Abstract
Dorso-ventral pigment patterning, characterized by a light ventrum and a dark dorsum, is one of the most widespread chromatic adaptations in vertebrate body coloration. In mammals, this countershading depends on differential expression of agouti-signaling protein (ASIP), which drives a switch of synthesis of one type of melanin to another within melanocytes. Teleost fish share countershading, but the pattern results from a differential distribution of multiple types of chromatophores, with black-brown melanophores most abundant in the dorsal body and reflective iridophores most abundant in the ventral body. We previously showed that Asip1 (a fish ortholog of mammalian ASIP) plays a role in patterning melanophores. This observation leads to the surprising hypothesis that agouti may control an evolutionarily conserved pigment pattern by regulating different mechanisms in mammals and fish. To test this hypothesis, we compared two ray-finned fishes: the teleost zebrafish and the non-teleost spotted gar (Lepisosteus oculatus). By examining the endogenous pattern of asip1 expression in gar, we demonstrate a dorso-ventral graded distribution of asip1 expression that is highest ventrally, similar to teleosts. Additionally, in the first reported experiments to generate zebrafish transgenic lines carrying a bacterial artificial chromosome (BAC) from spotted gar, we show that both transgenic zebrafish lines embryos replicate the endogenous asip1 expression pattern in adult zebrafish, showing that BAC transgenes from both species contain all of the regulatory elements required for regular asip1 expression within adult ray-finned fishes. These experiments provide evidence that the mechanism leading to an environmentally important pigment pattern was likely in place before the origin of teleosts.
Keywords: Agouti, pigmentation, BAC, Spotted gar
INTRODUTION
The origin of vertebrate pigment patterns – the differential pigmentation of skin, scales, hair, or feathers - is a classic problem in developmental biology. The extension and agouti genes are both key loci involved in the regulation of body pigmentation. The agouti locus encodes the Agouti-signaling protein (Asip), a ligand protein that competes with α-MSH in binding to the Mc1r receptor, which, in turn, is encoded by the extension locus. This competitive antagonism results, in amniotes, in melanocytes switching between the production of eumelanin (black/brown pigment) and pheomelanin (red/yellow pigment) (Lu et al., 1994). In amniotes, high Asip expression in the ventral region inhibits Mc1r signaling, which results in a pale belly coloration (phaeomelanin≫eumelanin) in contrast to the darker dorsal region (eumelanin≫phaeomelanin); the dorso-ventral pigment pattern gradient is adaptive and widely conserved in the animal kingdom, including fish (Vrieling et al., ‘94; Manceau et al., 2011). Curiously, this conserved pattern of dorsoventral pigment polarity occurs by a different mechanism in fish and mammals: in fish, the pattern appears by the distribution of different pigment cell types that produce chemically distinct pigments rather than by the switching of melanin types within the same kind of pigment cell as in mammals. In zebrafish, a teleost fish, the pigment pattern depends on the patterned distribution of three different pigment cell types: dark eumelanin-producing melanophores (homologous to the mammalian melanocytes, but without the ability to synthesize the lighter pheomelanin (Kottler et al., 2015), yellow to orange xanthophores, which produce pteridines, and silver shiny iridophores, which produce reflective plates of guanine crystals. These different pigment cell types derive from embryonic neural crest (Le Douarain and Kalcheim, ‘99; Dupin et al., 2000).
In adult zebrafish, melanophores are distributed in four horizontal stripes alternating with three white interstripes that are rich in iridophores and xanthophores; the ventrum is pale due to an abundance of iridophores and the absence of melanophores (Kelsh et al., 2009). Recent studies have characterized asip1 genes in several teleost fish species, showing that its high ventral expression is a conserved feature (Cerdá-Reverter et al., 2005; Klovins and Schiöth, 2005; Kurokawa et al., 2006; Guillot et al., 2012; Ceinos et al., 2015). Our previous experiments demonstrated a gradient of Asip1 expression with lowest expression levels in the dorsal region. Disruption of this expression gradient by gain-of-function approaches using ubiquitous promoters (Tol2-Eef1α1/asip1) interrupted pigment patterns by inhibiting dorsal melanogenesis and melanophore differentiation. Asip1 overexpression, however, had a minor impact on the pattern of stripes revealing the existence of two different pigment patterns in zebrafish: a dorsoventral pattern and stripe pattern, which are regulated by different mechanisms (Ceinos et al., 2015). Consequently, these results suggest that Asip-dependent countershading and its molecular machinery are conserved between mammals and teleost fish, but, rather surprisingly, the cellular basis for pigment pattern formation is different in the two groups.
We ask here the question: is the patterning mechanism found in teleosts specific to that taxon, perhaps arising as a feature of the teleost genome duplication (TGD) (Amores et al 1998; Postlethwait et al 1999; Taylor et al., 2003; Jallion et al., 2004), or alternatively, is it a more ancient feature shared by other groups of ray-finned fishes? To address this question, we investigated the regulation of asip1 in spotted gar, which represents a ray-finned lineage that diverged from teleosts before the TGD (Amores 2011).
To gain insight into the evolutionary conservation of these processes in ray-finned fish, we compare here the promoter regions of zebrafish and spotted gar (Lepisosteus oculatus). Gars diverged from the teleost lineage around 350 million years ago (Hoegg et al., 2004; Vandepoele et al., 2004; Hurley et al., 2007) just before the teleost specific genome duplication in a teleost ancestor (TGD), and gar is therefore considered a useful outgroup to the teleost lineage (Amores et al., 2011; Braasch et al., 2016). The use of a lineage closely related to teleosts, but diverging before the TSGD, facilitates connectivity, the sharing of biological understanding among ray-finned fish and the translation of knowledge to tetrapods including mammalian species (Braasch et al., 2016). Spotted gar can be reared in the laboratory and is thereby amenable for the study of spatial-temporal patterns of gene expression during development and the functional testing of hypotheses about the origin of gene activities in divergent fish lineages (Amores et al., 2011; Braasch et al., 2015, 2016).
Exploiting these advantages, we studied the functional evolution of the asip1 locus in ray-finned fish. We describe here the generation of bacterial artificial chromosome (BAC) transgenic (Tg) zebrafish lines, which harbor zebrafish or spotted gar BACs carrying the asip1 gene modified to drive enhanced green fluorescent protein (eGFP) expression. The resulting transgenic animals carrying asip1 BAC transgenes from either species showed eGFP expression in a pattern that mirrors that of endogenous asip1 gene expression in adult zebrafish and, also of spotted gar, and illustrate that the transgenic zebrafish is advantageous for identifying the conserved regulatory elements required for normal asip1 expression within adult fish. Thereby, our study is the first to generate zebrafish transgenic lines carrying a BAC clone from spotted gar. Additionally, we show a dorso-ventral gradient of asip1 expression in the skin of adult spotted gar with lower levels in the dorsum and higher the belly, similar to the expression gradient in zebrafish, and other teleosts.
MATERIALS AND METHODS
Fish
Zebrafish of TU strain (Tübingen, Nüsslein-Volhard Lab) were cultured as previously described (Westerfield, 2007) and staged according to Kimmel et al. (‘95). Ethical approval (Ref. Number: AGL2011-23581) for all studies was obtained from the Institutional Animal Care and Use Committee of the IIM-CSIC Institute in accordance with the National Advisory Committee for Laboratory Animal Research Guidelines licensed by the Spanish Authority (RD53/2013) and conformed to European animal directive (2010/63/UE) for the protection of experimental animals. Spotted gar (Lepisosteus oculatus) were reared and collected at the Institute of Neuroscience (University of Oregon). This work was approved by the University of Oregon institutional Animal Care and Use Committee (Animal Welfare Assurance Number A-3009-01, IACUC protocol 12-02RA).
Comparative genomic analysis and sequence conservation
To analyze the conserved sequences between zebrafish and spotted gar and identify any conserved non-coding elements (CNEs), repeat-masked sequences were aligned using MLAGAN alignment software in m-VISTA (Mayor et al., 2000; Brudno et al., 2003). Conserved sequences were predicted under 65% identity and 50bp window. Sequences analyzed were zebrafish masked sequence (http://www.ensembl.org/Danio_rerio/Info/Index) as reference genome and spotted gar masked sequence (http://www.ensembl.org/Lepisosteus_oculatus/Info/Index). The protein alignment was performed using the Expresso alignment with the T-coffee software. The predicted protein sequence was obtained from the Ensembl zebrafish genome database (ENSDART00000113083) and the GenBank database for spotted gar (XM_015365014.1). We used the web-based Genomicus synteny browser [http://www.dyogen.ens.fr/lcgi-bin/search.pl] and Ensembl genome browsers to assess the synteny between regions from zebrafish BAC clone (16N21) from DanioKey Library and spotted gar BAC clone (VMRC-56-161-M17) from BAC library VMRC-56 (Postlethwait Lab, Institute of Neuroscience, University of Oregon). Only genes annotated in these assembly versions were considered for our synteny comparison.
