R, Walker J. Fundamentals of physics. 6. New York: John Wiley & Sons, Inc.; 2000. [Google Scholar]
- 54.Sahin O, Magonov S, Su C, Quate CF, Solgaard O. An atomic force microscope tip designed to measure time-varying nanomechanical forces. Nat Nanotechnol. 2007;2(8):507–14. doi: 10.1038/nnano.2007.226. [DOI] [PubMed] [Google Scholar]
- 55.Leung L, Chan C, Baek S, Naguib H. Comparison of morphology and mechanical properties of PLGA bioscaffolds. Biomed Mater. 2008;3(2):025006. doi: 10.1088/1748-6041/3/2/025006. [DOI] [PubMed] [Google Scholar]
- 56.Yang Y, Kulangara K, Lam RTS, Dharmawan R, Leong KW. Effects of topographical and mechanical property alterations induced by oxygen plasma modification on stem cell behavior. ACS Nano. 2012;6(10):8591–8. doi: 10.1021/nn301713d. [DOI] [PubMed] [Google Scholar]
- 57.Kong HJ, Polte TR, Alsberg E, Mooney DJ. FRET measurements of cell-traction forces and nano-scale clustering of adhesion ligands varied by substrate stiffness. Proc Natl Acad Sci USA. 2005;102(12):4300–5. doi: 10.1073/pnas.0405873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo W, Frey MT, Burnham NA, Wang Y. Substrate rigidity regulates the formation and maintenance of tissues. Biophys J. 2006;90(6):2213–20. doi: 10.1529/biophysj.105.070144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khatiwala CB, Peyton SR, Putnam AJ. Intrinsic mechanical properties of the extracellular matrix affect the behavior of pre-osteoblastic MC3T3-E1 cells. Am J Physiol Cell Physiol. 2006;290(6):C1640–50. doi: 10.1152/ajpcell.00455.2005. [DOI] [PubMed] [Google Scholar]
- 60.Solon J, Levental I, Sengupta K, Georges PC, Janmey PA. Fibroblast adaptation and stiffness matching to soft elastic substrates. Biophys J. 2007;93(12):4453–61. doi: 10.1529/biophysj.106.101386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang H, Dembo M, Wang Y. Substrate flexibility regulates growth and apoptosis of normal but not transformed cells. Am J Physiol Cell Physiol. 2000;279(5):C1345–50. doi: 10.1152/ajpcell.2000.279.5.C1345. [DOI] [PubMed] [Google Scholar]
- 62.Engler AJ, Richert L, Wong JY, Picart C, Discher DE. Surface probe measurements of the elasticity of sectioned tissue, thin gels and polyelectrolyte multilayer films: correlations between substrate stiffness and cell adhesion. Surf Sci. 2004;570(1–2):142–54. [Google Scholar]
- 63.Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, et al. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil Cytoskeleton. 2005;60(1):24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- 64.Brown XQ, Ookawa K, Wong JY. Evaluation of polydimethylsiloxane scaffolds with physiologically-relevant elastic moduli: interplay of substrate mechanics and surface chemistry effects on vascular smooth muscle cell response. Biomaterials. 2005;26(16):3123–9. doi: 10.1016/j.biomaterials.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 65.Collin O, Tracqui P, Stephanou A, Usson Y, Clément-Lacroix J, Planus E. Spatiotemporal dynamics of actin-rich adhesion microdomains: influence of substrate flexibility. J Cell Sci. 2006;119(9):1914–25. doi: 10.1242/jcs.02838. [DOI] [PubMed] [Google Scholar]
- 66.Reinhart-King CA, Dembo M, Hammer DA. Cell-cell mechanical communication through compliant substrates. Biophys J. 2008;95(12):6044–51. doi: 10.1529/biophysj.107.127662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rowlands AS, George PA, Cooper-White JJ. Directing osteogenic and myogenic differentiation of MSCs: interplay of stiffness and adhesive ligand presentation. Am J Physiol Cell Physiol. 2008;295(4):C1037–44. doi: 10.1152/ajpcell.67.2008. [DOI] [PubMed] [Google Scholar]
- 68.Georges PC, Miller WJ, Meaney DF, Sawyer ES, Janmey PA. Matrices with compliance comparable to that of brain tissue select neuronal over glial growth in mixed cortical cultures. Biophys J. 2006;90(8):3012–8. doi: 10.1529/biophysj.105.073114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wong JY, Velasco A, Rajagopalan P, Pham Q. Directed movement of vascular smooth muscle cells on gradient-compliant hydrogels. Langmuir. 2003;19(5):1908–13. [Google Scholar]
- 70.Ghosh K, Pan Z, Guan E, Ge S, Liu Y, Nakamura T, et al. Cell adaptation to a physiologically relevant ECM mimic with different viscoelastic properties. Biomaterials. 2007;28(4):671–9. doi: 10.1016/j.biomaterials.2006.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mih JD, Marinkovic A, Liu F, Sharif AS, Tschumperlin DJ. Matrix stiffness reverses the effect of actomyosin tension on cell proliferation. J Cell Sci. 2012;125(24):5974–83. doi: 10.1242/jcs.108886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hsiong SX, Carampin P, Kong HJ, Lee KY, Mooney DJ. Differentiation stage alters matrix control of stem cells. J Biomed Mater Res A. 2008;85(1):145–56. doi: 10.1002/jbm.a.31521. [DOI] [PubMed] [Google Scholar]
- 73.Gu Y, Ji Y, Zhao Y, Liu Y, Ding F, Gu X, et al. The influence of substrate stiffness on the behavior and functions of Schwann cells in culture. Biomaterials. 2012;33(28):6672–81. doi: 10.1016/j.biomaterials.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 74.DiMilla PA, Stone JA, Quinn JA, Albelda SM, Lauffenburger DA. Maximal migration of human smooth-muscle cells on fibronectin and type-IV collagen occurs at an intermediate attachment strength. J Cell Biol. 1993;122(3):729–37. doi: 10.1083/jcb.122.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Peyton SR, Putnam AJ. Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J Cell Physiol. 2005;204(1):198–209. doi: 10.1002/jcp.20274. [DOI] [PubMed] [Google Scholar]
- 76.Calve S, Simon HG. Biochemical and mechanical environment cooperatively regulate skeletal muscle regeneration. FASEB J. 2012;26(6):2538–45. doi: 10.1096/fj.11-200162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Boontheekul T, Hill EE, Kong HJ, Mooney DJ. Regulating myoblast phenotype through controlled gel stiffness and degradation. Tissue Eng. 2007;13(7):1431–42. doi: 10.1089/ten.2006.0356. [DOI] [PubMed] [Google Scholar]
- 78.Saha K, Keung AJ, Irwin EF, Li Y, Little L, Schaffer DV, et al. Substrate modulus directs neural stem cell behavior. Biophys J. 2008;95(9):4426–38. doi: 10.1529/biophysj.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boonen KJM, Rosaria-Chak KY, Baaijens FPT, van der Schaft DWJ, Post MJ. Essential environmental cues from the satellite cell niche: optimizing proliferation and differentiation. Am J Physiol Cell Physiol. 2009;296(6):C1338–45. doi: 10.1152/ajpcell.00015.2009. [DOI] [PubMed] [Google Scholar]
- 80.Huang C, Butler PJ, Tong S, Muddana HS, Bao G, Zhang S. Substrate stiffness regulates cellular uptake of nanoparticles. Nano Lett. 2013;13(4):1611–5. doi: 10.1021/nl400033h. [DOI] [PubMed] [Google Scholar]
- 81.Balestrini JL, Chaudhry S, Sarrazy V, Koehler A, Hinz B. The mechanical memory of lung myofibroblasts. Integr Biol. 2012;4(4):410–21. doi: 10.1039/c2ib00149g. [DOI] [PubMed] [Google Scholar]
- 82.Yang C, Tibbitt MW, Basta L, Anseth KS. Mechanical memory and dosing influence stem cell fate. Nat Mater. 2014;13(6):645–52. doi: 10.1038/nmat3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee J, Abdeen AA, Kilian KA. Rewiring mesenchymal stem cell lineage specification by switching the biophysical microenvironment. Sci Rep. 2014;4:5188. doi: 10.1038/srep05188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li CX, Talele NP, Boo S, Koehler A, Knee-Walden E, Balestrini JL, et al. MicroRNA-21 preserves the fibrotic mechanical memory of mesenchymal stem cells. Nat Mater. 2016 doi: 10.1038/nmat4780. Epub 2016 Oct 31. [DOI] [PubMed] [Google Scholar]
