Abstract
Establishment of Kaposi’s sarcoma-associated herpesvirus (KSHV) latency following infection is a multistep process, during which polycomb proteins are recruited onto the KSHV genome, which is crucial for the genome-wide repression of lytic genes during latency. Strikingly, only a subset of lytic genes are expressed transiently in the early phase of infection prior to the binding of polycomb proteins onto the KSHV genome, which raises the question what restricts lytic gene expression in the first hours of infection. Here, we demonstrate that both CTCF and cohesin chromatin organizing factors are rapidly recruited to the viral genome prior to the binding of polycombs during de novo infection, but only cohesin is required for the genome-wide inhibition of lytic genes. We propose that cohesin is required for the establishment of KSHV latency by initiating the repression of lytic genes following infection, which is an essential step in persistent infection of humans.
Keywords: CTCF, cohesin, NIPBL, chromatin, KSHV, herpesvirus, polycomb proteins, viral latency, de novo infection, lytic gene expression
Introduction
Kaposi’s sarcoma-associated herpesvirus (KSHV) is a member of the gamma-2 herpesvirus (Rhadinovirus) subfamily (Chang et al., 1994). Persistent KSHV infection in humans can lead to the development of various KSHV-associated cancers such as Kaposi’s sarcoma, primary effusion lymphoma (PEL) or a subset of multicentric Castleman’s disease in immunocompromised individuals (Cesarman, 2011; Mesri et al., 2010). KSHV can establish latency in a number of different cell types (e.g. B cells, endothelial cells, fibroblasts), which is characterized by the inhibition of lytic viral genes and constitutive expression of latent viral genes (Bechtel et al., 2003). During latency the 165-kb DNA genome of KSHV adopts a stable chromatin structure similar to that of the host genome and persists in the nuclei of infected cells as a chromatinized, non-integrating episome, which replicates synchronously with the host genome (Ballestas et al., 1999; Renne et al., 1996; Tempera and Lieberman, 2010).
Following infection the KSHV genome undergoes a biphasic euchromatin-to-heterochromatin transition in infected cells before the establishment of latency (Toth et al., 2013b). The stepwise chromatinization of the viral DNA is regulated by chromatin modifying host factors that interact with the KSHV genome and deposit specific histone marks on it in a spatially and temporally ordered manner during infection (Gunther et al., 2014; Toth et al., 2013b). Two of the major cellular transcription repressors involved in the inhibition of lytic KSHV genes during latency are the Polycomb repressive complexes 1 and 2 (PRC1 and 2) (Gunther and Grundhoff, 2010; Simon and Kingston, 2009; Toth et al., 2013b; Toth et al., 2010). During de novo infection the PRCs are recruited to lytic viral promoters by the latent KSHV factor LANA 24 hours postinfection (hpi), which is required for the inhibition of the expression of lytic genes so that KSHV can establish latency (Toth et al., 2016). Strikingly, a limited number of lytic genes are known to be induced transiently during de novo KSHV infection despite neither PRC binding to the KSHV genome nor the viral DNA being heterochromatinized in the first 24 hours of KSHV infection (Gunther et al., 2014; Krishnan et al., 2004; Toth et al., 2013b). This implies that there must be transcription repressors other than PRCs, which can globally inhibit KSHV lytic gene expression immediately following de novo infection. However, nuclear factors that can be recruited onto the viral DNA prior to the heterochromatinization of the KSHV genome and can repress lytic gene expression during de novo infection have not yet been identified.
Several nuclear factors such as the 11-zinc finger protein CTCF and the cohesin complex have been shown to be involved in the regulation of the 3D chromatin structure of the host genome, which can influence gene expression (Kagey et al., 2010; Phillips and Corces, 2009). The cohesin complex is composed of the core subunits RAD21, SMC1, SMC3, STAG1, STAG2, and several transiently associated regulatory factors (Peters et al., 2008). The interaction of cohesin with chromatin is determined by several nuclear factors. While NIPBL and MAU2 proteins act as loading factors, which can recruit cohesin onto the chromosomes, the histone deacetylase 8 (HDAC8) functions as a recycling enzyme of acetylated cohesin factors releasing cohesin from chromatin. Both cohesin and NIPBL have been implicated not only in the regulation of chromosome segregation during cell division but also in transcriptional regulation where NIPBL can control gene transcription in a cohesin-independent manner (Schaaf et al., 2013a; Zuin et al., 2014). Importantly, several mutations in NIPBL and cohesin subunits have been linked to the developmental disorder Cornelia de Lange syndrome (CdLS), which is likely due to the misexpression of developmentally important genes rather than a defect in chromosome segregation (Strachan, 2005). In addition, CdLS patients have been shown to be more susceptible to infection indicating a role for cohesin factors in restricting microbial infection (Jyonouchi et al., 2013).
Recent studies revealed that CTCF and cohesin can act as restriction factors of KSHV lytic replication by suppressing lytic reactivation of KSHV in different cell types (Chen et al., 2012; Li et al., 2014). Here, we investigated the role CTCF and cohesin in the inhibition of the lytic cycle of KSHV during de novo infection. We found that CTCF, cohesin and NIBPL can rapidly bind to the KSHV genome within a few hours postinfection and both cohesin and NIBPL can inhibit the expression of lytic genes and KSHV replication following infection. We also show that the restriction of lytic viral replication after infection is mainly due to cohesin-mediated inhibition of the expression of the viral gene RTA, which is required for the induction of lytic genes and viral replication. Importantly, our data indicate that cohesin binds to the RTA promoter and can repress the expression of lytic genes prior to the recruitment of PRCs and heterochromatinization of the viral genome, suggesting that cohesins can function as global restriction factors of KSHV lytic gene expression and replication not only during latency but also following primary infection. Thus, we propose that cohesin factors are essential for the establishment of KSHV latency.
Materials and Methods
Cells, KSHV and de novo KSHV infection
293T, SLK, and HFF cells were maintained in DMEM medium supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin (P/S). The CdLS patient primary fibroblast cells designated as GM00045 and GM03478 were obtained from the Coriell Institute, which were cultured similarly to the primary fibroblast HFF. [Note: while NIPBL mutation has been identified in GM00045, it is not known which cohesin factor is mutated in GM03478 (Vrouwe et al., 2007)]. For de novo KSHV infection, we used the recombinant KSHV clone BAC16, which was produced using iSLK cell line that is described elsewhere (Myoung and Ganem, 2011; Toth et al., 2013b). The iSLK cell lines carrying WT KSHV (BAC16) or RTA KO KSHV (BAC16RTAstop) were maintained in DMEM medium supplemented with 10% FBS, P/S, 1 μg/ml puromycin, 250 μg/ml G418, and 1 mg/ml hygromycin. The construction of BAC16 and BAC16RTAstop viruses has been described previously (Brulois et al., 2012; Toth et al., 2012). KSHV was prepared from iSLKBAC16 and iSLKBAC16RTAstop cell cultures by treating the cells with 1 μg/ml doxycycline and 3 mM sodium butyrate for 84 hours. Media were collected from virus-producing cells, filtered through a 0.45 μM PES filter, and KSHV was concentrated by ultracentrifugation of the collected media by 24,000 rpm for 3 hours at 10°C. The concentrated virus was resuspended in DMEM and stored at −80 °C. The viral titer was calculated as described previously (Brulois et al., 2012; Toth et al., 2016). De novo infection was performed by spin-infection (2000 rpm, 45 min at 30°C). After infection the media was changed and the infected cells were harvested at the indicated time points.
