SUMMARY
The most abundant oxidants controlling bacterial colonization on mucosal barrier epithelia are hypochlorous acid (HOCl), hypobromous acid (HOBr), and hypothiocyanous acid (HOSCN). All three oxidants are highly antimicrobial but little is known about their relative efficacies, their respective cellular targets, or what specific responses they elicit in bacteria. To address these important questions, we directly tested the individual oxidants on the virulent Pseudomonas aeruginosa strain PA14. We discovered that HOCl and HOBr work almost interchangeably, impacting non-growing bacterial cultures more significantly than actively growing bacteria, and eliciting similar stress responses, including the heat shock response. HOSCN treatment is distinctly different, affecting primarily actively growing PA14, and evoking stress responses suggestive of membrane damage. What all three oxidants have in common, however, is their ability to cause substantial protein aggregation. This effect became particularly obvious in strains lacking polyphosphate, a newly recognized chemical chaperone. Treatment of PA14 with the FDA-approved anti-inflammatory drug mesalamine, which has recently been shown to attenuate polyP production in a wide range of bacteria, effectively decreased the resistance of PA14 towards all three oxidants, suggesting that we have discovered a novel, targetable defense system in P. aeruginosa.
Keywords: oxidative protein unfolding, polyphosphate, hypohalous acids, bacterial defense systems
ABBREVIATED SUMMARY
Accumulation of polyphosphate serves as the universal strategy of P. aeruginosa to respond to and resist the host-induced production of antimicrobial oxidants by reducing oxidative protein unfolding and aggregation. Impairment of polyphosphate production caused by the FDA-approved drug mesalamine sensitizes the pathogen towards all three physiological oxidants making mesalamine potentially an attractive new treatment option to improve the ability of the endogenous host defense systems to kill stress-resistant microbes.
INTRODUCTION
The natural ability of mammalian hosts to defend themselves against invading pathogens involves the production of antimicrobial oxidants, particularly hypochlorous acid (HOCl, bleach), hypobromous acid (HOBr) and hypothiocyanous acid (HOSCN) (Klebanoff et al., 2013). These antimicrobial hypohalous acids (HOX) are produced by human haloperoxidases, which include (i) myeloperoxidases (MPO) in activated neutrophils, monocytes, and tissue macrophages at sites of inflammation (Davies, 2011); (ii) lactoperoxidases (LPO) and salivary peroxidases, that are secreted from salivary and other mucosal glands and play important roles in controlling the oral microflora (Ashby, 2008); (iii) eosinophil peroxidases in eosinophil granulocytes (EPO) (Wang & Slungaard, 2006); and (iv) gastric peroxidases (GPO) found in cells of the fundic region in the stomach (Das et al., 1995). What all of these haloperoxidases have in common is their ability to catalyze the peroxide (H2O2)-mediated oxidation of halides (e.g., Cl−, Br−) and/or pseudohalides (e.g., SCN−), yielding the respective two-electron oxidants (Davies, 2011, Winterbourn et al., 2016). The levels of oxidants produced depend on the concentrations of the respective anions and on the peroxidases catalyzing the reaction. Previous reports indicated that MPO in the plasma produces almost equal amounts of HOCl and HOSCN, and only a minor (~5%) amount of HOBr (van Dalen et al., 1997). In contrast, LPO and GPO only oxidize SCN− under physiological conditions, whereas EPO oxidizes primarily SCN− and Br− (Hawkins et al., 2009, van Dalen et al., 1997).
All three oxidants are effective antimicrobials and considered more bactericidal/bacteriostatic than H2O2 (Love et al., 2016). HOCl, for instance, is known to rapidly oxidize a variety of amino acid side chains, causing extensive protein unfolding and aggregation (Dahl et al., 2015, Winter et al., 2008, Winterbourn, 2008). Not surprisingly, bacterial defense systems against HOCl include expression of transcription factors and genes involved in detoxification and reduction of side chain modifications (Gebendorfer et al., 2012, Gray et al., 2013, Parker et al., 2013), activation of stress specific molecular chaperones, such as Hsp33 and RidA that prevent protein aggregation (Muller et al., 2014, Winter et al., 2008), and conversion of ATP in inorganic polyphosphate, a chemical chaperone effective in stabilizing protein structures (Gray & Jakob, 2015, Gray et al., 2014). Absence of any one of these systems causes a significant decrease in HOCl stress resistance, suggesting that protein unfolding is a major contributor to the bactericidal activity of HOCl. Much less is known about the effects of and response to HOBr, which is also a strong oxidant that readily brominates tyrosine, tryptophan and nucleic acids (Davies et al., 2008, Klebanoff, 2005). Nevertheless, HOCl and HOBr are both associated with numerous diseases, including initiation and progression of various inflammatory diseases, such as atherosclerosis (Davies, 2011, Davies et al., 2008, Klebanoff, 2005). HOSCN appears to react with great specificity with thiols (e.g. GSH; protein cysteines) and selenogroups (Skaff et al., 2009, Skaff et al., 2012), resulting in a number of reversible oxidation products, including sulfenyl thiocyanates, sulfenic acids or disulfides (Barrett et al., 2012). High concentrations of HOSCN, however, result in non-reversible thiol products including sulfinic/sulfonic acids (Barrett & Hawkins, 2012). In contrast to HOCl/HOBr, the role of HOSCN in disease is still unclear and appears organism-specific, ranging from apparently protective functions in mammalian hosts to cytotoxic effects in bacteria (Pattison et al., 2012).
It is largely unknown how the three oxidants compare in bactericidal efficacies, what their specific in vivo targets are, and whether bacteria contain universal or individualized defense systems against these three different stressors. Finding answers to these important questions is especially pressing for patients suffering from cystic fibrosis, one of the most common fatal hereditary diseases, affecting about 30,000 people in the US (Cohen & Prince, 2012). Caused by a defect in the cystic fibrosis transmembrane conductance regulator (CFTR), patients experience frequent and reoccurring lower respiratory tract infections, chronic inflammation and progressive tissue damage in the lungs (Rao & Grigg, 2006). Analysis of the bronchoalveolar lavage fluid of CF patients revealed substantially altered levels of HOCl and HOSCN, presumably due to a combination of increased MPO activity caused by sustained airway neutrophilia, and a lack of secreted SCN− and glutathione (GSH), two CFTR-dependent compounds (Kogan et al., 2003, Lorentzen et al., 2011). HOCl levels were further increased in CF patients testing positive for P. aeruginosa infection, consistent with even higher numbers of activated neutrophils and further increased MPO concentrations during infections.
In this study, we now directly compared the antimicrobial efficacy of HOCl, HOBr and HOSCN on the pathogenic P. aeruginosa strain PA14. We determined the effects that the three oxidants exert on PA14, and identified common and oxidant-specific response and defense systems by RNAseq analysis. Despite their distinct reactivities, we discovered that all three oxidants cause extensive protein unfolding and aggregation. Stress-induced accumulation of polyphosphate appears to be the universal response to HOCl/HOBr and HOSCN treatment and serves as an effective strategy to prevent stress-induced protein aggregation and increase stress survival. Mesalamine, an FDA-approved drug recently discovered to reduce cellular polyP levels in a wide range of bacteria, including P. aeruginosa, effectively increases PA14’s sensitivity towards the antimicrobial oxidants, making it a potential treatment option for chronic P. aeruginosa infections.
RESULTS
The antimicrobial efficacies of HOCl, HOBr and HOSCN differ by growth conditions
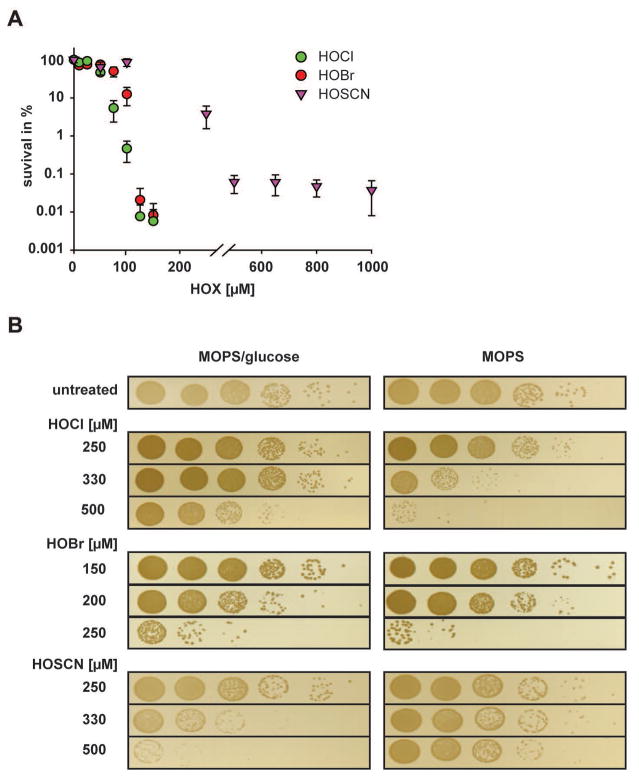
Most growth media are rich in components that react with and potentially quench the effective concentration of oxidants like HOCl and HOBr, making direct comparisons in relative bactericidal efficacy challenging. As a first approach, we therefore decided to treat the bacteria with the respective oxidants in phosphate-buffered saline (PBS), which is known not to react with any of the oxidants. We grew P. aeruginosa PA14 to mid-logarithmic phase in MOPS-glucose media, switched the media to PBS and treated the bacteria with various concentrations of the respective oxidants for 30 min. Next, we quenched the oxidants, serially diluted and spotted the bacteria onto LB plates and monitored their survival after 14 h of incubation. We found that HOCl and HOBr showed very comparable bactericidal activity with 100–125 μM of each oxidant being sufficient to reduce PA14 survival by about four orders of magnitude (Fig. 1A, Fig. S1). In contrast, HOSCN, when used at this concentration, reduced bacterial survival by less than one log, suggesting that HOSCN is much less bactericidal under these conditions (Fig. 1A, Fig. S1). Moreover, even at very high concentrations of HOSCN (i.e., 1 mM), a substantial fraction of bacteria still survived the treatment.