- 85.Trappmann B, Gautrot JE, Connelly JT, Strange DG, Li Y, Oyen ML, et al. Extracellular-matrix tethering regulates stem-cell fate. Nat Mater. 2012;11(7):642–9. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- 86.Houseman BT, Mrksich M. The microenvironment of immobilized Arg-Gly-Asp peptides is an important determinant of cell adhesion. Biomaterials. 2001;22(9):943–55. doi: 10.1016/s0142-9612(00)00259-3. [DOI] [PubMed] [Google Scholar]
- 87.Keselowsky BG, Collard DM, García AJ. Integrin binding specificity regulates biomaterial surface chemistry effects on cell differentiation. Proc Natl Acad Sci USA. 2005;102(17):5953–7. doi: 10.1073/pnas.0407356102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li B, Moshfegh C, Lin Z, Albuschies J, Vogel V. Mesenchymal stem cells exploit extracellular matrix as mechanotransducer. Sci Rep. 2013;3:2425. doi: 10.1038/srep02425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wen J, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H, et al. Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat Mater. 2014;13(10):979–87. doi: 10.1038/nmat4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lovett DB, Shekhar N, Nickerson JA, Roux KJ, Lele TP. Modulation of nuclear shape by substrate rigidity. Cell Mol Bioeng. 2013;6(2):230–8. doi: 10.1007/s12195-013-0270-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maloney JM, Walton EB, Bruce CM, van Vliet KJ. Influence of finite thickness and stiffness on cellular adhesion-induced deformation of compliant substrata. Phys Rev E. 2008;78(4):041923. doi: 10.1103/PhysRevE.78.041923. [DOI] [PubMed] [Google Scholar]
- 92.Merkel R, Kirchgeßner N, Cesa CM, Hoffmann B. Cell force microscopy on elastic layers of finite thickness. Biophys J. 2007;93(9):3314–23. doi: 10.1529/biophysj.107.111328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Buxboim A, Rajagopal K, Brown AEX, Discher DE. How deeply cells feel: methods for thin gels. J Phys Condens Matter. 2010;22(19):194116. doi: 10.1088/0953-8984/22/19/194116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Franck C, Maskarinec SA, Tirrell DA, Ravichandran G. Three-dimensional traction force microscopy: a new tool for quantifying cell-matrix interactions. PLoS One. 2011;6(3):e17833. doi: 10.1371/journal.pone.0017833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Roco MC. Nanotechnology: convergence with modern biology and medicine. Curr Opin Biotechnol. 2003;14(3):337–46. doi: 10.1016/s0958-1669(03)00068-5. [DOI] [PubMed] [Google Scholar]
- 96.Park J, Bauer S, Von der Mark K, Schmuki P. Nanosize and vitality: TiO2 nanotube diameter directs cell fate. Nano Lett. 2007;7(6):1686–91. doi: 10.1021/nl070678d. [DOI] [PubMed] [Google Scholar]
- 97.Wang K, Bruce A, Mezan R, Kadiyala A, Wang L, Dawson J, et al. Nanotopographical modulation of cell function through nuclear deformation. ACS Appl Mater Interfaces. 2016;8(8):5082–92. doi: 10.1021/acsami.5b10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lim JY, Hansen JC, Siedlecki CA, Runt J, Donahue HJ. Human foetal osteoblastic cell response to polymer-demixed nanotopographic interfaces. J R Soc Interface. 2005;2(2):97–108. doi: 10.1098/rsif.2004.0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lim JY, Hansen JC, Siedlecki CA, Hengstebeck RW, Cheng J, Winograd N, et al. Osteoblast adhesion on poly(L-lactic acid)/polystyrene demixed thin film blends: effect of nanotopography, surface chemistry, and wettability. Biomacromolecules. 2005;6(6):3319–27. doi: 10.1021/bm0503423. [DOI] [PubMed] [Google Scholar]
- 100.Dalby MJ, Riehle MO, Johnstone H, Affrossman S, Curtis ASG. In vitro reaction of endothelial cells to polymer demixed nanotopography. Biomaterials. 2002;23(14):2945–54. doi: 10.1016/s0142-9612(01)00424-0. [DOI] [PubMed] [Google Scholar]
- 101.Dalby MJ, Marshall GE, Johnstone HJH, Affrossman S, Riehle MO. Interactions of human blood and tissue cell types with 95-nm-high nanotopography. IEEE Trans Nanobioscience. 2002;1(1):18–23. doi: 10.1109/tnb.2002.806933. [DOI] [PubMed] [Google Scholar]
- 102.Frey MT, Tsai IY, Russell TP, Hanks SK, Wang YL. Cellular responses to substrate topography: role of myosin II and focal adhesion kinase. Biophys J. 2006;90(10):3774–82. doi: 10.1529/biophysj.105.074526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dalby MJ, Yarwood SJ, Riehle MO, Johnstone HJ, Affrossman S, Curtis AS. Increasing fibroblast response to materials using nanotopography: morphological and genetic measurements of cell response to 13-nm-high polymer demixed islands. Exp Cell Res. 2002;276(1):1–9. doi: 10.1006/excr.2002.5498. [DOI] [PubMed] [Google Scholar]
- 104.Csaderova L, Martines E, Seunarine K, Gadegaard N, Wilkinson CDW, Riehle MO. A biodegradable and biocompatible regular nanopattern for large-scale selective cell growth. Small. 2010;6(23):2755–61. doi: 10.1002/smll.201000193. [DOI] [PubMed] [Google Scholar]
- 105.Chen W, Villa-Diaz LG, Sun Y, Weng S, Kim JK, Lam RHW, et al. Nanotopography influences adhesion, spreading, and self-renewal of human embryonic stem cells. ACS Nano. 2012;6(5):4094–103. doi: 10.1021/nn3004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Thakar RG, Ho F, Huang NF, Liepmann D, Li S. Regulation of vascular smooth muscle cells by micropatterning. Biochem Biophys Res Commun. 2003;307(4):883–90. doi: 10.1016/s0006-291x(03)01285-3. [DOI] [PubMed] [Google Scholar]
- 107.Charest JL, Eliason MT, García AJ, King WP, Talin AA, Simmons BA. Polymer cell culture substrates with combined nanotopographical patterns and micropatterned chemical domains. J Vac Sci Technol B. 2005;23(6):3011–4. [Google Scholar]
- 108.Zhu B, Zhang Q, Lu Q, Xu Y, Yin J, Hu J, et al. Nanotopographical guidance of C6 glioma cell alignment and oriented growth. Biomaterials. 2004;25(18):4215–23. doi: 10.1016/j.biomaterials.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 109.Yim EKF, Reano RM, Pang SW, Yee AF, Chen CS, Leong KW. Nanopattern-induced changes in morphology and motility of smooth muscle cells. Biomaterials. 2005;26(26):5405–13. doi: 10.1016/j.biomaterials.2005.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gerecht S, Bettinger CJ, Zhang Z, Borenstein JT, Vunjak-Novakovic G, Langer R. The effect of actin disrupting agents on contact guidance of human embryonic stem cells. Biomaterials. 2007;28(28):4068–77. doi: 10.1016/j.biomaterials.2007.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bettinger CJ, Zhang Z, Gerecht S, Borenstein JT, Langer R. Enhancement of in vitro capillary tube formation by substrate nanotopography. Adv Mater. 2008;20(1):99–103. doi: 10.1002/adma.200702487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Teixeira AI, Abrams GA, Bertics PJ, Murphy CJ, Nealey PF. Epithelial contact guidance on well-defined micro- and nanostructured substrates. J Cell Sci. 2003;116(10):1881–92. doi: 10.1242/jcs.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ranucci CS, Moghe PV. Substrate microtopography can enhance cell adhesive and migratory responsiveness to matrix ligand density. J Biomed Mater Res. 2001;54(2):149–61. doi: 10.1002/1097-4636(200102)54:2<149::aid-jbm1>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 114.Prina-Mello A, Volkov Y, Kelleher D, Prendergast PJ. Comparative locomotory behavior of T lymphocytes versus T lymphoma cells on flat and grooved surfaces. Ann Biomed Eng. 2003;31(9):1106–13. doi: 10.1114/1.1603261. [DOI] [PubMed] [Google Scholar]
- 115.Brammer KS, Oh S, Gallagher JO, Jin S. Enhanced cellular mobility guided by TiO2 nanotube surfaces. Nano Lett. 2008;8(3):786–93. doi: 10.1021/nl072572o. [DOI] [PubMed] [Google Scholar]
- 116.Liliensiek SJ, Wood JA, Yong J, Auerbach R, Nealey PF, Murphy CJ. Modulation of human vascular endothelial cell behaviors by nanotopographic cues. Biomaterials. 2010;31(20):5418–26. doi: 10.1016/j.biomaterials.2010.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tan J, Saltzman WM. Topographical control of human neutrophil motility on micropatterned materials with various surface chemistry. Biomaterials. 2002;23(15):3215–25. doi: 10.1016/s0142-9612(02)00074-1. [DOI] [PubMed] [Google Scholar]
