Abstract
Type B coxsackieviruses (CVB) can cause myocarditis and dilated cardiomyopathy (DCM), a potentially-fatal sequela that has been correlated to the persistence of viral RNA. Herein, we demonstrate that cardiac RNA persistence can be established even after an inapparent primary infection. Using an inducible Cre/lox mouse model, we ask: (i) Does persistent CVB3 RNA cause ongoing immune activation? (ii) If T1IFN signaling into cardiomyocytes is ablated after RNA persistence is established, is there any change in the abundance of persistent CVB3 RNA and/or does cytopathic infectious virus re-emerge? (iii) Does this loss of T1IFN responsiveness by cardiomyocytes lead to the recurrence/exacerbation of myocarditis? Our findings suggest that persistent enteroviral RNAs probably do not contribute to ongoing myocardial disease, and are more likely to be the fading remnants of a recent, possibly sub-clinical, primary infection which may have set in motion the process that ultimately ends in DCM.
Keywords: Coxsackievirus, CVB3, myocarditis, cardiomyopathy, persistent RNA
Introduction
Myocarditis is a potentially very serious, and sometimes lethal, disease. Its exact prevalence is uncertain; in routine necropsies, it has been reported as an incidental finding in as many as ~10% of cases (Abelmann, 1966). Although other estimates are lower (e.g., ~1%, Gravanis and Sternby, 1991), even this remains striking, as it indicates that, at any given time, 1 in 100 people may have inflammatory lesions in their heart. This high prevalence of histologically-identifiable myocarditis far exceeds the frequency with which the clinical disease is diagnosed (Blauwet and Cooper, 2010), a discrepancy that is explained by the fact that most individuals with acute myocarditis remain relatively non-symptomatic; consistent with this, acute myocarditis was identified as the cause of death in 39 of 2560 serial autopsies, but it had been clinically suspected in only one of the cases (Carniel et al., 2004). Nevertheless, symptomless myocarditis can cause catastrophic dysfunction of the electrical pathways in the heart, particularly during exertion, and often explains the collapse and death of young and vigorous individuals (Bendig et al., 2001; Eckart et al., 2004; Ward, 1978). Indeed, in individuals <40 years of age, ~12% of sudden deaths have been attributed to myocarditis (Blauwet and Cooper, 2010). Furthermore, the disease can have long-term sequelae, including dilated cardiomyopathy (DCM), in which one or both ventricles dilate and decompensate, with resulting cardiac failure (O’Connell, 1987; Sole and Liu, 1993). The annual incidence of DCM is ~70 cases per million, and its prevalence is ~1/2500; in developed countries, it is the commonest indicator for heart transplantation (Maron et al., 2006; Rakar et al., 1997). Myocarditis has several different causes, non-infectious and infectious; the latter most frequently involves a virus infection, often by type B coxsackieviruses (CVB), and in particular by serotype 3 (CVB3) (Andreoletti et al., 2009; Blauwet and Cooper, 2010; Corsten et al., 2012; Esfandiarei and McManus, 2008; Magnani and Dec, 2006; Mahfoud et al., 2011; Marchant et al., 2008; Sagar et al., 2012). Patients with proven enteroviral myocarditis are at a vastly increased risk of developing DCM, which occurs in 10–20% of those cases, and a large study showed a strong correlation (p < 0.001) between prior coxsackievirus infection & DCM (Riecansky et al., 1989); thus, it is reasonable to argue a cause-and-effect relationship between coxsackievirus and many cases of DCM.
CVB3, which is a member of the picornavirus family and the human enterovirus B genus, has a positive-sense single-stranded RNA genome of ~7,500 bases. The virus infects mice and replicates to high titers, causing diseases – including acute myocarditis – that faithfully recapitulate those observed in humans. CVB RNA can persist in vivo; ~40–66% of heart biopsies from patients with healed myocarditis or DCM were reported positive for CVB RNA signal (Archard et al., 1991; Kuhl et al., 2005). A 1994 study of the hearts of CVB3-infected mice showed that infectious virus was rapidly cleared from all animals – by ~day 16 p.i., it was undetectable using a standard plaque assay – but viral RNA could be identified, in some mice, as late as 34 days p.i. (Rabausch-Starz et al., 1994). The mechanism that underpins this RNA persistence was identified in a landmark study by Chapman and colleagues (Kim et al., 2005), who isolated viral variants from mouse heart and from murine cardiomyocytes in culture, and showed that these variants could infect HeLa cells, but were not cytopathic and, therefore, were undetectable using a standard plaque assay. These non-cytopathic variants were identified in most at 21–28 days following infection with wt virus but, by 53 days p.i., they were found only in ~10% of animals. Following inoculation into mice, these variants could be identified in the heart, where they persisted but – mirroring their behavior in tissue culture – they did not trigger any detectable pathology. Analyses of the variants revealed 5′ terminal deletions (TD) varying from 7 to 49 bases, and transfection of HeLa cells with cloned RNA equivalents led to extremely limited RNA replication, no overt cytopathology, and production of very few infectious virions. That these deletions would impede viral RNA replication is consonant with the fact that the first ~100 bases of the 5′ noncoding region of CVB3 (and other enteroviruses) are folded into a structure that visually resembles a clover leaf, and plays a vital role in the syntheses of both positive and negative strands. Some time ago, our laboratory reported that deletion of as few as 32 bases at the 5′ end of the CVB3 cloverleaf had a profound negative impact on CVB3 replication, but had minimal impact on the molecule’s suitability as a template for translation (Hunziker et al., 2007), and more recent studies have shown that TD virus replication is ~105-fold less robust than that of the wt virus (Jaramillo et al., 2016; Leveque et al., 2017; Smithee et al., 2015). TD CVBs also have been identified in humans. A 39 year old male was hospitalized with fulminant myocarditis, and died 9 days after first experiencing severe symptoms (Oka et al., 2005). Autopsy implicated an enteroviral infection, and subsequent studies revealed the presence of TD variants of CVB2 (Chapman et al., 2008). Similar TDs (this time, of CVB3) were identified by deep sequencing of nucleic acids extracted from cardiac biopsies taken from a woman with a one month history of breathlessness, who was diagnosed as having DCM; ~85% of the 5′ noncoding sequences lacked 48 bases, ~15% lacked 15 bases, and ~1% were genetically-intact (Bouin et al., 2016).
Thus, we now have a reasonable understanding of how enteroviral RNA persists in the heart. However, the immunological and pathological consequences (if any) of this persistence is uncertain, and is the focus of this short communication. During normal enteroviral infection, production of type I interferons (T1IFNs) is induced when viral RNAs activate any one of several different cellular sensors. Might this also occur in response to persistent CVB3 RNA, potentially driving an ongoing innate response that could contribute to chronic myocarditis, and to the development or progression of DCM? Moreover, T1IFNs play a key role in controlling these viruses. Mice deficient in molecules that are involved in T1IFN production, such as MDA5 (Huhn et al., 2010; Wang et al., 2010), MAVS (Wang et al., 2010) TRIF (Abston et al., 2012; Mukherjee et al., 2011; Riad et al., 2011) and TLR3 (Abston et al., 2012; Abston et al., 2013; Negishi et al., 2008) are more susceptible to CVB infection, and if T1IFN signaling is blocked organism-wide, e.g., in T1IFNRKO mice, the animals rapidly succumb to this virus (Wessely et al., 2001). Furthermore, the exogenous administration of T1IFNs to mice can reduce CVB titers in several organs, including the heart (Deonarain et al., 2004; Wang et al., 2007), and recent data indicate that IFNβ may be an effective treatment of enteroviral myocarditis in humans (Kuehl et al., 2012). Since T1IFNs are so important in controlling wt CVB3, it seemed worthwhile to ask if T1IFN also might limit the abundance of persistent RNAs, whose existence appears to rely on ongoing – albeit slow – replication of TD viruses. Herein, we address these issues using a Cre/lox based approach that allows us to establish CVB3 RNA persistence in genetically-intact animals, and only thereafter to interrupt T1IFN signaling into cardiomyocytes. We ask: are there molecular signatures of T1IFN responses in hearts that carry persistent CVB3 RNA? After RNA persistence has taken hold, does removal of T1IFN responsiveness from cardiomyocytes have any effect on ISG expression in these hearts? Moreover, does this temporally-regulated ablation of T1IFN signaling result in increased levels of persistent RNA, or even lead to the re-emergence of cytopathic virus? And what is the effect, if any, on myocardial inflammatory disease?