Antibodies
The following antibodies were used in ChIPs and/or immunoblots: anti-histone H3 (Abcam ab1791), anti-H3K27me3 (Active Motif #39155), anti-H3K4me3 (Active Motif #39159), anti-EZH2 (Active Motif #39875), anti-RING1B (Abcam ab3832), anti-LANA (Advanced Biotechnologies #13-210-100), anti-CTCF (Millipore, #07-729), anti-RAD21 (Abcam, ab992), anti-SMC3 (Abcam, ab9263), anti-NIPBL (Bethyl Laboratories, A301-779A), anti-K8 (Abcam ab36617), anti-RNA polymerase II (RNAPII) (Abcam), and anti-actin (Abcam). Anti-K3 and anti-RTA antibodies were generous gifts from Drs. Jae U. Jung (University of Southern California) and Yoshihiro Izumiya (University of California, Davis).
Chromatin immunoprecipitation (ChIP) assay
KSHV-infected cells (2–4×106) were fixed with 1% formaldehyde for 10 min at RT followed by adding 2M glycine to the cells to stop crosslinking (final concentration of glycin 125 mM) for 5 min at RT. Cells were washed three times with cold PBS and then resuspended in cell lysis buffer (5 mM Tris-HCl, pH 8.0, 85 mM KCl, 0.5% NP40, 1x protease inhibitor cocktail (Roche)) and incubated on ice for 10 min. After centrifugation (5 min, 5000 rpm at 4°C), the pellet was resuspended in 1.2 ml of RIPA buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, pH 8.0, 140 mM NaCl, 0.1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100, 1 mM PMSF, 1x protease inhibitor cocktail), sonicated and centrifuged by 13000 rpm for 10 min at 4°C. Aliquots of the supernatant containing both cellular and viral chromatin were stored at −80°C.
To prepare input DNA, 20ul of chromatin was incubated in 100ul of TE buffer containing 50 μg/ml RNase A for 30 min at 37°C. The samples were then adjusted to contain 0.5% SDS and 0.5 mg/ml Proteinase K (Invitrogen) and incubated for 1 hr at 37°C. Formaldehyde crosslinks were reversed by adding sodium chloride (final concentration 300 mM) to the samples and incubated overnight at 65°C. DNA was extracted first by one volume of phenol/chloroform/isoamyl alcohol (25:24:1) saturated with 10 mM Tris, pH8.0 and 1 mM EDTA and then purified once by one volume of chloroform. DNA was precipitated by cold absolute ethanol, 10% 3 M sodium acetate, pH 5.2 and 1.5 μl of 15 mg/ml Glycogen Blue (Ambion) at −80°C at least for 1 hour following by wash with 70% ethanol. The input DNA was dried at RT and resuspended in 20 μl of water.
For ChIPs, chromatin containing 2–5 μg of DNA was first diluted in 500 μl of RIPA buffer and precleared by Sepharose A beads. Immunoprecipitation was carried out with 1–2 μg of antibodies overnight at 4°C. Next day, Protein-A/G agarose was added for 4 hours to pull down the DNA/protein complexes. ChIP was washed sequentially with RIPA buffer once briefly and once for 10 min followed by washing with LiCl buffer (10 mM Tris-HCl, pH 8.0, 1mM EDTA, pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate) once for 10 min and with TE buffer two times for 10 min. The ChIP-Protein A/G agarose complex was resuspended in 100 μl of TE buffer containing 50 μg/ml RNase A and incubated for 30 min at 37°C. The Proteinase K treatment, crosslink reversal and DNA purification was performed exactly as described for the preparation of input DNA. Both input and ChIP DNAs were measured by SYBR green-based qPCR (BioRad). Based on the standard curves for each primer pairs the enrichment of proteins and histone modifications on specific genomic regions were calculated as percentage of the immunoprecipitated DNA compared to input DNA. Each data points in ChIP figures were the averages of at least two independent ChIPs using independent chromatins. The following forward (Fw) and reverse (Rev) primers were used in ChIP-qPCR for HS1 (Fw-TTCCTATTTGCCAAGGCAGT and Rev- CTCTTCAGCCATCCCAAGAC), OSBP (Fw- GCTGCTGTTTCCGCCATTCATTTC and Rev- GCTGATACCAACCACCAATCCATGAG), and Neg (Fw- CAGGATCTCCGAGAATCAGC and Rev- GAGTTGGGAGAGCTGTCAGG). The sequences of the other primers used in ChIP-qPCR have been reported previously (Toth et al., 2012).
RNA purification and RT-qPCR
Total RNA was extracted from cells with Tri reagent (Sigma) according to the manufacturer’s protocol. Afterwards 1 μg of total RNA was treated with DNase I (Sigma), reverse transcribed by iScript cDNA Synthesis kit (Bio-Rad) and the cDNA was quantified by SYBR green-based qPCR using gene specific primers. The relative level of gene expression was calculated by the 2−ΔCt and the ΔΔCt methods, where 18S was used for normalization. The RT-qPCR graphs represent the average of at least three independent experiments. The sequences of the primers used in RT-qPCR have been reported previously (Toth et al., 2013b; Toth et al., 2016).
Total DNA purification and measuring relative viral copy number
KSHV-infected cells were lysed in RIPA buffer followed by sonication and then centrifugation to remove cell debris. Total DNA was purified from the supernatant by phenol-chloroform extraction and 10 ng of total DNA was analyzed in qPCR. The viral DNA was measured by qPCR using primers for ORF11 (Fw-GGCACCATACAGCTTCTACGA and Rev- CGTTTACTACTGCACACTGCA). The amount of viral DNA was normalized for the cellular DNA input, which was measured by qPCR specific for the HS1 genomic region. The viral DNA load change between different shRNA-treated KSHV infected cells was calculated by the ΔΔCt method where shcontrol samples were used as reference points.