Fig. 1. Effects of HOCl, HOBr and HOSCN on the survival of non-growing or exponentially growing PA14.
Wild-type PA14 was grown in MOPS-glucose media to mid-log phase (A600= 0.4–0.5) and shifted to either (A) PBS buffer or (B) MOPS media in the absence or presence of glucose. The indicated concentrations of HOCl, HOBr, or HOSCN were added. After 30 min incubation, excess HOX was quenched by the addition of 10 mM thiosulfate, and the cells were serially diluted with 0.85% NaCl, spot-titered onto LB agar plates, and incubated o/n at 37°C. The experiments were repeated at least three independent times. Data shown in (A) represent means ± s.e.m. A representative data set is shown in (B).
Surprisingly, we obtained very different results when we conducted the treatments in MOPS-glucose media. Under these conditions, HOBr was most effective in killing PA14, followed by HOSCN and HOCl (Fig. 1B, left panel). Explanations for this result included the possibility that components of the MOPS-glucose medium differentially quench the respective oxidants, or that cells grown in MOPS-glucose media might react with oxidants in ways that generate reaction products of differing bactericidal activities (e.g., chloramines, bromamines). Alternatively, however, we also considered the possibility that the three oxidants might act differently on metabolically active and inactive bacteria. To distinguish between these possibilities, we cultivated PA14 in MOPS-glucose, washed the cells, and, shortly before the stress treatment, shifted one part of the cells into fresh MOPS media with glucose (MOPS-glucose) to allow for continuous growth. The other part, we transferred into fresh MOPS media without glucose (MOPS), which resulted in an immediate growth arrest. We reasoned that this approach should allow us to keep the media composition constant while only varying the growth status of the bacteria. We found that HOSCN was substantially more bactericidal on actively growing cells, while HOCl and HOBr were both more effective in killing non-growing P. aeruginosa (Fig. 1B, compare left and right panel). For instance, in the presence of 0.5 mM HOSCN, cell survival in MOPS-glucose media was reduced by 4 orders of magnitude while no substantial killing was observed when the treatment was conducted in MOPS media. In contrast, HOCl and HOBr caused significant killing non-growing bacteria at concentrations that posed little threat to actively metabolizing bacteria (Fig. 1B). Importantly, we confirmed that glucose does not quench HOCl, suggesting that it is indeed the growth status of the cells that affects the toxicity of the oxidants. These results are fully consistent with our previous experiments conducted in PBS buffer, which showed that both HOCl/HOBr are significantly more effective than HOSCN in killing non-growing P. aeruginosa. They furthermore suggest that the three oxidants work by different mechanisms and/or affect different in vivo targets.
PA14 transcriptional changes in response to HOCl, HOBr and HOSCN treatment
Adaptation of gene expression is key for bacteria to quickly respond to changes in their environment. As a first step towards obtaining insights into the physiological consequences of HOCl, HOBr or HOSCN treatment, we tested gene expression changes in PA14 treated with sublethal concentrations of the respective oxidants. Since we wanted to keep the cultivation conditions the same and directly compare the in vivo effects of the three oxidants, we had to first identify concentrations of each oxidant that cause comparable growth defects without killing the cells. We settled on 0.5 mM HOCl, 0.15 mM HOBr or 0.25 mM HOSCN, which triggered a 1–1.5 h growth arrest (Fig. S2A). We isolated total RNA from the treated cells and conducted RNAseq analysis to monitor global changes in gene expression in PA14 in response to each of the three oxidants. All experiments were conducted in triplicates. For the transcriptome analysis, we included all genes annotated in the NCBI database and compared the expression values of the stress treated cells to non-stress treated controls. We set a false discovery rate (FDR) of <0.005 as a threshold for significance, and considered transcripts as upregulated when they showed a log2 fold change of >1.5 and downregulated when they showed a log2 fold change of < −1.5. A list of all expression data can be found in Tables S1A–C.
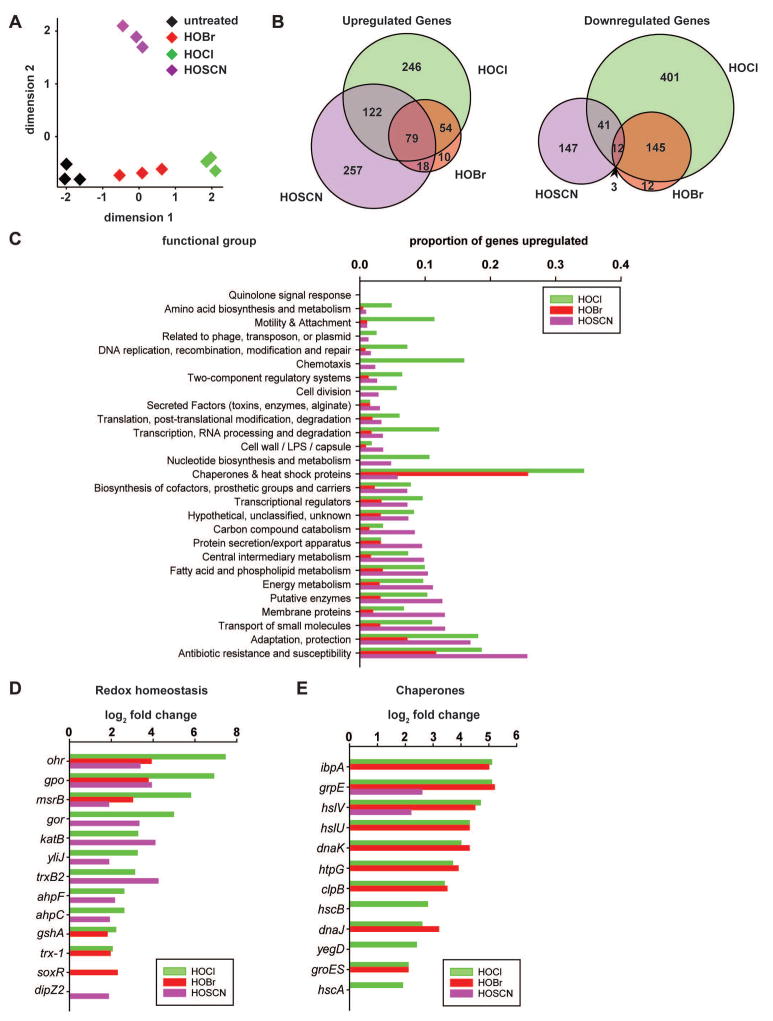
A multidimensional scaling plot of our data sets revealed that while gene expression changes in all three replicates treated with either HOCl or HOBr clustered in one dimension (Fig. 2A, green and red diamonds), treatment of PA14 with HOSCN led to distinctly different gene expression changes and clustered in two dimensions (Fig. 2A, purple diamonds). Closer analysis of all up- and downregulated genes in the three treatment groups revealed a >85% overlap between differentially expressed genes during HOBr and HOCl treatment while less than a 50% overlap existed between differentially expressed genes in PA14 treated with HOCl/HOBr and HOSCN (Fig. 2B). Closer analysis of the data revealed that all three stressors induced the expression of genes involved in oxidant detoxification (e.g., catalase, methionine sulfoxide reductase, alkyl hydroperoxide reductase) and restoration of redox homeostasis (e.g. glutathione peroxidase, glutathione reductase) (Fig. 2C, D; Table S1). Very similar results in regards to HOCl stress responses were previously reported for Bacillus subtilis, P. aeruginosa strain PAO1, Bacillus cereus ATCC14579 and E. coli MG1655 (Ceragioli et al., 2010, Chi et al., 2011, Small et al., 2007, Gray et al., 2013).
Fig. 2. Global gene expression changes in PA14 in response to HOCl, HOBr or HOSCN treatments.
Exponentially growing PA14 wild-type cells (A600= 0.4–0.5) were incubated with sublethal concentration of HOCl (0.5 mM), HOBr (0.15 mM), or HOSCN (0.25 mM) for 15 min. Incubation was stopped by the addition of ice-cold methanol. Reads were aligned to the P. aeruginosa UCBPP-PA14 reference genome (Accession number: NC_008463.1). (A) A multidimensional scaling plot revealed distinct clustering of transcriptomes between untreated PA14 (black) and PA14 treated with HOCl (green), HOBr (red), or HOSCN (purple). (B) Venn diagram shows the number of upregulated or downregulated genes upon treatment with HOCl (green), HOBr (red), or HOSCN (purple) and a potential overlap between treatments. (C) Proportion of upregulated genes with regards to functional groups defined in (Lee et al., 2006). (D) Log2-fold change in genes involved in redox homeostasis. (E) Log2-fold change in heat shock genes. Each condition was tested in triplicate. Significance was defined by a false discovery rate of <0.005 and absolute values of log2-fold change of >1.5.
We next ran Fisher’s Exact Tests on the 78 known regulons in PA14, and determined the false discovery rate (FDR) to identify differentially expressed genes that are statistically significantly enriched upon one or the other stress treatment (see Experimental Procedures for details). Surprisingly, only 6 regulons showed a FDR <0.05 in stress-treated versus untreated PA14, and only one of these regulons, MexT, responded comparably to all three antimicrobials (Table 1; Fig. S2B). MexT is a redox-sensitive LysR-type transcriptional regulator that modulates the expression of virulence factors and significantly contributes to the increased antibiotic resistance in P. aeruginosa (Fargier et al., 2012). It has recently been shown to induce expression of the MexEF-OprN multidrug efflux system, which exports electrophilic metabolites to restore redox homeostasis in the cell (Fargier et al., 2012). Our finding that all three oxidants induce the MexT regulon agrees with the redox-modifying properties of the three oxidants, and suggests that they activate MexT through thiol oxidation. Surprisingly, based on our FDR <0.05 criterium, we did not find the peroxide-sensitive OxyR regulon to be significantly induced by any of the three treatments (Fig. S2B), although OxyR has been previously shown to contribute to increased HOCl resistance in E. coli (Dukan & Touati, 1996).
Table 1.