- 118.Kim DH, Han K, Gupta K, Kwon KW, Suh KY, Levchenko A. Mechanosensitivity of fibroblast cell shape and movement to anisotropic substratum topography gradients. Biomaterials. 2009;30(29):5433–44. doi: 10.1016/j.biomaterials.2009.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lenhert S, Meier MB, Meyer U, Chi L, Wiesmann HP. Osteoblast alignment, elongation and migration on grooved polystyrene surfaces patterned by langmuir-blodgett lithography. Biomaterials. 2005;26(5):563–70. doi: 10.1016/j.biomaterials.2004.02.068. [DOI] [PubMed] [Google Scholar]
- 120.Sun X, Driscoll MK, Guven C, Das S, Parent CA, Fourkas JT, et al. Asymmetric nanotopography biases cytoskeletal dynamics and promotes unidirectional cell guidance. Proc Natl Acad Sci USA. 2015;112(41):12557–62. doi: 10.1073/pnas.1502970112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang PY, Thissen H, Tsai WB. The roles of RGD and grooved topography in the adhesion, morphology, and differentiation of C2C12 skeletal myoblasts. Biotechnol Bioeng. 2012;109(8):2104–15. doi: 10.1002/bit.24452. [DOI] [PubMed] [Google Scholar]
- 122.Patel S, Kurpinski K, Quigley R, Gao H, Hsiao BS, Poo MM, et al. Bioactive nanofibers: synergistic effects of nanotopography and chemical signaling on cell guidance. Nano Lett. 2007;7(7):2122–8. doi: 10.1021/nl071182z. [DOI] [PubMed] [Google Scholar]
- 123.Yang F, Murugan R, Wang S, Ramakrishna S. Electrospinning of nano/micro scale poly(L-lactic acid) aligned fibers and their potential in neural tissue engineering. Biomaterials. 2005;26(15):2603–10. doi: 10.1016/j.biomaterials.2004.06.051. [DOI] [PubMed] [Google Scholar]
- 124.Bryant DM, Mostov KE. From cells to organs: building polarized tissue. Nat Rev Mol Cell Biol. 2008;9(11):887–901. doi: 10.1038/nrm2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Petrie RJ, Doyle AD, Yamada KM. Random versus directionally persistent cell migration. Nat Rev Mol Cell Biol. 2009;10(8):538–49. doi: 10.1038/nrm2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Biggs MJP, Richards RG, Gadegaard N, Wilkinson CDW, Dalby MJ. Regulation of implant surface cell adhesion: characterization and quantification of S-phase primary osteoblast adhesions on biomimetic nanoscale substrates. J Orthop Res. 2007;25(2):273–82. doi: 10.1002/jor.20319. [DOI] [PubMed] [Google Scholar]
- 127.Wang S, Wang H, Jiao J, Chen K, Owens GE, Kamei K, et al. Three-dimensional nanostructured substrates toward efficient capture of circulating tumor cells. Angew Chem Int Ed. 2009;48(47):8970–3. doi: 10.1002/anie.200901668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang S, Liu K, Liu J, Yu Z, Xu X, Zhao L, et al. Highly efficient capture of circulating tumor cells by using nanostructured silicon substrates with integrated chaotic micromixers. Angew Chem Int Ed. 2011;50(13):3084–8. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu X, Chen L, Liu H, Yang G, Zhang P, Han D, et al. Bio-inspired soft polystyrene nanotube substrate for rapid and highly efficient breast cancer-cell capture. NPG Asia Mater. 2013;5:e63. [Google Scholar]
- 130.Chen W, Weng S, Zhang F, Allen S, Li X, Bao L, et al. Nanoroughened surfaces for efficient capture of circulating tumor cells without using capture antibodies. ACS Nano. 2013;7(1):566–75. doi: 10.1021/nn304719q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shi L, Wang K, Yang Y. Adhesion-based tumor cell capture using nanotopography. Colloids Surf B Biointerfaces. 2016;147:291–9. doi: 10.1016/j.colsurfb.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 132.Kulangara K, Adler AF, Wang H, Chellappan M, Hammett E, Yasuda R, et al. The effect of substrate topography on direct reprogramming of fibroblasts to induced neurons. Biomaterials. 2014;35(20):5327–36. doi: 10.1016/j.biomaterials.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Huang C, Ozdemir T, Xu L, Butler PJ, Siedlecki CA, Brown JL, et al. The role of substrate topography on the cellular uptake of nanoparticles. J Biomed Mater Res Part B. 2016;104(3):488–95. doi: 10.1002/jbm.b.33397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Iyer S, Gaikwad RM, Subba-Rao V, Woodworth CD, Sokolov I. Atomic force microscopy detects differences in the surface brush of normal and cancerous cells. Nat Nanotechnol. 2009;4(6):389–93. doi: 10.1038/nnano.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fischer KE, Alemán BJ, Tao SL, Hugh Daniels R, Li EM, Bünger MD, et al. Biomimetic nanowire coatings for next generation adhesive drug delivery systems. Nano Lett. 2009;9(2):716–20. doi: 10.1021/nl803219f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jeon H, Koo S, Reese WM, Loskill P, Grigoropoulos CP, Healy KE. Directing cell migration and organization via nanocrater-patterned cell-repellent interfaces. Nat Mater. 2015;14(9):918–23. doi: 10.1038/nmat4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Teo BKK, Goh KJ, Ng ZJ, Koo S, Yim EKF. Functional reconstruction of corneal endothelium using nanotopography for tissue-engineering applications. Acta Biomater. 2012;8(8):2941–52. doi: 10.1016/j.actbio.2012.04.020. [DOI] [PubMed] [Google Scholar]
- 138.Watari S, Hayashi K, Wood JA, Russell P, Nealey PF, Murphy CJ, et al. Modulation of osteogenic differentiation in hMSCs cells by submicron topographically- patterned ridges and grooves. Biomaterials. 2012;33(1):128–36. doi: 10.1016/j.biomaterials.2011.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wood JA, Ly I, Borjesson DL, Nealey PF, Russell P, Murphy CJ. The modulation of canine mesenchymal stem cells by nano-topographic cues. Exp Cell Res. 2012;318(19):2438–45. doi: 10.1016/j.yexcr.2012.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Janson IA, Kong YP, Putnam AJ. Nanotopographic substrates of poly(methyl methacrylate) do not strongly influence the osteogenic phenotype of mesenchymal stem cells in vitro. PLoS One. 2014;9(3):e90719. doi: 10.1371/journal.pone.0090719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Clements LR, Wang PY, Tsai WB, Thissen H, Voelcker NH. Electrochemistry-enabled fabrication of orthogonal nanotopography and surface chemistry gradients for high-throughput screening. Lab Chip. 2012;12(8):1480–6. doi: 10.1039/c2lc20732j. [DOI] [PubMed] [Google Scholar]
- 142.Yang J, Rose FRAJ, Gadegaard N, Alexander MR. A high-throughput assay of cell-surface interactions using topographical and chemical gradients. Adv Mater. 2009;21(3):300–4. [Google Scholar]
- 143.Ohara PT, Buck RC. Contact guidance in vitro: a light, transmission, and scanning electron microscopic study. Exp Cell Res. 1979;121(2):235–49. doi: 10.1016/0014-4827(79)90002-8. [DOI] [PubMed] [Google Scholar]
- 144.Kim DH, Lipke EA, Kim P, Cheong R, Thompson S, Delannoy M, et al. Nanoscale cues regulate the structure and function of macroscopic cardiac tissue constructs. Proc Natl Acad Sci USA. 2010;107(2):565–70. doi: 10.1073/pnas.0906504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ahn EH, Kim Y, Kshitiz, An SS, Afzal J, Lee S, et al. Spatial control of adult stem cell fate using nanotopographic cues. Biomaterials. 2014;35(8):2401–10. doi: 10.1016/j.biomaterials.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pan HA, Hung YC, Sui YP, Huang GS. Topographic control of the growth and function of cardiomyoblast H9c2 cells using nanodot arrays. Biomaterials. 2012;33(1):20–8. doi: 10.1016/j.biomaterials.2011.09.054. [DOI] [PubMed] [Google Scholar]