RNA isolation, RT-PCR and qRT-PCR
Animals were anesthetized with MS-222 and decapitated. Total RNA was extracted from adult fish (210 dpf zebrafish and 270 dpf spotted gar) from eye, gonads, liver, brain, gill, intestine, muscle, heart and dorsal and ventral skin. Furthermore, dorsal, stripes (1D, 1V, 2D and 2V in zebrafish; 2D, Medium and 2V in spotted gar) and belly skin samples of eight fish of both species were excised with a scalpel for qRT-PCR (see Fig. 3A, B). Tissue samples were frozen at −80ºC in RNAlater (Ambion, USA) until analysis. Total RNA was extracted with Direct-zol RNA miniprep kit (Zymo research) according to the manufacturer’s instructions. The Maxima First Strand cDNA Synthesis Kit (Fermentas) was used to synthetize cDNA from 100 ng of total RNA. Tissue expression patterns were analyzed by RT-PCR. A pair of specific primers was designed for spotted gar asip1 and spotted gar β-actin as internal control from the mRNA sequence published in Ensembl database (ENSLOCT00000008713 and ENSLOCT00000005654 respectively). Published zebrafish asip1 and zebrafish eEf1α primers were used for RT-PCR (McCurley and Callard, 2008; Ceinos et al., 2015). PCR reactions were performed in 25 μL volumes containing 1 μL of cDNA, 0′5 μL of each primer (10 μM), 12′5 μL of Dream Taq green PCR Master Mix (2×) and H2O nuclease free to 25 μL. PCR conditions were 94ºC for 5 min, 35 cycles of 94ºC for 30 sec, 60ºC for 30 sec, 72ºC for 2 min, and a final step of 72ºC for 7 min. PCR products were run on 1% agarose gels. Quantitative asip1 mRNA expression in the skin was determined by qRT-PCR. Each set of skin samples from zebrafish and spotted gar was set up in triplicate. The qRT-PCR reaction was carried out in 20 μL volumes, containing 1 μL of cDNA (diluted 1:10), 0′5 μL of each primer (10 μM), 12′5 μL of SYBR Green/ROX qPCR Master Mix (2×) (ThermoFisher) and H2O nuclease free to 20 μL. The thermal protocol was: 95ºC for 10 min and 40 cycles of 95ºC for 15 sec and 60ºC for 1 min. Primers are listed in Supplementary S1.
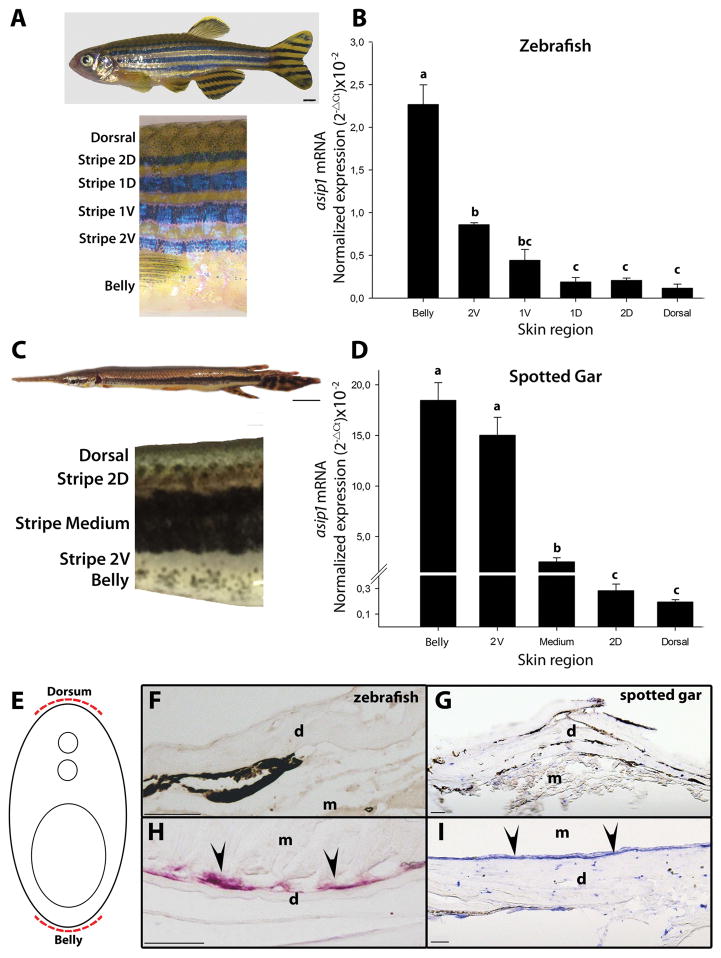
Figure 3.
Dorso-ventral gradient expression of asip1 in adult zebrafish (A) and spotted gar skin (C). Normalized gene expression levels of (B) zebrafish and (D) spotted gar asip1 in skin samples from belly to dorsum (locations of skin sample points are indicated in the picture). Shown are log 10-transformed ΔCt values of asip1 relative to eEf1α and β-actin, respectively. Data are the mean ±SEM from eight samples after triplicate PCR analysis. (E) Schematic representation of the rostro-caudal transverse sections. (F–I) Rostro-caudal transverse sections (12 μm thick) of paraffin embedded, in situ hybridization of asip1 expression in 210 dpf zebrafish (F,H) and 270dpf spotted gar (G,I). The sections in (H,I) shows the expression of asip1 transcripts in the dermis region of zebrafish (H) and spotted gar (I) ventral skin (black arrows). No aisp1 expression was found in the dermis region of dorsal skin of both species (C,D). Superscripts a, b, c and d indicate statistical differences (P<0.05) in gene expression levels among skin region (statistics data are similar if share at least one letter not sure what you mean). Scale bar: (A) 2 mm, (C) 1 cm, (F,G,H,I) 50 μm. d, dermis; m, muscle.
RNA in situ hybridization and confocal imaging
RNA in situ hybridization on zebrafish (210 dpf; 22mm) and spotted gar (270 dpf; 130 mm) paraffin sections were carried out following methods as described (Cerdá-Reverter et al., 2000; Rotllant et al., 2008). Fish (n=4; 2 of each fish species) were fixed in 4% paraformaldehyde and 0.1 M phosphate buffer, for 2 days at 4ºC, dehydrated and embedded in Paraplast (Sherwood, St Louis, MO, USA). Serial 12μm transverse anatomical sections (rostrocaudal direction) were cut using a rotary microtome. Sections were mounted on 3-aminopropyltriethoxylane (TESPA)-treated slides, air-dried at room temperature (RT) overnight and stored at 4ºC under dry conditions and used for hybridization within one month. RNA in situ hybridization was performed using digoxigenin-labeled antisense riboprobes as previously described (Rotllant et al., 2008). Antisense and sense riboprobes were made from linearized full-length zebrafish asip1 and partial-length spotted gar asip1. For cryostat sectioning, 150 dpf Tg(Drer.asip1-iTol2-eGFP-BAC) and Tg(Loc.asip1-iTol2-eGFP-BAC) fish were fixed in 4% paraformaldehyde overnight at 4ºC, washed with PBS, transferred to 15% sucrose, followed by 30% sucrose, and then embedded and frozen in OCT media. Serial 25μm rostro-caudal transverse sections were collected on Poly-Lysine slides (Thermo scientific) and allowed to dry. Slides were washed in PBS, mounted in mowiol and imaged on a Leica TCS SP5 confocal microscope.