Regulons determined to be significantly enriched for differentially expressed genes (FDR < 0.05) using one-sided Fisher’s Exact Tests.
| Regulon | FDR
|
||
|---|---|---|---|
| HOBr | HOCl | HOSCN | |
|
| |||
| mexT | 1.8 × 10−6 | 1.1 × 10−9 | 5.8 × 10−4 |
| lasR | 1.6 × 10−8 | 1.6 × 10−12 | n.s. |
| pchR | n.s. | 3.0 × 10−4 | n.s. |
| fur | n.s. | 0.03 | n.s. |
| rhlR | 1.1 × 10−4 | n.s. | n.s. |
| gacA | 0.015 | n.s. | n.s. |
The LasR regulon, which controls quorum sensing in P. aeruginosa was significantly down-regulated upon both HOCl and HOBr treatment but not by HOSCN. The remaining four regulons responded specifically to only one of the three oxidants; whereas both RhlR- and GacA regulons were significantly altered only upon HOBr treatment, Fur and PchR appeared to respond to HOCl treatment only (Table 1; Fig. S2B).
We also analyzed the expression levels of transcriptional regulators, whose regulons have not yet been identified in P. aeruginosa but which have previously shown to be HOCl-responsive in other organisms. Similar to our previous results, we found that HOCl, HOBr and HOSCN treatment elicits both overlapping and distinct responses in PA14. For instance, we found that the oxidative stress-sensing regulator OspR, which regulates glutathione peroxidase (Atichartpongkul et al., 2016), was strongly induced upon treatment with any of the three antimicrobials (Table S1). Expression of NemR (PA14_36300), a known HOCl-specific repressor in E. coli (Gray et al., 2013), was significantly upregulated in response to HOCl and HOSCN but not HOBr (Table S1). In contrast, the chlorine-specific transcription factor HypT/YjiE (PA14_71640) (Gebendorfer et al., 2012) was significantly induced only by HOCl, while RclR (PA14_07340), a third HOCl-responsive transcriptional activator in E. coli (Parker et al., 2013) was not significantly up-regulated in response to any of the three oxidants (Table S1).
By far the most significant differences in the proportion of upregulated genes, however, were found in the functional categories of molecular chaperones (Fig. 2C, E). While a total of 12 and 9 heat shock genes (e.g., ibpA, dnaK, groES, hslU/V) were significantly upregulated in response to HOCl and HOBr, respectively, only two (i.e., grpE, hslV) were upregulated in response to HOSCN (Fig. 2E). Moreover, the transcripts of both genes were about 6-fold less abundant in HOSCN-treated cells as compared to HOCl-treated cells. The HOCl-results were consistent with our previous microarray data in E. coli (Gray et al., 2013). Other functional categories that were specifically and significantly more upregulated in response to HOCl/HOBr as compared to HOSCN included genes involved in motility and attachment, chemotaxis, and nucleotide biosynthesis (Fig. 2C). This result fully agrees with previous observations in E. coli MG1655 and B. subtilis, where exposure to sublethal HOCl concentrations caused the induction of motility and chemotaxis-related genes (Chi et al., 2011, Gray et al., 2013). Interchangeably, a much higher proportion of genes involved in antibiotic resistance or encoding membrane proteins were upregulated in response to HOSCN as compared to HOCl and HOBr, suggesting that HOSCN might target membrane proteins (Fig. 2C). These data demonstrate that HOCl and HOBr work quite similarly in vivo, and elicit distinct gene expression changes when compared with the effects of HOSCN treatment.
Effects of HOCl, HOBr and HOSCN on the PA14 proteome
HOCl has previously been shown to induce extensive protein unfolding and aggregation (Winter et al., 2008). This effect of HOCl is presumably due to its ability to rapidly oxidize a variety of different amino acid side chains, thereby shifting the equilibrium of proteins more towards their aggregation-prone state (Dahl et al., 2015, Winter et al., 2008). HOCl-stress induced protein aggregation explains the upregulation of members of the heat shock regulon, whose expression is triggered by the accumulation of unfolded proteins (Arsene et al., 2000). Since bacterial HOCl-stress resistance is increased by preventing HOCl-induced protein aggregation, we previously concluded that protein unfolding is one mechanism by which HOCl kills bacteria (Winter et al., 2008). Based on the chemical similarity between HOCl and HOBr as well as the similarity by which cells react to the two stressors, it is likely that HOBr also triggers global protein aggregation. HOSCN, in contrast, is thought to be highly cysteine-specific, raising the question whether it kills bacteria by different means (Chandler et al., 2013, Skaff et al., 2009, Barrett et al., 2012).
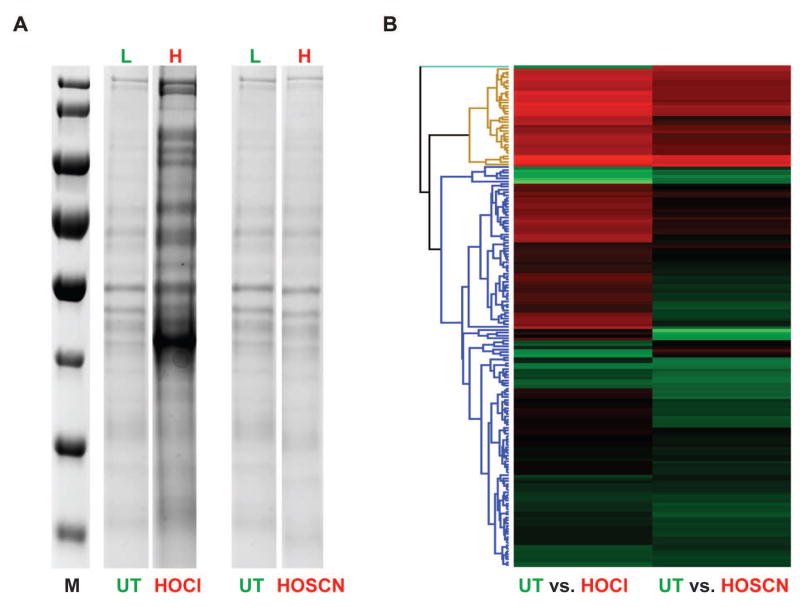
To directly compare the extent and targets of protein aggregation during our various oxidative stress treatments, we therefore treated wild-type PA14 with our previously established sublethal concentrations of HOCl (0.5 mM), HOBr (0.15 mM) or HOSCN (0.25 mM). After 30 min of treatment, we lysed the cells and analyzed the aggregated proteins by SDS-PAGE. While treatment with HOCl and HOBr triggered similar protein aggregation, affecting a large number of different PA14 proteins, we found much less protein aggregation when cells were treated with growth-delaying concentrations (i.e., 0.25 mM) of HOSCN (Fig. 3A, S3). This result was fully consistent with our RNAseq data that showed induction of the heat shock response when cells experienced sublethal doses of HOCl/HOBr but not HOSCN (Fig. 2E). Higher (i.e., lethal) concentrations of HOSCN, however, also triggered substantial protein aggregation, indicating that HOSCN either directly or indirectly exerts proteotoxic effects as well (Fig. S3).
Fig. 3. Effects of HOCl or HOSCN on the aggregation propensity of the PA14 wild-type proteome.
A SILAC experiment was performed to determine the distribution of aggregated proteins in PA14 upon treatment with either HOCl or HOSCN. PA14 wild-type cells were grown in MOPS-glucose medium supplemented with either 120 μg/ml isotopically light [12C6]Arg or heavy [13C6]Arg. PA14 cultures in light [12C6]Arg were either left untreated while cultures in heavy [13C6]Arg were exposed to either 0.5 mM HOCl or 0.25 mM HOSCN for 30 min. Cells were lysed, and the aggregated proteins were separated from the soluble fractions by centrifugation. (A) Insoluble proteins were analyzed by SDS-PAGE. (B) Equal volumes of insoluble proteins from light and heavy samples were mixed, and the proteins were identified by MS/MS analysis. A heat map was generated using the Hierarchical clustering function with Perseus. Row Trees were processed using the default settings with color red indicating those proteins that were enriched in the aggregates of HOCl (left) or HOSCN (right) treated cells.
Subsequent SILAC experiments on the protein aggregates on HOCl and HOSCN treated PA14 further confirmed these results and revealed a significantly increased aggregation of proteins in HOCl-treated bacteria as compared to HOSCN-treated PA14 (Fig 3B). However, due to the fact that PA14 produces endogenous arginine, the incorporation rate of 13C6-Arg was very low (6–8%). This caused our H/L ratios to be quite small, and made direct comparisons of the respective H/L ratios in HOCl and HOSCN treated bacteria challenging. As a first approach, we therefore decided to use the H/L ratio of the over 30 identified ribosomal proteins found in both of our aggregate preparations as internal reference points (see Fig S4 & Table S2, highlighted in orange). We then focused primarily on those annotated proteins, whose H/L ratio was substantially higher than that of ribosomal subunits. By using this analysis, we confirmed many of the proteins that have previously been identified to be aggregation prone in response to HOCl treatment (Winter et al., 2008), including GapA, EF-Tu and ClpX (see Fig S4 & Table S2, highlighted in blue). A few of these proteins were also identified among the aggregation-sensitive proteins in HOSCN-treated PA14. However, the H/L ratio was typically much lower. In addition, we found several proteins that appeared to be substantially more prone to HOSCN-mediated aggregation than to HOCl-mediated aggregation, suggesting that differences exist in the target proteins of the two oxidants. More detailed studies are needed in the future to reveal the features that make proteins particularly sensitive towards HOCl versus HOSCN treatment. A complete list of all proteins identified in the aggregates of HOCl and HOSCN-treated P. aeruginosa can be found in Table S2.
