Abstract
Pathogenic fungi must extend filamentous hyphae across solid surfaces to cause diseases of plants. However, the full inventory of genes which support this is incomplete and many may be currently concealed due to their essentiality for the hyphal growth form. During a random T-DNA mutagenesis screen performed on the pleomorphic wheat (Triticum aestivum) pathogen Zymoseptoria tritici, we acquired a mutant unable to extend hyphae specifically when on solid surfaces. In contrast “yeast-like” growth, and all other growth forms, were unaffected. The inability to extend surface hyphae resulted in a complete loss of virulence on plants. The affected gene encoded a predicted type 2 glycosyltransferase (ZtGT2). Analysis of >800 genomes from taxonomically diverse fungi highlighted a generally widespread, but discontinuous, distribution of ZtGT2 orthologues, and a complete absence of any similar proteins in non-filamentous ascomycete yeasts. Deletion mutants of the ZtGT2 orthologue in the taxonomically un-related fungus Fusarium graminearum were also severely impaired in hyphal growth and non-pathogenic on wheat ears. ZtGT2 expression increased during filamentous growth and electron microscopy on deletion mutants (ΔZtGT2) suggested the protein functions to maintain the outermost surface of the fungal cell wall. Despite this, adhesion to leaf surfaces was unaffected in ΔZtGT2 mutants and global RNAseq-based gene expression profiling highlighted that surface-sensing and protein secretion was also largely unaffected. However, ΔZtGT2 mutants constitutively overexpressed several transmembrane and secreted proteins, including an important LysM-domain chitin-binding virulence effector, Zt3LysM. ZtGT2 likely functions in the synthesis of a currently unknown, potentially minor but widespread, extracellular or outer cell wall polysaccharide which plays a key role in facilitating many interactions between plants and fungi by enabling hyphal growth on solid matrices.
Author summary
All plant-pathogenic fungi must grow hyphae across host tissues and cells to establish diseases. We have identified a single glycosyltransferase enzyme from the pleomorphic wheat pathogen Zymoseptoria tritici which functions specifically to enable hyphal growth on solid surfaces, and is therefore essential for fungal disease of wheat plants. ZtGT2 orthologues are present in most ascomycete filamentous fungi, and we show that the orthologous gene from the distantly related wheat ear infecting fungus, Fusarium graminearum, is also required for hyphal growth and virulence. Conversely the gene is completely absent from the genomes of most ascomycete yeast species, which do not form true hyphae. The data suggests that ZtGT2 orthologues may have played an important role in the evolution of pathogenic fungi, by enabling hyphal growth on solid surfaces. It is likely that this capability, which is also a requirement for the establishment of mutualistic interactions and for saprophytic growth, arose “inadvertently” from the need for fungi to adopt the characteristic filamentous lifestyle which enables them to seek out and access distal nutrient sources.
Introduction
Micro-organisms have evolved many different mechanisms to enable them to cause diseases of plants. Some of these mechanisms are pathogen species-host plant specific. For example, the evolution and deployment of suites of secreted “effector” proteins, allow pathogens to manipulate components of plant immunity to support infection [1]. The molecular interplay which underpins this exquisite control of host-pathogen interactions has become the focus of considerable research aimed at improving disease resistance in crop plants [2].
However, prior to engaging fully with plant immunity, fungal pathogens must first adhere to, recognise, respond to, and then grow on or through plant surfaces to initiate infection [3]. All known fungi infecting the aerial tissues of plants (flowers, leaves, stems) are referred to as “filamentous”, meaning that although they arrive on plant surfaces as either air- or water-borne spores, they subsequently transition into filamentous hyphae to grow over or through tissues and cells [4]. This discriminates filamentous fungi from the model ascomycete yeast species Saccharomyces cerevisiae and Schizosaccharomyces pombe and their close relatives, which are free living and do not form true hyphae. Many fungi which are pathogenic on animals can switch between yeast and hyphal growth forms (dimorphic), but most frequently in these cases the infectious module are often the spores, resulting in “yeast-like” infections [5–7]. In contrast, almost all (if not all) major plant infecting fungi use hyphal growth to initiate and/or complete infection. Therefore, hyphal growth over solid surfaces is a pre-requisite step for potentially all fungal diseases of plants.
Fungal hyphae are structurally supported by complex outer cell walls, which have multiple layered components of proteins and sugars and provide the strength and flexibility which enable growth, and sensing of growth surfaces [8, 9]. Major structural components of fungal cell walls include the complex polysaccharides chitin and beta glucans, which are present in most species. Both of these polysaccharides are also known to be triggers of plant defences, via their recognition by plant plasma membrane immune receptors [10]. However, fungal cell walls also contain various other polysaccharides including alpha glucans [11, 12] and other structurally undefined, but likely important molecules. Moreover, as fungi grow across and through solid surfaces they are also likely to deploy some form of extracellular matrix (ECM) to counteract surface-derived frictional and shear stresses [13, 14]. The importance of ECM is widely recognised for mammalian cells, which produce a matrix which contains a complex linear polysaccharide called Hyaluronan, as a key component [15, 16]. In contrast, whilst the presence of ECM surrounding fungal cells and hyphae has been detected via various staining and microscopy methods [17, 18], the precise polysaccharide components therein have only in rare cases been structurally defined or functionally characterised [19, 20]
The ascomycete filamentous fungus Zymoseptoria tritici (class Dothideomycete) is the causal agent of Septoria tritici blotch (STB) disease of wheat leaves, which represents one of the most economically important crop diseases of wheat worldwide [21, 22]. Z. tritici is also regarded as an emerging model fungus, due in part to its ability to grow in several different morphological states (or forms) depending on environmental conditions [23–25]. For this reason, Z. tritici is referred to as a pleomorphic fungus [26], able to grow in at least two different growth forms. One advantage of experimenting with pleomorphic (or dimorphic) fungi is that genes which are essential for life (or the recovery of stably transformed gene deletion strains) for only one growth form can still be characterised in viable cells growing in the alternative growth form(s). It is more problematic to define the functions of putative essential genes in fungi with only a single dominant growth form. Consistent with most plant pathogenic fungi, Z. tritici infection of wheat leaves begins with spores alighting, either through wind or rain splash, onto leaf surfaces. Initial infection then requires the spores to germinate and grow hyphae across the leaf surface and into stomatal pores [25, 27]. Once inside leaves, Z. tritici secretes a plethora of important effector proteins during hyphal growth [25], including a broadly conserved Lysin domain (LysM) effector protein which masks fungal chitin from plant immune receptors [28–32]. It is currently unknown what factors can trigger the up-regulation of effector production during phases of leaf infection.
This current study derives from a forward’s genetic screen aimed to identify novel virulence factors from Z. tritici, which contribute to its ability to cause plant disease. The data we present reports identification of a widely conserved but discontinuously distributed glycosyltransferase which plays a crucial role in pathogenicity of not only Z. tritici, but also a distinct and taxonomically un-related pathogen of wheat, Fusarium graminearum. We demonstrate its key role in enabling pathogenicity is to allow hyphal filaments to extend over solid surfaces. The distribution of orthologous genes within the fungal kingdom, suggests that this role in virulence likely evolved “inadvertently” from a more generic need for fungi to scavenge for nutrients through extending hyphal filaments over solid surfaces. The current study also identified an unexpected role for this protein in the regulation of pathogen effector gene expression.
Results and discussion
Isolation of a non-pathogenic Z. tritici T-DNA mutant unable to extend hyphae on solid surfaces
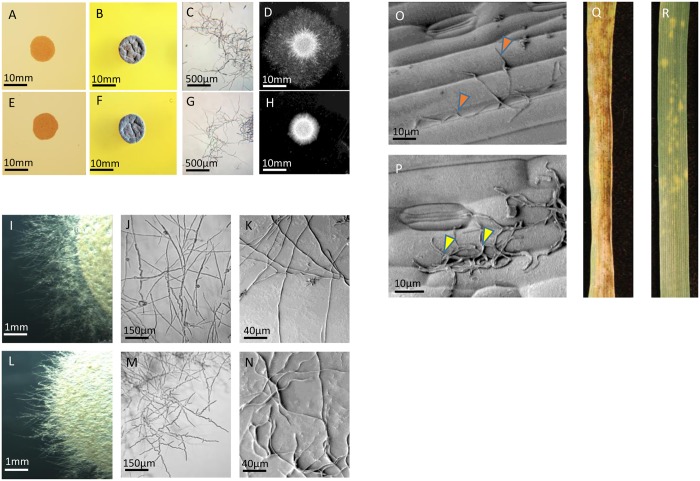
Z. tritici can grow in a “yeast-like” budding form or extend true hyphal filaments on agar plates depending on temperature and nutritional status. Fig 1 displays the growth phenotypes of the wild type (WT) fungus on different nutrient agar plates, at different temperatures, and in sterile liquid water. At 16°C on solid nutrient-rich Yeast extract peptone dextrose (YPD) agar, WT Z. tritici spores undergo yeast-like budding generating a mass of pink coloured spores (Fig 1A). On the same plates incubated at 25°C the fungus subsequently forms extensive networks of melanised aerial hypha over the top of the colony, with no direct contact with the underlying agar (Fig 1B). In nutrient limiting sterile shaking water flasks, spores germinate to form hyphal filaments in suspension (Fig 1C) and on solid water agar plates at 25°C spores germinate hyphae which extend radially from the spore droplet across the surrounding agar (1D). Fig 1E to 1H display the corresponding growth morphologies of a transposable DNA (T-DNA) tagged mutant of Z. tritici called 23–170 under the same conditions. The mutant had identical growth under all conditions (Fig 1E to 1G) except for growth on solid water agar, where hyphal growth was severely impaired (Fig 1H). Yeast-like growth of 23–170 on YPD agar occurred at a rate comparable to the WT strain and spore morphologies were macroscopically indistinguishable (S1 Fig). The filamentous growth defect of 23–170 on solid water agar was also seen at all tested agar strengths (S2 Fig) and during growth on Czapek-Dox and PDA agar, each of which can also support some level of filamentous growth by Z. tritici after protracted incubations (S3 Fig). Closer inspection of the hyphal filaments of both WT and 23–170 on water agar by both light microscopy and scanning electron microscopy (SEM) revealed that, in contrast to the mostly long and straight filaments produced by the WT strain (Fig 1L and 1K), the 23–170 filaments were both shorter and grew in a sinusoidal (“wavy”) manner (Fig 1L–1N). These data suggested that the mutation(s) present in 23–170 rendered it incapable of normally extending hyphae when in contact with a solid surface. Significantly, this phenotype was supported by wheat leaf infection data. In contrast to the WT strain which could rapidly elaborate hyphal filaments on the surfaces of wheat leaves (Fig 1O) and subsequently cause full disease (Fig 1Q), the 23–170 mutant displayed no clear hyphal growth on leaf surfaces (Fig 1P). This resulted in a dramatic loss of disease causing ability (Fig 1R). On occasions, some limited chlorosis was observed on leaves inoculated with 23–170, but these never progressed into necrotic lesions and no fungal sporulation was ever observed (Fig 1R).
Fig 1. Characterisation of a non-pathogenic T-DNA mutant of Z. tritici (23–170) unable to extend hyphae on solid surfaces.