Lentiviral shRNA knockdown during KSHV infection
The pLKO.1 lentiviral vector was used to express the CTCF, RAD21, SMC3, and NIPBL specific shRNAs. The lentivector shRNAs were co-transfected with packaging vectors into 293T cells and the media containing lentiviruses was collected 60 hours post-transfection. The lentivirus was concentrated by ultracentrifugation (24000 rpm, 2 hrs, 4°C). SLK cells were infected with shRNA lentiviruses in the presence of 8 μg/ml polybrene. 2 days after lentivirus infection, the cells were split, and infected with KSHV the following day.
Results
CTCF, cohesin, and NIPBL are rapidly recruited onto the KSHV genome during de novo infection
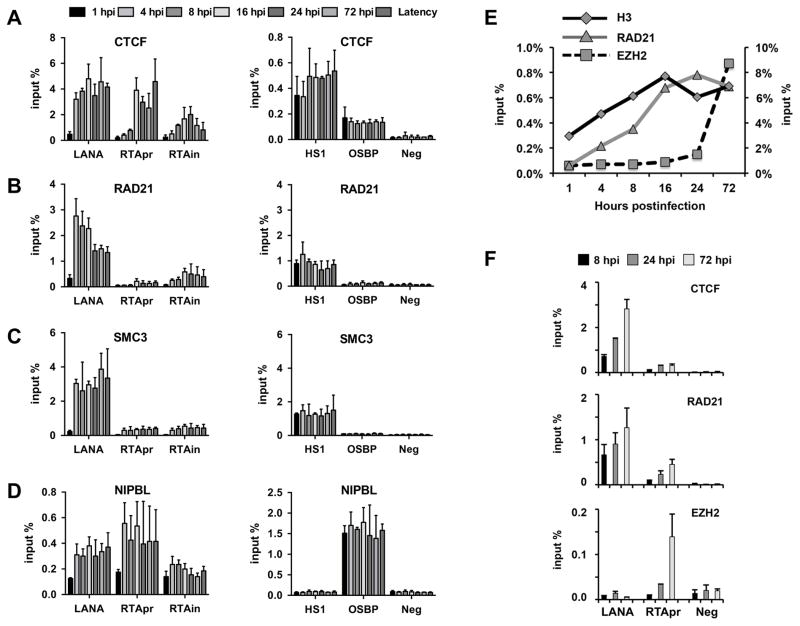
Repression of the replication and transcription activator gene (RTA) of KSHV following primary infection is essential for the establishment of KSHV latency in infected cells (Toth et al., 2013a; Toth et al., 2013b). To determine if CTCF and cohesin can play a role in the regulation of RTA gene expression following de novo infection, we first tested the binding of CTCF and cohesin factors RAD21 and SMC3 to the RTA locus compared to the promoter of the constitutively expressed latent gene LANA in the course of KSHV infection (Fig. 1A–1D). NIPBL, a cohesin-recruiting factor, with an additional cohesin-independent role in gene regulation, was also included in our investigation, which has not yet been indicated in controlling KSHV genes (Zuin et al., 2014). We infected SLK cells with KSHV for 1, 4, 8, 16, 24, and 72 hours and then performed ChIP assays at each time point to test the binding of CTCF, RAD21, SMC3, and NIPBL to the CTCF/cohesin-binding sites in the promoter (RTApr) and the intron of the RTA gene (RTAin), as well as in LANA promoter (LANA) (Fig. 1). Latently infected SLK cells were used as a reference point, in which CTCF and cohesin factors are highly enriched in the RTA locus and on the LANA promoter (Chen et al., 2012; Li et al., 2014; Stedman et al., 2008). The CTCF/cohesin-binding site in the erythroid-specific DNase I hypersensitive site (HS1) of the β-globin gene regulatory region and the NIPBL-binding site at the cellular OSBP gene were used as positive controls to show the comparable efficacy of CTCF, cohesin, and NIPBL ChIPs at each time point throughout the experiments, while an intergenic region of the host genome (Neg) was used as a negative control for all ChIPs. Our time course ChIP analyses revealed a gradual recruitment of CTCF, NIPBL, and the cohesin factors SMC3 and RAD21 to the RTA locus during de novo infection. In contrast, each of the tested host factors was already enriched at the LANA promoter as early as at 4 hpi as much as during latency. It can also be observed that the enrichment of cohesin is lower in the RTA locus compared to the LANA promoter, but it is still much higher relative to the control genomic region (Neg). Thus, we found a rapid but differential recruitment of CTCF, cohesin, and NIPBL to the RTA locus and the LANA promoter during de novo infection.
Figure 1. Time course ChIP assays for testing the binding of CTCF and cohesin factors on the viral genome during de novo KSHV infection.
SLK cells were infected with KSHV BAC16 and ChIP assays were performed at the indicated time points of infection. We determined the binding of CTCF (A), RAD21 (B), SMC3 (C), and NIPBL (D) on the LANA promoter, RTA promoter (RTApr), and in the intron of the RTA gene (RTAin). The host genomic regions HS1, OSBP, and Neg were used as controls in the analysis of ChIPs. (E) Time course ChIP assay to show the temporally ordered binding of histone H3, RAD21, and EZH2 onto RTA promoter during de novo KSHV infection of SLK cells. The Y-axis on the right is for the H3 ChIP. (F) Time course ChIP assay for binding of CTCF, RAD21, and EZH2 on the promoter of LANA and RTA during de novo KSHV infection of primary human fibroblast (HFF).
We also performed a comparative ChIP analysis for the binding of histone H3, RAD21 and the PRC2 subunit EZH2 on the RTA promoter at different time points of de novo KSHV infection (Fig. 1E). This time course ChIP experiment showed that RAD21 was recruited to the viral DNA early on during the chromatinization of the KSHV genome prior to the binding of EZH2 (Toth et al., 2013b; Toth et al., 2016). The rapid binding of CTCF and RAD21 to the viral DNA before the recruitment of EZH2 was also confirmed at the promoter of LANA and RTA in primary human fibroblast during de novo KSHV infection (Fig. 1F). The binding of CTCF, cohesin, and NIPBL to the viral DNA immediately following infection before PRC2 recruitment suggests that they can be involved in viral gene regulation and/or the organization of 3D structure of the KSHV genome during de novo infection, which can be critical for the establishment of viral latency.
