Abstract
B cell activation is dependent on a large increase in transcriptional output followed by focused expression on secreted immunoglobulin as the cell transitions to an antibody producing plasma cell. The rapid transcriptional induction is facilitated by the release of poised RNA pol II into productive elongation through assembly of the super elongation complex (SEC). We report that a SEC component, the Eleven nineteen Lysine-rich leukemia (ELL) family member 3 (ELL3) is dynamically up-regulated in mature and activated human B cells followed by suppression as B cells transition to plasma cells in part mediated by the transcription repressor PRDM1. Burkitt’s lymphoma and a sub-set of Diffuse Large B cell lymphoma cell lines abundantly express ELL3. Depletion of ELL3 in the germinal center derived lymphomas results in severe disruption of DNA replication and cell division along with increased DNA damage and cell death. This restricted utilization and survival dependence reveal a key step in B cell activation and indicate a potential therapeutic target against B cell lymphoma’s with a germinal center origin.
Keywords: ELL3, Transcription elongation, B-cell, Proliferation, Lymphoma
1. Introduction
Mature naïve B cells continuously circulate through the secondary lymphoid tissues and survey for antigens through B-cell receptor (BCR) recognition (1). Recognition is facilitated by antigen presentation, co-stimulation and cytokine stimulus provided by follicular T helper cells (2–9). Amongst the resulting fates is the germinal center (GC) reaction, which is key for development of long-term and high-affinity B cells (10–12). It involves rapid B cell proliferation and execution of somatic hypermutation and class switch recombination, which results in alteration of the BCR isotype and affinity, and selection to further differentiate into either long-lived plasma cells or memory B cells (13, 14).
Upon antigen encounter, previously quiescent naive lymphocytes increase their global transcriptional output (1). This rapid activation of gene expression in enabled by having poised RNA polymerase II (Pol II) but un-melted promoters (15). While crucial for productive transcriptional elongation, Pol II pause site release is the least studied rate-limiting step in lymphocyte activation. Our current understanding stems from its regulatory role in developmental genes, heat-shock-inducible genes, proto-oncogenes, retrovirus transcript production and leukaemogenesis (16–26). Productive elongation requires assembly of the Super Elongation Complex (SEC), which includes the heterodimeric kinase positive transcription elongation factor b (P-TEFb) composed of cyclin-dependent kinase 9 (CDK9) and Cyclin T, AFF family members (AFF1-4), YEATS domain containing protein family members (AF9 or ENL), ELL family members, and ELL associated factors (EAF) (26–29). These factors were identified through their purification with the mixed lineage leukemia (MLL) gene partner, Eleven nineteen Lysine-rich leukemia (ELL) (26). ELL was shown to increase the catalytic rate of transcription in vitro, initiate basal transcription, stabilize RNA Pol II, and recruit the remaining SEC members (28). ELL family members, ELL2 and ELL3, have similar elongation activity in SEC (30, 31). ELL3 was initially described in testis and was subsequently associated with priming future gene activation in embryonic stem cells, embryonic stem cell differentiation and promoting breast cancer (31–35). ELL2 is expressed in terminally differentiated plasma cells where it promotes secretion of immunoglobulin (36). This finding suggests that ELL family members may have functionally distinct roles during B cell differentiation. The presence and participation of ELL and ELL3 are poorly understood within the B cell lineage. In this study, we demonstrate the expression pattern ELL family members within the B-cell compartment and in B cell lymphoma, including a switch from ELL3 in fully matured and activated B cells to ELL2 upon plasma cell differentiation. Furthermore we establish that ELL3 is critical for proliferation and survival of B cell lymphoma cells.
2. Materials and Methods
2.1 Cell line culture
The CA46, Raji, Namalwa, Ramos, Mino, Jeko-1, Maver-1, Z138, U266, NCI-H929, Jurkat, and HEK-293T cell lines were purchased (ATCC, Manassas, VA). The 207 and 697 cell lines were provided by Dr. P.D. Burrows (University of Alabama Birmingham). The HBL2, Toledo, BJAB, OCI-Ly19, SU-DHL-4, and Pfeiffer cell lines were provided by Dr. J. Tao (H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL). The U2932 cell line was provided by Dr. I.S. Lossos (Sylvester Comprehensive Cancer Center, Miami, FL) (37). Cells were cultured in Hyclone RPMI 1640 or DMEM media supplemented with 10% FBS (GE Healthcare Life Sciences, Pittsburgh, PA) and 1% penicillin-streptomycin and for NCI-H929 only 55 μM 2-mercapotoethanol.
2.2 Primary cell stimulation
EasySep Human Naïve B-cell Enrichment Kit was utilized to negatively select B cells from peripheral blood mononuclear cells according to manufacturer’s protocol (STEMCELL Technologies Inc., Vancouver, Canada). In vitro stimulus was done at 1×106 cells/ml for 5 days with 100 U/ml IL-2 and 100 ng/ml IL-4 (PeproTech, Rocky Hill, NJ) for activation or 100 U/ml IL-2, 100 ng/ml IL-21, 5 μg/ml unlabeled goat anti-human IgM antibody (SouthernBioTech, Birmingham, AL), 10 ng/ml Histidine tagged CD40L, and 10 μg/ml polyHistidine antibody (R&D Systems Inc., Minneapolis, MN) for differentiation, as described previously (38).
2.3 ChIP-sequencing, data processing, and direct ChIP
PRDM1 enriched chromatin was prepared from a total of 2×107 cells using 5 μg of PRDM1 (C14A4) rabbit mAb (Cell Signaling Technology, Beverly, MA) as described previously (39). ChIP-seq was done on the U266 cell line using chromatin from 2 ×108 cells. Sequencing was performed by the Molecular Genomics Core Facility at the H. Lee Moffitt Cancer Center & Research Institute. 50 ng of PRDM1- or input DNA was used to generate sequencing libraries using the Illumina TruSeq Library Preparation Kit and sequenced on an Illumina HiScan SQ sequencer to generate approximately 15 million 50 base paired-end reads. The raw sequence data were de-multiplexed using the Illumina CASAVA 1.8.2 and aligned using BowTie (40). PRDM1 binding sites were identified using the MACS v1.4 peak-finding software and enriched for 50 or more mapped reads in peak located within 10 kb of a promoter, within a gene or within 2 kb of the 3′UTR and a False Discovery Rate of less than or equal to 5% (41). Data is deposited in GEO database under GSE102360. For direct-ChIP, PRDM1- or IgG-enriched DNA were analyzed by qPCR using primers described Supplemental Table I. Primers to HLA-DRA promoter were used as negative control for specificity. Ct values for each sample were linearized and the percentage over input calculated.
2.4 Immunoblotting
Immunoblotting procedure was as described previously (42). Primary antibodies include: ELL3 (1:300 dilution) (#H000080237-B02P lot WuLz 08310, and H00080237-B01P lot E1172, 08295 WuLz, Abnova, Taipei City, Taiwan), ELL2 (1:10,000 dilution) (A302-505A; Bethyl Laboratories Inc., Montgomery, TX), ELL (1:800 dilution) (#51044-1-AP, Proteintech Group, Chicago, IL), β-actin (1:12,000 dilution) (AC-15, Sigma Aldrich, St. Louis, MO), PARP (46D11), Phospho-Histone H2A.X (Ser139) (#2577), Cleaved Caspase-3 (Asp175) (#9661), Cyclin B1 (V152), Phospho-Cyclin B1 (Ser133) (9E3), p53 (1C12) and HA-Tag (C29F4), Cell Signaling Technology, Danvers, MA). Horse radish peroxidase conjugated secondary antibodies were purchased from GE Healthcare Life Sciences (Pittsburgh, PA), and IRDye conjugated secondary antibodies (IRDye®800CW, 926-32210 and IRDye®680RD, 926-68071) were purchased from LI-COR Biotechnologies (Lincoln, NE).