Materials & Methods
Ethics Statement
All animal experiments were approved by The Scripps Research Institute (TSRI) Institutional Animal Care and Use Committee and were carried out in accordance with the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals.
Mice strains, and Cre recombinase activation by tamoxifen
C57BL/6 mice were purchased from the TSRI rodent breeding colony. Most of the studies reported herein rely on a transgenic mouse line in which tamoxifen administration disrupts, specifically in cardiomyocytes, the expression of the T1IFN receptor (T1IFNR). We have previously described this line (Althof et al., 2014) but, in brief, it was generated by crossing two parental strains. The first encoded a tamoxifen-inducible form of Cre recombinase, MerCreMer, which is expressed under the control of the murine alpha myosin heavy chain (Myh6) promoter, limiting the protein’s expression to cardiomyocytes; consequently, we termed animals that carry this transgene cardiomyocyte-MerCreMer (CMMCM) mice. These mice were bred to the second line, IFNARflox/flox mice (Kamphuis et al., 2006), herein referred to as T1IFNRf/f mice, and backcrossed to develop CMMCM T1IFNRf/f mice. Intra-peritoneal (i.p.) administration of tamoxifen (Tam) to these mice leads to the rapid and near-complete deletion of the floxed sequence specifically in cardiomyocytes, with no deletion being observed in other tissues (Althof et al., 2014). In the present study, deletion was achieved following a single i.p. inoculation of 2mg of Tam. The success of the Tam-initiated deletion was determined using the following primers (both written 5′-3′): (JLWT1IFNR1) AAGCTCCTTGCTGCTATCTG and (JLWT1IFNR3) CACACCAGGCTTCTAATGTC, which together generate an 1144bp PCR band from floxed DNA, and a 309bp band from the deleted allele.
Virus, plaque assay, and quantitative PCR to determine CVB3 RNA load in the heart
The wt CVB3 used in these studies is a plaque purified isolate (designated H3) of the myocarditic Woodruff variant of CVB3. Viral stocks were generated from the plasmid pH 3, which encodes a full-length infectious clone of this virus (Knowlton et al., 1996). Mice were infected i.p. with the indicated quantity of wt CVB3 and, at 2, 6 and 8 days p.i., feces were collected, weighed, disrupted in 0.5 ml DMEM, briefly centrifuged to pellet debris, and the supernatant was used for virus titration. After sacrifice, organs were collected, and divided into segments for subsequent analyses. Plaque assays were performed on sub-confluent HeLa cell monolayers as described (Hunziker et al., 2007), and the virus titers (PFU/g) were calculated for each sample. Tissue destined for RNA analysis was immediately stored in RNAlater stabilization reagent (Ambion/LifeTechnologies). The tissue was weighed, and RNA was isolated using an RNeasy Mini Kit (QIAGEN), following the manufacturer’s instructions. 0.25 μg total RNA from the samples was reverse transcribed using SuperScript III Reverse Transcriptase (Life Technologies, CA, USA) according to the manufacturer’s protocol. For the data shown in Figure 1, two sets of primers were used, one termed “VP4”, the other “3D”. The VP4 primers were: (i) 5′GAGTGAAATCCTGCCGATTG3′, used as the genome-complementary primer for the RT reaction, and as the reverse primer in the subsequent PCR amplification; (ii) 5′AAAGACTGGGGCACATGAGA3′ as the forward PCR primer; and (iii) 5′CTGAATGCTAGCGGCAATTCC3′, as the FAM-TAMSP probe. The 3D primers were: (i) 5′TGGTCCATCTGATTGATTCG3′, used as the genome-complementary primer for the RT reaction, and as the reverse primer in the subsequent PCR amplification; (ii) 5′AGACAAAGGGGAGTGCTTCA3′ as the forward PCR primer; and (iii) 5′TTACTTGGACTAACGTCACATTCCT3′, as the FAM-TAMSP probe. For Figure 3, (i) the CVB3-specific genome complementary primer, used in the RT reaction and the PCR ampification, was 5′GAACGCTTTCTCCTTCAACC3′; (ii) the forward primer was 5′CACACTCCGATCAACAGTCA3′; and the FAM/TAMRA probe was 5′CGTGGCACACCAGCCATGTTT3′.
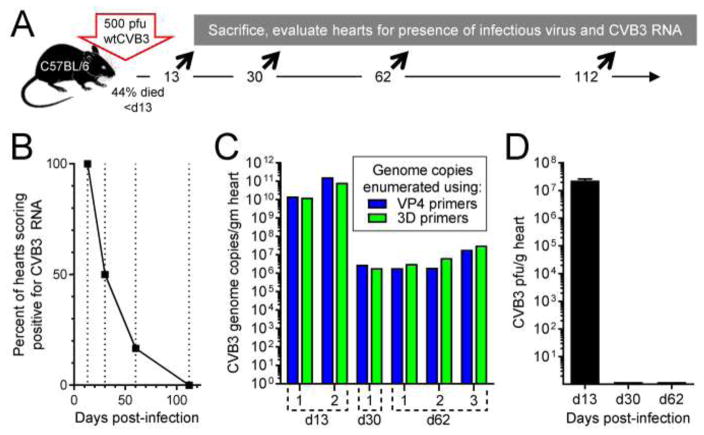
Figure 1. Near full-length CVB3 RNA transiently persists in the hearts of C57BL/6 mice.
As shown in panel A, C57BL/6 mice were infected with 500 pfu of CVB3 i.p, and were sacrificed 13 (two mice), 30 (two mice), 60 (three mice), or 112 (three mice) days later. RNA was extracted from the hearts, and CVB3 RNA levels were quantitated using qPCR. B. At each time point, the percentage of mice that were CVB3 RNA+ is shown. C. To determine if the viral RNA was near full-length at various times p.i., CVB3 genome copy numbers in six individual RNA+ mice were determined using two different primer sets, as described in the text. D. Infectious virus was detected, using a standard plaque assay, only on d13 p.i. The hearts of eleven mice were evaluated for the presence of infectious virus between days 30–112; all eleven scored negative.
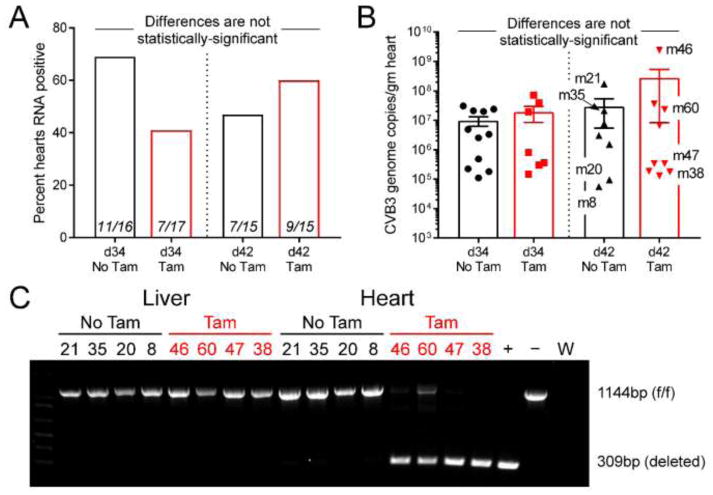
Figure 3. Deletion of the T1IFNR from cardiomyocytes at 30 days p.i. has no significant impact on the prevalence or the quantity of persistent CVB3 RNA.