- 147.You MH, Kwak MK, Kim DH, Kim K, Levchenko A, Kim DY, et al. Synergistically enhanced osteogenic differentiation of human mesenchymal stem cells by culture on nanostructured surfaces with induction media. Biomacromolecules. 2010;11(7):1856–62. doi: 10.1021/bm100374n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Crouch AS, Miller D, Luebke KJ, Hu W. Correlation of anisotropic cell behaviors with topographic aspect ratio. Biomaterials. 2009;30(8):1560–7. doi: 10.1016/j.biomaterials.2008.11.041. [DOI] [PubMed] [Google Scholar]
- 149.Hu W, Yim EKF, Reano RM, Leong KW, Pang SW. Effects of nanoimprinted patterns in tissue-culture polystyrene on cell behavior. J Vac Sci Technol B. 2005;23(6):2984–9. doi: 10.1116/1.2121729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Fraser SA, Ting YH, Mallon KS, Wendt AE, Murphy CJ, Nealey PF. Sub-micron and nanoscale feature depth modulates alignment of stromal fibroblasts and corneal epithelial cells in serum-rich and serum-free media. J Biomed Mater Res A. 2008;86A(3):725–35. doi: 10.1002/jbm.a.31519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Uttayarat P, Toworfe GK, Dietrich F, Lelkes PI, Composto RJ. Topographic guidance of endothelial cells on silicone surfaces with micro- to nano-grooves: orientation of actin filaments and focal adhesions. J Biomed Mater Res A. 2005;75A(3):668–80. doi: 10.1002/jbm.a.30478. [DOI] [PubMed] [Google Scholar]
- 152.Wong ST, Teo SK, Park S, Chiam KH, Yim EKF. Anisotropic rigidity sensing on grating topography directs human mesenchymal stem cell elongation. Biomech Model Mechanobiol. 2014;13(1):27–39. doi: 10.1007/s10237-013-0483-2. [DOI] [PubMed] [Google Scholar]
- 153.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276(5317):1425–8. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 154.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6(4):483–95. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 155.Dembo M, Wang Y. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys J. 1999;76(4):2307–16. doi: 10.1016/S0006-3495(99)77386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Calvo F, Ege N, Grande-Garcia A, Hooper S, Jenkins RP, Chaudhry SI, et al. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat Cell Biol. 2013;15(6):637–46. doi: 10.1038/ncb2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ma X, Schickel ME, Stevenson MD, Sarang-Sieminski AL, Gooch KJ, Ghadiali SN, et al. Fibers in the extracellular matrix enable long-range stress transmission between cells. Biophys J. 2013;104(7):1410–8. doi: 10.1016/j.bpj.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Harris A, Wild P, Stopak D. Silicone rubber substrata: a new wrinkle in the study of cell locomotion. Science. 1980;208(4440):177–9. doi: 10.1126/science.6987736. [DOI] [PubMed] [Google Scholar]
- 159.Chalut KJ, Kulangara K, Giacomelli MG, Wax A, Leong KW. Deformation of stem cell nuclei by nanotopographical cues. Soft Matter. 2010;6(8):1675–81. doi: 10.1039/B921206J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yim EKF, Darling EM, Kulangara K, Guilak F, Leong KW. Nanotopographyinduced changes in focal adhesions, cytoskeletal organization, and mechanical properties of human mesenchymal stem cells. Biomaterials. 2010;31(6):1299–306. doi: 10.1016/j.biomaterials.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tzvetkova-Chevolleau T, Stéphanou A, Fuard D, Ohayon J, Schiavone P, Tracqui P. The motility of normal and cancer cells in response to the combined influence of the substrate rigidity and anisotropic microstructure. Biomaterials. 2008;29(10):1541–51. doi: 10.1016/j.biomaterials.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 162.Forrest JA, Dalnoki-Veress K. The glass transition in thin polymer films. Adv Colloid Interface Sci. 2001;94(1–3):167–95. [Google Scholar]
- 163.Van Workum K, de Pablo JJ. Computer simulation of the mechanical properties of amorphous polymer nanostructures. Nano Lett. 2003;3(10):1405–10. [Google Scholar]
- 164.Stafford CM, Harrison C, Beers KL, Karim A, Amis EJ, Vanlandingham MR, et al. A buckling-based metrology for measuring the elastic moduli of polymeric thin films. Nat Mater. 2004;3(8):545–50. doi: 10.1038/nmat1175. [DOI] [PubMed] [Google Scholar]
- 165.Stafford CM, Vogt BD, Harrison C, Julthongpiput D, Huang R. Elastic moduli of ultrathin amorphous polymer films. Macromolecules. 2006;39(15):5095–9. [Google Scholar]
- 166.Fu J, Wang YK, Yang MT, Desai RA, Yu X, Liu Z, et al. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat Methods. 2010;7(9):733–6. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Park J, Kim HN, Kim DH, Levchenko A, Suh KY. Quantitative analysis of the combined effect of substrate rigidity and topographic guidance on cell morphology. IEEE Trans Nanobioscience. 2012;11(1):28–36. doi: 10.1109/TNB.2011.2165728. [DOI] [PubMed] [Google Scholar]
- 168.Balaban NQ, Schwarz US, Riveline D, Goichberg P, Tzur G, Sabanay I, et al. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat Cell Biol. 2001;3(5):466–72. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- 169.Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc Natl Acad Sci USA. 2003;100(4):1484–9. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.du Roure O, Saez A, Buguin A, Austin RH, Chavrier P, Siberzan P, et al. Force mapping in epithelial cell migration. Proc Natl Acad Sci USA. 2005;102(7):2390–5. doi: 10.1073/pnas.0408482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ghassemi S, Meacci G, Liu S, Gondarenko AA, Mathur A, Roca-Cusachs P, et al. Cells test substrate rigidity by local contractions on submicrometer pillars. Proc Natl Acad Sci USA. 2012;109(14):5328–33. doi: 10.1073/pnas.1119886109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yang M, Sniadecki NJ, Chen C. Geometric considerations of micro- to nanoscale elastomeric post arrays to study cellular traction forces. Adv Mater. 2007;19(20):3119–23. [Google Scholar]
- 173.Saez A, Ghibaudo M, Buguin A, Silberzan P, Ladoux B. Rigidity-driven growth and migration of epithelial cells on microstructured anisotropic substrates. Proc Natl Acad Sci USA. 2007;104(20):8281–6. doi: 10.1073/pnas.0702259104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sun Y, Yong KM, Villa-Diaz LG, Zhang X, Chen W, Philson R, et al. Hippo/YAP-mediated rigidity-dependent motor neuron differentiation of human pluripotent stem cells. Nat Mater. 2014;13(6):599–604. doi: 10.1038/nmat3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Han SJ, Bielawski KS, Ting LH, Rodriguez ML, Sniadecki NJ. Decoupling substrate stiffness, spread area, and micropost density: a close spatial relationship between traction forces and focal adhesions. Biophys J. 2012;103(4):640–8. doi: 10.1016/j.bpj.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Saez A, Buguin A, Silberzan P, Ladoux B. Is the mechanical activity of epithelial cells controlled by deformations or forces? Biophys J. 2005;89(6):L52–4. doi: 10.1529/biophysj.105.071217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110(6):673–87. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 178.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, et al. Cell migration: integrating signals from front to back. Science. 2003;302(5651):1704–9. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 179.Zaidel-Bar R, Cohen M, Addadi L, Geiger B. Hierarchical assembly of cell-matrix adhesion complexes. Biochem Soc Trans. 2004;32(3):416–20. doi: 10.1042/BST0320416. [DOI] [PubMed] [Google Scholar]