CTCF, cohesin, and NIPBL inhibit KSHV gene expression during de novo infection
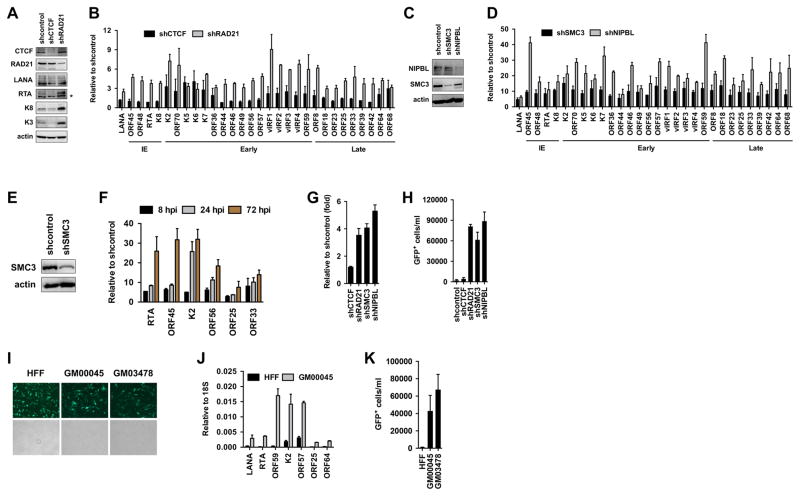
To test if CTCF, cohesin, or NIPBL is involved in the restriction of viral gene expression during de novo KSHV infection, we inhibited their expression by shRNAs in SLK cells during KSHV infection and analyzed the viral gene expression with real time quantitative PCR at 72 hours KSHV postinfection (Fig. 2A–2D). For this, SLK cells were first infected with lentiviruses expressing shRNAs for two days followed by KSHV infection for three days. Immunoblot analysis indicated that the lentiviral shRNAs could reduce the protein levels of CTCF, cohesin factors, and NIPBL in KSHV-infected cells (Fig. 2A and 2C), which led to increased lytic viral gene expression during de novo KSHV infection (Fig. 2B and 2D). Of note, shCTCF increased the expression of only a subset of the lytic genes during KSHV infection (e.g. K2, K5, K6, K7), while the shRNA depletion of RAD21, SMC3 or NIBPL resulted in the increased expression of each of the 29 lytic genes tested. The immunoblot analysis of shCTCF- and shRAD21-treated KSHV-infected cells also supported the results of the RT-qPCR assays showing increased viral protein production only in shRAD21-treated cells but not in shCTCF-treated cells during de novo KSHV infection (Fig. 2A). Since we found that cohesin factors were rapidly binding to the KSHV genome during de novo infection (Fig. 1), we tested whether cohesin can inhibit lytic gene expression in the early time points of KSHV infection. To this end, lentiviral shSMC3-treated SLK cells were infected with KSHV and lytic viral gene expression was analyzed at 8, 24, and 72 hours KSHV postinfection (Fig. 2E and 2F). We found that lytic gene expression was increased as early as 8 hpi when SMC3 was depleted. These data indicate that cohesin and NIPBL can act as potent restriction factors of KSHV lytic gene expression during de novo KSHV infection.
Figure 2. Cohesin factors are required for the restriction of lytic replication following KSHV infection.
(A) Immunoblot analysis of host and viral proteins in shCTCF- and shRAD21-treated SLK cells following KSHV infection. (B) Analysis of the effect of shCTCF and shRAD21 on viral gene expression at 72 hours KSHV postinfection. The classification of tested lytic viral genes based on their induction during KSHV lytic cycle is indicated such as IE (immediate early), early, and late. (C) Protein expression of the cohesin factors in shSMC3- and shNIPBL-treated SLK cells after KSHV infection. (D) Analysis of viral gene expression in shSMC3- and shNIPBL-treated cells at 72 hours KSHV postinfection using gene specific RT-qPCR. (E) Immunoblot showing the depletion of SMC3 expression in SLK cells upon shSMC3. (F) Time course RT-qPCR analysis of viral gene expression in shSMC3-treated SLK cells following KSHV infection. (G) The copy number of KSHV genome in different shRNA-treated SLK cells following KSHV infection was determined relative to shcontrol-treated cells at 72 hours KSHV postinfection. (H) KSHV titer was determined in the media of different shRNA-treated SLK cells at 72 hours KSHV postinfection. (I) Representative immunofluorescence images of KSHV BAC16-infected HFF and CdLS primary fibroblasts at 24 hpi. The KSHV clone BAC16 constitutively expresses GFP, which is used to detect infected cells. (J) RT-qPCR analysis of KSHV gene expression in infected HFF and CdLS fibroblasts at 24 hpi. (K) KSHV titer in the media collected from KSHV-infected HFF and CdLS fibroblasts at 72 hpi.
Cohesin and NIPBL inhibits KSHV lytic replication following de novo infection
KSHV infection of SLK cells results in establishment of viral latency, which is accompanied by the repression of lytic replication and virus production at 72 hpi (Toth et al., 2013b). To test if CTCF, cohesin or NIPBL plays a role in restricting KSHV lytic replication in SLK cells following de novo infection, we inhibited their expression by lentiviral shRNAs during KSHV infection and analyzed viral DNA replication and virus production at 72 hours KSHV postinfection (Fig. 2G and 2H). These experiments showed that shRAD21, shSMC3 and shNIBPL resulted in increased viral DNA replication and KSHV production compared to SLK cells that were treated with scrambled shRNA (shcontrol)- or shCTCF (Fig 2G and 2H).
CdLS patients who often carry genetic mutations in NIPBL or different cohesin factors have been shown to have increased susceptibility to viral and bacterial infections (Jyonouchi et al., 2013). Based on our finding that cohesin can inhibit lytic gene expression of KSHV following infection, we hypothesized that cells derived from CdLS patients could be defective in restricting KSHV replication following de novo infection. To address this question, we infected primary fibroblasts derived from two different CdLS patients (GM00045 and GM03478) with KSHV and examined viral gene expression and virus production in CdLS fibroblasts compared to KSHV-infected normal fibroblast (HFF) cells in which KSHV establishes latency (Fig. 2I–2K). While both HFF and CdLS fibroblasts could be efficiently infected with KSHV (Fig. 2I), viral gene expression and virus production were increased substantially only in infected CdLS fibroblasts (Fig. 2J and 2K). Collectively, these data indicate that cohesin and NIPBL can function as restriction factors to halt KSHV lytic gene expression and viral replication during de novo infection, which promotes the establishment of viral latency.