A. Thirty-four of the 63 hearts scored positive for viral RNA. The percentage of viral RNA+ mice in each group is shown, and the numerals indicate RNA+/total mice in each group. B. The amount of CVB3 RNA in the heart of each of the 34 RNA+ mice is shown, together with the group mean ± SE. Eight mice from the d42 groups (shown by mNN labels) were selected for further analysis. C. PCR evaluation of genomic DNA from all of the 8 selected mice confirms heart-specific and near-complete deletion of the floxed DNA fragment in Tam-treated animals.
The RT reactions were carried out in a thermocycler as follows: 65°C for 5 min, 50°C for 45 min, 70°C for 15 min. Samples were then treated with 1μl Ribonuclease H (Life Technologies, CA, USA) to remove RNA complementary to the cDNA. Next, Taqman quantitative real time PCR was performed using the CVB3-specific primers described above. PCR amplification was done using Platinum Quantitative PCR SuperMix-UDG ready to use cocktail (Life Technologies, CA, USA) as described by the manufacturer. Quantitative analysis of viral RNA was carried out using a BioRad iQ5 Real-Time PCR System in 96 well optical reaction plates heated to 50°C for 2 min to digest dUTP-containing contaminants, 95°C for 2 min to deactivate UNG and activate Platinum Taq DNA polymerase, followed by 40 cycles of: denaturation at 95°C for 15s and annealing and extension at 60°C for 30s. All samples were evaluated in triplicate amplification reactions. In order to assign a genome copy number to the cycle threshold value, a standard curve was generated: a known quantity of in vitro transcribed CVB genomic RNA was serially diluted, and all dilutions were subjected to the above reverse transcriptase and qPCR reactions. Additional control reactions were set up that omitted RT, and were invariably negative. Values are expressed as the average number of CVB genome copies per gram of tissue.
PCR array
The levels of interferon-related gene mRNAs in the hearts of mice carrying persistent CVB3 RNA were quantified by PCR array (Mouse Interferons & receptors PCR Array, PAMM-064Z, SA Biosciences, Frederick, MD), carried out in accordance with the manufacturer’s instructions. Data were uploaded to the Qiagen website (http://www.qiagen.com/us/shop/genes-and-pathways/data-analysis-center-overview-page/), where they were normalized to housekeeping genes and analyzed. The resulting data were downloaded in Excel format, and were analyzed using GraphPad Prism 7.
Histology
Segments of heart were fixed in neutral-buffered formalin, embedded in paraffin, and thin sections were prepared and stained with Masson’s trichrome.
Statistical analysis
Significance was determined (Prism 7, Graphpad, San Diego, CA) via one way ANOVA, unpaired, non-parametric t tests, or two way ANOVA where appropriate. Calculated p values <0.05 were considered significant.
Results
Near full-length CVB3 RNA transiently persists in the hearts of C57BL/6 mice
In most cases, when assessed by a regular plaque assay, infectious CVB3 is lost from the heart by ~2 weeks p.i., but CVB3 RNA can persist in the heart for many weeks thereafter; one early study demonstrated that viral RNA remained detectable in 66% of mice at 34 days p.i. (Rabausch-Starz et al., 1994). A subsequent analysis showed that 50% of mice were positive at 90 days p.i., and data extrapolation suggested that complete clearance might take 5–6 months (Reetoo et al., 2000). Because the primary aim of our study was to determine (i) the immunological consequences of CVB RNA persistence and (ii) the role of T1IFN signaling into cardiomyocytes during RNA persistence, it was important to select a time point after all mice have cleared infectious virus, but at which we could be confident that many of their hearts would score positive for RNA. Thus, we first assessed, in our own hands, the kinetics of CVB3 infection and RNA persistence in (and clearance from) the heart. As shown in Figure 1A, C57BL/6 mice were infected with wtCVB3 (500 pfu, i.p.), and surviving mice were sacrificed at days 13, 30, 62 or 112 p.i.; hearts were evaluated for the presence of viral RNA and infectious virus. Consistent with other reports, RNA was detected in all mice analyzed at d13, and at declining frequency thereafter (Figure 1B). For selected mice (two at d13, one at d30, three at d62), genome-sense RNA content was determined using two separate sets of primers, one set (VP4) detecting a region near the 5′ end of the viral open reading frame, and the other (3D) detecting a region close to the 3′ end (Figure 1C); in all cases, the numbers of RNA molecules identified were near-identical, consistent with the majority of the molecules being near full-length. Infectious virus was detected only in the hearts of d13 mice; using a standard plaque assay, no virus was detected in any RNA-containing heart at d30 and later (Figure 1D). We chose not to further characterize this persistent RNA because, as described in the Introduction, others have done so, demonstrating that it consists of TD viral materials. We wished instead to determine the biological consequences of RNA persistence and, to best do so, our subsequent experiments focused on the d30 time point, when infectious virus was absent, but persistent RNA was frequently found.
Deletion of T1IFNR from cardiomyocytes at 30 days p.i. does not lead to overt clinical disease, nor to reactivation of infectious virus
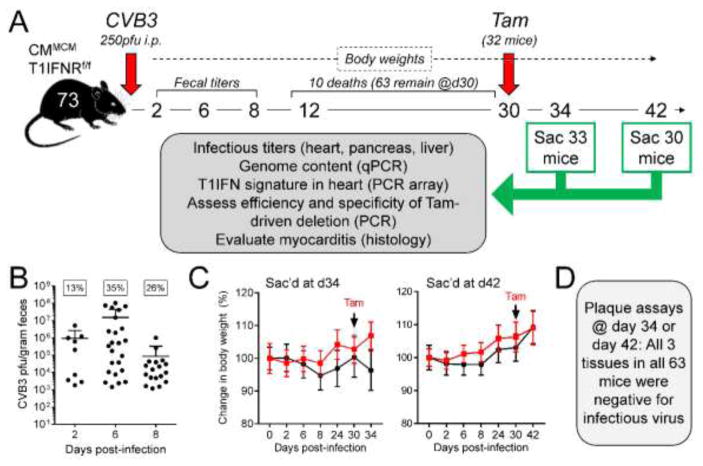
As diagrammed in Figure 2A, seventy-three genetically-intact MCM+ T1IFNRf/f mice received an i.p. injection of 250 pfu of CVB3. This slightly lower dose was selected to maximize mouse survival for at least 30 days. The early phase of the infection was monitored by determining fecal titers on days 2, 6 and 8 p.i., and general health was monitored by recording body weights. Ten of the mice died prior to d30, leaving a total of 63 mice to be used in our subsequent studies. On d30, 32 of these mice received a single injection of 2mg of tamoxifen. Four days later, 33 mice (17 that had received Tam, 16 that had not) were sacrificed, and the hearts, livers and pancreata were harvested. After a further 8 days (i.e., on d42) we sacrificed, and obtained organs from, the remaining 30 animals (15 of which had received Tam). The procedures outlined in the grey box in A were applied to all samples and, with the exception of virus titers, the related data are presented in Figure 3 and Figure 4. At d2 p.i., only 13% of the mice scored positive by fecal titer (Figure 2B), although higher proportions were positive on days 6 and 8. There was significant variability in fecal titers, and ~35% of the inoculated animals were negative at all three time points. Although it is tempting to conclude that the infection had not taken hold in these mice, this would not necessarily be correct (see below). The body weights of the mice remained relatively stable over the 30-day period, and Tam injection did not cause any detectable weight loss, or other overt signs of morbidity, over the subsequent 4 or 12 days (Figure 2C). One study of a patient with DCM revealed that, at ~1 month after the appearance of symptoms, ~1% of the persistent viral RNA was full-length (Bouin et al., 2016), causing us to hypothesize that Tam-mediated removal of T1IFN signaling might permit the re-emergence of full-length cytopathic virus; however, none of the 189 organ samples harvested at 34 or 42 days p.i. – not even those from Tam-treated animals – contained infectious CVB3 that could be detected by a regular plaque assay (Figure 2D).
