Abstract
Microbial expression of genes for resistance to heavy metals and metalloids is usually transcriptionally regulated by the toxic ions themselves. Arsenic is a ubiquitous, naturally occurring toxic metalloid widely distributed in soil and groundwater. Microbes biotransform both arsenate (As(V)) and arsenite (As(III)) into more toxic methylated metabolites methylarsenite (MAs(III)) and dimethylarsenite (DMAs(III)). Environmental arsenic is sensed by members of the ArsR/SmtB family. The arsR gene is autoregulated and is typically part of an operon that contains other ars genes involved in arsenic detoxification. To date every identified ArsR is regulated by inorganic As(III). Here we described a novel ArsR from Shewanella putrefaciens selective for MAs(III). SpArsR orthologs control expression of two MAs(III) resistance genes, arsP that encodes the ArsP MAs(III) efflux permease, and arsH encoding the ArsH MAs(III) oxidase. SpArsR has two conversed cysteine residues, Cys101 and Cys102. Mutation of either resulted in loss of MAs(III) binding, indicating that they form an MAs(III) binding site. SpArsR can be converted into an As(III)-responsive repressor by introduction of an additional cysteine that allows for 3-coordinate As(III) binding. Our results indicate that SpArsR evolved selectivity for MAs(III) over As(III) in order to control expression of genes for MAs(III) detoxification.
Graphical Abstract
A novel ArsR regulator of a bacterial arsenical resistance (ars) operon for resistance to toxic methylarsenite (MAs(III)) is described. The repressor is selective for MAs(III) over As(III). This structural model of the repressor shows the two-coordinate binding site with MAs(III). We propose that it evolved selectivity for MAs(III) over As(III) in order to control expression of genes for MAs(III) detoxification.
Introduction
Arsenic is a carcinogenic and toxic metalloid that is widely distributed in the earth’s crust, occurring naturally in soil, water and air. In addition, anthropogenic introduction of arsenic into the environment poses a significant threat to public health because of its toxicity, accumulation in the food chain and environmental persistence (Zhu et al., 2014). As a consequence, nearly every organism has genetic mechanisms for arsenic resistance (Liu et al., 2013). The majority of well-characterized resistances are to inorganic arsenic, including As(V) reduction (ArsC and ArrAB), As(III) oxidation (AoxAB), extrusion (ArsB and Acr3) and methylation (ArsM). These microbial biotranformations play key roles in the arsenic biogeological cycle (Zhu et al., 2014). Their genes are usually organized in ars operons, which are found either on the chromosome or on plasmids in Gram-positive bacteria, Gram-negative bacteria and archaea. Typically ars operons are regulated by an ArsR As(III)-responsive transcriptional repressor (Ordóñez et al., 2008, Qin et al., 2007, Wu & Rosen, 1993, Kang et al., 2016).
Recently a parallel biocycle for organoarsenicals has been identified (Li et al., 2016). Organisms with arsM genes methylate inorganic As(III) into the more toxic and potentially more carcinogenic MAs(III) (Qin et al., 2009, Qin et al., 2006, Chen et al., 2003, Drobna et al., 2005). In addition, MAs(V) is currently used in the U.S. and other countries as the herbicide MSMA (monosodium methanearsonate) on cotton fields or for post-emergence control of turf weeds and crabgrass (https://www.epa.gov/ingredients-used-pesticide-products/monosodium-methanearsonate-msma-organic-arsenical). Soil microbial such as Pseudomonas putida and Burkholderia MR1 reduce MAs(V) to MAs(III), which is subsequently degraded to the more mobile As(III) by other members of microbial communities such as Bacillus MD1. We propose that MAs(III) functions as an antibiotic to kill off other members of the microbial communities (Li et al., 2016, Yang & Rosen, 2016). In response to this environmental pressure, other bacteria have evolved mechanisms to detoxify MAs(III). To date, three MAs(III) resistance genes have been identified: arsI, arsH and arsP. ArsI is a C-As bond lyase, a member of the dioxygenase superfamily, that detoxifies MAs(III) by cleaving it into As(III) and H2CO (Yoshinaga & Rosen, 2014). ArsH is an NADPH-FMN dependent oxidoreductase that oxidizes MAs(III) to MAs(V) (Chen et al., 2015a). ArsP is a MAs(III) efflux permease, catalyzing MAs(III) extrusion from cells (Chen et al., 2015b).
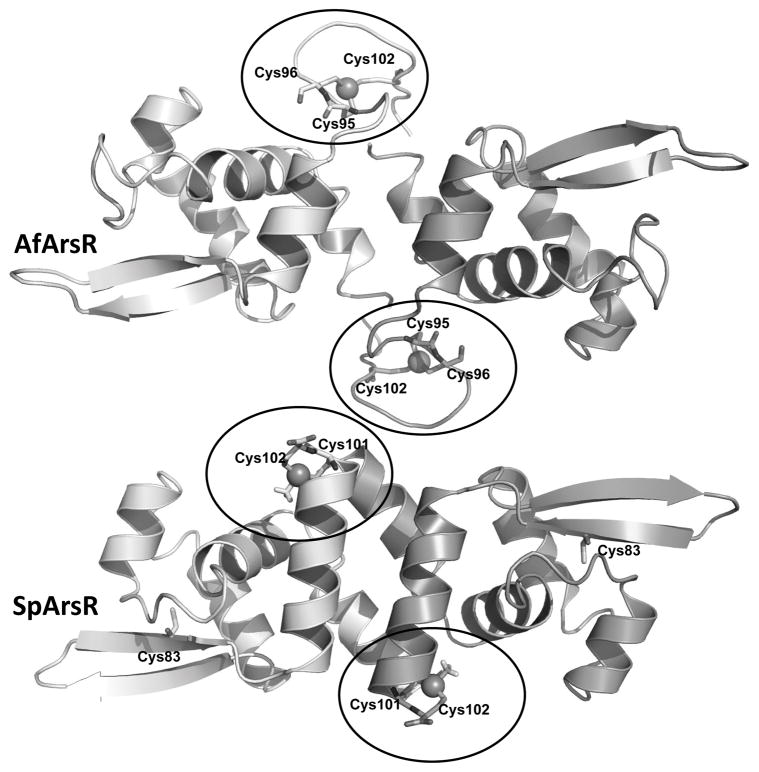
These organoarsenical detoxification genes are in ars operons regulated by a homodimeric As(III)-responsive ArsR repressor. Three different ArsRs have been identified that have three types of As(III) binding sites. The ArsR repressor encoded by Escherichia coli plasmid R773 binds to the promoter region of its respective ars operon in the absence of As(III) (Wu & Rosen, 1993). As(III) binds to a three-coordinate site in each monomer composed of three cysteine residues, Cys32, Cys34 and Cys37 (termed a Type 1 site), which are in an N-terminal α helix that forms part of the DNA binding site (Shi et al., 1996, Xu & Burke, 1996). As(III) binding is proposed to induce a conformational change in the DNA binding domain, resulting in dissociation of the repressor from the operator/promoter DNA and transcriptional derepression. Two other ArsR orthologs with different As(III) binding sites were identified in the ars operon of Acidithiobacillus ferrooxidans (Qin et al., 2007) and Corynebacterium glutamicum (Ordóñez et al., 2008), respectively. The Type 2 site of AfArsR is composed of three cysteine residues, Cys95, Cys96 and Cys102, which are located at the C-terminus of the repressor (Qin et al., 2007). Homology modeling indicates that the two As(III) binding sites (one in each monomer) are located at the ends of antiparallel C-terminal α helices in each monomer that form a dimerization domain. Binding of As(III) can be predicted to unwind the helix and cause a conformational change in the dimerization domain that results in derepression. The Type 3 As(III) binding site of CgArsR is composed of three cysteine residues, Cys15 and Cys16 from one subunit of the homodimer and Cys55 from the other subunit (Ordóñez et al., 2008). While it is located in a similar position to the Type 1 binding site, it is composed of different residues that form an inter-subunit binding site. Each ArsR has a high-affinity As(III) binding site composed of three cysteine residues at spatially distinct locations in their three-dimensional structures, suggesting that the Type 1, Type 2 and Type 3 ArsRs arose independently.