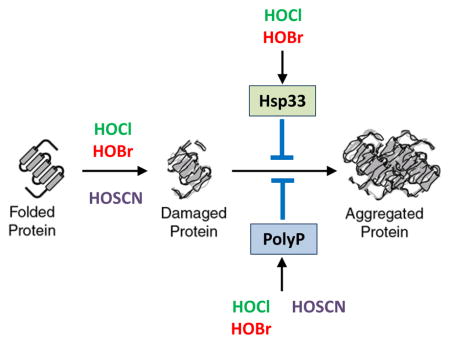
Polyphosphate – A universal oxidative stress defense system
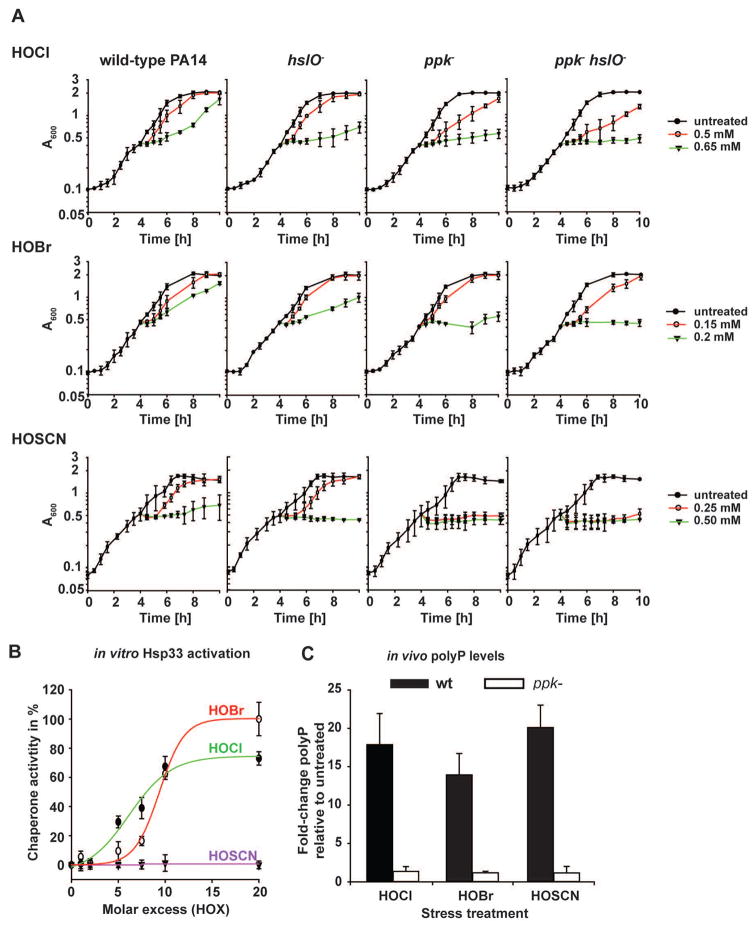
Previous work in E. coli and V. cholerae revealed both protein and non-protein based chaperone strategies that protect bacteria against oxidative stress-mediated protein aggregation (Dahl et al., 2015). These strategies include post-translational activation of the redox-regulated chaperone Hsp33 as well as the conversion of ATP into the chemical chaperone polyphosphate (polyP). Hsp33 is inactive under non-stress conditions but becomes rapidly activated as a molecular chaperone upon oxidative stress conditions that lead to widespread protein unfolding (Winter et al., 2008). Accumulation of polyP, a universal polymer, is at least in part achieved by the oxidant-mediated inactivation of the polyP degrading enzyme exopolyphosphatase (PPX) (Gray et al., 2014). Like activated Hsp33, polyP works as an effective protein–stabilizing chaperone, which binds unfolding proteins and prevents their in vivo aggregation. To test the extent to which Hsp33 and/or polyP participate in protecting P. aeruginosa against the three physiologically relevant oxidants, we constructed PA14 deletion mutants, lacking either the Hsp33 encoding gene hslO, the polyphosphate kinase (PPK) encoding gene ppk, or both hslO and ppk genes. As before, we exposed the cells to HOX-treatment in MOPS-glucose media, and either monitored growth in liquid culture after treatment (Fig. 4A) or their survival (Fig. S5). We found that the hslO deletion reproducibly increased the sensitivity of PA14 towards HOCl and HOBr treatment but not towards HOSCN stress (Fig. 4A; Fig. S5). This result was not completely unexpected given that 0.25 mM HOSCN did not appear to cause significant protein unfolding in PA14 wild-type cells, and hence would be predicted to fail to trigger unfolding of Hsp33. Indeed, in vitro activation studies using recombinantly expressed PA14 Hsp33 revealed that both HOBr and HOCl rapidly activated the chaperone function of PA14 Hsp33 whereas HOSCN treatment had no activating effect (Fig. 4B).
Fig. 4. Chaperones provide a defense mechanism against HOX-mediated killing.
(A) Logarithmically growing PA14 wild-type, ppk−, hslO− or ppk−hslO− cells in MOPS-glucose medium (A600= 0.4–0.5) were incubated with the indicated concentrations of either HOCl, HOBr or HOSCN. Growth was recorded every 30 min for 6 h post treatment. (B) To test the effects of HOX on the in vitro activation of PA14-Hsp33, 12 μM chemically denatured citrate synthase was diluted 160-fold into 40 mM HEPES, pH 7.5 at 30°C in the presence and absence of PA14 Hsp33 that was oxidized with the indicated molar excess of HOX. Light scattering was monitored with excitation and emission wavelength set to 360 nm. The light scattering signal of citrate synthase incubated in the presence of 20-fold molar excess of HOBr-activated PA14-Hsp33 was set to 100% activity while the light scattering signal of CS in the absence of any Hsp33 was set to 0% chaperone activity. (C) Exponentially growing PA14 wild-type and ppk− cells (A600= 0.4–0.5) were incubated with sublethal concentrations of either HOCl (0.5 mM), HOBr (0.15 mM), or HOSCN (0.25 mM) for 2.5 hours. Then, polyP was extracted, quantified, and the resulting amount was normalized to the respective protein amount in each sample. Experiments were performed independently at least three times. Data represent means ± s.d.
In contrast to the hslO deletion, we found that the deletion of the ppk gene dramatically increased the sensitivity of PA14 towards all three oxidants, HOCl, HOBr and HOSCN. For instance, we observed no recovery upon treatment of the ppk mutant with 0.25 mM HOSCN within the timeframe of the experiment (Fig. 4A), and survival was decreased by at least 2 orders of magnitude compared to wild-type PA14 (Fig. S5). However, whereas additional deletion of hslO did not cause any further increase in the HOSCN sensitivity of PA14, deletion of hslO increased the sensitivity towards HOCl and HOBr, suggesting that polyP and Hsp33 participate in protecting PA14 against HOCl and HOBr stress (Fig. 4A, Fig. S5).
Analysis of the polyP levels in HOCl, HOBr and HOSCN-treated wild-type PA14 confirmed that all three treatments cause an increase in polyP level (Fig. 4C). Since the respective polyP levels were quite similar, however, these results also excluded the possibility that polyP’s apparently more substantial influence on HOSCN resistance is due to higher amounts of polyP. Instead, our results suggest that polyP accumulation in wild-type PA14 is either sufficient to deal with HOSCN-mediated protein unfolding, explaining the lack of heat shock gene induction and failed activation of Hsp33, or that polyP prevents HOSCN-mediated damage of other cellular targets, such as DNA. In either case, our results suggested that we discovered a universal stress response system that protects PA14 against all three neutrophilic oxidants.
Absence of PolyP Leads to Massive Protein Aggregation in HOSCN-treated bacteria
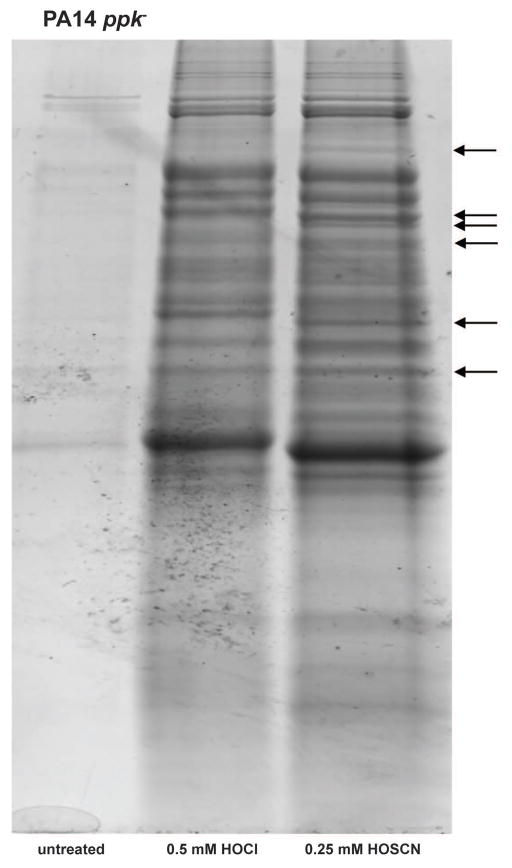
To gain insights into the mechanism(s) by which polyP protects PA14 against neutrophil-derived oxidants, we compared protein aggregation in the ppk deletion strain in response to HOCl and HOSCN. We reasoned that if polyP protected PA14 against HOSCN-mediated protein damage, absence of polyP would cause a dramatic increase in protein aggregation. In contrast, no change in the aggregation pattern would be expected if polyP’s effects were independent of protein unfolding. As before, we treated the ppk deletion strain with 0.25 mM HOSCN for 30-min. While this concentration did not cause any detectable protein aggregation in the wild-type strain (Fig. 3A–B, Fig. S3), in the ppk deletion strain it led to protein aggregation that was fully comparable to the massive protein aggregation triggered by a treatment with 0.5 mM HOCl (Fig. 5). In support of these results, we found that the ppk deletion strain now responds to HOSCN – treatment with the upregulation of the heat shock response, indicating that in the absence of polyP, cells try to compensate by increasing chaperone production (Fig. S6). Moreover, and confirming our earlier SILAC results, we again noted that several proteins were differentially affected by HOSCN and HOCl treatment, indicating that the two oxidants have slightly different in vivo targets (Fig. 5, arrows). In summary, these results demonstrate that HOSCN has the capacity to cause widespread protein aggregation and suggest that in wild-type PA14, endogenous polyP levels are sufficient to protect proteins against HOSCN-mediated protein damage.