(A-D) Typical growth characteristics of wild type (WT) Z. tritici on rich nutrient agar at 16°C (A); rich agar at 25°C (B); sterile liquid water (C) and solid water agar (D). (E-H) display the comparable growth morphologies of mutant 23–170. Note the severely impaired hyphal growth from the colony on solid water agar (H). (I-K) micrographs displaying hyphal morphology of wild type Z. tritici on solid water agar. (L-N) comparable micrographs displaying morphology of short, sinusoidal hyphae formed by 23–170. (O) SEM analysis of wild type fungal inoculated wheat leaf surfaces at 48 hpi. Arrows highlight surface growing hyphae. (P) SEM of 23–170 mutant cells at 48hpi on wheat leaf surfaces. Arrows highlight un-germinated fungal spores. (Q) Wheat leaf disease symptoms photographed 14 days after inoculation with wild type fungus. (R) Comparable levels of leaf disease caused by the 23–170 mutant at 14 dpi.
The 23–170 mutant phenotype results from inactivation of a gene encoding a predicted type 2 glycosyltransferase
Thermal asymmetric interlaced (TAIL)-PCR was used to identify the T-DNA insertion site in 23–170, with recovered sequences then subjected to a Blastn analysis against the fully sequenced genome of strain IPO323 [33] (http://genome.jgi.doe.gov/pages/search-for-genes.jsf?organism=Mycgr3). All sequenced PCR products mapped an identical T-DNA insertion site within the second intron of a gene model present on the antisense strand of Chromosome 1, between nucleotide positions 1786483-1788643 (Fig 2A). This gene model was well supported by RNA sequencing (RNAseq) raw read mapping data (Fig 2B), and the gene was annotated as encoding a type 2 glycosyltransferase (GT2) represented by Genbank ID XP_003857553.1 (Fig 2C). The T-DNA insertion likely resulted in a severely truncated protein (Fig 2D). To validate whether loss of the Z. tritici GT2 (ZtGT2) function was responsible for all the phenotypes seen for 23–170, the wild-type gene plus upstream and downstream regions was transformed back into strain 23–170 for complementation analysis. In addition, the ZtGT2 gene was also subject to independent targeted deletion in the WT fungus. Full hyphal growth on water agar and plant disease causing ability were restored by complementation of 23–170 with the native gene, and the targeted ΔZtGT2 mutants displayed all the original 23–170 mutant phenotypes (Fig 2D) including short “wavy” hyphal formation on the surface of all tested agar plates (S4 Fig). Thus, we conclude that the ZtGT2 protein plays an essential role in supporting the virulence of Z. tritici through enabling hyphal growth over solid surfaces.
Fig 2. All mutant phenotypes of 23–170 result from inactivation of a gene encoding a putative type 2 glycosyltransferase (ZtGT2).
(A) The gene model structure identified through TAIL-PCR analysis of the T-DNA insertion site in mutant 23–170. (B) RNAseq raw read mapping confirmed the predicted gene structure and highlights the position of the left border T-DNA insertion. (C) Functional annotation of the tagged gene highlighting the position of the T-DNA left border relative to the predicted type 2 glycosyltransferase catalytic domain. (D) The predicted amino acid sequence of ZtGT2 highlighting the protein region truncated by T-DNA insertion in brown font. The underlined region indicates the peptide sequence chosen for antibody generation (E) Complementation of the 23–170 mutant strain with the wild-type ZtGT2 gene (23–170::GT2comp) restores hyphal growth on solid surfaces and virulence on plants. Independent targeted deletion of ZtGT2 in the wild-type fungus (ΔZtGT2-19) results in the same aberrant hyphal growth and loss of virulence phenotypes shown for 23–170.
Orthologues of ZtGT2 are widely but discontinuously distributed within the fungal kingdom
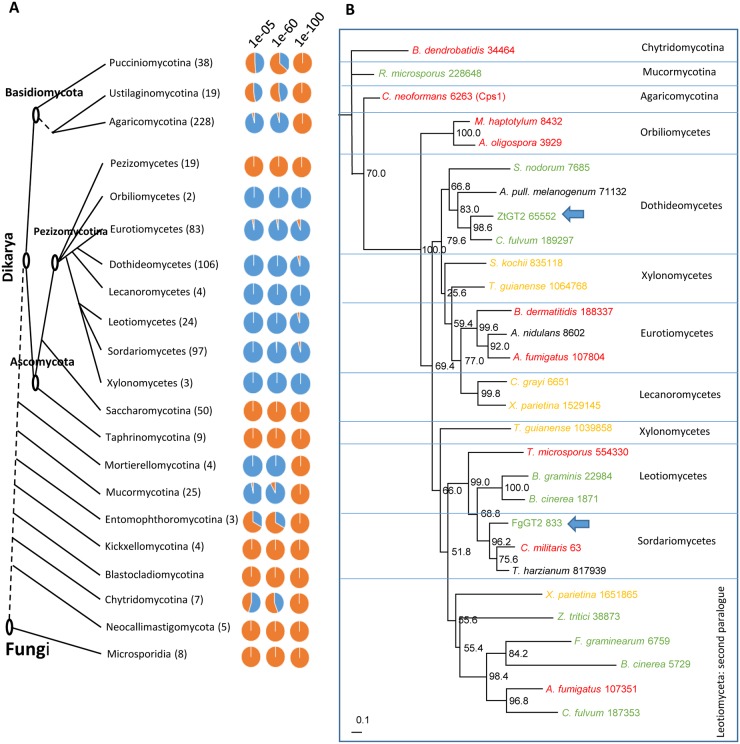
A Blastp analysis using the mature ZtGT2 protein sequence at expect (e-value) cut-off thresholds of 1.0e-100, 1.0e-60 and 1.0e-5 was performed on the predicted proteomes of 823 fungi present at the JGI Mycocosm portal [34] representing 20 different taxonomic classes or sub phylum (Fig 3A and S1 Table). Overall, proteins with varying degrees of similarity to ZtGT2, were identified in many of the species analysed including some of the earliest evolved fungal groups (Fig 3A and S1 Table). Phylogenetic analysis on a subset of transcripts representative of taxonomically distinct higher (ascomycete and basidiomycetes) and lower fungi, with parasitic, mutualistic or saprotrophic lifestyles (S2 Table), confirmed that orthologues of ZtGT2 were found in accordance with the fungal species tree and were present in some lower fungi (eg Chytrids and Mucor species) as well as many, but not all, basidiomycetes (Fig 3B). In contrast, most filamentous ascomycete fungi possess an orthologue of ZtGT2 irrespective of whether they were parasitic, mutualistic or saprotrophic. Numerous independent duplications were detected within basidiomycete species, but within ascomycetes, the 2nd Leotiomyceta paralogues grouped together, suggesting one duplication event there. (Fig 3B and S1 Table). A single paralogue of ZtGT2 was identified in Z. tritici (protein model 38873 in Fig 3B). The dramatic phenotypes observed for ΔZtGT2 mutants suggest this paralogue does not have an overlapping functionality. Interestingly no ascomycete species belonging to the class Pezizomycetes possessed orthologues, or any similar proteins whatsoever, to ZtGT2, highlighting a discontinuous distribution within this kingdom most likely arising from gene loss (Fig 3A and S1 Table). This distribution was further emphasised by the fact that no orthologues (or proteins with any similarity even at Blastp e-5) were identified in the genomes (proteomes) of all 59 ascomycete yeasts including all tested members of the Saccharomycotina and Taphrinomycotina, which include the model species Saccharomyces cerevisiae, Schizosaccharomyces pombe and the yeast-like human pathogenic Candida species (Fig 3A and S1 Table). The discontinuous distribution of proteins with similarity to ZtGT2 was also very evident within the tested Basidiomycetes (Fig 3A and S1 Table) with orthologues most frequently observed in the Agaricomycotina, represented by the gene model 6263 (Cps1) from Cryptococcus neoformans in Fig 3B.
Fig 3. ZtGT2 orthologues show a discontinuous distribution throughout the fungal tree of life but are present in saprophytes, mutualists and pathogens.
(A) Diagrammatic representation of the fungal tree of life adapted from the JGI MycoCosm genome portal (http://genome.jgi.doe.gov/programs/fungi/index.jsf). Taxonomic tree distances are only representative and do not accurately indicate evolutionary time. Each fungal taxonomic class or sub phylum is shown with brackets highlighting the number of species genomes present in each group analysed. Pie charts then display the number of species genomes which have a at least one similar protein to ZtGT2 at each of the indicated Blastp expect thresholds. Blue shading indicates the number of species with at least one similar proteins. Orange shading indicates the number of species with no similar proteins. (B) Maximum Likelihood phylogenetic tree displaying representative sequence orthologues to ZtGT2 from across the fungal tree of life. Node labels indicate percentage bootstrap support (500 replicates). Where similar genes were detected orthology is inferred by correspondence to the fungal species tree. Species names in green font indicate known plant pathogens, those in red font indicate known animal pathogens and those in yellow indicate those with mutualistic lifestyles. A true saprophytic lifestyle is included in the form of Aspergillus nidulans and other species in black font have currently ambiguous lifestyles. Numbers following species names represent the protein model Id retrievable from the JGI Mycocosm species genome portal.
The orthologous gene from the unrelated ascomycete wheat pathogen Fusarium graminearum is also important for hyphal growth and virulence on plants
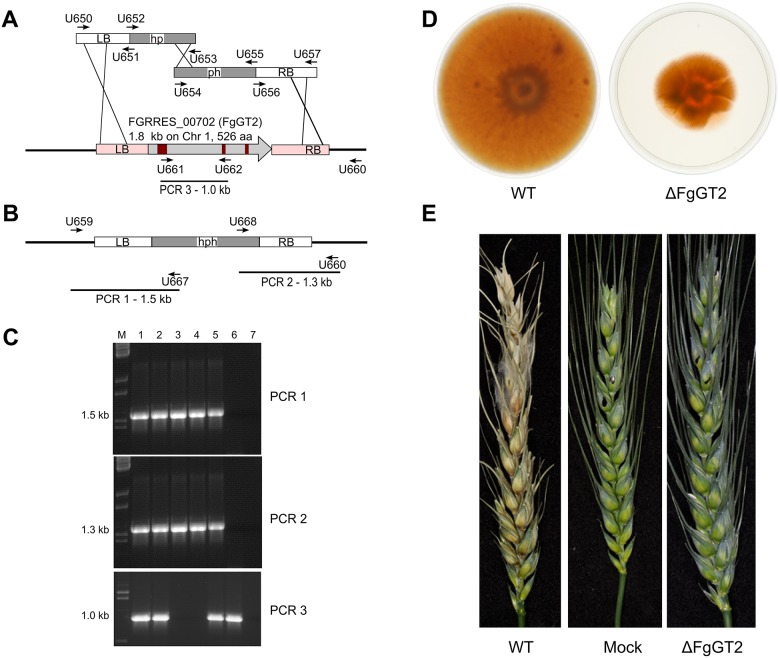
To test whether orthologues of ZtGT2 conferred the same function in other ascomycete fungi, we attempted to generate deletion strains in the wheat ear pathogen, Fusarium graminearum, a member of the taxonomically distant Sordariomycete class of fungi (Fig 3A and highlighted in Fig 3B). The gene selected for testing was FG00702.1 (http://fungi.ensembl.org/Fusarium_graminearum/Info/Index) with Blastp homology of 2.74e-142 to ZtGT2, encompassing 51% amino acid identity spanning 78.9% of the protein model. Phylogenetic analysis on transcripts confirmed the likely 1 to 1 orthology between ZtGT2 and FgGT2 (Fig 3B). F. graminearum is a rapidly growing filamentous fungus which unlike Z. tritici exhibits no known yeast-like growth form. Using standard protocols which select for hygromycin resistant transformants on solid agar [35], we were unable to recover any homokaryotic FgGT2 deletion strains, but instead recovered numerous heterokaryons. PCR analysis on these strains demonstrated that they possessed both a wild-type allele in addition to a deleted allele of FgGT2, and they grew hyphae at normal rates on agar plates (S5 Fig). It is known that attempts to delete “essential” genes in multinucleate fungi places strong selective pressure on the formation of such heterokaryons [36]. In view of this, and the phenotype described for ΔZtGT2 mutants, we modified the transformation protocol to perform selection in liquid medium instead of on solid agar (see Methods). This approach led to the acquisition of two independent homokaryotic deletion strains of FgGT2 (Fig 4C). Significantly both mutant strains were severely impaired in radial hyphal growth on solid agar (Fig 4D) and completely impaired in virulence towards wheat ears when either point inoculated via droplet, or sprayed across the entire wheat ear (Fig 4E). In addition to the radial growth defect seen on the surface of agar, the ΔFgGT2 mutants were also tested for ability to grow into (penetrate) agar plates through cellophane sheets [37]. This revealed that mutants were still capable of this type of invasive growth. However, this assay again demonstrated that subsequent radial growth on the agar surface was strongly affected (S6 Fig). This data highlights that invasive growth, which represents a key additional feature of F. graminearum infection of plants not known to occur in Z. tritici, does not require functional FgGT2 (S6 Fig).