In this study we identified a novel MAs(III)-responsive ArsR that regulates expression of the arsP and arsH MAs(III) resistance genes in Shewanella putrefaciens 200. SpArsR is induced most effectively by MAs(III), with little response to As(III). When aligned with other putative orthologs, SpArsR appears to have three conversed cysteine residues, Cys83, Cys101 and Cys102. However, only the latter two appear to be involved in MAs(III) induction. Substitutions of Cys101 or Cys102 with serine residues resulted in loss of MAs(III) binding affinity, while a C83S substitution had no effect. These results indicate that Cys101 or Cys102 are involved in MAs(III) binding. A homology model of SpArsR built on the structure of the CadC Cd(II)/Zn(II)/Pb(II)-responsive repressor show Cys101 and Cys102 forming a two-coordinate MAs(III) binding site. The structure is similar to AfArsR but lacks the third cysteine, Cys102, of the three-coordinate As(III) binding site in AfArsR. A C102S mutation converts AfArsR into a MAs(III)-responsive repressor that has a large reduction in As(III) binding affinity (Chen et al., 2014). Conversely, SpArsR can be converted into an As(III)-responsive repressor by introduction of the C-terminal sequence from AfArsR that includes Cys102. We postulate that SpArsR evolved selectivity for MAs(III) in order to respond to environmental MAs(III) produced by other soil microbes.
Results
Co-transcription of a novel arsR and MAs(III) resistance genes
Comparative genetic analysis of chromosomal ars operons from more than 40 bacterial ars operons revealed a non-canonical arsR gene that is potentially co-transcribed with either the arsP or arsH MAs(III) resistance determinants (representative sequences are shown in Fig. 1). In S. putrefaciens, SparsR (ADV53698) is linked to SparsP. The potential linkage of arsR suggests that the repressor may regulate expression of arsP and arsH genes in various ars operons. These ArsRs show a high degree of similarity to AfArsR (ACK80311) that has an As(III) binding site composed of three conserved cysteines in the C-terminus (Supplemental Fig. S1). However, these ArsRs do not have a cysteine residue corresponding to AfArsR residue 102, suggesting that they lack the third ligand required for high-affinity As(III) binding. One possibility is that another cysteine residue corresponding to Cys83 in SpArsR might be the third ligand. Another possibility is that the binding site has only two arsenic ligands, and so bind inorganic As(III) with low affinity. These alternatives were tested below.
Fig. 1. Genes for a novel type of arsR are linked with arsP or arsH in ars operons.
Shown are representative ars operons (accession numbers in parentheses) containing arsR and arsP or arsH genes (black fill). Vibrio splendidus 12B01 (AAMR00000000.1), Yersinia enterocolitica subsp (AM286415), Plesiomonas shigelloides 302–73 (LT575468.1), Escherichia coli IAI39 (NC_002695), Aeromonas veronii B565 (CP012504.1), Citrobacter freundii CFNIH1 (CP007557.1), Desulfobulbus propionicus DSM 2032 (NC_014972.1), Marinobacterium jannaschii (JHVJ00000000.1), Shewanella woodyi ATCC 51908 (NC_010506.1), Endozoicomonas elysicola (JOJP00000000.1), Photobacterium sp. SKA34 (GCF_000153325.1), Serratia liquefaciens FK01 (NC_021741.1), Edwardsiella tarda ATCC 23685 (ADGK00000000.1), Serratia plymuthica PRI-2C (CP015613.1), Shewanella putrefaciens 200 (NC_017566.1).
SpArsR has a MAs(III) binding site and is a MAs(III)-selective repressor
In small molecule thiolate-As(III) complexes arsenic is frequently three-coordinate. In ArsRs cysteine thiolates also form three-coordinate complexes with As(III) (Delnomdedieu et al., 1993, Delnomdedieu et al., 1994, Cullen et al., 1984). The location of three cysteine residues in the amino acid sequence and hence the location of the As(III) binding sites, of the three characterized ArsRs are distributed differently (Fig. 2A). In AfArsR, Cys95, Cys96 and Cys102 form the three-coordinate As(III) binding site. In a homology model of AfArsR built on the structure of the CadC Cd(III)-responsive repressor, the As(III) binding site can be visualized at the end of an antiparallel C-terminal helix that forms a dimerization domain in each monomer (Qin et al., 2007). MAs(III) is also an inducer and binds to the S3 site with even higher affinity than As(III) (Chen et al., 2012). In the absence of Cys102, the affinity for As(III) is greatly reduced, while the affinity for MAs(III) remains unchanged (Chen et al., 2014). In the homology model of SpArsR, Cys101 and Cys102 are superimposible with AfArsR Cys95 and Cys96. Cys83, which is found in putative orthologs but not in AfArsR, is located distant from Cys101 and Cys102 and would not be expected to contribute to inducer binding (Fig. 2B).
Fig. 2. Multiple alignment of SpArsR homologs and homology modeling.
