Abstract
Bacillus subtilis flagella are not only required for locomotion but also act as sensors that monitor environmental changes. Although how the signal transmission takes place is poorly understood, it has been shown that flagella play an important role in surface sensing by transmitting a mechanical signal to control the DegS-DegU two-component system. Here we report a role for flagella in the regulation of the K-state, which enables transformability and antibiotic tolerance (persistence). Mutations impairing flagellar synthesis are inferred to increase DegU-P, which inhibits the expression of ComK, the master regulator for the K-state, and reduces transformability. Tellingly, both deletion of the flagellin gene and straight filament (hagA233V) mutations increased DegU phosphorylation despite the fact that both mutants had wild type numbers of basal bodies and the flagellar motors were functional. We propose that higher viscous loads on flagellar motors result in lower DegU-P levels through an unknown signaling mechanism. This flagellar-load based mechanism ensures that cells in the motile subpopulation have a 10-fold enhanced likelihood of entering the K-state and taking up DNA from the environment. Further, our results suggest that the developmental states of motility and competence are related and most commonly occur in the same epigenetic cell type.
Keywords: flagella, K-state, hag, DegU-P, viscous load
Graphical Abstract
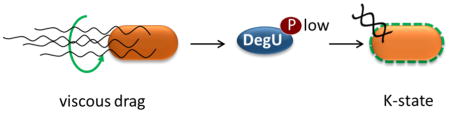
Viscous load on rotating flagella signals to lower the amount of DegU-P. This in turn increases the probability that Bacillus subtilis will enter the K-state.
Introduction
The most evident role for the rotary bacterial flagellum is in cell motility and chemotaxis (Berg & Brown, 1972, Adler, 1975, Purcell, 1977, Armitage, 1992). Remarkably, these molecular machines can also serve a sensory function, by detecting surface interactions to initiate the expression of biofilm genes (Belas, 2014, Ellison & Brun, 2015). The earliest evidence for a flagellar mechanosensory role is in the control of swarming in Vibrio parahaemolyticus (Belas et al., 1986, McCarter et al., 1988). In Vibrio cholerae, proper motor rotation is critical for permanent cell attachment, perhaps because flagellar interaction with a surface results in hyperpolarization of the membrane, an event that is correlated with attachment (Van Dellen et al., 2008). In Pseudomonas aeruginosa, the reversal frequency of flagella rotation plays a role in biofilm formation (Caiazza & O’Toole, 2004, Caiazza et al., 2007), while in Caulobacter crescentus, inhibition of flagella rotation initiates adhesion to a surface by stimulating holdfast synthesis (Li et al., 2012). Recent work in Escherichia coli suggests that within the motor, it is the stator (the torque-generating protein complex) that functions as a mechanosensor (Lele et al., 2013, Tipping et al., 2013, Che et al., 2014). Upon sensing an increase in the viscous load, the stator-complex recruits additional stator-units (Lele et al., 2013, Tipping et al., 2013). Although the mechanisms are unknown at present, stator-remodeling might be one way in which some bacterial species adapt to a surface-associated lifestyle.
In the Gram-positive model organism Bacillus subtilis, flagellar defects appear to cause the accumulation of the phosphorylated transcription factor DegU-P, resulting in the transcriptional up-regulation of poly-γ-glutamate synthesis (Cairns et al., 2013, Chan et al., 2014). It has been proposed that in the wild-type, interaction with a surface perturbs flagellar rotation and up-regulates genes needed for biofilm formation, including those dependent on DegU-P (Cairns et al., 2013). During the process of biofilm formation, the protein EpsE is synthesized, acting both as an essential enzyme for the production of the extracellular matrix and as a clutch, disengaging the flagellum from its motor and contributing to the transition from a motile to a sessile state (Blair et al., 2008, Guttenplan et al., 2010). Thus, flagellar rotation is intimately related to the formation of the B. subtilis biofilm at multiple levels.
B. subtilis contains typical ion-driven flagella, consisting of three architectural domains: the basal body, the hook and the filament (Berg, 2003, Chevance & Hughes, 2008, Minamino et al., 2008, Guttenplan et al., 2013, Mukherjee & Kearns, 2014). The filament is helical and is composed of repeating units of the protein flagellin, encoded by hag. The hook, a distinct structure, is located at the base of the filament and is also attached to the basal body, located in the cell envelope. The motor included in the basal body contains two functional entities; the rotor and the proton-conducting stator complex built by MotA and MotB (Block & Berg, 1984, Kojima & Blair, 2004). Protonation of the Asp24 residue in the B. subtilis MotB is thought to cause conformational changes in the cytoplasmic region of MotA, generating a torque at the rotor interface (the FliG ring) (Kojima & Blair, 2001, Cairns et al., 2013, Chan et al., 2014). Torque is then imparted to the filament through the rod, causing the flagellum to rotate.
While previous work in B. subtilis highlighted the role of flagella as mechanosensors for surface contact (Cairns et al., 2013) and for the induction of poly-γ-glutamate synthesis (Chan et al., 2014), we report a previously unrecognized effect of flagellar signaling in the regulation of the K-state. The K-state refers to a physiological condition in which ComK is expressed, along with about 100 genes dependent for their transcription on ComK (van Sinderen et al., 1995, Berka et al., 2002, Hamoen et al., 2002, Ogura et al., 2002).. Among the proteins encoded by these genes are those that enable the uptake and processing of transforming DNA; K-state cells are thus genetically competent for transformation. In addition, K-state cells are arrested for growth and division and are tolerant of a number of antibiotics (Hahn et al., 2015). Importantly, the K-state is bistably expressed, turning on in a subpopulation of cells. We show that mutations impairing flagella synthesis are less transformable due to the decreased basal expression of comK The lowering of the basal transcription rate, caused by the accumulation of DegU-P, results in a decreased frequency of transitions to the bistably expressed K-state (Mirouze et al., 2012). In addition to confirming that flagella function suppresses DegU-P, we show that the generation of this signal is dependent on the viscous drag experienced by the flagellar motor. We propose that increased viscous loads on the motor depress DegU phosphorylation, perhaps as a consequence of motor remodeling. This mechanism ensures that K-state development takes place predominantly in the motile sub-population of B. subtilis cells.
Results
comK transcription is low in the absence of flagella
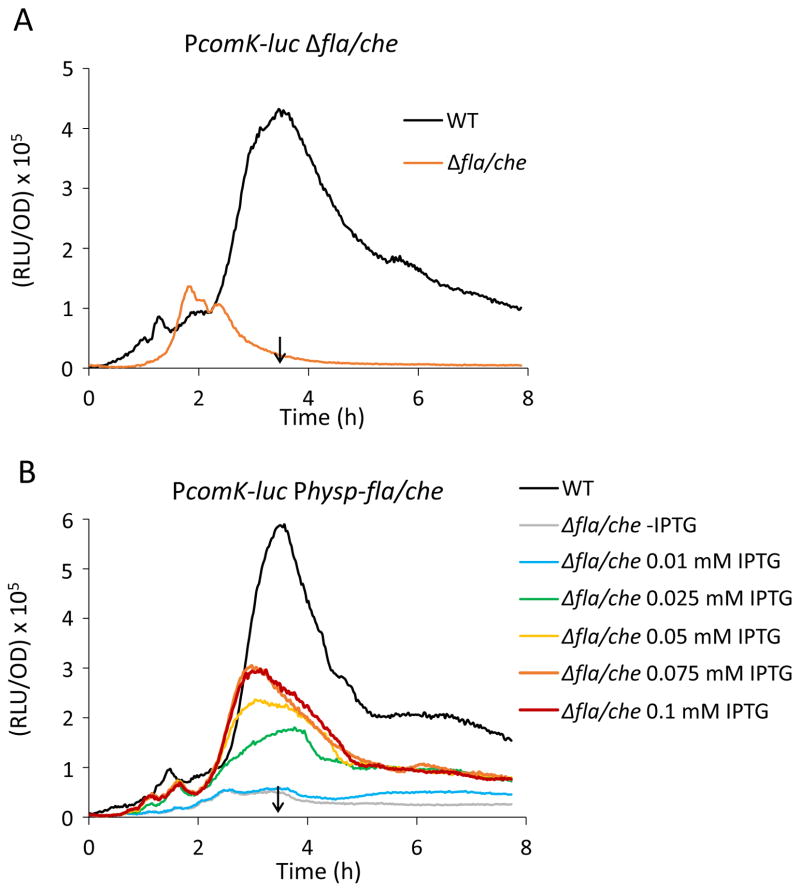
We observed that a deletion of the entire fla/che operon reduces the transcription rate from the PcomK promoter about 10-fold, shown using a promoter fusion to firefly luciferase as a reporter (Fig. 1A). The fla/che operon encodes many genes required for the synthesis of flagella and in its absence, motors, hooks and filaments are not produced (Zuberi et al., 1990, Marquez-Magana & Chamberlin, 1994, Mukherjee & Kearns, 2014). Additionally, we replaced the native promoter of the fla/che operon by an isopropyl-β-D-thiogalactoside (IPTG)-inducible Phyperspank promoter and measured comK expression in the absence or presence of different IPTG concentrations (Fig. 1B). comK expression was reduced in the absence of induction and increased in the presence of IPTG in a dose-dependent manner until it reached a maximum at 0.1 mM IPTG. In these experiments, carried out in the undomesticated ancestral strain 3610, comK expression peaked at the time of entry into the stationary phase (T0) at intermediate concentrations of IPTG, showing that the normal growth stage regulation of comK is unperturbed by the fla/che deletion.