Fig 4. The ZtGT2 orthologue from the wheat ear blight pathogen Fusarium graminearum (FgGT2) is required for hyphal growth on solid surfaces and virulence.
The F. graminearum gene FGRRES_00702 (http://fungi.ensembl.org/Fusarium_graminearum/Info/Index) orthologous to ZtGT2 was functionally characterized by gene deletion using a selection step in liquid rather than solid medium. (A) Genomic left border and right border regions (white bars) were amplified with primers and fused to parts of the hph hygromycin resistance gene. Fused PCR fragments were used in a split-marker strategy to replace FGRRES_00702. Horizontal black bars represent genomic areas outside the replacement construct, vertical black bars represent 3 introns. (B) Anticipated diagnostic PCR for successful gene replacement of FGRRES_00702. (C) Results of diagnostic PCR and expected sizes indicated in (A) and (B). Loadings are: M—λ DNA-BstEII digest, 1–5 transformants MU424 to MU428, WT, no DNA. MU426 and MU427 have the ΔFGRRES_00702 null allele and lost the 1.0 kb GT2 fragment. (D) Severely reduced growth for the FgGT2 null allele mutants on the surface of potato dextrose agar plates after 8 days incubation compared to the wild-type PH-1 strain. (E) Wheat ears inoculated with FgGT2 null mutant and wild-type strain 13 days post-inoculation. Spore-droplet inoculated spikelets are marked with black dots. For the mock inoculation, only water was used.
ZtGT2 transcripts accumulate during hyphal growth and mutant cells display outer cell wall abnormalities
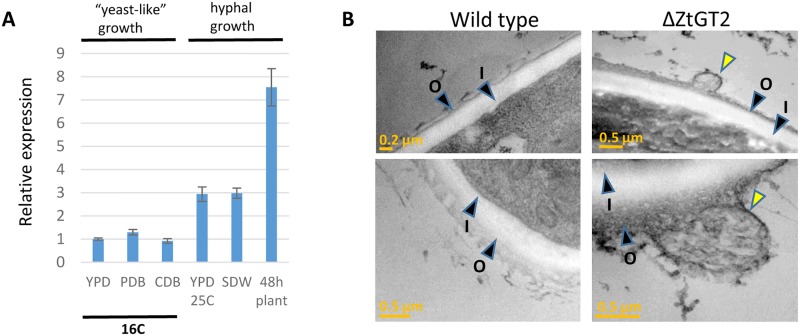
Quantitative Real-Time PCR (qPCR) gene expression profiling demonstrated that ZtGT2 was expressed most strongly under conditions which favour hyphal growth of Z. tritici, including elevated temperatures, low nutrients and early (48h) growth on wheat leaf surfaces (Fig 5A). To ascertain the likely protein localisation of ZtGT2, we generated a peptide antibody. Western analysis demonstrated that this antibody cross reacted with a specific ~52kDa protein present in wild type cells but absent from ΔZtGT2 mutants (S7 Fig). This protein was detected in the low speed cell wall pellet of fungal cells extracted in detergent-free buffer. In contrast, no protein was present in the soluble fraction, and little to none in the high speed microsomal fraction. This suggested that ZtGT2 may be present in, or attached to, the fungal cell wall (S7 Fig).
Fig 5. ZtGT2 transcripts accumulate under conditions which stimulate hyphal growth and mutant cells display an abnormal outer cell wall structure.
(A) Real-Time qPCR analysis on ZtGT2 expression. Overall expression is relatively low in three different liquid culture media (Yeast extract peptone dextrose broth (YPD); Potato dextrose broth (PDB) and Czapek-Dox (CDB) broth) grown at 16°C. Transcript levels are increased during growth in YPD at 25°C and during growth in liquid water (24h) which both stimulate more hyphal growth. Highest transcript accumulation overall was observed during hyphal growth on wheat leaf surfaces at 48 hpi. Data illustrate the results of three replicate experiments with standard error bars deriving from the combined data. (B) Transmission electron micrographs display abnormal structures on the outer cell wall of ΔZtGT2 mutants (highlighted with yellow arrowhead). These structures were observed only in ΔZtGT2 mutants and seen in ~50% of all sections analysed. The outer (O) and inner (I) cell wall regions are highlighted with black arrowheads.
Analysis of cell wall ultrastructure by TEM, following collection of yeast-like spores from the surfaces of solid agar plates, revealed the frequent presence of outer wall surface irregularities, which appeared as bulges, protrusions or as breakages (Fig 5B and S8 Fig). These were observed in over 50% of all sections examined from mutant cells and were never seen in sections of WT cells, suggesting that ZtGT2 plays a role in generating or maintaining an outer wall component. We then sought to determine whether there were any differences in monosaccharide levels (glucose, mannose and galactose) of the alcohol insoluble polysaccharides deriving from either the cell wall or culture filtrates of ΔZtGT2 relative to WT strains. We were unable to determine any quantitative differences in these monosaccharides deriving from total cell walls (S9A Fig). Comparable analysis of the ethanol precipitated culture filtrates revealed an unexpected increase in monosaccharides from ΔZtGT2 relative to WT (S9B Fig). Thus, overall the results were inconclusive but we detected no depletion of materials due to loss of ZtGT2. This may suggest that the product of ZtGT2 is a relatively minor component of either the fungal cell wall or extracellular matrix. In this case its overall contribution to the total monosaccharide composition may be masked by the more abundant chitin, beta- and alpha- glucans. It is also possible that loss of ΔZtGT2 function could induce changes, such as the alterations seen in outer cell wall structure (Fig 5B and S8 Fig) that could result in / from changes in the levels of other polysaccharides, influenced indirectly by loss of ZtGT2 function.
We also developed assays to determine whether the outer wall irregularities gave rise to changes in the ability of conidia to adhere to leaf surfaces (S10 Fig), or to plastic (hydrophobic) surfaces (S10 Fig). Like most fungi which are pathogenic on leaves of plants, the spores of Z. tritici adhere very strongly and rapidly to hydrophobic surfaces such as those posed by waxy leaf surfaces [3]. No change in the ability of ΔZtGT2 mutant spores to adhere to either of these hydrophobic surfaces was detected (S10 Fig) demonstrating that the macroscopic differences observed between the outer walls of each strain did not compromise surface attachment. Similarly, ΔZtGT2 mutant spores displayed no altered sensitivity to chemical de-stabilisation of chitin synthesis (Calcoflour white), beta glucan function (Caspofungin), temperature (30°C), oxidative (H2O2) or osmotic (sorbitol) stress (S11 Fig), suggesting that major cell wall and membrane components were not strongly affected by gene loss.
ZtΔGT2 mutants retain ability to respond to plant surfaces, but constitutively overexpress chitin-binding effectors and transmembrane proteins
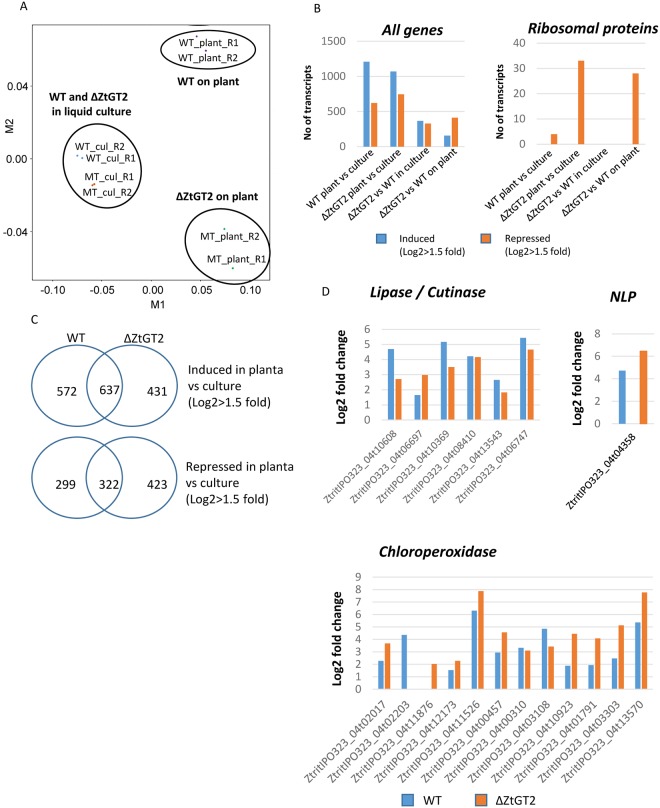
Based upon the abnormalities seen on the outer wall surfaces of ΔZtGT2 mutants, and their failure to germinate hyphae on leaf surfaces, we speculated that leaf surface-sensing may be compromised in the mutants. To test this, we performed RNAseq based whole genome expression profiling of early leaf infection compared with growth in liquid culture. Materials were sampled as shown in S12 Fig to allow several pairwise analyses on the fungal and plant transcriptomes. Global Principle Components Analysis (PCA) on the fungal datasets highlighted that, relative to growth of WT and ΔZtGT2 strains in liquid culture, 48 h growth on leaf surfaces distinguished their respective transcriptomes the most (Fig 6A and S3 Table). Much of this effect was attributed to relative growth rates on wheat leaf surfaces, as illustrated by the repression of many ribosomal protein encoding genes in ΔZtGT2 on the leaf surface. This feature was not observed when comparing the two strains transcriptomes in liquid culture, which further emphasised the key role of ZtGT2 in regulating contact-dependent fungal growth (Fig 6B). Despite this, many genes normally expressed by the WT fungus on leaves relative to liquid culture, were also similarly expressed by ΔZtGT2 (Fig 6C and S3 Table). These included the characteristic early transcriptional up-regulation of suites of secreted protein encoding genes including Lipases, Cutinases, Necrosis-inducing proteins (NLP) and Chloroperoxidases [24, 38–40] (Fig 6D). These data highlight that whilst hyphal growth is rapidly impaired, ΔZtGT2 can still sense and react transcriptionally to the leaf surface environment. The fungal leaf infection data also highlighted several protein families which are transcriptionally upregulated by the WT fungus on leaves, but which are not by ΔZtGT2. One example being several CFEM domain containing membrane-spanning proteins [41] which were strongly negatively affected in ΔZtGT2, both in liquid culture and even more so on leaf surfaces (S13 Fig). This data suggests these genes may play key roles in regulating processes associated with elongating hyphae during normal plant infection by the WT fungus.
Fig 6. Global RNAseq expression profiling highlights that ΔZtGT2 mutants are rapidly arrested on leaf surfaces but still retain many early transcriptional responses.