- 180.Galbraith CG, Yamada KM, Sheetz MP. The relationship between force and focal complex development. J Cell Biol. 2002;159(4):695–705. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Besser A, Safran SA. Force-induced adsorption and anisotropic growth of focal adhesions. Biophys J. 2006;90(10):3469–84. doi: 10.1529/biophysj.105.074377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Riveline D, Zamir E, Balaban NQ, Schwarz US, Ishizaki T, Narumiya S, et al. Focal contacts as mechanosensors. J Cell Biol. 2001;153(6):1175–86. doi: 10.1083/jcb.153.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Nobes CD, Hall A. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81(1):53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 184.DeMali KA, Burridge K. Coupling membrane protrusion and cell adhesion. J Cell Sci. 2003;116(12):2389–97. doi: 10.1242/jcs.00605. [DOI] [PubMed] [Google Scholar]
- 185.Choi CK, Vicente-Manzanares M, Zareno J, Whitmore LA, Mogilner A, Horwitz AR. Actin and α-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat Cell Biol. 2008;10(9):1039–50. doi: 10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Plotnikov SV, Pasapera AM, Sabass B, Waterman CM. Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell. 2012;151(7):1513–27. doi: 10.1016/j.cell.2012.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Coyer SR, Singh A, Dumbauld DW, Calderwood DA, Craig SW, Delamarche E, et al. Nanopatterning reveals an ECM area threshold for focal adhesion assembly and force transmission that is regulated by integrin activation and cytoskeleton tension. J Cell Sci. 2012;125(21):5110–23. doi: 10.1242/jcs.108035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Stricker J, Aratyn-Schaus Y, Oakes PW, Gardel ML. Spatiotemporal constraints on the force-dependent growth of focal adhesions. Biophys J. 2011;100(12):2883–93. doi: 10.1016/j.bpj.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Cary LA, Chang J, Guan J. Stimulation of cell migration by overexpression of focal adhesion kinase and its association with Src and Fyn. J Cell Sci. 1996;109(Pt 7):1787–94. doi: 10.1242/jcs.109.7.1787. [DOI] [PubMed] [Google Scholar]
- 190.Xu B, Song G, Ju Y, Li X, Song Y, Watanabe S. RhoA/ROCK, cytoskeletal dynamics, and focal adhesion kinase are required for mechanical stretch-induced tenogenic differentiation of human mesenchymal stem cells. J Cell Physiol. 2012;227(6):2722–9. doi: 10.1002/jcp.23016. [DOI] [PubMed] [Google Scholar]
- 191.Salasznyk RM, Klees RF, Williams WA, Boskey A, Plopper GE. Focal adhesion kinase signaling pathways regulate the osteogenic differentiation of human mesenchymal stem cells. Exp Cell Res. 2007;313(1):22–37. doi: 10.1016/j.yexcr.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wang H, Dembo M, Hanks SK, Wang Y. Focal adhesion kinase is involved in mechanosensing during fibroblast migration. Proc Natl Acad Sci USA. 2001;98(20):11295–300. doi: 10.1073/pnas.201201198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Pasapera AM, Schneider IC, Rericha E, Schlaepfer DD, Waterman CM. Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J Cell Biol. 2010;188(6):877–90. doi: 10.1083/jcb.200906012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Provenzano PP, Inman DR, Eliceiri KW, Keely PJ. Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK-ERK linkage. Oncogene. 2009;28(49):4326–43. doi: 10.1038/onc.2009.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Humphrey JD, Dufresne ER, Schwartz MA. Mechanotransduction and extracellular matrix homeostasis. Nat Rev Mol Cell Biol. 2014;15(12):802–12. doi: 10.1038/nrm3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Teo BK, Wong ST, Lim CK, Kung TY, Yap CH, Ramagopal Y, et al. Nanotopography modulates mechanotransduction of stem cells and induces differentiation through focal adhesion kinase. ACS Nano. 2013;7(6):4785–98. doi: 10.1021/nn304966z. [DOI] [PubMed] [Google Scholar]
- 197.Kulangara K, Yang Y, Yang J, Leong KW. Nanotopography as modulator of human mesenchymal stem cell function. Biomaterials. 2012;33(20):4998–5003. doi: 10.1016/j.biomaterials.2012.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol. 2009;10(1):21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 199.Burridge K, Wennerberg K. Rho and Rac take center stage. Cell. 2004;116(2):167–79. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- 200.Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C, Brown RA. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat Rev Mol Cell Biol. 2002;3(5):349–63. doi: 10.1038/nrm809. [DOI] [PubMed] [Google Scholar]
- 201.Treiser MD, Yang EH, Gordonov S, Cohen DM, Androulakis IP, Kohn J, et al. Cytoskeleton-based forecasting of stem cell lineage fates. Proc Natl Acad Sci USA. 2010;107(2):610–5. doi: 10.1073/pnas.0909597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Murphy WL, McDevitt TC, Engler AJ. Materials as stem cell regulators. Nat Mater. 2014;13(6):547–57. doi: 10.1038/nmat3937. Erratum in: Nat Mater 2014;13(7):756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wang N, Tolić-Nørrelykke IM, Chen J, Mijailovich SM, Butler JP, Fredberg JJ, et al. Cell prestress. I. Stiffness and prestress are closely associated in adherent contractile cells. Am J Physiol. 2002;282(3):C606–16. doi: 10.1152/ajpcell.00269.2001. [DOI] [PubMed] [Google Scholar]
- 204.Kilian KA, Bugarija B, Lahn BT, Mrksich M. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc Natl Acad Sci USA. 2010;107(11):4872–7. doi: 10.1073/pnas.0903269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wang JH, Lin JS. Cell traction force and measurement methods. Biomech Model Mechanobiol. 2007;6(6):361–71. doi: 10.1007/s10237-006-0068-4. [DOI] [PubMed] [Google Scholar]
- 206.Ghibaudo M, Saez A, Trichet L, Xayaphoummine A, Browaeys J, Silberzan P, et al. Traction forces and rigidity sensing regulate cell functions. Soft Matter. 2008;4(9):1836–43. [Google Scholar]
- 207.Lo C, Wang H, Dembo M, Wang Y. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79(1):144–52. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Klein EA, Yin L, Kothapalli D, Castagnino P, Byfield FJ, Xu T, et al. Cell-cycle control by physiological matrix elasticity and in vivo tissue stiffening. Curr Biol. 2009;19(18):1511–8. doi: 10.1016/j.cub.2009.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Jay PY, Pham PA, Wong SA, Elson EL. A mechanical function of myosin II in cell motility. J Cell Sci. 1995;108(Pt 1):387–93. doi: 10.1242/jcs.108.1.387. [DOI] [PubMed] [Google Scholar]
- 210.Keung AJ, de Juan-Pardo EM, Schaffer DV, Kumar S. Rho GTPases mediate the mechanosensitive lineage commitment of neural stem cells. Stem Cells. 2011;29(11):1886–97. doi: 10.1002/stem.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wang K, He X, Linthicum W, Mezan R, Wang L, Rojanasakul Y, et al. Carbon nanotubes induced fibrogenesis on nanostructured substrates. Environ Sci Nano. 2017 doi: 10.1039/C6EN00402D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Ankam S, Lim CK, Yim EK. Actomyosin contractility plays a role in MAP2 expression during nanotopography-directed neuronal differentiation of human embryonic stem cells. Biomaterials. 2015;47:20–8. doi: 10.1016/j.biomaterials.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 213.Kulangara K, Yang J, Chellappan M, Yang Y, Leong KW. Nanotopography alters nuclear protein expression, proliferation and differentiation of human mesenchymal stem/stromal cells. PLoS One. 2014;9(12):e114698. doi: 10.1371/journal.pone.0114698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci USA. 1997;94(3):849–54. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wormer DB, Davis KA, Henderson JH, Turner CE. The focal adhesion-localized CdGAP regulates matrix rigidity sensing and durotaxis. PLoS One. 2014;9(3):e91815. doi: 10.1371/journal.pone.0091815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Thomas CH, Collier JH, Sfeir CS, Healy KE. Engineering gene expression and protein synthesis by modulation of nuclear shape. Proc Natl Acad Sci USA. 2002;99(4):1972–7. doi: 10.1073/pnas.032668799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.McBride SH, Knothe Tate ML. Modulation of stem cell shape and fate A: the role of density and seeding protocol on nucleus shape and gene expression. Tissue Eng Part A. 2008;14(9):1561–72. doi: 10.1089/ten.tea.2008.0112. [DOI] [PubMed] [Google Scholar]