2.5 Quantitative mRNA analysis
RNA was isolated using the E.Z.N. A. Total RNA Kit I (Omega Bio-Tek, Norcross, GA), cDNA was prepared with the qScript cDNA synthesis Kit (Quanta Biosciences Inc., Gaithersburg, MD) and diluted one to eleven with purified water. 3 μl of the diluted cDNA sample was analyzed in duplicate at primer set specific annealing temperatures. Expression was analyzed using the ΔΔCt method, with 18S as a normalization gene (43). Primer sequences are described in Supplemental Table I.
2.6 DNA constructs
The −587 to +343 ELL3 proximal promoter region was PCR cloned into pCR2.1 (Invitrogen Life technologies, Grand Island, NY) from human genomic DNA. The XhoI-KpnI fragment was subcloned into pGL3-basic (Promega, Madison, MI), generating pGL3-ELL3-WT. Mutations in the PRDM1 binding sites were generated by substitution mutagenesis. pGL3-ELL3-Mut I eliminates the −239 to −229 PRDM1 site, substituting 5′-AACTTTCACTG-3′ with 5′-AgagcTCACTG-3′ and generating a SacI site. pGL3-ELL3-Mut II eliminates the +14 to +24 PRDM1 site, substituting 5′-AGCTTTCACTT-3′ with 5′-AGCggTacCTT-3′, and generating a KpnI site. pGL3-ELL3-Mut I & II was created through SacII-XhoI fragment subcloning from the single mutant constructs. Transfections and analysis were described previously and utilized 15 μg of promoter constructs, 5 μg of empty pcDNA3.1 or pcDNA3.1-HA-PRDM1α and 0.5 μg pRL-TK internal control construct (39, 42). The ELL3-expression plasmid was created from Raji cell cDNA, PCR amplified and cloned into pCR2.1. The KpnI/EcoRV fragment was subcloned into KpnI/PmeI fragment of the previously described pcDNA3.1-HA-PRDM1α construct (44). Utilized primers are described in Supplemental Table I. All vectors were verified by sequencing.
2.7 siRNA and lentiviral shRNA constructs
siRNA knockdown was performed using the ELL3-specific siRNA SMARTpool (E-014601-00-0005) and the non-targeting control Accell siRNA (D-001910-01-50) (GE Dharmacon, Lafayette, CO) according to the manufacturers protocol. shRNA knockdown was done using shRNA vectors MISSSION® TRC2 pLKO.5-puro ELL3shRNA (ELL3sh-1, TRCN0000289149; and ELL3sh-2, TRCN0000296220), and MISSSION® TRC2 pLKO.5-puro Non-Mammalian control shRNA (NTsh; SHC202; Sigma Aldrich, St. Louis, MO). mCherry tagged shRNA vectors were generated by replacing the puromycin resistance gene in the above mentioned constructs with the mCherry gene from the pLVmCherry vector (Addgene, Cambridge, MA). Lentiviral particles were produced with jetPRIME transfection reagent (Polyplus transfection, Illkirch, France) in HEK-293T using 3rd generation lentiviral packaging mixture (Applied Biological Materials Inc., Richmond, Canada). Transduction was performed at 5×107 cells/ml for 2 h at 1500xg, room temperature (RT) in the presence of 0.6 μg/ml polybrene (Merck Millipore, Billerica, MA).
2.8 Proliferation and DNA replication
Bromodeoxyuridine (BrdU) incorporation was performed at day 5 post transduction on exponentially growing shRNA transduced cells at (2×105 cells/ml) for 30 min. Four biological replicates were generated over an 8 h period at 2 h intervals for two independent experiments and stained simultaneously with anti-Brdu-FITC (BD Biosciences, San Jose, CA) and 4′,6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI) (Sigma Aldrich, St. Louis, MO) according to manufacturer’s protocol. Flow cytometric detection was performed at the Flow Cytometry Core Facility at the H. Lee Moffitt Cancer Center & Research Institute using the LSRII (BD Biosciences, San Jose, CA). Cell cycle distribution was assessed in mCherry+DAPI+ cells using ModFit (Verity Software House, Topsham, ME) and incorporated BrdU levels was assessed in mCherry+DAPI+FITC+ cells using FlowJo (FlowJo, LLC, Ashland, OR).
2.9 Microscopy
Imaging was performed at the Analytic Microscopy Core Facility at the H. Lee Moffitt Cancer Center & Research Institute. To assess cell area 3 wells were plated per condition on a glass bottom 12-well plate (MatTek, Ashland, MA) at 5×105 cells/ml and 4 images were taken per well on a Zeiss inverted microscope (Carl Zeiss Microscopy, LLC, Thornwood, NY). Image studio software (LICOR, Inc, Lincoln, NE) was used to determine the cell area and categorization done using Excel analysis toolpak (Microsoft, Redmond, WA).
3. Results
3.1 ELL3 and ELL2 are differentially expressed in the B cell compartment
ELL2 is highly expressed in plasma cells where it contributes to secretion of immunoglobulin (36). Given the potential significance of ELL proteins in B cell function, we assessed the mRNA levels of all three family members (Figure 1A). As expected, ELL2 was strongly expressed in the two Multiple Myeloma (MM) plasma cell lines, U266 and NCI-H929, but not the two Burkitt’s Lymphoma (BL) B cell lines, CA46 and Raji. Low, but detectable levels of ELL were found in all four cell lines. In direct contrast, ELL3 expression was observed in both BL cell lines but not the MM cell lines. Our re-analysis of microarray expression from Longo et al. on purified human tonsillar B cell sub-populations, revealed ELL2 mRNA selectively in the terminally differentiated plasma cell population (Figure 1B) (45). Consistent with its known role in plasma cell differentiation, PRDM1 was also exclusively detected in plasma cells (46, 47). In contrast, ELL3 mRNA was highly expressed in primary GC B cells and to a lesser extent in the naïve and memory B cells, while ELL mRNA was minimally expressed across the B-cell compartment. In a previous study of mRNA processing in plasma cells, Benson et al. demonstrated selective ELL2 mRNA expression in the murine MCP11 plasmacytoma cell line and murine plasma cells while ELL3 mRNA was most abundantly detected in the murine splenic B cells and the murine A20 GC B cell line (48). These findings indicate that the expression pattern of ELL family members are highly conserved between human and mouse.
Figure 1. Differential expression of ELL family members in B cell lines and primary B cells.
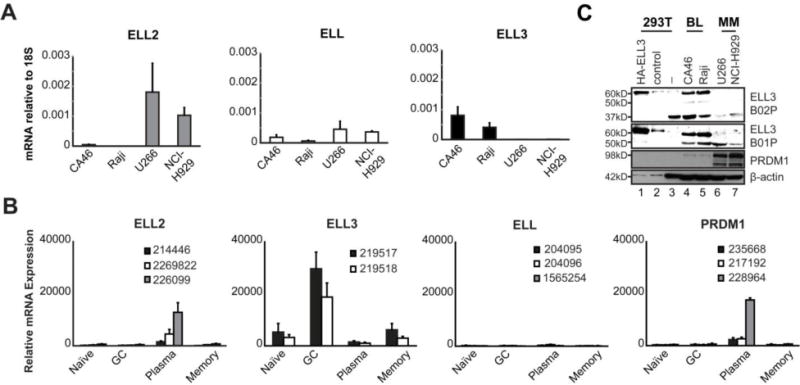
A. The relative mRNA quantitation of ELL family members in BL and MM cell lines. Data is the average of 3 independent experiments; errors bars represent standard deviation (SD). B. Expression levels of all ELL family members were obtained from GSE12366 (45). Depicted values are an average of 3 microarray hybridizations on cell sorted primary human B-cell subpopulations; error bars represent SD. Probe ID number is indicated in each graph. C. ELL3 immunoblot using two independent antibodies detects expression in BL lines but not MM lines. Cell lines are indicated above each lane. Whole cell lysates from 0.5×106 cells were used for each except HEK-293T lysates were diluted 1:50.