(A) Global clustering analysis of fungal RNAseq data from growth of wild type and ΔZtGT2 mutants in liquid culture, and at 48h after leaf inoculation. Key to data point labelling- WT_plant_R1 indicates wild type fungus on plant surface 1st replication; MT_cul_R2 indicates ΔZtGT2 mutant in YPD culture broth 2nd replicate sample. (B) Rapid arrest of growth on wheat leaf surfaces is indicated by dramatically reduced expression of many genes encoding ribosomal protein in ΔZtGT2. (C) Many genes induced and repressed by the WT fungus following leaf inoculation retain the same dynamics in ΔZtGT2 mutants. (D) Many classes of early expressed genes encoding secreted proteins are up-regulated to equivalent levels in both the WT and ΔZtGT2 mutant strains indicating surface sensing is still retained. All data is expressed relative to expression levels detected in the equivalent fungal liquid culture. (NLP = necrosis and ethylene inducing protein).
The most strongly up-regulated genes in ΔZtGT2 growing in liquid culture encoded secreted or membrane bound proteins. Table 1 highlights that amongst the “top 30” genes upregulated in the ΔZtGT2 mutant relative to WT, twenty-eight fell into these categories. Seventeen of these 28 were also upregulated in the WT fungus at 48h on leaf surfaces relative to its growth in liquid culture (Table 1). This suggested that loss of ZtGT2 function stimulated some transcriptional changes to occur which are commonly induced early by the fungus on wheat leaf surfaces. Amongst this set of genes was one encoding a functionally validated effector protein, Zt3LysM (Gene Id ZtritIPO323_04t03143 in Table 1 and S14 and S15 Figs), which binds to chitin fragments and mediates the evasion of plant chitin-triggered immunity [31, 32]. This suggests that the loss of normal outer cell wall structure in ΔZtGT2 mutants may mimic changes which usually occur following inoculation onto plants, and that this may serve to trigger increased expression of the Zt3LysM effector.
Table 1. The Top30 most upregulated genes in ΔZtGT2 vs WT fungus growing in liquid medium.
| *Gene_id new Ensembl | **Original Id Ensembl | FPKM WT | FPKM ΔZtGT2 | Log2 fold change ΔZtGT2/WT | Blastp annotation (NCBI) | Signal peptide? | Transmembrane? | log2 fold WT 48h plant / culture |
|---|---|---|---|---|---|---|---|---|
| ZtritIPO323_04t13800 | Mycgr3G47776 | 2.38361 | 187.623 | 6.29855 | GPR1 FUN34 -class plasma membrane | No | YES | 3.69719 |
| ZtritIPO323_04t06606 | Mycgr3G103427 | 36.5046 | 2328.37 | 5.9951 | Tandem Repeat Protein 4 (TRP4)*** | Yes | No | 4.56836 |
| ZtritIPO323_04t08955 | Mycgr3G109137 | 50.7599 | 2372.23 | 5.54641 | hypothetical protein MYCGRDRAFT_109137 | Yes | No | 4.48632 |
| ZtritIPO323_04t03143 | Mycgr3G111221 | 21.2326 | 742.174 | 5.12741 | 3xLysM domain effector (3LysM) | Yes | No | 5.30617 |
| ZtritIPO323_04t10723 | Mycgr3G104794 | 327.83 | 10093.8 | 4.94438 | hypothetical protein MYCGRDRAFT_104794 | Yes | No | 1.90893 |
| ZtritIPO323_04t01868 | Mycgr3G89984 | 9.07987 | 277.305 | 4.93266 | hypothetical protein MYCGRDRAFT_89984 | No | YES | 2.02453 |
| ZtritIPO323_04t07933 | Mycgr3G91855 | 5.51628 | 168.164 | 4.93003 | aspergillopepsin-2 precursor like | Yes | No | No change |
| ZtritIPO323_04t01956 | Mycgr3G67250 | 4.68437 | 124.825 | 4.73591 | peroxidase catalase like | Yes | No | 1.71448 |
| ZtritIPO323_04t00231 | Mycgr3G34306 | 5.01835 | 132.068 | 4.71792 | Aspartic protease pep1 like | Yes | No | 3.47866 |
| ZtritIPO323_04t13637 | Mycgr3G95831 | 20.0937 | 519.83 | 4.69323 | hypothetical protein MYCGRDRAFT_95831 | Yes | No | No change |
| ZtritIPO323_04t04294 | Mycgr3G111636 | 235.826 | 5976.16 | 4.66342 | Ecp2-3 effector homologue | Yes | No | 1.9105 |
| ZtritIPO323_04t08501 | Mycgr3G104000 | 38.9405 | 950.309 | 4.60905 | hypothetical protein MYCGRDRAFT_104000 | Yes | No | 6.39787 |
| ZtritIPO323_04t10384 | Mycgr3G72646 | 6.27375 | 148.704 | 4.56697 | alpha 1,3 glucan synthase | No | YES | 1.76247 |
| ZtritIPO323_04t11777 | Mycgr3G110220 | 32.942 | 745.202 | 4.49963 | hypothetical protein MYCGRDRAFT_110220 | Yes | No | 2.4987 |
| ZtritIPO323_04t04149 | Mycgr3G77689 | 2.1705 | 43.3503 | 4.31994 | carboxypeptidase S1 | Yes | No | 5.99527 |
| ZtritIPO323_04t03806 | Mycgr3G97031 | 19.0535 | 376.75 | 4.30548 | hypothetical protein MYCGRDRAFT_97031 | Yes | No | No change |
| ZtritIPO323_04t09707 | none | 28.0532 | 550.981 | 4.29576 | none | Yes | No | No change |
| ZtritIPO323_04t13185 | Mycgr3G48129 | 91.2925 | 1728.88 | 4.2432 | Cerato-platanin / hydrophobin | Yes | No | No change |
| ZtritIPO323_04t11297 | Mycgr3G12051 | 1.65444 | 28.5554 | 4.10935 | galactose oxidase | Yes | No | 4.3809 |
| ZtritIPO323_04t09426 | Mycgr3G71659 | 4.53293 | 77.3318 | 4.09255 | NAD(P)-binding Rossmann-fold containing | No | No | No change |
| ZtritIPO323_04t07171 | Mycgr3G108399 | 38.5037 | 644.32 | 4.06471 | hypothetical protein MYCGRDRAFT_108399 | No | No | No change |
| ZtritIPO323_04t03262 | Mycgr3G96651 | 1.87684 | 31.0977 | 4.05043 | hypothetical protein MYCGRDRAFT_96651 | No | No | No change |
| ZtritIPO323_04t04165 | none | 9.77584 | 156.07 | 3.99683 | hypothetical protein TI39_contig4278g00011 | Yes | No | 5.34383 |
| ZtritIPO323_04t00584 | none | 77.6554 | 1221.01 | 3.97484 | hypothetical protein TI39_contig279g00027 | No | No | No change |
| ZtritIPO323_04t09555 | Mycgr3G92998 | 25.6961 | 402.709 | 3.97012 | hypothetical protein MYCGRDRAFT_92998 | Yes | No | 1.87831 |
| ZtritIPO323_04t06766 | Mycgr3G69117 | 4.18014 | 64.4432 | 3.9464 | MFS monosaccharide transporter like | No | YES | No change |
| ZtritIPO323_04t11928 | Mycgr3G19274 | 257.455 | 3908.59 | 3.92425 | major allergen alt a1 | Yes | No | No change |
| ZtritIPO323_04t13136 | Mycgr3G46937 | 8.05712 | 121.448 | 3.91393 | major facilitator superfamily transporter like | No | YES | No change |
| ZtritIPO323_04t12624 | Mycgr3G105419 | 276.522 | 4164.69 | 3.91274 | small threonine-rich | Yes | No | -1.58721 |
| ZtritIPO323_04t13161 | Mycgr3G101121 | 2.95145 | 42.827 | 3.85902 | general substrate transporter | No | YES | 5.83776 |
*- sequences available from https://doi.org/10.6084/m9.figshare.4753708.v1 [42].
** sequences available from http://fungi.ensembl.org/Zymoseptoria_tritici/Info/Index.
*** previously described in [43].
Also notable amongst the genes shown in Table 1 was a strong up-regulation (>25 fold levels in the WT strain) of a transcript predicted to encode an alpha-1,3-glucan synthase in the ΔZtGT2 mutant (ZtritIPO323_04t10384 in Table 1). In the filamentous fungal pathogen of animals Aspergillus fumigatus, alpha-1,3-glucan represents a major constituent of the outer cell wall and is required for full virulence [11, 12]. In contrast to the putative Z. tritici alpha-1,3-glucan synthase gene, no other genes likely to be involved in the biosynthesis of either chitin or beta-glucans were differentially expressed in the mutant relative to the WT. Hence from the data here provided it may be that loss of the outer wall integrity in ΔZtGT2 triggers a specific compensatory activation of alpha glucan synthesis.
Analysis of the wheat leaf transcriptome at 48h hpi with ΔZtGT2 and the WT fungus also revealed many differentially expressed genes (S4 Table). Prominent amongst these were plant defence-associated genes including those encoding pathogenesis-related (PR) proteins, receptor-like kinases (RLK) and WRKY transcription factors, which were all more highly expressed in leaves infected with the WT fungus than those infected with ΔZtGT2 (S16 Fig). This likely reflects the comparable lack of hyphal growth by the ΔZtGT2 mutants, relative to that by the WT fungus, over the growth period. In support of this, independent qPCR assessments on wheat defence gene expression incorporating mock-inoculated leaves (no fungus) indicated that the lower induction of plant defence genes by ΔZtGT2 was unlikely to be a consequence of loss of a specific elicitor activity (S16 Fig).
Conclusions
All fungal pathogens of plants require the ability to grow hyphae over solid surfaces posed by host tissues and cells to initiate or complete infections. The present study identified a widely (but not universally) conserved glycosyltransferase (GT2) as a likely key facilitator of this process. The pleomorphic growth characteristics of Zymoseptoria tritci enabled the identification of the specific growth state in which ZtGT2 plays a crucial role, that being hyphal growth when in physical contact with solid matrices. It is likely for fungi which exhibit a predominantly hyphal growth state on solid agar, that gene deletion mutants of ZtGT2 orthologues may be difficult to recover using standard protocols. This could be interpreted as either them being “essential” genes in filamentous fungi, or that hyphal growth is so severely affected in mutants that they cannot be recovered. In support of the latter interpretation was our requirement to perform selection in a liquid, rather than solid growth medium, to obtain homokaryotic ΔFgGT2 deletion mutants in the highly filamentous fungus, Fusarium graminearum. This highlights that the terminology “essential gene” should be used with caution and in consideration of the technical approaches used, when interpreting failed attempts to obtain specific gene deletion mutants from filamentous fungi. In contrast, the fact that yeast-like growth of Z. tritici ΔZtGT2 mutants occurs un-impeded on solid agar, highlights the utility of this species for characterising genes which may be “essential” for hyphal growth in fungi. The ability for Z. tritici to undergo “yeast-like” budding growth in the absence of ZtGT2, agrees well with the observation that all true free-living ascomycete budding yeasts, particularly in the classes Saccharomycotina and Taphrinomycotina, lack any similar proteins. This suggests that ZtGT2 orthologues may be dispensable for the budding growth form of ascomycete fungi.