Figure 1.
Low comK expression in the absence of flagella synthesis. (A) The rate of comK expression is inhibited in the fla/che deletion strain (BD7763) compared to its isogenic wild-type equivalent (BD6439). (B) Dose-dependent suppression of comK expression by flagellar synthesis. Transcription rates from the PcomK-luc reporter in a strain with an IPTG inducible fla/che operon in the native locus. Strain BD7695 (Physp-fla/che-operon PcomK-luc) was grown in the absence and presence of different concentrations of IPTG (as indicated). PcomK-luc expression in the wild-type (BD6439) is shown for comparison. All strains used for this experiment were built in the undomesticated background NCIB 3610. The vertical arrows in each panel point to the time of entry into stationary phase (T0), determined from optical density measurements during growth in the plate reader.
The inactivation of hag or of motB also lowers comK expression
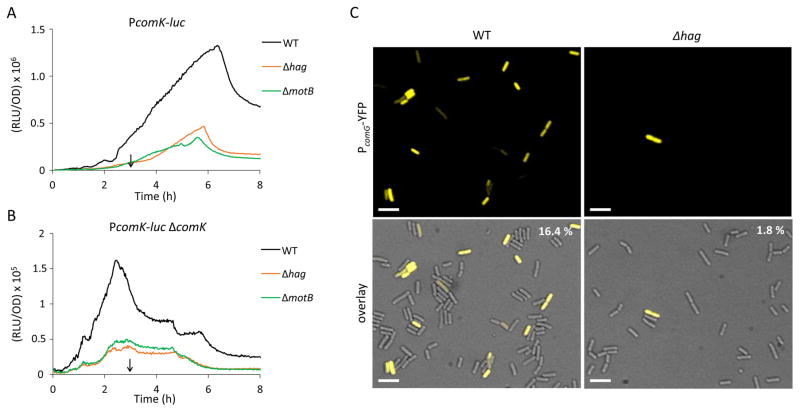
To determine whether the absence of individual flagellar components mimics that of the entire fla/che operon, we tested strains that were deleted for genes required for the most downstream flagellar components, hag or motB, which encode the flagellar filament and a stator protein that powers filament rotation. Both of these strains exhibited reductions in comK expression indistinguishable from that of the fla/che deletant (Fig. 2A). Consistent with this, the hag mutant (Table 1) was less transformable than the wild-type strain. Because the K-state is normally expressed in a subpopulation (Maamar & Dubnau, 2005, Suel et al., 2006, Maamar et al., 2007), we used a fusion of the comG promoter to yellow fluorescent protein (YFP) to determine whether fewer cells expressed this ComK-dependent promoter in the hag strain. Microscopic enumeration of PcomG-yfp expressing cells in the mutant and wild-type populations demonstrated about 10-fold fewer K-state cells in the hag strain than in the wild-type (Fig. 2C). In contrast, a non-polar deletion of cheA, a chemotaxis gene embedded in the fla/che operon, had no effect on K-state expression (not shown), suggesting that the chemotactic response is not needed for regulation. We conclude that the probability of transitions to the K-state is augmented by a signal that requires fully assembled and rotating flagella.
Figure 2.
Deletion of hag and motB lower the expression of comK and a Δhag mutation lowers the frequency of entry to the K-state. The effects of hag and motB knockouts on PcomK-luc expression in the presence (A) or absence (B) of ComK are shown. The following strains were used in these experiments: wild-type PcomK-luc (BD4773), Δhag PcomK-luc (BD7636), ΔmotB PcomK-luc (BD7466), ΔcomK PcomK-luc (BD4893), Δhag ΔcomK PcomK-luc (BD7261) and ΔmotB ΔcomK PcomK-luc (BD7488). The expression profiles in A and B are plotted on different scales. The vertical arrows in panels A and B point to T0. (C) Single-cell expression and microscopic enumeration of PcomG-yfp expressing cells in Δhag (BD7262) and wild-type (BD5783) populations. The indicated strains were grown to the time of maximum K-state expression (T2) and samples were taken for microscopy. Representative images are shown. In the upper right corner are the average percentages of K-state cells determined by counting at least 1000 cells for each strain, done in duplicate. Scale bar is 5μm.
Table 1.
Transformation frequencies
| strain | Relative transformation frequencya |
|---|---|
|
| |
| IS75 | 2.1x10−01 ± 0.093 |
| Δhag (BD7309) | 2.3x10−02 ± 0.006 |
Transforming DNA was incubated with competent cells for 30 min before plating for leucine prototrophy. The mean and standard deviation of three experiments is shown.
Deletion of hag, like that of motB, is inferred to raise the level of DegU-P
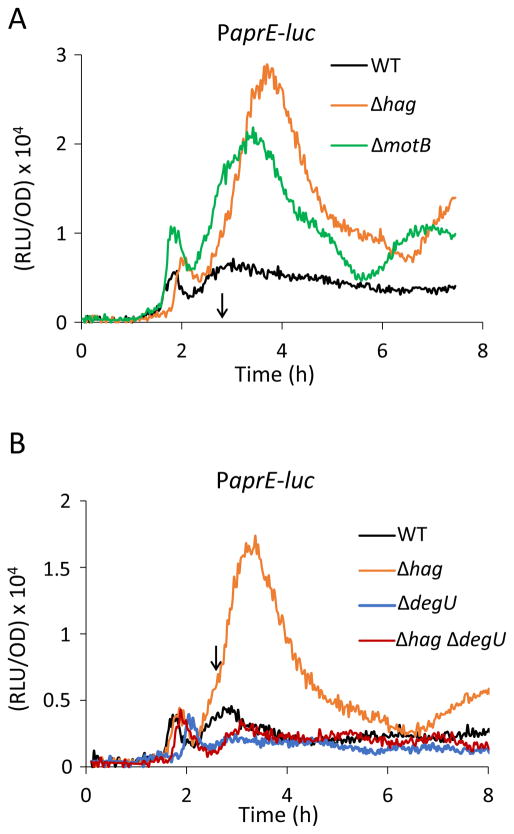
It has been shown in B. subtilis that flagella are not only important for motility, but act as mechanosensors to regulate exoprotease and poly-γ-glutamate synthesis on the level of gene expression by controlling the level of the phosphorylated response regulator protein, DegU-P (Cairns et al., 2013, Chan et al., 2014). It was proposed that the inhibition of flagellar rotation or of proton flux through MotB somehow signaled an increase in DegU-P, resulting in increased synthesis of poly-γ-glutamate in motB mutants (Cairns et al., 2013). Based on these findings, we suspected that an increase in DegU-P might be responsible for the phenotypes reported above, although it would seem that the elimination of hag may not lead to the cessation of hook rotation or to an interruption of proton flow, because the hook and stator assemblies would remain intact. To assess the level of DegU-P in the hag strain, we used DegU-P dependent promoter fusions as well as mutations in degU and its cognate histidine kinase, degS. Transcription of the alkaline protease subtilisin, encoded by aprE, is activated by high DegU-P (Dahl et al., 1992, Verhamme et al., 2007, Murray et al., 2009). To measure aprE transcription in the hag knockout, we used a promoter fusion of aprE to the luciferase gene and followed the expression of the PaprE during growth in hag and wild-type cells. As shown in Fig. 3, the inactivation of hag increased the expression of aprE, as shown previously for the motB deletion (Cairns et al., 2013). To inquire whether this increase depended on DegU, the degU gene was disrupted in the hag background. Although inactivation of degU did not affect the expression of aprE in the hag+ strain, aprE expression in the hag degU strain reverted back to the wild-type level (Fig. 3), confirming that DegU mediates the increase in aprE transcription caused by deletion of hag.
Figure 3.
DegU mediates the increase in aprE transcription caused by deletion of hag. (A) Increased expression of the PaprE-luc reporter in the hag and motB knock out strains. (B) aprE expression can be completely suppressed by a degU deletion in the hag background. The strains used were as follows: wild-type PaprE-luc (BD6904), Δhag PaprE-luc (BD7639), ΔmotB PaprE-luc (BD7719), ΔdegU PaprE-luc (BD7084) and Δhag ΔdegU PaprE-luc (BD7371). The vertical arrows in each panel point to T0.