3.2 Abundant ELL3 protein expression in B cells
We next investigated if the ELL3 mRNA data was reflected at the protein level. Several ELL3 reactive antibodies are commercially available, however validation is minimal. To establish specificity, ELL3 was depleted in BL cell lines with a siRNA cocktail. The ELL3 B02P antibody recognized a protein band of the expected 60kD molecular weight in the un-transduced and control cells that was eliminated in the ELL3-depleted cells (Supplemental Figure 1). In both BL cell lines, two additional protein bands of unknown origin (50kD and 37kD) were also detected but not affected by ELL3-depletion. Over-expressed HA-tagged ELL3 was also detected at approximately 60kD by the B02P antibody and a second ELL3 antibody, B01P (Figure 1C). Both ELL3 antibodies specifically recognized ELL3 in the BL cell lines, CA46 and Raji but not in the MM cell lines (Figure 1C). These findings indicate that the ELL3 protein expression is observed in B cells prior to plasma cell differentiation.
3.3 ELL3 expression is primarily restricted to mature and activated B cells
To assess the differential expression of ELL family members across the B lineage, we profiled their expression in a panel of B cell lines at different stages of activation or differentiation (Figure 2A and Figure 1A Data in Brief (49)). ELL3 protein expression is robust in 8 of the 10 cell lines that represent the GC, including all of the BL lines and 3 of 5 Diffuse Large B cell Lymphoma (DLBCL) lines. Similarly, we found that ELL3 mRNA levels are the highest in these B cell lines. Low expression of ELL3 was observed in the Mantle Cell Lymphoma (MCL) naïve B cell line, Jeko-1, and to a lesser extent in the Mino MCL cell line. The MM cell lines have undetectable levels of ELL3 protein and mRNA. ELL protein and mRNA were at similar levels in most cell lines, but displayed comparatively low protein expression in the B cell Precursor Acute Lymphoblastic Leukemia (BCP-ALL) premature B cell line, 697, and low mRNA in the DLBCL line Toledo. The ubiquitous expression of ELL is consistent with prior findings that ELL is crucial for assembling the SEC and the little elongation complex (LEC) (28, 50). As expected, ELL2 protein and mRNA levels were most abundant in MM cell lines. However, we also observed ELL2 protein expression in one MCL and one DLBCL cell line. These results in immortalized cell lines broadly suggest that ELL expression is ubiquitous, while the family members ELL3 and ELL2 predominate at distinct stages of B cell differentiation. These findings further imply a switch from ELL3 expression to ELL2 occurring during B cell differentiation.
Figure 2. Profile of ELL family member expression during B cell differentiation and in lymphoma cell lines.
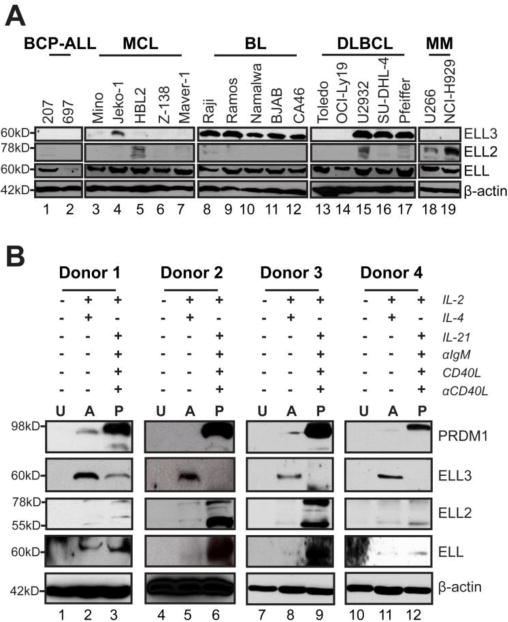
A. Immunoblot of ELL family members in B cell lymphoma cell lines. Cell line labels are BCP-ALL (B cell Precursor Acute Lymphoblastic Leukemia) MCL (Mantle Cell Lymphoma), BL (Burkitts’ Lymphoma), DLBCL (Diffuse Large B Cell Lymphoma) and MM (Multiple Myeloma). B. Immunoblot of ELL family members and PRDM1 in human primary naïve B cells un-stimulated (U), stimulate into activated B cells (A), and stimulated into plasmablasts (P). Data is from 4 representative healthy human donors.
3.4 ELL3 preceded the expression of ELL2 in terminally differentiated plasma cells
We next directly assessed ELL family member protein expression during ex vivo primary B cell activation and differentiation. Peripheral naïve B cells from healthy human donors had undetectable levels of the ELL family members (Figure 2B). Upon B cell activation, extensive ELL3 up-regulation as well as modest ELL expression was observed, while ELL2 levels remained minimal. In contrast, primary B cells stimulated to differentiate into plasma cells had minimal ELL3, while ELL2 was up-regulated. Additionally, ELL expression further increased upon differentiation. Robust up-regulation of PRDM1 in this population confirmed differentiation toward the plasma cell phenotype (Figure 2B). In support of this observation, we re-analyzed the global gene expression data from Barwick et al. on in vivo LPS stimulated adoptively transferred mouse B cells Figure 1B Data in Brief (49, 51)). As expected, ELL3 levels predominated during early CD138− B cell divisions. ELL3 expression diminished as B cells progressed towards the plasma cell phenotype and was extinguished in the CD138+ plasma cells.
This inversely coincided with PRDM1 expression. Furthermore, ELL2 expression was induced in plasma cells. Indicating that as in humans, murine B cells exhibit a switch from ELL3 to ELL2 during differentiation and suggests that these two ELL family members may have distinct roles within the B cell compartment.
3.5 PRDM1 directly associates and represses the ELL3 promoter
Terminal plasma cell differentiation is driven by transcriptional reprogramming mediated by the transcriptional repressor PRDM1 (46). Our lab and others have previously demonstrated that PRDM1 directly suppresses components of activated B cells, suggesting that PRDM1 might have a key role in regulating ELL family expression (47, 52, 53). In Figures 1A, 2B and Figure 1C, the expression of PRDM1 is shown to coincide with loss of ELL3 expression. To directly assess the functional role of PRDM1, we analyzed microarray expression data of human tonsilar GC B cells transfected with a PRDM1 expression plasmid which we previously used to demonstrate a role for PRDM1 in Epstein Barr Virus (EBV) pathogenesis (54). ELL3 expression was reduced by 19.1% and 38.5% in two independent donors upon expression of PRDM1 (Table 1). This effect on ELL3 paralleled the impact on well characterized targets of PRDM1, including CIITA and BCL6. We utilized ChIP-Seq in the U266 MM cell line for PRDM1 associations and identified a predominant PRDM1 association centered 180 base pairs upstream of the ELL3 transcription start site (Figure 3A). The association was confirmed by direct ChIP in both PRDM1-positive MM cell lines (Figure 3B). We identified two consensus PRDM1 binding sites at the ELL3 locus, termed site I (−239 to −229) and site II (+14 to +24). Both sites are located in the proximal promoter region within the area defined by ChIP-seq. To assess the effect of PRDM1 on ELL3 promoter activity, a DNA fragment containing both sites was cloned into a luciferase-reporter construct (Figure 3C). When transfected into Raji BL cells the ELL3 promoter demonstrated a high level of basal activity that was repressed by approximately 50% when co-transfected with a PRDM1 expression construct (Figure 3D). This degree of repression is consistent with known PRDM1 regulated genes including CIITA, LMO2 and HGAL (52, 55). The PRDM1-mediated repression was lost by simultaneous mutation of both sites without altering the basal promoter activity. However, mutation of either site independently did not impair PRDM1 mediated repression. These results indicate that the ELL3 promoter is suppressed by direct PRDM1 binding and either site is sufficient to mediate transcriptional repression.