Isolation of BAC clones containing the Asip1 locus and BAC transgene construction
The zebrafish BAC clone (16N21, 214 Kb) from Danio Key Library, which includes the asip1 locus, was ordered from Source BioScience (Nottingham, UK). Spotted gar BAC clone (VMRC-56-161-M17, 130 Kb) from BAC library VMRC-56 (Braasch et al., 2016) was identified by PCR screening with a pair of asip1-specific primers that were designed using the asip1 gene sequence in the spotted gar Ensembl database (accession no. ENSLOCG00000007193). BAC transgene construction was performed as previously described (Suster et al., 2011). We used homologous recombination in E. coli to introduce the iTol2-amp cassette from the pCR8GW-iTol2-amp plasmid (Suster et al., 2011) into the BAC plasmid backbone, which contains the inverted minimal cis-sequences required for Tol2 transposition. The insertion of the reporter gene eGFP into the Drer.asip1 and Loc.asip1 locus of Drer.asip1-iTol2-BAC clone and Loc.asip1-iTol2-BAC clone, respectively, was carried out by homologous recombination. Briefly, the eGFP-Kan reporter gen cassette was amplified from eGFP-pA-FRT-Kan-FRT plasmid by PCR, together with a sequence of 50bp homologies to the Drer.asip1 or Loc.asip1 translation start site. After transformation of Drer.asip1-iTol2 or Loc.asip1-iTol2 BAC clone-containing cells with the PCR product, homologous recombination took place between the PCR product and the Drer.asip1-iTol2 or Loc.asip1-iTol2 BAC clone resulting in integration of the vector insert into the BAC clone, placing the eGFP reporter gene under control of the Drer.asip1 or Loc.asip1 regulatory regions within the clone (Drer.asip1-iTol2-eGFP-BAC or Loc.asip-iTol2-eGFP-BAC). Primers used are listed in Supplementary S1.
Generation of BAC-transgenic zebrafish and analysis of transgene expression
BACs functionalized with iTol2kan were prepared in a concentration of 200 ng/μL together with Tol2 transposase mRNA in a concentration of 200 ng/μL and Phenol red solution (0,1%). Around 2nL of this mixture was microinjected into the cytoplasm of zebrafish embryos at the one- or two-cell stage. A dissection microscope (MZ8, Leica) equipped with a MPPI-2 pressure injector (ASI systems) was used for microinjection. To observe pigment cells, double transgenic zebrafish lines (Tg(Drer.asip1-iTol2-eGFP-BAC;sox10:mRFP) and Tg(Loc.asip1-iTol2-eGFP-BAC;sox10:mRFP)) were obtained by setting up crosses between the asip1 BAC reporter gene transgenic lines and Tg(sox10:mRFP) (M. Levesque; obtained from the Nüsslein-Volhard Lab). For fluorescent microscope imaging, transgenic zebrafish of 3-days post-fertilization (dpf) (n=50), 30 dpf (n=20) and 100 dpf (n=15) were anesthetized with tricaine methasulfonate (MS-222-, Sigma-Aldrich) and photographed with a Leica M165FC stereomicroscope equipped with a Leica DFC310FX camera. Confocal imaging was carried out in a Leica TCS SP5 confocal microscope.
RESULTS
Genomic and peptide comparison
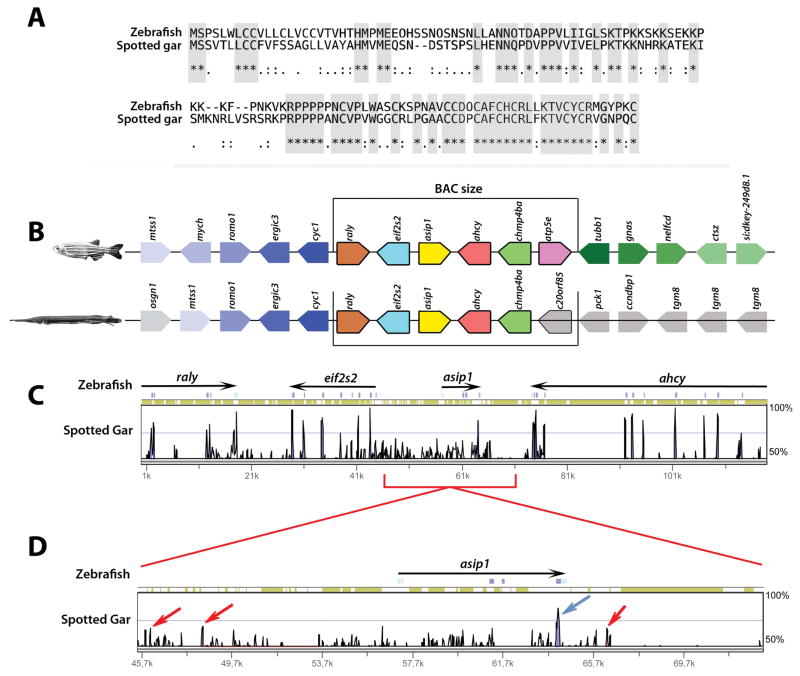
We determined the organization of the asip1 locus in gar and compared it to zebrafish. In both gar and zebrafish, the asip1 gene has three coding exons distributed over regions of about 3.6 and 7.4 kb, respectively. Peptide alignment shows an identity level of 47% and a similarity of 62% between zebrafish and spotted gar Asip1 (Fig. 1A; Supplementary S2B). Comparative mapping of asip1 regions in zebrafish and spotted gar shows high conserved genomic organization (Fig. 1B). Sequence conservation analysis of the genomic region upstream and downstream of asip1 was performed to learn if any non-coding elements are conserved between zebrafish and spotted gar. MLAGAN alignments of genomic sequence spanning 6:49873114 to 6:49991996 for Zebrafish (Zv9), and 18:14032011 to 18:14456756 for gar (LepOcu1) show few conserved non-coding regions (red arrows) and coding regions (blue arrows) (Fig. 1C,D).
Figure 1.
Identification of zebrafish and spotted gar BACs likely to recapitulate asip1 expression. (A) T-coffee asip1 peptide sequence alignment of Drer.asip1 (ENSDART00000113083.2) and Loc.asip1. (ENSLOCT00000008713.1). Grey boxes show conserved amino acid residues. (B) Genomic organization and gene synteny comparison for genes annotated from zebrafish BAC clone (16N21) from DanioKey Library and spotted gar BAC clone (VMRC-56-161-M17) from BAC library VMRC-56 (Postlethwait Lab; Institute of Neuroscience, University of Oregon). Picture style was adapted from Genomicus [http://www.dyogen.ens.fr/genomicus-57.01/cgi-bin/search.pl]. Genes are depicted by colored polygons and transcriptional orientation is indicated at the angled end of each gene. Gene names are indicated above. Orthologs across the two species are depicted in the same colors. (C, D) Distribution of the spotted gar conserved coding and non-coding regions. Vista browser representation of the genomic region surrounding the spotted gar asip1 gene (LepOcu1, chr18:14032011-14456756), using Zv9 zebrafish genome (chr6:49873114-49991996) as reference. Conservation is represented as vertical peaks. Conserved non-coding elements (CNEs) and coding elements with possible enhancer activity are marked as red and blue bars, respectively.
Expression of asip1 in spotted gar compared to zebrafish
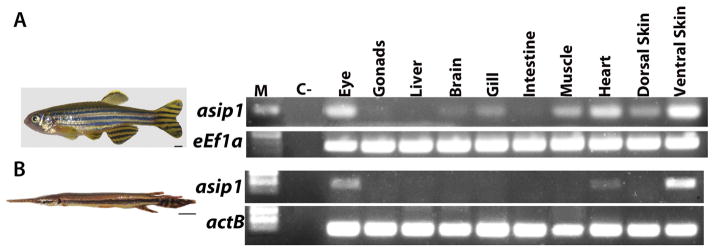
To investigate the role of Asip1 in establishing the spotted gar dorso-ventral pigment pattern, we first compared the tissue expression distribution of asip1 expression as detected by RT-PCR in zebrafish (Fig. 2A) and spotted gar (Fig. 2B). In 270 dpf adult gar, high levels of asip1 mRNA were found only in the eye and ventral skin and were present at lower levels in the heart (Fig. 2B). In 11 dpf spotted gar larvae, asip1 expression was not found in the skin (data not shown). Detecting asip1 expression particularly in the ventral skin of zebrafish and adult gar (Fig. 2A and 2B), was consistent with a possible role in adult pigment pattern formation, but did not have the quantitative resolution to decide whether that expression might show a spatial difference. To test this question, we investigated whether there were dorso-ventral differences in expression levels by qRT-PCR. We measured asip1 transcript levels in skin from different dorso-ventral positions, specifically, in the belly, each body stripe (2V, 1V, D and 2D) and dorsal skin regions for zebrafish (Fig. 3A) and belly, each body stripe (2V, Medium and 2D) and dorsal skin regions for spotted gar (Fig. 3C). Figure 3B shows the expression levels of asip1 in skin samples obtained along the dorso-ventral axis in wild-type adult zebrafish. The highest levels of expression of asip1 were found on the belly; however, asip1 transcripts showed a gradual decrease from the belly to the dorsal midline of the fish. Figure 3D shows the expression levels of asip1 in skin samples obtained along the dorso-ventral axis in adult gar. Equivalent to what is found in zebrafish, the highest levels of expression of asip1 in gar were also found in the belly; and asip1 transcripts similarly showed a gradual decrease from the belly to the dorsal midline of the fish. Additionally, we use mRNA in situ hybridization to examine whether asip1 mRNA was dorsoventrally expressed. In adult fish, we only saw detectable asip1 expression in the ventral skin area in both species (Fig. 3E,F,G,H,I). This expression analysis suggests that asip1 may have a conserved function among ray-finned fish and that function might be conserved with mammals, which also suggests that the regulation and the asip1-dependent mechanism might be conserved among ray-finned fish and mammals.