High DegU-P in flagellar mutants represses the basal expression of comK
We next addressed which step in the comK signaling pathway is affected by the absence of flagella. Central to K-state decision-making is a positive feedback in which ComK directly activates its own promoter (van Sinderen & Venema, 1994, Maamar & Dubnau, 2005, Smits et al., 2005, Gamba et al., 2015). The decision to enter the K-state is governed by the basal level expression from PcomK, defined as expression in a ΔcomK mutant, in which the feedback is interrupted (Leisner et al., 2007, Mirouze et al., 2012). Basal expression normally rises during growth to a maximum at T0 and then decreases. Because a threshold basal concentration of ComK is needed to activate the positive feedback on PcomK, the probability that a cell will have enough ComK to induce transcription from PcomK increases and then decreases as cells enter the stationary phase of growth. This rise and fall in the basal rate of comK expression is due to the gradually increasing level of the phosphorylated master response regulator protein Spo0A-P during growth, because Spo0A-P activates and then represses comK transcription by directly binding to a series of operator sites in PcomK (Mirouze et al., 2012), Thus, within a temporal window of opportunity set by the rate of Spo0A-P accumulation, some cells in the population (~15% in the domesticated (laboratory) strains of B. subtilis) achieve a level of ComK sufficient to activate the positive feedback loop by binding to its own promoter. These cells switch to the K-state and become competent for transformation. Fig. 2B shows, using a PcomK-luc reporter in a ΔcomK strain, that the basal expression is lowered by the deletion of hag. This result suggests that the effect of the hag deletion is independent of ComK and its positive feedback loop. A motB mutant exhibits a similarly decreased basal level expression of comK (Fig. 2B). We conclude, based on the data in Figs. 1A and B, that when flagellar synthesis is perturbed, cells will have a lower chance to reach the threshold for ComK to activate its own expression and fewer cells will be able to enter the K-state and become competent for transformation. This result is consistent with the data in Fig. 2, which show a decreased frequency of K-state cells in the fla/che and hag deletants.
It has been shown that unphosphorylated DegU binds to the comK promoter (Hamoen et al., 2000). This binding increases the affinity of ComK for its own promoter, thus lowering the threshold of ComK concentration needed to activate positive auto-regulation. To determine whether high DegU-P can inhibit the basal expression of comK, we used degU32, a degUh allele that increases the amount of DegU-P (Henner et al., 1988, Dahl et al., 1992). This mutation caused the reduced expression of PcomK-luc in both wild-type and ΔcomK backgrounds (Fig. S1A and B), consistent with previous reports (Steinmetz et al., 1976, Msadek et al., 1990, Dahl et al., 1992). It is not known if this negative effect is exerted directly on PcomK, but it is consistent with the possibility that the effect of flagellar mutations on K-state expression is mediated by DegU-P.
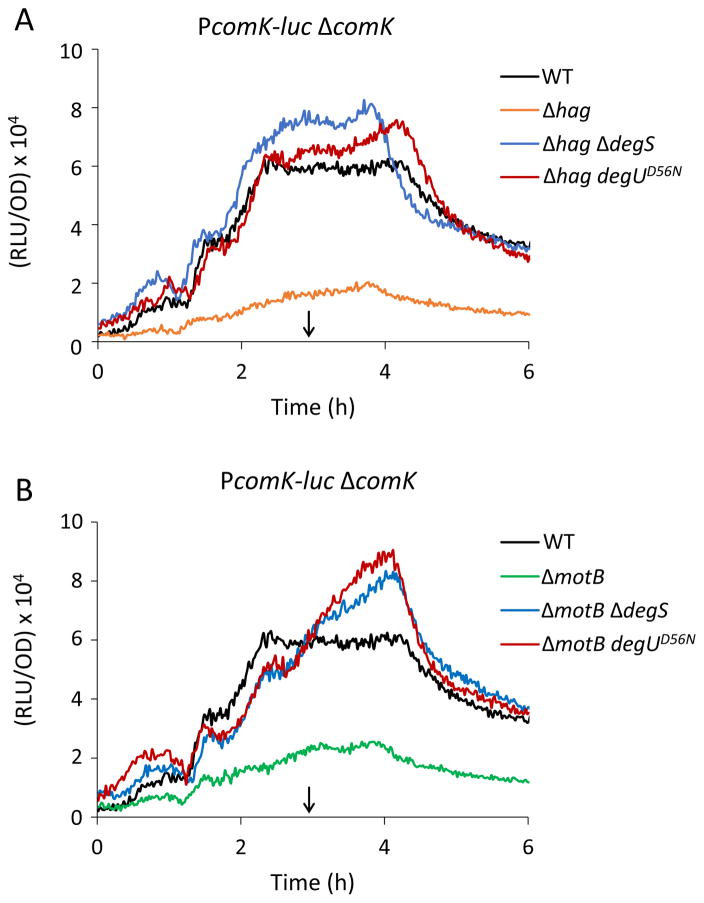
To further investigate the effects of hag inactivation on the basal expression of comK expression, we utilized two mutations that prevent the phosphorylation of DegU; a knockout of degS and a degUD56N mutation, which alters the site of phosphorylation (Msadek et al., 1990, Dahl et al., 1991). The PcomK-luc reporter was used in a comK mutant strain so that all the expression from this promoter reflects the basal expression (Fig. 4A). Importantly, both the D56N mutation and inactivation of degS bypass the effect of the hag deletion to levels of basal expression slightly higher than in the hag+ strain. This suggests that excess phosphorylation of DegU is responsible for the inhibition observed in the hag strain, while in the hag+ strain, the basal expression is probably limited by DegU-P. The degS and DegUD56N mutations similarly bypassed the deletion of motB (Fig. 4B).
Figure 4.
DegU phosphorylation represses the basal expression of comK in motB and hag knockouts. The expression curves show PcomK-luc expression in a comK deletion background. The degUD56N allele as well as the degS deletion bypass Δhag (A) and ΔmotB (B) for expression from PcomK. Strains used were as follows: ΔcomK PcomK-luc (BD4893), Δhag ΔcomK PcomK-luc (BD7261), ΔmotB ΔcomK PcomK-luc (BD7488), Δhag ΔdegS ΔcomK PcomK-luc (BD7491), ΔmotB ΔdegS ΔcomK PcomK-luc (BD7490), Δhag degUD56NΔ comK PcomK-luc (BD7475) and ΔmotB degUD56N ΔcomK PcomK-luc (BD7492). The vertical arrows point to T0.
It has been reported that the accumulation of DegU-P induces an increased level of Spo0A-P (Marlow et al., 2014). Because excess Spo0A-P will repress the basal expression of comK by direct binding (Mirouze et al., 2012), the effect of flagellar mutants could be mediated via an effect of DegU-P on the level of Spo0A-P. However, Fig. S2 shows that the deletion of motB decreases the residual basal expression of comK in a spo0A deletion strain and that this decrease is suppressed by the DegUD56N allele. Collectively, these findings show that a deletion of motB results in an increase in the level of DegU-P that represses the basal expression of comK and that this effect is probably not mediated by Spo0A-P. We do not know if DegU-P, like DegU, binds directly to the promoter region of comK.
K-state cells predominantly express low levels of DegU-P
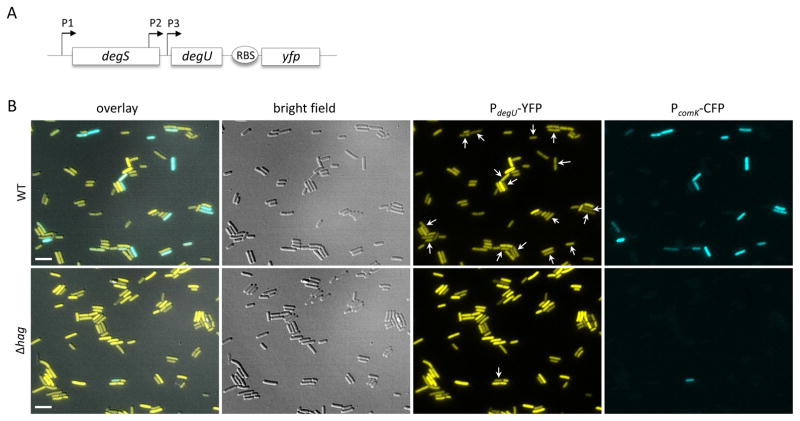
To explore the relationship between DegU-P and the K-state on a single-cell level, we used a strain expressing a promoter fusion of comK to cfp as well as a degU-yfp promoter fusion placed downstream from the native degU gene (Fig. 5A). degU is transcribed from three promoters: P1 is in front of the degSU operon, while P2 and P3 transcribe degU (Fig. 5A). Importantly, DegU-P auto-regulates its own expression by positive feedback at P3 (Ogura & Tsukahara, 2010), so that a DegU-YFP fusion can be used as a readout to infer the level of DegU-P.
Figure 5.