Table 1. Microarray signal intensity in tonsillar GC B cells transiently transfected with either control or PRDM1.
Expression levels of ELL3 and the known PRDM1-silenced genes in purified human tonsillar GC B cells transfected with either a PRDM1 expression construct (pcDNA3.1-PRDM1α) or an empty vector (pcDNA3.1). The microarray data set source is GSE27670 (54). Depicted values are fold change over control of from 2 independent donors. Probe ID number is indicated in each graph.
| Probe ID | Gene symbol | Donor 1 | Donor 2 | ||
|---|---|---|---|---|---|
| +pcDNA3.1 | +pcDNA3.1-PRDM1α | +pcDNA3.1 | +pcDNA3.1-PRDM1α | ||
| 217192 | PRDM1 | 64.8 | 7095.7 | 44.8 | 3364.0 |
| 219518 | ELL3 | 3462.6 | 2802.4 | 2782.3 | 1712.3 |
| 203140 | BCL6 | 3144.1 | 2261.8 | 3632.7 | 2312.2 |
| 205101 | CIITA | 490.5 | 400.5 | 642.7 | 110.5 |
Figure 3. PRDM1 mediates repression of ELL3 promoter activity.
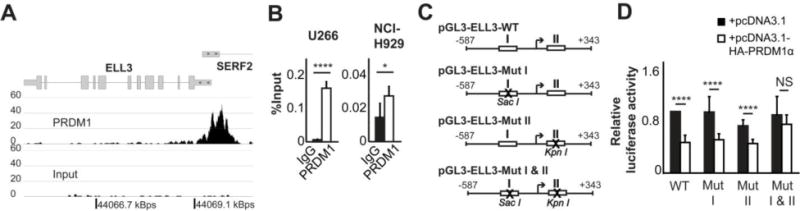
A. Plot of PRDM1 ChIP-seq reads at the ELL3 locus in the U266 MM cell line. B. ChIP-qPCR assessment of PRDM1 binding at ELL3 in MM lines. Data is represented as the average of at least 4 independent experiments. C. Schematic depiction of the cloned 930 nt ELL3 promoter constructs. The two PRDM1 sites are indicated with I and II. D. Promoter activity of ELL3 promoter constructs co-transfected with either control or PRDM1 expression vector. Data represents the average of 6 independent experiments; error bars represent SD.*p<0.05, ****p<0.0001 (two-tailed t-test).
3.6 ELL3 is necessary for proliferation and cell cycle progression
To assess ELL3 function, we transiently depleted ELL3 in Namalwa BL cells using two independent ELL3-shRNAs. Depletion of ELL3 protein and mRNA expression was highly efficient and was not accompanied by compensatory up-regulation of ELL or ELL2 (Figure 4A and 4B). Unexpectedly, PRDM1 expression was somewhat increased in ELL3-depleted cells (Figure 2B Data in Brief (49)). To establish if the increase of PRDM1 caused reactivation of latent EBV into lytic replication (56), we assessed expression of the immediate early- (BZLF1), early- (BMRF1) and late- (BLLF1) lytic replication factors (Figure 2C Data in Brief (49)). All genes remained un-affected indicating no change in EBV status. We also assessed if induction of PRDM1 caused B cell differentiation by repressing B cell factors BCL6, PAX5, MYC and secretion of immunoglobulin (Figure 2D and 2E Data in Brief (49)). All genes remained unaffected indicating that ELL3 depletion does not cause any detectable changes in differentiation state.
Figure 4. ELL3 depletion inhibits B cell proliferation and cell cycle progression.
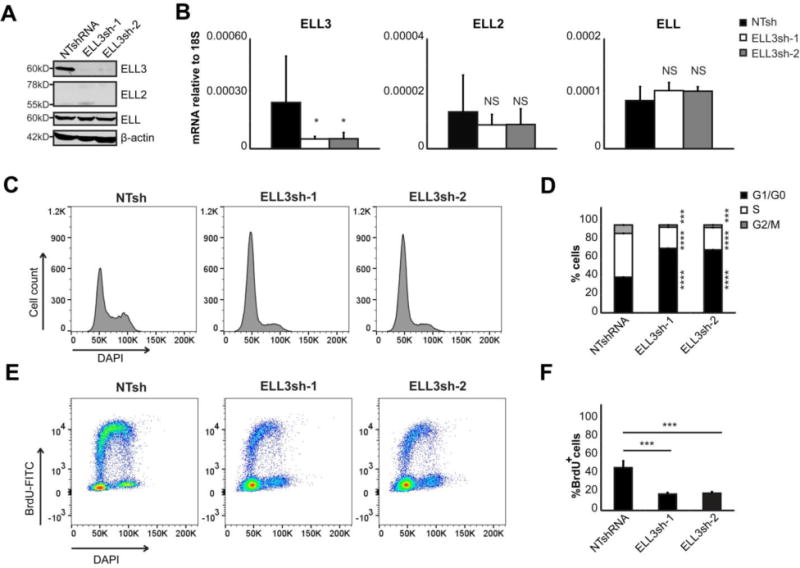
Namalwa cells were transduced with either NTsh, ELL3sh-1, or ELL3sh-2. A. Immunoblot of ELL3, ELL2 and ELL. The equivalent of 0.5×106 whole cell lysates were assayed. Data depicts representative images of 3 independent experiments. B. The relative mRNA quantitation of all ELL family members after ELL3 depletion. Data is presented as the average of 5 independent experiments; errors bars represent SD. *p<0.05; NS is not significant (two-tailed t-test). C. Representative total DNA content profile after ELL3 depletion measured by DAPI staining and flow cytometry. D. Cell cycle distribution after ELL3 depletion. Cells stained with DAPI for DNA content and gated on mCherry+ as marker of shRNA transduced cells. 150,000 cells were modeled with ModFit. Depicted are percentages at each stage of the cell cycle. E. Representative dual color fluorescence density plot of DNA-DAPI and BrdU-FITC from cells pulsed with BrdU after ELL3 depletion. F. DNA replication detected by BrdU pulse labeling (30 min) and modeled for at least 190,000 cells. Data is depicted as percentage of total mCherry+ cells. Data in panels C-F are from 2 biological independent experiments with 4 replicates each. All assays were performed on day 5 post-transduction. Errors bars represent SD in percentage. *p<0.05, **p<0.01, ***p<0.001, ****p<0.00001 (two-tailed t-test).
We observed less cell growth upon ELL3 depletion in BL cell lines and were unable to establish stable cell lines in the absence of ELL3. Similar results were observed in the Raji BL cell line (data not shown). This finding suggests that ELL3 deficiency may compromise cell cycle progression or viability. We addressed this by assessing total DNA content by flow cytometry and observed an altered cell cycle distribution in ELL3-depleted conditions (Figure 4C and 4D). Based on total DAPI levels, we determined the cell cycle distribution. Approximately 42% of control cells were in G1/G0 and this was elevated to approximately 75% after ELL3 depletion (black bars). Similarly, the S-phase population decreased in ELL3-depleted cells to 24% (ELL3sh-1) and 23% (ELL3sh-2), compared to 52% in control cells (white bars). Furthermore, the G2/M population peaked at 12.6% in control cells while remaining below 4% in ELL3-depleted cells (grey bars). We next directly assessed DNA replication with BrdU pulse labeling and observed compromised levels in ELL3-depleted conditions (Figure 4E and 4F). Only 20% of ELL3-depleted cells incorporated BrdU compared to approximately 50% of the control cells, indicating that DNA replication was compromised in ELL3-depleted cells. In combination, these findings suggest that ELL3-depleted cells arrest in G1/G0 and are limited in the ability to replicate DNA.