Inhibition of cohesin or NIPBL modulates the levels of histone modifications on viral promoters during de novo KSHV infection
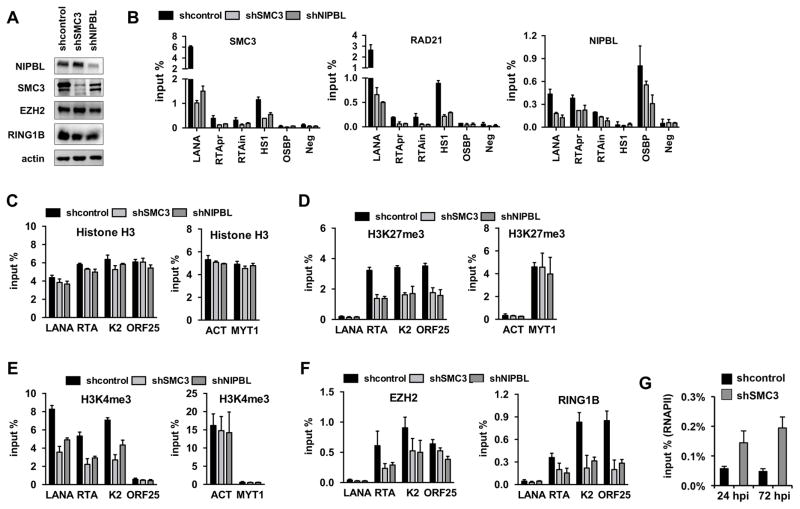
To investigate the function of cohesin factors in the chromatinization of the KSHV genome during de novo infection, we analyzed the epigenetic changes on the viral genome upon the inhibition of cohesin factors. To this end, SLK cells were first infected with lentiviruses expressing either shSMC3 or shNIPBL for two days, which was then followed by KSHV infection for three days. Immunoblots showed efficient and specific depletion of both SMC3 and NIPBL in KSHV-infected shSMC3- and shNIPBL-treated SLK cells while the expression of the H3K27me3 histone methyltransferase EZH2 of PRC2, and RING1B of PRC1 was not affected (Fig. 3A). ChIP assays also confirmed the depletion of SMC3 and NIPBL, which was reflected by their loss of binding on their viral and host target DNA regions (Fig. 3B). Both shSMC3 and shNIPBL resulted in reduced binding of RAD21 as well. This could be due to the fact that these proteins form a complex on their DNA target sites, which can be unstable in the absence of any of the cohesin components. In addition, we analyzed the deposition of histone H3, H3K27me3, H3K4me3, and the binding of EZH2 and RING1B to the promoter of LANA (latent), RTA (immediate early), K2 (early), and ORF25 (late) genes in shSMC3- and shNIPBL-treated cells at 72 hours KSHV postinfection (Fig. 3C–3F). These ChIP experiments revealed that while shSMC3 or shNIPBL did not alter significantly the deposition of histone H3 on the promoters of LANA, K2, and ORF25 during de novo KSHV infection (Fig. 3B), the enrichment of H3K27me3 and H3K4me3 as well as the binding of polycomb proteins on the viral promoters were reduced (Fig. 3C–3E). In contrast, SMC3 or NIPBL depletion had little effect on histone modifications associated with the cellular actin (ACT) and MYT1 promoters indicating target specific affect of shSMC3 and shNIPBL on the viral genome. These results show that the loss of cohesin or NIPBL during KSHV infection can cause insufficient binding of chromatin regulatory factors to viral promoters resulting in reduced deposition of histone marks on the viral genome. However, RNAPII ChIP showed that the binding of RNAPII was increased on the RTA promoter in shSMC3-treated SLK cells during de novo KSHV infection (Fig. 3G). This is in agreement with previous studies, which showed that cohesin is critical for maintaining the histone modification pattern in the RTA locus and can also affect RNAPII-binding on the RTA promoter in PEL cells (Chen et al., 2012). Our data suggests that cohesin and NIPBL can play a role in the establishment of viral chromatin with proper levels of histone modifications on the KSHV genome following de novo infection.
Figure 3. Effect of cohesin depletion on the deposition of histone marks onto the KSHV genome during de novo infection.
(A) Immunoblot showing the expression of host factors indicated on the left in lentiviral shRNA-treated SLK cells. (B) ChIP assay for SMC3, RAD21, and NIPBL on the indicated viral and host genomic sites in different cohesin factor depleted KSHV-infected SLK cells at 72 hpi. (C) Histone H3 ChIP, (D) H3K27me3 ChIP, (E) H3K4me3 ChIP as well as (F) EZH2 and RING1B ChIPs on viral and host genomic sites in shSMC3 and shNIPBL SLK cells at 72 hours KSHV postinfection. (G) RNAPII ChIP on RTA promoter in shcontrol- or shSMC3-treated SLK cells that were infected with KSHV for 24 or 72 hours. A two-tailed student’s t-test was applied to compare the H3 levels on the viral genomic regions between shcontrol and shSMC3 as well as between shcontrol and shNIPBL. For LANA, K2, and ORF25 p>0.05 while for RTA p<0.05.
Inhibition of the KSHV lytic cycle by cohesin is partly through the repression of RTA following de novo infection
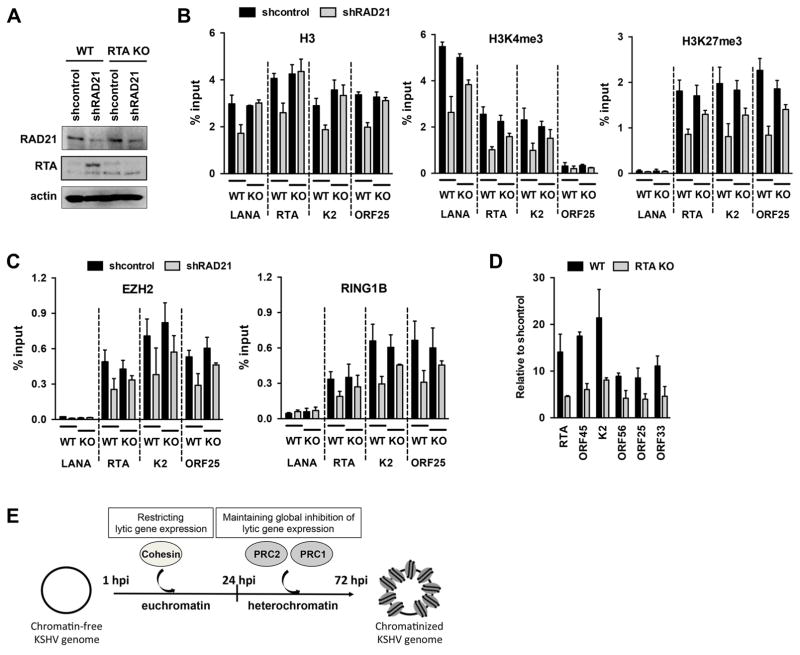
RTA is a potent viral transcription factor whose expression in latently KSHV-infected cells can induce the dissociation of polycomb proteins and the reduction of their repressive histone marks on the promoters of lytic genes resulting in the expression of lytic genes (Guito and Lukac, 2012; Toth et al., 2013a; Toth et al., 2013b; Toth et al., 2010). To test if chromatin changes and increased lytic gene expression in cohesin-depleted cells are due to RTA induction, we performed ChIP and RT-qPCR assays using RTA knockout (RTA KO) KSHV (Fig. 4). We infected shRAD21-treated SLK cells with RTA KO KSHV or wild-type (WT) KSHV and analyzed the enrichment of histone H3, H3K27me3, H3K4me3, EZH2, and RING1B on select viral promoters by ChIP assays at 72 hours KSHV postinfection (Fig. 4A–4C). Our ChIP experiments revealed that shRAD21 significantly reduced the level of histone H3 deposition on the promoters of RTA and ORF25 only in WT KSHV-infected but not in RTA KO KSHV-infected cells (Fig. 4B). In contrast, both the enrichment of histone marks (H3K4me3 and H3K27me3) and the binding of polycomb proteins (EZH2 and RING1B) were reduced on viral promoters in both WT and RTA KO KSHV infected cells upon shRAD21 (Fig. 4B and 4C). However, strikingly, the deposition of histone marks and polycomb proteins was decreased to a lesser extent on viral promoters in RTA KO KSHV-infected cells compared to WT KSHV-infected cells in shRAD21 cells (Fig. 4B and 4C). This finding indicates that shRAD21-induced chromatin changes on the KSHV genome during de novo infection are partly due to the induction of RTA and RTA-mediated viral DNA replication. In agreement with this, we found that lytic viral gene expression was reduced in RTA KO KSHV-infected cells as expected because RTA is required for lytic gene expression, but viral lytic gene transcription was still induced by several fold in shRAD21-treated compared to the shcontrol-treated RTA KO KSHV-infected cells (Fig. 4D). These findings suggest that the cohesin complex can repress KSHV lytic cycle not only via inhibiting RTA expression but also cohesin can exert a global inhibition on lytic gene expression by modulating the chromatinization of the viral DNA early on during de novo KSHV infection prior to the recruitment of Polycomb Repressive Complexes onto the KSHV genome (Fig. 4E).