K-state cells have low DegU-P. (A) The diagram explains the construct used for panel B. YFP with its own ribosomal binding site was placed downstream of the degS degU operon, so that it is expressed from the three native promoters. The P3 promoter is activated by DegU-P (Ogura & Tsukahara, 2010). This strain (BD8556) also carries PcomK-cfp (blue) to monitor expression of the K-state. (B) The top row shows expression of YFP (yellow) and CFP (blue) in strain BD8556 while the bottom row shows expression from its isogenic equivalent (BD8557) that also carries a hag deletion. In the PdegU-YFP images, the comK expressing cells are indicated by arrows. In the Δhag strain, 39 % of the cells were scored visually as expressing high level of YFP, whereas in the wild-type strain shown in the top row, 3.8 % were high expressers. More than 2000 cells were scored for each strain. Cultures were grown to the time of maximum competence (T2). Representative images are shown. Scale bar is 5 μm.
In this strain, the DegU-YFP signal was distributed heterogeneously across the population as reported previously (Veening et al., 2008) (Fig. 5B, top row of images). ComK expression was observed to be negatively correlated with DegU-YFP expression, consistent with a role for DegU-P in down-regulation of the K-state. In this strain 3.8 % of the cells were scored visually as high expressing for YFP. Inactivation of hag resulted in an increase in the percentage of high YFP-expressing cells to 39 % (Fig. 5B, bottom row). At least 2000 cells were scored for each strain. This increase can be explained by autoregulation at P3. The activation of DegU phosphorylation caused by interference with flagellar function leads to an amount of DegU-P that exceeds the threshold for auto-activation in many cells. This would imply that the positive auto-regulation at the P3 promoter is at least partly responsible for the heterogeneous expression of DegU. As expected, the frequency of K-state cells in the Δhag population is lower than in the wild-type background, consistent with Fig. 2. These single cell data lead to two important conclusions. First, as predicted from the ensemble measurements transitions to the K-state occur preferentially in cells with low levels of DegU-P. Second, the elimination of hag causes an increase in DegU-P and a decrease in the frequency of K-state cells.
The hag strain has functional motors
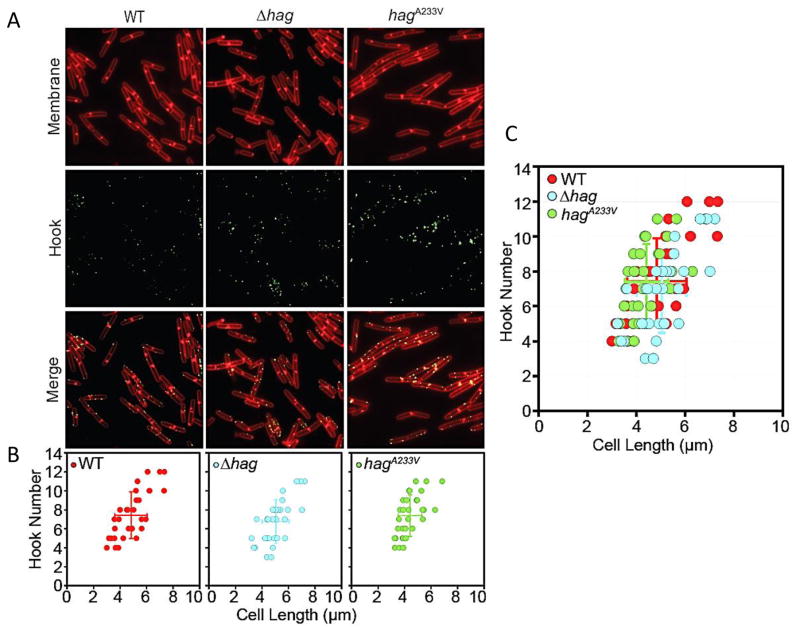
What is the initial signal that leads to suppression of the accumulation of DegU-P? We can consider three possibilities for this primary signal; rotation, proton flux and viscous load on the flagellar filament. In their important study, Cairns et al. (2013) postulated that restriction of flagellar rotation or possibly of proton flux could activate the DegS-DegU signal transduction system. In support of these suggestions, a mutation in the conserved aspartate of MotB, essential for protonation and torque generation phenocopies the motB deletion. However, as noted above, if the primary signal is rotation or proton flux, it is not immediately obvious why a hag mutation would have the same phenotype as a motB knockout. It is known that in E. coli, elimination of the filament leaves intact hooks that still rotate driven by ion flux, whereas the motB mutant flagella would rotate only due to Brownian motion (Suzuki & Komeda, 1981, Yuan & Berg, 2008). If the motor continues to function in hag deletions in B. subtilis as in E. coli, it would support a role for viscous load in generating the signal. However, if the signal strength is determined by the sum of the loads experienced by all the flagella in a cell, it is important to determine if the numbers of hooks, representative of the total number of basal bodies per cell, are the same in the wild-type and the hag mutant strains. To enable staining of hooks, we genetically altered the hook protein FlgE, by insertion of a cysteine (T123C) as previously described by Courtney et al. (2012) and swapped the mutation into the native locus by allelic exchange. The flgET123C allele was swapped into the native locus and hooks were fluorescently labeled and counted in the wild-type and the hag mutant strains (Fig. 6). The average number of hooks was calculated in relation to the cell length because hook numbers increase with increasing cell length (Guttenplan et al., 2013) (Fig. 6B and C). hag cells synthesized an average of 7 hooks per cell close to the number for the wild-type. Like the wild-type, the number of hooks appear to increase proportionally with cell length in the hag mutant.
Figure 6.
Basal body numbers are unaltered in Δhag and hagA233V mutants. (A) Fluorescence microscopy of strains with the indicated genotypes stained for membranes with FM4-64 (red) and flagellar hooks (green). The following strains were used: wild-type (DK3455), Δhag (BD8396) and hagA233V (DK3456). (B) Graphical representation of flagellar hooks as determined by counting the number of FlgET123C puncta per cell per cell length using Imaris image analysis software. Individual cell data presented as a scatter plot. Average of length and spot number indicated as a larger solid dot with standard deviations that emerge from the solid dot as horizontal and vertical lines. N = 28 cells. Red symbols indicate wild-type (DK3455), blue symbols indicate Δhag (BD8396) and green symbols indicate hagA233V (DK3456). (C) Overlay of hook scatter plots from (B).
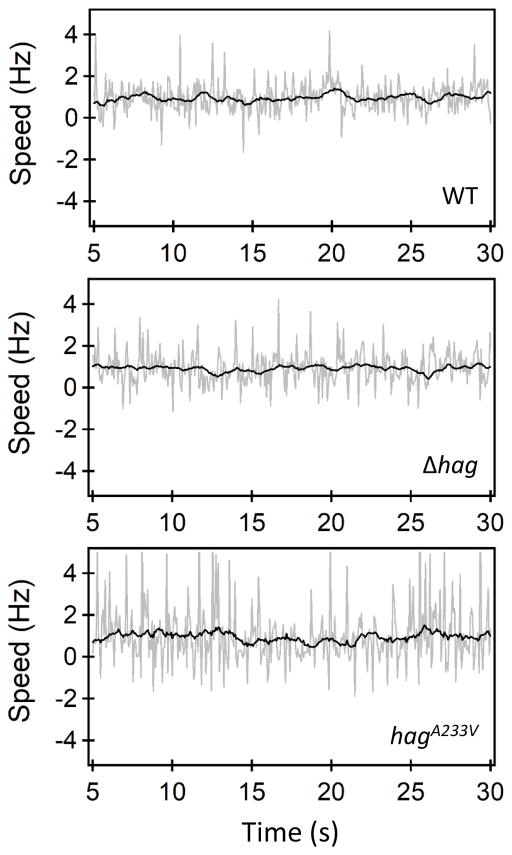
To test motor functions in the hag strain, hook rotation was compared in wild-type and hag strains. To enable tethering of cells to a surface, we used the genetically altered hook protein FlgET123C. Swim assays confirmed the wild-type function of the mutated flgE allele (data not shown). Finally, a hag deletion was inserted into this flgET123C background. Tethered cell assays were carried out on hag and wild-type cells attached to maleimide-coated surfaces as previously described (Blair et al., 2008). Cell rotation was digitally recorded over a time period up to 100s and quantitatively analyzed using ImageJ and Matlab codes (Lele et al., 2013). Motors in the hag deletion strain were functional; cells were observed to tether and rotate in a manner similar to motors in the wild-type strain (Fig. 7, Movies S1, S2). The average motor speed in the hag knockout strain was 0.7 +/− 0.2 Hz and the average torque was 341 +/− 68 pN-nm. The average motor speed in the wild-type cells was 0.8 +/− 0.2 Hz and the average torque was 353 +/− 117 (Table 2). These data suggest that flagellar motors in the hag knockouts and the wild type strains generate similar torque and that these strains contain similar numbers of basal bodies, suggesting that proton flow or rotation may not be the primary signals. We propose that the primary signal is load, sensed by helical filaments rotating in viscous media.
Figure 7.