Fig. 5. HOCl and HOSCN cause widespread protein aggregation in PA14 lacking polyP.
Exponentially growing PA14 ppk− cells in MOPS-glucose medium (A600= 0.4–0.5) were either left untreated or incubated with HOCl (0.5 mM) or HOSCN (0.25 mM) for 30 min. Incubation was stopped by the addition of 10 mM thiosulfate. Cells were lysed and aggregated proteins were separated from the soluble fraction by centrifugation. The insoluble proteins were analyzed by SDS-PAGE. Arrows indicate changes in the aggregation pattern between HOCl- and HOSCN-treated samples. Experiments were performed independently at least three times.
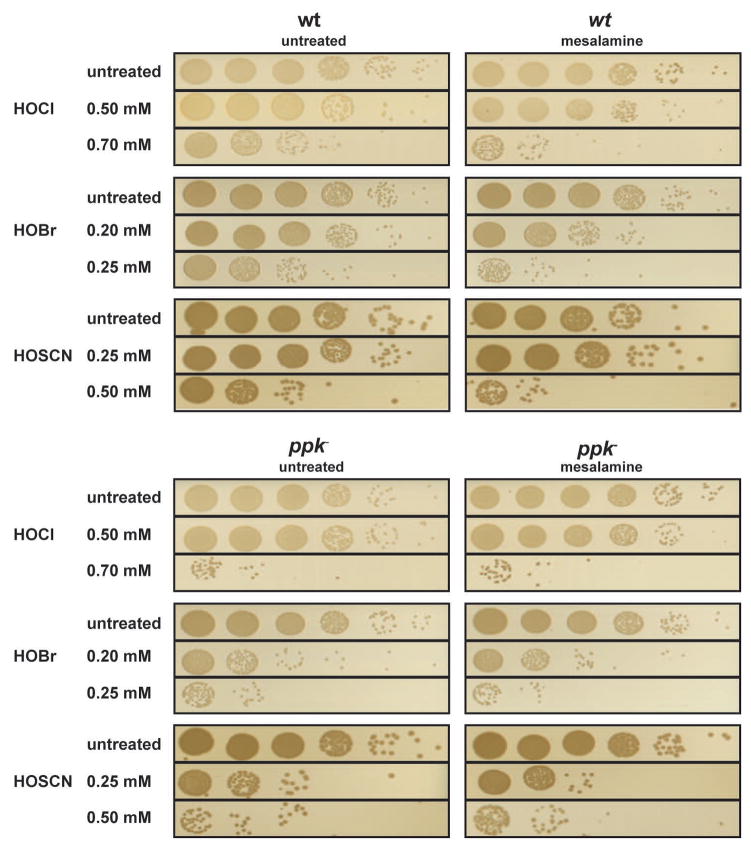
Mesalamine increases PA14 sensitivity towards all three oxidants
Our data revealed that accumulation of polyP serves as an efficient posttranslational stress response system that protects Pseudomonas against the deleterious effects of all three major neutrophilic oxidants. We recently identified mesalamine, the gold standard in treating patients with mild to moderate ulcerative colitis, as a potent inhibitor of bacterial polyP production in a wide range of bacteria, including P. aeruginosa PA14 (Dahl et al., 2017). We now wondered whether mesalamine treatment could be used to sensitize PA14 towards the three physiological oxidants, making it potentially a powerful drug to combat P. aeruginosa infections within chronically inflamed environments. To test this idea, we treated logarithmically growing PA14 wild-type and ppk deletion mutants for 120 min with mesalamine before we exposed the cells to our 30-min HOCl, HOBr or HOSCN stress. As before, we monitored and compared cell survival. As shown in Figure 6, mesalamine-treated wild-type PA14 was almost as sensitive towards all three oxidants as the ppk deletion strain. Consistent with our previous findings (Dahl et al., 2017), mesalamine did not alter the sensitivity of the ppk deletion strain. These results demonstrate that a decrease in polyphosphate levels sensitizes P. aeruginosa towards HOCl, HOBr and HOSCN stress, and suggest that mesalamine might be an appropriate option in the treatment of PA14 infections.
Fig. 6. Mesalamine-treatment increases HOX sensitivity of wild-type PA14 by targeting polyP homeostasis.
Exponentially growing PA14 wild-type and ppk− cells in MOPS-glucose medium (A600= 0.4–0.5) were either left untreated or pretreated with 0.5 mM mesalamine for 120 min. Then, treatment with the indicated concentrations of HOCl, HOBr or HOSCN was conducted as before. After 30 min of incubation, excess HOX was quenched by the addition of 10 mM thiosulfate. Cells were serially diluted with 0.85% NaCl, spot-titered onto LB agar plates, and incubated overnight at 37°C. The experiments were repeated at least three independent times.
DISCUSSION
The accumulation of reactive oxygen and chlorine species (RO/CS) is highly toxic, causing membrane alterations, protein unfolding and DNA damage. It is therefore not surprising that organisms developed systems that utilize increased RO/CS production and secretion as a strategy to defend themselves against their enemies. One example for such a system comes in the form of the mammalian innate immune defense, where neutrophils produce and release high levels of HOCl, HOBr, and HOSCN to kill invading pathogens and control bacterial colonization (Davies et al., 2008). To investigate what mechanisms bacteria use for their defense, we compared the effects and consequences of HOCl, HOBr and HOSCN treatment on P. aeruginosa, a microorganism known to persist within chronically inflamed environments that are enriched for neutrophil-derived oxidants (Rada, 2017). We reasoned that by identifying and subsequently targeting such bacteria-specific defense mechanisms, we would have the potential to increase the efficacy of the host to kill colonizing bacteria. This might potentially provide a novel approach to combat chronic P. aeruginosa infections.
We determined the bacteriotoxic concentrations for HOCl, HOBr and HOSCN, and compared their bactericidal effects on both actively growing and dormant cells. We found that HOCl and HOBr are most toxic for non-growing bacteria, cultivated in either PBS or MOPS buffer. These results are consistent with the high reactivity of these oxidants, and suggest that some of the organic compounds that are produced and secreted by actively metabolizing bacteria likely react with and partially detoxify the oxidants. In contrast, HOSCN was significantly more bactericidal for actively growing PA14 than for non-metabolizing cells, suggesting that HOSCN treatment affects cellular systems involved in metabolism and/or growth. Previous studies conducted in bacteria and mammalian cell cultures suggested several mechanisms by which HOSCN might affect metabolism (Love et al., 2016). For instance, redox proteomic studies revealed active site cysteines in glycolytic enzymes as one of the major in vivo oxidation targets of HOSCN, together with proteins involved in redox signaling, structure maintenance, and protein folding (Love et al., 2016). Moreover, HOSCN has been shown to inhibit glucose and amino acid uptake in a broad spectrum of bacteria, most likely by targeting membrane-bound transport proteins for these nutrients (Chandler & Day, 2015). These results are consistent with our RNAseq data, which revealed significant HOSCN-specific upregulation of genes encoding for membrane proteins and transporters. They furthermore revealed that HOCl/HOBr and HOSCN exert distinctly different effects on bacteria and, when produced in combination, as is the case in healthy individuals, should be effective in targeting both actively growing and dormant bacteria. Analysis of the defense strategies in P. aeruginosa revealed at least one cellular system, which significantly promotes bacterial resistance towards all three oxidants. We found that in response to HOCl/HOBr and HOSCN, PA14 produces high levels of polyphosphate, which confers considerable resistance against all three oxidants. PolyP is a universally conserved, energy-rich polymer, which was recently shown to bind unfolded proteins and prevent their non-specific aggregation, suggesting it has chaperone-like activities (Gray et al., 2014). At first, this result was surprising since HOSCN treatment, in contrast to HOCl, did not cause any detectable protein aggregation or upregulation of the heat shock response in wild-type PA14, suggesting that HOSCN does not affect protein homeostasis. However, when we conducted these experiments in strains lacking PPK, the kinase responsible for polyP synthesis, we fou nd that HOSCN treatment triggered massive amounts of protein aggregation, and led to increased expression of heat-shock regulated genes. Since wild-type PA14 produced approximately the same levels of polyP in response to HOCl/HOBr and HOSCN treatment, we concluded that polyP production is more effective in protecting PA14 against HOSCN-mediated protein unfolding as compared to HOCl-mediated unfolding. These results make sense when one considers the reactivity of HOCl/HOBr and HOSCN. HOCl/HOBr are highly reactive towards a number of different amino acid side chains, whose oxidation rapidly cause protein unfolding both in vitro and in vivo (Winter et al., 2008). In contrast, HOSCN, which is highly thiol specific (Skaff et al., 2009), will presumably react first with surface-exposed cysteine residues, whose oxidation is unlikely to cause rapid protein unfolding. Cellular levels of polyP increase in both circumstances since polyP accumulation is in part mediated by the oxidative modification of a surface-exposed cysteine in the exopolyphosphatase PPX (Gray et al., 2014). It is therefore possible that polyP production in response to HOCl treatment is simply not fast enough to compete with the sudden onset of protein unfolding, a conclusion that would also explain why cells require the extremely fast activation of the redox-regulated chaperone Hsp33 for additional protection. HOSCN might either work much slower, and hence give polyP the time to accumulate and stabilize unfolding proteins, or might affect only a few crucially important redox-sensitive proteins, such as cysteine containing chaperones (Winter et al., 2005), whose inactivation would trigger more widespread protein aggregation. Either of these scenarios is consistent with our finding that HOCl and HOSCN target a set of only partially overlapping cellular proteins for aggregation.
In summary, our results suggest that we have identified a protection mechanism that would become crucial for PA14 when present in an environment of chronic inflammation, such as it is found in the airways of patients suffering from cystic fibrosis. Successful clearance of chronic P. aeruginosa infection is often hampered by the high antibiotic resistance of P. aeruginosa (Lister et al., 2009). We have now discovered that mesalamine, which decreases endogenous polyP levels in PA14, substantially increases the sensitivity of PA14 to neutrophilic oxidants. The fact that PA14 does not require polyP for growth under non-stress conditions likely reduces their chance to develop resistance. Our studies therefore serve as a first step towards alternative treatment strategies aimed to improve the ability of the endogenous host defense systems to kill stress-resistant microbes.