The basidiomycete pathogen of mammals, Cryptococcus neoformans, is a dimorphic fungus pathogenic in the yeast form, but which can also grow as true hyphae during sexual reproduction [44]. C. neoformans possesses a ZtGT2 orthologue named CPS1 (Blastp homology of 9.05e-90 to ZtGT2 with 47.3% identity covering 73.5% of the protein), shown in Fig 3B. This protein is predicted to function as a putative capsule polysaccharide synthase and is essential for full virulence of the yeast form against mammals [45, 46]. In contrast to this, the yeast-like non-pathogenic growth form of Z. tritici is unaffected in ΔZtGT2 mutants, and it is instead the infectious hyphal growth form which is affected, and then only when in physical contact with surfaces. This re-emphasises a key distinguishing feature of infection of plants and animals by ascomycete and basidiomycete fungi which can replicate in either yeast-like or filamentous forms. The polysaccharide produced by CPS1 has been suggested to be similar to Hyaluronic acid, the key and ubiquitous extracellular matrix component of mammals, buts it’s precise structure has not yet been determined. The morphological abnormalities we detected in the ΔZtGT2 mutant suggests that ZtGT2 plays a key role in maintaining outer cell wall integrity, although there is currently no evidence to suggest that Z. tritici (or any filamentous fungus), forms anything functionally equivalent to the unique C. neoformans yeast capsule [47]. Efforts to detect quantitative changes in cell wall or culture filtrate monosaccharides (deriving from precipitated polysaccharide) between WT and ΔZtGT2 mutants of Z. tritici were inconsistent. Neither of the analyses performed identified decreased levels of monosaccharides deriving from polysaccharides from ΔZtGT2 mutants. For total cell walls, the levels of glucose, mannose and galactose were unchanged. Conversely the culture filtrate of ΔZtGT2 mutants unexpectedly gave increased levels. As fungal cell walls possess chitin and beta glucan as some major components, it is possible that other functionally important, but more minor constituents might be beyond detection if only using comparisons between gene deletion strains with wild-type strains. It is also possible that such components may have been lost in preparation without prior knowledge of their physicochemical properties. However, identifying differences via the approach of comparing WT strains with gene deletion mutants in this case was also likely complicated by the fact that our transcriptome data demonstrated that ΔZtGT2 strains overexpress a conserved alpha-1,3- glucan synthase to ~25 fold higher levels than WT strains (Table 1). So its possible that the increased levels of polysaccharides derived from culture filtrates of ΔZtGT2 mutants might be a consequence of other polysaccharides being over-produced to compensate for the loss of ZtGT2. It may also be that the irregularities seen in the outer cell wall surface may have also contributed to the increased levels of monosaccharides detected. This requires further study. However, future approaches aimed to identify and structurally characterise the polysaccharide produced by ZtGT2 and/or its orthologues may be better addressed using different approaches to circumvent these potential complications.
At this point it is unclear why some filamentous fungi lack orthologues, or proteins with any similarity whatsoever, to ZtGT2. For example, the model basidiomycete plant pathogens Puccinia graminis and Ustilago maydis have no similar proteins to ZtGT2, but they do grow hyphae over plant surfaces (including wheat leaves for the former). As presence / absence of proteins with similarity to ZtGT2 appears discontinuous, even within the ascomycete kingdom, it is possible that some fungal species use a different protein, or mechanism, to provide the equivalent functionality. This might again be reflected in the chemical composition of the outer cell wall surfaces in these fungi. However, for ZtGT2 and its close orthologues, some conclusions can be made. Firstly, the gene is present in almost all analysed ascomycete plant pathogens and in several opportunistic animal pathogens (for example Aspergillus fumigatus). However, it is also frequently observed to be present in many saprophytic fungi including those which perform the primary events in leaf litter decay. Thus, we speculate that the importance of this gene for the evolution of fungal pathogenesis likely derived “inadvertently” from an ancient requirement for fungi to evolve the hyphal growth form, to extend filaments over solid surfaces for purposes such as nutrient acquisition. By acquiring this basic functionality, fungi also acquired a key competency which subsequently enabled them to develop parasitic (as well as potentially mutualistic) interactions with many plants and animals. Based upon all the available data it is likely that ZtGT2 synthesises or modifies a potentially widespread and essential extracellular matrix or outer cell wall polysaccharide component, which may be only a minor constituent (Fig 7). This may function to alleviate surface friction and shear stresses normally imposed on rapid hyphal tip growth over solid matrices [13, 14]. This currently structurally undefined, polysaccharide may be functionally important for many plant (and perhaps animal) pathogens and may therefore represent a viable target for future widespread control of fungal diseases. This study also emphasises the tractability of Z. tritici as a model organism for isolating genes which may be essential for contact-dependent filamentous fungal growth.
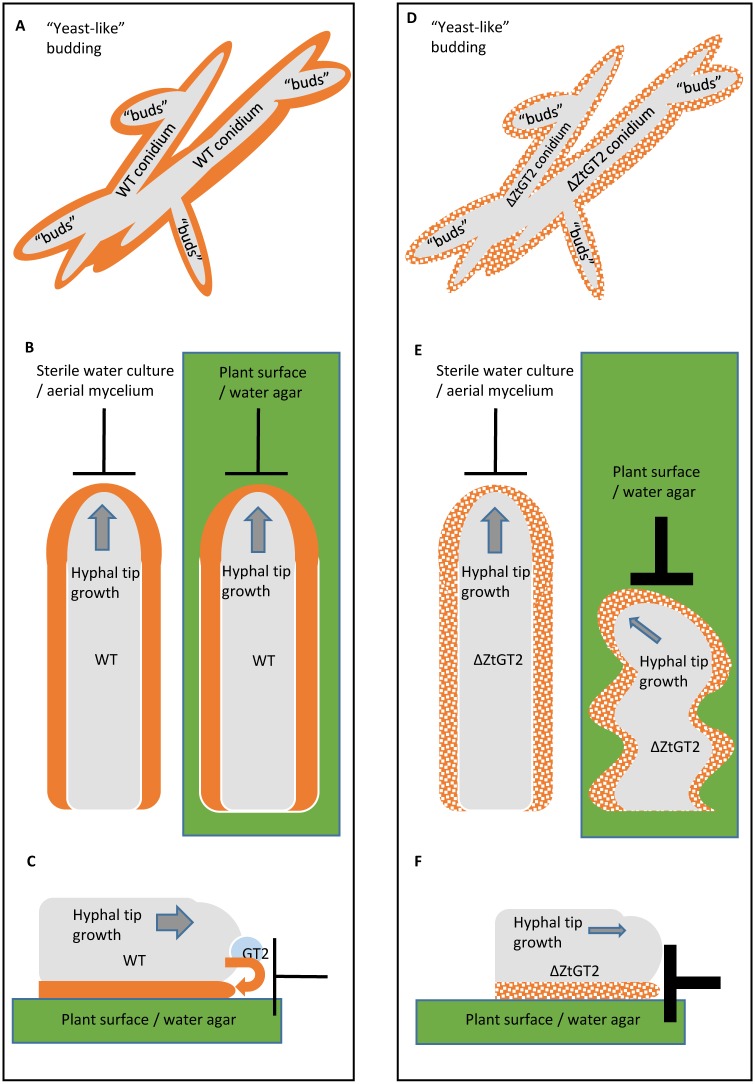
Fig 7. Model for the function of ZtGT2 in overcoming surface-derived frictional (opposing) forces.
The phenotype of ΔZtGT2 mutants, the predicted protein localisation, homology to C. neoformans polysaccharide synthase CPS1, and alterations to the outer cell wall, suggest that ZtGT2 is involved in the production of an unknown polysaccharide which acts as an important component of the outer cell wall or extracellular matrix (ECM) of fungal cells (indicated by solid orange colouring surrounding grey cells in wild type fungus depicted in A to C). The cells missing this component are depicted by hatched shading surrounding ΔZtGT2 mutants in D to F. In budding spores (conidia) of Z. tritici this component is functionally dispensable and both wild type and ΔZtGT2 mutants grow at comparable rates (A and D). However, for Z. tritici hyphae this feature may provide the fluidity which enables rapid extension in a forward’s direction in the face of the counter frictional forces of the leaf (or other) surfaces (B and C). These counter forces resulting from frictional or other stresses are depicted as cross hatches throughout (B, C, E and F). ΔZtGT2 lacks a functional version of the outer cell wall, or ECM, polysaccharide so whilst mutant cells can grow normally as hyphae in liquid suspensions (where they encounter no frictional counter forces- left panel of E) they are unable to effectively counter these forces on solid surfaces (right panel of E and F). This gives rise to short, occasionally sinusoidal (or aborted) hyphae on agar and a loss of virulence on plants.
Methods
Plant and fungal materials
The fully genome sequenced Z. tritici isolate IPO323 was used in all experiments (http://genome.jgi-psf.org/Mycgr3/Mycgr3.home.html). For all experiments, fungal spores were initially harvested from 5 day old cultures growing (budding) on Yeast extract peptone dextrose (YPD) plates (Oxoid Ltd., Hampshire, UK) at 16°C. For RNAseq and qRT-PCR based gene expression analysis, duplicate or triplicate flasks containing 40 ml YPD broth (Oxoid Ltd., Hampshire, UK) were inoculated for 96h at 18°C (or other specified temperatures) on an orbital shaking incubator at 120 rpm. After this time, fungal materials were collected via vacuum filtration and snap frozen in liquid N2 until RNA extraction. Wheat seedling infection assays (cultivar Riband) was performed as described previously [48]. Briefly the adaxial surfaces of leaves were inoculated with spore suspensions collected from YPD cultures, which were washed and re-suspended in dH20 +0.01% Tween 20 to a density of 1x106 spores / ml. After the indicated time, inoculated leaves were excised and snap frozen in liquid N2 until RNA extraction. Photographs displaying disease levels on leaves were taken twenty-one days after inoculation of the second leaf of three-week-old seedlings. All plant materials for RNAseq were prepared from duplicate (two independent) experiments performed three weeks apart. A total of six leaves were taken from six independent inoculated wheat seedlings and used to prepare a single total RNA preparation. This constituted a single biological replicate sample.
Generation of T-DNA insertional mutants and gene deletion and complementation strains for ZtGT2
The T-DNA mutagenesis screen of Z. tritici was previously described [49]. TAIL PCR was carried out on fungal genomic DNA isolated from the 23–170 mutant according to previous methods [49, 50]. For targeted disruption of the ZtGT2 gene (JGI protein model 65552; NCBI Reference Sequence: XP_003857553.1) two regions (flanks) of approximately 1000 bp of fungal genomic DNA were amplified by PCR. Flank1 was then cloned into vector pCHYG using SacI and KpnI (primers P1 and P2 -S5 Table) and Flank 2 using PstI and HindIII (P3 and P4) and the resulting plasmid was transformed into A. tumefaciens strain Agl-1 via the freeze-thaw method [51]. For targeted gene deletion, a modified ΔKu70 strain of IPO323 was used [52]. To generate the complementation strain 23–170::GT2comp the entire open reading frame of ZtGT2 plus upstream and downstream sequences was amplified by PCR on genomic DNA (primers P5 and P6). The amplicon was cloned using SacI and KpnI into vector pCGEN for fungal transformation of strain 23–170 and selection used 100 μg/ml Geneticin (Sigma St Louis). Agrobacterium-mediated transformation of Z. tritici was performed as previously described [24, 53]. Targeted mutants were confirmed by PCR on genomic DNA. All oligonucleotide primer sequences are provided in S5 Table.
Identification of proteins with similarity to ZtGT2 in other fungi
The mature protein sequence of ZtGT2 (Genbank XP_003857553.1) was used for a Blastp analysis against the contents of the JGI MycoCosm fungal genome portal (http://genome.jgi.doe.gov/programs/fungi/index.jsf) in July 2017. Blastp searches at expect value homology cut-offs of 1e-100; 1e-60 and 1e-05 were performed for each taxonomic class / sub phylum. Species which returned one or more hits at each cut-off were included as a positive (harbouring a protein with the indicated level of similarity). Species returning no hits at any cut-off were deemed negative (lacking any similar proteins). Data was then presented in the form of pie charts for each taxonomic class/ sub phylum displaying number of species genomes having or lacking similar proteins at each e value cut-off, as a proportion of the total number of species genomes analysed in each category.