A hag deletant and a straight filament mutant have functional motors. Rotational speeds of individual tethered cells belonging to the wild-type strain (top panel), the Δhag strain (middle panel) and the straight mutant (bottom panel) are indicated. The gray lines indicate raw data and the black curves represent filtered values. A majority of the cells predominantly rotated in the CCW direction (positive speeds). A small fraction (< 10%) of the cells exhibited CW rotation. Strains used for this experiment were: wild-type flgET123C (BD8207), Δhag flgET123C (BD8263) and hagA233V-T209C (BD7757).
Table 2.
Motor measurements
| wild type (BD8207), n=11 | hag (BD8263), n=17 | hagA233V (BD7757), n=15 | |
|---|---|---|---|
|
| |||
| Cell length (μm) | 3.3 ± 0.2 | 4.6 ± 0.4 | 4.1 ± 0.2 |
| Cell width (μm) | 1.4 ± 0.1 | 0.9 ± 0.1 | 1.5 ± 0.1 |
| Speed (Hz) | 0.8 ± 0.2 | 0.7 ± 0.2 | 0.4 ± 0.1 |
| Torque (pN-nm) | 353 ± 117 | 341 ± 68 | 248 ± 38 |
The standard errors have been indicated.
Straight filaments actively rotate and cause an increase in DegU-P levels
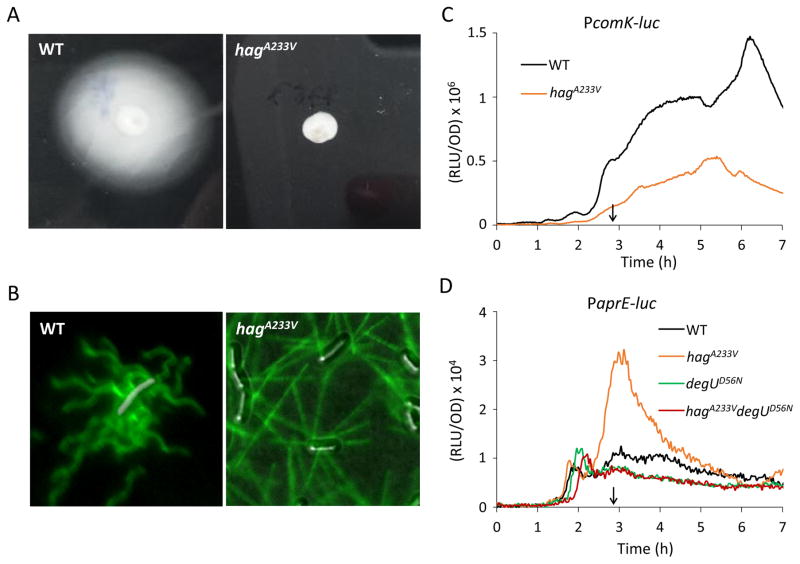
The load on a straight filament is lower than that on a helical flagellum rotating with the same angular velocity. To test whether the signal affecting the K-state is generated by load, we used a HagA233V substitution that results in straight filaments (Martinez et al., 1968). This substitution was inserted into the hag gene and swapped into the native locus. To enable visualization of the flagella by Alexa488-maleimide labeling, a T209C mutation was also introduced (Blair et al., 2008). The non-motile phenotype of this strain, previously described by (Martinez et al., 1968), was confirmed on a swim agar plate (Fig. 8A). Wild-type and straight flagella stained with the fluorescent dye are shown in Fig. 8B, verifying the straight morphology of the mutant flagella.
Figure 8.
Cells with straight filaments have increased DegU-P levels. (A) The nonmotile phenotype of a hagA233V mutant. The wild-type (IS75) and the hagA233V mutant (BD7599) were centrally inoculated on LB fortified with 0.3% agar. After 18 h incubation at 30°C, the plates were photographed against a black background. (B) Fluorescence microscopy of flagella filaments (HagT209C). HagT209C was stained with a maleimide reactive dye (green) in the wild-type strain (BD7816, hagT209C) and the hagA233V mutant (BD7757, hagT209C-A233V). Strains were grown to mid log phase in LB. (C) hagA233V decreases comK expression. Effect of hagA233V (BD7626) on PcomK-luc expression compared to wild-type (BD4773). (D) degUD56N suppresses high aprE expression in hagA233V. Expression from PaprE-luc in hagA233V is higher than wild-type expression. The degUD56N allele bypasses hagA233V for expression from PaprE. Strains used were: wild-type PaprE-luc (BD6904), hagA233V PaprE-luc (BD8007), degUD56N PaprE-luc (BD7423), and hagA233V degUD56N PaprE-luc (BD8011). The vertical arrows in panel C and D indicate T0.
The levels of DegU-P were inferred in the hagA233V strain, by testing the transcription of comK and aprE (Fig. 8C and D). comK expression decreased, whereas aprE expression increased in the hagA233V strain compared to wild-type. Thus, the results obtained for hagA233V resemble those of the hag and motB deletions for DegU-P dependent gene expression. Furthermore, a bypass experiment with the non-phosphorylatable degUD56N allele confirmed that DegU-P is needed to activate aprE expression in the hagA233V strain (Fig. 8D). These results show that the straight filament mutant produces high DegU-P and consequently lowers K-state expression, consistent with the hypothesis that the cells sense viscous drag to regulate DegU phosphorylation.
To determine whether the straight filament strain has functional motors, tethering experiments were performed, as described above for the hag mutant. Motor speeds, shown in Fig. 7, reveal that the straight filaments rotate and that functional motors are assembled (see also Movie S3). The average motor speed in the straight filament mutant was 0.4 +/− 0.1 Hz and the average torque was 248 +/− 38 pN-nm (Table 2). All measurements were at high viscous loads since the tethered cell represents a considerable load. Cells carrying the hagA233V allele synthesized an average of 7.5 hooks per cells, close to the result for the wild-type (Fig. 6). No significant differences were observed between the hagA233V mutant and wild-type in the cell length/hook number correlation (Fig. 6B and C). We conclude from these data, that disruption of motor functions in the straight flagella mutant is not the reason for low K-state expression and that the signal for K-state regulation is viscous drag on the flagellar filament.
Discussion
Our observations indicated a reduction in the K-state transition probability when the entire fla/che operon was deleted. Data also suggested that K-state cells carried lower levels of DegU-P. In a hag deletion strain and in a straight filament mutant, the normal DegU-P-suppressing signal was abrogated and DegU-P was elevated. Experiments confirmed that the flagellar motors were functional in these mutants. Considering that flagellar hooks spin at much higher speeds accompanied by a significant proton flux (Yuan & Berg, 2008), it is unlikely that the elevation in DegU-P is due to a cessation in either motor-rotation or the flux. Instead, given that the viscous loads in the two mutants are lower compared to the loads on motors in the wild type, it appears that motor operation at higher loads is necessary to suppress DegU-P. Presumably, the loads encountered by motors in freely swimming wild type cells are adequate for the suppression of DegU-P and any reduction in the load increases DegU-P. Our data further suggest that high DegU-P levels reduce the basal expression of comK, ultimately decreasing the number of cells in a population that exceed a threshold needed to activate positive auto-regulation at PcomK. These observations together strongly support a heuristic model in which the viscous drag on flagellar filaments mediates transition to the K-state.
We do not know if the phosphorylated protein binds directly to repress the basal expression of comK. In addition to ComK and DegU, it is known that AbrB, Rok and CodY bind to PcomK (Hamoen et al., 2003, Smits et al., 2007) and it is possible that DegU-P acts via an effect on one of these proteins or in some other manner. Unphosphorylated DegU helps ComK bind to its own promoter, thereby activating positive auto-regulation (Hamoen et al., 2000). However, it is unlikely that the depletion of DegU, due to increased phosphorylation, inhibits K-state expression. If DegU were to be depleted considerably due to phosphorylation, it would likely increase the level of ComK required to initiate auto-regulation rather than lowering the basal expression. High Spo0A-P is known to repress the basal expression from PcomK (Mirouze et al., 2012) and it has been reported that elevated DegU-P increases the concentration of Spo0A-P (Marlow et al., 2014) However, our results show that DegU-P is able to lower the residual basal expression in a spo0A knockout strain, making it less plausible that DegU-P action on PcomK is mediated by Spo0A-P.
The number of stator-units bound to individual flagellar motors increases with viscous loads (Lele et al., 2013, Tipping et al., 2013). Our recent work suggested that this likely occurs due to a load-induced increase in the torque delivered by individual units (Chawla et al., Sci Rep, in press). It is possible then, that the modulation of stator-assembly or the torque by the viscous load facilitates transitions to the K-state, Stalling of flagellar motors by locking filaments with anti-Hag antibodies was observed to elevate DegU-P levels (Cairns et al., 2013). This appears to be at odds with the proposed mechanism since mechanical stalling results in maximum loads on individual motors in E. coli (Berry & Berg, 1997). However, it might be that the treatment with antibodies or motor-stall causes the dissociation of stators in B. subtilis, leading to a concomitant increase in the level of DegU-P. Although our study has focused on the regulation of the K-state by flagellar motors, it is tempting to suggest that the viscous load on the motor also regulates other forms of gene expression, such as the production of poly-γ-glutamate, exoprotease and biofilm formation (Osera et al., 2009, Cairns et al., 2013, Chan et al., 2014). Indeed, DegU directly or indirectly regulates well over 150 genes (Ogura et al., 2001, Mader et al., 2002, Kobayashi, 2007).