Figure 2. Deletion of T1IFNR from cardiomyocytes at 30 days p.i. does not lead to overt clinical disease, nor to reactivation of infectious virus.
The experimental approach is summarized in panel A; 73 CMMCM T1IFNRf/f mice were infected with 250 pfu of CVB3, and were monitored as shown. B. Fecal samples from all 73 mice were analyzed at the three indicated time points. The box above each time point indicates the percentage of mice that scored positive at that time point and, for each positive sample at each time point, the infectious titer is shown. As stated in the text, ~35% of animals were negative at all time points. C. Percentage changes in body weights across the course of infection are shown, for all four groups of mice (± Tam at d30; sacrificed on d34 or d42. Weights of the mice in the Tam groups are shown in red). none of the animals suffered significant weight loss, and there was no statistically-significant difference among the groups. D. All 189 tissues (heart, liver and pancreas from 63 mice) were titrated for the presence of cytopathic virus; none were positive.
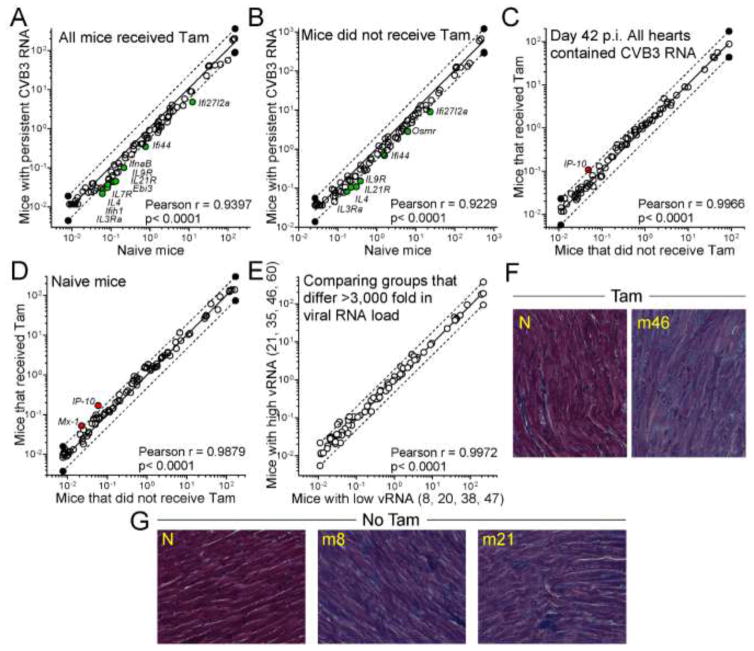
Figure 4. Neither the persistence of CVB3 RNA nor the disruption of T1IFNR expression has a marked impact on IFN-related mRNA abundance or cardiac inflammation.
A – E. PCR array was used to quantify the expression of interferon-related mRNAs in the cardiac RNAs from the selected mice, and from control CMMCM T1IFNRf/f mice. For all comparisons, a linear regression analysis was carried out (solid line in each graph) and 2-fold changes are delimited by dashed lines. Each symbol indicates the relative expression of an individual gene in the two groups shown on the x and y axes. A reduction in mRNA content (y lower than x) is indicated by a green symbol; an increase (y greater than x) is shown by a red symbol. All such changes are annotated with the gene name. None of the changes were flagged as statistically-significant. A. Viral RNA+ Tam-treated mice (y axis) were compared to control Tam-treated mice (x axis), to determine if persistent viral RNA is associated with an interferon signature. B. A similar comparison, this time of mice that did not receive Tam. C. Using the 8 selected RNA+ mice, gene expression in Tam-receiving animals (in which T1IFN signaling is disrupted in cardiomyocytes; y axis) was compared to that in untreated animals (x axis). D. The impact of Tam on gene expression was determined in uninfected mice. E. The effect of viral RNA load was determined by comparing four mice with high load (mean 6.6 × 108 genome copies/g heart, y axis) with four mice with low RNA load (mean 2.1 × 105 genome copies/g heart, x axis) F, G. Histological sections are shown from the indicated d42 mice, and from naïve (N) mice ± Tam (Masson’s trichrome stain).
Deletion of the T1IFNR from cardiomyocytes at 30 days p.i. has no significant impact on the prevalence or the quantity of persistent CVB3 RNA
Although no infectious virus was detected in any mouse heart by plaque assay, work from other labs, and the data in Figure 1, indicated a high probability that some of the mice would harbor persistent CVB3 RNA. Thus, the hearts of all 63 mice were evaluated by qPCR. As shown in Figure 3A, ~54% of the hearts (34/63) contained viral RNA (>105 copies/gram of tissue). This is consistent with Figure 1, and with data from other laboratories, but we considered the possibility that, in these genetically-intact animals, the RNA-negative mice might not be truly negative, and instead might have viral RNA that was being held at a below-detectable level, perhaps by the action of T1IFNs. If that hypothesis were valid, one would predict that the eradication of T1IFN signaling would increase the frequency at which viral RNA was detected. The data in Figure 3A show that this was not the case: although there was some variability among the four groups, none of the differences reached the level of statistical significance and, more specifically, the Tam-mediated abrogation of T1IFN signaling (red bars) did not significantly increase the frequency with which CVB3 RNA was detected. This suggests that, at 30 days p.i., T1IFN is not actively suppressing the levels of CVB3 RNA in mouse hearts to below detectable levels. Individual assessment of the 34 RNA-positive animals allows us to extend this conclusion. If T1IFN signaling played a role in limiting the level of persistent CVB3 RNA, one would predict that, among all RNA-positive animals, Tam-treated mice should have higher levels of viral RNA than their untreated counterparts. However, when analyses were limited to the RNA-positive mice (Figure 3B), there was no statistically-significant difference in genomic RNA levels between Tam-treated and untreated groups. To exclude the possibility that Tam had been ineffective, PCR analyses were carried out on genomic DNA isolated from hearts and livers of eight d42 mice, four of which had received Tam; these mice are individually labeled in Figure 3B. Near-complete deletion of the floxed DNA was observed in the hearts of all four Tam-treated animals, while their liver DNA was intact, demonstrating the cardiac specificity of the CMMCM model (Figure 3C); as expected, no deleted DNA was present in mice that had not received Tam. Thus, Tam administration successfully ablated the floxed DNA in cardiomyocytes but, despite this, there was no increase in infectious virus (Figure 2) or in the frequency or abundance of CVB3 RNA (Figure 3). Taken together, these data indicate that T1IFNs do not play a major part in regulating the prevalence/abundance of viral RNAs, after they have become persistent. Finally, several mice that carried persistent RNA (m8, m35, m38, m60) had had negative fecal titers on all three occasions, demonstrating that fecal titers – perhaps surprisingly, for an enteric infection – are not an infallible means by which to determine the existence, or the extent, of an enteroviral infection.