Phylogenetics analysis of ZtGT2 orthologues
Data from the protein similarity search was used for selection of sequences for phylogenetics analysis. For Maximum Likelihood gene tree analysis, the best Blastp hits were selected from 2–3 species representative of the different taxonomic subdivisions or classes from the fungal tree of life present within the JGI Mycocosm Genome portal [34]. For select species, including Z. tritici and F. graminearum, the next best Blastp hit was also included. To produce the most accurate nucleotide alignments, coding sequences were trimmed from each transcript to the conserved Type 2 glycosyltransferase domain as identified by the NCBI conserved domain database displayed in the Blastp output. The accession for the conserved domain sequence was cd06434. Genes were then aligned using MAFFT v.7.308 [54, 55], in Geneious 10.0.9 [56]. For phylogenetic reconstruction, the GTR+I+G nucleotide substitution model was selected by AIC in jModeltest 2.1.10 [57, 58]. The Maximum Likelihood phylogeny was reconstructed using PhyML [59], with the substitution model selected in jModeltest; starting tree with optimised topology, length and rate parameters; topology searching by the best of NNI and SPR; and 500 bootstraps.
Gene deletion of the ZtGT2 homologue (FgGT2) in Fusarium graminearum
The FgGT2 gene, FGRRES_00702 (http://fungi.ensembl.org/Fusarium_graminearum/Info/Index), was deleted in F. graminearum wild-type strain PH-1 (NRRL 31084) for which the complete genome sequence is available [60]. A PCR-based split-marker gene deletion strategy was chosen [35]. The DNA flanks (1000 bp 5’- and 997 bp 3’-sequence) of the FgGT2 gene were first amplified by PCR using primers (S5 Table) pairs U650/U651 and U656/U657 and cloned into the plasmid vector pGEM-T (Promega) using the Gibson assembly Master Mix kit (New England BioLabs Inc.) according to the manufacturer’s instructions to generate vectors pMU421 and pMU422, respectively. Specific PCR was carried out in 25 μl volumes, containing 50 ng of DNA, 1 U of Expand High Fidelity Taq-Pwo polymerase mixture (Boehringer Mannheim), 10 pmol of each primer and 0.25 mM each deoxynucleoside triphosphate, in a standard buffer for 35 cycles with the following cycling parameters: denaturation at 94°C for 30 s; annealing at 54°C for 30 s; and DNA synthesis at 72°C for 1 min. For transformation both split-marker constructs contained in pMU421 and pMU422 were quantitatively amplified by PCR using HotStar TAQ polymerase (Qiagen) following the manufacturer’s instructions. The concentration of the PCR products was adjusted to 2 μg μl-1 and 5 μl of each construct was mixed and transformed into 1x108 protoplasts of F. graminearum strain as previously described [61, 62]. Recovery of transformants was accomplished in liquid TB3 medium containing hygromycin (75 μg/ml) to allow selection of mutants which might otherwise be impaired in hyphal growth on selective solid agar. 0.2 ml transformation mix was added to 10 ml TB3 liquid medium containing 75 μg/ml hygromycin contained in 50 ml Falcon tubes. Cultures were grown for further 10 days in a shaking incubator set to 28 C and 180 rpm. Hygromycin resistant transformants were then transferred to PDA agar plates containing hygromycin (10 μg/ml) for further analysis. Fungal genomic DNA was extracted from transformants grown in 10 ml potato dextrose medium in the presence of hygromycin (10 μg/ml) as described [63].
In the two isolated gene replacement mutants MU426 and MU427 two diagnostic PCR fragments of 1.5kb and 1.3 kb size are detectable using oligomer pairs U659/U667 (PCR 1) and U660/U668 (PCR 2) respectively (Fig 4). In both mutants MU426 and MU427, the FgGT2 gene is absent (PCR 3 in Fig 4). Plant infection and pathogenicity tests on wheat (Triticum aestivum) plants of cultivar Bobwhite were grown and infected by point and whole-wheat spike spray inoculation with spore solutions as previously described [64]. Each experiment was performed in triplicate with similar results.
Transmission electron microscopy (TEM)
Fungal cells were collected from the surfaces of YPD agar plates using a sterile loop. Samples were then high pressure frozen using a Leica Microsystems EM HPM100 and stored in liquid nitrogen before staining with 1% osmium and freeze substitution using acetone in a Leica Microsystems EM AFS. Following freeze substitution, the samples were stored at -20C for 24 hours then 4C for 24 hours before resin infiltration. Samples were infiltrated with an acetone:Spurr resin series and polymerised at 60C overnight. 70nm thin sections were cut using a Leica Microsystems UC7 microtome and collected on copper grids coated with formvar and carbon. Micrographs were collected using a JEOL 2011 transmission electron microscope at 200kV and a Gatan Ultrascan CCD camera.
Scanning electron microscopy (SEM)
Approximately 5mm square regions were cut from samples and attached to aluminium stubs using a 50:50 mixture of graphite:TissueTek. The samples were plunge frozen in liquid nitrogen and transferred to the GATAN ALTO 2100 cryo prep system. Samples were etched and coated in a thin layer of gold. Micrographs were collected using a JEOL 6360 scanning electron microscope at 5kV.
RNA sequencing and differential gene expression analysis
Total RNA was isolated from frozen materials using the TRIZOL procedure (Invitrogen). Library preparation and sequencing was performed at the Earlham Institute, Norwich Research Park, Norwich, UK. No pre-processing of the reads took place. Hisat2 (v2.0.4) was used to map the reads to Zymoseptoria tritici IPO323 (Ensembl fungi v30) or Triticum aestivum Chinese Spring (Ensembl plants v32) using default settings. Cuffdiff (v2.2.1) was used to produce FPKM and differential testing of counts with a classic-fpkm library normalisation method, pooled dispersion estimation method, and bias correction with effective length correction, for wheat using Ensembl plants v32 annotation but for fungi using a custom gff3 annotation file https://doi.org/10.6084/m9.figshare.4753708.v1 [42] to count against only these models. CummeRbund (v2.8.2) was used to produce the PCA plots in R (v3.1.2). Annotations were added to the cuffdiff output using Blast2Go (v3.3.5) with Blast2Go GO database 05/2016 and default filtering settings, using input from InterPro (v58.0) and Timelogic DeCypher with the NCBI NR database (23/06/16) using an e-value threshold of 1e-2. For robust differential expression analysis, only genes with fold changes of Log2 >1.5 (Padj 0.05) were considered further.
Quantitative RT-PCR analysis
Total RNA was isolated from freeze-dried, fungal material collected from the stated liquid culture or from fungal infected leaf tissues, was prepared using the TRIZOL procedure (Invitrogen). Total RNA was used for all RT-PCR and Real-Time RT-PCR analyses. First-strand cDNA was synthesised from total RNA using the SuperScript III First_Strand Synthesis System for RT-PCR (Invitrogen). A 5 μg aliquot of total RNA primed with oligo(dT)20 was used in a 20 μl reaction, following the suppliers instructions. The resulting cDNA was analysed by Real-Time RT-PCR using a QuantiTect SYBR Green PCR Kit (Qiagen), following the supplier’s instruction. A 0.5 μl aliquot of cDNA was used in each 20 μl PCR reaction, with an annealing temperature of 60°C. Primers were used at a final concentration of 0.25 μM. Real-Time RT-PCR reactions were run for 40 cycles and analysed using an ABI 7500 Real Time PCR System. The relative expression of each fungal or plant gene was determined by normalisation with the constitutively expressed Z. tritici beta-tubulin gene [48] or with the TaCdc48 gene for T. aestivum [31]. All oligonucleotides used are listed in S5 Table.
Supporting information
Wild type or 23–170 mutant spore suspensions at 104 spores / ml in water was inoculated (5 μl) onto a YPD plate. The inoculated region was photographed at 24h intervals for a macroscopic analysis (upper panels). Middle panels display the typical characteristic spore morphologies of each strain grown on YPD agar coated slides for 24 hours. Lower panels show spores under higher magnification stained with the chitin binding fluorophore calcoflour white.
(TIF)
A spore suspension of 104 spores / ml in water was inoculated (5 μl) onto a water agar plate at the indicated agar concentration. Plates were incubated at RT then photographed after 10 days.
(TIF)
A spore suspension of 104 spores / ml water was inoculated (5 μl) onto the surface of the indicated agar plate. Plates were incubated at RT then photographed after 21 days.
(TIF)
A spore suspension of 104 spores / ml water of wild-type or ΔZtGT2 was inoculated (5 μl) onto the surface of the indicated agar plate. Plates were incubated at RT then photographed after 21 days (Left panels). Hyphal morphology radiating from the colony edge was studied by light microscopy (right panels).
(TIF)
Gene replacement strategy for the ZtGT2 orthologue from F. graminearum with gene Id FG00702.1 (http://fungi.ensembl.org/Fusarium_graminearum/Info/Index) (A) Genomic left border and right border regions (white bars) were amplified with primers and fused to parts of the hph hygromycin resistance gene. Fused PCR fragments were used in a split-marker strategy to replace FGRRES_00702. Horizontal black bars represent genomic areas outside the replacement construct, vertical black bars represent 3 introns. (B) Anticipated diagnostic PCR for successful gene replacement of FGRRES_0702. (C) Results of diagnostic PCR and expected sizes indicated in (A) and (B). Transformants S#11, S#19 and S#24 have the ΔFGRRES_00702 null allele but also retain a wild-type gene (heterokaryons). This was observed for all hygromycin resistant strains originating from 4 independent transformations. M—λ DNA-BstEII digest; WT—wild type. (D) Growth of F. graminearum heterokaryotic strains carrying both a WT and FGRRES_00702 null allele on potato dextrose agar after 72 hours incubation. No alteration in hyphal growth was observed. Top left to bottom right: WT, FG transformants S#10, 11, 15, 19, 24.
(TIF)
Penetration of cellophane membranes by F. graminearum strains. Fungal spores were plated and grown for 2 d at 22°C on top of cellophane membrane on the surface of PDA plates (images labelled “Before”). The cellophane with the fungal colonies was removed and plates were incubated for an additional two days to determine whether fungal growth occurred on the plates, indicating penetration of the cellophane disk (images labelled “After”). The ΔFgMAP1 mutant strain was used as a control strain shown previously to be defective in cellophane and plant penetration [64].
(TIF)
Fungal mycelium grown at 25°C in liquid YPD was subjected to crude protein fractionation and western blot analysis with an anti-ZtGT2 peptide antibody. (A) The antiserum detects a ~56kDa protein found only in the cell wall fraction of wild type fungal cells (B) The specific binding to this protein in wild type cells is confirmed by a peptide competition experiment.
(TIF)
Arrows highlight the different surface structures observed in the mutants.
(TIF)
Fungal strains were grown in shaking culture flasks until saturation and then separated by filtration. AIR was generated from cell walls, samples were hydrolysed and monosaccharaides levels (glucose, mannose and galactose) were quantified (A). Culture filtrates were ethanol precipitated and then analysed the same way (B). Data shown derives from three biological replicates and in each case and monosaccharide levels are expressed in milligrams / gram of total dry weight (mg /g. dwt).
(TIF)
Conidial suspensions were left to adhere to either plastic petri dishes (upper panels) before extensive washing and counting of remaining adhered spores (from micrograph images taken before and after washing). Lower panels- cell suspensions were applied to surface of wheat leaves or to glass coverslips and allowed to adhere. Leaves or coverslips were then washed by vortexing and the resulting suspensions plated out on YPD agar + 100 μg/ml G418 antibiotic. Plates were incubated for 6 days and colonies were counted from photographic images. A decrease in the number of colony forming units (cfus) retrieved from leaf surfaces relative to glass coverslips was taken as indicative of adhesion. There were no statistical differences between wild-type and ΔZtGT2 strains from all assays.