Among the most interesting unanswered questions concern the biological roles of this signal transduction mechanism. Dynamic signaling by flagellar load may occur during switching between motile and sessile states in planktonic cells, when cells encounter a surface and initiate the formation of biofilms and during swarming, when viscous drag is likely to be greater than in free-swimming cells.
Experimental procedures
Strains and growth conditions
All strains are listed in Table S1. The B. subtilis strains used in this work are derived from the laboratory wild type strain IS75, a derivative of strain 168, or the undomesticated strain NCIB3610ΔcomI. The comI mutation abolishes ComI activity (Konkol et al., 2013), removing a block in DNA uptake. Constructs were introduced into IS75 by transformation (Albano et al., 1987) and into NCIB3610 by transduction using bacteriophage SPP1 (Cozy & Kearns, 2010). Bacterial growth was at 37°C. Antibiotic selections were carried out on Luria Broth (LB) agar plates (10 g/L Tryptone, 5 g/L Yeast Extract and 5 g/L NaCl) containing ampicillin (100 μg ml−1), spectinomycin (100 μg ml−1), erythromycin (5 μg ml−1), kanamycin (5 μg ml−1), tetracycline (25 μg ml−1) or chloramphenicol (5 μg ml−1). In some cases selection was for erythromycin (1 μg ml−1) plus lincomycin (20 μg ml−1). Solid media were solidified by the addition of 1.5% agar. Transformation frequencies were determined using genomic DNA isolated from a leucine prototroph. Isopropyl beta-D-thiogalactopyranoside (IPTG, Sigma) was added to the medium at the indicated concentrations when appropriate.
Strain construction
To construct the following point mutations: degUD56N, hagT209C, hagA233V, hagT209C/A233V and flgET123C at their native loci, we used the pMiniMAD2 cloning strategy, as described previously (Cozy & Kearns, 2010, Mukherjee et al., 2013). Plasmids are listed in Table S2.
All oligonucleotides were provided by Eton Biosciences (Union, NJ) and are listed in Table S3. All plasmid constructs and all resulting Bacillus strains were confirmed by sequencing performed by Eton Biosciences. A fragment containing the degUD56N mutation was amplified from genomic DNA containing the degUD56N mutation (Msadek et al., 1990) using the primers degUfwd and CD47-degUrev and cloned into the HindIII and KpnI sites on plasmid pMiniMAD2. The resulting plasmid, pMiniMAD2-degUD56N (pED1839), was used to create the strain BD7394.
A fragment containing the hag open reading frame was amplified from IS75 genomic DNA by using the primers CD60-hagfwd and CD61-hagrev and cloned into the BamHI site on plasmid pMiniMAD2 using the In-Fusion HD cloning kit (Clontech). The resulting plasmid pED1933 was mutagenized using the Change-IT multiple mutation site-directed mutagenesis kit (Affymetrix), per the manufacturer’s instructions and the primers CD62-hagA233V and CD78-hagT209C, to construct plasmids with the following pointmutations in hag: pED1907 (hagA233V), pED1927 (hagT209C) and pED1925 (hagA233V-T209C). The plasmids were used to transform IS75 to create the strains BD7599 (hagA233V), BD7816 (hagT209C) and BD7757 (hagA233V-T209C).
To construct the flgET123C allele, a fragment was amplified from genomic DNA containing the flgET123C mutation using the primers CD126A-flgEfwd and CD127A-flgErev and cloned into the BamHI site on plasmid pMiniMAD2 as described above. The plasmid pMiniMAD2-flgET123C (pED2004) was used to create BD8207.
To create a transcriptional promoter fusion of degU to mYpet (yellow fluorescent protein, Ohashi et al. (2007)), the mYpet coding sequence isolated from pDP429 via a SalI and SphI digest and a DNA fragment encoding the C-terminal portions of degU amplified with CD149-degU-yfpfwd and CD150-degU-yfprev were cloned into the EcoRI and SalI sites of the vector pUS19 (Benson & Haldenwang, 1993). The resulting plasmid pED2117 was transformed into B. subtilis to create a transcriptional promoter fusion of PdegU to the mYpet gene which has its own ribosomal binding site (BD8556).
Motility assay
Swimming analysis were performed as described before (Guttenplan et al., 2013). Swim agar plates containing 25 ml of LB fortified with 0.3% Bacto agar were prepared the night before use and each plate was toothpick inoculated from an overnight colony and scored for motility after 18h at 30°C.
Microscopy
For PcomG-yfp and degU-yfp PcomK-cfp microscopy, cells were grown at 37°C in competence media until they reached the maximum of competence expression at T2 (2h after transition from exponential to stationary phase of growth). One microliter of the cell culture was spotted on an agarose pad (1% agarose made up in 0.5 x TAE buffer). Cells were imaged on an upright Nikon Eclipse 90i microscope outfitted with an Orca ER Digital Camera (Hamamatsu) with a Nikon Tirf 1.45 NA Plan Neoflur 100 oil immersion objective. Semrock Optical filter sets were used for fluorescence detection. The Volocity software package 6.3 was used for image acquisition.
For fluorescent microscopy of flagella filaments, cells were grown at 37°C in LB broth until they reached an OD600 of 0.5–0.8. 1 ml of broth culture was harvested and resuspended in 50 μl of PBS containing 5 μg/ml Alexa Fluor 488 C5 maleimide (Molecular Probes), and incubated for 5 min at room temperature. Cells were then washed with 1 ml of PBS and resuspended to an OD600 of 10. Samples were observed by spotting 1 ul of the suspension on an agarose pad (see above). Images were collected using a Nikon Eclipse Ti microscope equipped with an Orca Flash 4.0 Digital camera (Hamamatsu) with a Nikon TIRF 1.45 NA Plan Neoflur 100 oil immersion objective. NIS-Elements AR (v 4.40, Nikon) software was used for image acquisition and data analysis.
For fluorescent microscopy of flagella hooks, cells were grown at 37°C in LB broth until they reached an OD600 of 0.5–0.8. 1.0 ml of broth culture was harvested and resuspended in 30 μl of PBS containing 5μg/ml Alexa Fluor 488 C5 maleimide (Molecular Probes), and incubated for 5 min at room temperature. Cells were then washed with 1 ml of PBS, pelleted, and supernatant removed. Membranes were stained by resuspension in 30 μl of PBS containing 5 μg/ml FM4-64 and incubated for 5 min at room temperature. Cells were washed in 1 ml of PBS, and pelleted again. Samples were finally resuspended to an OD600 of 10 in PBS. Samples were observed by spotting 4 μl of the suspension on a glass microscope slide and immobilized with a poly-L-lysine-treated glass coverslip.
Conventional fluorescence microscopy was performed with a Nikon 80i microscope with a phase contrast objective Nikon Plan Apo 100X and an Excite 120 metal halide lamp. FM4-64 membrane stains were visualized with a C-FL HYQ Texas Red Filter Cube (excitation filter 532–587 nm, barrier filter 590 nm). Alexa Fluor 488 C5 maleimide fluorescent signals were visualized using a C-FL HYQ FITC Filter Cube (FITC, excitation filter 460–500 nm, barrier filter 515–550 nm). Images were captured with a Photometrics Coolsnap HQ2 camera in black and white, false colored, and superimposed using Metamorph image software.
Super Resolution fluorescence microscopy was performed using the structured illumination OMX 3D-SIM Super Resolution system at Indiana University Bloomington Light Microscopy Imaging Center. Super Resolution microscopy was performed using a 1.4NA Olympus 100x oil objective. FM4-64 was visualized using laser line 561nm and emission filter 609–654nm, and Alexa Fluor 488 was visualized using laser line 488nm and emission filter 500–550 nm. Images were captured by Photometrics Cascade II EMCCD camera, and processed by SoftWoRx software (Applied Precision).
To count hooks, images reconstructed with SoftWoRx were processed in Imaris (Bitplane) to measure cell length and determine the number of FlgET123C foci on the surface of each cell. Cell length was measured by hand using the ‘Slice’ measurement tool. The ‘Spots’ feature labelled each FlgET123C foci by identifying spots of 0.1 μM in the 488 wavelength range. We then verified by eye that the labelling identified bona fide foci on the cell surface. Number of foci was then recorded and plotted against cell length.