Figure 2.
Spatial expression of asip1 gene in zebrafish (A) and spotted gar (B). RT-PCR analysis of tissue specific expression pattern of asip1 in adults. Scale bar: (A)2 mm, (B) 1 cm.
Generation and molecular analysis of BAC transgenic zebrafish
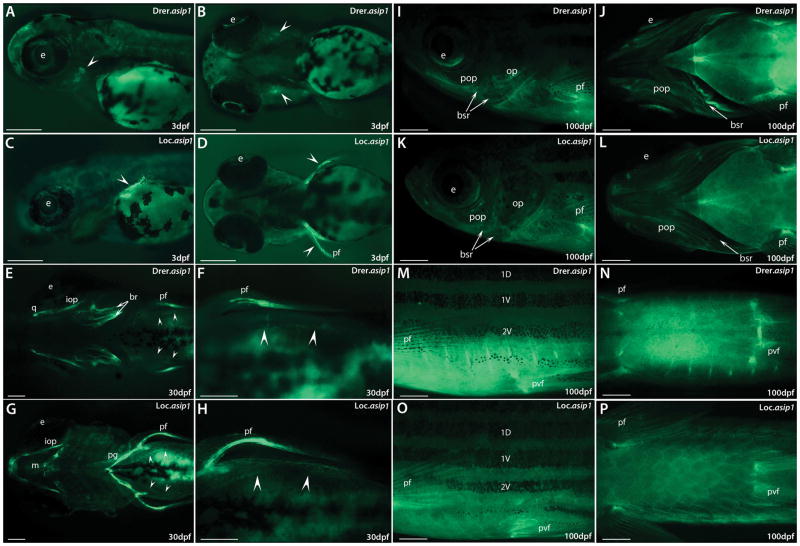
Having shown that asip1 was expressed with dorso-ventral asymmetry in the skin of both zebrafish (Fig. 3B) and gar (Fig. 3D), we generated transgenic zebrafish lines to characterize conserved regulatory elements required for the evolutionarily conserved dorso-ventral asip1 expression pattern found in zebrafish and spotted gar. We first tried conventional reporter transgene approaches generating several constructs of different sizes up to 13kb of the promoter sequence upstream of the zebrafish asip1 TSS (Transcription Starting Site) (data not shown). Injection of these reporter constructs did not show eGFP expression, suggesting that the regulatory elements of asip1 are not located in the regions used in the reporter assay. Therefore, we created four independent BAC transgenic zebrafish lines, containing either a BAC from a zebrafish or a spotted gar library carrying the asip1 gene modified to drive enhanced green fluorescent protein (eGFP) (two lines per BAC/species). In stable transgenic lines, eGFP expression was first detectable in 3 dpf embryos, when the developing opercle was the initial site of expression for Drer.asip1-iTol2-eGFP-BAC (Fig. 4A–B, White arrows), which mimicked the already well described expression pattern of the endogenous asip1 mRNAs in zebrafish (Ceinos et al., 2015) and in the developing pectoral fins for the spotted gar BAC (Fig. 4C–D, white arrows). At 30 dpf, eGFP expression was still present in the opercle, dermal branchiostegal rays, quadrate dermal bone and pectoral fin folds for Drer.asip1-iTol2-eGFP-BAC (Fig. 4E–F). In the Loc.asip1-iTol2-eGFP-BAC transgenic line, eGFP was mainly expressed in the pectoral fin folds, interopercular bone and pectoral girdle (Fig. 4G–H). Furthermore, eGFP was also detectable in ventral skin area (Fig. 4F–H, white arrows) in both BAC transgenic lines. At 100 dpf, strong eGFP expression was seen in the ventral skin area, in the base of pectoral and pelvic fins, and eGFP expression was still detectable in the pharyngeal region in both BAC transgenic lines (Fig. 4I–P). eGFP expression in adult zebrafish (150 dpf) monitored by direct observation under confocal fluorescence was strongly detected in the ventral skin region and peritoneal membrane, while low expression levels were found in the dorsal skin region in both BAC transgenic lines (Fig. 5A–D). Therefore, at 100–150 dpf, both transgenic zebrafish lines containing either a BAC from a zebrafish or a spotted gar library showed highly similar eGFP expression patterns recapitulating the spatial pattern of endogenous asip1 transcript in adult zebrafish (Fig. 3C–F).
Figure 4.
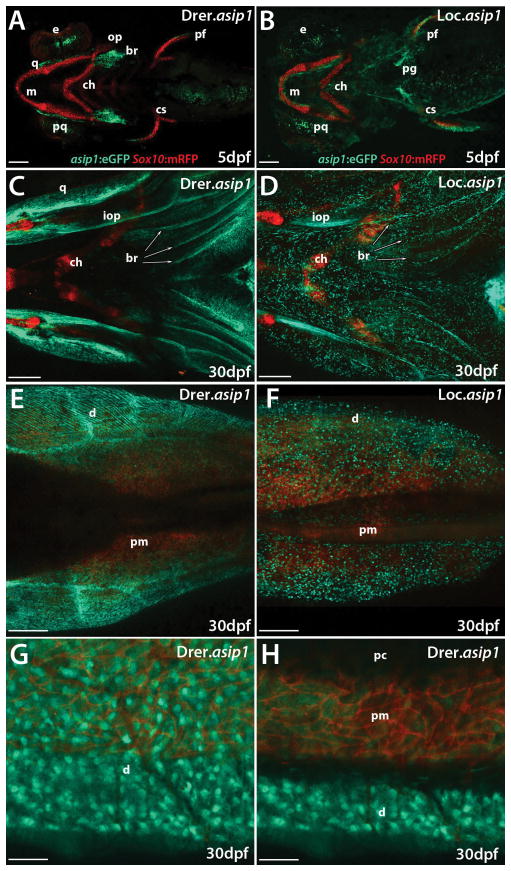
BAC transgenes exhibit endogenous asip1-like expression in most tissues. Shown are representative eGFP fluorescence images in Drer.asip1-iTol2-eGFP-BAC (A,B,E,F,I,J,M,N)) and Loc.asip1-iTol2-eGFP-BAC (C,D,G,H,K,L,O,P) stable transgenic zebrafish lines. (A,C,I,K,M,O) are lateral views anterior to the left and (B,D,E,F,G,H,J,L,N,P) ventral views anterior to the left. eGFP fluorescence is first detected at 3dpf (A,B,C,D). (A, B) lateral and ventral views of eGFP expression in larval heads of Drer.asip1-iTol2-eGFP-BAC transgenic zebrafish showing specific expression restricted to the developing opercle. (C,D) lateral and ventral views of eGFP expression in larval heads of Loc.asip1-iTol2-eGFP-BAC transgenic zebrafish showing expression limited to the pectoral fins. (E–H) ventral views of eGFP expression in 30dph larvae, showing increased expression in different pharyngeal bone structures (opercle, dermal branchiostegal rays, quadrate dermal bone and pectoral fin folds) and in ventral skin area (white arrows) in Drer.asip1-iTol2-eGFP-BAC (E,F) and in the pectoral fin folds, interopercular bone, pectoral girdle and ventral skin area (white arrows) in Loc.asip1-iTol2-eGFP-BAC transgenic zebrafish line (G–H). (I–P) eGFP expression in 100dph fish, showing increased expression in the ventral skin region and the base of pectoral and pelvic fins of both BAC transgenic lines (M–P), eGFP was also detectable in different bone structures of pharyngeal region in both BAC Transgenic lines (I–L). Scale bars: (A–H) 200 μm; (I–P) 1mm. br, brachiostegals; e, eye; iop, interopercular bone; m, Meckel’s cartilage; op, operculum; pf, pectoral fin; pop, preopercle; pvf, pelvic fin; pg, pectoral girdle q, quadrate; 1D, 1V, 2V stripes.
Figure 5.