(TIF)
Wild type; 23–170 mutant; ΔZtGT2 and 23–170::GT2comp spore suspensions of initially 106 spores / ml (or three successive 3-fold dilutions in water), were inoculated (5 μl) onto a YPD agar plates containing the added stress agent. Plates were then photographed after growth at 16°C or 30°C for six days.
(TIF)
(TIF)
Seven genes in this category show significantly reduced expression in ΔZtGT2 mutants growing both in liquid culture and on leaf surfaces.
(TIF)
(TIF)
An independent experiment was performed growing the wild type and ΔZtGT2 fungus in either YPD or PDB broth for 5 days. Real-Time qRT-PCR was then used to measure relative expression of the Zt3LysM effector. Data was normalised to the expression of the Z. tritici beta tubulin gene and presented as fold change relative to gene expression by the WT fungus in YPD broth.
(TIF)
(A) indicates the number of transcripts with higher expression in wheat leaves inoculated with WT fungus than with ΔZtGT2. (B) qRT-PCR validation of expression of the two indicated wheat PR genes. Data is normalised to expression of the wheat cdc48 gene.
(TIF)
(XLSX)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
Acknowledgments
The authors would like to thanks members of the Rothamsted Horticultural team. The authors also acknowledge use of equipment and advice provided by members of the Rothamsted BioImaging suite. Experiments involving licenced pathogen and transgenic isolates were conducted in a biological containment facility under DEFRA licence 101948/198285/4.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Rothamsted Research receives grant-aided support from the Biotechnology and Biological Sciences Research Council (BBSRC), UK, as part of the Institute Strategic Programme grants 20:20® wheat [BB/J/00426X/1] and Designing Future Wheat [BB/P016855/1]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jones J.D.G. and Dangl J.L., The plant immune system. Nature, 2006. 444 (7117): p. 323–329. doi: 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- 2.de Wit P.J.G.M., et al. , Fungal effector proteins: past, present and future. Molecular Plant Pathology, 2009. 10(6): p. 735–747. doi: 10.1111/j.1364-3703.2009.00591.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tucker S.L. and Talbot N.J., Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annual Review of Phytopathology, 2001. 39: p. 385–417. doi: 10.1146/annurev.phyto.39.1.385 [DOI] [PubMed] [Google Scholar]
- 4.Steinberg G., Hyphal growth: a tale of motors, lipids, and the Spitzenkorper. Eukaryotic Cell, 2007. 6(3): p. 351–360. doi: 10.1128/EC.00381-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyce K.J. and Andrianopoulos A., Fungal dimorphism: the switch from hyphae to yeast is a specialized morphogenetic adaptation allowing colonization of a host. Fems Microbiology Reviews, 2015. 39(6): p. 797–811. doi: 10.1093/femsre/fuv035 [DOI] [PubMed] [Google Scholar]
- 6.Lengeler K.B., et al. , Signal transduction cascades regulating fungal development and virulence. Microbiology and Molecular Biology Reviews, 2000. 64(4): p. 746–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nemecek J.C., Wuthrich M., and Klein B.S., Global control of dimorphism and virulence in fungi. Science, 2006. 312(5773): p. 583–588. doi: 10.1126/science.1124105 [DOI] [PubMed] [Google Scholar]
- 8.Bartnicki-Garcia S. CELL WALL CHEMISTRY MORPHOGENESIS AND TAXONOMY OF FUNGI. Annual Review of Microbiology, 1968. 22: p. 87–108. doi: 10.1146/annurev.mi.22.100168.000511 [DOI] [PubMed] [Google Scholar]
- 9.Latge J.P., The cell wall: a carbohydrate armour for the fungal cell. Molecular Microbiology, 2007. 66(2): p. 279–290. doi: 10.1111/j.1365-2958.2007.05872.x [DOI] [PubMed] [Google Scholar]
- 10.Silipo A., et al. , Glyco-conjugates as elicitors or suppressors of plant innate immunity. Glycobiology, 2010. 20(4): p. 406–419. doi: 10.1093/glycob/cwp201 [DOI] [PubMed] [Google Scholar]
- 11.Beauvais A., et al. , Deletion of the alpha-(1,3)-Glucan Synthase Genes Induces a Restructuring of the Conidial Cell Wall Responsible for the Avirulence of Aspergillus fumigatus. Plos Pathogens, 2013. 9(11). e1003716 doi: 10.1371/journal.ppat.1003716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshimi A., et al. , Functional Analysis of the alpha-1,3-Glucan Synthase Genes agsA and agsB in Aspergillus nidulans: AgsB Is the Major alpha-1,3-Glucan Synthase in This Fungus. Plos One, 2013. 8(1). e54893 doi: 10.1371/journal.pone.0054893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goriely A. and Tabor M., Mathematical modeling of hyphal tip growth. Fungal Biology Reviews, 2008. 22(2): p. 77–83. [Google Scholar]
- 14.Money N., Insights on the mechanics of hyphal growth. Fungal Biology Reviews, 2008. 22: p. 71–76. [Google Scholar]
- 15.Toole B.P., Hyaluronan: From extracellular glue to pericellular cue. Nature Reviews Cancer, 2004. 4(7): p. 528–539. doi: 10.1038/nrc1391 [DOI] [PubMed] [Google Scholar]
- 16.Torronen K., et al. , Tissue distribution and subcellular localization of hyaluronan synthase isoenzymes. Histochemistry and Cell Biology, 2014. 141(1): p. 17–31. doi: 10.1007/s00418-013-1143-4 [DOI] [PubMed] [Google Scholar]
- 17.Beauvais A., et al. , An extracellular matrix glues together the aerial-grown hyphae of Aspergillus fumigatus. Cellular Microbiology, 2007. 9(6): p. 1588–1600. doi: 10.1111/j.1462-5822.2007.00895.x [DOI] [PubMed] [Google Scholar]
- 18.Duncan K.E. and Howard R.J., Cytological analysis of wheat infection by the leaf blotch pathogen Mycosphaerella graminicola. Mycological Research, 2000. 104: p. 1074–1082. [Google Scholar]
- 19.Beauvais A., et al. , Aspergillus Cell Wall and Biofilm. Mycopathologia, 2014. 178(5–6): p. 371–377. doi: 10.1007/s11046-014-9766-0 [DOI] [PubMed] [Google Scholar]
- 20.Gravelat F.N., et al. , Aspergillus Galactosaminogalactan Mediates Adherence to Host Constituents and Conceals Hyphal beta-Glucan from the Immune System. Plos Pathogens, 2013. 9(8). e1003575 doi: 10.1371/journal.ppat.1003575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dean R., et al. , The Top 10 fungal pathogens in molecular plant pathology. Molecular Plant Pathology, 2012. 13(4): p. 414–430. doi: 10.1111/j.1364-3703.2011.00783.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fones H. and Gurr S., The impact of Septoria tritici Blotch disease on wheat: An EU perspective. Fungal Genetics and Biology, 2015. 79: p. 3–7. doi: 10.1016/j.fgb.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mehrabi R., et al. , MgHog1 regulates dimorphism and pathogenicity in the fungal wheat pathogen Mycosphaerella graminicola. Molecular Plant-Microbe Interactions, 2006. 19(11): p. 1262–1269. doi: 10.1094/MPMI-19-1262 [DOI] [PubMed] [Google Scholar]
- 24.Motteram J., et al. , Molecular Characterization and Functional Analysis of MgNLP, the Sole NPP1 Domain-Containing Protein, from the Fungal Wheat Leaf Pathogen Mycosphaerella graminicola. Molecular Plant-Microbe Interactions, 2009. 22(7): p. 790–799. doi: 10.1094/MPMI-22-7-0790 [DOI] [PubMed] [Google Scholar]
- 25.Steinberg G., Cell biology of Zymoseptoria tritici: Pathogen cell organization and wheat infection. Fungal Genetics and Biology, 2015. 79: p. 17–23. doi: 10.1016/j.fgb.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossman A.Y., et al. , Recommended names for pleomorphic genera in Dothideomycetes. Ima Fungus, 2015. 6(2): p. 507–523. doi: 10.5598/imafungus.2015.06.02.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kema G.H.J., et al. , Histology of the pathogenesis of Mycosphaerella graminicola in wheat. Phytopathology, 1996. 86(7): p. 777–786. [Google Scholar]
- 28.Bolton M.D., et al. , The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Molecular Microbiology, 2008. 69(1): p. 119–136. doi: 10.1111/j.1365-2958.2008.06270.x [DOI] [PubMed] [Google Scholar]
- 29.Sanchez-Vallet A., et al. , Fungal effector Ecp6 outcompetes host immune receptor for chitin binding through intrachain LysM dimerization. Elife, 2013. 2 e00790 doi: 10.7554/eLife.00790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Jonge R., et al. , Conserved Fungal LysM Effector Ecp6 Prevents Chitin-Triggered Immunity in Plants. Science, 2010. 329(5994): p. 953–955. doi: 10.1126/science.1190859 [DOI] [PubMed] [Google Scholar]
- 31.Lee W.S., et al. , Mycosphaerella graminicola LysM Effector-Mediated Stealth Pathogenesis Subverts Recognition Through Both CERK1 and CEBiP Homologues in Wheat. Molecular Plant-Microbe Interactions, 2014. 27(3): p. 236–243. doi: 10.1094/MPMI-07-13-0201-R [DOI] [PubMed] [Google Scholar]
- 32.Marshall R., et al. , Analysis of Two in Planta Expressed LysM Effector Homologs from the Fungus Mycosphaerella graminicola Reveals Novel Functional Properties and Varying Contributions to Virulence on Wheat. Plant Physiology, 2011. 156(2): p. 756–769. doi: 10.1104/pp.111.176347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goodwin S.B., et al. , Finished Genome of the Fungal Wheat Pathogen Mycosphaerella graminicola Reveals Dispensome Structure, Chromosome Plasticity, and Stealth Pathogenesis. Plos Genetics, 2011. 7(6). e1002070 doi: 10.1371/journal.pgen.1002070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grigoriev IV, N R., Haridas S, Kuo A, Ohm R, Otillar R, Riley R, Salamov A, Zhao X, Korzeniewski F, Smirnova T, Nordberg H, Dubchak I, Shabalov I., MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Research, 2014. 42: p. 699–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu J.H., et al. , Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genetics and Biology, 2004. 41(11): p. 973–981. doi: 10.1016/j.fgb.2004.08.001 [DOI] [PubMed] [Google Scholar]
- 36.Weld R.J., et al. , Approaches to functional genomics in filamentous fungi. Cell Research, 2006. 16(1): p. 31–44. doi: 10.1038/sj.cr.7310006 [DOI] [PubMed] [Google Scholar]
- 37.Lopez-Berges M.S., et al. , A Nitrogen Response Pathway Regulates Virulence Functions in Fusarium oxysporum via the Protein Kinase TOR and the bZIP Protein MeaB. Plant Cell, 2010. 22(7): p. 2459–2475. doi: 10.1105/tpc.110.075937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kellner R., et al. , Expression Profiling of the Wheat Pathogen Zymoseptoria tritici Reveals Genomic Patterns of Transcription and Host-Specific Regulatory Programs. Genome Biology and Evolution, 2014. 6(6): p. 1353–1365. doi: 10.1093/gbe/evu101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palma-Guerrero J., et al. , Comparative transcriptomic analyses of Zymoseptoria tritici strains show complex lifestyle transitions and intraspecific variability in transcription profiles. Molecular Plant Pathology, 2016. 17(6): p. 845–859. doi: 10.1111/mpp.12333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rudd J.J., et al. , Transcriptome and Metabolite Profiling of the Infection Cycle of Zymoseptoria tritici on Wheat Reveals a Biphasic Interaction with Plant Immunity Involving Differential Pathogen Chromosomal Contributions and a Variation on the Hemibiotrophic Lifestyle Definition. Plant Physiology, 2015. 167(3): p. 1158–1185. doi: 10.1104/pp.114.255927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kulkarni R.D., Kelkar H.S., and Dean R.A., An eight-cysteine-containing CFEM domain unique to a group of fungal membrane proteins. Trends in Biochemical Sciences, 2003. 28(3): p. 118–121. doi: 10.1016/S0968-0004(03)00025-2 [DOI] [PubMed] [Google Scholar]
- 42.King, R., Zym_tritici_RRes_v4.0_RK_public.gff.figshare. 2017.