A: Representative ArsR orthologs (accession numbers in parentheses) are from: S. putrefaciens (ADV53698); Enterovibrio calviensis (WP_017007765); Sinorhizobium meliloti 1021 (NP_385183); A. ferrooxidans (ACK80311); Propionibacterium avidum (WP_063279264); C. glutamicum CgArsR1 (CAF21518); R773 ArsR (CAA34168); Synechocystis sp. PCC 7942 SmtB (CAA45872) and plasmid pI258 CadC (P20047). The multiple alignment was calculated with CLUSTAL W. B: Homology modeling of As(III)-bound AfArsR and MAs(III)-bound SpArsR were performed as described in Experimental Procedures. The cysteines are shown in sticks. The arsenic atoms are shown as spheres.
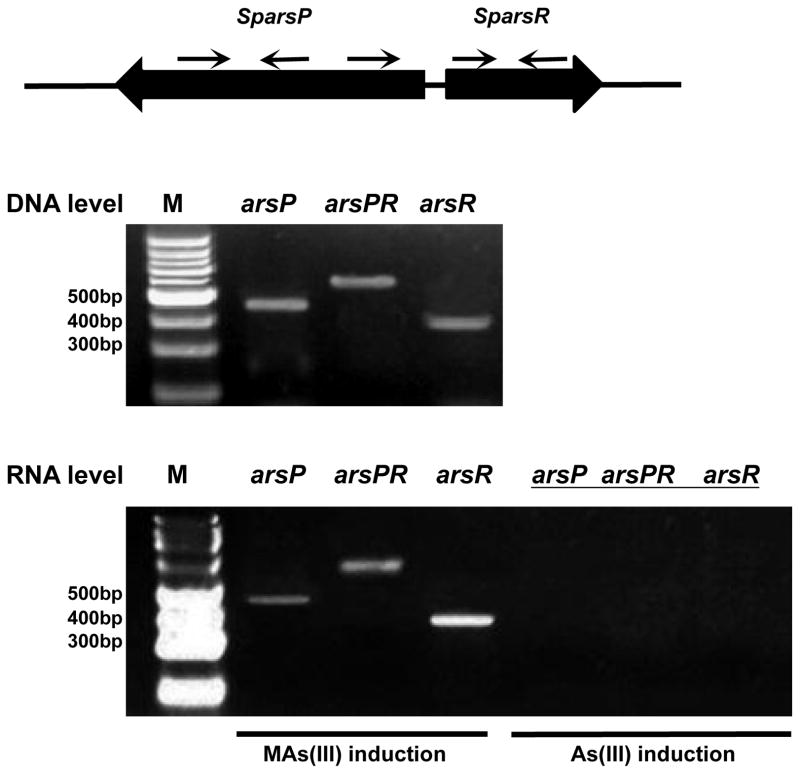
From the homology model, SpArsR should not be able to form a three-coordinate As(III) binding site but should still be able to bind MAs(III). To test this idea, we examined the expression and regulation of the arsP and arsR genes. Qualitative reverse transcription-PCR (RT-PCR) was used to assess SparsR and SparsP expression following exposure to As(III) or MAs(III). There was no detectable expression of either SparsR or SparsP expression following exposure to As(III) (Fig. 3). In contrast, both were expressed at high levels following exposure to MAs(III). In this experiment arsR and arsP were co-transcribed, demonstrating that SpArsR controls expression of both genes. These results show that SparsR is induced by MAs(III) and not As(III) and regulates a gene for MAs(III) detoxification.
Fig. 3. SparsR and SparsP are linked transcriptionally by MAs(III) induction.
The SparsR and SparsP are transcribed in opposite directions. Shown is the location of the primers for RT-PCR analysis (Table S1) of the ars transcripts. Total RNA isolated from S. putrefaciens 200 in exponential phase (A600nm = 1.2) in the presence of 2 μM MAs(III) or 20 μM As(III) were used as templates in reverse transcriptase reactions to generate cDNA and then amplified with the indicated primers. PCR products were resolved on a 1% agarose gel stained with ethidium bromide. Genomic DNA from S. putrefaciens was used as a positive control for each primer set.
The MAs(III) binding site in SpArsR
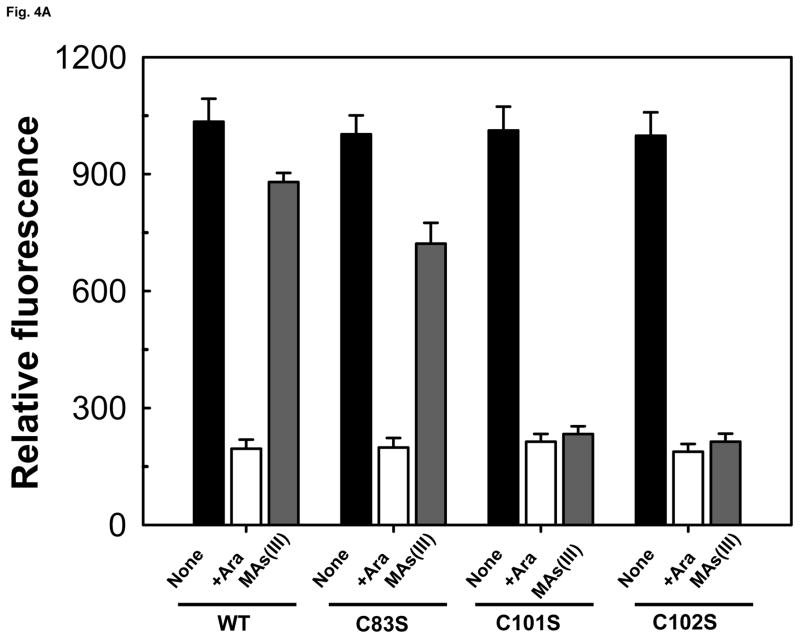
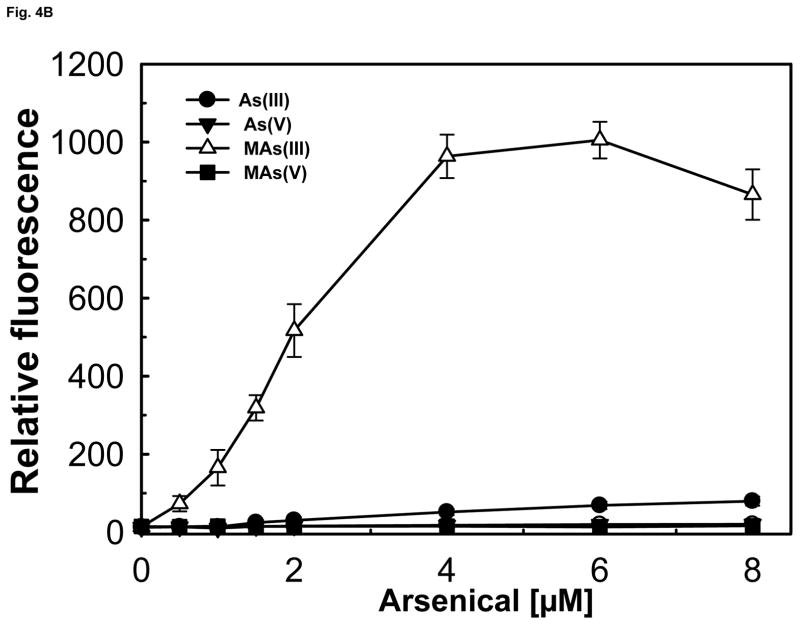
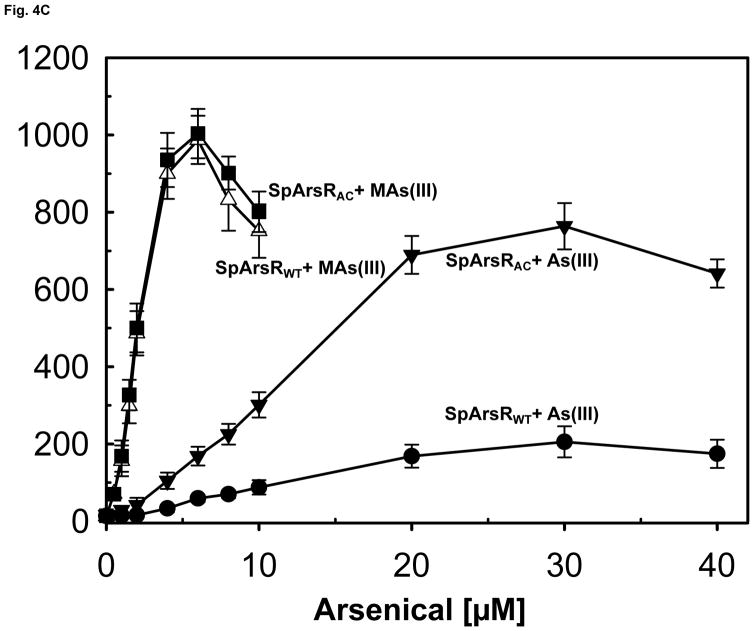
We have previously reported construction of a two-plasmid biosensor utilizing wild type AfArsR to measure As(III) and MAs(III) binding affinity (Chen et al., 2012). The objective of this study was to analyze whether SpArsR was selective for MAs(III) binding. A similar biosensor was constructed with the SparsR gene and SparsP promoter (Fig. S2A). The biosensors incorporate SparsR gene under control of the arabinose promoter in one plasmid, pBAD-SparsR, and the SparsP promoter (ParsP) controlling expression of gfp on a second plasmid, pACYC184-ParsP-gfp. As the concentration of arabinose is increased, AraC becomes a positive regulatory protein that drives expression of SparsR, and consequently expression of gfp from pACYC184-ParsP-gfp is decreased. To increase sensitivity, the arsenic-hypersensitive E. coli strain AW3110 (ars:cam), which is unable to extrude As(III) (Carlin et al., 1995), was utilized as host for the plasmids. Expression of gfp was constitutive when SparsR was repressed by growth on glucose (Fig. S2B) and could be quantified by spectrofluorometry (Fig. S2C). When the repressors were induced by