Figure 4. Reduced deposition of histone marks on the KSHV genome accompanied by lytic gene expression during RTA KO KSHV infection in RAD21 depleted cells.
(A) Immunoblot of RAD21 and RTA in RAD21 depleted WT KSHV and RTA KO KSHV infected SLK cells. (B) ChIP assay for histone H3 and histone marks on different viral genes in RAD21 depleted WT KSHV and RTA KO KSHV infected SLK cells at 72 hpi. A two-tailed student’s t-test was applied to compare H3 levels on the viral genomic regions between shcontrol and shRAD21 in both WT and RTA KO KSHV-infected cells. In WT KSHV-infected cells the H3 changes were not significant (p>0.05) on LANA and K2 promoters while it is significant at RTA and ORF25 (p<0.05). In RTA KO KSHV-infected cells none of the tested viral promoters showed significant H3 differences (p>0.3). (C) ChIP assay for polycomb proteins as described in panel B. (D) RT-qPCR analysis of viral gene expression at 72 hours WT KSHV or RTA KO KSHV postinfection in RAD21 depleted cells. (E) Working model showing that cohesin can play a role in the genome-wide restriction of lytic gene expression early on during de novo KSHV infection by binding to the KSHV genome prior to the recruitment of the Polycomb Repressive Complexes.
Discussion
One of the critical steps in KSHV pathogenesis is the establishment of viral latency following primary infection, which is characterized by the inhibition of lytic gene expression and lytic viral replication (Giffin and Damania, 2014). In this study, we investigated the role of CTCF and cohesin factors in the establishment of viral latency following de novo KSHV infection. We found that although both CTCF and cohesin are rapidly recruited to the viral genome during infection, only cohesin is critical for the establishment of KSHV latency (Figs. 1 and 2). Despite that CTCF has been demonstrated to co-localize with cohesin at many sites on the KSHV genome, several studies showed that they have distinct roles in the regulation of viral gene expression (Chen et al., 2012; Li et al., 2014; Stedman et al., 2008; Tempera and Lieberman, 2010). While cohesin inhibition can reactivate KSHV from latency, CTCF inhibition cannot (Chen et al., 2012). In this study we showed that cohesin knockdown during de novo KSHV infection reduced the binding of polycomb proteins to the promoters of lytic genes and the deposition of histone modifications on the KSHV genome, which was accompanied by increased RNAPII-binding to the RTA promoter, lytic gene expression, viral DNA replication, and virus production in infected cells (Figs. 2 and 3). We also found that RTA, which is known to be required for KSHV lytic replication by antagonizing PRC2-mediated lytic gene inhibition, remains expressed upon cohesin knockdown following KSHV infection (Fig. 2), which can explain the incomplete heterochromatinization of the incoming viral DNA and continuous lytic replication (Toth et al., 2013a; Toth et al., 2013b; Toth et al., 2010). Strikingly, cohesin depletion also compromises the proper chromatinization of the KSHV genome in RTA KO KSHV infected cells, which is associated with increased lytic gene expression compared to shcontrol-treated RTA KO KSHV infected cells (Fig. 4). Based on these findings, we propose that cohesin functions as a global restriction host factor of lytic KSHV replication during de novo infection by regulating the proper chromatinization of the incoming viral DNA, which is critical for suppressing the expression of lytic genes so that KSHV can establish latency. The importance of cohesin in restricting KSHV replication was also supported by our observation that KSHV can replicate in fibroblasts derived from CdLS patients following de novo infection. CdLS patients carry mutations in different cohesin factors, which can cause development disorders while also making CdLS patients more susceptible to viral infections (Jyonouchi et al., 2013; Strachan, 2005). While cohesin controls a number of immune functions, which can be defective when cohesin is mutated, our results are also in line with other studies suggesting a role for cohesin in the direct regulation of KSHV replication in infected cells (Chen et al., 2012; Degner-Leisso and Feeney, 2010; Feng and Barnes, 2013; Li et al., 2014; Rawlings, 2017).