Neither the persistence of CVB3 RNA nor the disruption of T1IFNR expression has a marked impact on IFN-related mRNA abundance or cardiac inflammation
Others have shown that, when present in abnormal amounts in the heart, single-stranded DNA derived from endogenous retroelements can drive T1IFN-dependent myocarditis (Stetson et al., 2008). This, together with the abundant evidence (see Introduction) that T1IFNs play a key role during CVB infection, led us to hypothesize that persistent CVB RNA – even at the levels generated by slowly-replicating deletion mutants – might stimulate the innate immune system, possibly causing chronic inflammation and contributing to the development of chronic sequelae including DCM. Therefore, using a PCR array approach, we determined if the expression of 84 IFN-related genes in the hearts of the eight selected viral RNA+ mice was altered by (i) the presence of persistent CVB3 RNA and/or (ii) the presence or absence of T1IFN on cardiomyocytes. We found that the expression of these genes in CVB3 RNA+ hearts was very similar to that observed in RNA-negative hearts, regardless of whether or not the mice had (Figure 4A) or had not (Figure 4B) received Tam. A small number of mRNAs (green symbols) were present at marginally (~2-fold) lower levels in the viral RNA-carrying hearts than in the control hearts, but none of the differences reached the level of statistical significance. The Pearson correlation coefficients and their related p values were extremely strong for both panels A and B, indicating that, with or without Tam administration, persistent CVB3 RNA had little impact on the expression of IFN-related genes. Moreover, the similarity between the two panels suggested that ablation of the T1IFNR had minimal effect on the abundance of IFN-related mRNAs, and this was confirmed by the direct comparison (Figure 4C) of RNA-positive mice that had (y axis) or had not (x axis) received Tam. Most mRNAs were unaffected by the deletion, arguing strongly that, at d30 p.i. and thereafter, there is no ongoing T1IFN-driven response in these hearts. Only one mRNA, encoding IP10 (aka Cxcl10) was slightly higher (2.1-fold) in the hearts of Tam-treated mice (Figure 4C). This was potentially interesting, as others have reported that this chemokine is induced by CVB3, and inhibits viral replication by recruiting natural killer cells to the heart (Yuan et al., 2009); however, the small change in IP10 mRNA content that we observed appears to be a side-effect of Tam treatment, because it also was present following Tam administration to naive animals (Figure 4D). Our final comparison was between mice that carry high viral RNA loads and those that carry low loads (Figure 4E); although the mean viral load in the y-axis group was >3,000-fold higher than the x-axis group, there was no significant difference in IFN-related gene expression. In summary, there is no indication that the persistence of viral RNA in the heart at d42 p.i. is accompanied by an ongoing innate immune response that (i) up-regulates the cardiac expression IFN-responsive genes, or (ii) is affected by T1IFN signaling in cardiomyocytes. Despite the absence of evidence of an ongoing viral RNA-driven innate response, we thought it prudent to carry out a histological evaluation of the hearts of these mice, and representative images are presented in Figure 4F and G. We found no indication of ongoing myocarditis, even though some of these hearts (e.g., m46) contained >109 CVB3-related RNA molecules per gram of tissue.
Discussion
Many viruses have long been known to establish persistent/latent infections; examples include hepatitis B virus and several herpesviruses. In contrast, until relatively recently, enteroviruses were thought to cause only acute infections, and to be completely eradicated with ~2 weeks of the primary infection. However, over the past decade it has become clear that these viruses can persist, in immunocompetent hosts, for at least several weeks, albeit in a form that is not detectable using a standard plaque assay. These TD viruses are non-cytopathic, and their inoculation in vivo does not initiate detectable disease. However, even if unable to kill infected cells, or to trigger disease de novo, in principle these TD viruses could exacerbate existing disease, e.g. by prolonging immunopathogenic responses to the original infection.
Our study had several goals: first, all studies of TD viruses to date have employed hosts with an intact immune system. We wished to assess the impact, if any, of disrupting the innate immune response of cardiomyocytes after CVB3 RNA persistence was established: would the viral RNA load increase? Would standard (cytopathic) infectious virus re-emerge? Second, it has been proposed that these persistent TD viruses, or the RNAs, might contribute to the gradual development of DCM. We wished to determine if, at >30 days following infection, there is an interferon signature that correlates with the persistence of viral RNA. Third, bringing these threads together, would the ablation of T1IFN signaling into cardiomyocytes have any impact on T1IFN signatures, and/or myocarditis, at >4 weeks p.i.? A total of 73 mice were infected with wtCVB3, and 63 survived until sacrificed. ~50% of those mice received Tam at d30 p.i., and were sacrificed 4 or 12 days later. Consistent with other studies, none of the hearts (or liver or pancreata) contained infectious virus, judged by the criterion of a standard plaque assay. However, others have reported the presence of full-length CVB3 RNA in a cardiac biopsy taken from a patient who reported ~1 month of symptoms (Bouin et al., 2016), so we tested the hypothesis that ablation of T1IFN signaling during the RNA persistence phase might permit the re-emergence of any full-length (and presumably cytopathic) viruses. This did not happen; plaque assays remained negative at both 4 and 12 days after Tam administration, suggesting that, at d30 after acute infection, T1IFNs are not actively suppressing the replication of full-length viruses in cardiomyocytes. Although our work focused on cardiomyocytes, it is important to note that tissue culture studies of cardiac fibroblasts have shown that these cells, too, can be infected with CVB3 and thereafter produce various cytokines (Lindner et al., 2014).
Having demonstrated that no cytopathic virus was detectable, even after Tam administration, we next evaluated the frequencies with which persistent CVB3 RNA was detected in the various mouse groups. We note, in passing, that it would have been interesting to determine if CVB RNA persistence is more readily established in cardiomyocytes that are unable to respond to T1IFNs, but that experiment is not feasible, because Tam-treatment prior to wt CVB3 infection almost invariably results in death by ~12 days p.i. (Althof et al., 2014). Thus, we relied on the approach taken herein: wt cardiovirulent CVB3 was administered to genetically-intact mice, most of which can survive the acute infection, and only when the acute phase was over did we ablate T1IFN signaling in some of the survivors. Under those circumstances, 54% of the hearts (34/63) scored positive for viral RNA, but there was no statistically-significant difference in frequency between Tam-treated and untreated groups. At first sight, it appears promising to compare the frequencies at d34 with those at d42 (Figure 3A). In mice that had remained genetically-intact (black bars), the frequency of RNA+ mice dropped between d34 and d42, consistent with the notion that the RNA (and TD viruses) is/are cleared by the immune response in a time-dependent manner (see Figure 1, and the work of other labs). Conversely, in Tam-treated mice (red bars), the proportion of RNA+ hearts at d42 p.i (9/15, 60%) is higher than the proportion at d34 (7/17, 41%), raising the possibility that ablation of T1IFN signaling in cardiomyocytes at d30 had permitted the increased replication of TD viruses, and the consequent gradual increase in mice that scored positive for viral RNA. However, such a conclusion is unsupportable for at least two reasons. First, at d34, one would predict that the frequency of RNA+ hearts would be higher in Tam-treated than in non-treated mice; the opposite is the case. Second, statistical comparison of the four groups (ANOVA) revealed no significant difference; this was true even of a t-test limited to the d34 and d42 Tam-treated groups (p>0.3). Moreover, we found no significant difference in the quantities of viral RNAs in individual Tam-treated versus untreated mice (Figure 3B) supporting our conclusion that, at d30 p.i., T1IFNs are not actively regulating the abundance of persistent CVB3 RNAs. This conclusion has a broader implication: it suggests that, despite the presence of readily-detectable virus-related RNAs at d30 p.i., T1IFNs may be relatively quiescent at this time point. However, that inference is based on the lack of impact of T1IFNs on CVB3 (i.e., on infectious virus or viral RNA load), so we next used PCR arrays to evaluate the expression of host interferon-related mRNAs. No significant IFN signature was detected in RNA+ mice, regardless of time point (d34 or d42) and of whether or not the animals had received Tam (Figure 4A–D). Near-identical gene expression patterns were present in mice that differed >3,000-fold in viral RNA load (Figure 4E), and there was no recrudescence of myocarditis, even in the mice with the highest RNA load (Figure 4F). Others have shown that cardiomyocyte-specific transgenic over-expression of MDA5 – a protein considered to play a key role in sensing picornaviral RNA – results in increased baseline expression of antiviral cytokines in the heart, and to cellular infiltration, even in the absence of infection (Philip et al., 2013), suggesting that the MDA5-mediated innate response balances on a hair-trigger; in that light, the failure of persistent CVB3 RNA to induce a detectable inflammatory response is even more surprising. In summary, our data suggest that persistent cardiac CVB3 RNA does not cause a substantial ongoing innate response in the heart; this is consistent with our finding that removal of the T1IFNR from cardiomyocytes has no significant impact on the levels of viral, or of IFN-related, RNAs. We therefore propose that the viral RNAs that remain at 30d p.i. most probably play no role in ongoing pathogenesis, and are merely the last remnants of the preceding infection. Indeed, we speculate that the inability of these RNAs (and of TD viruses) to trigger an innate response may provide them with a selective advantage, explaining both their emergence, and their ability to persist for several weeks.