3.7 ELL3 depletion results in DNA damage and manifests as morphological aberrations
A diminished DNA replication and cell cycle progression suggest a compromised genomic integrity (57). We assessed the presence of DNA damage in ELL3-depleted conditions and observed increased levels of the DNA damage marker phosphorylated H2AX (γH2AX) (Figure 5A). This increase was not accompanied by an increase of the DNA damage checkpoint gene p53 (Figure 5A) (57). In addition, ELL3-depleted B cells undergo morphological changes consistent with loss of genomic integrity. Measurement of cell granularity and size was done by the forward scatter (FSC) and side scatter (SSC) flow cytometric parameters. ELL3 depletion increased both parameters and the cells appeared visually larger (Figure 5B and 5C).
Figure 5. ELL3 depletion results in morphological aberrations.
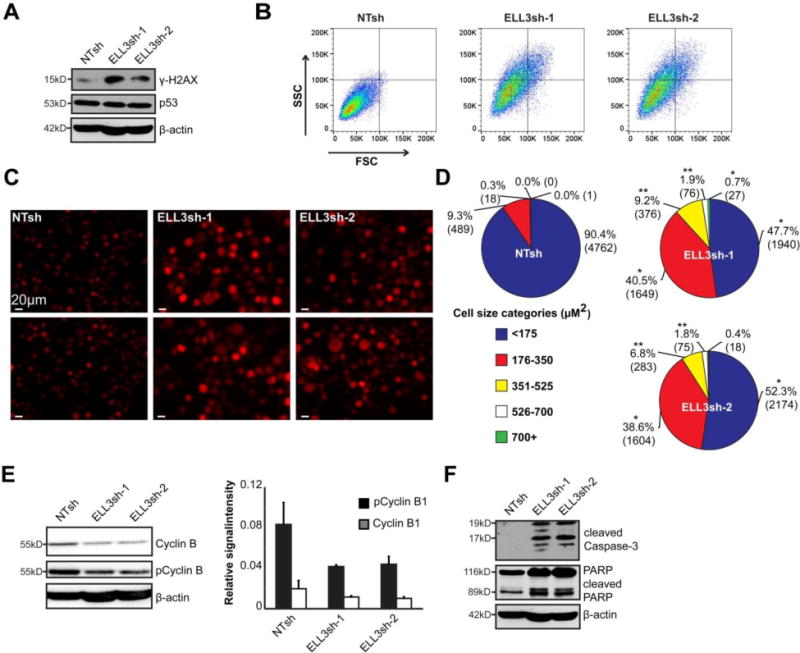
Namalwa cells were transduced with NTsh, ELL3sh-1, or ELL3sh-2 and assayed at day 5 post-transduction. A. Immunoblot of Ser139 phosphorylated H2AX and total p53. Data depicts representative images of 3 independent experiments. B. Cell size and granularity were assessed by flow cytometric detection of Forward Scatter (FSC) and Side Scatter (SSC) signals in mCherry+ cells. Data is representative of 2 independent experiments. C. Imaging of mCherry+ shRNA transduced cells at day 6 post transduction indicates alterations in cell size. Two representative images are shown per condition. D. Quantitation of cell area was determined from at least 4000 mCherry+ imaged cells per condition and categorized into 5 size bins. Data is presented as the percentage of total cells per cell size category across 3 independent experiments. Total number of cells indicated in parentheses. *p<0.05, **p<0.01 (two-tailed t-test). E. Immunoblot of Cyclin B1 and Ser133 phosphorylated Cyclin B1 levels. Fluorescence intensity of Cyclin B1 and pCyclin B1 was quantified and normalized to β-actin. Bar graph on right represents average fluorescence intensity of 3 independent experiments. F. Immunoblot of cleaved Caspase-3, and its substrate PARP. Data depicts representative images of 3 independent experiments. All assessments were done at day 5 post transduction unless otherwise stated.
Measurement and categorization of cell sizes indicate that while 90% of control cells were in the smallest cell size range of <175 μM2, comparatively only 48% and 52% of ELL3-depleted populations were within this category (Figure 5D). Overall, ELL3-depleted cells had a significantly higher representation within the 176–350 μM2 size range, at 40% (ELL3sh-1) and 39% (ELL3sh-2), compared to only 9% of control cells. In addition, while only 0.3% of the control cells were larger than 350 μM2, 10–12% of the ELL3-depleted cells were found in size ranges, representing cells more than double the normal cell volume. These findings were accompanied by the observation that ELL3-depleted cells exhibit mitotic aberrations over time, which include failure to divide, increased nuclear size, multiple nuclei and cells with aberrant cytokinesis (Figure 2A Data in Brief (49)). Together these findings suggest that ELL3 depletion dramatically perturbs cell growth and division.
The M-phase promoting factor (MPF) Cyclin B1-CDK1 complex is the key initiator of mitosis (58, 59). To assess if ELL3-depletion affected this complex, we probed for both total and phosphorylated Cyclin B1 (pCyclinB1) levels. In ELL3-depleted cells, both total and pCyclin B1 levels were diminished (Figure 5F). The Cyclin B1-CDK1 complex also has a well established role in regulating cell viability through survivin-mediated regulation of apoptosis (60, 61). Given the depleted levels of Cyclin B1, we assessed if the apoptosis pathway was induced. Both ELL3-depleted conditions displayed higher levels of cleaved caspase-3 and cleavage of its target, poly ADP ribose polymerase (PARP), indicating activation of apoptosis (Figure 5F). Collectively, these findings indicate that depletion of ELL3 compromises mitotic regulators resulting in mitotic disarray and loss of survival.
4. Discussion
In this report, we have established ELL3 as a factor that is up-regulated in B cells and subsequently down-regulated by the expression of PRDM1 during terminal plasma cell differentiation. This finding is consistent with ELL3 participation in the release of paused RNA Pol II required for activation of B cells (1, 15, 62). The expression of ELL3 is required for proliferation and survival of B cells and ELL3 expression lymphoma cell lines. Interestingly, over-expression of ELL in a HEK-293 cell model also impaired viability (63). This suggests that a regulated expression level of the ELL family members is required for proper cell proliferation and viability.
ELL family members were initially described to function as components of the SEC (29). Here, our findings illustrate that expression of ELL3 precedes ELL2 during B cell differentiation, a finding that is consistent with the existence of distinct SEC-like complexes. Known to contain P-TEFb with various combinations of the transcriptional elongation factors of the AFF family, YEATS domain containing protein family members, and ELL family members, SEC-like complexes are thought to have target specificity (64). An examination of murine embryonic stem cells revealed that ELL3 is preferentially bound at transcription enhancers. In addition the study indicated that ELL3 functions prior to ELL2 during development to establish gene promoter occupancy and allow the ELL2/SEC transcription program (32). Whether ELL3 and ELL2 have a similar linked activity in B cells or independently regulate specific genes remains to be investigated. However our data indicates that ELL3 and ELL2 are not co-expressed in B cells and thus unlikely to be present at the same promoter. Although not concurrently expressed in B cells, ELL3 and ELL2 are each observed in the presence of ELL. This finding is consistent with previous observations that even when all three proteins are over-expressed concurrently the complexes contain ELL with either ELL2 or ELL3 but not both (27). The presence of other SEC elongation factors is yet to be determined, however we postulate that differential usage of ELL3 and ELL2 may contribute to target specificity of SEC-like complexes in the B cell compartment (28). In addition to promoting elongation, it remains to be determined if the presence of ELL3 also alters RNA splicing as proposed for ELL2 at the immunoglobulin locus (36, 48, 65).