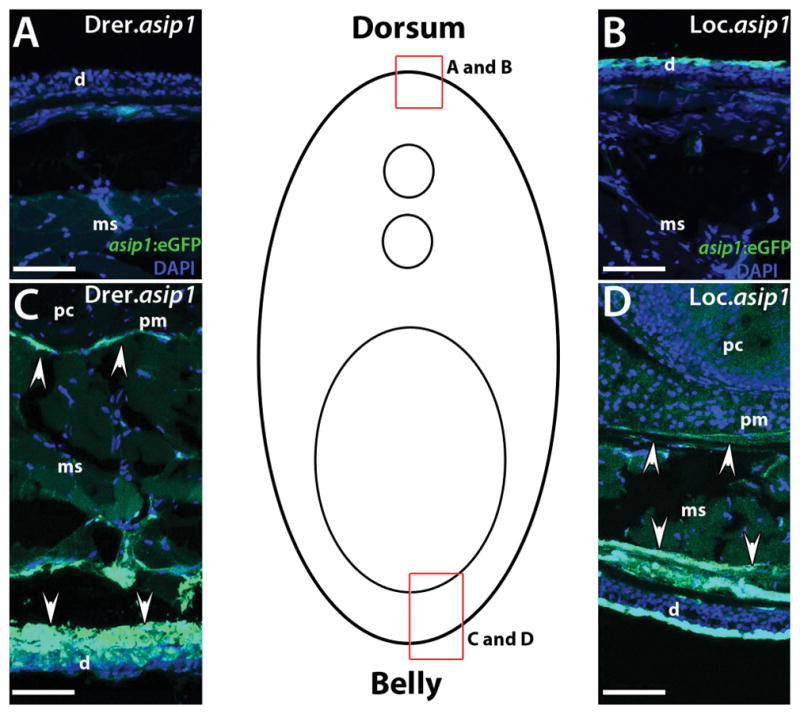
Comparison of eGFP expression in rostro-caudal transverse sections (25 μm) from 150dpf Drer.asip1-iTol2-eGFP-BAC (A,C) and the Loc.asip1-iTol2-eGFP-BAC (B,D) transgenic zebrafish. Drer.asip1-iTol2-eGFP-BAC and Loc.asip1-iTol2-eGFP-BAC zebrafish show strong eGFP expression in the ventral skin region (dermis) and peritoneal membrane (C,D). Variegated eGFP fluorescence is seen in Loc.asip1-iTol2-eGFP-BAC dorsal dermis area (B). Cells that do not express eGFP are revealed with DAPI staining (blue color). Scale bars: 50 μm. d, dermis; ms, muscle; pc, peritoneal cavity; pm, peritoneal membrane.
To further characterize cells expressing asip1 in the developing opercle, dermal branchiostegal rays, quadrate dermal bone, pectoral fins and ventral skin, we crossed BAC transgenic lines to a sox10:mRFP line, which marks neural crest derivatives with membrane-bound mRFP (Fadeev et al. 2015) (Fig. 6). Interestingly, although both asip1 and sox10 transgenes were expressed in the pharyngeal bones and pectoral fins, they were expressed in different cell types (Fig. 6A–F, revealing that the asip1 gene (eGFP positive cells) was not expressed in the pre- and post-migratory neural crest cells (mRFP positive cells) in both zebrafish or spotted gar BAC transgenic lines. Figure 6G–H shows the sox10:mRFP positive cell localization in the ventral skin region (E–G) and dorso-ventral sagittal section (H) in Drer.asip1-iTol2-eGFP-BAC 30dph zebrafish transgenic line. mRFP-labeled neural crest derivatives were predominantly present in the peritoneal membrane region where a reflecting iridophore layer is located. In contrast, eGFP-labeled asip1 cells in the peritoneal membrane region were present, but remained mostly in the ventral dermis area (Fig. 6H). This revealed that asip1 gene was not detected in pre- and post-migratory neural crest cells and their derivatives, which strongly suggests that asip1 is not expressed by the pigment cells itself but by surrounding dermal cells. Therefore, we show that the asip1 gene is specifically expressed in fish dermal fibroblasts (Supplementary S3).
Figure 6.
Confocal image projections of live Tg(Drer.asip1-iTol2-eGFP-BAC/sox10:mRFP) (A,C,E,G,H) and Tg(Loc.asip1-iTol2-eGFP-BAC/sox10:mRFP) (B,D,F) 5dpf larvae and 30dpf juvenile zebrafish. (A,B,C,D) Ventral view of a live 5 dpf larvae, anterior is to the left. sox10:mRFP specifically labels the pre- and post-migratory neural crest cells. (A,B) mRFP strongly labels the cartilages of maxillary, mandibular arches and pectoral fins. (A) Drer.asip1-iTol2-eGFP-BAC labels the developing opercle and dermal branchiostegal rays. (B) Loc.asip1-iTol2-eGFP-BAC labels the pectoral gridle. (A,B) The pectoral fin is labeled by both fluorescent proteins at 5 dpf. (C,D) Ventral view of a live 30dpf larvae heads, anterior is to the left. sox10:mRFP labels the cartilages of maxillary and mandibular arches. Drer.asip1-iTol2-eGFP-BAC labels different pharyngeal bone structures (interopercle, dermal branchiostegal rays, quadrate dermal bone) (C), and and Loc.asip1-iTol2-eGFP-BAC also labels some pharyngeal bone structures (interopercle and dermal branchiostegal rays) (D). (E,F) Ventral dermis region was strongly labeled by Drer.asip1-iTol2-eGFP-BAC and Loc.asip1-iTol2-eGFP-BAC. (G) Magnified view of ventral skin region from (E), showing dermis, peritoneal membrane and peritoneal cavity region. (H) Fluorescent dorso-ventral sagittal projection image of ventral skin area of 30 dpf juvenile showing clear sox10:mRFP expression in peritoneal membrane region where a reflecting iridophore layer is located. The peritoneal membrane region is also positive for Drer.asip1-iTol2-eGFP-BAC. However, the dermis region is exclusively labeled by Drer.asip1-iTol2-eGFP-BAC. Scale bar: (A,B) 100 μm, (C,D,E, F) 200 μm, (H,J) 50 μm. br, brachiostegals; ch, ceratohyal; cs, coraco scapular cartilage; d, dermis; e, eye; iop, interopercular bone; m, Meckel’s cartilage; ms, muscle; o, opercle; pc, peritoneal cavity; pf, pectoral fin; pg, pectoral gridle; pm, peritoneal membrane; pq, palatoquadrate; q, quadrate; sk, skin.
DISCUSSION
It is well known that the dorso-ventral countershading pigment pattern, which is one of the most common pigmentary adaptations in vertebrates, is driven by differential expression of agouti-signaling protein Asip, with high ventral Asip expression driving pale belly color (Manceau et al., 2011; Ceinos et al., 2015). In mammals, pigment pattern formation depends on the regulation of the production of different melanin types and levels in melanocytes (reviewed in Lin and Fisher, 2007), whereas in teleost fish, it results from a patterned distribution of multiple types of chromatophores (Hirata et al., 2005; Kelsh et al., 2009; Irion et al., 2016; Parichy & Spiewak, 2015). It would be reasonable to hypothesize that the conserved expression of Asip reflects a conserved role in pigment pattern formation in vertebrates, and specifically that the pale ventrum of vertebrates depends upon elevated levels of Asip expression.
Whereas in mammals, ASIP influences the type of melanin produced, in teleost fish it is likely to affect the ratios between different cell-types, i.e. promoting development of iridophores and suppressing development of melanophores (Guillot et al., 2012; Ceinos et al., 2015). Thus, rather unexpectedly, Asip appears to control an evolutionarily conserved pattern by regulating different cellular mechanisms in mammals and in teleost fish. To test of this model, we analyzed and compared the agouti locus in zebrafish and spotted gar using a BAC transgenic approach. As mentioned above, within ray-finned fishes, spotted gar is a member of holostean linage that diverged from the teleost linage before the TGD; consequently, the spotted gar genome provides an outgroup to the teleost lineage with a genome duplication status at the same level as mammals and thus spotted gar provides information regarding vertebrate genome evolution (Amores et al., 2011; Braasch et al., 2016). We report here the cloning, genomic organization and spatial and temporal expression of the asip1 gene in spotted gar, note that Asip is member of a larger gene family, and that the mammalian gene should be called Asip1 as well (see Braasch and Postlethwait 2011) although we follow Kurokawa’s et al. 2006 gene nomenclature to avoid confusion (see Supplementary S2A).