EXPERIMENTAL PROCEDURES
Bacterial strains, plasmids and primers
All strains, plasmids and primers used in this study are listed in Table S3. A clean deletion of hslO, ppK or hslO/ppK in P. aeruginosa strain PA14 was generated as previously described (Hmelo et al., 2015). In brief, a 500 bp upstream and a 500 bp downstream region of the PA14 hslO and ppk gene were amplified by PCR, using the respective primer pairs (Table S3). Sowing overlap extension PCR was used to amplify the constructs hslOupdwSOE and ppkupdwSOE. The plasmids pEX18GmhslOupdwSOE and pEX18GmppkupdwSOE were generated by integrating the constructs into the multicloning-site (MCS) of the allelic exchange vector pEX18Gm using BamHI and HindIII restriction sites (Table S3). Plasmids were verified by PCR using MCS specific primers M13F-21 and M13R. For subsequent puddle mating with the respective PA14 strain, the plasmid constructs were transformed into the donor strain E. coli S17.1 (λ pir+) using standard chemical transformation. Transformed E. coli cells were plated onto LB agar plates supplemented with 10 μg/ml gentamicin and grown overnight at 37°C. The same day, an overnight culture of the recipient strain PA14 was prepared. The following day, an equal volume of fresh LB was added to the PA14 overnight culture, and the PA14 strain was incubated at 42°C for 3 h and until the E. coli donor strain had reached A600 = 0.5–0.6. 1.5 ml of the E. coli donor strain and 0.5 ml of the PA14 recipient strain were mixed and centrifuged for 5 min at 10,000g at room temperature. Cells were pelleted, resuspended in 50 μl LB medium and transferred onto an LB plate and overnight incubation at 30°C. The bacterial lawn was scraped from the plate and resuspended in 1 ml of sterile PBS. Then 10 μl, 100 μl and 500 μl aliquots of the resuspension were plated onto Vogel-Bonner Minimal Medium plates supplemented with 60 μg/ml gentamicin and the plates were incubated for 48 h at 37°C. For the counter-selection, an isolated colony from the VBMM plate was picked, streaked onto no-salt LB-plates supplemented with 15% (w/v) sucrose, and incubated for 36–48 h at 30°C. Colonies were PCR verified to confirm the deletion of the PA14 hslO and ppk genes using the respective sequencing primers (Table S3).
Preparation of HOCl, HOBr and HOSCN stock solutions
HOCl was prepared by diluting concentrated NaOCl into 40 mM potassium phosphate buffer (KPi), pH 7.5, and the concentration was determined in 10 mM NaOH using ε292 nm 350 M−1 cm−1 (Morris, 1966). HOBr was generated by mixing equal volumes of 40 mM HOCl and 45 mM KBr, and the concentration was determined in 10 mM NaOH using ε329 nm 332 M−1 cm−1 (Hawkins et al., 2009). HOSCN was generated enzymatically as described previously with modifications (5). In brief, 26 mM NaSCN and 200 units ml−1 lactoperoxidase (Sigma) were added to 2 mM aliquots of H2O2 in 40 mM KPi buffer, pH 7.5 over 5 min, followed by the addition of 500 units ml−1 catalase. The solution was centrifuged at 14,000 g for 7.5 min at 4°C using a 10-kDa cutoff filter to remove proteins. The concentration of HOSCN was determined by monitoring the loss of signal at 412 nm (ε412 nm 14,150 M−1 cm−1), which occurs when HOSCN interacts with 5-thio-2-nitrobenzoic acid (Chandler et al., 2013).
Growth conditions and bacterial survival assays
Unless specified otherwise, all strains were grown aerobically at 37°C in lysogenic broth (LB) or potassium morpholinopropanesulfonate (MOPS) minimal medium (Teknova, Inc.) containing 0.2% glucose, 1.32 mM potassium phosphate and 10 μM thiamine. For oxidative stress treatment, PA14 wild-type and the respective mutant strains were grown until A600 = 0.4–0.5 was reached. Then, the cells were washed with and resuspended in pre-warmed (i) phosphate buffer saline (PBS), pH 7.5, (ii) MOPS-glucose media, or (iii) MOPS media, and treated with the indicated concentrations of HOCl, HOBr, and HOSCN at either 30°C (PBS) or 37°C (MOPS-glucose; MOPS), respectively. Growth was recorded every 30 to 60 min for 10 h by measuring the optical density at 600 nm. To test for cell survival, 0.5 ml of bacteria were harvested by centrifugation at the indicated time points after normalizing them to their A600, washed with MOPS medium containing 10 mM Na2S2O3 to quench excess HOX, diluted into 0.85% NaCl and spot-titered onto LB agar using a Precision XS Microplate Sample Processor (Bio-Tek). The LB plates were incubated overnight at 37°C. To test the effects of mesalamine (Sigma Aldrich) treatment on the oxidative stress resistance of PA14 wild-type and ppk deletion strain, 500 μM mesalamine dissolved in MOPS media was added to the cultures 120 min before the stress treatment. Stress treatment with the respective oxidants and survival analysis was conducted as described above.
RNASeq library construction and sequence analysis
Wild-type P. aeruginosa PA14 was cultivated in MOPS-glucose medium at 37°C until OD600 of 0.4–0.5 was reached. Then, cells were treated with either 0.5 mM HOCl, 0.15 mM HOBr, or 0.25 mM HOSCN. Cells equivalent to 1 ml with an A600 = 0.4 were harvested either before (control) or 15 min after the treatment onto 1 ml of ice-cold methanol (−80°C) to stop transcription. After centrifugation and removal of the supernatant, total RNA was prepared from three biological replicates of untreated and HOX-treated PA14 using the Ambion RiboPure-Bacteria Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The samples were DNase I treated, followed by depletion of rRNA using the Illumina Ribo Zero Kit (Illumina) for Gram-negative bacteria. 50 base single end sequencing was performed on an Illumina HiSeq 2500 by the University of Michigan DNA Sequencing Core. Reads were aligned to the P. aeruginosa UCBPP-PA14 reference genome (Accession number: NC_008463.1) using bwa v. 0.7.8-r455 (Li & Durbin, 2009). Alignment files were generated using samtools (Li et al., 2009). Subsequent analyses were performed using the statistical software package R. The R packages edgeR and limma were used for read count normalization and determining differential expression, respectively (Robinson et al., 2010, Ritchie et al., 2015). The false discovery rate (fdr) was calculated via the Benjamini-Hochberg procedure (Benjamini & Hochberg, 1995). The threshold chosen for calling a gene differentially expressed was fdr ≤ 0.005 and an absolute value of log2(fold change) ≥ 1.5. Enrichment of differentially expressed genes in each of 78 regulons was tested using one-sided Fisher’s Exact Tests. P-values were adjusted for false discovery rate using the method of Benjamini and Hochberg (Benjamini & Hochberg, 1995). Information on P. aeruginosa regulons was taken from (Galan-Vasquez et al., 2011), with additional information on the OxyR regulon supplemented from a recently published study (Wei et al., 2012). Regulons determined to be significantly enriched for differentially expressed genes (FDR < 0.05) were presented as heatmaps using the heatmap function from the R package NMF. Log2(Fold change) values of each treatment group compared to the untreated control group were used for clustering genes by their Euclidean distance with complete linkage.
In vivo aggregation assays
Preparation of cellular insoluble protein fractions was conducted according to (Tomoyasu et al., 2001) with slight modifications. In brief, wild-type P. aeruginosa PA14 and mutant strains were grown in MOPS-glucose medium at 37°C to an OD600 ~0.4–0.5 and treated with the indicated concentrations of HOCl, HOBr and HOSCN for 30 min. Cells equivalent to 4 ml of OD600 = 1 were harvested by centrifugation and resuspended in 40 μl Buffer A (10 mM KPi, pH 6.5, 1 mM EDTA, 20% [w/v] sucrose, 1 mg/ml lysozyme, 50 U/ml benzonase), followed by a 30 min incubation on ice. The samples were then frozen at −80°C. After thawing the samples on ice and addition of 160 μl Buffer B (10 mM KPi, pH 6.5, 1 mM EDTA), ~100 μl 0.5 mm glass beads (BioSpec Products) were added, and the microfuge tubes were shaken for 30 min at 1,400 rpm, 8°C, to lyse the cells completely. The samples were centrifuged for 20 min at 16,100 × g and 4°C to separate protein aggregates from the soluble fraction. The pellets were washed once with Buffer B, subsequently with Buffer C (Buffer B plus 2% Nonidet P-40 [ICN Biomedicals]) to dissolve membrane proteins, and finally with Buffer B. The insoluble fractions were resuspended in SDS-Laemmli buffer and visualized by SDS-PAGE.
Analysis of in vivo protein aggregation upon HOX treatment and identification of protein aggregates by SILAC
Wild-type PA14 was cultivated in MOPS-glucose medium supplemented with either 120 μg/ml heavy [13C6]Arg or the isotopically light [12C6]Arg at 37°C until OD600 of 0.4–0.5 was reached. Then, cells were treated with either 0.5 mM HOCl or 0.25 mM HOSCN for 30 min. Cells equivalent to 4 ml of OD600 = 1 were harvested by centrifugation and resuspended in 40 μl Buffer A (10 mM KPi, pH 6.5, 1 mM EDTA, 20% [w/v] sucrose, 1 mg/ml lysozyme, 50 U/ml benzonase), followed by a 30 min incubation on ice. Preparation of cellular insoluble protein fraction was performed as described in the previous section. Aggregated proteins were dissolved in DAB buffer (200 mM Tris/HCl, pH 7.5, 6 M Urea, 10 mM EDTA, 0.5% SDS) and mixed in a 1:1 ratio. Protein mixtures were then processed by SDS-PAGE using a 10% Bis-Tris NuPAGE gel (Invitrogen). Protein bands were cut out and in-gel digestion was processed on each using a robot (ProGest, DigiLab) with the following protocol: (i) washed with 25 mM ammonium bicarbonate followed by acetonitrile; (ii) reduced with 10 mM dithiothreitol at 60 C followed by alkylation with 50 mM iodacetamideat room temperature; (iii) digested with trypsin (Promega) at 37 C for 4 h; and quenched with formic acid, lyophilized and reconstituted in 0.1% trifluoroacetic acid. Samples were analyzed by nano-LC-MS/MS (MS Bioworks). Half of each digested sample was analyzed by nano LC-MS/MS with a Waters NanoAcquity HPLC system interfaced to a ThermoFisher Fusion Lumos. Peptides were loaded on a trapping column and eluted over a 75 μm analytical column at 350nL/min; both columns were packed with Luna C18 resin (Phenomenex). A 2 h gradient was employed. The mass spectrometer was operated in data-dependent mode, with the Orbitrap operating at 60,000 FWHM and 17,500 FWHM for MS and MS/MS respectively. The instrument was run with a 3s cycle for MS and MS/MS.