We have also shown that PRDM1, a master regulator of B cell differentiation into plasma cells, binds the ELL3 promoter in intact cells and directly represses the cloned ELL3 promoter. The cloned promoter was not completely inactivated by PRDM1 suggesting that additional regulatory factors, such as IRF4, may contribute to silencing. PRDM1 has previously been reported to have activating functions in mouse plasma blast (66). There ELL2 was reported as a PRDM1 activated gene, suggesting that the ELL3-to-ELL2 switch can be attributed to both the repressive and activating functions of PRDM1. Direct analysis of ELL3 transcriptional activation has not been reported. However, studies of the Early B-cell Factor 1 (EBF1) in vivo binding sites in murine B cells reveals association at the ELL3 loci and EBF1 expression correlates with ELL3 transcript levels (67, 68). EBF1, which is known for its roles in lineage specification and activation of B cells may serve as a pioneer factor to activate future ELL3 expression during B cell activation.
ELL3 has limited expression in normal human tissues with predominant presence in lymph nodes and spleen (69). Previous studies in human breast cancer cells revealed ELL3 expression and implicated it in tumor cell proliferation and survival mechanisms (33, 35). Our studies now show that malignantly transformed GC B cell lines express abundant ELL3 but not ELL2. At the protein level, all BL and three out of five DLBCL cell lines tested expressed abundant ELL3 levels, indicating a possible dichotomy within DLBCL for ELL3 function. This finding is consistent with the heterogeneity observed within DLBCL. Several molecular subtypes of DLBCL are defined based on differing origin, host responses, and genetic heterogeneity, including inactivating mutations of PRDM1 (70–74). Interestingly ELL3 expression in the DLBCL lines did not correlate with the known classifications. Further exploration of this dichotomy may be a useful marker in future studies to distinguish DLBCL subtypes and responses.
5. Conclusions
In summary, this study establishes ELL3 as a B-cell specific factor whose expression precedes terminal differentiation. ELL3 is abundant in multiple germinal center derived B-cell lymphoma lines and its expression maintains their capacity to proliferate and survive. These findings suggest that ELL3 may be a viable therapeutic target in treatment of ELL3-positive B-cell lymphomas. Finally, this study provides new insights into the mechanisms underlying the burst of transcriptional activity required for normal B cell activation and function.
Supplementary Material
Highlights.
The transcriptional elongation factor ELL3 is up-regulated in activated B cells.
Two ELL family members are non-concurrently expressed at specific B cell stages.
The expression of ELL3 precedes expression of ELL2 during B cell differentiation.
ELL3 plays a critical role in proliferation and survival of B cell models.
PRDM1 (Blimp-1) silences expression of ELL3 in plasma cells.
Acknowledgments
The authors thank the Molecular Genomics-, Flow Cytometry- and Analytic Microscopy Cores at the Moffitt Cancer Center & Research Institute for their support and technical assistance.
Funding details
This work was supported by the National Cancer Institute of the National Institutes of Health Grants R01CA164641 to KLW and T32CA115308 to JW and JAR. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- ELL
Eleven-nineteen Lysine-rich Leukemia
- Pol II
RNA polymerase II
- SEC
Super Elongation Complex
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
The authors declare that there are no conflicts of interest.
References
- 1.Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 2.Ferguson SE, Han S, Kelsoe G, Thompson CB. CD28 is required for germinal center formation. J Immunol. 1996;156:4576–4581. [PubMed] [Google Scholar]
- 3.Kim CH, Rott LS, Clark-Lewis I, Campbell DJ, Wu L, Butcher EC. Subspecialization of CXCR5+ T cells: B helper activity is focused in a germinal center-localized subset of CXCR5+ T cells. J Exp Med. 2001;193:1373–1381. doi: 10.1084/jem.193.12.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chtanova T, Tangye SG, Newton R, Frank N, Hodge MR, Rolph MS, Mackay CR. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. J Immunol. 2004;173:68–78. doi: 10.4049/jimmunol.173.1.68. [DOI] [PubMed] [Google Scholar]
- 5.Xu J, Foy TM, Laman JD, Elliott EA, Dunn JJ, Waldschmidt TJ, Elsemore J, Noelle RJ, Flavell RA. Mice deficient for the CD40 ligand. Immunity. 1994;1:423–431. doi: 10.1016/1074-7613(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 6.Kawabe T, Naka T, Yoshida K, Tanaka T, Fujiwara H, Suematsu S, Yoshida N, Kishimoto T, Kikutani H. The immune responses in CD40-deficient mice: impaired immunoglobulin class switching and germinal center formation. Immunity. 1994;1:167–178. doi: 10.1016/1074-7613(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 7.Reinhardt RL, Liang HE, Locksley RM. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat Immunol. 2009;10:385–393. doi: 10.1038/ni.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howard M, Farrar J, Hilfiker M, Johnson B, Takatsu K, Hamaoka T, Paul WE. Identification of a T cell-derived b cell growth factor distinct from interleukin 2. J Exp Med. 1982;155:914–923. doi: 10.1084/jem.155.3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozaki K, Spolski R, Feng CG, Qi CF, Cheng J, Sher A, Morse HC, 3rd, Liu C, Schwartzberg PL, Leonard WJ. A critical role for IL-21 in regulating immunoglobulin production. Science. 2002;298:1630–1634. doi: 10.1126/science.1077002. [DOI] [PubMed] [Google Scholar]
- 10.Jacob J, Kassir R, Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. I. The architecture and dynamics of responding cell populations. J Exp Med. 1991;173:1165–1175. doi: 10.1084/jem.173.5.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu YJ, Zhang J, Lane PJ, Chan EY, MacLennan IC. Sites of specific B cell activation in primary and secondary responses to T cell-dependent and T cell-independent antigens. Eur J Immunol. 1991;21:2951–2962. doi: 10.1002/eji.1830211209. [DOI] [PubMed] [Google Scholar]
- 12.Inamine A, Takahashi Y, Baba N, Miyake K, Tokuhisa T, Takemori T, Abe R. Two waves of memory B-cell generation in the primary immune response. Int Immunol. 2005;17:581–589. doi: 10.1093/intimm/dxh241. [DOI] [PubMed] [Google Scholar]
- 13.MacLennan IC. Germinal centers. Annu Rev Immunol. 1994;12:117–139. doi: 10.1146/annurev.iy.12.040194.001001. [DOI] [PubMed] [Google Scholar]