- 43.Rudd J.J., et al. , Identification and characterisation of Mycosphaerella graminicola secreted or surface-associated proteins with variable intragenic coding repeats. Fungal Genetics and Biology, 2010. 47(1): p. 19–32. doi: 10.1016/j.fgb.2009.10.009 [DOI] [PubMed] [Google Scholar]
- 44.Sia R.A., Lengeler K.B., and Heitman J., Diploid strains of the pathogenic basidiomycete Cryptococcus neoformans are thermally dimorphic. Fungal Genetics and Biology, 2000. 29(3): p. 153–163. doi: 10.1006/fgbi.2000.1192 [DOI] [PubMed] [Google Scholar]
- 45.Chang Y.C., et al. , CPS1, a homolog of the Streptococcus pneumoniae type 3 polysaccharide synthase gene, is important for the pathobiology of Cryptococcus neoformans. Infection and Immunity, 2006. 74(7): p. 3930–3938. doi: 10.1128/IAI.00089-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jong A., et al. , Identification and characterization of CPS1 as a hyaluronic acid synthase contributing to the pathogenesis of Cryptococcus neoformans infection. Eukaryotic Cell, 2007. 6(8): p. 1486–1496. doi: 10.1128/EC.00120-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bose I., et al. , A yeast under cover: the capsule of Cryptococcus neoformans. Eukaryotic Cell, 2003. 2(4): p. 655–663. doi: 10.1128/EC.2.4.655-663.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keon J., et al. , Transcriptional adaptation of Mycosphaerella graminicola to programmed cell death (PCD) of its susceptible wheat host. Molecular Plant-Microbe Interactions, 2007. 20(2): p. 178–193. doi: 10.1094/MPMI-20-2-0178 [DOI] [PubMed] [Google Scholar]
- 49.Motteram J., et al. , Aberrant protein N-glycosylation impacts upon infection-related growth transitions of the haploid plant-pathogenic fungus Mycosphaerella graminicola. Molecular Microbiology, 2011. 81(2): p. 415–433. doi: 10.1111/j.1365-2958.2011.07701.x [DOI] [PubMed] [Google Scholar]
- 50.Liu Y.G., et al. , EFFICIENT ISOLATION AND MAPPING OF ARABIDOPSIS-THALIANA T-DNA INSERT JUNCTIONS BY THERMAL ASYMMETRIC INTERLACED PCR. Plant Journal, 1995. 8(3): p. 457–463. [DOI] [PubMed] [Google Scholar]
- 51.An G., Binary Vectors In: Plant Molecular Biology Manual A3. Gelvin S. B. & Schilperroort R. A. (eds). Dordrecht: Kluwer Academic Press, 1988: p. 1–19. [Google Scholar]
- 52.Bowler J., et al. , New capabilities for Mycosphaerella graminicola research. Molecular Plant Pathology, 2010. 11(5): p. 691–704. doi: 10.1111/j.1364-3703.2010.00629.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zwiers L.H. and De Waard M.A., Efficient Agrobacterium tumefaciens-mediated gene disruption in the phytopathogen Mycosphaerella graminicola. Current Genetics, 2001. 39(5–6): p. 388–393. [DOI] [PubMed] [Google Scholar]
- 54.Katoh K., Misawa K., Kuma K. I. and Miyata T. Mafft: A novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Research 2002. 30: p. 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katoh K. S a.D.M.., Mafft multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution 2013. 30: p. 772–780. doi: 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Mentjies P. and Drummond A. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. 2012. 28: p. 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Darriba D., Taboada G. L., Doallo R. and Posada D. Jmodeltest 2: More models, new heuristics and parallel computing. Nature Methods, 2012. 9: p. 772–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Posada D., Jmodeltest: Phylogenetic model averaging. Molecular Biology and Evolution 2008. 25: p. 1253–1256. doi: 10.1093/molbev/msn083 [DOI] [PubMed] [Google Scholar]
- 59.Guindon S. G. a.O., A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic biology 2003. 52: p. 696–704. [DOI] [PubMed] [Google Scholar]
- 60.King R., et al. , The completed genome sequence of the pathogenic ascomycete fungus Fusarium graminearum. Bmc Genomics, 2015. 16 544 doi: 10.1186/s12864-015-1756-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hohn T.M. and Desjardins A.E., ISOLATION AND GENE DISRUPTION OF THE TOX5 GENE ENCODING TRICHODIENE SYNTHASE IN GIBBERELLA-PULICARIS. Molecular Plant-Microbe Interactions, 1992. 5(3): p. 249–256. [DOI] [PubMed] [Google Scholar]
- 62.Proctor R.H., Hohn T.M., and McCormick S.P., Restoration of wild-type virulence to Tri5 disruption mutants of Gibberella zeae via gene reversion and mutant complementation. Microbiology-Uk, 1997. 143: p. 2583–2591. [DOI] [PubMed] [Google Scholar]
- 63.Fernandez-Ortuno D., Atkins S.L., and Fraaije B.A., The use of a CYP51C gene based PCR-RFLP assay for simultaneous detection and identification of Fusarium avenaceum and F. tricinctum in wheat. International Journal of Food Microbiology, 2011. 145(1): p. 370–374. doi: 10.1016/j.ijfoodmicro.2011.01.011 [DOI] [PubMed] [Google Scholar]
- 64.Urban M., et al. , The Fusarium graminearum MAP1 gene is essential for pathogenicity and development of perithecia. Molecular Plant Pathology, 2003. 4(5): p. 347–359. doi: 10.1046/j.1364-3703.2003.00183.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Wild type or 23–170 mutant spore suspensions at 104 spores / ml in water was inoculated (5 μl) onto a YPD plate. The inoculated region was photographed at 24h intervals for a macroscopic analysis (upper panels). Middle panels display the typical characteristic spore morphologies of each strain grown on YPD agar coated slides for 24 hours. Lower panels show spores under higher magnification stained with the chitin binding fluorophore calcoflour white.
(TIF)
A spore suspension of 104 spores / ml in water was inoculated (5 μl) onto a water agar plate at the indicated agar concentration. Plates were incubated at RT then photographed after 10 days.
(TIF)
A spore suspension of 104 spores / ml water was inoculated (5 μl) onto the surface of the indicated agar plate. Plates were incubated at RT then photographed after 21 days.
(TIF)
A spore suspension of 104 spores / ml water of wild-type or ΔZtGT2 was inoculated (5 μl) onto the surface of the indicated agar plate. Plates were incubated at RT then photographed after 21 days (Left panels). Hyphal morphology radiating from the colony edge was studied by light microscopy (right panels).
(TIF)
Gene replacement strategy for the ZtGT2 orthologue from F. graminearum with gene Id FG00702.1 (http://fungi.ensembl.org/Fusarium_graminearum/Info/Index) (A) Genomic left border and right border regions (white bars) were amplified with primers and fused to parts of the hph hygromycin resistance gene. Fused PCR fragments were used in a split-marker strategy to replace FGRRES_00702. Horizontal black bars represent genomic areas outside the replacement construct, vertical black bars represent 3 introns. (B) Anticipated diagnostic PCR for successful gene replacement of FGRRES_0702. (C) Results of diagnostic PCR and expected sizes indicated in (A) and (B). Transformants S#11, S#19 and S#24 have the ΔFGRRES_00702 null allele but also retain a wild-type gene (heterokaryons). This was observed for all hygromycin resistant strains originating from 4 independent transformations. M—λ DNA-BstEII digest; WT—wild type. (D) Growth of F. graminearum heterokaryotic strains carrying both a WT and FGRRES_00702 null allele on potato dextrose agar after 72 hours incubation. No alteration in hyphal growth was observed. Top left to bottom right: WT, FG transformants S#10, 11, 15, 19, 24.
(TIF)
Penetration of cellophane membranes by F. graminearum strains. Fungal spores were plated and grown for 2 d at 22°C on top of cellophane membrane on the surface of PDA plates (images labelled “Before”). The cellophane with the fungal colonies was removed and plates were incubated for an additional two days to determine whether fungal growth occurred on the plates, indicating penetration of the cellophane disk (images labelled “After”). The ΔFgMAP1 mutant strain was used as a control strain shown previously to be defective in cellophane and plant penetration [64].
(TIF)
Fungal mycelium grown at 25°C in liquid YPD was subjected to crude protein fractionation and western blot analysis with an anti-ZtGT2 peptide antibody. (A) The antiserum detects a ~56kDa protein found only in the cell wall fraction of wild type fungal cells (B) The specific binding to this protein in wild type cells is confirmed by a peptide competition experiment.
(TIF)
Arrows highlight the different surface structures observed in the mutants.
(TIF)
Fungal strains were grown in shaking culture flasks until saturation and then separated by filtration. AIR was generated from cell walls, samples were hydrolysed and monosaccharaides levels (glucose, mannose and galactose) were quantified (A). Culture filtrates were ethanol precipitated and then analysed the same way (B). Data shown derives from three biological replicates and in each case and monosaccharide levels are expressed in milligrams / gram of total dry weight (mg /g. dwt).
(TIF)
Conidial suspensions were left to adhere to either plastic petri dishes (upper panels) before extensive washing and counting of remaining adhered spores (from micrograph images taken before and after washing). Lower panels- cell suspensions were applied to surface of wheat leaves or to glass coverslips and allowed to adhere. Leaves or coverslips were then washed by vortexing and the resulting suspensions plated out on YPD agar + 100 μg/ml G418 antibiotic. Plates were incubated for 6 days and colonies were counted from photographic images. A decrease in the number of colony forming units (cfus) retrieved from leaf surfaces relative to glass coverslips was taken as indicative of adhesion. There were no statistical differences between wild-type and ΔZtGT2 strains from all assays.
(TIF)
Wild type; 23–170 mutant; ΔZtGT2 and 23–170::GT2comp spore suspensions of initially 106 spores / ml (or three successive 3-fold dilutions in water), were inoculated (5 μl) onto a YPD agar plates containing the added stress agent. Plates were then photographed after growth at 16°C or 30°C for six days.
(TIF)
(TIF)
Seven genes in this category show significantly reduced expression in ΔZtGT2 mutants growing both in liquid culture and on leaf surfaces.
(TIF)
(TIF)
An independent experiment was performed growing the wild type and ΔZtGT2 fungus in either YPD or PDB broth for 5 days. Real-Time qRT-PCR was then used to measure relative expression of the Zt3LysM effector. Data was normalised to the expression of the Z. tritici beta tubulin gene and presented as fold change relative to gene expression by the WT fungus in YPD broth.
(TIF)
(A) indicates the number of transcripts with higher expression in wheat leaves inoculated with WT fungus than with ΔZtGT2. (B) qRT-PCR validation of expression of the two indicated wheat PR genes. Data is normalised to expression of the wheat cdc48 gene.
(TIF)
(XLSX)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.