- 218.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Physical plasticity of the nucleus in stem cell differentiation. Proc Natl Acad Sci USA. 2007;104(40):15619–24. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Sims JR, Karp S, Ingber DE. Altering the cellular mechanical force balance results in integrated changes in cell, cytoskeletal and nuclear shape. J Cell Sci. 1992;103(Pt 4):1215–22. doi: 10.1242/jcs.103.4.1215. [DOI] [PubMed] [Google Scholar]
- 220.Dahl KN, Ribeiro AJ, Lammerding J. Nuclear shape, mechanics, and mechanotransduction. Circ Res. 2008;102(11):1307–18. doi: 10.1161/CIRCRESAHA.108.173989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Dalby MJ, Riehle MO, Yarwood SJ, Wilkinson CDW, Curtis ASG. Nucleus alignment and cell signaling in fibroblasts: response to a micro-grooved topography. Exp Cell Res. 2003;284(2):274–80. doi: 10.1016/s0014-4827(02)00053-8. [DOI] [PubMed] [Google Scholar]
- 222.Roca-Cusachs P, Alcaraz J, Sunyer R, Samitier J, Farré R, Navajas D. Micropatterning of single endothelial cell shape reveals a tight coupling between nuclear volume in G1 and proliferation. Biophys J. 2008;94(12):4984–95. doi: 10.1529/biophysj.107.116863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol. 2006;7(4):265–75. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 224.Yang Y, Kulangara K, Sia J, Wang L, Leong KW. Engineering of a microfluidic cell culture platform embedded with nanoscale features. Lab Chip. 2011;11(9):1638–46. doi: 10.1039/c0lc00736f. [DOI] [PubMed] [Google Scholar]
- 225.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474(7350):179–83. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 226.Wada K, Itoga K, Okano T, Yonemura S, Sasaki H. Hippo pathway regulation by cell morphology and stress fibers. Development. 2011;138(18):3907–14. doi: 10.1242/dev.070987. [DOI] [PubMed] [Google Scholar]
- 227.Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011;13(8):877–83. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, et al. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 2013;154(5):1047–59. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 229.Liu F, Lagares D, Choi KM, Stopfer L, Marinkovic A, Vrbanac V, et al. Mechanosignaling through YAP and TAZ drives fibroblast activation and fibrosis. Am J Physiol Lung C. 2015;308(4):L344–57. doi: 10.1152/ajplung.00300.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Tremblay AM, Camargo FD. Hippo signaling in mammalian stem cells. Semin Cell Dev Biol. 2012;23(7):818–26. doi: 10.1016/j.semcdb.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 231.Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 2010;24(11):1106–18. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Mosqueira D, Pagliari S, Uto K, Ebara M, Romanazzo S, Escobedo-Lucea C, et al. Hippo pathway effectors control cardiac progenitor cell fate by acting as dynamic sensors of substrate mechanics and nanostructure. ACS Nano. 2014;8(3):2033–47. doi: 10.1021/nn4058984. [DOI] [PubMed] [Google Scholar]
- 233.Zhao B, Li L, Wang L, Wang C, Yu J, Guan K. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012;26(1):54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Song L, Wang K, Li Y, Yang Y. Nanotopography promoted neuronal differentiation of human induced pluripotent stem cells. Colloids Surf B. 2016;148:49–58. doi: 10.1016/j.colsurfb.2016.08.041. [DOI] [PubMed] [Google Scholar]
- 235.Musah S, Wrighton PJ, Zaltsman Y, Zhong X, Zorn S, Parlato MB, et al. Substratum-induced differentiation of human pluripotent stem cells reveals the coactivator YAP is a potent regulator of neuronal specification. Proc Natl Acad Sci USA. 2014;111(38):13805–10. doi: 10.1073/pnas.1415330111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Biggs MJP, Richards RG, Gadegaard N, Wilkinson CDW, Dalby MJ. The effects of nanoscale pits on primary human osteoblast adhesion formation and cellular spreading. J Mater Sci–Mater Med. 2007;18(2):399–404. doi: 10.1007/s10856-006-0705-6. [DOI] [PubMed] [Google Scholar]
- 237.Gray DS, Tien J, Chen CS. Repositioning of cells by mechanotaxis on surfaces with micropatterned Young’s modulus. J Biomed Mater Res A. 2003;66A(3):605–14. doi: 10.1002/jbm.a.10585. [DOI] [PubMed] [Google Scholar]
- 238.Trichet L, Le Digabel J, Hawkins RJ, Vedula SR, Gupta M, Ribrault C, et al. Evidence of a large-scale mechanosensing mechanism for cellular adaptation to substrate stiffness. Proc Natl Acad Sci USA. 2012;109(18):6933–8. doi: 10.1073/pnas.1117810109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Sunyer R, Jin AJ, Nossal R, Sackett DL. Fabrication of hydrogels with steep stiffness gradients for studying cell mechanical response. PLoS One. 2012;7(10):e46107. doi: 10.1371/journal.pone.0046107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Zaari N, Rajagopalan P, Kim SK, Engler AJ, Wong JY. Photopolymerization in microfluidic gradient generators: microscale control of substrate compliance to manipulate cell response. Adv Mater. 2004;16(23–24):2133–7. [Google Scholar]
- 241.Park J, Kim DH, Kim HN, Wang CJ, Kwak MK, Hur E, et al. Directed migration of cancer cells guided by the graded texture of the underlying matrix. Nat Mater. 2016;15(7):792–801. doi: 10.1038/nmat4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Kim DH, Seo CH, Han K, Kwon KW, Levchenko A, Suh KY. Guided cell migration on microtextured substrates with variable local density and anisotropy. Adv Funct Mater. 2009;19(10):1579–86. doi: 10.1002/adfm.200990041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Khung YL, Barritt G, Voelcker NH. Using continuous porous silicon gradients to study the influence of surface topography on the behaviour of neuroblastoma cells. Exp Cell Res. 2008;314(4):789–800. doi: 10.1016/j.yexcr.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 244.Arnold M, Hirschfeld-Warneken VC, Lohmueller T, Heil P, Blüemmel J, Cavalcanti-Adam EA, et al. Induction of cell polarization and migration by a gradient of nanoscale variations in adhesive ligand spacing. Nano Lett. 2008;8(7):2063–9. doi: 10.1021/nl801483w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Arnold M, Cavalcanti-Adam EA, Glass R, Blüemmel J, Eck W, Kantlehner M, et al. Activation of integrin function by nanopatterned adhesive interfaces. Chemphyschem. 2004;5(3):383–8. doi: 10.1002/cphc.200301014. [DOI] [PubMed] [Google Scholar]
- 246.Arnold M, Schwieder M, Blüemmel J, Cavalcanti-Adam EA, López-Garcia M, Kessler H, et al. Cell interactions with hierarchically structured nanopatterned adhesive surfaces. Soft Matter. 2009;5(1):72–7. doi: 10.1039/B815634D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Xiong J, Stehle T, Diefenbach B, Zhang R, Dunker R, Scott DL, et al. Crystal structure of the extracellular segment of integrin αVβ3. Science. 2001;294(5541):339–45. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Gautrot JE, Malmström J, Sundh M, Margadant C, Sonnenberg A, Sutherland DS. The nanoscale geometrical maturation of focal adhesions controls stem cell differentiation and mechanotransduction. Nano Lett. 2014;14(7):3945–52. doi: 10.1021/nl501248y. [DOI] [PubMed] [Google Scholar]
- 249.Bischofs IB, Safran SA, Schwarz US. Elastic interactions of active cells with soft materials. Phys Rev E. 2004;69(2):021911. doi: 10.1103/PhysRevE.69.021911. [DOI] [PubMed] [Google Scholar]