- 14.Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 15.Kouzine F, Wojtowicz D, Yamane A, Resch W, Kieffer-Kwon KR, Bandle R, Nelson S, Nakahashi H, Awasthi P, Feigenbaum L, Menoni H, Hoeijmakers J, Vermeulen W, Ge H, Przytycka TM, Levens D, Casellas R. Global regulation of promoter melting in naive lymphocytes. Cell. 2013;153:988–999. doi: 10.1016/j.cell.2013.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin C, Garrett AS, De Kumar B, Smith ER, Gogol M, Seidel C, Krumlauf R, Shilatifard A. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC) Genes Dev. 2011;25:1486–1498. doi: 10.1101/gad.2059211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eissenberg JC, Ma J, Gerber MA, Christensen A, Kennison JA, Shilatifard A. dELL is an essential RNA polymerase II elongation factor with a general role in development. Proc Natl Acad Sci U S A. 2002;99:9894–9899. doi: 10.1073/pnas.152193699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerber M, Ma J, Dean K, Eissenberg JC, Shilatifard A. Drosophila ELL is associated with actively elongating RNA polymerase II on transcriptionally active sites in vivo. Embo j. 2001;20:6104–6114. doi: 10.1093/emboj/20.21.6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith ER, Winter B, Eissenberg JC, Shilatifard A. Regulation of the transcriptional activity of poised RNA polymerase II by the elongation factor ELL. Proc Natl Acad Sci U S A. 2008;105:8575–8579. doi: 10.1073/pnas.0804379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 24.Kao SY, Calman AF, Luciw PA, Peterlin BM. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- 25.Laspia MF, Rice AP, Mathews MB. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell. 1989;59:283–292. doi: 10.1016/0092-8674(89)90290-0. [DOI] [PubMed] [Google Scholar]
- 26.Thirman MJ, Levitan DA, Kobayashi H, Simon MC, Rowley JD. Cloning of ELL, a gene that fuses to MLL in a t(11;19)(q23;p13.1) in acute myeloid leukemia. Proc Natl Acad Sci U S A. 1994;91:12110–12114. doi: 10.1073/pnas.91.25.12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Byun JS, Fufa TD, Wakano C, Fernandez A, Haggerty CM, Sung MH, Gardner K. ELL facilitates RNA polymerase II pause site entry and release. Nat Commun. 2012;3:633. doi: 10.1038/ncomms1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shilatifard A, Conaway RC, Conaway JW. The RNA polymerase II elongation complex. Annu Rev Biochem. 2003;72:693–715. doi: 10.1146/annurev.biochem.72.121801.161551. [DOI] [PubMed] [Google Scholar]
- 30.Shilatifard A, Duan DR, Haque D, Florence C, Schubach WH, Conaway JW, Conaway RC. ELL2, a new member of an ELL family of RNA polymerase II elongation factors. Proc Natl Acad Sci U S A. 1997;94:3639–3643. doi: 10.1073/pnas.94.8.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller T, Williams K, Johnstone RW, Shilatifard A. Identification, cloning, expression, and biochemical characterization of the testis-specific RNA polymerase II elongation factor ELL3. J Biol Chem. 2000;275:32052–32056. doi: 10.1074/jbc.M005175200. [DOI] [PubMed] [Google Scholar]
- 32.Lin C, Garruss AS, Luo Z, Guo F, Shilatifard A. The RNA Pol II elongation factor Ell3 marks enhancers in ES cells and primes future gene activation. Cell. 2013;152:144–156. doi: 10.1016/j.cell.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahn HJ, Kim KS, Shin KW, Lim KH, Kim JO, Lee JY, Kim J, Park JH, Yang KM, Baek KH, Ko JJ, Park KS. Ell3 stabilizes p53 following CDDP treatment via its effects on ubiquitin-dependent and -independent proteasomal degradation pathways in breast cancer cells. Oncotarget. 2015;6:44523–44537. doi: 10.18632/oncotarget.5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahn HJ, Cha Y, Moon SH, Jung JE, Park KS. Ell3 enhances differentiation of mouse embryonic stem cells by regulating epithelial-mesenchymal transition and apoptosis. PLoS One. 2012;7:e40293. doi: 10.1371/journal.pone.0040293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahn HJ, Kim G, Park KS. Ell3 stimulates proliferation, drug resistance, and cancer stem cell properties of breast cancer cells via a MEK/ERK-dependent signaling pathway. Biochem Biophys Res Commun. 2013;437:557–564. doi: 10.1016/j.bbrc.2013.06.114. [DOI] [PubMed] [Google Scholar]
- 36.Martincic K, Alkan SA, Cheatle A, Borghesi L, Milcarek C. Transcription elongation factor ELL2 directs immunoglobulin secretion in plasma cells by stimulating altered RNA processing. Nat Immunol. 2009;10:1102–1109. doi: 10.1038/ni.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cubedo E, Gentles AJ, Huang C, Natkunam Y, Bhatt S, Lu X, Jiang X, Romero-Camarero I, Freud A, Zhao S, Bacchi CE, Martinez-Climent JA, Sanchez-Garcia I, Melnick A, Lossos IS. Identification of LMO2 transcriptome and interactome in diffuse large B-cell lymphoma. Blood. 2012;119:5478–5491. doi: 10.1182/blood-2012-01-403154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ettinger R, Sims GP, Fairhurst AM, Robbins R, da Silva YS, Spolski R, Leonard WJ, Lipsky PE. IL-21 induces differentiation of human naive and memory B cells into antibody-secreting plasma cells. J Immunol. 2005;175:7867–7879. doi: 10.4049/jimmunol.175.12.7867. [DOI] [PubMed] [Google Scholar]
- 39.Desai S, Bolick SC, Maurin M, Wright KL. PU.1 regulates positive regulatory domain I-binding factor 1/Blimp-1 transcription in lymphoma cells. J Immunol. 2009;183:5778–5787. doi: 10.4049/jimmunol.0901120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith MA, Maurin M, Cho HI, Becknell B, Freud AG, Yu J, Wei S, Djeu J, Celis E, Caligiuri MA, Wright KL. PRDM1/Blimp-1 controls effector cytokine production in human NK cells. J Immunol. 2010;185:6058–6067. doi: 10.4049/jimmunol.1001682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 44.Ren B, Chee KJ, Kim TH, Maniatis T. PRDI-BF1/Blimp-1 repression is mediated by corepressors of the Groucho family of proteins. Genes Dev. 1999;13:125–137. doi: 10.1101/gad.13.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Longo NS, Lugar PL, Yavuz S, Zhang W, Krijger PH, Russ DE, Jima DD, Dave SS, Grammer AC, Lipsky PE. Analysis of somatic hypermutation in X-linked hyper-IgM syndrome shows specific deficiencies in mutational targeting. Blood. 2009;113:3706–3715. doi: 10.1182/blood-2008-10-183632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 47.Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, Giltnane JM, Yang L, Zhao H, Calame K, Staudt LM. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 48.Benson MJ, Aijo T, Chang X, Gagnon J, Pape UJ, Anantharaman V, Aravind L, Pursiheimo JP, Oberdoerffer S, Liu XS, Lahesmaa R, Lahdesmaki H, Rao A. Heterogeneous nuclear ribonucleoprotein L-like (hnRNPLL) and elongation factor, RNA polymerase II, 2 (ELL2) are regulators of mRNA processing in plasma cells. Proc Natl Acad Sci U S A. 2012;109:16252–16257. doi: 10.1073/pnas.1214414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.[data set]; Alexander LEMM, Watters J, Reusch JA, Maurin M, Nepon-Sixt BS, Vrzalikova K, Alexandrow MG, Murray PG, Wright KL. Data supporting the functional role of ELL3 in B cell lymphoma cell line cells. 2017 doi: 10.1016/j.dib.2017.09.042. Submitted to Journal - Data in Brief. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu D, Smith ER, Garruss AS, Mohaghegh N, Varberg JM, Lin C, Jackson J, Gao X, Saraf A, Florens L, Washburn MP, Eissenberg JC, Shilatifard A. The little elongation complex functions at initiation and elongation phases of snRNA gene transcription. Mol Cell. 2013;51:493–505. doi: 10.1016/j.molcel.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barwick BG, Scharer CD, Bally AP, Boss JM. Plasma cell differentiation is coupled to division-dependent DNA hypomethylation and gene regulation. Nat Immunol. 2016;17:1216–1225. doi: 10.1038/ni.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cubedo E, Maurin M, Jiang X, Lossos IS, Wright KL. PRDM1/Blimp1 downregulates expression of germinal center genes LMO2 and HGAL. Febs j. 2011;278:3065–3075. doi: 10.1111/j.1742-4658.2011.08227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martins G, Calame K. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu Rev Immunol. 2008;26:133–169. doi: 10.1146/annurev.immunol.26.021607.090241. [DOI] [PubMed] [Google Scholar]