Tethering
Overnight cultures were diluted in fresh TB (10 g/L tryptone; 5 g/L NaCl) at 30°C and grown to an OD600 of 0.5–0.6. Cells were washed twice in motility buffer containing lactate (0.055 M NaCl, 10 mM EDTA, 10 mM potassium phosphate, pH 7.0) and sheared 70 times to shorten the filaments (Ford et al., 2017). These strains carried cysteine groups in the flagellin or hook proteins (hagT209C/flgET123C) in order to facilitate tethering to maleimide-coated coverslips (MicroSurfaces, Inc.). Cells were allowed to tether for 10 minutes and observed with a 20 X objective on an inverted microscope (Nikon Ti-E). Cell-geometries and rotational-speeds were quantitatively tracked with custom-written MATLAB codes (Lele et al., 2016).
Luciferase assays
For the detection of luciferase activity, strains were first grown in LB medium to an OD600 of 2. Cells were then centrifuged and resuspended in fresh competence medium (Albano et al., 1987), adjusting all the cultures to an OD600 of 2. These pre-cultures were then diluted 20-fold in fresh competence medium and 200 μl was distributed in each of two wells in a 96-well black plate (Corning Incorporated Costar®). 10 μl of luciferin was added to each well to reach a final concentration of 1.5 mg/ml (4.7 mM). The cultures were incubated at 37°C with agitation in a PerkinElmer Envision 2104 Multilabel Reader equipped with an enhanced sensitivity photomultiplier for luminometry. The temperature of the clear plastic lid was maintained at 38°C to avoid condensation. Relative Luminescence Units (RLU) and OD600 were measured at 1 min intervals after two 30 second shaking steps. The data were processed using a script written in MATLAB, exported to Excel and plotted as RLU/OD600 versus time from the beginning of growth.
Supplementary Material
Acknowledgments
This work was supported by NIH grant GM057720 (DD), NIH GM093030 to DBK and by funds from the Texas A&M Engineering Experiment Station (PL). We thank K. Lemon and A. Grossman for the gift of the pKL184 plasmid.
References
- Adler J. Chemotaxis in bacteria. Annu Rev Biochem. 1975;44:341–356. doi: 10.1146/annurev.bi.44.070175.002013. [DOI] [PubMed] [Google Scholar]
- Albano M, Hahn J, Dubnau D. Expression of competence genes in Bacillus subtilis. J Bacteriol. 1987;169:3110–3117. doi: 10.1128/jb.169.7.3110-3117.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage JP. Bacterial motility and chemotaxis. Sci Prog. 1992;76:451–477. [PubMed] [Google Scholar]
- Belas R. Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol. 2014;22:517–527. doi: 10.1016/j.tim.2014.05.002. [DOI] [PubMed] [Google Scholar]
- Belas R, Simon M, Silverman M. Regulation of lateral flagella gene transcription in Vibrio parahaemolyticus. J Bacteriol. 1986;167:210–218. doi: 10.1128/jb.167.1.210-218.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson AK, Haldenwang WG. Regulation of sigma B levels and activity in Bacillus subtilis. J Bacteriol. 1993;175:2347–2356. doi: 10.1128/jb.175.8.2347-2356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- Berg HC, Brown DA. Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature. 1972;239:500–504. doi: 10.1038/239500a0. [DOI] [PubMed] [Google Scholar]
- Berka RM, Hahn J, Albano M, Draskovic I, Persuh M, Cui X, Sloma A, Widner W, Dubnau D. Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol Microbiol. 2002;43:1331–1345. doi: 10.1046/j.1365-2958.2002.02833.x. [DOI] [PubMed] [Google Scholar]
- Berry RM, Berg HC. Absence of a barrier to backwards rotation of the bacterial flagellar motor demonstrated with optical tweezers. Proc Natl Acad Sci U S A. 1997;94:14433–14437. doi: 10.1073/pnas.94.26.14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science. 2008;320:1636–1638. doi: 10.1126/science.1157877. [DOI] [PubMed] [Google Scholar]
- Block SM, Berg HC. Successive incorporation of force-generating units in the bacterial rotary motor. Nature. 1984;309:470–472. doi: 10.1038/309470a0. [DOI] [PubMed] [Google Scholar]
- Caiazza NC, Merritt JH, Brothers KM, O’Toole GA. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol. 2007;189:3603–3612. doi: 10.1128/JB.01685-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazza NC, O’Toole GA. SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J Bacteriol. 2004;186:4476–4485. doi: 10.1128/JB.186.14.4476-4485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns LS, V, Marlow L, Bissett E, Ostrowski A, Stanley-Wall NR. A mechanical signal transmitted by the flagellum controls signalling in Bacillus subtilis. Mol Microbiol. 2013;90:6–21. doi: 10.1111/mmi.12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JM, Guttenplan SB, Kearns DB. Defects in the flagellar motor increase synthesis of poly-gamma-glutamate in Bacillus subtilis. J Bacteriol. 2014;196:740–753. doi: 10.1128/JB.01217-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che YS, Nakamura S, Morimoto YV, Kami-Ike N, Namba K, Minamino T. Load-sensitive coupling of proton translocation and torque generation in the bacterial flagellar motor. Mol Microbiol. 2014;91:175–184. doi: 10.1111/mmi.12453. [DOI] [PubMed] [Google Scholar]
- Chevance FF, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol. 2008;6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney CR, Cozy LM, Kearns DB. Molecular characterization of the flagellar hook in Bacillus subtilis. J Bacteriol. 2012;194:4619–4629. doi: 10.1128/JB.00444-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozy LM, Kearns DB. Gene position in a long operon governs motility development in Bacillus subtilis. Mol Microbiol. 2010;76:273–285. doi: 10.1111/j.1365-2958.2010.07112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl MK, Msadek T, Kunst F, Rapoport G. Mutational analysis of the Bacillus subtilis DegU regulator and its phosphorylation by the DegS protein kinase. J Bacteriol. 1991;173:2539–2547. doi: 10.1128/jb.173.8.2539-2547.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl MK, Msadek T, Kunst F, Rapoport G. The phosphorylation state of the DegU response regulator acts as a molecular switch allowing either degradative enzyme synthesis or expression of genetic competence in Bacillus subtilis. J Biol Chem. 1992;267:14509–14514. [PubMed] [Google Scholar]
- Ellison C, Brun YV. Mechanosensing: a regulation sensation. Curr Biol. 2015;25:R113–115. doi: 10.1016/j.cub.2014.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford KM, Chawla R, Lele PP. Biophysical Characterization of Flagellar Motor Functions. J Vis Exp. 2017 doi: 10.3791/55240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamba P, Jonker MJ, Hamoen LW. A Novel Feedback Loop That Controls Bimodal Expression of Genetic Competence. PLoS Genet. 2015;11:e1005047. doi: 10.1371/journal.pgen.1005047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttenplan SB, Blair KM, Kearns DB. The EpsE flagellar clutch is bifunctional and synergizes with EPS biosynthesis to promote Bacillus subtilis biofilm formation. PLoS Genet. 2010;6:e1001243. doi: 10.1371/journal.pgen.1001243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttenplan SB, Shaw S, Kearns DB. The cell biology of peritrichous flagella in Bacillus subtilis. Mol Microbiol. 2013;87:211–229. doi: 10.1111/mmi.12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J, Tanner AW, Carabetta VJ, Cristea IM, Dubnau D. ComGA-RelA interaction and persistence in the Bacillus subtilis K-state. Mol Microbiol. 2015;97:454–471. doi: 10.1111/mmi.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamoen LW, Smits WK, de Jong A, Holsappel S, Kuipers OP. Improving the predictive value of the competence transcription factor (ComK) binding site in Bacillus subtilis using a genomic approach. Nucleic Acids Res. 2002;30:5517–5528. doi: 10.1093/nar/gkf698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamoen LW, Van Werkhoven AF, Venema G, Dubnau D. The pleiotropic response regulator DegU functions as a priming protein in competence development in Bacillus subtilis. Proc Natl Acad Sci U S A. 2000;97:9246–9251. doi: 10.1073/pnas.160010597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamoen LW, Venema G, Kuipers OP. Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology. 2003;149:9–17. doi: 10.1099/mic.0.26003-0. [DOI] [PubMed] [Google Scholar]
- Henner DJ, Yang M, Ferrari E. Localization of Bacillus subtilis sacU(Hy) mutations to two linked genes with similarities to the conserved procaryotic family of two-component signalling systems. J Bacteriol. 1988;170:5102–5109. doi: 10.1128/jb.170.11.5102-5109.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K. Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis. Mol Microbiol. 2007;66:395–409. doi: 10.1111/j.1365-2958.2007.05923.x. [DOI] [PubMed] [Google Scholar]
- Kojima S, Blair DF. Conformational change in the stator of the bacterial flagellar motor. Biochemistry. 2001;40:13041–13050. doi: 10.1021/bi011263o. [DOI] [PubMed] [Google Scholar]