The initial infections were monitored, in all mice, by three fecal titers taken on days 2, 6 and 8 p.i. Of the 63 mice that survived to d30, 34 were CVB RNA+ at day 34 or 42 and, remarkably, almost 1/3rd of these RNA+ mice (10/34) had scored negative by fecal titers on all three occasions (days 2, 6 & 8 p.i.), and during subsequent days they did not suffer weight loss indicative of a severe infection; we speculate that these animals may represent a parallel to the well-recognized phenomenon of human DCM patients who have enterovirus RNA+ cardiac biopsies without any history suggestive of acute viral myocarditis. In a recent study of 23 DCM patients (N’Guyen et al., 2017), 10 individuals were classified as having “explained” DCM (eDCM), and the remaining 13 were considered to have idiopathic DCM (iDCM). Cardiac biopsies from both DCM groups, and samples from two control groups without DCM, were evaluated for the presence of nucleic acids of six different viruses; in all four groups, >80% of hearts contained genomes from at least one virus. Parvovirus genomes were especially frequent, being observed in 60–80% of hearts, with no significant difference among the groups, indicating that these genomes may be passenger/bystander materials. In contrast, enteroviral genomes were identified only in the iDCM group (3/13 hearts), and these samples also displayed footprints of an inflammatory response. Thus, that study strengthens the case that enteroviruses may cause a significant proportion of DCM cases that currently are considered idiopathic. It is tempting, from those data, to infer that the persistent EV genomes in those three DCM patients may have contributed directly to the development of their disease. However, the EV RNA load in two of the three patients was extremely low, and could be detected only using a highly-sensitive technique (PCR assays coupled to microarray hybridization analyses). Using standard real-time PCR, viral genome was quantifiable only in one of the three patients, and we show here that, even at that “high” level of persistent CVB3 RNA, no ongoing T1IFN response exists, at least in mice (Figure 4). This begs the question: in humans, does persistent enteroviral RNA have an effect on disease progression, or might it be merely a marker of the prior infection? In mice, persistent CVB RNA, and TD viruses, are readily identified at 1 month p.i., but the frequency with which they can be found declines dramatically as time passes. In humans, the exact date of infection is rarely known but, in both of the reported cases of TD CVB in the heart (Bouin et al., 2016; Chapman et al., 2008), the cardiac tissues were obtained within 2–4 weeks of the appearance of clinical symptoms, raising the possibility that these TD variants are evidence of a recent primary infection. Moreover, the TD variants are non-cytopathic in tissue culture and, to our knowledge, they have never been shown to cause disease in vivo. Thus, as noted above, we favor the notion that TD viral variants, and the related persistent RNA, are not themselves pathogenic, and instead are the last vestiges of a fading infection; as such, they may be useful for diagnostic purposes, as they allow the correct etiological assignment of cases that otherwise would be considered idiopathic. If this contention is valid, it follows that DCM must develop soon after the primary enteroviral infection – otherwise, persistent viral RNA would not be detected in biopsies of DCM patients. In that light, how rapidly does DCM develop? Around 1/3rd of DCM cases are genetic in origin (Kimura, 2016), and several strains of transgenic mice have been established to model some of these defects. Muscle LIM protein (MLP) null mice are engineered by a deletion of MLP, an actin-associated cytoskeletal protein. In these mice, functional analyses are normal at 2 weeks of age but, thereafter, chamber dilation and contractile dysfunction develop very rapidly (within ~2 weeks, see Knoll et al., 2002). DCM also develops quickly (in ~6 weeks) in knock-in mice expressing an autosomal dominant cardiac troponin mutation (Ramratnam et al., 2016). What of enterovirus-related DCM: how quickly can it develop? Cardiomyocyte-specific transgenic expression of a CVB variant that could replicate, but could not generate infectious particles, led to histological changes consistent with DCM in 4- to 6-week-old animals (Wessely et al., 1998). A second transgenic model exploited the CVB3 2A protease, which can cleave a key cardiomyocyte protein, dystrophin (Badorff et al., 2000). A tamoxifen-inducible version of the 2A protease was expressed specifically in cardiomyocytes, and DCM occurred within 22 days of Tam administration (Xiong et al., 2007). Taken together, these observations indicate that the interval between CVB infection and DCM may be measured in weeks, rather than in months or years. Moreover, we show herein that mice with no overt signs of infection frequently had viral RNA in their hearts. This explains the scenario – not uncommon in human cases of DCM – of a patient who presents without a prior history suggestive of viral myocarditis, but who tests positive for enteroviral RNA in the heart; he/she may have had subclinical infection in the recent past, explaining the existence of the residual viral RNAs.
Highlights.
CVB3 RNA persists in mouse hearts for weeks after clearance of infectious virus
Although abundant, this RNA triggers neither T1IFN responses nor local inflammation
After RNA persistence was established, T1IFNR was ablated in vivo from cardiomyocytes
This had no effect on RNA levels, and infectious virus did not emerge
Acknowledgments
This work was supported by NIH grants R01AI114615 and R01AI110621 to JLW. This is manuscript number 29567 from the Scripps Research Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abelmann WH. Myocarditis. N Engl J Med. 1966;275:832–834. doi: 10.1056/NEJM196610132751509. [DOI] [PubMed] [Google Scholar]
- Abston ED, Coronado MJ, Bucek A, Bedja D, Shin J, Kim JB, Kim E, Gabrielson KL, Georgakopoulos D, Mitzner W, Fairweather D. Th2 regulation of viral myocarditis in mice: different roles for TLR3 versus TRIF in progression to chronic disease. Clin Dev Immunol. 2012;2012:129486. doi: 10.1155/2012/129486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abston ED, Coronado MJ, Bucek A, Onyimba JA, Brandt JE, Frisancho JA, Kim E, Bedja D, Sung YK, Radtke AJ, Gabrielson KL, Mitzner W, Fairweather D. TLR3 deficiency induces chronic inflammatory cardiomyopathy in resistant mice following coxsackievirus B3 infection: role for IL-4. Am J Physiol Regul Integr Comp Physiol. 2013;304:R267–R277. doi: 10.1152/ajpregu.00516.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Althof N, Harkins S, Kemball CC, Flynn CT, Alirezaei M, Whitton JL. In vivo ablation of type I interferon receptor from cardiomyocytes delays coxsackieviral clearance and accelerates myocardial disease. J Virol. 2014;88:5087–5099. doi: 10.1128/JVI.00184-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreoletti L, Leveque N, Boulagnon C, Brasselet C, Fornes P. Viral causes of human myocarditis. Arch Cardiovasc Dis. 2009;102:559–568. doi: 10.1016/j.acvd.2009.04.010. [DOI] [PubMed] [Google Scholar]