addition of 0.2% arabinose, gfp was repressed, and the cells were not fluorescent. The biosensor with wild type SpArsR exhibited little gfp expression upon addition of 20 μM sodium arsenite, about the same as controls with As(V) or MAs(V). In contrast, elevated gfp expression was detected when exposed to 3 μM MAs(III). This result indicates that SpArsR is highly selective for MAs(III) binding and derepresses gfp expression. SpArsR has three cysteines found in putative orthologs: Cys83, Cys101 and Cys102 (Fig. S1). A C83S mutant retained responsiveness to addition of MAs(III), which exhibited derepressed gfp fluorescent, showing that Cys83 is not required for MAs(III) response. In either the C101S or C102S mutant, gfp was repressed and cells were not fluorescent (Fig. 4A). This indicates that both Cys101 and Cys102 are involved in MAs(III) binding. The response of biosensor with SparsR to arsenicals was quantified (Fig. 4B). Cells with the wild type SparsR biosensor responded only to MAs(III), and cellular gfp derepression was proportional to the concentration of MAs(III). Cells with the wild type SpArsR biosensor were unresponsive to MAs(V) or As(V) and exhibited only a very low response to As(III).
Fig. 4. Binding of MAs(III) to SpArsR involves specific cysteine residues.
Expression of the gfp reporter gene was assayed as described under Experimental Procedures. A) Mutagenesis of Cys83, Cys101 and Cys102 and their contribution to MAs(III) binding. Cells were grown in M9 medium for 14 h. Cells of E. coli strain AW3110(DE3) bearing plasmids with wild type SpArsR, C83S, C101S or the C102S mutant in trans with reporter plasmid pACYC184-ParsP-gfp were grown without arabinose (solid black bars ((left)), 0.2% arabinose (open bars (middle)), or 0.2% arabinose and 2 μM MAs(III) (solid grey bars (right)). B) Comparison of the response of the bacterial biosensor to inorganic and organic arsenicals. Additions: (●), As(III); (▼), As(V); (△), MAs(III); (■), MAs(V).
C) SpArsR can be converted to an As(III)-responsive ArsR by introduction of an additional cysteine residue from AfArsR. SpArsR with addition of the C-terminus of AfArsR containing AfArsR Cys102 (SpArsRAC) responds to As(III) four-fold better than the wild type. As(III) and MAs(III) were assayed at the indicated concentrations. Fluorescence intensities of cell suspensions were quantified using a Photon Technology International spectrofluorometer with an excitation wavelength of 470 nm and emission wavelength of 510 nm. The data are the mean ± SE (n = 3).
AfArsR has three sulfur thiolates from Cys95, Cys96 and Cys102 that form an S3 binding site for As(III). Cys101 and Cys102 in SpArsR correspond to Cys95 and Cys96 of AfArsR (Fig. 2), but SpArsR lacks a residue corresponding to AfArsR Cys102 and responds with low affinity. Would SpArsR respond to As(III) with higher affinity if it had a third cysteine residue corresponding to AfArsR Cys102? A hybrid SpArsR-AfArsR repressor was constructed by replacing the last 3 residues of SpArsR with the last 22 residues of AfArsR, which includes Cys102. This hybrid repressor, termed SpArsRAC, responds to As(III) more than 4-fold better than wild type SpArsR without affecting responsiveness to MAs(III) (Fig. 4C), indicating that SpArsR could be converted into an As(III)-responsive repressor by introduction of an additional cysteine to form a 3-coordinate As(III) binding site.