- 250.Bischofs IB, Schwarz US. Cell organization in soft media due to active mechanosensing. Proc Natl Acad Sci USA. 2003;100(16):9274–9. doi: 10.1073/pnas.1233544100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Dalby MJ, Biggs MJ, Gadegaard N, Kalna G, Wilkinson CD, Curtis AS. Nanotopographical stimulation of mechanotransduction and changes in interphase centromere positioning. J Cell Biochem. 2007;100(2):326–38. doi: 10.1002/jcb.21058. [DOI] [PubMed] [Google Scholar]
- 252.Kilian KA, Mrksich M. Directing stem cell fate by controlling the affinity and density of ligand–receptor interactions at the biomaterials interface. Angew Chem Int Ed. 2012;51(20):4891–5. doi: 10.1002/anie.201108746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Razafiarison T, Silván U, Meier D, Snedeker JG. Surface-driven collagen self-assembly affects early osteogenic stem cell signaling. Adv Healthc Mater. 2016;5(12):1481–92. doi: 10.1002/adhm.201600128. [DOI] [PubMed] [Google Scholar]
- 254.Siegel RW. Creating nanophase materials. Sci Am. 1996;275(6):74–9. [Google Scholar]
- 255.Webster TJ, Schadler LS, Siegel RW, Bizios R. Mechanisms of enhanced osteoblast adhesion on nanophase alumina involve vitronectin. Tissue Eng. 2004;7(3):291–301. doi: 10.1089/10763270152044152. [DOI] [PubMed] [Google Scholar]
- 256.Puckett SD, Lee PP, Ciombor DM, Aaron RK, Webster TJ. Nanotextured titanium surfaces for enhancing skin growth on transcutaneous osseointegrated devices. Acta Biomater. 2010;6(6):2352–62. doi: 10.1016/j.actbio.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 257.Yang Y, Liu D, Xie Y, Lee LJ, Tomasko DL. Low-temperature fusion of polymeric nanostructures using carbon dioxide. Adv Mater. 2007;19(2):251–4. [Google Scholar]
- 258.Yang Y, Cheng MMC, Hu X, Liu D, Goyette RJ, Lee LJ, et al. Low-pressure carbon dioxide enhanced polymer chain mobility below the bulk glass transition temperature. Macromolecules. 2007;40(4):1108–11. [Google Scholar]
- 259.den Braber ET, de Ruijter JE, Ginsel LA, von Recum AF, Jansen JA. Orientation of ECM protein deposition, fibroblast cytoskeleton, and attachment complex components on silicone microgrooved surfaces. J Biomed Mater Res. 1998;40(2):291–300. doi: 10.1002/(sici)1097-4636(199805)40:2<291::aid-jbm14>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 260.Andersson AS, Brink J, Lidberg U, Sutherland DS. Influence of systematically varied nanoscale topography on the morphology of epithelial cells. IEEE Trans Nanobioscience. 2003;2(2):49–57. doi: 10.1109/tnb.2003.813934. [DOI] [PubMed] [Google Scholar]
- 261.Choudhary S, Haberstroh KM, Webster TJ. Enhanced functions of vascular cells on nanostructured Ti for improved stent applications. Tissue Eng. 2007;13(7):1421–30. doi: 10.1089/ten.2006.0376. [DOI] [PubMed] [Google Scholar]
- 262.Koh LB, Rodriguez I, Venkatraman SS. Conformational behavior of fibrinogen on topographically modified polymer surfaces. Phys Chem Chem Phys. 2010;12(35):10301–8. doi: 10.1039/c001747g. [DOI] [PubMed] [Google Scholar]
- 263.Norde W, Horbett TA, Brash JL. Proteins at interfaces III: introductory overview. In: Horbett T, Brash JL, Norde W, editors. Proteins at interfaces III state of the art. Washington, DC: American Chemical Society; 2012. pp. 1–34. [Google Scholar]
- 264.Chirasatitsin S, Engler AJ. Detecting cell-adhesive sites in extracellular matrix using force spectroscopy mapping. J Phys Condens Matter. 2010;22(19):194102. doi: 10.1088/0953-8984/22/19/194102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Berning S, Willig KI, Steffens H, Dibaj P, Hell SW. Nanoscopy in a living mouse brain. Science. 2012;335(6068):551. doi: 10.1126/science.1215369. [DOI] [PubMed] [Google Scholar]
- 266.Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang M, et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466(7303):263–6. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Heil P, Spatz JP. Lateral shear forces applied to cells with single elastic micropillars to influence focal adhesion dynamics. J Phys Condens Matter. 2010;22(19):194108. doi: 10.1088/0953-8984/22/19/194108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Thompson MT, Berg MC, Tobias IS, Lichter JA, Rubner MF, Van Vliet KJ. Biochemical functionalization of polymeric cell substrata can alter mechanical compliance. Biomacromolecules. 2006;7(6):1990–5. doi: 10.1021/bm060146b. [DOI] [PubMed] [Google Scholar]
- 269.Lee M, Kang DK, Yang HK, Park KH, Choe SY, Kang C, et al. Protein nanoarray on Prolinker™ surface constructed by atomic force microscopy dip-pen nanolithography for analysis of protein interaction. Proteomics. 2006;6(4):1094–103. doi: 10.1002/pmic.200500392. [DOI] [PubMed] [Google Scholar]
- 270.Sahin O, Erina N. High-resolution and large dynamic range nanomechanical mapping in tapping-mode atomic force microscopy. Nanotechnology. 2008;19(44):445717. doi: 10.1088/0957-4484/19/44/445717. [DOI] [PubMed] [Google Scholar]
- 271.Ankam S, Suryana M, Chan LY, Moe AA, Teo BK, Law JB, et al. Substrate topography and size determine the fate of human embryonic stem cells to neuronal or glial lineage. Acta Biomater. 2013;9(1):4535–45. doi: 10.1016/j.actbio.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 272.Tan KK, Tann JY, Sathe SR, Goh SH, Ma D, Goh EL, et al. Enhanced differentiation of neural progenitor cells into neurons of the mesencephalic dopaminergic subtype on topographical patterns. Biomaterials. 2015;43:32–43. doi: 10.1016/j.biomaterials.2014.11.036. [DOI] [PubMed] [Google Scholar]
- 273.Unadkat HV, Hulsman M, Cornelissen K, Papenburg BJ, Truckenmuller RK, Carpenter AE, et al. An algorithm-based topographical biomaterials library to instruct cell fate. Proc Natl Acad Sci USA. 2011;108(40):16565–70. doi: 10.1073/pnas.1109861108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Gu Y, Zhu J, Xue C, Li Z, Ding F, Yang Y, et al. Chitosan/silk fibroin-based, Schwann cell-derived extracellular matrix-modified scaffolds for bridging rat sciatic nerve gaps. Biomaterials. 2014;35(7):2253–63. doi: 10.1016/j.biomaterials.2013.11.087. [DOI] [PubMed] [Google Scholar]
- 275.Liu X, Zhang F, Wang Q, Gao J, Meng J, Wang S, et al. Platelet-inspired multiscaled cytophilic interfaces with high specificity and efficiency toward point-of-care cancer diagnosis. Small. 2014;10(22):4677–83. doi: 10.1002/smll.201401530. [DOI] [PubMed] [Google Scholar]
- 276.Zhao L, Mei S, Chu P, Zhang Y, Wu Z. The influence of hierarchical hybrid micro/nano-textured titanium surface with titania nanotubes on osteoblast functions. Biomaterials. 2010;31(19):5072–82. doi: 10.1016/j.biomaterials.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 277.Kubo K, Tsukimura N, Iwasa F, Ueno T, Saruwatari L, Aita H, et al. Cellular behavior on TiO2 nanonodular structures in a micro-to-nanoscale hierarchy model. Biomaterials. 2009;30(29):5319–29. doi: 10.1016/j.biomaterials.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 278.Gittens R, McLachlan T, Olivares-Navarrete R, Cai Y, Berner S, Tannenbaum R, et al. The effects of combined micron-/submicron-scale surface roughness and nanoscale features on cell proliferation and differentiation. Biomaterials. 2011;32(13):3395–403. doi: 10.1016/j.biomaterials.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Tsukimura N, Yamada M, Iwasa F, Minamikawa H, Att W, Ueno T, et al. Synergistic effects of UV photofunctionalization and micro-nano hybrid topography on the biological properties of titanium. Biomaterials. 2011;32(19):4358–68. doi: 10.1016/j.biomaterials.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 280.Tocce EJ, Smirnov VK, Kibalov DS, Liliensiek SJ, Murphy CJ, Nealey PF. The ability of corneal epithelial cells to recognize high aspect ratio nanostructures. Biomaterials. 2010;31(14):4064–72. doi: 10.1016/j.biomaterials.2010.01.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Jia Z, Xiu P, Li M, Xu X, Shi Y, Cheng Y, et al. Bioinspired anchoring AgNPs onto micro-nanoporous TiO2 orthopedic coatings: trap-killing of bacteria, surface-regulated osteoblast functions and host responses. Biomaterials. 2016;75:203–22. doi: 10.1016/j.biomaterials.2015.10.035. [DOI] [PubMed] [Google Scholar]