- Archard LC, Bowles NE, Cunningham L, Freeke CA, Olsen EG, Rose ML, Meany B, Why HJ, Richardson PJ. Molecular probes for detection of persisting enterovirus infection of human heart and their prognostic value. Eur Heart J. 1991;12(Suppl D):56–59. doi: 10.1093/eurheartj/12.suppl_d.56. [DOI] [PubMed] [Google Scholar]
- Badorff C, Berkely N, Mehrotra S, Talhouk JW, Rhoads RE, Knowlton KU. Enteroviral protease 2A directly cleaves dystrophin and is inhibited by a dystrophin-based substrate analogue. J Biol Chem. 2000;275:11191–11197. doi: 10.1074/jbc.275.15.11191. [DOI] [PubMed] [Google Scholar]
- Bendig JWA, O’Brien PS, Muir P, Porter HJ, Caul EO. Enterovirus sequences resembling coxsackievirus A2 detected in stool and spleen from a girl with fatal myocarditis. J Med Virol. 2001;64:482–486. doi: 10.1002/jmv.1075. [DOI] [PubMed] [Google Scholar]
- Blauwet LA, Cooper LT. Myocarditis. Prog Cardiovasc Dis. 2010;52:274–288. doi: 10.1016/j.pcad.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouin A, Nguyen Y, Wehbe M, Renois F, Fornes P, Bani-Sadr F, Metz D, Andreoletti L. Major Persistent 5′ Terminally Deleted Coxsackievirus B3 Populations in Human Endomyocardial Tissues. Emerg Infect Dis. 2016;22:1488–1490. doi: 10.3201/eid2208.160186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carniel E, Sinagra G, Bussani R, Di LA, Pinamonti B, Lardieri G, Silvestri F. Fatal myocarditis: morphologic and clinical features. Ital Heart J. 2004;5:702–706. [PubMed] [Google Scholar]
- Chapman NM, Kim KS, Drescher KM, Oka K, Tracy S. 5′ terminal deletions in the genome of a coxsackievirus B2 strain occurred naturally in human heart. Virology. 2008;375:480–491. doi: 10.1016/j.virol.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsten MF, Schroen B, Heymans S. Inflammation in viral myocarditis: friend or foe? Trends Mol Med. 2012;18:426–437. doi: 10.1016/j.molmed.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Deonarain R, Cerullo D, Fuse K, Liu PP, Fish EN. Protective role for interferon-β in coxsackievirus B3 infection. Circulation. 2004;110:3540–3543. doi: 10.1161/01.CIR.0000136824.73458.20. [DOI] [PubMed] [Google Scholar]
- Eckart RE, Scoville SL, Campbell CL, Shry EA, Stajduhar KC, Potter RN, Pearse LA, Virmani R. Sudden death in young adults: a 25-year review of autopsies in military recruits. Ann Intern Med. 2004;141:829–834. doi: 10.7326/0003-4819-141-11-200412070-00005. [DOI] [PubMed] [Google Scholar]
- Esfandiarei M, McManus BM. Molecular biology and pathogenesis of viral myocarditis. Annu Rev Pathol. 2008;3:127–155. doi: 10.1146/annurev.pathmechdis.3.121806.151534. [DOI] [PubMed] [Google Scholar]
- Gravanis MB, Sternby NH. Incidence of myocarditis. A 10-year autopsy study from Malmo, Sweden. Arch Pathol Lab Med. 1991;115:390–392. [PubMed] [Google Scholar]
- Huhn MH, McCartney SA, Lind K, Svedin E, Colonna M, Flodstrom-Tullberg M. Melanoma differentiation-associated protein-5 (MDA-5) limits early viral replication but is not essential for the induction of type 1 interferons after Coxsackievirus infection. Virology. 2010;401:42–48. doi: 10.1016/j.virol.2010.02.010. [DOI] [PubMed] [Google Scholar]
- Hunziker IP, Cornell CT, Whitton JL. Deletions within the 5′UTR of coxsackievirus B3: consequences for virus translation and replication. Virology. 2007;360:120–128. doi: 10.1016/j.virol.2006.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo L, Smithee S, Tracy S, Chapman NM. Domain I of the 5′ non-translated genomic region in coxsackievirus B3 RNA is not required for productive replication. Virology. 2016;496:127–130. doi: 10.1016/j.virol.2016.05.021. [DOI] [PubMed] [Google Scholar]
- Kamphuis E, Junt T, Waibler Z, Forster R, Kalinke U. Type I interferons directly regulate lymphocyte recirculation and cause transient blood lymphopenia. Blood. 2006;108:3253–3261. doi: 10.1182/blood-2006-06-027599. [DOI] [PubMed] [Google Scholar]
- Kim KS, Tracy S, Tapprich W, Bailey J, Lee CK, Kim K, Barry WH, Chapman NM. 5′-Terminal Deletions Occur in Coxsackievirus B3 during Replication in Murine Hearts and Cardiac Myocyte Cultures and Correlate with Encapsidation of Negative-Strand Viral RNA. J Virol. 2005;79:7024–7041. doi: 10.1128/JVI.79.11.7024-7041.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A. Molecular genetics and pathogenesis of cardiomyopathy. J Hum Genet. 2016;61:41–50. doi: 10.1038/jhg.2015.83. [DOI] [PubMed] [Google Scholar]
- Knoll R, Hoshijima M, Hoffman HM, Person V, Lorenzen-Schmidt I, Bang ML, Hayashi T, Shiga N, Yasukawa H, Schaper W, McKenna W, Yokoyama M, Schork NJ, Omens JH, McCulloch AD, Kimura A, Gregorio CC, Poller W, Schaper J, Schultheiss HP, Chien KR. The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell. 2002;111:943–955. doi: 10.1016/s0092-8674(02)01226-6. [DOI] [PubMed] [Google Scholar]
- Knowlton KU, Jeon ES, Berkley N, Wessely R, Huber SA. A mutation in the puff region of VP2 attenuates the myocarditic phenotype of an infectious cDNA of the Woodruff variant of coxsackievirus B3. J Virol. 1996;70:7811–7818. doi: 10.1128/jvi.70.11.7811-7818.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehl U, Lassner D, von Schlippenbach J, Poller W, Schultheiss HP. Interferon-beta improves survival in enterovirus-associated cardiomyopathy. J Am Coll Cardiol. 2012;60:1295–1296. doi: 10.1016/j.jacc.2012.06.026. [DOI] [PubMed] [Google Scholar]
- Kuhl U, Pauschinger M, Noutsias M, Seeberg B, Bock T, Lassner D, Poller W, Kandolf R, Schultheiss HP. High prevalence of viral genomes and multiple viral infections in the myocardium of adults with “idiopathic” left ventricular dysfunction. Circulation. 2005;111:887–893. doi: 10.1161/01.CIR.0000155616.07901.35. [DOI] [PubMed] [Google Scholar]
- Leveque N, Garcia M, Bouin A, Nguyen JHC, Tran GP, Andreoletti L, Semler BL. Functional Consequences of RNA 5′-Terminal Deletions on Coxsackievirus B3 RNA Replication and Ribonucleoprotein Complex Formation. J Virol. 2017:91. doi: 10.1128/JVI.00423-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner D, Li J, Savvatis K, Klingel K, Blankenberg S, Tschope C, Westermann D. Cardiac fibroblasts aggravate viral myocarditis: cell specific coxsackievirus B3 replication. Mediators Inflamm. 2014;2014:519528. doi: 10.1155/2014/519528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani JW, Dec GW. Myocarditis: current trends in diagnosis and treatment. Circulation. 2006;113:876–890. doi: 10.1161/CIRCULATIONAHA.105.584532. [DOI] [PubMed] [Google Scholar]
- Mahfoud F, Gartner B, Kindermann M, Ukena C, Gadomski K, Klingel K, Kandolf R, Bohm M, Kindermann I. Virus serology in patients with suspected myocarditis: utility or futility? Eur Heart J. 2011;32:897–903. doi: 10.1093/eurheartj/ehq493. [DOI] [PubMed] [Google Scholar]
- Marchant D, Si X, Luo H, McManus B, Yang D. The impact of CVB3 infection on host cell biology. Curr Top Microbiol Immunol. 2008;323:177–198. doi: 10.1007/978-3-540-75546-3_8. [DOI] [PubMed] [Google Scholar]