Phylogenetic relatedness of ArsR proteins
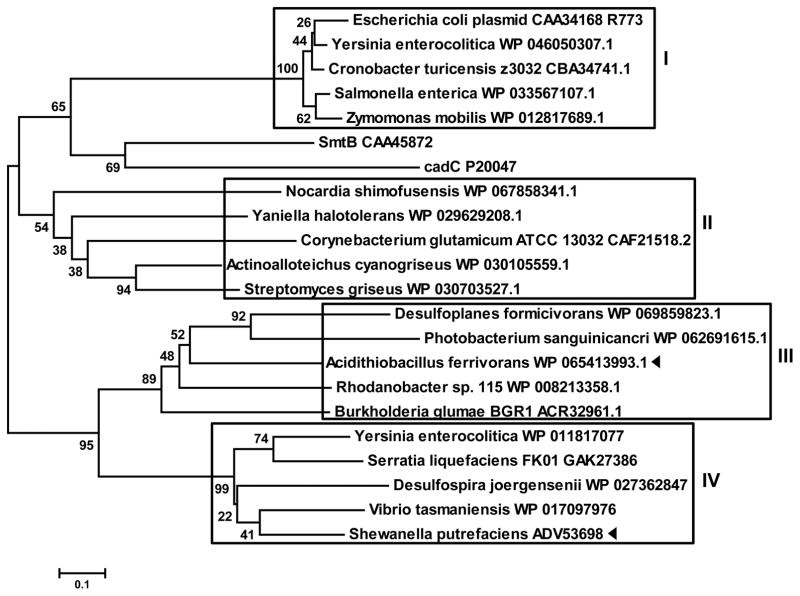
To examine the evolutionary history of SpArsR-like MAs(III)-responsive repressors, a phylogenetic analysis of ArsR sequences from 20 bacterial species was conducted (Fig. 5). The ArsR repressors cluster in four groups. ArsRs similar to E. coli plasmid R773 ArsR with the arsenic binding site “CVCXXC” cluster in Group I. ArsRs similar to C. glutamicum ArsR cluster in Group II. ArsRs similar to A. ferrooxidans ArsR cluster in Group III. SpArsR-like repressors cluster in Group IV. Group III and Group IV are more closely related to each other than to the other two groups, suggesting that AfArsR and SpArsR have a more recent common ancestor than R773 ArsR and CgArsR.
Fig. 5. Evolutionary relationships of SpArsR with ArsRs from members of other bacterial species.
A neighbor-joining phylogenetic tree shows that there are four type of ArsRs (boxes) with different placement of As(III)- or MAs(III)-binding cysteine residues. SpArsR and AfArsR, are indicated by black triangles.
Discussion
The ArsR/SmtB family of metal responsive repressors includes members that respond to transition metals, heavy metals, and metalloids. As of June 2017 more than 3000 members in archaea, prokaryotic genomes and plasmids have been identified in the NCBI database. Recently genes such as arsH and arsP for detoxification of organic arsenicals including MAs(III) have been identified. MAs(III) is considerably more toxic than As(III), and may be evolved as a primordial antibiotic (Li et al., 2016). In many ars operons a novel arsR is linked with arsP or arsH genes. The arsR and arsP genes are found in both anaerobes and aerobes, while arsH genes appear to only be in aerobes. It is possible that SpArsR and ArsP co-evolved before the atmosphere became oxidizing, and ArsH was acquired afterwards. Most of these arsR genes are located divergently, which is not unusual. Bidirectional ars operons have been found to be controlled by single ArsR that provide differential response to changes in arsenic stress, including the ars operons from A. ferrooxidans (Qin et al., 2007), C. glutamicum ATCC13032 (Ordóñez et al., 2008), Microbacterium sp. strain A33 (Achour-Rokbani et al., 2010) and pR478 of Serratia marsescens (Ryan & Colleran, 2002).
In S. putrefaciens 200, the arsenic ars cluster consists of 19 genes, which are regulated by three different arsR genes (Chen & Rosen, 2016). One, termed SparsR (accession number: ADV53698), is novel in its gene product has a two-cysteine arsenic binding motif and lacks a third ligand for binding of inorganic As(III) compared to characterized ArsRs such as R773 ArsR, CgArsR and AfArsR. This gene is adjacent to a divergently transcribed arsP gene, which suggested that it might regulate arsP transcription. In the related Shewanella sp. strain ANA-3, a similar arsR gene with this novel cysteine motif is also adjacent to an arsP gene that responds poorly or not at all to inorganic As(III) (Murphy & Saltikov, 2009). A similar As(III) non-responsive arsR gene (WP_003518036) was observed in Agrobacterium tumefaciens 5A (WP_003518036) (Kang et al., 2016). The results of phylogenetic analysis of ArsR orthologs with different arsenic binding sites suggest that the number and location of cysteine residues in the primary sequence reflects the evolution of the binding site for MAs(III). SpArsR clustered in a group that has only two conserved cysteines residues and is closely related to the A. ferrooxidans ArsR group that has three conserved cysteines, two of which correspond to those in the SpArsR group (Fig. S1). These results suggest that these SpArsR-like repressors may not respond to inorganic As(III) physiologically and are not involved in detoxification of As(III). Considering that they are adjacent to arsP or arsH genes, we considered the possibility that they are induced primarily by MAs(III), and regulate MAs(III) detoxification. Transcriptional analysis demonstrated that SparsR is co-transcribed with the neighboring gene SparsP when induced with MAs(III) but not As(III). These results show that the occurrence of multiple arsR genes and ars operons is not necessarily redundancy, but instead allows for coping with different arsenical stresses, in this case MAs(III) exposure. Why would organisms evolve a two-coordinate ArsR when three-coordinate ArsRs respond to MAs(IIII) with higher affinity than As(III)? One possibility is that a two-coordinate ArsR that does not respond to As(III) allows the organism to control expression of organoarsenical detoxification genes only when organoarsenicals are present. If environmental As(III) induces genes for detoxification of organoarsenicals when those species are not present, the cells would waste cellular resources synthesizing unnecessary proteins. Thus, arsenic-resistant microorganisms benefit by having multiple arsenic-resistance operons.