- 282.Moffa M, Sciancalepore AG, Passione LG, Pisignano D. Combined nano- and micro-scale topographic cues for engineered vascular constructs by electrospinning and imprinted micro-patterns. Small. 2014;10(12):2439–50. doi: 10.1002/smll.201303179. [DOI] [PubMed] [Google Scholar]
- 283.López-Bosque MJ, Tejeda-Montes E, Cazorla M, Linacero J, Atienza Y, Smith KH, et al. Fabrication of hierarchical micro-nanotopographies for cell attachment studies. Nanotechnology. 2013;24(25):255305. doi: 10.1088/0957-4484/24/25/255305. [DOI] [PubMed] [Google Scholar]
- 284.Kim J, Bae WG, Choung HW, Lim KT, Seonwoo H, Jeong HE, et al. Multiscale patterned transplantable stem cell patches for bone tissue regeneration. Biomaterials. 2014;35(33):9058–67. doi: 10.1016/j.biomaterials.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 285.Yang K, Jung H, Lee HR, Lee JS, Kim SR, Song KY, et al. Multiscale, hierarchically patterned topography for directing human neural stem cells into functional neurons. ACS Nano. 2014;8(8):7809–22. doi: 10.1021/nn501182f. [DOI] [PubMed] [Google Scholar]
- 286.Bao L, Cheng X, Huang X, Guo L, Pang S, Yee A. Nanoimprinting over topography and multilayer three-dimensional printing. J Vac Sci Technol B. 2002;20(6):2881–6. [Google Scholar]
- 287.Eliason MT, Charest JL, Simmons BA, García AJ, King WP. Nanoimprint fabrication of polymer cell substrates with combined microscale and nanoscale topography. J Vac Sci Technol B. 2007;25(4):L31–4. [Google Scholar]
- 288.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294(5547):1708–12. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 289.Bryant SJ, Chowdhury TT, Lee DA, Bader DL, Anseth KS. Crosslinking density influences chondrocyte metabolism in dynamically loaded photocrosslinked poly(ethylene glycol) hydrogels. Ann Biomed Eng. 2004;32(3):407–17. doi: 10.1023/b:abme.0000017535.00602.ca. [DOI] [PubMed] [Google Scholar]
- 290.Park Y, Lutolf MP, Hubbell JA, Hunziker EB, Wong M. Bovine primary chondrocyte culture in synthetic matrix metalloproteinase-sensitive poly(ethylene glycol)-based hydrogels as a scaffold for cartilage repair. Tissue Eng. 2004;10(3–4):515–22. doi: 10.1089/107632704323061870. [DOI] [PubMed] [Google Scholar]
- 291.Fouchard J, Bimbard C, Bufi N, Durand-Smet P, Proag A, Richert A, et al. Three-dimensional cell body shape dictates the onset of traction force generation and growth of focal adhesions. Proc Natl Acad Sci USA. 2014;111(36):13075–80. doi: 10.1073/pnas.1411785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Hogrebe NJ, Gooch KJ. Direct influence of culture dimensionality on human mesenchymal stem cell differentiation at various matrix stiffnesses using a fibrous self-assembling peptide hydrogel. J Biomed Mater Res A. 2016;104(9):2356–68. doi: 10.1002/jbm.a.35755. [DOI] [PubMed] [Google Scholar]
- 293.Fischbach C, Chen R, Matsumoto T, Schmelzle T, Brugge JS, Polverini PJ, et al. Engineering tumors with 3D scaffolds. Nat Methods. 2007;4(10):855–60. doi: 10.1038/nmeth1085. [DOI] [PubMed] [Google Scholar]
- 194.Aljitawi OS, Li D, Xiao Y, Zhang D, Ramachandran K, Stehno-Bittel L, et al. A novel three-dimensional stromal-based model for in vitro chemotherapy sensitivity testing of leukemia cells. Leuk Lymphoma. 2014;55(2):378–91. doi: 10.3109/10428194.2013.793323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Talukdar S, Kundu SC. A non-mulberry silk fibroin protein based 3D in vitro tumor model for evaluation of anticancer drug activity. Adv Funct Mater. 2012;22(22):4778–88. [Google Scholar]
- 296.Bruce A, Evans R, Mezan R, Shi L, Moses BS, Martin KH, et al. Three-dimensional microfluidic tri-culture model of the bone marrow microenvironment for study of acute lymphoblastic leukemia. PLoS One. 2015;10(10):e0140506. doi: 10.1371/journal.pone.0140506. Erratum in: PLoS One 2015;10(12):e0146203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 2005;23(1):47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 298.Sun Y, Jallerat Q, Szymanski JM, Feinberg AW. Conformal nanopatterning of extracellular matrix proteins onto topographically complex surfaces. Nat Methods. 2015;12(2):134–6. doi: 10.1038/nmeth.3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Perschmann N, Hellmann JK, Frischknecht F, Spatz JP. Induction of malaria parasite migration by synthetically tunable microenvironments. Nano Lett. 2011;11(10):4468–74. doi: 10.1021/nl202788r. [DOI] [PubMed] [Google Scholar]
- 300.Aydin D, Louban I, Perschmann N, Blümmel J, Lohmüller T, Cavalcanti-Adam EA, et al. Polymeric substrates with tunable elasticity and nanoscopically controlled biomolecule presentation. Langmuir. 2010;26(19):15472–80. doi: 10.1021/la103065x. [DOI] [PubMed] [Google Scholar]
- 301.Li S, Wang X, Cao B, Ye K, Li Z, Ding J. Effects of nanoscale spatial arrangement of arginine-glycine-aspartate peptides on dedifferentiation of chondrocytes. Nano Lett. 2015;15(11):7755–65. doi: 10.1021/acs.nanolett.5b04043. [DOI] [PubMed] [Google Scholar]
- 302.Kruss S, Erpenbeck L, Schön MP, Spatz JP. Circular, nanostructured and biofunctionalized hydrogel microchannels for dynamic cell adhesion studies. Lab Chip. 2012;12(18):3285–9. doi: 10.1039/c2lc40611j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Chaudhuri O, Koshy ST, Branco da Cunha C, Shin JW, Verbeke CS, Allison KH, et al. Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat Mater. 2014;13(10):970–8. doi: 10.1038/nmat4009. [DOI] [PubMed] [Google Scholar]
- 304.Madl CM, Katz LM, Heilshorn SC. Bio-orthogonally crosslinked, engineered protein hydrogels with tunable mechanics and biochemistry for cell encapsulation. Adv Funct Mater. 2016;26(21):3612–20. doi: 10.1002/adfm.201505329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.White ES. Lung extracellular matrix and fibroblast function. Ann Am Thorac Soc. 2015;12(Suppl 1):S30–3. doi: 10.1513/AnnalsATS.201406-240MG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Kisseleva T, Brenner DA. Mechanisms of fibrogenesis. Exp Biol Med. 2008;233(2):109–22. doi: 10.3181/0707-MR-190. [DOI] [PubMed] [Google Scholar]
- 307.Burdick JA, Murphy WL. Moving from static to dynamic complexity in hydrogel design. Nat Commun. 2012;3:1269. doi: 10.1038/ncomms2271. [DOI] [PubMed] [Google Scholar]
- 308.Tibbitt MW, Anseth KS. Dynamic microenvironments: the fourth dimension. Sci Transl Med. 2012;4(160):160ps24. doi: 10.1126/scitranslmed.3004804. [DOI] [PubMed] [Google Scholar]
- 309.Guvendiren M, Burdick JA. Stiffening hydrogels to probe short- and long-term cellular responses to dynamic mechanics. Nat Commun. 2012;3:792. doi: 10.1038/ncomms1792. [DOI] [PubMed] [Google Scholar]
- 310.Young JL, Engler AJ. Hydrogels with time-dependent material properties enhance cardiomyocyte differentiation in vitro. Biomaterials. 2011;32(4):1002–9. doi: 10.1016/j.biomaterials.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Khetan S, Guvendiren M, Legant WR, Cohen DM, Chen CS, Burdick JA. Degradation-mediated cellular traction directs stem cell fate in covalently crosslinked three-dimensional hydrogels. Nat Mater. 2013;12(5):458–65. doi: 10.1038/nmat3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Le DM, Kulangara K, Adler AF, Leong KW, Ashby VS. Dynamic topographical control of mesenchymal stem cells by culture on responsive poly(ε-caprolactone) surfaces. Adv Mater. 2011;23(29):3278–83. doi: 10.1002/adma.201100821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science. 2009;324(5923):59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.