SILAC data were processed with MaxQuant 1.5.8.3 (Cox and Mann, 2008). Second peptide and Match between runs were enabled otherwise the default parameters were used. The multiplicity was set to 2, with Arg6 configured as the heavy label. Data were searched against UniProt Pseudomonas aeruginosa UCBPP-PA14 (2/14/2017). The MaxQuant output was further processed using Perseus. This involved filtering out contaminant and reverse protein identifications. The heat map in Fig. 3B was generated using the Hierarchical clustering function with Perseus, Row Trees were processed using the default settings.
Protein Expression, purification and in vitro oxidation of PA14 Hsp33
The hslO gene from genomic P. aeruginosa PA14 was cloned into a pET11a vector using BamHI and HindIII restriction sites. The plasmid was transformed into the E. coli expression strain BL21 (DE3) hslO::kan. PA14 Hsp33 was expressed and purified as described for E. coli Hsp33 (Graumann et al., 2001). Reduced, chaperone-inactive PA14 Hsp33 (PA14-Hsp33red) and HOX-treated PA14-Hsp33 (PA14 Hsp33HOX) were prepared as previously described for E. coli Hsp33 using the indicated concentrations of HOCl, HOBr or HOSCN (Winter et al., 2008).
Chaperone Activity Assays
Chaperone activity measurements with chemically denatured citrate synthase from porcine heart (CS, Sigma-Aldrich) were conducted as previously described (Ilbert et al., 2007). In short, CS was denatured to a final concentration of 12 μM in 6.0 M Gdn*HCl, 40 mM HEPES, pH 7.5 overnight at room temperature. To initiate aggregation of CS, the unfolded enzyme was diluted 1:160 into 40 mM HEPES, pH 7.5 at 30°C in the absence or presence of various concentrations of reduced or oxidized Hsp33. Light scattering was monitored using a Hitachi F4500 fluorimeter equipped with a thermostated cell holder and stirrer. Excitation and emission wavelengths were set to 360 nm, and the excitation and emission slit widths were set to 2.5 nm.
Extraction and quantification of polyphosphate (polyP)
Changes in intracellular polyP levels upon treatment with HOCl, HOBr, or HOSCN were quantified as previously described (Dahl et al., 2017) with minor modifications. PA14 wild-type and PA14 ppk strains were grown aerobically in 50 ml MOPS glucose medium and split into 10 ml cultures in new flasks when the culture reached an optical density at 600nm of 0.4 – 0.5. Then, 0.5 mM HOCl, 0.15 mM HOBr or 0.25mM HOSCN were added and cells sufficient to yield 200 μg total cellular protein were harvested by centrifugation after incubation for 2.5 h and lysed by incubation for 10 min at 95°C in 0.25 ml GITC Lysis Buffer (4 M guanidine isothiocyanate, 50 mM Tris-HCl, pH 7.0). Protein content of each sample was determined using the Bradford assay (Bio-Rad). PolyP was extracted by sequential addition of 15 μl 10% sodium dodecyl sulfate, 0.5 ml 95% ethanol, and 5 μl glassmilk (0.1 g/ml acid-washed silicon dioxide in GITC Lysis Buffer). This mixture was applied (1 min @ 3,000 × g) to silica membrane spin columns (Econospin™, Epoch Life Science, Inc.) and rinsed twice with 0.75 ml NW Buffer (5 mM Tris-HCl, pH 7.5, 50 mM NaCl, 5 mM EDTA, 50% v/v ethanol). After centrifuging once more to dry the membrane, polyP was eluted by adding 50 μl 50 mM Tris-HCl, pH 8, incubating for 15 min at room temperature, and centrifuging to collect eluate. PolyP extracts were incubated for 1 hour at 37°C with 250 μM highly-pure ADP (Cell Technology), 50 mM HEPES (pH 7.5), 50 mM ammonium sulfate, 5 mM MgCl2 in the presence or absence of 50 nM E. coli PPK. The resulting ATP was quantified with QuantiLum® Recombinant Luciferase (Promega). The reactions contained 50 mM Tricine buffer, pH 7.8, 10 mM MgSO4, 0.2 mM EDTA, 0.2 mM sodium azide, 1 mM DTT, 100 μM luciferin, 25 nM luciferase, and measurements were conducted in a FLUOstar Omega microplate reader (BMG Labtech). Values obtained from samples incubated in the absence of PPK were subtracted from the values of samples incubated in the presence of PPK and normalized to the values obtained for the untreated samples of each strain in order to calculate the fold-change in polyP accumulation in each sample.
Quantitative real-time PCR
Gene expression analysis by RT-PCR was performed upon HOSCN treatment of PA14 wild-type and PA14 ppk strains. Briefly, cells were grown aerobically to mid-log phase in MOPS glucose at 37°C. HOSCN was added to a final concentration of 0.25 mM and incubated for 20 min. RNA was prepared using the NucleoSpin RNA kit (Macherey&Nagel) and DNA-free™ kit (Ambion). PrimeScript 1st strand cDNA Synthesis Kit (Takara) was used to generate cDNA, and RT-PCRs were set up with SYBR(R) GreenER™ qRT-PCR mix (Invitrogen) and a Mastercycler(R) realplex2 real-time PCR system (Eppendorf). Expression ratios were calculated compared with expression of each gene in untreated PA14 and PA14 ppk cultures, respectively, by the ΔΔCT method (Pfaffl, 2001) and normalized to expression of rrsD (encoding 16S rRNA), expression of which did not change under the conditions tested. Primers used for RT-PCR analysis are listed in Table S3.
Supplementary Material
Acknowledgments
FUNDING INFORMATION
This work was funded by the National Institute of Health grants GM065318 and GM116582 to U.J.; J.-U. D. was supported by a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft (DA 1697/1-1).
We thank Dao Ngyuen (McGill University) for providing us with protocols, plasmids and strains to construct the PA14 mutant strains. Wild-type PA14 was kindly provided by B. Boles. We are grateful to M. Gray for helpful discussions. We thank the University of Michigan DNA Sequencing Core for RNA sample processing and sequencing. The authors declare no conflict of interest.
Footnotes
AUTHOR CONTRIBUTIONS
B.G. and J.-U.D. designed and carried out experiments. J.S. evaluated the RNAseq analysis. B.G., J.-U.D. and U.J. conceived the study, interpreted the results, and wrote the paper.