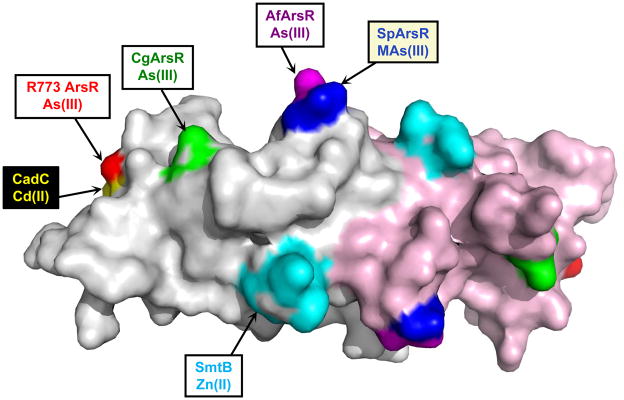
Members of the ArsR/SmtB family share the same overall structure of a winged helix DNA-binding dimmer (Bushweller et al., 1992). Upon this common backbone a variety of different metal/metalloid binding sites have evolved independently from each other and in different spatial locations (Ordóñez et al., 2008, Qin et al., 2007), demonstrating the plasticity of the regulatory sites of these transcriptional repressors (Fig. 6). Metal binding sites can be located either near the DNA binding domain, such as found in R773 ArsR or CadC, or near the C-terminal dimer interface, as in SmtB, CmtR, or AfArsR. For As(III), Sb(III) or Cd(II), the binding sites are composed of three or four cysteine residues. The metal binding sites appear to have evolved independently but have similarities. In plasmid R773, the S3 As(III) binding site is composed of Cys32, Cys34 and Cys37. In CadC, the S4 Cd(II) binding site is composed of Cys7 and Cys11 from the N-terminus of one monomer and Cys58 and Cys60 in the DNA binding site of the other monomer (Ye et al., 2005). CadC Cys58 and Cys60 are congruent with Cys32 and Cys34 of R773 ArsR, suggesting that two binding sites may have evolved from a common ancestor. Like CadC, CgArsR, the three-coordinate As(III) binding site consists of Cys15 and Cys16 from one subunit and Cys55 in the DNA binding site of the other monomer. CadC has two metal binding sites. In addition to the Cd(II)-binding regulatory site, there is a structural Zn(II) binding that is composed of two residues from the C-terminus of each monomer. Similarly, the S3 As(III) binding site of A. ferrooxidans is composed of residues Cys95, Cys96 and Cys102 in the C-terminal dimerization domain. The two C-terminal binding sites do not appear to be evolutionarily related, but their similar locations show that binding sites evolve convergently. In contrast, the MAs(III) binding site of SpArsR appears to have evolved from an ancestor common to the AfArsR As(III) binding site. The SpArsR MAs(III) binding site appears to be the result of a loss-of-function mutation where the third As(III) ligand is no longer present. In reality, though, it is a gain-of-function mutation to provide specificity for MAs(III) that allows for transcriptional regulation of MAs(III) detoxification mechanisms such as the ArsP MAs(III) efflux permease or the ArsH MAs(III) oxidase.
Fig. 6. Location of metal(loid) binding sites in ArsR/SmtB repressors.
Metal(loid) binding sites in members of ArsR/SmtB family of repressor proteins are shown on a surface model of the CadC aporepressor structure by coloring CadC residues corresponding to each binding site as identified from the structure-based alignment (Fig. 2A). The S3 As(III) binding site of the R773 ArsR (red) formed within each monomer overlaps with the corresponding S4 Cd(II) binding site of CadC (yellow) formed between the N-terminus of one subunit and the DNA binding domain of the other subunit. The S3 binding sites of CgArsR1 (green) include residues in the DNA binding site. The Zn(II) binding sites of CadC and SmtB (cyan) are formed between the antiparallel C-terminal α6 helices. The S3 As(III)-binding site of AfArsR (purple) and the S2 MAs(III)-binding site of SpArsR (blue) differ by a single cysteine residues. The variety of the location of metal(lloid) binding sites distributed over the surface of the respective repressors demonstrates the plasticity of evolutionary solutions to similar environmental stresses.
Experimental procedures
Strains, plasmids, medium, and reagents
Escherichia coli Stellar™ (Clontech Laboratories, Mountain View, CA) (F–, endA1, supE44, thi-1, recA1, relA1, gyrA96 phoA, Φ80d lacZΔ M15, Δ(lacZYA-argF)U169, Δ(mrrhsdRMS-mcrBC), ΔmcrA, λ–) (ref) was used for plasmid DNA construction and replication. E. coli AW3110(DE3) (Δars::cam F−IN(rrn-rrnE) (Carlin et al., 1995) bearing two plasmids, pACYC184-ParsP-gfp and pBAD-SparsR was constructed for use as an MAs(III) biosensor. For most experiments cultures of E. coli bearing the indicated plasmids were grown aerobically in Luria-Bertani (LB) medium or M9 medium at 37 °C supplemented with 100 μg/ml ampicillin or 34 μg/ml chloramphenicol, as required (Sambrook et al., 1989). S. putrefaciens 200 was a gift from Flynn Picardal, Indiana University. Unless otherwise noted, cultures of S. putrefaciens were grown aerobically in Luria–Bertani (LB) medium or M9 medium at 30 °C with shaking. Bacterial growth was monitored by measuring the optical density at 600 nm (OD600). Unless otherwise indicated, all reagents were purchased from Sigma-Aldrich. MAs(V) was obtained from Chem Service, Inc., West Chester, PA,. MAs(III) were reduced by the method of Reay and Asher (Reay & Asher, 1977).
Plasmid construction
For expression of SpArsR from S. putrefaciens in E. coli, plasmid pBAD-SpArsR was constructed in which the SparsR gene is under the control of the arabinose promoter and has the sequence for a C-terminal His tag. The SparsR gene was cloned from S. putrefaciens genomic DNA isolated using a QIAamp DNA Mini Kit (QIAGEN, Valencia, CA) with primers SparsRF (NcoI) and SparsRR (SalI) (The sequences of the primers used in this study are listed in Supplemental Table S1, with restriction sites underlined). The PCR fragment was gel purified and digested using NcoI and SalI, then cloned into vector plasmid pBAD/myc-HisA that had been digested with NcoI and SalI, generating plasmid pBAD-SparsR. To analyze SpArsR binding affinity with As(III) and MAs(III), a biosensor with gfp reporter was constructed (Chen et al., 2012). The gfp gene was obtained from pGreen vector, which was digested by SalI. The gfp reporter is under control of the SparsP (ADV53697) promoter, which was cloned from S. putrefaciens genomic DNA using primers ParsPF (BamHI) and ParsPR (SalI). The two fragments were gel purified, digested with restriction enzymes and ligated into vector plasmid pACYC184 that had been digested with the same restriction enzymes, generating plasmid pACYC184-ParsP-gfp. To convert SpArsR to an As(III) responsive ArsR, hybrid pBAD-SpArsRAC was generated by fusing the sequence for the C-terminus of AfArsR (His97-Q118) to sequence for C-truncated SpArsR (1–102). The fragment of SparsR was amplified using primers HSpArsR102F and HSpArsR102R. The fragment of AfarsR was amplified with primers HAfArsR97F and HAfArsR97R using plasmid pBAD-AfarsR as a template (Qin et al., 2007). The two fragments were gel purified, digested with restriction enzymes and ligated into vector plasmid pBAD. The final constructs were further confirmed by DNA sequencing (Sequetech, Mountain View, CA).