Latent infection is considered to be the default pathway of KSHV infection, but establishment of latency is still not a well understood process (Bechtel et al., 2003). Previous studies showed that chromatinization of the viral genome and the expression of lytic genes are regulated in a biphasic manner during de novo infection (Gunther et al., 2014; Krishnan et al., 2004; Toth et al., 2013b; Toth et al., 2016). First, KSHV acquires a transcriptionally active chromatin following infection, which is associated with the transient expression of a subset of lytic genes. This is followed by the recruitment of PRC2 and PRC1 by the latent KSHV factor LANA and establishment of transcriptionally repressive chromatin on the promoters of lytic genes after 24 hpi resulting in the silencing of lytic genes (Toth et al., 2013b; Toth et al., 2016). The reason that there is no full-blown lytic gene expression and lytic replication following KSHV infection despite the lack of heterochromatin on the KSHV genome in the first 24 hours of infection is still an enigma. Since cohesin binds to the KSHV DNA prior to the establishment of heterochromatin on the viral episome, and cohesin depletion can markedly increase lytic gene expression as early as 8 hours KSHV postinfection, we propose that cohesin is involved in the genome-wide repression of lytic genes prior to the recruitment of polycomb proteins during de novo infection (Fig. 4E). Based on our data cohesin can repress RTA expression by rapidly binding to the RTA locus during de novo KSHV infection thereby blocking RTA-mediated lytic infection. Previous studies showed that (1) cohesin rapidly dissociates from the KSHV genome during lytic reactivation, (2) RAD21 knockdown during latency induces lytic replication, and (3) RAD21 overexpression can repress KSHV lytic genes, which are consistent with the notion that cohesin can act as a global repressor of KSHV lytic genes (Chen et al., 2012; Kang and Lieberman, 2009; Li et al., 2014). Cohesin factors have been reported to interact with polycomb proteins and we found that cohesin inhibition reduced the binding of EZH2 and the level of H3K27me3 on lytic promoters during KSHV infection (Schaaf et al., 2013b; Stelloh et al., 2016; Strubbe et al., 2011). However, we do not consider cohesin as an EZH2 recruiting factor per se on the KSHV genome. Despite the binding of cohesin to the KSHV genome within 24 hpi, EZH2 is not recruited and also, cohesin inhibition affects the deposition of several other histone marks besides H3K27me3 indicating that cohesin globally affects the KSHV epigenome (Fig. 3) and (Chen et al., 2012). Because PRCs are known to be responsible for maintaining transcription repression rather than initiating gene silencing, it is conceivable that cohesin rapidly binding to the incoming KSHV DNA initiates the repression of lytic genes, which are then preserved by PRCs recruited after 24 hpi during de novo infection (Di Croce and Helin, 2013; Toth et al., 2013b; Toth et al., 2016).
Cohesin factors have been shown to play a role both in repression and activation of gene transcription by controlling interaction between promoters and enhancers, regulating the processivity of RNA polymerase II or modulating the recruitment of transcription factors to gene regulatory regions (Peters et al., 2008; Schaaf et al., 2013a; Stelloh et al., 2016). There is also evidence for the role of cohesin in intrachromosomal looping within the KSHV genome, which regulates the 3D structure of the KSHV genome affecting viral gene expression (Kang et al., 2011; Tempera and Lieberman, 2010). Mutation of CTCF/cohesin-binding sites in the KSHV genome, or the inhibition of cohesin function, disrupts KSHV chromosome conformation, reduces the level of histone marks in the RTA locus, and increases lytic gene expression in latently infected primary effusion lymphoma cells (Chen et al., 2017; Chen et al., 2012; Kang et al., 2011; Stedman et al., 2008). Similarly, we also found that cohesin inhibition reduces the deposition of histone marks on the KSHV genome during de novo infection, which is also associated with upregulated lytic gene expression. It is possible that cohesin may also be involved in the coordination of correct KSHV chromosome conformation during infection, which is crucial for the deposition of the proper histone modification pattern on the viral chromatin that is required for the establishment of latency. Additional studies will be required to demonstrate the role of cohesin in the regulation of the KSHV chromatin architecture during de novo infection and how cohesin-initiated silencing of lytic genes can be linked to the maintenance of lytic gene repression during latency.
In summary, we identified cohesin as one of the early host restriction factors, which can block lytic replication of KSHV immediately following primary infection by restricting the expression of lytic genes including RTA that is required for the full-blown lytic replication of the virus. We envision that there are many other host transcription factors, which can be involved in the downregulation of lytic genes of KSHV early on following de novo infection. The identification of these factors will help us better understand how KSHV establishes viral latency, which is an essential step to persistent infection of humans.
Highlights.
Rapid binding of CTCF and cohesin to the KSHV DNA during de novo infection.
KSHV replicates in cohesin depleted cells.
Cohesin is required for the inhibition of KSHV lytic replication following primary infection, which is essential for the establishment of viral latency.
Acknowledgments
We thank Jae U. Jung (University of Southern California) for generously providing K3 antibody and Yoshihiro Izumiya (University of California, Davis) for RTA antibody. This work was supported by the NIH grants R21AI119597, and R03DE025562 as well as the Cornelia de Lange Syndrome Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ballestas ME, Chatis PA, Kaye KM. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science (New York, N Y. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- Bechtel JT, Liang Y, Hvidding J, Ganem D. Host range of Kaposi’s sarcoma-associated herpesvirus in cultured cells. Journal of virology. 2003;77:6474–6481. doi: 10.1128/JVI.77.11.6474-6481.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brulois KF, Chang H, Lee AS, Ensser A, Wong LY, Toth Z, Lee SH, Lee HR, Myoung J, Ganem D, Oh TK, Kim JF, Gao SJ, Jung JU. Construction and manipulation of a new Kaposi’s sarcoma-associated herpesvirus bacterial artificial chromosome clone. Journal of virology. 2012;86:9708–9720. doi: 10.1128/JVI.01019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesarman E. Gammaherpesvirus and lymphoproliferative disorders in immunocompromised patients. Cancer Lett. 2011;305:163–174. doi: 10.1016/j.canlet.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science (New York, N Y. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- Chen HS, De Leo A, Wang Z, Kerekovic A, Hills R, Lieberman PM. BET-Inhibitors Disrupt Rad21-Dependent Conformational Control of KSHV Latency. PLoS pathogens. 2017;13:e1006100. doi: 10.1371/journal.ppat.1006100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HS, Wikramasinghe P, Showe L, Lieberman PM. Cohesins repress Kaposi’s sarcoma-associated herpesvirus immediate early gene transcription during latency. Journal of virology. 2012;86:9454–9464. doi: 10.1128/JVI.00787-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner-Leisso SC, Feeney AJ. Epigenetic and 3-dimensional regulation of V(D)J rearrangement of immunoglobulin genes. Semin Immunol. 