- Maron BJ, Towbin JA, Thiene G, Antzelevitch C, Corrado D, Arnett D, Moss AJ, Seidman CE, Young JB American Heart A, Council on Clinical Cardiology HF, Transplantation C, Quality of C, Outcomes R, Functional G, Translational Biology Interdisciplinary Working G, Council on E, Prevention. Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation. 2006;113:1807–1816. doi: 10.1161/CIRCULATIONAHA.106.174287. [DOI] [PubMed] [Google Scholar]
- Mukherjee A, Morosky SA, Delorme-Axford E, Dybdahl-Sissoko N, Oberste MS, Wang T, Coyne CB. The coxsackievirus B 3C protease cleaves MAVS and TRIF to attenuate host type I interferon and apoptotic signaling. PLoS Pathog. 2011;7:e1001311. doi: 10.1371/journal.ppat.1001311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- N’Guyen Y, Lesaffre F, Metz D, Tassan S, Saade Y, Boulagnon C, Fornes P, Renois F, Andreoletti L. Enterovirus but not Parvovirus B19 is associated with idiopathic dilated cardiomyopathy and endomyocardial CD3, CD68, or HLA-DR expression. J Med Virol. 2017;89:55–63. doi: 10.1002/jmv.24600. [DOI] [PubMed] [Google Scholar]
- Negishi H, Osawa T, Ogami K, Ouyang X, Sakaguchi S, Koshiba R, Yanai H, Seko Y, Shitara H, Bishop K, Yonekawa H, Tamura T, Kaisho T, Taya C, Taniguchi T, Honda K. A critical link between Toll-like receptor 3 and type II interferon signaling pathways in antiviral innate immunity. Proc Natl Acad Sci U S A. 2008;105:20446–20451. doi: 10.1073/pnas.0810372105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell JB. The role of myocarditis in end-stage dilated cardiomyopathy. Tex Heart Inst J. 1987;14:268–275. [PMC free article] [PubMed] [Google Scholar]
- Oka K, Oohira K, Yatabe Y, Tanaka T, Kurano K, Kosugi R, Murata M, Hakozaki H, Nishikawa T, Tsutsumi Y. Fulminant myocarditis demonstrating uncommon morphology--a report of two autopsy cases. Virchows Arch. 2005;446:259–264. doi: 10.1007/s00428-004-1173-3. [DOI] [PubMed] [Google Scholar]
- Philip J, Xu Z, Bowles NE, Vallejo JG. Cardiac-Specific Overexpression of Melanoma Differentiation Associated Gene-5 Protects Mice from Lethal Viral Myocarditis. Circ Heart Fail. 2013;6:326–334. doi: 10.1161/CIRCHEARTFAILURE.112.969402. [DOI] [PubMed] [Google Scholar]
- Rabausch-Starz I, Schwaiger A, Grunewald K, Muller-Hermelink HK, Neu N. Persistence of virus and viral genome in myocardium after coxsackievirus B3-induced murine myocarditis. Clin Exp Immunol. 1994;96:69–74. doi: 10.1111/j.1365-2249.1994.tb06232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakar S, Sinagra G, Di Lenarda A, Poletti A, Bussani R, Silvestri F, Camerini F. Epidemiology of dilated cardiomyopathy. A prospective post-mortem study of 5252 necropsies The Heart Muscle Disease Study Group. Eur Heart J. 1997;18:117–123. doi: 10.1093/oxfordjournals.eurheartj.a015092. [DOI] [PubMed] [Google Scholar]
- Ramratnam M, Salama G, Sharma RK, Wang DW, Smith SH, Banerjee SK, Huang XN, Gifford LM, Pruce ML, Gabris BE, Saba S, Shroff SG, Ahmad F. Gene-Targeted Mice with the Human Troponin T R141W Mutation Develop Dilated Cardiomyopathy with Calcium Desensitization. PLoS One. 2016;11:e0167681. doi: 10.1371/journal.pone.0167681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reetoo KN, Osman SA, Illavia SJ, Cameron-Wilson CL, Banatvala JE, Muir P. Quantitative analysis of viral RNA kinetics in coxsackievirus B3-induced murine myocarditis: biphasic pattern of clearance following acute infection, with persistence of residual viral RNA throughout and beyond the inflammatory phase of disease. Journal of General Virology. 2000;81:2755–2762. doi: 10.1099/0022-1317-81-11-2755. [DOI] [PubMed] [Google Scholar]
- Riad A, Westermann D, Zietsch C, Savvatis K, Becher PM, Bereswill S, Heimesaat MM, Lettau O, Lassner D, Dorner A, Poller W, Busch M, Felix SB, Schultheiss HP, Tschope C. TRIF is a critical survival factor in viral cardiomyopathy. J Immunol. 2011;186:2561–2570. doi: 10.4049/jimmunol.1002029. [DOI] [PubMed] [Google Scholar]
- Riecansky I, Schreinerova Z, Egnerova A, Petrovicova A, Bzduchova O. Incidence of Coxsackie virus infection in patients with dilated cardiomyopathy. Cor Vasa. 1989;31:225–230. [PubMed] [Google Scholar]
- Sagar S, Liu PP, Cooper LT., Jr Myocarditis. Lancet. 2012;379:738–747. doi: 10.1016/S0140-6736(11)60648-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smithee S, Tracy S, Chapman NM. Mutational Disruption of cis-Acting Replication Element 2C in Coxsackievirus B3 Leads to 5′-Terminal Genomic Deletions. J Virol. 2015;89:11761–11772. doi: 10.1128/JVI.01308-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sole MJ, Liu P. Viral myocarditis: a paradigm for understanding the pathogenesis and treatment of dilated cardiomyopathy. J Am Coll Cardiol. 1993;22:99A–105A. doi: 10.1016/0735-1097(93)90470-l. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JP, Cerny A, Asher DR, Kurt-Jones EA, Bronson RT, Finberg RW. MDA5 and MAVS mediate type I IFN responses to Coxsackie B virus. J Virol. 2010;84:254–260. doi: 10.1128/JVI.00631-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YX, da C V, Vincelette J, White K, Velichko S, Xu Y, Gross C, Fitch RM, Halks-Miller M, Larsen BR, Yajima T, Knowlton KU, Vergona R, Sullivan ME, Croze E. Antiviral and myocyte protective effects of murine interferon-β and -α2 in coxsackievirus B3-induced myocarditis and epicarditis in Balb/c mice. Am J Physiol Heart Circ Physiol. 2007;293:H69–H76. doi: 10.1152/ajpheart.00154.2007. [DOI] [PubMed] [Google Scholar]
- Ward C. Severe arrhythmias in Coxsackievirus B3 myopericarditis. Arch Dis Child. 1978;53:174–176. doi: 10.1136/adc.53.2.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessely R, Klingel K, Knowlton KU, Kandolf R. Cardioselective infection with coxsackievirus B3 requires intact type I interferon signaling: implications for mortality and early viral replication. Circulation. 2001;103:756–761. doi: 10.1161/01.cir.103.5.756. [DOI] [PubMed] [Google Scholar]
- Wessely R, Klingel K, Santana LF, Dalton N, Hongo M, Jonathan LW, Kandolf R, Knowlton KU. Transgenic expression of replication-restricted enteroviral genomes in heart muscle induces defective excitation-contraction coupling and dilated cardiomyopathy. J Clin Invest. 1998;102:1444–1453. doi: 10.1172/JCI1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong D, Yajima T, Lim BK, Stenbit A, Dublin A, Dalton ND, Summers-Torres D, Molkentin JD, Duplain H, Wessely R, Chen J, Knowlton KU. Inducible cardiac-restricted expression of enteroviral protease 2A is sufficient to induce dilated cardiomyopathy. Circulation. 2007;115:94–102. doi: 10.1161/CIRCULATIONAHA.106.631093. [DOI] [PubMed] [Google Scholar]
- Yuan J, Liu Z, Lim T, Zhang H, He J, Walker E, Shier C, Wang Y, Su Y, Sall A, McManus B, Yang D. CXCL10 inhibits viral replication through recruitment of natural killer cells in coxsackievirus B3-induced myocarditis. Circ Res. 2009;104:628–638. doi: 10.1161/CIRCRESAHA.108.192179. [DOI] [PubMed] [Google Scholar]