Reverse transcription polymerase chain reaction (RT-PCR)
Total RNA from S. putrefaciens 200 was isolated using the TRIZOL reagent (Life Sciences, Thermo Fisher Scientific, Grand Island, NY) according to the manufacturer’s protocol. DNA was removed from total RNA preparation using TURBO DNA free Kit (Ambion, Thermo Fisher Scientific, Grand Island, NY). cDNA synthesis was carried out with 0.5 mg of total RNA using an AccuScript High Fidelity 1st strand cDNA synthesis kit (Agilent, Santa Clara, CA). For RT-PCR total RNA isolated from exponentially growing S. putrefaciens 200 in the absence or presence of 20 μM As(III) or 2 μM MAs(III) was used as template in a reverse transcriptase reaction to generate cDNAs that were then amplified with the indicated primer sets (Supplemental Table S1). The primers covering arsR and arsP with overlap were purchased from Integrated DNA technologies (IDT) (Coralville, lA). PCR products were resolved on 1% agarose gels using standard methods and stained with ethidium bromide.
Mutagenesis of the SpArsR gene
SparsR mutations were generated by site-directed mutagenesis using a Quick Change mutagenesis kit (Stratagene, La Jolla, CA). The mutagenic oligonucleotides used for both strands and the respective changes introduced (underlined) are listed in (Supplemental Table S1). The codons for residues Cys83, Cys101 and Cys102 were changed to serine codons, generating three different single-cysteine SpArsR mutants. Each mutation was confirmed by DNA sequencing (Sequetech, Mountain View, CA).
Phylogenetic analysis
Multiple alignment of ArsR homolog sequence was performed using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). ArsR sequences with conserved cysteines were selected for phylogenetic analysis. Acquisition of sequences was performed by searching a list of reference organisms or from the National Center for Biotechnology Information (NCBI) protein database using a BLASTP search (Johnson et al., 2008). Phylogenetic analysis was performed to infer the evolutionary relationship among the arsenic repressor of various organisms. The phylogenetic tree was constructed using the Neighbor-Joining method using MEGA 6.0.1 (Tamura et al., 2013). The statistical significance of the branch pattern was estimated by conducting a 1000 bootstrap.
Assay of MAs(III) binding in vivo
Transcriptional activity of the biosensor was estimated from arsenical-responsive expression of gfp (Chen et al., 2014). Cultures of the biosensor (E. coli strain AW3110 bearing plasmids pBAD-SpArsR and pACYC184-ParsP-gfp) were grown to mid-exponential phase in M9 medium at 37 °C with 100 μg/mL ampicillin and 34 μg/mL chloramphenicol with shaking. Glucose (0.2 %) was added for constitutive expression of gfp. SparsR gene was induced by addition of 0.2 % arabinose for 5 h. Derepression was produced by simultaneous addition of arabinose and arsenicals for 5 h. Cell densities were normalized by dilution or suspension to the same A600 nm, and expression of gfp was assayed from the fluorescence of cells using a Photon Technology International spectrofluorometer with an excitation wavelength of 470 nm and emission wavelength of 510 nm.
Construction of homology models
Homology modeling and computational studies were undertaken to explore the structural similarities and differences of arsenic binding site in ArsR orthologs. AfArsR, CgArsR, R773ArsR and SpArsR homology models were built using the crystal structure of ArsR/SmtB family proteins available in the protein data bank. All models were built as the homodimer. AfArsR from I10 to S105, CgArsR from G21 to L112, R773 ArsR from P6 to K102 and SpArsR from V13 to Cys102 were built by PDBs 2JSC (Banci et al., 2007), 1R1T (Eicken et al., 2003), 3F6V (Chang, C., Xu, X., Zheng, H., Savchenko, A., Edwards, A and Joachimiak, A, unpublished) and 3JTH (Nishi et al., 2010), respectively, as the templates. The homology models were constructed using a fully automated protein structure homology modeling server SWISS-MODEL (Kiefer et al., 2009) (http://swissmodel.expasy.org/). Model quality was estimated based on the QMEAN scoring function. The As(III)-bound AfArsR model was constructed as previously reported (Qin et al., 2007). MAs(III)-bound SpArsR was built by manually docking MAs(III) to the cysteine pairs using COOT (Emsley & Cowtan, 2004). PyMOL v1.3 was used to visualize the structural models (DeLano, 2001) (https://www.pymol.org/citing).
Supplementary Material
Acknowledgments
This work was supported by NIH grant R01 GM55425. The authors certify that they have no conflicts of interest to declare. We thank Flynn Picardal, Indiana University, for the gift of S. putrefaciens 200.
Abbreviations
- MAs(III)
Methylarsenite
- MAs(V)
methylarsenate
- DMAs(V)
dimethylarsenate
- MSMA
monosodium methylarsenate
References
- Achour-Rokbani A, Cordi A, Poupin P, Bauda P, Billard P. Characterization of the ars gene cluster from extremely arsenic-resistant Microbacterium sp. strain A33. Appl Environ Microbiol. 2010;76:948–955. doi: 10.1128/AEM.01738-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banci L, Bertini I, Cantini F, Ciofi-Baffoni S, Cavet JS, Dennison C, Graham A, Harvie DR, Robinson NJ. NMR structural analysis of cadmium-sensing by winged helix repressor CmtR. J Biol Chem. 2007;282:30181–30188. doi: 10.1074/jbc.M701119200. [DOI] [PubMed] [Google Scholar]
- Bushweller JH, Åslund F, Wuthrich K, Holmgren A. Structural and functional characterization of the mutant Escherichia coli glutaredoxin (C14S) and its mixed disulfide with glutathione. Biochemistry. 1992;31:9288–9293. doi: 10.1021/bi00153a023. [DOI] [PubMed] [Google Scholar]
- Carlin A, Shi W, Dey S, Rosen BP. The ars operon of Escherichia coli confers arsenical and antimonial resistance. J Bacteriol. 1995;177:981–986. doi: 10.1128/jb.177.4.981-986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GQ, Zhou L, Styblo M, Walton F, Jing Y, Weinberg R, Chen Z, Waxman S. Methylated metabolites of arsenic trioxide are more potent than arsenic trioxide as apoptotic but not differentiation inducers in leukemia and lymphoma cells. Cancer Res. 2003;63:1853–1859. [PubMed] [Google Scholar]