To characterize conserved regulatory elements required for the evolutionary conserved asip1 expression in the adult fish, we report the generation and expression analysis of two independent BAC transgenic zebrafish lines, containing a zebrafish and/or spotted gar BAC carrying the asip1 gene modified to drive enhanced green fluorescent protein (eGFP). Our gar asip1 transgenic zebrafish line is the first one ever to introduce a holostean BAC clone into the zebrafish genome. Because asip1 transcriptional control is complex, we decided to use BACs, which is likely to avoid potential problems associated with “standard” reporter transgene approaches, including positional effects (Giraldo and Montoliu, 2001; Suster et al., 2011) and the isolation of isolated and individualized elements that may not recapitulate the complete endogenous cooperative interactions between regulatory elements of a larger genomic region.
The spotted gar asip1 gene shows a similar genomic organization present in other vertebrate species (Bultman et al., ‘92; Kwon et al., ‘94; Cerdá-Reverter et al., 2005; Guillot et al., 2012; Agullerio et al., 2014). It consists of three coding exons that encode a putative peptide of 127 amino acids. Multiple sequence alignments indicated that the deduced amino acid sequence of spotted gar Asip1 was also highly conserved compared to orthologs from other vertebrate lineages (Supplementary S2B).
Spatial expression analysis revealed that the asip1 gene in adult spotted gar is expressed in diverse tissues as shown in other vertebrate species (Ceinos et al., 2015; Cerdá-Reverter et al., 2005; Girardot et al., 2005; Guillot et al., 2012; Agulleiro et al., 2014), therefore suggesting other possible roles of the asip1 gene in addition to its role in pigmentation, such as energy balance (Miller et al., ‘93; Smith et al., 2003; Guillot et al., 2016). We also show conservation of a dorso-ventral gradient of asip1 expression in the skin of adult spotted gar with lower levels in the dorsum and higher levels in the belly. This expression pattern correlates with the countershading coloration in which the dorsal side (upper side) of the animal is darker than its ventral (lower) side. In contrast to the high expression level of asip1 found in adult spotted gar skin, our data give no evidence of asip1 expression in 11 dpf pre-metamorphic spotted gar larvae skin (data not shown). Therefore, our results reveal that asip1 is important for adult spotted gar pigment pattern formation with a reduced or non-existent effect in embryonic and larval pigment pattern as it has been described in other fish species like zebrafish (Ceinos et al., 2015).
Asip transcriptional control is complex because the gene uses at least two alternatives promoters in different mammalian species (Bultman et al., ‘92; Vrieling et al., 1994; Girardot et al., 2005; Fontanesi et al., 2010; Oribe et al., 2012). Our previous attempts to characterize the conserved regulatory elements of asip1 in zebrafish by conventional reporter transgene assays did not show eGFP expression, which suggests that essential regulatory elements of asip1 must be located further from the locus. Alternatively, BAC transgenesis showed that 214 Kb and 130 Kb genomic sequences including the asip1 locus of zebrafish or spotted gar, respectively were capable of recapitulating the endogenous dorso-ventral expression pattern of asip1 in adult zebrafish and similar to the dorso-ventral asip1 expression pattern in adult spotted gar. Therefore, our BAC transgenesis approach identified large genomic regions carrying divergent sequences but a conserved function in adult ray-finned fish. These results confirm the complex regulation of the Asip gene and show the conserved evolutionary roots of this complex regulation among ray-finned fish. Surprisingly, our observations also indicate that this conserved evolutionary asip1 expression pattern in adult zebrafish and spotted gar is not found in larvae and/or pre-metamorphic fish, raising the possibility that different regulatory elements could be involved in the larval control of asip1 gene expression in both species. Further analyses are necessary to fully characterize the specific regulatory elements (enhancers) that control the spatial and temporal gene expression of asip1 and its sequence and function conservation.
In most mammalian species ASIP is a protein secreted by dermal papilla cells (Bultman et al., ‘92; Miller et al., ‘93) that acts as a paracrine factor for melanocyte pigment production (Millar et al., ‘95). In adult teleost fish, Asip1 is produced mainly by cells in the ventral skin and possibly acts nearby (in a paracrine fashion) to reduce melanophore number and melanophore differentiation in the ventrum, but has no effect on larvae and/or pre-metamorphic pigment pattern (Ceinos et al., 2015), likely because asip1 in zebrafish larvae is not expressed in the skin.
Using transgenic reporter lines expressing eGFP and mRFP under the asip1 regulatory machinery and sox10 promoters, respectively, we demonstrated here that, although both transgenes were expressed in the pharyngeal bones, pectoral fins, and peritoneal membrane, they were expressed in distinctive cell types. asip1/green was expressed in dermal fibroblast, however, sox10/red was expressed in neural crest derivatives, including craniofacial chondrocytes and pigment cells, suggesting that asip1 transgenes (eGFP positive cells) and thus endogenous asip1 was not expressed in the pre- and post-migratory neural crest cells (sox10:mRFP positive cells).
In conclusion, we demonstrate for the first time the existence a dorso-ventral gradient of asip1 expression in the skin of adult spotted gar with a high level in the ventrum and lower levels in the dorsum, conserved with the dorso-ventral gradient in zebrafish and other teleosts. We describe the generation of bacterial artificial chromosome (BAC) transgenic (Tg) zebrafish lines that harbor zebrafish or spotted gar BACs carrying the asip1 gene modified to drive enhanced green fluorescent protein (eGFP). The resulting transgenic lines showed eGFP expression in a pattern that mirrors that of endogenous asip1 gene expression in adult zebrafish and spotted gar and illustrated that these transgenic zebrafish lines are a promising tool for future identification of the regulatory elements required for regular dorso-ventrally gradients of asip1 expression in adult fish. The development and the pigment pattern of both transgenic lines were totally normal. Therefore, our study provides new molecular genetic tools for functional investigations of the regulatory elements that control spatial and temporal pattern of asip1 expression and its role in pigment pattern formation. The work emphasizes the need to characterize asip1 transcriptional regulation to further elucidate the conserved functions of ASIP protein among bony vertebrates despite the different cellular mechanisms of pigment pattern formation in mammals and fish.
Supplementary Material
Gene-specific primer sequences used for RT-PCR and qRT-PCR analysis. Dre (Danio rerio), Loc (Lepisosteus oculatus).
(A) Two different nomenclatures for the Agouti gene family have been proposed: the earlier Kurokawa nomenclature and the recently proposed Braasch & Postlethwait nomenclature. In order to avoid confusion, we have used the conventional Kurokawa nomenclature. (B) Alignment of agouti-signaling protein (ASIP) and agouti-related protein (AGRP) amino acid sequences. Gar sequences are in bold letters. Dashes were introduced to improve alignment. Black boxes show amino acid residues conserved in all sequences. Green boxes show residues only conserved in Asip1 sequences. Red boxes indicate residues only conserved in Agrp1 sequences. Blue boxes show residues of the short tail present in all AGRP sequences. Lines joining cysteine residues indicate putative disulfide bonds forming the cysteine domain. Arrow shows conserved motif for AGRP post-transcriptional processing.
Image projections of rostro-caudal transverse sections of ventral skin region (40 μm) from Drer.asip1-iTol2-eGFP-BAC. Expression of eGFP (green) together with 4,6-diamidino-2-phenylindole (DAPI) (blue) was analyzed in 1 year old fish. Stereomicroscope image projection of the ventral region of skin (A, B) shows the different layers of the skin (epidermis and dermis) around ventral scales with DAPI staining (A) and eGFP fluorescence (B). eGFP labels a thin layer of fibroblast within the dermis layer of skin next to the scales. Confocal image projection of the ventral skin (C) shows the different layers of the skin (epidermis and dermis) in a region without scales. eGFP labels dermal fibroblasts within the dermis layer of skin. Scale bars: (A, B) 50 μm, (C) 25 μm.
Acknowledgments
This work was funded by the Spanish Economy and Competitiveness Ministry project AGL2011-23581 and AGL2014-52473R to JR; NIH grants R01 OD011116 (alias R01 RR020833) and R24 OD01119004 to JHP; a Feodor Lynen Fellowship from the Alexander von Humboldt Foundation and the Volkswagen Foundation Initiative Evolutionary Biology, grant I/84 815 to IB.