- 54.Vrzalikova K, Vockerodt M, Leonard S, Bell A, Wei W, Schrader A, Wright KL, Kube D, Rowe M, Woodman CB, Murray PG. Down-regulation of BLIMP1alpha by the EBV oncogene, LMP-1, disrupts the plasma cell differentiation program and prevents viral replication in B cells: implications for the pathogenesis of EBV-associated B-cell lymphomas. Blood. 2011;117:5907–5917. doi: 10.1182/blood-2010-09-307710. [data set] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ghosh N, Gyory I, Wright G, Wood J, Wright KL. Positive regulatory domain I binding factor 1 silences class II transactivator expression in multiple myeloma cells. J Biol Chem. 2001;276:15264–15268. doi: 10.1074/jbc.M100862200. [DOI] [PubMed] [Google Scholar]
- 56.Reusch JA, Nawandar DM, Wright KL, Kenney SC, Mertz JE. Cellular differentiation regulator BLIMP1 induces Epstein-Barr virus lytic reactivation in epithelial and B cells by activating transcription from both the R and Z promoters. J Virol. 2015;89:1731–1743. doi: 10.1128/JVI.02781-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bartek J, Lukas J. Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr Opin Cell Biol. 2001;13:738–747. doi: 10.1016/s0955-0674(00)00280-5. [DOI] [PubMed] [Google Scholar]
- 58.Fung TK, Poon RY. A roller coaster ride with the mitotic cyclins. Semin Cell Dev Biol. 2005;16:335–342. doi: 10.1016/j.semcdb.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 59.Hagting A, Jackman M, Simpson K, Pines J. Translocation of cyclin B1 to the nucleus at prophase requires a phosphorylation-dependent nuclear import signal. Curr Biol. 1999;9:680–689. doi: 10.1016/s0960-9822(99)80308-x. [DOI] [PubMed] [Google Scholar]
- 60.O’Connor DS, Grossman D, Plescia J, Li F, Zhang H, Villa A, Tognin S, Marchisio PC, Altieri DC. Regulation of apoptosis at cell division by p34cdc2 phosphorylation of survivin. Proc Natl Acad Sci U S A. 2000;97:13103–13107. doi: 10.1073/pnas.240390697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li F, Ambrosini G, Chu EY, Plescia J, Tognin S, Marchisio PC, Altieri DC. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- 62.Sprent J. Lifespans of naive, memory and effector lymphocytes. Curr Opin Immunol. 1993;5:433–438. doi: 10.1016/0952-7915(93)90065-z. [DOI] [PubMed] [Google Scholar]
- 63.Johnstone RW, Gerber M, Landewe T, Tollefson A, Wold WS, Shilatifard A. Functional analysis of the leukemia protein ELL: evidence for a role in the regulation of cell growth and survival. Mol Cell Biol. 2001;21:1672–1681. doi: 10.1128/MCB.21.5.1672-1681.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luo Z, Lin C, Guest E, Garrett AS, Mohaghegh N, Swanson S, Marshall S, Florens L, Washburn MP, Shilatifard A. The super elongation complex family of RNA polymerase II elongation factors: gene target specificity and transcriptional output. Mol Cell Biol. 2012;32:2608–2617. doi: 10.1128/MCB.00182-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Park KS, Bayles I, Szlachta-McGinn A, Paul J, Boiko J, Santos P, Liu J, Wang Z, Borghesi L, Milcarek C. Transcription elongation factor ELL2 drives Ig secretory-specific mRNA production and the unfolded protein response. J Immunol. 2014;193:4663–4674. doi: 10.4049/jimmunol.1401608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Minnich M, Tagoh H, Bonelt P, Axelsson E, Fischer M, Cebolla B, Tarakhovsky A, Nutt SL, Jaritz M, Busslinger M. Multifunctional role of the transcription factor Blimp-1 in coordinating plasma cell differentiation. Nat Immunol. 2016;17:331–343. doi: 10.1038/ni.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hagman J, Belanger C, Travis A, Turck CW, Grosschedl R. Cloning and functional characterization of early B-cell factor, a regulator of lymphocyte-specific gene expression. Genes Dev. 1993;7:760–773. doi: 10.1101/gad.7.5.760. [DOI] [PubMed] [Google Scholar]
- 68.Treiber T, Mandel EM, Pott S, Gyory I, Firner S, Liu ET, Grosschedl R. Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription- independent poising of chromatin. Immunity. 2010;32:714–725. doi: 10.1016/j.immuni.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 69.Wilhelm M, Schlegl J, Hahne H, Gholami AM, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, Mathieson T, Lemeer S, Schnatbaum K, Reimer U, Wenschuh H, Mollenhauer M, Slotta-Huspenina J, Boese JH, Bantscheff M, Gerstmair A, Faerber F, Kuster B. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 70.Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, Hurt EM, Zhao H, Averett L, Yang L, Wilson WH, Jaffe ES, Simon R, Klausner RD, Powell J, Duffey PL, Longo DL, Greiner TC, Weisenburger DD, Sanger WG, Dave BJ, Lynch JC, Vose J, Armitage JO, Montserrat E, Lopez-Guillermo A, Grogan TM, Miller TP, LeBlanc M, Ott G, Kvaloy S, Delabie J, Holte H, Krajci P, Stokke T, Staudt LM. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 71.Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2003;100:9991–9996. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Monti S, Savage KJ, Kutok JL, Feuerhake F, Kurtin P, Mihm M, Wu B, Pasqualucci L, Neuberg D, Aguiar RC, Dal Cin P, Ladd C, Pinkus GS, Salles G, Harris NL, Dalla-Favera R, Habermann TM, Aster JC, Golub TR, Shipp MA. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood. 2005;105:1851–1861. doi: 10.1182/blood-2004-07-2947. [DOI] [PubMed] [Google Scholar]
- 73.Mandelbaum J, Bhagat G, Tang H, Mo T, Brahmachary M, Shen Q, Chadburn A, Rajewsky K, Tarakhovsky A, Pasqualucci L, Dalla-Favera R. BLIMP1 is a tumor suppressor gene frequently disrupted in activated B cell-like diffuse large B cell lymphoma. Cancer Cell. 2010;18:568–579. doi: 10.1016/j.ccr.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pasqualucci L, Compagno M, Houldsworth J, Monti S, Grunn A, Nandula SV, Aster JC, Murty VV, Shipp MA, Dalla-Favera R. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. J Exp Med. 2006;203:311–317. doi: 10.1084/jem.20052204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tam W, Gomez M, Chadburn A, Lee JW, Chan WC, Knowles DM. Mutational analysis of PRDM1 indicates a tumor-suppressor role in diffuse large B-cell lymphomas. Blood. 2006;107:4090–4100. doi: 10.1182/blood-2005-09-3778. [DOI] [PubMed] [Google Scholar]
- 76.Ryan JL, Fan H, Glaser SL, Schichman SA, Raab-Traub N, Gulley ML. Epstein-Barr virus quantitation by real-time PCR targeting multiple gene segments: a novel approach to screen for the virus in paraffin-embedded tissue and plasma. J Mol Diagn. 2004;6:378–385. doi: 10.1016/S1525-1578(10)60535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hilscher C, Vahrson W, Dittmer DP. Faster quantitative real-time PCR protocols may lose sensitivity and show increased variability. Nucleic Acids Res. 2005;33:e182. doi: 10.1093/nar/gni181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu X, McCarthy PJ, Wang Z, Gorlen DA, Mertz JE. Shutoff of BZLF1 gene expression is necessary for immortalization of primary B cells by Epstein-Barr virus. J Virol. 2012;86:8086–8096. doi: 10.1128/JVI.00234-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