- Chen J, Bhattacharjee H, Rosen BP. ArsH is an organoarsenical oxidase that confers resistance to trivalent forms of the herbicide monosodium methylarsenate and the poultry growth promoter roxarsone. Mol Microbiol. 2015a;96:1042–1052. doi: 10.1111/mmi.12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Madegowda M, Bhattacharjee H, Rosen BP. ArsP: a methylarsenite efflux permease. Mol Microbiol. 2015b;98:625–635. doi: 10.1111/mmi.13145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Rosen BP. Organoarsenical biotransformations by Shewanella putrefaciens. Environ Sci Technol. 2016;50:7956–7963. doi: 10.1021/acs.est.6b00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Sun S, Li CZ, Zhu YG, Rosen BP. Biosensor for organoarsenical herbicides and growth promoters. Environ Sci Technol. 2014;48:1141–1147. doi: 10.1021/es4038319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhu YG, Rosen BP. A novel biosensor selective for organoarsenicals. Appl Environ Microbiol. 2012;78:7145–7147. doi: 10.1128/AEM.01721-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen WR, McBride BC, Reglinski J. The reaction of methylarsenicals with thiols: Some biological implications. J Inorg Biochem. 1984;21:179–194. [Google Scholar]
- DeLano WL. The PyMOL user’s manual. DeLano Scientific; San Carlos, CA: 2001. [Google Scholar]
- Delnomdedieu M, Basti MM, Otvos JD, Thomas DJ. Transfer of arsenite from glutathione to dithiols: a model of interaction. Chem Res Toxicol. 1993;6:598–602. doi: 10.1021/tx00035a002. [DOI] [PubMed] [Google Scholar]
- Delnomdedieu M, Basti MM, Otvos JD, Thomas DJ. Reduction and binding of arsenate and dimethylarsinate by glutathione: a magnetic resonance study. Chem Biol Interact. 1994;90:139–155. doi: 10.1016/0009-2797(94)90099-x. [DOI] [PubMed] [Google Scholar]
- Drobna Z, Waters SB, Devesa V, Harmon AW, Thomas DJ, Styblo M. Metabolism and toxicity of arsenic in human urothelial cells expressing rat arsenic (+3 oxidation state)-methyltransferase. Toxicol Appl Pharmacol. 2005;207:147–159. doi: 10.1016/j.taap.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eicken C, Pennella MA, Chen X, Koshlap KM, VanZile ML, Sacchettini JC, Giedroc DP. A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins. J Mol Biol. 2003;333:683–695. doi: 10.1016/j.jmb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang YS, Brame K, Jetter J, Bothner BB, Wang G, Thiyagarajan S, McDermott TR. Regulatory activities of four ArsR proteins in Agrobacterium tumefaciens 5A. Appl Environ Microbiol. 2016;82:3471–3480. doi: 10.1128/AEM.00262-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37:D387–392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Pawitwar SS, Rosen BP. The organoarsenical biocycle and the primordial antibiotic methylarsenite. Metallomics. 2016;8:1047–1055. doi: 10.1039/c6mt00168h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Rensing C, Rosen BP. Resistance pathways for metalloids and toxic metals. In: Culotta V, Scott RA, editors. Metals in Cells. Hoboken, NJ: Wiley & Sons, Inc; 2013. pp. 429–442. [Google Scholar]
- Murphy JN, Saltikov CW. The ArsR repressor mediates arsenite-dependent regulation of arsenate respiration and detoxification operons of Shewanella sp. strain ANA-3. J Bacteriol. 2009;191:6722–6731. doi: 10.1128/JB.00801-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi K, Lee HJ, Park SY, Bae SJ, Lee SE, Adams PD, Rhee JH, Kim JS. Crystal structure of the transcriptional activator HlyU from Vibrio vulnificus CMCP6. FEBS Lett. 2010;584:1097–1102. doi: 10.1016/j.febslet.2010.02.052. [DOI] [PubMed] [Google Scholar]
- Ordóñez E, Thiyagarajan S, Cook JD, Stemmler TL, Gil JA, Mateos LM, Rosen BP. Evolution of metal(loid) binding sites in transcriptional regulators. J Biol Chem. 2008;283:25706–25714. doi: 10.1074/jbc.M803209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Fu HL, Ye J, Bencze KZ, Stemmler TL, Rawlings DE, Rosen BP. Convergent evolution of a new arsenic binding site in the ArsR/SmtB family of metalloregulators. J Biol Chem. 2007;282:34346–34355. doi: 10.1074/jbc.M706565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Lehr CR, Yuan C, Le XC, McDermott TR, Rosen BP. Biotransformation of arsenic by a Yellowstone thermoacidophilic eukaryotic alga. Proc Natl Acad Sci U S A. 2009;106:5213–5217. doi: 10.1073/pnas.0900238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Rosen BP, Zhang Y, Wang G, Franke S, Rensing C. Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc Natl Acad Sci U S A. 2006;103:2075–2080. doi: 10.1073/pnas.0506836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reay PF, Asher CJ. Preparation and purification of 74As-labeled arsenate and arsenite for use in biological experiments. Anal Biochem. 1977;78:557–560. doi: 10.1016/0003-2697(77)90117-8. [DOI] [PubMed] [Google Scholar]
- Ryan D, Colleran E. Arsenical resistance in the IncHI2 plasmids. Plasmid. 2002;47:234–240. doi: 10.1016/s0147-619x(02)00012-4. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning, a laboratory manual. Cold Spring Harbor Laboratory; New York: 1989. [Google Scholar]
- Shi W, Dong J, Scott RA, Ksenzenko MY, Rosen BP. The role of arsenic-thiol interactions in metalloregulation of the ars operon. J Biol Chem. 1996;271:9291–9297. doi: 10.1074/jbc.271.16.9291. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Rosen BP. Metalloregulated expression of the ars operon. J Biol Chem. 1993;268:52–58. [PubMed] [Google Scholar]
- Xu X, Burke SP. Roles of active site residues and the NH2-terminal domain in the catalysis and substrate binding of human Cdc25. J Biol Chem. 1996;271:5118–5124. doi: 10.1074/jbc.271.9.5118. [DOI] [PubMed] [Google Scholar]
- Yang HC, Rosen BP. New mechanisms of bacterial arsenic resistance. Biomed J. 2016;39:5–13. doi: 10.1016/j.bj.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Kandegedara A, Martin P, Rosen BP. Crystal structure of the Staphylococcus aureus pI258 CadC Cd(II)/Pb(II)/Zn(II)-responsive repressor. J Bacteriol. 2005;187:4214–4221. doi: 10.1128/JB.187.12.4214-4221.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga M, Rosen BP. A C-As lyase for degradation of environmental organoarsenical herbicides and animal husbandry growth promoters. Proc Natl Acad Sci U S A. 2014;111:7701–7706. doi: 10.1073/pnas.1403057111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YG, Yoshinaga M, Zhao FJ, Rosen BP. Earth abides arsenic biotransformations. Annu Rev Earth and Planet Sci. 2014;42:443–467. doi: 10.1146/annurev-earth-060313-054942. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.