We thank Christiane Nüsslein-Volhard from Max-Planck Institute (Germany) for providing the sox10:mRFP transgenic line. Also, we would also like to thank Inés Pazos Garridos (CACTI, University of Vigo, Spain) for her assistance with confocal imaging and to Fátima Adrio for her support with cryostat sectioning. We thank Carlos Gómez-Marin, Charlotte Taylor and Julia Ganz for help with BAC recombineering and the University of Oregon Aquatic Animal Care Services for supporting gar husbandry. This work was funded by the Spanish Economy and Competitiveness Ministry projects AGL2011-23581 and AGL2014-52473R to JR. Partial funding was obtained from AGL2013-46448-C3-3-R to JMCR. L. Cal was supported by pre-doctoral fellowship FPI funded by Spanish Economy and Competitiveness Ministry (AGL2011-23581) and by pre-doctoral fellowship of the Spanish Personnel Research Training Programe funded by Spanish Economy and Competitiveness Ministry (EEBB-C-14-00467). We thank NIH grants R01 OD011116 (alias R01 RR020833) and R24 OD01119004 to JHP, and a Feodor Lynen Fellowship from the Alexander von Humboldt Foundation and the Volkswagen Foundation Initiative Evolutionary Biology, grant I/84 815 to IB. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- Agulleiro MJ, Cortés R, Leal E, Ríos D, Sánchez E, Cerdá-Reverter JM. Characterization, tissue distribution and regulation by fasting of the agouti family of peptides in the sea bass (Dicentrarchus labrax) Gen Comp Endocrinol. 2014;205:251–259. doi: 10.1016/j.ygcen.2014.02.009. [DOI] [PubMed] [Google Scholar]
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, Westerfield M, Ekker M, Postlethwait JH. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–4. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Amores A, Catchen J, Ferrara A, Fontenot Q, Postlethwait JH. Genome evolution and meiotic maps by massively parallel DNA sequencing: spotted gar, an outgroup for the teleost genome duplication. Genetics. 2011;188:799–808. doi: 10.1534/genetics.111.127324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch I, Gehrke AR, Smith JJ, Kawasaki K, Manousaki T, Pasquier J, Amores A, Desvinges T, Batzel P, Catchen J, Berlin AM, Campbell MS, Barrell D, Martin KJ, Mulley JF, Ravi V, Lee AP, Nakamura T, Chalopin D, Fan S, Wcisel D, Cañestro C, Sydes J, Beaudry FEG, Sun Y, Hertel J, Beam MJ, Fasold M, Ishiyama M, Johnson J, Kehr S, Lara M, Letaw JH, Litman GW, Litman RT, Mikami M, Ota T, Saha NR, Williams L, Stadler PF, Wang H, Taylor JS, Fontenot Q, Ferrara A, Searle SMJ, Aken B, Yandell M, Schneider I, Yoder JA, Volff JN, Meyer A, Amemiya CT, Venkatesh B, Holland PWH, Guiguen Y, Bobe J, Shubin NH, Palma FD, Alföldi J, Lindblad-Toh K, Postlethwait JH. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet. 2016;48:427–437. doi: 10.1038/ng.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch I, Peterson SM, Desvignes T, McCluskey BM, Batzel P, Postlethwait JH. A New Model Armi: Emerging Fish Models to Study the Genomics of Vertebrate Evo-Devo. J Exp Zool Part B Mol Dev Evol. 2015;324:316–341. doi: 10.1002/jez.b.22589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch I, Postlethwait JH. The teleost agouti-related protein 2 gene is an ohnolog gone missing from the tetrapod genome. Proc Natl Acad Sci U S A. 2011;108:E47–E48. doi: 10.1073/pnas.1101594108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Green ED, Sidow A, Batzoglou S. LAGAN and Multi-LAGAN: Efficient Tools for Large-Scale Multiple Alignment of Genomic DNA. Genome Res. 2003;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman SJ, Michaud EJ, Woychik RP NISC Comparative Sequencing Program. Molecular characterization of the mouse agouti locus. Cell. 1992;71:1195–1204. doi: 10.1016/s0092-8674(05)80067-4. [DOI] [PubMed] [Google Scholar]
- Ceinos RM, Guillot R, Kelsh RN, Cerdá-Reverter JM, Rotllant J. Pigment patterns in adult fish result from superimposition of two largely independent pigmentation mechanisms. Pigment Cell Melanoma Res. 2015;28:196–209. doi: 10.1111/pcmr.12335. [DOI] [PubMed] [Google Scholar]
- Cerdá-Reverter JM, Haitina T, Schiöth HB, Peter RE. Gene Structure of the Goldfish Agouti-Signaling Protein: A Putative Role in the Dorsal-Ventral Pigment Pattern of Fish. Endocrinology. 2005;146:1597–1610. doi: 10.1210/en.2004-1346. [DOI] [PubMed] [Google Scholar]
- Cerdá-Reverter JM, Martínez-Rodríguez G, Anglade I, Kah O, Zanuy S. Peptide YY (PYY) and fish pancreatic peptide Y (PY) expression in the brain of the sea bass (Dicentrarchus labrax) as revealed by in situ hybridization. J Comp Neurol. 2000;426:197–208. doi: 10.1002/1096-9861(20001016)426:2<197::aid-cne3>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Le Douarin N, Kalcheim C. Developmental and Cell Biology Series. 2. Cambridge: Cambridge University Press; 1999. The Neural Crest. [Google Scholar]
- Dupin E, Creuzet S, Le Douarin NM. The Contribution of the Neural Crest to the Vertebrate Body. Madame Curie Biosci Database [Internet] 2000 doi: 10.1007/978-0-387-46954-6_6. [DOI] [PubMed] [Google Scholar]
- Fadeev A, Krauss J, Frohnhöfer HG, Irion U, Nüsslein-Volhard C. Tight Junction Protein 1a regulates pigment cell organization during zebrafish colour patterning. eLife. 2015;4:e06545. doi: 10.7554/eLife.06545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanesi L, Forestier L, Allain D, Scotti E, Beretti F, Deretz-Picoulet S, Pecchioli E, Vernesi C, Robinson TJ, Malaney JL, Russo V, Oulmouden A. Characterization of the rabbit agouti signaling protein (ASIP) gene: transcripts and phylogenetic analyses and identification of the causative mutation of the nonagouti black coat colour. Genomics. 2010;95:166–175. doi: 10.1016/j.ygeno.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Giraldo P, Montoliu L. Size matters: use of YACs, BACs and PACs in transgenic animals. Transgenic Res. 2001;10:83–103. doi: 10.1023/a:1008918913249. [DOI] [PubMed] [Google Scholar]
- Girardot M, Martin J, Guibert S, Leveziel H, Julien R, Oulmouden A. Widespread expression of the bovine Agouti gene results from at least three alternative promoters. Pigment Cell Res. 2005;18:34–41. doi: 10.1111/j.1600-0749.2004.00195.x. [DOI] [PubMed] [Google Scholar]
- Guillot R, Ceinos RM, Cal R, Rotllant J, Cerdá-Reverter JM. Transient ectopic overexpression of agouti-signalling protein 1 (Asip1) induces pigment anomalies in flatfish. PLoS One. 2012;7:e48526. doi: 10.1371/journal.pone.0048526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot R, Cortés R, Navarro S, Mischitelli M, García-Herranz V, Sánchez E, Cal L, Navarro JC, Míguez JM, Afanasyev S, Krasnov A, Cone RD, Rotllant J, Cerdá-Reverter JM. Behind melanocortin antagonist overexpression in the zebrafish brain: A behavioral and transcriptomic approach. Horm Behav. 2016;82:87–100. doi: 10.1016/j.yhbeh.2016.04.011. [DOI] [PubMed] [Google Scholar]
- Hirata M, Nakamura K-I, Kondo S. Pigment cell distributions in different tissues of the zebrafish, with special reference to the striped pigment pattern. Dev Dyn. 2005;234:293–300. doi: 10.1002/dvdy.20513. [DOI] [PubMed] [Google Scholar]
- Hoegg S, Brinkmann H, Taylor JS, Meyer A. Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. J Mol Evol. 2004;59:190–203. doi: 10.1007/s00239-004-2613-z. [DOI] [PubMed] [Google Scholar]