- Kojima S, Blair DF. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry. 2004;43:26–34. doi: 10.1021/bi035405l. [DOI] [PubMed] [Google Scholar]
- Konkol MA, Blair KM, Kearns DB. Plasmid-encoded ComI inhibits competence in the ancestral 3610 strain of Bacillus subtilis. J Bacteriol. 2013;195:4085–4093. doi: 10.1128/JB.00696-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisner M, Stingl K, Radler JO, Maier B. Basal expression rate of comK sets a ‘switching-window’ into the K-state of Bacillus subtilis. Mol Microbiol. 2007;63:1806–1816. doi: 10.1111/j.1365-2958.2007.05628.x. [DOI] [PubMed] [Google Scholar]
- Lele PP, Hosu BG, Berg HC. Dynamics of mechanosensing in the bacterial flagellar motor. Proc Natl Acad Sci U S A. 2013;110:11839–11844. doi: 10.1073/pnas.1305885110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lele PP, Roland T, Shrivastava A, Chen Y, Berg HC. The flagellar motor of Caulobacter crescentus generates more torque when a cell swims backward. Nat Phys. 2016;12:175–178. doi: 10.1038/nphys3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Brown PJ, Tang JX, Xu J, Quardokus EM, Fuqua C, Brun YV. Surface contact stimulates the just-in-time deployment of bacterial adhesins. Mol Microbiol. 2012;83:41–51. doi: 10.1111/j.1365-2958.2011.07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamar H, Dubnau D. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol Microbiol. 2005;56:615–624. doi: 10.1111/j.1365-2958.2005.04592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamar H, Raj A, Dubnau D. Noise in gene expression determines cell fate in Bacillus subtilis. Science. 2007;317:526–529. doi: 10.1126/science.1140818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader U, Antelmann H, Buder T, Dahl MK, Hecker M, Homuth G. Bacillus subtilis functional genomics: genome-wide analysis of the DegS-DegU regulon by transcriptomics and proteomics. Mol Genet Genomics. 2002;268:455–467. doi: 10.1007/s00438-002-0774-2. [DOI] [PubMed] [Google Scholar]
- Marlow VL, Porter M, Hobley L, Kiley TB, Swedlow JR, Davidson FA, Stanley-Wall NR. Phosphorylated DegU manipulates cell fate differentiation in the Bacillus subtilis biofilm. J Bacteriol. 2014;196:16–27. doi: 10.1128/JB.00930-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez-Magana LM, Chamberlin MJ. Characterization of the sigD transcription unit of Bacillus subtilis. J Bacteriol. 1994;176:2427–2434. doi: 10.1128/jb.176.8.2427-2434.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez RJ, Ichiki AT, Lundh NP, Tronick SR. A single amino acid substitution responsible for altered flagellar morphology. J Mol Biol. 1968;34:559–564. doi: 10.1016/0022-2836(68)90180-0. [DOI] [PubMed] [Google Scholar]
- McCarter L, Hilmen M, Silverman M. Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell. 1988;54:345–351. doi: 10.1016/0092-8674(88)90197-3. [DOI] [PubMed] [Google Scholar]
- Minamino T, Imada K, Namba K. Molecular motors of the bacterial flagella. Curr Opin Struct Biol. 2008;18:693–701. doi: 10.1016/j.sbi.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Mirouze N, Desai Y, Raj A, Dubnau D. Spo0A~P imposes a temporal gate for the bimodal expression of competence in Bacillus subtilis. PLoS Genet. 2012;8:e1002586. doi: 10.1371/journal.pgen.1002586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Msadek T, Kunst F, Henner D, Klier A, Rapoport G, Dedonder R. Signal transduction pathway controlling synthesis of a class of degradative enzymes in Bacillus subtilis: expression of the regulatory genes and analysis of mutations in degS and degU. J Bacteriol. 1990;172:824–834. doi: 10.1128/jb.172.2.824-834.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Babitzke P, Kearns DB. FliW and FliS function independently to control cytoplasmic flagellin levels in Bacillus subtilis. J Bacteriol. 2013;195:297–306. doi: 10.1128/JB.01654-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Kearns DB. The structure and regulation of flagella in Bacillus subtilis. Annu Rev Genet. 2014;48:319–340. doi: 10.1146/annurev-genet-120213-092406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray EJ, Kiley TB, Stanley-Wall NR. A pivotal role for the response regulator DegU in controlling multicellular behaviour. Microbiology. 2009;155:1–8. doi: 10.1099/mic.0.023903-0. [DOI] [PubMed] [Google Scholar]
- Ogura M, Tsukahara K. Autoregulation of the Bacillus subtilis response regulator gene degU is coupled with the proteolysis of DegU-P by ClpCP. Mol Microbiol. 2010;75:1244–1259. doi: 10.1111/j.1365-2958.2010.07047.x. [DOI] [PubMed] [Google Scholar]
- Ogura M, Yamaguchi H, Kobayashi K, Ogasawara N, Fujita Y, Tanaka T. Whole-genome analysis of genes regulated by the Bacillus subtilis competence transcription factor ComK. J Bacteriol. 2002;184:2344–2351. doi: 10.1128/JB.184.9.2344-2351.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura M, Yamaguchi H, Yoshida K, Fujita Y, Tanaka T. DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: an approach to comprehensive analysis of B.subtilis two-component regulatory systems. Nucleic Acids Res. 2001;29:3804–3813. doi: 10.1093/nar/29.18.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi T, Galiacy SD, Briscoe G, Erickson HP. An experimental study of GFP-based FRET, with application to intrinsically unstructured proteins. Protein Sci. 2007;16:1429–1438. doi: 10.1110/ps.072845607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osera C, Amati G, Calvio C, Galizzi A. SwrAA activates poly-gamma-glutamate synthesis in addition to swarming in Bacillus subtilis. Microbiology. 2009;155:2282–2287. doi: 10.1099/mic.0.026435-0. [DOI] [PubMed] [Google Scholar]
- Purcell EM. Life at low Reynolds number. American Journal of Physics. 1977;45:3–11. [Google Scholar]
- Smits WK, Eschevins CC, Susanna KA, Bron S, Kuipers OP, Hamoen LW. Stripping Bacillus: ComK auto-stimulation is responsible for the bistable response in competence development. Mol Microbiol. 2005;56:604–614. doi: 10.1111/j.1365-2958.2005.04488.x. [DOI] [PubMed] [Google Scholar]
- Smits WK, Hoa TT, Hamoen LW, Kuipers OP, Dubnau D. Antirepression as a second mechanism of transcriptional activation by a minor groove binding protein. Mol Microbiol. 2007;64:368–381. doi: 10.1111/j.1365-2958.2007.05662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M, Kunst F, Dedonder R. Mapping of mutations affecting synthesis of exocellular enzymes in Bacillus subtilis. Identity of the sacUh, amyB and pap mutations. Mol Gen Genet. 1976;148:281–285. doi: 10.1007/BF00332902. [DOI] [PubMed] [Google Scholar]
- Suel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 2006;440:545–550. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Komeda Y. Incomplete flagellar structures in Escherichia coli mutants. J Bacteriol. 1981;145:1036–1041. doi: 10.1128/jb.145.2.1036-1041.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipping MJ, Delalez NJ, Lim R, Berry RM, Armitage JP. Load-dependent assembly of the bacterial flagellar motor. MBio. 2013:4. doi: 10.1128/mBio.00551-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dellen KL, Houot L, Watnick PI. Genetic analysis of Vibrio cholerae monolayer formation reveals a key role for DeltaPsi in the transition to permanent attachment. J Bacteriol. 2008;190:8185–8196. doi: 10.1128/JB.00948-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Sinderen D, Luttinger A, Kong L, Dubnau D, Venema G, Hamoen L. comK encodes the competence transcription factor, the key regulatory protein for competence development in Bacillus subtilis. Mol Microbiol. 1995;15:455–462. doi: 10.1111/j.1365-2958.1995.tb02259.x. [DOI] [PubMed] [Google Scholar]
- van Sinderen D, Venema G. comK acts as an autoregulatory control switch in the signal transduction route to competence in Bacillus subtilis. J Bacteriol. 1994;176:5762–5770. doi: 10.1128/jb.176.18.5762-5770.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening JW, Igoshin OA, Eijlander RT, Nijland R, Hamoen LW, Kuipers OP. Transient heterogeneity in extracellular protease production by Bacillus subtilis. Mol Syst Biol. 2008;4:184. doi: 10.1038/msb.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhamme DT, Kiley TB, Stanley-Wall NR. DegU co-ordinates multicellular behaviour exhibited by Bacillus subtilis. Mol Microbiol. 2007;65:554–568. doi: 10.1111/j.1365-2958.2007.05810.x. [DOI] [PubMed] [Google Scholar]
- Yuan J, Berg HC. Resurrection of the flagellar rotary motor near zero load. Proc Natl Acad Sci U S A. 2008;105:1182–1185. doi: 10.1073/pnas.0711539105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuberi AR, Ying CW, Weinreich MR, Ordal GW. Transcriptional organization of a cloned chemotaxis locus of Bacillus subtilis. J Bacteriol. 1990;172:1870–1876. doi: 10.1128/jb.172.4.1870-1876.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.