2010;22:346–352. doi: 10.1016/j.smim.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol. 2013;20:1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- Feng D, Barnes BJ. Bioinformatics analysis of the factors controlling type I IFN gene expression in autoimmune disease and virus-induced immunity. Front Immunol. 2013;4:291. doi: 10.3389/fimmu.2013.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giffin L, Damania B. KSHV: pathways to tumorigenesis and persistent infection. Advances in virus research. 2014;88:111–159. doi: 10.1016/B978-0-12-800098-4.00002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guito J, Lukac DM. KSHV Rta Promoter Specification and Viral Reactivation. Front Microbiol. 2012;3:30. doi: 10.3389/fmicb.2012.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther T, Grundhoff A. The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PLoS pathogens. 2010;6:e1000935. doi: 10.1371/journal.ppat.1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther T, Schreiner S, Dobner T, Tessmer U, Grundhoff A. Influence of ND10 components on epigenetic determinants of early KSHV latency establishment. PLoS pathogens. 2014;10:e1004274. doi: 10.1371/journal.ppat.1004274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jyonouchi S, Orange J, Sullivan KE, Krantz I, Deardorff M. Immunologic features of Cornelia de Lange syndrome. Pediatrics. 2013;132:e484–489. doi: 10.1542/peds.2012-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Lieberman PM. Cell cycle control of Kaposi’s sarcoma-associated herpesvirus latency transcription by CTCF-cohesin interactions. Journal of virology. 2009;83:6199–6210. doi: 10.1128/JVI.00052-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Wiedmer A, Yuan Y, Robertson E, Lieberman PM. Coordination of KSHV latent and lytic gene control by CTCF-cohesin mediated chromosome conformation. PLoS pathogens. 2011;7:e1002140. doi: 10.1371/journal.ppat.1002140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan HH, Naranatt PP, Smith MS, Zeng L, Bloomer C, Chandran B. Concurrent expression of latent and a limited number of lytic genes with immune modulation and antiapoptotic function by Kaposi’s sarcoma-associated herpesvirus early during infection of primary endothelial and fibroblast cells and subsequent decline of lytic gene expression. Journal of virology. 2004;78:3601–3620. doi: 10.1128/JVI.78.7.3601-3620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DJ, Verma D, Mosbruger T, Swaminathan S. CTCF and Rad21 act as host cell restriction factors for Kaposi’s sarcoma-associated herpesvirus (KSHV) lytic replication by modulating viral gene transcription. PLoS pathogens. 2014;10:e1003880. doi: 10.1371/journal.ppat.1003880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesri EA, Cesarman E, Boshoff C. Kaposi’s sarcoma and its associated herpesvirus. Nat Rev Cancer. 2010;10:707–719. doi: 10.1038/nrc2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myoung J, Ganem D. Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: maintenance of tight latency with efficient reactivation upon induction. Journal of virological methods. 2011;174:12–21. doi: 10.1016/j.jviromet.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Tedeschi A, Schmitz J. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008;22:3089–3114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings JS. Roles of SMC Complexes During T Lymphocyte Development and Function. Adv Protein Chem Struct Biol. 2017;106:17–42. doi: 10.1016/bs.apcsb.2016.08.001. [DOI] [PubMed] [Google Scholar]
- Renne R, Lagunoff M, Zhong W, Ganem D. The size and conformation of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) DNA in infected cells and virions. Journal of virology. 1996;70:8151–8154. doi: 10.1128/jvi.70.11.8151-8154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf CA, Kwak H, Koenig A, Misulovin Z, Gohara DW, Watson A, Zhou Y, Lis JT, Dorsett D. Genome-wide control of RNA polymerase II activity by cohesin. PLoS Genet. 2013a;9:e1003382. doi: 10.1371/journal.pgen.1003382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf CA, Misulovin Z, Gause M, Koenig A, Gohara DW, Watson A, Dorsett D. Cohesin and polycomb proteins functionally interact to control transcription at silenced and active genes. PLoS Genet. 2013b;9:e1003560. doi: 10.1371/journal.pgen.1003560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. Embo J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelloh C, Reimer MH, Pulakanti K, Blinka S, Peterson J, Pinello L, Jia S, Roumiantsev S, Hessner MJ, Milanovich S, Yuan GC, Rao S. The cohesin-associated protein Wapal is required for proper Polycomb-mediated gene silencing. Epigenetics Chromatin. 2016;9:14. doi: 10.1186/s13072-016-0063-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strachan T. Cornelia de Lange Syndrome and the link between chromosomal function, DNA repair and developmental gene regulation. Curr Opin Genet Dev. 2005;15:258–264. doi: 10.1016/j.gde.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Strubbe G, Popp C, Schmidt A, Pauli A, Ringrose L, Beisel C, Paro R. Polycomb purification by in vivo biotinylation tagging reveals cohesin and Trithorax group proteins as interaction partners. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5572–5577. doi: 10.1073/pnas.1007916108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempera I, Lieberman PM. Chromatin organization of gammaherpesvirus latent genomes. Biochimica et biophysica acta. 2010;1799:236–245. doi: 10.1016/j.bbagrm.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth Z, Brulois K, Jung JU. The chromatin landscape of Kaposi’s sarcoma-associated herpesvirus. Viruses. 2013a;5:1346–1373. doi: 10.3390/v5051346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth Z, Brulois K, Lee HR, Izumiya Y, Tepper C, Kung HJ, Jung JU. Biphasic Euchromatin-to-Heterochromatin Transition on the KSHV Genome Following De Novo Infection. PLoS pathogens. 2013b;9:e1003813. doi: 10.1371/journal.ppat.1003813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth Z, Brulois KF, Wong LY, Lee HR, Chung B, Jung JU. Negative elongation factor-mediated suppression of RNA polymerase II elongation of Kaposi’s sarcoma-associated herpesvirus lytic gene expression. Journal of virology. 2012;86:9696–9707. doi: 10.1128/JVI.01012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth Z, Maglinte DT, Lee SH, Lee HR, Wong LY, Brulois KF, Lee S, Buckley JD, Laird PW, Marquez VE, Jung JU. Epigenetic analysis of KSHV latent and lytic genomes. PLoS pathogens. 2010;6:e1001013. doi: 10.1371/journal.ppat.1001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth Z, Papp B, Brulois K, Choi YJ, Gao SJ, Jung JU. LANA-Mediated Recruitment of Host Polycomb Repressive Complexes onto the KSHV Genome during De Novo Infection. PLoS pathogens. 2016;12:e1005878. doi: 10.1371/journal.ppat.1005878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrouwe MG, Elghalbzouri-Maghrani E, Meijers M, Schouten P, Godthelp BC, Bhuiyan ZA, Redeker EJ, Mannens MM, Mullenders LH, Pastink A, Darroudi F. Increased DNA damage sensitivity of Cornelia de Lange syndrome cells: evidence for impaired recombinational repair. Hum Mol Genet. 2007;16:1478–1487. doi: 10.1093/hmg/ddm098. [DOI] [PubMed] [Google Scholar]
- Zuin J, Franke V, van Ijcken WF, van der Sloot A, Krantz ID, van der Reijden MI, Nakato R, Lenhard B, Wendt KS. A cohesin-independent role for NIPBL at promoters provides insights in CdLS. PLoS Genet. 2014;10:e1004153. doi: 10.1371/journal.pgen.1004153. [DOI] [PMC free article] [PubMed] [Google Scholar]