- Hurley Ia, Mueller RL, Dunn Ka, Schmidt EJ, Friedman M, Ho RK, Prince VE, Yang Z, Thomas MG, Coates MI. A new time-scale for ray-finned fish evolution. Proc R Soc B Biol Sci. 2007;274:489–498. doi: 10.1098/rspb.2006.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion U, Singh AP, Nüsslein-Volhard C. The Developmental Genetics of Vertebrate Color Pattern Formation: Lessons from zebrafish. 2016;117:141–169. doi: 10.1016/bs.ctdb.2015.12.012. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, Nicaud S, Jaffe D, Fisher S, Lutfalla G, Dossat C, Segurens B, Dasilva C, Salanoubat M, Levy M, Boudet N, Castellano S, Anthouard V, Jubin C, Castelli V, Katinka M, Vacherie B, Biémont C, Skalli Z, Cattolico L, Poulain J, De Berardinis V, Cruaud C, Duprat S, Brottier P, Coutanceau JP, Gouzy J, Parra G, Lardier G, Chapple C, McKernan KJ, McEwan P, Bosak S, Kellis M, Volff JN, Guigó R, Zody MC, Mesirov J, Lindblad-Toh K, Birren B, Nusbaum C, Kahn D, Robinson-Rechavi M, Laudet V, Schachter V, Quétier F, Saurin W, Scarpelli C, Wincker P, Lander ES, Weissenbach J, Roest Crollius H. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431(7011):946–57. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Kelsh RN, Harris ML, Colanesi S, Erickson CA. Stripes and belly-spots - a review of pigment cell morphogenesis in vertebrates. Semin Cell Dev Biol. 2009;20:90–104. doi: 10.1016/j.semcdb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Klovins J, Schiöth HB. Agouti-related proteins (AGRPs) and agouti-signaling peptide (ASIP) in fish and chicken. Ann N Y Acad Sci. 2005;1040:363–367. doi: 10.1196/annals.1327.063. [DOI] [PubMed] [Google Scholar]
- Kottler VA, Künstner A, Schartl M. Pheomelanin in fish? Pigment Cell Melanoma Res. 2015;28:355–356. doi: 10.1111/pcmr.12359. [DOI] [PubMed] [Google Scholar]
- Kurokawa T, Murashita K, Uji S. Characterization and tissue distribution of multiple agouti-family genes in pufferfish, Takifugu rubripes. Peptides. 2006;27:3165–3175. doi: 10.1016/j.peptides.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Kwon HY, Bultman SJ, Löffler C, Chen WJ, Furdon PJ, Powell JG, Usala AL, Wilkison W, Hansmann I. Molecular structure and chromosomal mapping of the human homolog of the agouti gene. Proc Natl Acad Sci U S A. 1994;91:9760–9764. doi: 10.1073/pnas.91.21.9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JY, Fisher DE. Melanocyte biology and skin pigmentation. Nature. 2007;445:843–850. doi: 10.1038/nature05660. [DOI] [PubMed] [Google Scholar]
- Lu D, Willard D, Patel IR, Kadwell S, Overton L, Kost T, Luther M, Chen W, Woychik RP, Wilkison WO. Agouti protein is an antagonist of the melanocyte-stimulating-hormone receptor. Nature. 1994;371:799–802. doi: 10.1038/371799a0. [DOI] [PubMed] [Google Scholar]
- Manceau M, Domingues VS, Mallarino R, Hoekstra HE. The developmental role of Agouti in color pattern evolution. Science. 2011;331:1062–1065. doi: 10.1126/science.1200684. [DOI] [PubMed] [Google Scholar]
- Mayor C, Brudno M, Schartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- McCurley AT, Callard GV. Characterization of housekeeping genes in zebrafish: male-female differences and effects of tissue type, developmental stage and chemical treatment. BMC Mol Biol. 2008;9:102. doi: 10.1186/1471-2199-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar SE, Miller MW, Stevens ME, Barsh GS. Expression and transgenic studies of the mouse agouti gene provide insight into the mechanisms by which mammalian coat color patterns are generated. Development. 1995;121:3223–3232. doi: 10.1242/dev.121.10.3223. [DOI] [PubMed] [Google Scholar]
- Miller MW, Duhl DM, Vrieling H, Cordes SP, Ollmann MM, Winkes BM, Barsh GS. Cloning of the mouse agouti gene predicts a secreted protein ubiquitously expressed in mice carrying the lethal yellow mutation. Genes Dev. 1993;7:454–467. doi: 10.1101/gad.7.3.454. [DOI] [PubMed] [Google Scholar]
- Ollmann MM, Wilson BD, Yang YK, Kerns Ja, Chen Y, Gantz I, Barsh GS. Antagonism of central melanocortin receptors in vitro and in vivo by agouti-related protein. Science. 1997;278:135–138. doi: 10.1126/science.278.5335.135. [DOI] [PubMed] [Google Scholar]
- Oribe E, Fukao A, Yoshihara C, Mendori M, Rosal KG, Takahashi S, Takeuchi S. Conserved distal promoter of the agouti signaling protein (ASIP) gene controls sexual dichromatism in chickens. Gen Comp Endocrinol. 2012;177:231–237. doi: 10.1016/j.ygcen.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Parichy DM, Spiewak JE. Origins of adult pigmentation: diversity in pigment stem cell lineages and implications for pattern evolution. Pigment Cell Melanoma Res. 2015 Jan;28(1):31–50. doi: 10.1111/pcmr.12332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait J, Amores A, Force A, Yan YL. The zebrafish genome. Methods Cell Biol. 1999;60:149–63. [PubMed] [Google Scholar]
- Rotllant J, Liu D, Yan YL, Postlethwait JH, Westerfield M, Du SJ. Sparc (Osteonectin) functions in morphogenesis of the pharyngeal skeleton and inner ear. Matrix Biol. 2008;27:561–572. doi: 10.1016/j.matbio.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SR, Gawronska-Kozak B, Janderová L, Nguyen T, Murrel A, Stephens JM, Mynatt RL. Agouti Expression in Human Adipose Tissue. Diabetes. 2003;52:2914–2922. doi: 10.2337/diabetes.52.12.2914. [DOI] [PubMed] [Google Scholar]
- Suster ML, Abe G, Schouw A, Kawakami K. Transposon-mediated BAC transgenesis in zebrafish. Nat Protoc. 2011;6:1998–2021. doi: 10.1038/nprot.2011.416. [DOI] [PubMed] [Google Scholar]
- Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y. Genome duplication, a trait shared by 22,000 species of ray-finned fish. Genome Res. 2003;13(3):382–390. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, De Vos W, Taylor JS, Meyer A, Van de Peer Y. Major events in the genome evolution of vertebrates: paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proc Natl Acad Sci U S A. 2004;101:1638–1643. doi: 10.1073/pnas.0307968100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieling H, Duhl DM, Millar SE, Miller Ka, Barsh GS. Differences in dorsal and ventral pigmentation result from regional expression of the mouse agouti gene. Proc Natl Acad Sci U S A. 1994;91:5667–5671. doi: 10.1073/pnas.91.12.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. A Guide for the Laboratory Use of Zebrafish (Danio rerio) 5. Eugene: University of Oregon Press; 2007. The Zebrafish Book. [Google Scholar]
- Zhang C, Song Y, Thompson Da, Madonna Ma, Millhauser GL, Toro S, Varga Z, Westerfield M, Gamse J, Chen W, Cone RD. Pineal-specific agouti protein regulates teleost background adaptation. Proc Natl Acad Sci U S A. 2010;107:20164–20171. doi: 10.1073/pnas.1014941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene-specific primer sequences used for RT-PCR and qRT-PCR analysis. Dre (Danio rerio), Loc (Lepisosteus oculatus).
(A) Two different nomenclatures for the Agouti gene family have been proposed: the earlier Kurokawa nomenclature and the recently proposed Braasch & Postlethwait nomenclature. In order to avoid confusion, we have used the conventional Kurokawa nomenclature. (B) Alignment of agouti-signaling protein (ASIP) and agouti-related protein (AGRP) amino acid sequences. Gar sequences are in bold letters. Dashes were introduced to improve alignment. Black boxes show amino acid residues conserved in all sequences. Green boxes show residues only conserved in Asip1 sequences. Red boxes indicate residues only conserved in Agrp1 sequences. Blue boxes show residues of the short tail present in all AGRP sequences. Lines joining cysteine residues indicate putative disulfide bonds forming the cysteine domain. Arrow shows conserved motif for AGRP post-transcriptional processing.
Image projections of rostro-caudal transverse sections of ventral skin region (40 μm) from Drer.asip1-iTol2-eGFP-BAC. Expression of eGFP (green) together with 4,6-diamidino-2-phenylindole (DAPI) (blue) was analyzed in 1 year old fish. Stereomicroscope image projection of the ventral region of skin (A, B) shows the different layers of the skin (epidermis and dermis) around ventral scales with DAPI staining (A) and eGFP fluorescence (B). eGFP labels a thin layer of fibroblast within the dermis layer of skin next to the scales. Confocal image projection of the ventral skin (C) shows the different layers of the skin (epidermis and dermis) in a region without scales. eGFP labels dermal fibroblasts within the dermis layer of skin. Scale bars: (A, B) 50 μm, (C) 25 μm.