References
- Arsene F, Tomoyasu T, Bukau B. The heat shock response of Escherichia coli. Int J Food Microbiol. 2000;55:3–9. doi: 10.1016/s0168-1605(00)00206-3. [DOI] [PubMed] [Google Scholar]
- Ashby MT. Inorganic chemistry of defensive peroxidases in the human oral cavity. J Dent Res. 2008;87:900–914. doi: 10.1177/154405910808701003. [DOI] [PubMed] [Google Scholar]
- Atichartpongkul S, Vattanaviboon P, Wisitkamol R, Jaroensuk J, Mongkolsuk S, Fuangthong M. Regulation of Organic Hydroperoxide Stress Response by Two OhrR Homologs in Pseudomonas aeruginosa. PLoS One. 2016;11:e0161982. doi: 10.1371/journal.pone.0161982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett TJ, Hawkins CL. Hypothiocyanous acid: benign or deadly? Chem Res Toxicol. 2012;25:263–273. doi: 10.1021/tx200219s. [DOI] [PubMed] [Google Scholar]
- Barrett TJ, Pattison DI, Leonard SE, Carroll KS, Davies MJ, Hawkins CL. Inactivation of thiol-dependent enzymes by hypothiocyanous acid: role of sulfenyl thiocyanate and sulfenic acid intermediates. Free Radic Biol Med. 2012;52:1075–1085. doi: 10.1016/j.freeradbiomed.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- Ceragioli M, Mols M, Moezelaar R, Ghelardi E, Senesi S, Abee T. Comparative transcriptomic and phenotypic analysis of the responses of Bacillus cereus to various disinfectant treatments. Appl Environ Microbiol. 2010;76:3352–3360. doi: 10.1128/AEM.03003-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JD, Day BJ. Biochemical mechanisms and therapeutic potential of pseudohalide thiocyanate in human health. Free Radic Res. 2015;49:695–710. doi: 10.3109/10715762.2014.1003372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JD, Nichols DP, Nick JA, Hondal RJ, Day BJ. Selective metabolism of hypothiocyanous acid by mammalian thioredoxin reductase promotes lung innate immunity and antioxidant defense. J Biol Chem. 2013;288:18421–18428. doi: 10.1074/jbc.M113.468090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi BK, Gronau K, Mader U, Hessling B, Becher D, Antelmann H. S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics. Mol Cell Proteomics. 2011;10:M111009506. doi: 10.1074/mcp.M111.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen TS, Prince A. Cystic fibrosis: a mucosal immunodeficiency syndrome. Nat Med. 2012;18:509–519. doi: 10.1038/nm.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Dahl JU, Gray MJ, Bazopoulou D, Beaufay F, Lempart J, Koenigsknecht MJ, Wang Y, Baker JR, Hasler WL, Young VB, Sun D, Jakob U. The anti-inflammatory drug mesalamine targets bacterial polyphosphate accumulation. Nat Microbiol. 2017;2:16267. doi: 10.1038/nmicrobiol.2016.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JU, Gray MJ, Jakob U. Protein quality control under oxidative stress conditions. J Mol Biol. 2015;427:1549–1563. doi: 10.1016/j.jmb.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D, De PK, Banerjee RK. Thiocyanate, a plausible physiological electron donor of gastric peroxidase. Biochem J. 1995;305(Pt 1):59–64. doi: 10.1042/bj3050059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MJ. Myeloperoxidase-derived oxidation: mechanisms of biological damage and its prevention. J Clin Biochem Nutr. 2011;48:8–19. doi: 10.3164/jcbn.11-006FR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MJ, Hawkins CL, Pattison DI, Rees MD. Mammalian heme peroxidases: from molecular mechanisms to health implications. Antioxid Redox Signal. 2008;10:1199–1234. doi: 10.1089/ars.2007.1927. [DOI] [PubMed] [Google Scholar]
- Dukan S, Touati D. Hypochlorous acid stress in Escherichia coli: resistance, DNA damage, and comparison with hydrogen peroxide stress. J Bacteriol. 1996;178:6145–6150. doi: 10.1128/jb.178.21.6145-6150.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fargier E, Mac Aogain M, Mooij MJ, Woods DF, Morrissey JP, Dobson AD, Adams F, O’Gara C. MexT functions as a redox-responsive regulator modulating disulfide stress resistance in Pseudomonas aeruginosa. J Bacteriol. 2012;194:3502–3511. doi: 10.1128/JB.06632-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan-Vasquez E, Luna B, Martinez-Antonio A. The Regulatory Network of Pseudomonas aeruginosa. Microb Inform Exp. 2011;1:3. doi: 10.1186/2042-5783-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebendorfer KM, Drazic A, Le Y, Gundlach J, Bepperling A, Kastenmuller A, Ganzinger KA, Braun N, Franzmann TM, Winter J. Identification of a hypochlorite-specific transcription factor from Escherichia coli. J Biol Chem. 2012;287:6892–6903. doi: 10.1074/jbc.M111.287219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann J, Lilie H, Tang X, Tucker KA, Hoffmann JH, Vijayalakshmi J, Saper M, Bardwell JC, Jakob U. Activation of the redox-regulated molecular chaperone Hsp33--a two-step mechanism. Structure. 2001;9:377–387. doi: 10.1016/s0969-2126(01)00599-8. [DOI] [PubMed] [Google Scholar]
- Gray MJ, Jakob U. Oxidative stress protection by polyphosphate--new roles for an old player. Curr Opin Microbiol. 2015;24:1–6. doi: 10.1016/j.mib.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MJ, Wholey WY, Parker BW, Kim M, Jakob U. NemR is a bleach-sensing transcription factor. J Biol Chem. 2013;288:13789–13798. doi: 10.1074/jbc.M113.454421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MJ, Wholey WY, Wagner NO, Cremers CM, Mueller-Schickert A, Hock NT, Krieger AG, Smith EM, Bender RA, Bardwell JC, Jakob U. Polyphosphate is a primordial chaperone. Mol Cell. 2014;53:689–699. doi: 10.1016/j.molcel.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins CL, Morgan PE, Davies MJ. Quantification of protein modification by oxidants. Free Radic Biol Med. 2009;46:965–988. doi: 10.1016/j.freeradbiomed.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Hmelo LR, Borlee BR, Almblad H, Love ME, Randall TE, Tseng BS, Lin C, Irie Y, Storek KM, Yang JJ, Siehnel RJ, Howell PL, Singh PK, Tolker-Nielsen T, Parsek MR, Schweizer HP, Harrison JJ. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat Protoc. 2015;10:1820–1841. doi: 10.1038/nprot.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilbert M, Horst J, Ahrens S, Winter J, Graf PC, Lilie H, Jakob U. The redox-switch domain of Hsp33 functions as dual stress sensor. Nat Struct Mol Biol. 2007;14:556–563. doi: 10.1038/nsmb1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebanoff SJ. Myeloperoxidase: friend and foe. J Leukoc Biol. 2005;77:598–625. doi: 10.1189/jlb.1204697. [DOI] [PubMed] [Google Scholar]
- Klebanoff SJ, Kettle AJ, Rosen H, Winterbourn CC, Nauseef WM. Myeloperoxidase: a front-line defender against phagocytosed microorganisms. J Leukoc Biol. 2013;93:185–198. doi: 10.1189/jlb.0712349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogan I, Ramjeesingh M, Li C, Kidd JF, Wang Y, Leslie EM, Cole SP, Bear CE. CFTR directly mediates nucleotide-regulated glutathione flux. EMBO J. 2003;22:1981–1989. doi: 10.1093/emboj/cdg194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DG, Urbach JM, Wu G, Liberati NT, Feinbaum RL, Miyata S, Diggins LT, He J, Saucier M, Deziel E, Friedman L, Li L, Grills G, Montgomery K, Kucherlapati R, Rahme LG, Ausubel FM. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 2006;7:R90. doi: 10.1186/gb-2006-7-10-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev. 2009;22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentzen D, Durairaj L, Pezzulo AA, Nakano Y, Launspach J, Stoltz DA, Zamba G, McCray PB, Jr, Zabner J, Welsh MJ, Nauseef WM, Banfi B. Concentration of the antibacterial precursor thiocyanate in cystic fibrosis airway secretions. Free Radic Biol Med. 2011;50:1144–1150. doi: 10.1016/j.freeradbiomed.2011.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love DT, Barrett TJ, White MY, Cordwell SJ, Davies MJ, Hawkins CL. Cellular targets of the myeloperoxidase-derived oxidant hypothiocyanous acid (HOSCN) and its role in the inhibition of glycolysis in macrophages. Free Radic Biol Med. 2016;94:88–98. doi: 10.1016/j.freeradbiomed.2016.02.016. [DOI] [PubMed] [Google Scholar]
- Morris JC. The Acid Ionization Constant of HOCl from 5 to 35°. The Journal of Physical Chemistry. 1966;70:3798–3805. [Google Scholar]
- Muller A, Langklotz S, Lupilova N, Kuhlmann K, Bandow JE, Leichert LI. Activation of RidA chaperone function by N-chlorination. Nat Commun. 2014;5:5804. doi: 10.1038/ncomms6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker BW, Schwessinger EA, Jakob U, Gray MJ. The RclR protein is a reactive chlorine-specific transcription factor in Escherichia coli. J Biol Chem. 2013;288:32574–32584. doi: 10.1074/jbc.M113.503516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattison DI, Davies MJ, Hawkins CL. Reactions and reactivity of myeloperoxidase-derived oxidants: differential biological effects of hypochlorous and hypothiocyanous acids. Free Radic Res. 2012;46:975–995. doi: 10.3109/10715762.2012.667566. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada B. Interactions between Neutrophils and Pseudomonas aeruginosa in Cystic Fibrosis. Pathogens. 2017:6. doi: 10.3390/pathogens6010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Grigg J. New insights into pulmonary inflammation in cystic fibrosis. Arch Dis Child. 2006;91:786–788. doi: 10.1136/adc.2004.069419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaff O, Pattison DI, Davies MJ. Hypothiocyanous acid reactivity with low-molecular-mass and protein thiols: absolute rate constants and assessment of biological relevance. Biochem J. 2009;422:111–117. doi: 10.1042/BJ20090276. [DOI] [PubMed] [Google Scholar]
- Skaff O, Pattison DI, Morgan PE, Bachana R, Jain VK, Priyadarsini KI, Davies MJ. Selenium-containing amino acids are targets for myeloperoxidase-derived hypothiocyanous acid: determination of absolute rate constants and implications for biological damage. Biochem J. 2012;441:305–316. doi: 10.1042/BJ20101762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small DA, Chang W, Toghrol F, Bentley WE. Toxicogenomic analysis of sodium hypochlorite antimicrobial mechanisms in Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2007;74:176–185. doi: 10.1007/s00253-006-0644-7. [DOI] [PubMed] [Google Scholar]
- Tomoyasu T, Mogk A, Langen H, Goloubinoff P, Bukau B. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol Microbiol. 2001;40:397–413. doi: 10.1046/j.1365-2958.2001.02383.x. [DOI] [PubMed] [Google Scholar]
- van Dalen CJ, Whitehouse MW, Winterbourn CC, Kettle AJ. Thiocyanate and chloride as competing substrates for myeloperoxidase. Biochem J. 1997;327(Pt 2):487–492. doi: 10.1042/bj3270487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Slungaard A. Role of eosinophil peroxidase in host defense and disease pathology. Arch Biochem Biophys. 2006;445:256–260. doi: 10.1016/j.abb.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Wei Q, Minh PN, Dotsch A, Hildebrand F, Panmanee W, Elfarash A, Schulz S, Plaisance S, Charlier D, Hassett D, Haussler S, Cornelis P. Global regulation of gene expression by OxyR in an important human opportunistic pathogen. Nucleic Acids Res. 2012;40:4320–4333. doi: 10.1093/nar/gks017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Ilbert M, Graf PC, Ozcelik D, Jakob U. Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell. 2008;135:691–701. doi: 10.1016/j.cell.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Linke K, Jatzek A, Jakob U. Severe oxidative stress causes inactivation of DnaK and activation of the redox-regulated chaperone Hsp33. Mol Cell. 2005;17:381–392. doi: 10.1016/j.molcel.2004.12.027. [DOI] [PubMed] [Google Scholar]
- Winterbourn CC. Reconciling the chemistry and biology of reactive oxygen species. Nat Chem Biol. 2008;4:278–286. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- Winterbourn CC, Kettle AJ, Hampton MB. Reactive Oxygen Species and Neutrophil Function. Annu Rev Biochem. 2016;85:765–792. doi: 10.1146/annurev-biochem-060815-014442. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.